Abstract
Acute promyelocytic leukemia is a myeloid disorder that is characterized by the specific t(15;17) variant in ~98% of cases. The typical hypergranular and microgranular or hypogranular types exist, and are frequently associated with disseminated intravascular coagulopathy. Rare cases of promyelocytic leukemia-retinoic acid receptor α (PML-RARA) fusion without the reciprocal RARA-PML have been reported in cytogenetically normal samples. Conversely, fluorescence in situ hybridization (FISH) analysis has revealed a cryptic insertion of the RARA gene into the PML gene on chromosome 15. The current study reports a unique case with a normal karyotype and molecular evidence of the PML-RARA short isoform 3-fusion transcript, with FISH analysis revealing two fusion signals on the two copies of chromosome 15, but absence of the reciprocal on the two copies of chromosome 17. This finding raised the hypothesis of chromosome 15 uniparental isodysomy as consequence of normal chromosome 15 loss and duplication of the rearranged chromosome, as supported by polymorphic loci molecular analysis. The clinical, cytogenetic and molecular characterization of this case are presented and discussed in the present study.
Keywords: acute promyelocytic leukemia variant, cryptic promyelocytic leukemia-retinoic acid receptor α, fluorescence in situ hybridization, quenching loop-mediated isothermal amplification, uniparental disomy
Introduction
The diagnostic hallmark of acute promyelocytic leukemia (APL) is the reciprocal translocation t(15;17)(q24;q21), leading to the disruption of the promyelocytic leukemia (PML) and retinoic acid receptor α (RARA) genes, resulting in PML-RARA and RARA-PML fusion products in ~98% of cases (1–7). The PML-RARA fusion transcript from der(15)t(15;17) serves a key role in leukemogenesis, inhibiting the differentiation and promoting the survival of myeloid precursor cells (8). Three regions of the PML locus are primarily involved in the t(15;17) translocation breakpoint cluster regions (bcrs): intron 6 (bcr1), exon 6 (bcr2) and intron 3 (bcr3), whereas RARA breakpoints always occur in intron 2. As a consequence, there are three possible PML-RARA isoforms, referred to the as long (bcr1), variant (bcr2) and short (bcr3) isoforms (9).
Assessment of PML-RARA formation, or variant RARA gene rearrangements by means of conventional karyotyping, fluorescence in situ hybridization (FISH) or reverse transcription-polymerase chain reaction (RT-PCR), is required for the diagnosis of APL (10). In rare cytogenetically normal cases, FISH or molecular methods demonstrate the presence of the PML-RARA fusion gene without the reciprocal RARA-PML, resulting from a submicroscopic insertion of RARA into PML. Since this cryptic insertion has rarely been reported, no prognostic significance has been clearly established (9,11–22); however, a prompt diagnosis and the administration of targeted therapies, including all-trans retinoic acid (ATRA) and arsenic trioxide (ATO), are essential to improve the outcome in these patients (21). Due to the use of contemporary targeted therapy, APL has become a highly curable disease with complete remission rates of >95% and cure rates of >80% (23–27).
To the best of our knowledge, the current case is the first reported with two PML-RARA FISH fusion signals present on the two copies of chromosome 15, as result of a cryptic insertion of RARA into PML and chromosome 15 uniparental isodisomy (iUPD), likely due to loss of the normal chromosome 15 and duplication of the rearranged one (28,29). Written informed consent was obtained from the patient.
