Abstract
Infection of the colon with the Gram-positive bacterium Clostridium difficile is potentially life threatening, especially in elderly people and in patients who have dysbiosis of the gut microbiota following antimicrobial drug exposure. C. difficile is the leading cause of health-care-associated infective diarrhoea. The life cycle of C. difficile is influenced by antimicrobial agents, the host immune system, and the host microbiota and its associated metabolites. The primary mediators of inflammation in C. difficile infection (CDI) are large clostridial toxins, toxin A (TcdA) and toxin B (TcdB), and, in some bacterial strains, the binary toxin CDT. The toxins trigger a complex cascade of host cellular responses to cause diarrhoea, inflammation and tissue necrosis — the major symptoms of CDI. The factors responsible for the epidemic of some C. difficile strains are poorly understood. Recurrent infections are common and can be debilitating. Toxin detection for diagnosis is important for accurate epidemiological study, and for optimal management and prevention strategies. Infections are commonly treated with specific antimicrobial agents, but faecal microbiota transplants have shown promise for recurrent infections. Future biotherapies for C. difficile infections are likely to involve defined combinations of key gut microbiota.
Clostridium difficile is a Gram-positive obligate anaerobic bacterium that was originally identified as part of the flora of healthy infants in 1935, described as an “actively motile, heavy-bodied rod with elongated subterminal or nearly terminal spores” (REF. 1) (FIG. 1). At that time, the strain was named Bacillus difficilis to reflect the difficulty experienced in isolating and culturing it. Despite the organism being present as a commensal bacterium in neonates, researchers noted that it could induce disease in animals, probably due to the production of a secreted toxin1. Subsequent work established the high molecular weight clostridial toxins toxin A (TcdA) and/or toxin B (TcdB) as the main virulence (disease-causing) factors of C. difficile2,3. A hallmark feature of C. difficile that sets it apart from other species in the class Clostridia is its ability to decarboxylate parahydroxyphenylacetic acid to produce p-cresol, which gives C. difficile its characteristic tar-like or pig-like smell4.
Figure 1. Clostridium difficile.
a | Typical image of Clostridium difficile colonies on a blood agar plate. b | Phase-contrast microscopy image of a C. difficile culture with vegetative cells (elongated rods), phase-dark spores (subterminal dark spots) and phase-bright spores (bright ellipsoids). Inset, Gram stain of culture. c | Scanning electron micrograph of C. difficile spores. d | Endoscopic picture of pseudomembranous colitis caused by C. difficile. Healthy colon tissue is pink, pseudomembranes resulting from C. difficile infection are yellow.
It was not until the 1970s that a detailed characterization of the bacterium, then called C. difficile, revealed its involvement in human disease5. This disease became widely known as C. difficile-associated disease/diarrhoea (CDAD); more recently, the term C. difficile infection (CDI) is preferred. In the early 2000s, an increase in severe cases of CDI was noted in Canada, the United States and Europe6, which was attributed to the emergence of certain epidemic types of C. difficile7,8. The first complete genome sequence for C. difficile9, together with the development of tools for the genetic manipulation of C. difficile10,11, has greatly stimulated research on the bacterium. C. difficile is now recognized as the leading cause of health-care-associated infective diarrhoea and is increasingly being linked to community-acquired cases of colitis12. C. difficile can be found in the intestinal tracts of both humans and animals, but its spores are also ubiquitous in the environment and can be isolated from food13. Importantly, people with an adequate immune response will either eliminate the infection and/or become asymptomatic carriers14. In 2013, it was proposed that C. difficile should be reclassified as Peptoclostridium difficile on the basis of a detailed phylogenetic analysis15, and this has been adopted by the US National Center for Biotechnology Information. However, considering the public awareness of the disease, the large body of scientific literature using C. difficile and the lack of formal acceptance of this proposal, we will refer to the organism as C. difficile throughout this Primer, and to the disease it causes as CDI. We describe key aspects of the epidemiology of C. difficile infections, the mechanisms underlying the disease, strategies for diagnosis, prevention and management, and summarize the impact that CDIs have on patients and society.
Epidemiology
Molecular typing (BOX 1) is the characterization of organisms beyond the species level, enabling clustering of individual bacterial isolates in a meaningful manner16. Typing is crucial for epidemiological studies and facilitates effective infection prevention and disease management. Different types of C. difficile are known; here, we will refer to PCR ribotypes when relevant, a typing system that is based on a banding pattern obtained from PCR amplifying ribosomal 16S–23S intergenic spacer sequences17. Certain PCR ribotypes of C. difficile (such as PCR ribotype 010) are non-pathogenic as they lack the toxin genes. Epidemic PCR ribotypes are distinguished from non-epidemic types by their frequent occurrence in multiple settings across several countries.
Box 1. Molecular typing of Clostridium difficile.
Epidemiological studies are dependent on standardized typing methods. Many typing methods have been developed for Clostridium difficile that evaluate either phenotypic or genotypic traits17 (see the figure). Given their high reproducibility, typability (ability to type a strain unambiguously) and discriminatory power, genotyping methods have become standard for typing of C. difficile. Band-based typing methods, such as restriction enzyme analysis (REA), pulsed-field gel electrophoresis (PFGE) and PCR ribotyping, are common. Historically, REA and PFGE have been the methods of choice in North America, whereas PCR ribotyping has primarily been used in Europe. As a result, epidemic strains are often indicated with multiple typing designations247. For instance, PCR ribotype 027 strains that have caused outbreaks globally8 have been classified as REA group BI and PFGE type NAP1. Similarly, PCR ribotype 078 strains are known as REA group BK and PFGE type NAP7/NAP8. Global surveillance is becoming key for health-care management, and efforts have been made to harmonize the different typing schemes. Capillary gel-based electrophoresis ribotyping has been standardized and validated in a collaborative effort of the European Centre for Disease Control and Prevention, the US Centers for Disease Control and Prevention and the Public Health Agency of Canada, and is likely to become commonplace throughout the world248. In addition, sequence-based methods, such as multilocus sequence typing (MLST) and whole-genome sequencing (WGS; specifically single-nucleotide polymorphism (SNP) typing), have gained interest specifically to study evolutionary relationships between various C. difficile strains (phylogeny)17. Overall, different lineages can be discriminated and several different PCR ribotypes have been shown to be closely related to the epidemic types using these sequence-based methods98,230. WGS is also used to study transmission and outbreaks25,231, although this approach can be costly and is predominantly performed retrospectively. Relatedness of strains in an outbreak setting is more commonly performed using multilocus variable-number tandem repeat analysis17.

Owing to a lack of systematic surveillance, no comprehensive data are available on circulating C. difficile types before 2003. After 2003, large increases in incidence and mortality rates in North America — and subsequently in several European countries — were observed, which were associated with PCR ribotype 027 (REF. 8) and, to a lesser extent, PCR ribotype 078 (REF. 7). Such increases have also been observed in Australia, Asia and Central America after 2008 (REF. 18). It should be noted, however, that CDI and CDI-related epidemics are not limited to these types. Ribotypes 001, 002 and 014/020 frequently cause CDI clusters in the United States and Europe19,20. Furthermore, outbreaks have also been reported for strains of other PCR ribotypes, such as 017 (REF. 12), 018 (REF. 21), 106 (REF. 12), 176 (REF. 22) and 244 (REF. 23). Epidemiological data on C. difficile in Africa and the Middle East are sparse12.
CDI is historically regarded as a nosocomial infection; antibiotic exposure (either prophylactic or as treatment) during hospitalization is the foremost risk factor for CDI. However, C. difficile is increasingly being recognized as a cause of community-associated diarrhoea. Indeed, PCR ribotypes of strains isolated from patients with community-associated CDI have a large overlap with strains cultured from patients with health-care-associated CDI in an endemic setting, suggesting a common source (or sources) of C. difficile24,25. The incidence of community-associated CDI is estimated at 30 to 120 cases per 100,000 persons per year in the United States26. The incidence of community-associated CDI in the Netherlands is estimated at 390 to 730 per 100,000 person years, similar to the incidence of Campylobacter spp. infection and higher than the incidence of Salmonella spp. infection27. More than 30% of patients who develop community-associated CDI do not have typical risk factors for CDI, such as antibiotic treatment or recent hospitalization (see below)28,29.
The US Centers for Disease Control and Prevention (CDC) estimated that almost 500,000 patients had CDI, with 29,000 attributable deaths in the United States in 2011 (REF. 26). However, there is uncertainty regarding the precision of these estimates given the accuracy of the testing methodology, although data have been adjusted based on the frequency of nucleic acid amplification test use. Between 2001 and 2010, the incidence of CDI among hospitalized adults in the United States approximately doubled according to International Classification of Diseases (ICD)-9 discharge diagnoses30. Again, ascertainment bias secondary to awareness (frequency of requesting) and to diagnostic methods could be relevant here.
The first European Centre for Disease Prevention and Control (ECDC) point-prevalence survey in 2011 and 2012 estimated that ~124,000 patients developed health-care-associated CDI within the European Union each year31. However, this surveillance was performed with a variety of diagnostic tests, and CDI was often only tested upon physician request, resulting in considerable underestimation of the true CDI incidence; on average, ~80 cases of CDI are missed per hospital per annum in Europe32. A pilot study supported by the ECDC using standardized CDI surveillance performed in 37 hospitals in 14 European countries demonstrated large differences in the incidence of CDI per hospital and per country33. The incidence of health-care-associated CDI by hospital was in the range of 4.2–131.8 per 10,000 discharges (median 16.4 per 10,000 discharges) and 0.6–18.5 per 10,000 patient days (median 3.7 per 10,000 patient days). In some countries, a high incidence of PCR ribotype 027 strains has been noted, but other countries demonstrated a decrease in the incidence of these strains, most probably as a consequence of effective management strategies (see below)32.
The all-cause mortality associated with CDI due to non-epidemic PCR ribotypes has been reported to be ~15–20% within a month of diagnosis, and approximately half of the deaths are directly attributed to CDI34. Given the multiple comorbidities (such as respiratory disease and renal failure) typically present in patients with CDI, mortality is often related to one or more of these conditions.
C. difficile is a human pathogen but can also infect and cause disease in animals that can enter the food chain; however, the relevance of foodborne transmission in human disease is unclear13. Since the beginning of 2000, C. difficile has been reported as a major cause of neonatal enteritis in piglets. The predominant strains found in piglets (belonging to PCR ribotype 078) are similar to isolates from some human patients with CDI and to isolates from asymptomatic farm workers, suggesting zoonotic transmission13,35. As a consequence of increasing pressure from spores present in the environment and animals, humans might become colonized more frequently. One meta-analysis revealed an increased rate (>8% of admitted patients) of asymptomatic colonization by toxigenic isolates in patients at the time of admission to the hospital36. It should be noted, however, that patients who have been admitted to hospital typically have a much higher colonization rate than people in the community, because of exposure to a hospital environment, comorbidities and/or antibiotic use.
Mechanisms/pathophysiology
C. difficile lifecycle
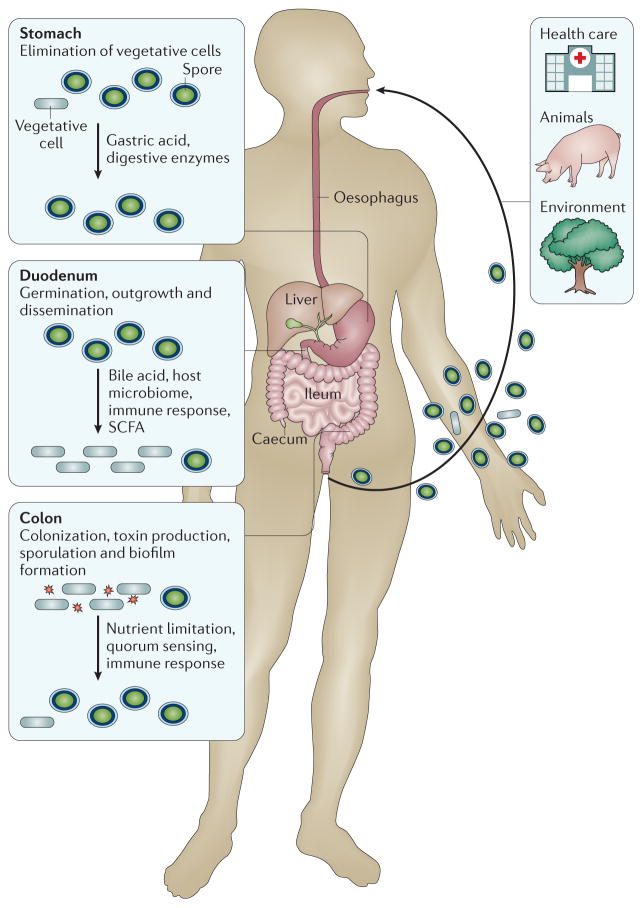
C. difficile is transmitted via the oral–faecal route (FIG. 2). Spores are dormant cells that are highly resistant to environmental conditions37, including some disinfectants and many antimicrobials (BOX 2), which generally target metabolically active cells. Spores are thought to be the infectious vehicle given that vegetative (metabolically active) cells of obligate anaerobic bacteria are unlikely to survive the oxygenated environment outside the host or the acidic environment of the stomach. Indeed, an asporogenic strain of C. difficile is unable to persist in the environment or transmit between hosts38.
Figure 2. Stages of the Clostridium difficile life cycle in the human gastrointestinal tract.
Three sources of infection (health care, animal and environmental) are indicated. A range of host factors influence the Clostridium difficile life cycle, as well as the relative numbers of spores and vegetative (metabolically active) cells in the gut. Note that passage through the stomach eliminates most vegetative cells (but spores survive), and spores germinate and grow out in the duodenum. In the caecum and colon, C. difficile starts producing spores again, and vegetative cells are excreted by the patient during infection. Toxin is produced in the colon. As C. difficile is an obligate anaerobic bacterium, transmission occurs primarily via spores. SCFA, short-chain fatty acid (such as butyrate).