Case report
Patient presentation
A 73-year-old female Caucasian patient was admitted to the Humanitas Clinical and Research Center (Milan, Italy) in January 2016 with monocytosis, anemia and thrombocytopenia incidentally diagnosed during a knee replacement surgery. The patient's medical history revealed β-thalassemia minor trait, obesity, hypertension, mild fasting hyperglycemia and toxic multinodular goiter. In 1982, the patient had undergone a bilateral hysteroannessiectomy to remove fibroids, and in 2010 the patient had undergone a cholecystectomy due to cholelithiasis. During the admission, the peripheral blood examination revealed a hemoglobin count of 8.8 g/dl (normal range, 12–16 g/dl) and a platelet count of 4.7×1010/l (normal range, 13.0–40.0×1010/l), as well as a white blood cell count of 5.24×109/l (normal range, 4–10×109/l). A peripheral blood cell smear revealed prominent leukocytosis with blast cells accounting for 94% of all nucleated cells, characterized by hypogranular bilobed nuclei (French-American-British classification M3 variant) (30). Peripheral blood flow cytometric analysis revealed positivity for cluster of differentiation (CD) 13, CD33, myeloperoxidase, CD2 and CD9, and a negative result for human leukocyte antigen-antigen D related, CD117, CD15, CD4, CD19, CD14, CD10, CD3 and CD34. The patient was clinically diagnosed with APL. The karyotype, as determined from the peripheral blood, was 46,XX. As Q-LAMP revealed positivity for the PML-RARA transcript, FISH was performed and two fusion signals on the two copies of chromosome 15 were observed.
Treatment
According to the ATRA and idarubicin (AIDA) protocol, the patient was treated with induction chemotherapy and received 45 mg/m2 ATRA twice a day and 12 mg/m2/day idarubicin for 3 days (one cycle). The patient developed differentiation syndrome symptoms and disseminated intravascular coagulation with intracranial bleeding. On day 20 post-therapy, the patient was in clinical remission. Molecular analysis using microsatellites and performed on a peripheral blood sample supported the hypothesis of chromosome 15 iUPD.
On day 48 post-therapy, cytogenetic, FISH and RT-PCR analyses were performed, with normal results. The patient received consolidation therapy with 1,000 mg/m2/day cytarabine for 5 days, 5 mg/m2/day idarubicin for 5 days and 45 mg/m2 ATRA twice a day for 15 days, and is presently under maintenance therapy. During the clinical course no substantial difference compared with classical APL patients was observed.
Cytogenetic analysis
Cytogenetic analysis was performed on peripheral blood samples incubated for 24 h according to standard procedures (31). A total of 27 spontaneous quinacrine-banded metaphases were analyzed and the karyotype described according to the International System for Human Cytogenetic Nomenclature 2013 criteria (32). Following induction therapy on day 48, the karyotype was obtained from a bone marrow sample following a 24-h incubation period. A total of 25 spontaneous metaphases were analyzed.
FISH analysis
FISH was performed according to the manufacturer's protocol on metaphase and interphase nuclei using the commercially available SureFISH PML-RARA dual-color dual-fusion DNA probe, specific for the PML (15q24; spectrum red) and RARA (17q21; spectrum green) loci (Agilent Technologies, Inc., Santa Clara, CA, USA). A total of 15 metaphases and 100 nuclei were scored for the peripheral blood and the bone marrow samples.
Molecular methods
The presence of the PML-RARA transcript was evaluated using a commercial kit (DiaSorin, Saluggia, Italy) based on the non-PCR quenching loop-mediated isothermal amplification (Q-LAMP) method modified to introduce fluorescent oligonucleotides and a new polymerase with RNA reverse transcription and DNA amplification activity, as previously described by Spinelli et al (33). To investigate the iUPD hypothesis, the analysis of short tandem repeats (STR) present on 15 different alleles comprising the Penta E locus on chromosome 15 was performed on 0.5 ng of DNA at diagnosis and on day 20 post-therapy using a commercial kit (PowerPlex® 16 System; Promega Corporation, Madison, WI, USA) according to the manufacturer's protocol.
Results
Cytogenetics and FISH results
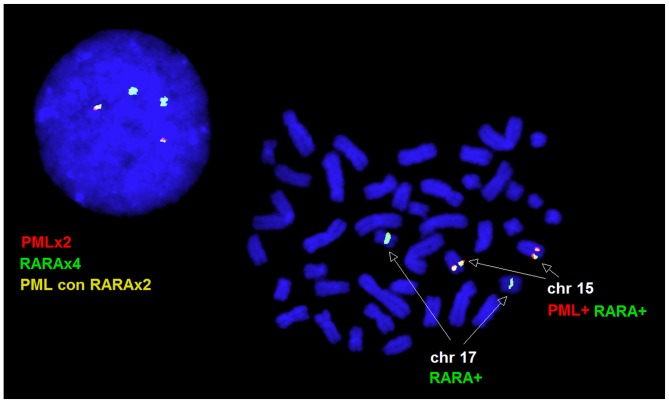
A 46,XX karyotype was observed in the peripheral blood and bone marrow samples. In the peripheral blood sample, interphase FISH revealed normal cells with two red PML and two green RARA signals in 12% of the nuclei, and a variant fusion pattern characterized by two green RARA and two yellow PML-RARA fusion signals in 88% of nuclei. All the metaphase cells analyzed by FISH exhibited two PML/RARA fusion signals, one on each copy of chromosome 15, and two normal RARA signals on the two copies of chromosome 17, consistent with the interphase FISH pattern (Fig. 1). A normal FISH pattern was observed on the bone marrow specimen following induction therapy.
Figure 1.
Fluoresence in situ hybridization pattern observed on metaphase cells and nuclei. PML, promyelocytic leukemia; RARA, retinoic acid receptor α; chr, chromosome; con, fused with.
Molecular results
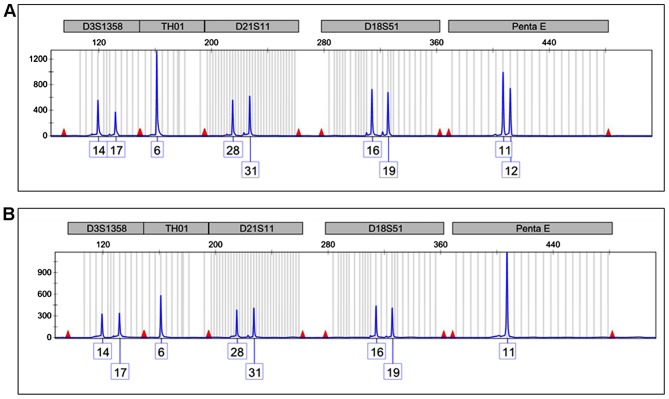
The Q-LAMP assay assessed the presence of the bcr3/short form PML-RARA fusion transcript. The STR analysis of the Penta E locus on chromosome 15 revealed the presence of two peaks of 11 and 12 STR repeats in the remission sample (Fig. 2A) demonstrating that two chromosomes with varying STR numbers were present in the patient's normal cells. Conversely, only the 11 repeat signal was present in the diagnostic sample, representing the APL cells (Fig. 2B). This latter feature is consistent with the presence of only one chromosome 15 or with the presence of two identical copies of chromosome 15 (iUPD).
Figure 2.
Short tandem repeat analysis of 5 representative loci out of 15 studied located on 5 differing chromosomes. (A) The Penta E locus (chromosome 15) in the remission sample exhibits peaks 11 and 12 compatible with the presence of two varied copies of chromosome 15. (B) The sample at the onset of the disease exhibits only a double-sized 11 peak, suggesting that only one chromosome 15 or two identical copies are present.
Discussion
Approximately 9% of APL patients do not harbor the classic t(15;17) translocation; however, certain patients still express the PML-RARA fusion gene. These cases are considered to have a ‘cryptic’ transcript resulting from sub-microscopic insertions of PML or RARA or more complex rearrangements, thus escaping detection with conventional cytogenetic analysis. In these rare cases, the cryptic transcript is usually detected by RT-PCR. As ATRA and ATO are targeted therapies against the action of the PML-RARA protein, patients who lack the classic translocation but present the fusion product may also benefit from these therapies (14,15,20,21). There have been a number of previous studies describing patients with morphological features of APL and a normal karyotype who are FISH-negative for t(15;17), but RT-PCR-positive for PML-RARA. The treatment with ATRA reveals similar good responses and favorable prognoses compared with those observed in patients harboring the classic t(15;17) variant (14,15,20,21).