Box 2. Antibiotic resistance of Clostridium difficile.
Most antimicrobial compounds target metabolically active cells and have limited or no activity against dormant cells, such as spores. This intrinsic resistance of spores ensures that Clostridium difficile can persist in the presence of antibiotics or in the host immune system249,250. C. difficile also demonstrates extensive acquired antimicrobial resistance9,251. Interestingly, C. difficile vegetative cells are sensitive to teicoplanin and vancomycin despite harbouring a genomic region that resembles a vanG glycopeptide resistance cluster9,252. This cluster can confer vancomycin resistance to a heterologous host, but why it is not functional in C. difficile is unclear253. The mobile genome of C. difficile (which consists of transposons, insertion sequences and (pro)phages) probably contributes to antibiotic resistance because these elements commonly contain resistance determinants9,254. For example, the Tn6218 element of C. difficile contains a cfr-like gene that can confer resistance to peptidyl transferase inhibitors such as linezolid255 and the Tn5397 element carries a tetracycline resistance determinant256. Reduced susceptibility or resistance to the commonly used antibiotics vancomycin, metronidazole and fidaxomicin has been noted257–260. Although this is cause for concern, the clinical relevance of resistance to these antibiotics so far is limited. However, C. difficile antibiotic resistance is only part of the reason why C. difficile has been classified as an Urgent Antibiotic Resistance Threat by the US Centers for Disease Control and Prevention261. Other major factors include the bacterium affecting people treated by antibiotics for other infections, the ageing of the general population and the emergence of epidemic types, such as PCR ribotype 027. At least in this ribotype, epidemic lineages are associated with resistance against fluoroquinolones8; although this class of antibiotics is not used for the treatment of C. difficile infections, the antibiotics can select for C. difficile when used to treat other infections. Fluoroquinolone resistance is also common in other PCR ribotypes262.
Germination of spores is dependent on sensing primary bile acids from the liver, such as taurocholate, by the germinant receptor CspC and is inhibited by secondary bile acids in the colon37,39,40. In addition, glycine can act as a germinant through an uncharacterized mechanism41. A proteolytic cascade then leads to the degradation of the spore peptidoglycan, release of calcium dipicolinic acid and rehydration of the spore, ultimately resulting in outgrowth of the cells37,40.
The propensity of spores to outgrow and colonize the intestine is greatly influenced by the host microbiota and its associated metabolome39,42. For example, antibiotic-induced shifts in the microbiota can generate an environment conducive to C. difficile infection43. Mucolytic enzymes, such as cell surface protein Cwp84 (REF. 44), are secreted by the bacterium and degrade the colonic mucosa. Bacterial cell surface-associated proteins have been shown to affect the adhesion of the bacterium to colon epithelial cells in vitro45–48; mutations in genes encoding these proteins, or in the genes encoding proteins for their processing, generally attenuate virulence49,50. The expression of at least a subset of colonization factors by the bacterium, such as cell surface protein Cwp84 and surface layer protein A (SlpA), is stimulated in the presence of the antibiotics ampicillin and clindamycin51.
Before germination into vegetative cells, spores are also capable of adhering to colon cells52. C. difficile is a motile bacterium, and the switch between the sessile and motile phase is regulated by the secondary messenger cyclic di-GMP53–55. C. difficile is also capable — at least in vitro — of forming robust biofilms56–58. Cell–cell signalling contributes to both colonization and virulence factor expression59,60. Host recognition of C. difficile is mediated via pattern recognition receptors and myeloid differentiation primary response 88 (MYD88)- and nucleotide-binding oligomerization domain-containing protein 1 (NOD1)-dependent pathways61,62 (FIG. 3). Specifically, SlpA can activate a Toll-like receptor 4 (TLR4)-dependent response63, flagellin can stimulate TLR5 (REF. 64) and NOD1 is probably activated via peptidoglycan-derived compounds65. The first line of host defence against C. difficile is the production of antimicrobial compounds such as lysozyme and cationic antimicrobial peptides66,67. Interestingly, the bactericidal α-defensins, but not β-defensin 1 or cathelicidin antimicrobial peptide (also known as LL-37), can also inhibit the activity of the TcdB (see below) through a mechanism that involves direct binding to the toxin66. Innate lymphoid cells, specifically class 1, are implicated in resistance against CDI68.
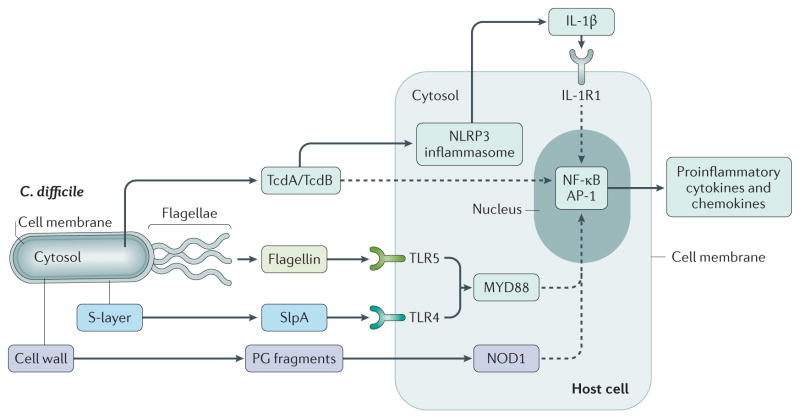
Figure 3. Innate immune response of host cells to Clostridium difficile.
Clostridium difficile elicits the innate immune response via at least four different effectors, leading to the induction of proinflammatory cytokines and chemokines via nuclear factor-κB (NF-κB) and transcription factor AP-1. The large clostridial toxins toxin A (TcdA) and TcdB act via NLRP3 (NOD-, LRR- and pyrin domain-containing 3) inflammasome-dependent and -independent pathways. Surface layer protein A (SlpA) and flagellin act via myeloid differentiation primary response 88 (MYD88)-dependent pathways through Toll-like receptor 4 (TLR4) and TLR5, respectively. The nucleotide-binding oligomerization domain-containing protein 1 (NOD1)-dependent pathway of induction most probably detects fragments of peptidoglycan (PG) derived from the cell wall of C. difficile. Dashed lines indicate indirect effects. IL-1R1, IL-1 receptor type 1.
Resistance of the bacterium to host-produced antimicrobial compounds is multifactorial69 and mediated by, for example, the extracytoplasmic function (ECF) sigma factor CsfV70, the site-1 protease PrsW71, the proteins encoded by the cpr locus72 and modulation of cell wall charge73.
The large clostridial toxins TcdA and TcdB
Regulation of expression
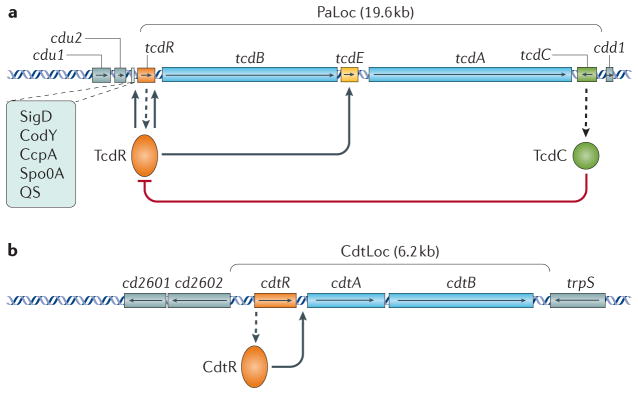
Although several virulence factors contribute to the retention of C. difficile within the gastrointestinal tract74,75, the symptoms of CDI correlate with the presence of a toxin-encoding pathogenicity locus (PaLoc) in the bacterial genome76,77. In most strains, the PaLoc is located at the same site in the chromosome77,78. The PaLoc in most pathogenic C. difficile strains encodes two large homologous toxins (TcdA and TcdB) and three proteins that seem to regulate toxin production and secretion (TcdR, TcdE and TcdC)74,75,78 (FIG. 4a).
Figure 4. Regulation of the Clostridium difficile toxins.
a | Schematic representation of the pathogenicity locus (PaLoc) and the flanking regions with regulatory interactions of Clostridium difficile. Boxes with arrows indicate open reading frames with the direction of the arrows showing the direction of transcription. Toxin genes tcdA and tcdB are in blue, regulatory genes are in orange (positive) and green (negative); tcdE is in yellow and genes located outside the PaLoc are in grey. Dashed arrows indicate the production of protein from a gene transcript. Other regulators (Sigma D (SigD), the nutritional repressor CodY (known as GTP-sensing transcriptional pleiotropic repressor CodY), catabolite control protein A (CcpA), Stage 0 sporulation protein A (Spo0A) and quorum sensing (QS)) that affect toxin gene transcription (boxed) mostly act via expression of the tcdR gene. The TcdR protein is involved in the initiation of the production of toxin A (TcdA) and TcdB. b | Schematic representation of the binary toxin locus (CdtLoc) and flanking regions with regulatory interactions. Boxes with arrows indicate open reading frames with arrows showing the direction of transcription. The cdtA and cdtB toxin genes are in blue, the regulatory gene cdtR is in orange and genes located outside the CdtLoc are in grey.
TcdR is a member of the ECF family of alternative sigma factors and is critical for the initiation of TcdA and TcdB production in C. difficile79,80. TcdC is thought to encode an anti-sigma factor that negatively regulates toxin production81. Epidemic ribotype 027 strains carry a nonsense mutation within the tcdC gene, leading to the suggestion that derepression of the toxin genes by the inactivation of TcdC might contribute to the increased virulence of these strains82. However, despite many studies aimed at defining the role of TcdC in toxin production and virulence, conflicting findings have been reported and the functional role of this protein remains unclear75.
TcdE has homology with bacteriophage holin proteins, which are involved in the release of progeny phages from infected bacterial cells83. The role of TcdE is also poorly understood, although some evidence suggests that it facilitates toxin (TcdA and TcdB) secretion78,84,85. TcdA and TcdB do not possess any recognizable signal or export sequences, suggesting that they might be exported from the bacterial cell by lysis or a non-classic secretion pathway, possibly involving TcdE84,85.
The synthesis of TcdR and the subsequent activation of tcdA and tcdB expression is influenced by many environmental stimuli, including short-chain fatty acids such as butyric acid that are common in the gut and sub-inhibitory concentrations of certain antimicrobials that may be relevant in the context of disease79,86–89. Amino acids such as proline, cysteine and certain branched chain amino acids in the local environment of the bacterium repress toxin production through the action of the global transcriptional regulator CodY (known as GTP-sensing transcriptional pleiotropic repressor CodY)90. The presence of glucose or other rapidly metabolizable carbon sources in the local environment of the bacterium also inhibits the production of TcdA and TcdB via the carbon catabolite control protein A (CcpA)91,92. The sigma factor SigD, which is associated with the expression of motility genes, promotes toxin gene expression by binding to a SigD-dependent promoter sequence upstream of tcdR93,94. The master regulator of sporulation in both Bacillus and Clostridium species, Spo0A, can also regulate toxin production in C. difficile, but only in some strains95. Specifically, Spo0A negatively regulates TcdA and TcdB production in epidemic type PCR ribotype 027 strains but not in others that have been tested95–97. These results highlight the heterogeneous nature of C. difficile isolates and the need to study strains belonging to distinct evolutionary lineages95,98. Finally, growth signals and cell density play an important part in toxin regulation59,60. Cell–cell signalling is partially dependent on an accessory gene regulator quorum signalling system, which is mediated by a novel thiolactone quorum signalling peptide59,60. Overall, C. difficile toxin synthesis is closely connected to the metabolic state of the bacterium and its environment99.
Mechanism of action
Structural and functional studies have provided insight into the mechanisms of action of all of the C. difficile toxins, particularly TcdA and TcdB. Once secreted, TcdA and TcdB bind and enter the colonic epithelium to cause inflammatory chemokine and cytokine production, an influx of neutrophils, disruption of tight junctions, fluid secretion and epithelial cell death100 (FIGS 5,6). Given the homology between the two proteins, it is notable that TcdA and TcdB have markedly different functions in animal toxicity models. Historically, TcdA has been viewed as the more potent enterotoxin, as administration of purified TcdA into the intestines of rabbits and rodents causes tissue necrosis and an intense infiltration of immune cells101–103. High levels of TcdB in identical experiments failed to induce these effects, although it should be noted that most of these studies were conducted in an ileal loop model and, accordingly, represent only the response of the small intestine. In studies involving human colonic tissue, TcdB seems to be a potent inflammatory toxin104,105, whereas TcdA is weaker104. These data suggest that the differential toxin responses might partially stem from differences in receptor tropism and highlight the importance of conducting mechanistic studies within the colon, which is the site of bacterial outgrowth in the host.