The increasing number of reported cases with cryptic rearrangements supports the requirement for varied integrated diagnostic approaches in order to recognize the presence of fusion products that respond to targeted therapy. In particular, the current case demonstrated the importance of FISH evaluation not only on interphase nuclei, but also on metaphase cells, which allowed the identification of the underlying mechanism leading to the rearrangement. Q-LAMP revealed the presence of the PML-RARA transcript, but only FISH detected the presence of the double insertion of RARA into each copy of the PML gene, supporting the hypothesis of chromosome 15 iUPD, which was then confirmed using microsatellite analysis.
Acquired UPD (aUPD), most frequently segmental, has been reported in 15–20% of patients affected by acute myeloid leukemia, resulting in gene dosage alterations and homozygosity for mutated genes that can provide a proliferative advantage or increased chemoresistance (34,35). Chromosomes 11 and 13 have the highest number of aUPDs, but aUPDs on Xq, 1p, 2p, 2q, 6p, 9p, 17p, 17q, 19q and 21q have also been reported (34,35). To the best of our knowledge, no aUPD has been reported for chromosome 15; however, it is well established that chromosome 15 is imprinted and constitutional UPD causes specific syndromes (28,29).
Numerical somatic UPDs may occur due to mitotic errors, including non-disjunction or loss of a chromosome due to anaphase lag followed by duplication of the remaining chromosome (28,29). The chromosome 15 iUPD observed in the current case may be the result of an insertion of RARA into PML, loss of the normal chromosome 15 and monosomy rescue by duplication of the rearranged one or trisomy consequent to non-disjunction followed by loss of the homolog, as reported in constitutional cases (Fig. 3) (28,29). These two mechanisms may be possible in acute myeloid leukemia, and chromosome 15 trisomy, despite being rare, is more frequently reported (36).
Figure 3.
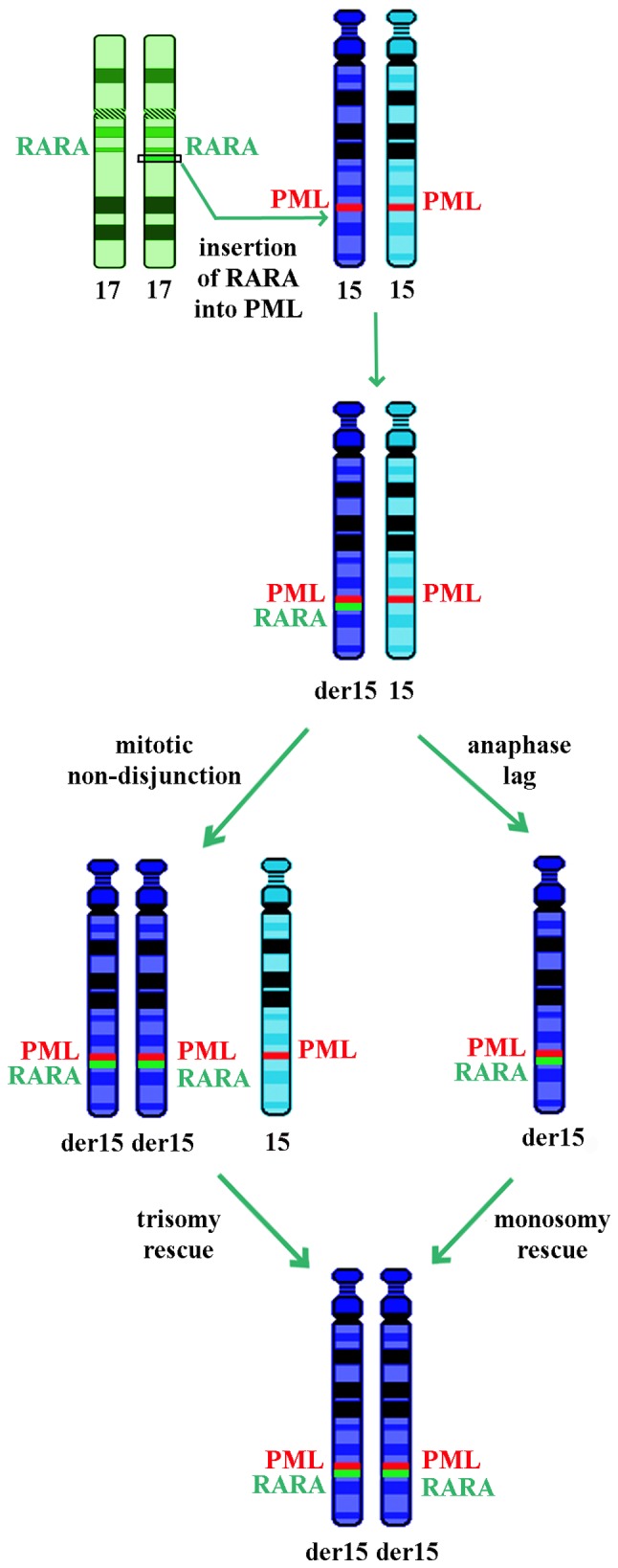
Potential underlying mechanisms leading to uniparental isodisomy of the derivative chromosome 15. RARA, retinoic acid receptor α; PML, promyelocytic leukemia.
The presence of iUPD in the present case apparently did not affect the good outcome of therapy and the prognosis. However, the consequent loss of heterozygosity for the entire chromosome may have introduced homozygosity for coding polymorphisms with variable functional activity, including those in the drug metabolizing enzymes, non-coding regulatory polymorphisms that result in differential expression of alleles and haploinsufficiency or overexpression of important proteins (37–39). A close follow-up is required for this patient in order to monitor the possible prognostic negative effect due to the iUPD.
References
- 1.Lo Coco F, Nervi C, Avvisati G, Mandelli F. Acute promyelocytic leukemia: A curable disease. Leukemia. 1998;12:1866–1880. doi: 10.1038/sj.leu.2401230. [DOI] [PubMed] [Google Scholar]
- 2.Kadam PR, Merchant AA, Advani SH. Cytogenetic findings in patients with acute promyelocytic leukemia and a case of cml blast crisis with promyelocytic proliferation. Cancer Genet Cytogenet. 1990;50:109–117. doi: 10.1016/0165-4608(90)90244-5. [DOI] [PubMed] [Google Scholar]
- 3.Bapna A, Nair R, Tapan KS, Nair CN, Kadam P, Gladstone B, Advani SH. All-trans-retinoic acid (ATRA): Pediatric acute promyelocytic leukemia. Pediatr Hematol Oncol. 1998;15:243–248. doi: 10.3109/08880019809028791. [DOI] [PubMed] [Google Scholar]
- 4.Advani SH, Nair R, Bapna A, Gladstone B, Kadam P, Saikia TK, Parekh PM, Gopal R, Nair CN. Acute promyelocytic leukemia: All-trans retinoic acid (ATRA) along with chemotherapy is superior to ATRA alone. Am J Hematol. 1999;60:87–93. doi: 10.1002/(SICI)1096-8652(199902)60:2<87::AID-AJH1>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 5.Burnett AK, Grimwade D, Solomon E, Wheatley K, Goldstone AH. Presenting white blood cell count and kinetics of molecular remission predict prognosis in acute promyelocytic leukemia treated with all-trans retinoic acid: Result of the Randomized MRC Trial. Blood. 1999;93:4131–4143. [PubMed] [Google Scholar]
- 6.Amare PS, Baisane C, Saikia T, Nair R, Gawade H, Advani S. Fluorescence in situ hybridization: A highly efficient technique of molecular diagnosis and prediction for disease course in patients with myeloid leukemias. Cancer Genet Cytogenet. 2001;131:125–134. doi: 10.1016/S0165-4608(01)00504-0. [DOI] [PubMed] [Google Scholar]
- 7.Brockman SR, Paternoster SF, Ketterling RP, Dewald GW. New highly sensitive fluorescence in situ hybridization method to detect PML/RARA fusion in acute promyelocytic leukemia. Cancer Genet Cytogenet. 2003;145:144–151. doi: 10.1016/S0165-4608(03)00061-X. [DOI] [PubMed] [Google Scholar]
- 8.Mistry AR, Pedersen EW, Solomon E, Grimwade D. The molecular pathogenesis of acute promyelocytic leukaemia: Implications for the clinical management of the disease. Blood Rev. 2003;17:71–97. doi: 10.1016/S0268-960X(02)00075-9. [DOI] [PubMed] [Google Scholar]