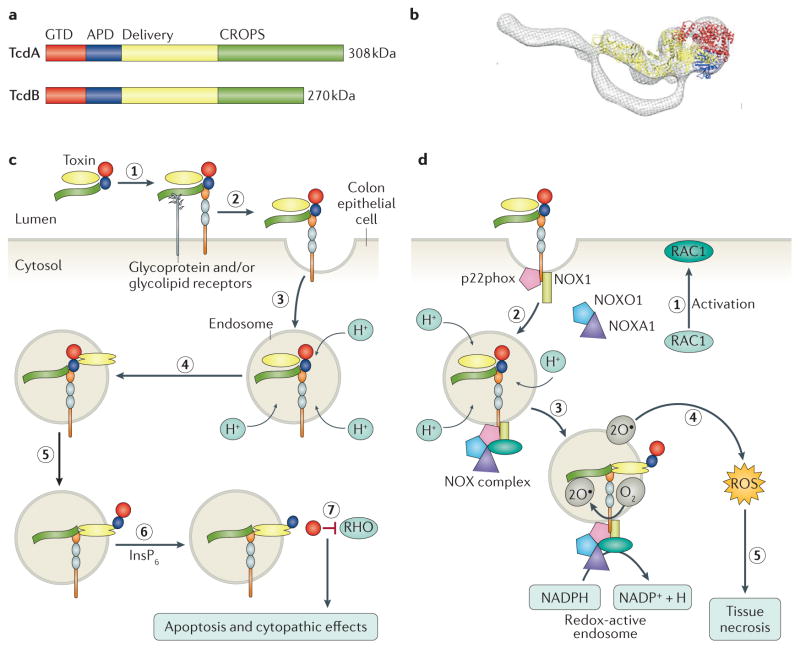
Figure 5. Structure and function of the large clostridial toxins.
a | Schematic of the toxin A (TcdA) and TcdB primary structures, highlighting the four functional domains: the glucosyltransferase domain (GTD; red), the autoprotease domain (APD; blue), the delivery domain (yellow) and the combined repetitive oligopeptides (CROPS) domain (green) that binds carbohydrates on the host cell surface to facilitate bacterial entry. b | Overlay of an electron microscopy reconstruction of the structure of TcdA with the X-ray crystal structure of TcdA lacking the CROPS domain (Protein Data Bank code: 4R04 ). Colour-coding reflects the domain structure in part a. c | The discrete structural and functional domains of the toxins contribute to a multi-step mechanism of intoxication. Toxins bind to one or more receptors (carbohydrate and/or protein) on the cell surface (step 1) and are internalized by receptor-mediated endocytosis (step 2). As the endosome matures, the V-ATPase contributes to a decrease in pH (step 3). The acidic pH causes a conformational change in the toxin delivery domain, resulting in pore formation (step 4) and the translocation of the APD and GTD into the cytosol (step 5). Inositol hexakisphosphate (InsP6) binds and activates the APD, resulting in the release of the GTD (step 6), which can inactivate RHO family proteins (step 7) to cause apoptosis and cytopathic ‘rounding’ effects. d | At concentrations >0.1 nM, TcdB can promote RAS-related C3 botulinum toxin substrate 1 (RAC1) activation (step 1) and complex formation between p22phox (also known as cytochrome b-245 light chain), NADPH oxidase 1 (NOX1), NADPH oxidase activator 1 (NOXA1), NADPH oxidase organizer 1 (NOXO1) and RAC1 on the endosomal membrane to form the NOX complex (step 2). The fully assembled NOX complex generates superoxide by transferring an electron from NADPH to molecular oxygen (step 3). Superoxide generation leads to the production of reactive oxygen species (ROS), which — at high levels — promote necrosis by causing mitochondrial damage, lipid peroxidation and protein oxidation (step 5).
Figure 6. Histopathology of Clostridium difficile infection in a mouse model.
Histopathological analysis of haematoxylin and eosin-stained caecal and colonic tissues collected from mice infected with a wild-type PCR ribotype 027 strain (TcdA+ TcdB+), infected with an isogenic double tcdA and tcdB mutant (TcdA− TcdB−) or mock-infected with phosphate buffered saline (Mock). Note that both wild-type and double-mutant strains contain an intact binary toxin locus. Arrows indicate major histological differences; oedema and polymorphonuclear cell influx into the lamina propria (black arrows), erosion of crypts and goblet cell loss (yellow arrows) and hyperplasia (white arrows).
TcdA and TcdB have four functional domains100,106: an amino-terminal glucosyltransferase domain (GTD), an autoprotease domain (APD), a pore-forming and delivery domain, and the combined repetitive oligopeptides (CROPS) domain, which extends from around residue 1,830 to the carboxyl terminus (FIG. 5a,b). A combination of electron microscopy and X-ray crystallography studies has revealed the structural organization of these domains in TcdA and suggests that the structure of TcdB is similar106,107. TcdA and TcdB enter cells via receptor-mediated endocytosis108. Historically, receptor binding has been associated with a CROPS domain located at the C termini of TcdA and TcdB109. The CROPS domain is capable of binding carbohydrates, which is consistent with the model wherein TcdA engages glycosylated receptors110. Evidence supporting the idea that the TcdA CROPS domain contributes to receptor binding includes the observations that antibodies against the TcdA CROPS domain can block intoxication111 and that excess TcdA CROPS domain can compete with TcdA holotoxin for cell binding112. TcdA binds a variety of carbohydrates, and although multiple glycolipids and glycosylated proteins have been proposed as receptors109, the specific receptors used to bind human epithelial cells remain unknown. However, accumulating evidence suggests that domains other than CROPS participate in receptor binding. Indeed, TcdA and TcdB lacking CROPS domains are still capable of intoxicating cells113,114, and the homologous toxin TpeL from Clostridium perfringens lacks a CROPS domain entirely115. Recently, two protein receptors have been reported for TcdB: poliovirus receptor-like protein 3 (PVRL3; also called nectin 3)116 and chondroitin sulfate proteoglycan 4 (CSPG4)117. PVRL3 is highly expressed on the surface of human colon epithelial cells and co-localizes with TcdB in tissue resected from a C. difficile-infected individual116, suggesting that PVRL3 could serve as the initial receptor that TcdB encounters in the context of infection. CSPG4 is highly expressed in the intestinal subepithelial myofibroblasts of mouse and human intestines118, suggesting that this receptor could be engaged after initial damage to the colonic epithelium117. Both CSPG4 and PVRL3 bind outside the CROPS domain116,117. It is conceivable that additional alternative receptors for TcdB exist.
Following receptor binding and endocytosis, acidification of the endosome is thought to trigger a structural change in the delivery domain, facilitating pore formation and translocation of the GTD into the cytosol109 (FIG. 5c). The APDs share sequence homology with the cysteine protease of the MARTX family of toxins119,120, but one publication has shown that the catalytic dyad of TcdA and TcdB serves to coordinate a zinc ion that is essential for function106. Activation of the APD by eukaryotic inositol hexakisphosphate (InsP6) results in the release of the GTD into the cell, enabling access to cytosolic substrates119,120. Host S-nitrosylation at the conserved cysteine of the APD can inactivate the protease activity in an InsP6-dependent manner121. It has been hypothesized that the autoprotease and translocation efficiency of the epidemic PCR ribotype 027 strain contribute to its increased virulence122. The GTD transfers glucose from UDP glucose onto RHO family GTPases such as RHO, RAC1 and cell division control protein 42 (CDC42)123,124. These modifications cause a cytopathic effect resulting from rearrangement of the actin cytoskeleton and can lead to apoptosis125 (FIG. 5c). At higher concentrations, TcdB is also capable of coordinating the assembly of the NADPH oxidase complex on endosomes126. The resulting production of reactive oxygen species leads to cell death by a necrotic mechanism126 (FIG. 5d). Both mechanisms might be important in the context of disease; cytopathic effects promote inflammation and disruption of the tight junctions, whereas TcdB-induced necrosis contributes to the colonic tissue damage observed in severe cases of CDI.
Knowledge of the mechanisms of action of C. difficile toxins has served as a foundation for several preclinical studies aimed at identifying small-molecule inhibitors of intoxication, particularly those that are already approved for other uses127. A recent screen for inhibitors of TcdB-induced cytopathic effects identified compounds that act by inhibiting toxin binding to cells, endosomal maturation or glucosyltransferase function128. Another screen, conducted using an activity-based probe for inhibitors of the APD, identified ebselen, a FDA-approved compound, which reduced tissue pathology in a mouse infection model129. Notably, ebselen has also been reported to block NADPH oxidase 1 activity130, a function that could contribute to a decrease in toxin-induced production of reactive oxygen species. Similarly, N-acetylcysteine, an FDA-approved antioxidant, prevented TcdB-induced tissue damage in a colonic explant model126.
Pathology
Although the roles of TcdA and TcdB in the context of CDI have been difficult to assess, recent progress has come through advances in the genetic manipulation of C. difficile. Four studies have been conducted in both hamster and mouse models of infection. All studies indicate that TcdB is capable of inducing the phenotypes of disease in the absence of TcdA, but differ on the interpretation of the role of TcdA in the absence of TcdB on the survival of animals2,3,131,132. Histological examination of colonic and caecal tissue from mice infected with TcdB-positive C. difficile strains (either wild-type TcdA+ TcdB+ or TcdA− TcdB+ mutants) showed severe gut damage associated with eroded and often absent crypts, mucosal ulceration and goblet cell loss132. Polymorphonuclear cell influx into the lamina propria, enterocyte hyperplasia and severe submucosal oedema associated with haemorrhage were also observed in these tissues. TcdB-negative strains (TcdA+ TcdB−) caused less tissue damage that was confined to mild oedema and polymorphonuclear cell influx132. Tissue damage was strictly dependent on TcdB or TcdA given that tissues from mice infected with a strain that did not produce TcdA or TcdB (TcdA− TcdB−) resembled those of mock-infected control animals132 (FIG. 6).
Consistent with the finding that TcdB is independently capable of causing disease, a considerable number of clinical C. difficile isolates express only TcdB133. The prevalence of these strains, which include PCR ribotype 017, has been increasing, sometimes to epidemic proportions134. Recently, the first strain with an intact tcdA gene, but no tcdB gene, in a different genomic context than the PaLoc has been characterized78. This work has raised the hypothesis that the single toxin-encoding loci might have fused to form the typical two-toxin locus (PaLoc), which is the most common form currently detected in clinical isolates. The study also suggests a conserved relationship between the presence of toxin genes and holin genes, and demonstrates that the PaLoc does not always encode a tcdC homologue78. However, it should be noted that confirmed clinical cases of CDI caused by strains that only produce TcdA are extremely limited78.
Binary toxin CDT
Regulation of expression
C. difficile transferase (CDT; or binary toxin) is a third toxin produced by some C. difficile strains, including the epidemic PCR ribotypes 027 and 078. CDT has received increased attention in recent years because of its increasing prevalence in isolates of both human and animal origin135. CDT is encoded by two genes, cdtA and cdtB, that are located in an operon on the binary toxin locus (CdtLoc)136,137 (FIG. 3b). In CDT-negative strains, this locus contains an ~2 kb deletion138. The CdtLoc also harbours a response regulator gene, cdtR, upstream of the cdtAB operon139. CdtR is an orphan LytTR-like positive transcriptional regulator of the cdt operon and CDT production. The cognate sensor histidine kinase that interacts with the orphan histidine kinase CdtR has yet to be identified139. A truncation in the C. difficile PCR ribotype 078 CdtR does not abrogate CDT expression, suggesting that full-length CdtR is not essential for expression140. In contrast to the PaLoc, the environmental signals that regulate the expression of the CdtLoc genes are not known.
Mechanism of action
Recent studies have provided insight into the mechanisms of action of CDT, although the role of this toxin in disease pathogenesis remains unclear. CDT belongs to the binary ADP-ribosylating toxin family and consists of two components: an enzymatic component (CDTa) and a binding/translocation component (CDTb). CDTa has ADP ribosyltransferase activity, whereas CDTb facilitates the passage of the enzymatic component to the cell cytosol135 (FIG. 7). CDTa leads to the complete destruction of the actin cytoskeleton and, ultimately, cell death135,141.
Figure 7. Mechanism of action of Clostridium difficile transferase (binary toxin).
Clostridium difficile transferase (CDT) is a binary toxin consisting of the CDTa ADP-ribosyltransferase (red) and the CDTb protein (yellow and green). The monomeric form of CDTb binds to the lipolysis-stimulated lipoprotein receptor (LSR)263, which is found in many tissues, including the gut. CDTb undergoes proteolytic activation and oligomerizes to form a heptameric prepore, which facilitates the binding of CDTa to the prepore–receptor complex. This complex enters cells by endocytosis and as the endosome matures, the V-ATPase contributes to a decrease in pH. The low pH of the endosome triggers pore formation and the translocation of CDTa into the cell. Once in the cytosol CDTa, ribosylates actin at arginine 177, resulting in a dual effect whereby G-actin polymerization is inhibited and F-actin depolymerization is favoured, which leads to the complete destruction of the actin cytoskeleton and, ultimately, cell death135,141. Inset, structure of CTDa (Protein Data Bank code: 2WN7 ).
Pathology
Despite a thorough understanding of the mechanism of action of CDT on intoxicated cells, the role of this toxin in disease pathogenesis is unclear. Experimental data have suggested that CDT results in the formation of microtubule-based protrusions on epithelial cells that might increase the adherence and colonization of C. difficile142. Importantly, the increasing presence of CDT in clinically relevant strain types commonly associated with severe CDI, such as PCR ribotypes 027 and 078, and the isolation of TcdA− TcdB− CDT+ strains suggest that this toxin is likely to play an important but as yet undefined part in CDI143,144.
Experimental models
Multiple experimental models have been developed as a proxy for CDI and its treatment in humans145–147. The most common models are the female golden (Syrian) hamster model, which is exquisitely sensitive to toxin and is primarily suited to study acute disease, and the mouse model. The mouse model mimics certain aspects of human disease that are difficult to assess in hamsters; for instance, mice can be colonized asymptomatically, they exhibit differential sensitivity towards different PCR ribotypes and they can experience relapsing disease. For these reasons, the model is suitable to study colonization, transmission and persistence phenotypes38,148,149. A piglet model is of particular interest to study C. difficile strains that are problematic in both animal and human populations13,35,150. In vitro gut models have been developed to study interactions of C. difficile with therapeutics in the context of a complex microbiome13,151,152. Each model is greatly influenced by variables such as qualitative and quantitative differences in inoculum and the choice of C. difficile strain147. Furthermore, non-animal models (for example, the in vitro gut models) may not fully reflect the interaction with the host, but some have been shown to be more reflective of human CDI than animal models147.