- 9.Park TS, Kim JS, Song J, Lee KA, Yoon S, Suh B, Lee JH, Lee HJ, Kim JK, Choi JR. Acute promyelocytic leukemia with insertion of PML exon 7a and partial deletion of exon 3 of RARA: A novel variant transcript related to aggressive course and not detected with real-time polymerase chain reaction analysis. Cancer Genet Cytogenet. 2009;188:103–107. doi: 10.1016/j.cancergencyto.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 10.Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, Thiele J, Vardiman JW, editors. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. 4th Edition. IARC; Lyon: 2008. [Google Scholar]
- 11.Tirado CA, Jahn JA, Scheerle J, Eid M, Meister RJ, Christie RJ, Croft CD, Wallingford S, Heritage DW, Mowrey PN, Meloni-Ehrig AM. Variant acute promyelocytic leukemia translocation (15;17) originating from two subsequent balanced translocations involving the same chromosomes 15 and 17 while preserving the PML/RARA fusion. Cancer Genet Cytogenet. 2005;161:70–73. doi: 10.1016/j.cancergencyto.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 12.Wang HY, Ding J, Vasef MA, Wilson KF. A bcr3/short form PML-RARalpha transcript in an acute promyelocytic leukemia resulted from a derivative chromosome 17 due to submicroscopic insertion of the PML gene into the RARalpha locus. Am J Clin Pathol. 2009;131:64–71. doi: 10.1309/AJCPE0L7CIJZBIFR. [DOI] [PubMed] [Google Scholar]
- 13.Goldschmidt N, Yehuda-Gafni O, Abeliovich D, Slyusarevsky E, Rund D. Interstitial insertion of RARα gene into PML gene in a patient with acute promyelocytic leukemia (APL) lacking the classic t(15;17) Hematology. 2010;15:332–337. doi: 10.1179/102453310X12647083621083. [DOI] [PubMed] [Google Scholar]
- 14.Kim MJ, Cho SY, Kim MH, Lee JJ, Kang SY, Cho EH, Huh J, Yoon HJ, Park TS, Lee WI, et al. FISH-negative cryptic PML-RARA rearrangement detected by long-distance polymerase chain reaction and sequencing analyses: A case study and review of the literature. Cancer Genet Cytogenet. 2010;203:278–283. doi: 10.1016/j.cancergencyto.2010.08.026. [DOI] [PubMed] [Google Scholar]
- 15.Lewis C, Patel V, Abhyankar S, Zhang D, Ketterling RP, McClure RF, Persons DL. Microgranular variant of acute promyelocytic leukemia with normal conventional cytogenetics, negative PML/RARA FISH and positive PML/RARA transcripts by RT-PCR. Cancer Genet. 2011;204:522–523. doi: 10.1016/j.cancergen.2011.09.001. [DOI] [PubMed] [Google Scholar]
- 16.Welch JS, Westervelt P, Ding L, Larson DE, Klco JM, Kulkarni S, Wallis J, Chen K, Payton JE, Fulton RS, et al. Use of whole-genome sequencing to diagnose a cryptic fusion oncogene. JAMA. 2011;305:1577–1584. doi: 10.1001/jama.2011.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Amare PK, Baisane C, Nair R, Menon H, Banavali S, Kabre S, Gujral S, Subramaniam P. Characterization of cryptic rearrangements, deletion, complex variants of PML, RARA in acute promyelocytic leukemia. Indian J Hum Genet. 2011;17:54–58. doi: 10.4103/0971-6866.86174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koshy J, Qian YW, Bhagwath G, Willis M, Kelley TW, Papenhausen P. Microarray, gene sequencing, and reverse transcriptase-polymerase chain reaction analyses of a cryptic PML-RARA translocation. Cancer Genet. 2012;205:537–540. doi: 10.1016/j.cancergen.2012.07.017. [DOI] [PubMed] [Google Scholar]
- 19.Gruver AM, Rogers HJ, Cook JR, Ballif BC, Schultz RA, Batanian JR, Fesler MJ, Tubbs RR. Modified array-based comparative genomic hybridization detects cryptic and variant PML-RARA rearrangements in acute promyelocytic leukemia lacking classic translocations. Diagn Mol Pathol. 2013;22:10–21. doi: 10.1097/PDM.0b013e31825b8326. [DOI] [PubMed] [Google Scholar]
- 20.Blanco EM, Curry CV, Lu XY, Sarabia SF, Redell MS, Lopez-Terrada DH, Roy A. Cytogenetically cryptic and FISH-negative PML/RARA rearrangement in acute promyelocytic leukemia detected only by PCR: An exceedingly rare phenomenon. Cancer Genet. 2014;207:48–49. doi: 10.1016/j.cancergen.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 21.Fan H, Ortega V, Fanasch HM, Wang Y, Holder KN, Higgins RA, Mendiola C, Mohamed G, Vadlamudi K, Velagaleti G. PML-RARA fusion resulting from a cryptic insertion of RARA gene into PML gene without the reciprocal RARA-PML fusion: Clinical, cytogenetic and molecular characterization and prognosis. Eur J Haematol. 2014;93:354–358. doi: 10.1111/ejh.12326. [DOI] [PubMed] [Google Scholar]
- 22.Shepshelovich D, Oniashvili N, Parnes D, Klein A, Muchtar E, Yeshaya J, Aviram A, Rabizadeh E, Raanani P. Acute promyelocytic leukemia with isochromosome 17q and cryptic PML-RARA successfully treated with all-trans retinoic acid and arsenic trioxide. Cancer Genet. 2015;208:575–579. doi: 10.1016/j.cancergen.2015.08.001. [DOI] [PubMed] [Google Scholar]
- 23.Tallman MS, Andersen JW, Schiffer CA, Appelbaum FR, Feusner JH, Woods WG, Ogden A, Weinstein H, Shepherd L, Willman C, et al. All-trans retinoic acid in acute promyelocytic leukemia: long-term outcome and prognostic factor analysis from the North American Intergroup protocol. Blood. 2002;100:4298–4302. doi: 10.1182/blood-2002-02-0632. [DOI] [PubMed] [Google Scholar]
- 24.Adès L, Guerci A, Raffoux E, Sanz M, Chevallier P, Lapusan S, Recher C, Thomas X, Rayon C, Castaigne S, et al. Very long-term outcome of acute promyelocytic leukemia after treatment with all-trans retinoic acid and chemotherapy: The European APL Group experience. Blood. 2010;115:1690–1696. doi: 10.1182/blood-2009-07-233387. [DOI] [PubMed] [Google Scholar]
- 25.Lo-Coco F, Avvisati G, Vignetti M, Breccia M, Gallo E, Rambaldi A, Paoloni F, Fioritoni G, Ferrara F, Specchia G, et al. Front-line treatment of acute promyelocytic leukemia with AIDA induction followed by risk-adapted consolidation for adults younger than 61 years: Results of the AIDA-2000 trial of the GIMEMA Group. Blood. 2010;116:3171–3179. doi: 10.1182/blood-2010-03-276196. [DOI] [PubMed] [Google Scholar]
- 26.Iland HJ, Bradstock K, Supple SG, Catalano A, Collins M, Hertzberg M, Browett P, Grigg A, Firkin F, Hugman A, et al. All-trans-retinoic acid, idarubicin, and IV arsenic trioxide as initial therapy in acute promyelocytic leukemia (APML4) Blood. 2012;120:1570–1580, quiz 1752. doi: 10.1182/blood-2012-02-410746. [DOI] [PubMed] [Google Scholar]
- 27.Burnett AK, Russel NH, Hills RK, Bowen D, Kell J, Knapper S, Morgan YG, Lok J, Grech A, Jones G, et al. Arsenic trioxide and all-trans retinoic acid treatment for acute promyelocytic leukaemia in all risk groups (AML17): Results of a randomised, controlled, phase 3 trial. Lancet Oncol. 2015;16:1295–1305. doi: 10.1016/S1470-2045(15)00193-X. [DOI] [PubMed] [Google Scholar]