Diagnosis, screening and prevention
Symptoms and risk factors
The clinical symptoms associated with CDI range from mild, self-limiting diarrhoea to fulminant colitis, and can include pseudomembranous colitis (FIG. 1d), toxic megacolon (severe dilatation of the colon), bowel perforation and sepsis, and/or multiple organ dysfunction syndrome14,132,153. Given that the characteristics (for example, age, use of medication or underlying comorbidities) of patients who can acquire CDI are highly variable, there is considerable variation in the possible severity assessments for this disease, which is reflected in the differing criteria used in guidelines154,155. Severe diarrhoea associated with C. difficile is often accompanied by a typical endoscopic picture of pseudomembranous colitis with haemorrhage and deep ulcerations. Toxic megacolon is considered the most serious disease entity and is characterized by systemic toxicity and high mortality. Extra-intestinal manifestations of CDI (including bacteraemia) are extremely rare, and this emphasizes that it is the localized effects of toxins, associated with depleted intestinal microbiota, that cause the range of signs and symptoms of CDI. C. difficile toxins in sera from patients with CDI can be detected with an ultrasensitive cell-based assay156, but studies are required to assess the relationship between severe CDI and levels of toxaemia.
Known risk factors for CDI are previous hospitalization, underlying disease, advanced age (>65 years) and, most importantly, the use of antibiotics. All antibiotic classes can be associated with CDI, but clindamycin, cephalosporins and fluoroquinolones are the most frequently cited157. Antibiotic-induced dysbiosis of the protective intestinal microbiota often underlies C. difficile outgrowth and toxin production39,43. Thus, even low-risk antibiotics (such as trimethoprim and piperacillin-tazobactam) can predispose the patient to CDI, especially when two or more courses of (different) antibiotics are prescribed; the cumulative damage to the intestinal microbiota could be sufficient to enable C. difficile to proliferate.
In addition to the antibiotic class, the number of administered antibiotics, dose and duration of therapy have been identified as risk factors for CDI. Given that disruption of the intestinal flora persists for >3 months after antibiotic therapy, patients can remain susceptible to CDI development long after ending the treatment28. Acid suppression by proton pump inhibitors (commonly used for dyspepsia, peptic ulcer disease and gastroesophageal reflux disease) has frequently been associated with CDI158,159 but the precise role of proton pump inhibitors in CDI (and a causal relationship) remains unclear160,161. Of all patients with antibiotic-associated diarrhoea, 20–30% is caused by C. difficile162. A differential diagnosis could consider a role for Staphylococcus aureus, C. perfringens, Clostridium sordellii or Klebsiella oxytoca as causative agents of antibiotic-associated diarrhoea163.
Diagnosis
The mainstay for diagnosing CDI is the presence of clinical symptoms plus a well-chosen laboratory assay. The diagnostic tests for C. difficile can be divided into tests for C. difficile products (glutamate dehydrogenase (GDH), aromatic fatty acids, TcdA and/or TcdB); culture methods for the detection of toxin-producing C. difficile (toxigenic culture); and nucleic acid amplification tests for C. difficile genes (detecting 16S rRNA, toxin genes or the gene encoding GDH)162. The test selection is important to differentiate between patients with CDI and asymptomatic carriers34,164. Tests that detect toxins are specific to CDI, whereas those that detect (a component of) the bacterium might indicate colonization rather than disease34,164.
Exclusive reliance on molecular tests for CDI diagnosis without tests for toxins is likely to result in overdiagnosis and overtreatment34,165. Owing to large variations in sensitivity and specificity of various diagnostic tests, the European Society of Clinical Microbiology and Infectious Diseases recommends using a two-step algorithm, including a test for the presence of C. difficile and one to detect free toxins in the faeces166. As tests remain positive for toxins during treatment and toxins can even be found after successful treatment167, regular monitoring using toxin tests as follow up for treatment is not advised. If free toxins are absent, CDI is highly unlikely (FIG. 8). Importantly, C. difficile toxin assays vary markedly in their sensitivity34,164,168. If C. difficile is present but the toxin test result is negative, CDI cannot be definitively excluded. Patients could be (asymptomatically) colonized by C. difficile with diarrhoea due to an alternative cause or could be experiencing CDI with toxin levels below the lower limit of detection of the assay used. For these patients, clinical evaluation is required to decide whether treatment for CDI is warranted.
Figure 8. Diagnosis and treatment options for Clostridium difficile infections.
When a patient is suspected of having Clostridium difficile infection (CDI), the recommended option is to detect toxins of C. difficile in the stool. Several diagnostic algorithms have been condensed into this figure264. Treatment options indicated here are based on reports by Leffler and Lamont14, and Debast et al.155. For moderate CDI, metronidazole is given orally, in severe cases intravenously. Hospitalization refers to admission as a result of CDI (not as a result of comorbidities; patients might already be hospitalized). Note that recurrence after clinical cure (resolution of symptoms) can be observed. Faecal microbiota transplant is an effective but non-standard form of treatment (shown by a dashed line). *Fidaxomicin is a treatment option if the risk of recurrence is high, but not for complicated CDI.
An alternative diagnostic algorithm uses a coupled enzyme immune assay that simultaneously detects both GDH and TcdA or TcdB169. As the sensitivity of the toxin component is unclear, samples that are GDH-positive but toxin-negative could undergo reflex testing using a nucleic acid amplification test to determine whether a toxigenic C. difficile strain is present. Ideally, every laboratory should also have the opportunity to isolate C. difficile from faecal samples. Isolation enables toxigenic culture and susceptibility testing, and offers the ability to perform molecular typing that is required for surveillance (BOX 1). Many countries have implemented reference laboratories for this purpose33. The most recent US guidelines on testing were published in 2010 (by the Society for Healthcare Epidemiology of America and the Infectious Diseases Society of America); the guidelines did not make a firm recommendation regarding the routine diagnosis of CDI154.
Screening
There is considerable variation between and within countries in the diagnostic algorithm applied but also the frequency of testing (when to test). A study involving ~500 hospitals in 20 countries across Europe revealed that 23% of diarrhoeal samples with a positive CDI test result (at a reference laboratory) were initially missed owing to a lack of clinical suspicion32. Hence, restricting the testing of samples to those for which a physician has requested CDI testing will lead to underdiagnosis. All stool samples from hospitalized patients with diarrhoea should be tested unless a plausible alternative explanation (such as laxative use or diagnosis of inflammatory bowel disease) is available155,164,170 or if the patient is <2 years of age. Indeed, C. difficile is commonly found in the faeces of healthy infants, and there is no general agreement on how to define CDI in children or when to test for CDI in children, especially with respect to age, underlying disease and use of antibiotics1,171. However, CDI can occasionally occur in infants and young children, and can cause microscopic intestinal lesions with symptoms other than diarrhoea. The American Academy of Pediatrics has recently recommended testing for CDI only if age-specific clinical criteria are met172.
As C. difficile is increasingly recognized as a causative agent of community-associated diarrhoea, one can consider testing all diarrhoeal samples from community patients, although it can be cost-prohibitive to test a large number of samples. Application of specific algorithms, such as testing those with diarrhoea who have previously been hospitalized or used antibiotics, results in recognition of only 61% of patients with CDI in the community28. The introduction of multiplex molecular tests for enteric pathogens makes it easier to implement routine testing (screening) for CDI, although positive tests of C. difficile should be followed by a stool toxin detection test (two-step algorithm). In a European, multicentre, quarterly point-prevalence study of community-acquired diarrhoea using molecular tests, C. difficile was found among 709 samples to be the third most frequently occurring pathogen, after enteropathogenic Escherichia coli and Campylobacter spp.173. In that study, children were also included (potentially identifying colonized rather than infected individuals) and a large variation per country was observed173. C. difficile occurred more frequently in the >60-year age group than in other age groups. Application of molecular diagnostics in a case–control study of patients with diarrhoea attending a general practitioner in the Netherlands revealed that C. difficile was present in 4.2% of 1,515 cases and 0.8% of 1,195 healthy controls174. This number is considerably higher than the prevalence of Salmonella spp. and Shigella spp., and confirmed that C. difficile is frequently found in general practice24.
Asymptomatic carriage
For both health-care- and community-associated infection, it is necessary to differentiate between colonization and disease34,164,165. Carriage and colonization are often deemed to be interchangeable terms175,176. Indeed, the criteria used to determine asymptomatic carriage and/or colonization vary markedly between studies. A clarification of terminology is required, as a single C. difficile-positive faecal sample could indicate colonization, transient carriage or ‘pass through’ (REF. 177). We prefer to use the term ‘carriage’ to refer to persistent colonization.
Most studies use just one single culture and, therefore, evaluate passage or transient colonization, defined here as asymptomatic colonization. Asymptomatic C. difficile colonization is common in health-care facilities and in the community, and can be attributable to toxin-positive or toxin-negative strains36,178–180. In hospitals, the prevalence of asymptomatic colonization varies from 7% to 18%, depending on the length of stay, exposure to antibiotics and to C. difficile (infection pressure), underlying disease and possibly use of acid-suppressive medication. Indeed, the environment is most contaminated in rooms of patients with CDI, less so in rooms of patients asymptomatically colonized with C. difficile and least in patients who are not colonized181. During outbreaks, colonization rates may further increase to >50%179. Asymptomatic colonization in the community is lower than in the health-care setting and is in the range of 2–4%. Several studies report high carriage rates (25–35%) in children in the first year; the rate drops (to 15%) for ages 1–8 years182–184. This variation is probably associated with the unstable intestinal microbiota in the first 2–3 years of life, which enables C. difficile to remain established in the intestinal tract185. Interestingly, high levels of free toxin can also be detected in faecal samples of young children without diarrhoeal symptoms, therefore, further reinforcing the need for information on the effects of toxins on intestinal epithelial cells in children.
Asymptomatic colonization has been considered a protective factor against the acquisition of new C. difficile isolates. A frequently cited meta-analysis of four studies from 1994 showed that patients colonized with C. difficile actually had a lower risk of subsequently developing CDI, but no distinction was made between colonization by toxigenic versus non-toxigenic strains186. A recent meta-analysis included studies in which patients were colonized with toxigenic strains at hospital admission only36. It should be noted that in this study, unlike the previous study186, patients with previous CDI were not excluded and this could have confounded the results. Overall, the patients who were colonized had an increased risk of developing CDI (relative risk 5.86; 95% confidence interval 4.21–8.16). By contrast, patients asymptomatically colonized by non-toxigenic strains do not seem to have an increased risk or are even protected from progressing to CDI. This concept was recently tested in humans; patients who could be colonized by a non-toxigenic C. difficile strain after receiving standard of care treatment for CDI had significantly reduced CDI recurrence rates187. However, further explorations of this approach should consider that non-toxinogenic strains can become toxinogenic by horizontal gene transfer188, although it is unknown whether this process also occurs in a human host.
Asymptomatically colonized patients can shed spores into their environment and consequently to other patients. As early as 1992, it was recognized that nosocomial acquisition of a C. difficile strain was preceded by introduction of that strain to the ward by an asymptomatically colonized patient189. On the basis of an epidemiological model of C. difficile transmission in health-care settings, admission of colonized patients was shown to play an important part in sustaining ward-based transmission190. This observation needs confirmation as it could have major implications for infection control and prevention measures.
Management
Infection control
Several reviews and guidelines for controlling CDI have been published154,155,157,191–195. Although some differences exist, most of these guidelines have similar recommendations (BOX 3). If a patient is suspected of having CDI, rapid diagnostic testing should be performed to enable treatment initiation as soon as possible. Spores are highly infectious and problematic in health-care settings as they are able to persist on surfaces and are resistant to many disinfectants and alcohol-based hand washes37. Up to 1 × 107 spores per gram of faeces are present in patients with CDI, representing considerable potential for environmental contamination. Consequently, treatment should always be combined with patient isolation to prevent spread of C. difficile or other enteropathogenic microorganisms.
Box 3. Infection control and prevention of Clostridium difficile infection.
Ensure rapid diagnostic testing of patients with diarrhoeal illness acquired in the hospital or associated with antimicrobial therapy
A hospital-based infection control programme combined with active surveillance can help to decrease the incidence of Clostridium difficile infection (CDI); locally defined thresholds or triggers for the addition of enhanced control measures are needed
Antibiotic stewardship, including restriction of specific high-risk antimicrobials (such as clindamycin, cephalosporins and fluoroquinolones), is recommended to reduce the risk of CDI
Patient isolation and contact precautions (including hand hygiene with soap and water) should be maintained until at least the diarrhoea has resolved
If isolation in a single room is not possible, alternatives are segregation within wards and/or cohorting of cases
Disinfection of environmental surfaces is recommended using chlorine-releasing agents as a minimum in clinical areas with CDI cases
Educate health-care personnel, cleaning staff and visitors on contact precautions to minimize the transmission of spores
In a health-care setting, transmission of C. difficile spores occurs primarily via the contaminated hands of health-care workers, but contact with a contaminated environment, contaminated utensils or medical devices has also been implicated; C. difficile spores have been identified in rooms of patients who have tested negative. Environmental decontamination of clinical areas, ideally using chlorine-releasing agents or a sporicidal product, is recommended; however, in practice, compliance with cleaning protocols is often suboptimal154. Newer alternatives for environmental decontamination have been introduced, notably gaseous hydrogen peroxide and, more recently, UV decontamination196. The former is particular effective at killing C. difficile spores, but the cost-effectiveness of these approaches is unclear.
Antimicrobial therapy and surgery
The currently available antibiotics that are recommended for treatment of CDI are metronidazole, vancomycin and fidaxomicin (FIG. 8). Stopping the inciting antibiotics and clinical observation can treat very mild CDI that has been induced by antibiotics155. Patients with mild-to-moderate CDI can be treated with oral metronidazole whereas oral vancomycin is recommended for severe or complicated infections154,155,193. Two multinational randomized controlled trials included patients managed with either vancomycin or metronidazole197. Metronidazole was inferior to vancomycin on an intent-to-treat basis (clinical cure rates 72.7% versus 81.1%). Also, in a post-hoc multivariate analysis, vancomycin, treatment-naive status and mild-to-moderate CDI severity predicted treatment success.
Concomitant antibiotics are associated with reduced clinical cure, increased recurrence rates and longer time to cessation of diarrhoea198. Consequently, additional measures to curb CDI include discontinuation of unnecessary antimicrobial therapy (that is, for the presenting infection), as well as avoidance of anti-motility medications and reviewing proton pump inhibitor use155. Patients with complicated CDI with ileus (paralysed bowel) or toxic megacolon, in whom oral antibiotics cannot reach the disease site, can be treated with vancomycin delivered per rectum plus intravenous metronidazole.
Fulminant CDI is a highly lethal disease with mortality rates of up to 80%. These patients often require a total abdominal colectomy, but there is no established management protocol. Alternatively, a diverting loop ileostomy and colonic lavage might be associated with reduced morbidity and mortality199. Surgical therapy should be considered in patients with toxic megacolon, clinical signs of sepsis and severe organ dysfunction, acute abdomen and severe ileus. A white blood cell count of ≥15,000–20,000 per μl and elevated serum lactate (>5.0 mM) might serve as markers of disease severity154,155,200.
Recurrent infection
After treatment of an initial episode of CDI, the chance of a recurrence within 8 weeks is 15–25%; for a patient with 1–2 previous recurrences, the risk of further recurrences is 40–65%201. Recurrences are associated with an impaired immune response to C. difficile toxins and/or alteration of the colonic microbiota. Fewer recurrences occur after treatment with oral fidaxomicin (13%) than with vancomycin (25%)202,203. However, after a first recurrence, optimal treatment options are less clear, but fidaxomicin might be effective198,202,203. Despite these shortcomings, fidaxomicin is generally used for treating a first recurrence of CDI, unless disease has progressed from non-severe to severe (FIG. 8). Given that the main strength of fidaxomicin is prevention of recurrent infections (except for those due to PCR ribotype 027 that generally respond less well to antibiotics), clinical prediction markers for recognizing patients at risk for recurrent CDI could be helpful. Multiple risk factors for recurrent CDI have been suggested in the literature, but as not all of these are evident at the time of treatment initiation, it is difficult to select appropriate parameters. Age >65 years, concomitant antibiotics, renal failure, history of previous CDI, possibly continued use of antacid medications and initial disease severity155,204 are associated with increased risk of recurrence.
Bacteriotherapy and faecal transplantation
Limited evidence supports the use of probiotics to decrease recurrence of CDI, and no effective immunotherapy is currently available205. Faecal microbiota transplantation (FMT) is a highly effective rescue treatment and should be considered in patients who have had >2 recurrences, as the efficacy of antibiotics in these patients is ~30%. A randomized controlled trial of FMT revealed that it is effective (~81%) in treating multiple recurrent CDI206 (FIG. 9). FMT is best reserved for patients with multiple recurrences of CDI who have failed to respond to other treatment options. Importantly, FMT is a non-standardized procedure, and the long-term consequences of altering a patient’s gut microbiota are unknown. Several national guidelines have, therefore, been developed to standardize FMT, including donor screening and selection207–209. Results from a preliminary study among patients with relapsing CDI revealed that administration of FMT using frozen encapsulated inoculum from unrelated donors also resulted in a good outcome210. It is likely that future research will define mixtures of selected microorganisms, designed according to the microorganisms’ roles in the microbiota against CDI, as ‘pharmacological’ alternatives to FMT. For example, a mixture of 33 bacteria has been shown to be effective in two patients with CDI211, although the selection of the isolates was not based on microbiota studies. Rectal bacteriotherapy with a mixture of 12 bacteria from healthy donors resolved CDI and prevented recurrence within 30 days in 64% of the patients212. A combination of four bacterial species selected on the basis of strongest association with resistance to CDI, protected mice from infection, most probably indirectly through an effect on bile acid metabolism42. As noted for FMT, long-term safety data on rectal bacteriotherapy will be needed, given the far-reaching effects of gut microbiota in health and disease.
Figure 9. Faecal microbiota transplant.
In faecal microbiota transplant (FMT), faecal material from a healthy donor is collected. The material is processed (blending, filtration) into pills or a solution. As part of this process, a check for the presence of pathogenic and multidrug-resistant organisms is performed. The processed material can be stored before (one-off) administration by nasoduodenal infusion, colonoscopic infusion or rectal enema for solution formulations or orally for pill formulations. Antibiotic treatment generally precedes the administration of the FMT to reduce Clostridium difficile levels. Alongside FMT, efforts are ongoing to standardize bacteriotherapy. On the basis of microbiome and metabolome analyses, signatures of resistance to colonization by C. difficile are identified. After collecting faecal material from healthy donors, species identified in these microbiome signatures or believed to be responsible for the metabolomic signature are cultured. Defined mixtures of these strains are tested for safety and ability to confer colonization resistance in preclinical trials and subsequently validated in clinical studies. Coloured bars indicate diversity of the microbiota, which is severely reduced in the patient with C. difficile infection (CDI) compared with the healthy subject.
Novel therapeutics
Several therapeutics targeting different stages of CDI are currently in clinical development (TABLE 1). In brief, these consist of treatments to restore a complex microbiota (SER-109)213, to prevent off-target effects of antibiotic treatment on the intestinal microbiota (SYN-004), to neutralize C. difficile toxins (including monoclonal antibodies)214 or to inhibit C. difficile proliferation (SMT19969, LFF571, surotomycin, cadazolid, PolyCAb and CRS3123).
Table 1.
Selected agents for the treatment and prevention of CDI in clinical trials
| Agent (manufacturer) | Indication | Notes | Clinical trial identifier |
|---|---|---|---|
| Phase III | |||
| Actoxumab and bezlotoxumab alone or in combination (Merck) | Prevention of recurrent CDI | Antitoxin A (MK-3415) and antitoxin B (MK-6072) monoclonal antibodies given intravenously as adjuncts to standard treatment | NCT01241552, NCT01513239 |
| Surotomycin (Merck) | Treatment | Cyclic lipopeptide antibiotic related to daptomycin but administered orally | NCT01598311, NCT01597505 |
| Cadazolid (Actelion) | Treatment | Hybrid antibiotic molecule, consisting of fluoroquinolone and oxazolidinone moieties, for oral administration | NCT01987895, NCT01983683 |
| Cdiffense (Sanofi Pasteur) | Prevention | Vaccine containing toxoids of TcdA and TcdB from C. difficile | NCT01887912 |
| Phase II | |||
| IC84 vaccine (Valneva) | Prevention | Vaccine consisting of recombinant protein of the two truncated toxins TcdA and TcdB from C. difficile | NCT02316470 |
| LFF571 (Novartis) | Treatment of moderate CDI | Semi-synthetic thiopeptide | NCT01232595 |
| SER-109 (Seres Therapeutics) | Treatment of recurrent CDI | Oral microbiome therapeutic (mixture of bacterial spores) granted orphan drug designation by the FDA | NCT02437500 |
| SMT19969 (Summit Pharmaceuticals) | Treatment | Oral non-absorbable antibiotic with a narrow spectrum of activity and high selectivity for C. difficile | NCT02092935 |
| C. difficile vaccine (Pfizer) | Prevention | Bivalent toxin vaccine | NCT02561195, NCT02117570 |
| SYN-004 (Synthetic Biologics) | Prevention | Class A β-lactamase designed to protect gut microbiota from the action of systemically administered β-lactam antibiotics that might otherwise predispose for CDI | NCT02563106 |
| VP20621 (Shire) | Prevention of recurrent CDI | Orally administered non-toxigenic C. difficile | NCT01259726 |
| Phase I | |||
| PolyCAb (Micropharm) | Treatment of severe CDI | Polyclonal antibodies against C. difficile given intravenously | Not available |
| CRS3123/REP3123 (National Institute of Allergy and Infectious Diseases) | Treatment | Methionyl-tRNA synthetase inhibitor oral antibiotic | NCT02106338, NCT01551004 |
CDI, Clostridium difficile infection; Tcd, C. difficile toxin.
Quality of life
Economic burden of CDI
The burden of health-care-associated CDI can be expressed in terms of mortality, recurrence, (additional) length of hospital stay or economic cost215–217. Economic analyses of health-care-associated CDI have shown that direct health-care cost, and costs due to increased length of stay, were the main cost drivers. An integrative review showed a wide variation in the difference in length of stay between people with and those without CDI (2.8–16.1 days), which was attributed to differences in design and data collection218. Overall, people with CDI stay longer in hospital than people without CDI despite this variation.
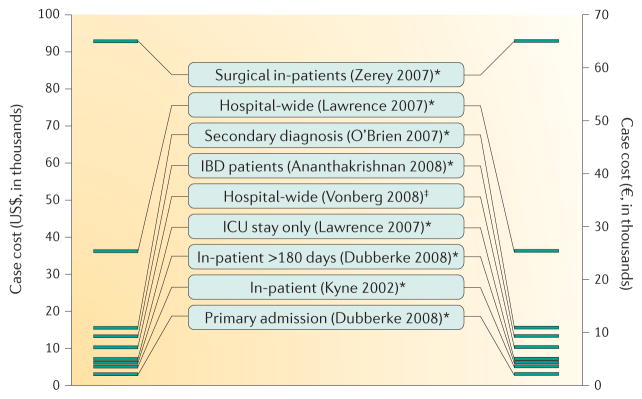
A systematic review of the effects of CDI in Europe showed that the median length of stay for patients with CDI was in the range of 8–27 days, with an additional length of stay (due to CDI) of between 2.8 days and 18 days217. The incremental per-case cost of CDI in this study was UK£4,577 in Ireland and £8,843 in Germany, after standardization to 2010 prices217. Others have estimated the incremental per-case cost of CDI, after standardization to 2008 prices, to be US$2,871–90,644 (REFS 216,219) (FIG. 10). A recent meta-analysis identified a total of 45 studies (mostly from North America) that measured the economic impact of CDI. For hospitalized patients, attributable mean CDI costs ranged from $8,911 to $30,049. However, the authors noted that costing methods were heterogeneous, making inter-study and setting comparisons difficult. Standardization of such measurements would be helpful, although differences between health-care systems remain a barrier when comparing financial costs220.
Figure 10. Cost per case of Clostridium difficile infection.
Data are from Ghantoji et al.219 (indicated with *) and Vonberg et al.216 (indicated with‡); surnames of first authors and years indicate the original studies described in these publications. Conversion between US$ and € is based on 2008 exchange rates. Note that several studies have estimated the cost of Clostridium difficile infections more recently220,265,266. IBD, inflammatory bowel disease; ICU, intensive care unit.
The total direct cost of CDI to the European Union in 2006 was estimated at €3 billion per year6. Assuming a 3% annual inflation rate, this approximates to >€4 billion in 2015. Estimates for the economic burden of CDI in the United States and Canada are in excess of US$1 billion221 and CAN$280 million222, respectively. These figures do not include the indirect socioeconomic costs (see below). Only for Canada does the estimate include a parameter for community-associated CDI in addition to health-care-associated CDI222; so, the total burden in the United States and European Union probably exceeds the numbers given above.
Patient-reported quality of life
The stark mortality rates associated with CDI emphasize the serious consequences of this disease. Furthermore, given that CDI is characterized by diarrhoea, relatively frequent symptomatic recurrences and often altered bowel habit for possibly weeks or months following cessation of acute symptoms, it is perhaps not surprising that patients regularly report that this disease is one of the worst that they have experienced223. As patients are typically older and have comorbidities, the additional burden of CDI can affect both their dignity and ability to cope. Despite these well-recognized effects of CDI, very few data are available to formally measure how the disease affects an individual’s health status, functionality and quality of life. Two recent studies have begun to explore these under-reported issues.
A prospective study of 66 outpatients with CDI used the RAND Short-Form 36 (SF-36) Health Survey and concluded that CDI significantly decreased overall quality of life but that a more specific health-related questionnaire is needed. The Patient-Reported Outcome Measurement Information System (PROMIS; http://www.nihpromis.org) is a large, US National Institutes of Health-funded database of questions to measure patient-reported health status for physical, mental and social well-being. PROMIS has recently been explored in a prospective, observational, multicentre study as a potential way to evaluate self-reported health status in patients with CDI224. Patients (n = 95) with active CDI (58%) or who were hospitalized (42%) had worse scores with regard to bowel function, nausea and abdominal pain compared with controls (P < 0.001). Those with recurrent CDI had worse anxiety scores than any other group (patients with first occurrence of CDI and controls; P < 0.001). The authors concluded that the 18 patient-reported health status questions were discriminatory for active CDI and primary versus recurrent CDI. These questions might be suitable for measuring short-term and long-term differences in patient-reported health status in people with CDI. CDI can also have long-lasting effects on families, but there is no systematic evaluation of these effects.
More work is needed to optimize these measurements and to determine which interventions are associated with the best improvements in outcomes for both patients and relatives.