- 28.Ledbetter DH, Engel E. Uniparental disomy in humans: Development of an imprinting map and its implications for prenatal diagnosis. Hum Mol Genet 4 Spec. 1995:1757–1764. doi: 10.1093/hmg/4.suppl_1.1757. [DOI] [PubMed] [Google Scholar]
- 29.Liher T. Cytogenetic contribution to uniparental disomy (UPD) Mol Cytogenet. 2010;3:8. doi: 10.1186/1755-8166-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bennett JM, Catovsky D, Daniel MT, Flandrin G, Galton DA, Gralnick HR, Sultan C. Proposals for the classification of the acute leukaemias. French-American-British (FAB) co-operative group. Br J Haematol. 1976;33:451–481. doi: 10.1111/j.1365-2141.1976.tb03563.x. [DOI] [PubMed] [Google Scholar]
- 31.Barch MJ, Knutsen T, Spurbeck JL, editors. The AGT cytogenetics laboratory manual. 3rd edition. Lippincott-Raven Publishers; Philadelphia: 1997. [Google Scholar]
- 32.Shaffer LG, McGowan-Jordan J, Schmid, editors. ISCN 2013 An International System for Human Cytogenetic Nomenclature. Karger Medical and Scientific Publishers; Basel: 2013. [Google Scholar]
- 33.Spinelli O, Rambaldi A, Rigo F, Zanghì P, D'Agostini E, Amicarelli G, Colotta F, Divona M, Ciardi C, Coco FL, et al. Simple, rapid and accurate molecular diagnosis of acute promyelocytic leukemia by loop mediated amplification technology. Oncoscience. 2014;2:50–58. doi: 10.18632/oncoscience.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Raghavan M, Smith LL, Lillington DM, Chaplin T, Kakkas I, Molloy G, Chelala C, Cazier JB, Cavenagh JD, Fitzgibbon J, et al. Segmental uniparental disomy is a commonly acquired genetic event in relapsed acute myeloid leukemia. Blood. 2008;112:814–821. doi: 10.1182/blood-2008-01-132431. [DOI] [PubMed] [Google Scholar]
- 35.Gupta M, Raghavan M, Gale R, Chelala C, Allen C, Molloy G, Chaplin T, Linch DC, Cazier JB, Young BD. Novel regions of acquired uniparental disomy discovered in acute myeloid leukemia. Genes Chromosomes Cancer. 2008;47:729–739. doi: 10.1002/gcc.20573. [DOI] [PubMed] [Google Scholar]
- 36.Mitelman F, Johansson B, Mertens F, editors. http://cgap.nci.nih.gov/Chromosomes/Mitelman. Mitelman Database of Chromosome Aberrations and Gene Fusions in Cancer. 2016 2016 Oct 6; [Google Scholar]
- 37.Stebbing J, Bower M, Syed N, Smith P, Yu V, Crook T. Epigenetics: An emerging technology in the diagnosis and treatment of cancer. Pharmacogenomics. 2006;7:747–757. doi: 10.2217/14622416.7.5.747. [DOI] [PubMed] [Google Scholar]
- 38.Schwahn B, Rozen R. Polymorphisms in the methylenetetrahydrofolate reductase gene: Clinical consequences. Am J Pharmacogenomics. 2001;1:189–201. doi: 10.2165/00129785-200101030-00004. [DOI] [PubMed] [Google Scholar]
- 39.Milani L, Gupta M, Andersen M, Dhar S, Fryknäs M, Isaksson A, Larsson R, Syvänen AC. Allelic imbalance in gene expression as a guide to cis-acting regulatory single nucleotide polymorphisms in cancer cells. Nucleic Acids Res. 2007;35:e34. doi: 10.1093/nar/gkl1152. [DOI] [PMC free article] [PubMed] [Google Scholar]