Outlook
Outstanding research questions
Substantial progress has been made in our understanding of C. difficile physiology and pathogenesis. Studies have provided insight into the workings of the pathogen and highlighted aspects of its biology that differ from the situation in other studied bacteria. For instance, the order, activation and function of sporulation sigma factors of C. difficile deviate from what is known for the best-studied Gram-positive spore former, Bacillus subtilis225. Perhaps this difference should not come as a surprise, as the last common ancestor of bacilli and clostridia dates back ~2.7 billion years, only shortly after the divergence of Gram-positive and Gram-negative bacteria (3.2 billion years ago)226. It is conceivable that more detailed investigations of the molecular biology of clostridia in general, and C. difficile in particular, will reveal unexpected features.
Laboratory investigations under controlled conditions have clearly demonstrated that the production of enterotoxins is regulated in a complex manner and integrates signals from both bacterial metabolism and the culture conditions99. It is uncertain, however, whether this process reflects infection within the host. Mutants that are used to assess these effects in the laboratory might display reduced virulence as a result of reduced fitness. Although the role of enterotoxins in disease is established, it remains unknown how toxin production is triggered in vivo, and how or when the toxins are secreted into the gastrointestinal tract. Similarly, it is unknown what triggers sporulation during infection. Both toxins (production and activity) and spores have been implicated in epidemicity82,122,227. However, strong evidence supporting this link is lacking and the ability of C. difficile to cause epidemics is likely to be a multifactorial process that involves a delicate balance of factors that affect virulence and transmissibility74,75. It should be noted that increased virulence might not correlate with transmissibility, which might be favoured when hosts remain relatively healthy75.
Epidemic types of C. difficile have received considerable attention as a result of their higher mortality and morbidity in comparison to other, non-epidemic, types7,8,12,18,82. Often, enhanced infection control measures are taken when transmission of epidemic C. difficile types is detected, which has probably contributed to the decline of PCR ribotype 027 in different countries in 2014 (REF. 32). But is the increased vigilance towards these strains warranted? First, epidemiological analyses ignore the fact that not all strains of the same PCR ribotype exhibit the same characteristics, as has been demonstrated for sporulation228. Second, other PCR ribotypes can also cause outbreaks of CDI20,21,229. Third, with the advent of sequence-based typing methods, it is becoming clear that strains with different PCR ribotypes can be highly related98,230. For instance, strains of PCR ribotypes 244 and 176, which are related to PCR ribotype 027, can cause outbreaks of severe CDI22,23. PCR ribotype 244 seems to be primarily implicated in community-associated CDI, indicating that highly related strains might emerge as epidemic types in different settings231. Finally, new types could emerge. Thus, care should be taken to not generate an unjustified bias towards certain strains in epidemiological vigilance.
Colonization and pathogenesis
What determines whether C. difficile successfully establishes an infection is an important question. The host microbiota and its associated metabolites greatly influence the ability of C. difficile spores to germinate and colonize the gut39,43. However, niche-specific competition232 or collaboration (for instance, in a multispecies biofilm56,58) might also contribute. Most of the metagenomic studies have focused on bacterial species, whereas the contribution of fungal species and viruses (bacteriophages) is poorly explored. Notably, population groups with relatively unstable microbiomes (that is, infants185 and the elderly233) are most commonly colonized by C. difficile.
Other host factors might also play a part. Failure to mount a protective immune response is associated with disease progression; patients with an adequate response could either eliminate the infection or become asymptomatic carriers14. Host-dependent expression levels of toxin or colonization factor receptors could also be an important determinant for disease development. It has been postulated that infants remain asymptomatic in the presence of high levels of toxins due to the absence of receptors in their still-developing gastrointestinal tract; however, there is no evidence yet to support this hypothesis. Detailed studies of the interaction of C. difficile with the host are necessary to delineate the contribution of host factors to colonization and pathogenesis.
Clinical needs
Although antimicrobial resistance of C. difficile is not a major issue in the clinic, novel narrow-spectrum therapeutics are needed. The primary reason is that current (broad-spectrum) antibiotics with activity against C. difficile, such as metronidazole and vancomycin, can concurrently predispose patients to CDI (and possibly recurrent CDI) owing to their effects on other members of the microbiota234. Furthermore, some antibiotics are not cost-effective as first-line treatment options. It is possible that specific prophylactic elimination of C. difficile in high-risk groups could reduce the risk of CDI without altering the host microbiota. It is of interest that bacteriocins and viruses (bacteriophages) seem to be able to target C. difficile specifically232,235,236.
FMT is an excellent potential alternative to antibiotic therapy206, but long-term safety, public acceptance and relative lack of standardized donor material are limiting broad application. Steps have been taken to generate standardized formulations of FMT or bacteria42,149,211,212. However, it remains to be established whether a single species of bacteria or mixtures of different bacteria are effective for all (or most) patients. It is likely that bacterial or metabolic signatures need to be identified that confer broad protection or activity against CDI (FIG. 9).
Antivirulence strategies237 might become valuable additions or alternatives to the current therapeutic spectrum. Neutralizing antitoxin antibodies have shown clinical promise214,238 (TABLE 1), and toxin activity has also been targeted using small molecules128,129. Interference with quorum sensing or colonization factors has been underexplored so far, although it is clear that these factors can also be targeted by antibodies239,240. Small-molecule inhibition of extracellular protein processing or function could prove a viable strategy to reduce colonization or transmission of CDI.
A final issue is the clinical need for accurate prediction models155. Accurate risk assessment tools that can be applied in real time would be beneficial to target diagnostic methods and for optimized treatment of those at risk of severe or complicated CDI, or recurrence241–244. Although several tools have been developed, there is considerable room for simplification and improvements in predictive values to make these tools applicable at the bedside.
The burden of CDI
Current conservative estimates of the economic burden of CDI are based on estimates of incidence and an incremental — per (hospitalized) patient — cost. These estimates fail to take into account the changes in demographics that are projected over the coming years. Age is a risk factor for CDI, although it is unclear whether this is an independent factor or caused by underlying confounders (such as immune senescence, comorbidities, need to stay at long-term care facilities and additional required health care). The European Union has projected that the demographic old-age dependency ratio (the ratio of those aged >65 years old to those aged 15–64 years) will increase from 27.8 to 50.1% between 2013 and 2060 (REF. 245). Similarly, US-based population projections foresee an increase in the percentage of people aged >65 years of 13.7–20.9% between 2012 and 2050 (REF. 246). On the basis of these projections, the impact of CDI is expected to become considerable in coming years.
Epidemiological evidence suggests that an increasing number of CDI cases is linked to populations that are generally considered to be at low risk for CDI. These community-associated infections generally affect a different demographic (younger patients) who have frequently not been exposed to antibiotics. Further studies are required to determine risk factors for community-associated CDI. As many cases of community-acquired CDI go undetected24,27,29, more studies are required to determine its contribution to the total burden of CDI at national and international levels.
Finally, C. difficile is increasingly recognized in veterinary medicine, with highly variable disease entities in different species of animals. No information is available to date on the economic burden of CDI in the food (animal) production industry.
Acknowledgments
This work was partially supported by a Vidi fellowship from the Netherlands Organization for Scientific Research and a Gisela Thier Fellowship from the Leiden University Medical Center to W.K.S.; a National Institutes of Health Award AI95755 and a Burroughs Wellcome Investigator in the Pathogenesis of Infectious Disease Award to D.B.L.; and a Future Fellowship FT120100779 of the Australian Research Council to D.L. The authors thank the ESCMID Study Group for Clostridium difficile (ESGCD) for their advice and suggestions and apologize to the authors whose work could not be cited due to restrictions imposed by the format of this Primer. W. Knetsch is acknowledged for drafting the illustration used in Box 1.
Footnotes
Author contributions
Introduction (W.K.S.); Epidemiology (M.H.W., E.J.K. and W.K.S.); Mechanisms/pathophysiology (D.B.L., D.L. and W.K.S.); Diagnosis, screening and prevention (M.H.W., E.J.K. and W.K.S.); Management (M.H.W., E.J.K. and W.K.S.); Quality of life (M.H.W., E.J.K. and W.K.S.); Outlook (W.K.S. and E.J.K.); overview of the Primer (W.K.S.).
Competing interests statement
W.K.S. has performed research for Cubist. D.L. has performed research for Immuron and Adenium Biotech. D.B.L. has performed research for MedImmune and Merck. M.H.W. has received consulting fees from Abbott, Actelion Pharmaceuticals, Astellas, AstraZeneca, Bayer, Cerexa, Cubist, Durata, The European Tissue Symposium, The Medicines Company, MedImmune, Merck, Motif Biosciences, Nabriva, Optimer, Paratek, Pfizer, Roche, Sanofi Pasteur, Seres Therapeutics, Summit Pharmaceuticals, and Synthetic Biologics. M.H.W. has also received lecture fees from Abbott, Alere, Astellas, AstraZeneca, Pfizer and Hoffmann La Roche, and received grant support from Abbott, Actelion, Astellas, bioMérieux, Cubist, Da Volterra, The European Tissue Symposium, Merck and Summit Pharmaceuticals. E.J.K. has performed research for Cubist, Novartis and Qiagen, and has participated in advisory forums of Astellas, Optimer, Actelion, Pfizer, Sanofi Pasteur and Seres Therapeutics. These companies had no role in the writing of this Primer.
References
- 1.Hall IC, O’Toole E. Intestinal flora in new-born infants: with a description of a new pathogenic anaerobe, Bacillus difficilis. Am J Child Dis. 1935;49:390–402. [Google Scholar]
- 2.Lyras D, et al. Toxin B is essential for virulence of Clostridium difficile. Nature. 2009;458:1176–1179. doi: 10.1038/nature07822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kuehne SA, et al. The role of toxin A and toxin B in Clostridium difficile infection. Nature. 2010;467:711–713. doi: 10.1038/nature09397. [DOI] [PubMed] [Google Scholar]
- 4.Hafiz S, Oakley CL. Clostridium difficile: isolation and characteristics. J Med Microbiol. 1976;9:129–136. doi: 10.1099/00222615-9-2-129. [DOI] [PubMed] [Google Scholar]
- 5.Bartlett JG. Clostridium difficile: history of its role as an enteric pathogen and the current state of knowledge about the organism. Clin Infect Dis. 1994;18:S265–S272. doi: 10.1093/clinids/18.supplement_4.s265. An exceptional overview of the early experiments demonstrating the involvement of C. difficile in (antibiotic-associated) colitis. [DOI] [PubMed] [Google Scholar]
- 6.Kuijper EJ, Coignard B, Tüll P. Emergence of Clostridium difficile-associated disease in North America and Europe. Clin Microbiol Infect. 2006;12(Suppl 6):2–18. doi: 10.1111/j.1469-0691.2006.01580.x. [DOI] [PubMed] [Google Scholar]
- 7.Goorhuis A, et al. Emergence of Clostridium difficile infection due to a new hypervirulent strain, polymerase chain reaction ribotype 078. Clin Infect Dis. 2008;47:1162–1170. doi: 10.1086/592257. [DOI] [PubMed] [Google Scholar]
- 8.He M, et al. Emergence and global spread of epidemic healthcare-associated Clostridium difficile. Nat Genet. 2013;45:109–113. doi: 10.1038/ng.2478. A large-scale whole-genome sequencing study that was the first to demonstrate the potential of the technique to trace the emergence of epidemic strains and relatedness between isolates. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sebaihia M, et al. The multidrug-resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome. Nat Genet. 2006;38:779–786. doi: 10.1038/ng1830. [DOI] [PubMed] [Google Scholar]
- 10.Minton N, et al. The development of Clostridium difficile genetic systems. Anaerobe. 2004;10:75–84. doi: 10.1016/j.anaerobe.2003.11.003. [DOI] [PubMed] [Google Scholar]
- 11.Kuehne SA, Heap JT, Cooksley CM, Cartman ST, Minton NP. ClosTron-mediated engineering of Clostridium. Methods Mol Biol. 2011;765:389–407. doi: 10.1007/978-1-61779-197-0_23. [DOI] [PubMed] [Google Scholar]
- 12.Freeman J, et al. The changing epidemiology of Clostridium difficile infections. Clin Microbiol Rev. 2010;23:529–549. doi: 10.1128/CMR.00082-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hensgens MP, et al. Clostridium difficile infection in the community: a zoonotic disease? Clin Microbiol Infect. 2012;18:635–645. doi: 10.1111/j.1469-0691.2012.03853.x. [DOI] [PubMed] [Google Scholar]
- 14.Leffler DA, LaMont JT. Clostridium difficile infection. N Engl J Med. 2015;372:1539–1548. doi: 10.1056/NEJMra1403772. [DOI] [PubMed] [Google Scholar]
- 15.Yutin N, Galperin MY. A genomic update on clostridial phylogeny: Gram-negative spore formers and other misplaced clostridia. Environ Microbiol. 2013;15:2631–2641. doi: 10.1111/1462-2920.12173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nadon CA, et al. Development and application of MLVA methods as a tool for inter-laboratory surveillance. Euro Surveill. 2013;18:20565. doi: 10.2807/1560-7917.es2013.18.35.20565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Knetsch CW, et al. Current application and future perspectives of molecular typing methods to study Clostridium difficile infections. Euro Surveill. 2013;18:20381. doi: 10.2807/ese.18.04.20381-en. An updated review of typing methods for C. difficile. [DOI] [PubMed] [Google Scholar]
- 18.Clements AC, Magalhaes RJ, Tatem AJ, Paterson DL, Riley TV. Clostridium difficile PCR ribotype 027: assessing the risks of further worldwide spread. Lancet Infect Dis. 1992;10:395–404. doi: 10.1016/S1473-3099(10)70080-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tickler IA, et al. Strain types and antimicrobial resistance patterns of Clostridium difficile isolates from the United States, 2011 to 2013. Antimicrob Agents Chemother. 2014;58:4214–4218. doi: 10.1128/AAC.02775-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bauer MP, et al. Clostridium difficile infection in Europe: a hospital-based survey. Lancet. 2011;377:63–73. doi: 10.1016/S0140-6736(10)61266-4. The first pan-European study of the epidemiology of CDI. [DOI] [PubMed] [Google Scholar]
- 21.Baldan R, et al. Clostridium difficile PCR ribotype 018, a successful epidemic genotype. J Clin Microbiol. 2015;53:2575–2580. doi: 10.1128/JCM.00533-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pituch H, et al. Hospital-based Clostridium difficile infection surveillance reveals high proportions of PCR ribotypes 027 and 176 in different areas of Poland, 2011 to 2013. Euro Surveill. 2015;20:30025. doi: 10.2807/1560-7917.ES.2015.20.38.30025. [DOI] [PubMed] [Google Scholar]
- 23.Lim SK, et al. Emergence of a ribotype 244 strain of Clostridium difficile associated with severe disease and related to the epidemic ribotype 027 strain. Clin Infect Dis. 2014;58:1723–1730. doi: 10.1093/cid/ciu203. [DOI] [PubMed] [Google Scholar]
- 24.Hensgens MP, et al. Diarrhoea in general practice: when should a Clostridium difficile infection be considered? Results of a nested case–control study. Clin Microbiol Infect. 2014;20:O1067–O1074. doi: 10.1111/1469-0691.12758. [DOI] [PubMed] [Google Scholar]
- 25.Eyre DW, et al. Diverse sources of C difficile infection identified on whole-genome sequencing. N Engl J Med. 2013;369:1195–1205. doi: 10.1056/NEJMoa1216064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lessa FC, et al. Burden of Clostridium difficile infection in the United States. N Engl J Med. 2015;372:825–834. doi: 10.1056/NEJMoa1408913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bouwknegt M, Van DS, Kuijper E. Burden of Clostridium difficile infection in the United States. N Engl J Med. 2015;372:2368. doi: 10.1056/NEJMc1505190. [DOI] [PubMed] [Google Scholar]
- 28.Hensgens MP, et al. Clostridium difficile infection in an endemic setting in the Netherlands. Eur J Clin Microbiol Infect Dis. 2011;30:587–593. doi: 10.1007/s10096-010-1127-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilcox MH, Mooney L, Bendall R, Settle CD, Fawley WN. A case–control study of community-associated Clostridium difficile infection. J Antimicrob Chemother. 2008;62:388–396. doi: 10.1093/jac/dkn163. [DOI] [PubMed] [Google Scholar]
- 30.Reveles KR, Lee GC, Boyd NK, Frei CR. The rise in Clostridium difficile infection incidence among hospitalized adults in the United States: 2001–2010. Am J Infect Control. 2014;42:1028–1032. doi: 10.1016/j.ajic.2014.06.011. [DOI] [PubMed] [Google Scholar]
- 31.European Centre for Disease Prevention and Control. Point prevalence survey of healthcare-associated infections and antimicrobial use in European acute care hospitals 2011–2012. ECDU. 2013 online http://www.ecdc.europa.eu/en/publications/Publications/healthcare-associated-infections-antimicrobial-use-PPS.pdf.
- 32.Davies KA, et al. Underdiagnosis of Clostridium difficile across Europe: the European, multicentre, prospective, biannual, point-prevalence study of Clostridium difficile infection in hospitalised patients with diarrhoea (EUCLID) Lancet Infect Dis. 2014;14:1208–1219. doi: 10.1016/S1473-3099(14)70991-0. A large pan-European study demonstrating the extent of missed diagnoses of CDI. [DOI] [PubMed] [Google Scholar]
- 33.European Centre for Disease Prevention and Control. European surveillance of Clostridium difficile infections. Surveillance protocol version 2.1. ECDU [online] 2015 http://ecdc.europa.eu/en/publications/Publications/Clostridium-difficile-infections-surveillance-protocol-version-2.1.pdf.
- 34.Planche TD, et al. Differences in outcome according to Clostridium difficile testing method: a prospective multicentre diagnostic validation study of C. difficile infection. Lancet Infect Dis. 2013;13:936–945. doi: 10.1016/S1473-3099(13)70200-7. A large multicentre study demonstrating the importance of toxin detection as part of a diagnostic algorithm for CDI. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Knetsch CW, et al. Whole genome sequencing reveals potential spread of Clostridium difficile between humans and farm animals in the Netherlands, 2002 to 2011. Euro Surveill. 2014;19:20954. doi: 10.2807/1560-7917.es2014.19.45.20954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zacharioudakis IM, Zervou FN, Pliakos EE, Ziakas PD, Mylonakis E. Colonization with toxinogenic C difficile upon hospital admission, and risk of infection: a systematic review and meta-analysis. Am J Gastroenterol. 2015;110:381–390. doi: 10.1038/ajg.2015.22. [DOI] [PubMed] [Google Scholar]
- 37.Paredes-Sabja D, Shen A, Sorg JA. Clostridium difficile spore biology: sporulation, germination, and spore structural proteins. Trends Microbiol. 2014;22:406–416. doi: 10.1016/j.tim.2014.04.003. A comprehensive overview of C. difficile sporulation, including the role of bile acids in germination. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deakin LJ, et al. The Clostridium difficile spo0A gene is a persistence and transmission factor. Infect Immun. 2012;80:2704–2711. doi: 10.1128/IAI.00147-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Theriot CM, Young VB. Interactions between the gastrointestinal microbiome and Clostridium difficile. Annu Rev Microbiol. 2015;69:445–461. doi: 10.1146/annurev-micro-091014-104115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bhattacharjee D, et al. Reexamining the germination phenotypes of several Clostridium difficile strains suggests another role for the CspC germinant receptor. J Bacteriol. 2015;198:777–786. doi: 10.1128/JB.00908-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sorg JA, Sonenshein AL. Bile salts and glycine as cogerminants for Clostridium difficile spores. J Bacteriol. 2008;190:2505–2512. doi: 10.1128/JB.01765-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Buffie CG, et al. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature. 2015;517:205–208. doi: 10.1038/nature13828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Theriot CM, Bowman AA, Young VB. Antibiotic-induced alterations of the gut microbiota alter secondary bile acid production and allow for Clostridium difficile spore germination and outgrowth in the large intestine. mSphere. 2016;1:e00045–15. doi: 10.1128/mSphere.00045-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Janoir C, Pechine S, Grosdidier C, Collignon A. Cwp84, a surface-associated protein of Clostridium difficile, is a cysteine protease with degrading activity on extracellular matrix proteins. J Bacteriol. 2007;189:7174–7180. doi: 10.1128/JB.00578-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Merrigan MM, et al. Surface-layer protein A (SlpA) is a major contributor to host-cell adherence of Clostridium difficile. PLoS ONE. 2013;8:e78404. doi: 10.1371/journal.pone.0078404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tasteyre A, Barc MC, Collignon A, Boureau H, Karjalainen T. Role of FliC and FliD flagellar proteins of Clostridium difficile in adherence and gut colonization. Infect Immun. 2001;69:7937–7940. doi: 10.1128/IAI.69.12.7937-7940.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Spigaglia P, et al. Surface-layer (S-layer) of human and animal Clostridium difficile strains and their behaviour in adherence to epithelial cells and intestinal colonization. J Med Microbiol. 2013;62:1386–1393. doi: 10.1099/jmm.0.056556-0. [DOI] [PubMed] [Google Scholar]
- 48.Lin YP, Kuo CJ, Koleci X, McDonough SP, Chang YF. Manganese binds to Clostridium difficile Fbp68 and is essential for fibronectin binding. J Biol Chem. 2011;286:3957–3969. doi: 10.1074/jbc.M110.184523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kovacs-Simon A, et al. Lipoprotein CD0873 is a novel adhesin of Clostridium difficile. J Infect Dis. 2014;210:274–284. doi: 10.1093/infdis/jiu070. [DOI] [PubMed] [Google Scholar]
- 50.Tulli L, et al. CbpA: a novel surface exposed adhesin of Clostridium difficile targeting human collagen. Cell Microbiol. 2013;15:1674–1687. doi: 10.1111/cmi.12139. [DOI] [PubMed] [Google Scholar]
- 51.Deneve C, Delomenie C, Barc MC, Collignon A, Janoir C. Antibiotics involved in Clostridium difficile-associated disease increase colonization factor gene expression. J Med Microbiol. 2008;57:732–738. doi: 10.1099/jmm.0.47676-0. [DOI] [PubMed] [Google Scholar]
- 52.Paredes-Sabja D, Sarker MR. Adherence of Clostridium difficile spores to Caco-2 cells in culture. J Med Microbiol. 2012;61:1208–1218. doi: 10.1099/jmm.0.043687-0. [DOI] [PubMed] [Google Scholar]
- 53.Bordeleau E, Burrus V. Cyclic-di-GMP signaling in the Gram-positive pathogen Clostridium difficile. Curr Genet. 2015;61:497–502. doi: 10.1007/s00294-015-0484-z. [DOI] [PubMed] [Google Scholar]
- 54.Purcell EB, McKee RW, McBride SM, Waters CM, Tamayo R. Cyclic diguanylate inversely regulates motility and aggregation in Clostridium difficile. J Bacteriol. 2012;194:3307–3316. doi: 10.1128/JB.00100-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Peltier J, et al. Cyclic diGMP regulates production of sortase substrates of Clostridium difficile and their surface exposure through ZmpI protease-mediated cleavage. J Biol Chem. 2015;290:24453–24469. doi: 10.1074/jbc.M115.665091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Crowther GS, et al. Comparison of planktonic and biofilm-associated communities of Clostridium difficile and indigenous gut microbiota in a triple-stage chemostat gut model. J Antimicrob Chemother. 2014;69:2137–2147. doi: 10.1093/jac/dku116. [DOI] [PubMed] [Google Scholar]
- 57.Dapa T, Unnikrishnan M. Biofilm formation by Clostridium difficile. Gut Microbes. 2013;4:397–402. doi: 10.4161/gmic.25862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Semenyuk EG, et al. Analysis of bacterial communities during C difficile infection in the mouse. Infect Immun. 2015;83:4383–4391. doi: 10.1128/IAI.00145-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Darkoh C, DuPont HL, Norris SJ, Kaplan HB. Toxin synthesis by Clostridium difficile is regulated through quorum signaling. mBio. 2015;6:e02569. doi: 10.1128/mBio.02569-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Martin MJ, et al. The agr locus regulates virulence and colonization genes in Clostridium difficile 027. J Bacteriol. 2013;195:3672–3681. doi: 10.1128/JB.00473-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sun X, Hirota SA. The roles of host and pathogen factors and the innate immune response in the pathogenesis of Clostridium difficile infection. Mol Immunol. 2015;63:193–202. doi: 10.1016/j.molimm.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cowardin CA, Petri WA., Jr Host recognition of Clostridium difficile and the innate immune response. Anaerobe. 2014;30:205–209. doi: 10.1016/j.anaerobe.2014.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ryan A, et al. A role for TLR4 in Clostridium difficile infection and the recognition of surface layer proteins. PLoS Pathog. 2011;7:e1002076. doi: 10.1371/journal.ppat.1002076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yoshino Y, et al. Clostridium difficile flagellin stimulates Toll-like receptor 5, and toxin B promotes flagellin-induced chemokine production via TLR5. Life Sci. 2013;92:211–217. doi: 10.1016/j.lfs.2012.11.017. [DOI] [PubMed] [Google Scholar]
- 65.Hasegawa M, et al. Nucleotide-binding oligomerization domain 1 mediates recognition of Clostridium difficile and induces neutrophil recruitment and protection against the pathogen. J Immunol. 2011;186:4872–4880. doi: 10.4049/jimmunol.1003761. [DOI] [PubMed] [Google Scholar]
- 66.Giesemann T, Guttenberg G, Aktories K. Human α-defensins inhibit Clostridium difficile toxin B. Gastroenterology. 2008;134:2049–2058. doi: 10.1053/j.gastro.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 67.Hing TC, et al. The antimicrobial peptide cathelicidin modulates Clostridium difficile-associated colitis and toxin A-mediated enteritis in mice. Gut. 2013;62:1295–1305. doi: 10.1136/gutjnl-2012-302180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Abt MC, et al. Innate immune defenses mediated by two ILC subsets are critical for protection against acute Clostridium difficile infection. Cell Host Microbe. 2015;18:27–37. doi: 10.1016/j.chom.2015.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.McQuade R, Roxas B, Viswanathan VK, Vedantam G. Clostridium difficile clinical isolates exhibit variable susceptibility and proteome alterations upon exposure to mammalian cationic antimicrobial peptides. Anaerobe. 2012;18:614–620. doi: 10.1016/j.anaerobe.2012.09.004. [DOI] [PubMed] [Google Scholar]
- 70.Ho TD, et al. Clostridium difficile extracytoplasmic function σ factor σV regulates lysozyme resistance and is necessary for pathogenesis in the hamster model of infection. Infect Immun. 2014;82:2345–2355. doi: 10.1128/IAI.01483-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ho TD, Ellermeier CD. PrsW is required for colonization, resistance to antimicrobial peptides, and expression of extracytoplasmic function sigma factors in Clostridium difficile. Infect Immun. 2011;79:3229–3238. doi: 10.1128/IAI.00019-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Suarez JM, Edwards AN, McBride SM. The Clostridium difficile cpr locus is regulated by a noncontiguous two-component system in response to type A and B lantibiotics. J Bacteriol. 2013;195:2621–2631. doi: 10.1128/JB.00166-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.McBride SM, Sonenshein AL. The dlt operon confers resistance to cationic antimicrobial peptides in Clostridium difficile. Microbiology. 2011;157:1457–1465. doi: 10.1099/mic.0.045997-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Awad MM, Johanesen PA, Carter GP, Rose E, Lyras D. Clostridium difficile virulence factors: Insights into an anaerobic spore-forming pathogen. Gut Microbes. 2014;5:579–593. doi: 10.4161/19490976.2014.969632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Smits WK. Hype or hypervirulence: a reflection on problematic C difficile strains. Virulence. 2013;4:592–596. doi: 10.4161/viru.26297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hammond GA, Johnson JL. The toxigenic element of Clostridium difficile strain VPI 10463. Microb Pathog. 1995;19:203–213. doi: 10.1016/s0882-4010(95)90263-5. [DOI] [PubMed] [Google Scholar]
- 77.Braun V, Hundsberger T, Leukel P, Sauerborn M, von Eichel-Streiber C. Definition of the single integration site of the pathogenicity locus in Clostridium difficile. Gene. 1996;181:29–38. doi: 10.1016/s0378-1119(96)00398-8. [DOI] [PubMed] [Google Scholar]
- 78.Monot M, et al. Clostridium difficile: new insights into the evolution of the pathogenicity locus. Sci Rep. 2015;5:15023. doi: 10.1038/srep15023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mani N, et al. Environmental response and autoregulation of Clostridium difficile TxeR, a sigma factor for toxin gene expression. J Bacteriol. 2002;184:5971–5978. doi: 10.1128/JB.184.21.5971-5978.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mani N, Dupuy B. Regulation of toxin synthesis in Clostridium difficile by an alternative RNA polymerase sigma factor. Proc Natl Acad Sci USA. 2001;98:5844–5849. doi: 10.1073/pnas.101126598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dupuy B, Govind R, Antunes A, Matamouros S. Clostridium difficile toxin synthesis is negatively regulated by TcdC. J Med Microbiol. 2008;57:685–689. doi: 10.1099/jmm.0.47775-0. [DOI] [PubMed] [Google Scholar]
- 82.Warny M, et al. Toxin production by an emerging strain of Clostridium difficile associated with outbreaks of severe disease in North America and Europe. Lancet. 2005;366:1079–1084. doi: 10.1016/S0140-6736(05)67420-X. [DOI] [PubMed] [Google Scholar]
- 83.Tan KS, Wee BY, Song KP. Evidence for holin function of tcdE gene in the pathogenicity of Clostridium difficile. J Med Microbiol. 2001;50:613–619. doi: 10.1099/0022-1317-50-7-613. [DOI] [PubMed] [Google Scholar]
- 84.Govind R, Dupuy B. Secretion of Clostridium difficile toxins A. and B requires the holin-like protein TcdE. PLoS Pathog. 2012;8:e1002727. doi: 10.1371/journal.ppat.1002727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Olling A, et al. Release of TcdA and TcdB from Clostridium difficile cdi 630 is not affected by functional inactivation of the tcdE gene. Microb Pathog. 2012;52:92–100. doi: 10.1016/j.micpath.2011.10.009. [DOI] [PubMed] [Google Scholar]
- 86.Karlsson S, et al. Expression of Clostridium difficile toxins A and B and their sigma factor TcdD is controlled by temperature. Infect Immun. 2003;71:1784–1793. doi: 10.1128/IAI.71.4.1784-1793.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Karlsson S, Lindberg A, Norin E, Burman LG, Akerlund T. Toxins, butyric acid, and other short-chain fatty acids are coordinately expressed and down-regulated by cysteine in Clostridium difficile. Infect Immun. 2000;68:5881–5888. doi: 10.1128/iai.68.10.5881-5888.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Aldape MJ, Packham AE, Nute DW, Bryant AE, Stevens DL. Effects of ciprofloxacin on the expression and production of exotoxins by Clostridium difficile. J Med Microbiol. 2013;62:741–747. doi: 10.1099/jmm.0.056218-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chilton CH, et al. Co-amoxiclav induces proliferation and cytotoxin production of Clostridium difficile ribotype 027 in a human gut model. J Antimicrob Chemother. 2012;67:951–954. doi: 10.1093/jac/dkr584. [DOI] [PubMed] [Google Scholar]
- 90.Dineen SS, McBride SM, Sonenshein AL. Integration of metabolism and virulence by Clostridium difficile CodY. J Bacteriol. 2010;192:5350–5362. doi: 10.1128/JB.00341-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Antunes A, et al. Global transcriptional control by glucose and carbon regulator CcpA in Clostridium difficile. Nucleic Acids Res. 2012;40:10701–10718. doi: 10.1093/nar/gks864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Dupuy B, Sonenshein AL. Regulated transcription of Clostridium difficile toxin genes. Mol Microbiol. 1998;27:107–120. doi: 10.1046/j.1365-2958.1998.00663.x. [DOI] [PubMed] [Google Scholar]
- 93.McKee RW, Mangalea MR, Purcell EB, Borchardt EK, Tamayo R. The second messenger cyclic Di-GMP regulates Clostridium difficile toxin production by controlling expression of sigD. J Bacteriol. 2013;195:5174–5185. doi: 10.1128/JB.00501-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.El Meouche I, et al. Characterization of the SigD regulon of C difficile and its positive control of toxin production through the regulation of tcdR. PLoS ONE. 2013;8:e83748. doi: 10.1371/journal.pone.0083748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mackin KE, Carter GP, Howarth P, Rood JI, Lyras D. Spo0A differentially regulates toxin production in evolutionarily diverse strains of Clostridium difficile. PLoS ONE. 2013;8:e79666. doi: 10.1371/journal.pone.0079666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Rosenbusch KE, Bakker D, Kuijper EJ, Smits WK. C difficile 630Δerm Spo0A regulates sporulation, but does not contribute to toxin production, by direct high-affinity binding to target DNA. PLoS ONE. 2012;7:e48608. doi: 10.1371/journal.pone.0048608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pettit LJ, et al. Functional genomics reveals that Clostridium difficile Spo0A coordinates sporulation, virulence and metabolism. BMC Genomics. 2014;15:160. doi: 10.1186/1471-2164-15-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Knetsch CW, et al. Comparative analysis of an expanded Clostridium difficile reference strain collection reveals genetic diversity and evolution through six lineages. Infect Genet Evol. 2012;12:1577–1585. doi: 10.1016/j.meegid.2012.06.003. [DOI] [PubMed] [Google Scholar]
- 99.Bouillaut L, Dubois T, Sonenshein AL, Dupuy B. Integration of metabolism and virulence in Clostridium difficile. Res Microbiol. 2015;166:375–383. doi: 10.1016/j.resmic.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Shen A. Clostridium difficile toxins: mediators of inflammation. J Innate Immun. 2012;4:149–158. doi: 10.1159/000332946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lyerly DM, Saum KE, MacDonald DK, Wilkins TD. Effects of Clostridium difficile toxins given intragastrically to animals. Infect Immun. 1985;47:349–352. doi: 10.1128/iai.47.2.349-352.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mitchell TJ, et al. Effect of toxin A and B of Clostridium difficile on rabbit ileum and colon. Gut. 1986;27:78–85. doi: 10.1136/gut.27.1.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Triadafilopoulos G, Pothoulakis C, O’Brien MJ, LaMont JT. Differential effects of Clostridium difficile toxins A and B on rabbit ileum. Gastroenterology. 1987;93:273–279. doi: 10.1016/0016-5085(87)91014-6. [DOI] [PubMed] [Google Scholar]
- 104.Riegler M, et al. Clostridium difficile toxin B is more potent than toxin A in damaging human colonic epithelium in vitro. J Clin Invest. 1995;95:2004–2011. doi: 10.1172/JCI117885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Savidge TC, et al. Clostridium difficile toxin B is an inflammatory enterotoxin in human intestine. Gastroenterology. 2003;125:413–420. doi: 10.1016/s0016-5085(03)00902-8. [DOI] [PubMed] [Google Scholar]
- 106.Chumbler NM, et al. Crystal structure of Clostridium difficile toxin A. Nat Microbiol. 2016;1:15002. doi: 10.1038/nmicrobiol.2015.2. A structural study that provided insight into the mode of action of autoproteolytic activity and allosteric activation of the large clostridial toxins. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Pruitt RN, et al. Structural determinants of Clostridium difficile toxin A glucosyltransferase activity. J Biol Chem. 2012;287:8013–8020. doi: 10.1074/jbc.M111.298414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Papatheodorou P, Zamboglou C, Genisyuerek S, Guttenberg G, Aktories K. Clostridial glucosylating toxins enter cells via clathrin-mediated endocytosis. PLoS ONE. 2010;5:e10673. doi: 10.1371/journal.pone.0010673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Pruitt RN, Lacy DB. Toward a structural understanding of Clostridium difficile toxins A and B. Front Cell Infect Microbiol. 2012;2:28. doi: 10.3389/fcimb.2012.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Greco A, et al. Carbohydrate recognition by Clostridium difficile toxin A. Nat Struct Mol Biol. 2006;13:460–461. doi: 10.1038/nsmb1084. [DOI] [PubMed] [Google Scholar]
- 111.Murase T, et al. Structural basis for antibody recognition in the receptor-binding domains of toxins A and B from Clostridium difficile. J Biol Chem. 2014;289:2331–2343. doi: 10.1074/jbc.M113.505917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Sauerborn M, Leukel P, von Eichel-Streiber C. The C-terminal ligand-binding domain of Clostridium difficile toxin A (TcdA) abrogates TcdA-specific binding to cells and prevents mouse lethality. FEMS Microbiol Lett. 1997;155:45–54. doi: 10.1111/j.1574-6968.1997.tb12684.x. [DOI] [PubMed] [Google Scholar]
- 113.Genisyuerek S, et al. Structural determinants for membrane insertion, pore formation and translocation of Clostridium difficile toxin B. Mol Microbiol. 2011;79:1643–1654. doi: 10.1111/j.1365-2958.2011.07549.x. [DOI] [PubMed] [Google Scholar]
- 114.Olling A, et al. The repetitive oligopeptide sequences modulate cytopathic potency but are not crucial for cellular uptake of Clostridium difficile toxin A. PLoS ONE. 2011;6:e17623. doi: 10.1371/journal.pone.0017623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Schorch B, et al. LRP1 is a receptor for Clostridium perfringens TpeL toxin indicating a two-receptor model of clostridial glycosylating toxins. Proc Natl Acad Sci USA. 2014;111:6431–6436. doi: 10.1073/pnas.1323790111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.LaFrance ME, et al. Identification of an epithelial cell receptor responsible for Clostridium difficile TcdB-induced cytotoxicity. Proc Natl Acad Sci USA. 2015;112:7073–7078. doi: 10.1073/pnas.1500791112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Yuan P, et al. Chondroitin sulfate proteoglycan 4 functions as the cellular receptor for Clostridium difficile toxin B. Cell Res. 2015;25:157–168. doi: 10.1038/cr.2014.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Terada N, et al. Immunohistochemical study of NG2 chondroitin sulfate proteoglycan expression in the small and large intestines. Histochem Cell Biol. 2006;126:483–490. doi: 10.1007/s00418-006-0184-3. [DOI] [PubMed] [Google Scholar]
- 119.Shen A, et al. Defining an allosteric circuit in the cysteine protease domain of Clostridium difficile toxins. Nat Struct Mol Biol. 2011;18:364–371. doi: 10.1038/nsmb.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Egerer M, Giesemann T, Jank T, Satchell KJ, Aktories K. Auto-catalytic cleavage of Clostridium difficile toxins A and B depends on cysteine protease activity. J Biol Chem. 2015;282:25314–25321. doi: 10.1074/jbc.M703062200. [DOI] [PubMed] [Google Scholar]
- 121.Savidge TC, et al. Host S-nitrosylation inhibits clostridial small molecule-activated glucosylating toxins. Nat Med. 2011;17:1136–1141. doi: 10.1038/nm.2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Lanis JM, Hightower LD, Shen A, Ballard JD. TcdB from hypervirulent Clostridium difficile exhibits increased efficiency of autoprocessing. Mol Microbiol. 2012;84:66–76. doi: 10.1111/j.1365-2958.2012.08009.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Just I, et al. The enterotoxin from Clostridium difficile (ToxA) monoglucosylates the Rho proteins. J Biol Chem. 1995;270:13932–13936. doi: 10.1074/jbc.270.23.13932. [DOI] [PubMed] [Google Scholar]
- 124.Just I, et al. Glucosylation of Rho proteins by Clostridium difficile toxin B. Nature. 1995;375:500–503. doi: 10.1038/375500a0. [DOI] [PubMed] [Google Scholar]
- 125.Brito GA, et al. Mechanism of Clostridium difficile toxin A-induced apoptosis in T84 cells. J Infect Dis. 2002;186:1438–1447. doi: 10.1086/344729. [DOI] [PubMed] [Google Scholar]
- 126.Farrow MA, et al. Clostridium difficile toxin B-induced necrosis is mediated by the host epithelial cell NADPH oxidase complex. Proc Natl Acad Sci USA. 2013;110:18674–18679. doi: 10.1073/pnas.1313658110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Slater LH, et al. Identification of novel host-targeted compounds that protect from anthrax lethal toxin-induced cell death. ACS Chem Biol. 2013;8:812–822. doi: 10.1021/cb300555n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Tam J, et al. Small molecule inhibitors of Clostridium difficile toxin B-induced cellular damage. Chem Biol. 2015;22:175–185. doi: 10.1016/j.chembiol.2014.12.010. [DOI] [PubMed] [Google Scholar]
- 129.Bender KO, et al. A small-molecule antivirulence agent for treating Clostridium difficile infection. Sci Transl Med. 2015;7:306ra148. doi: 10.1126/scitranslmed.aac9103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Smith SM, et al. Ebselen and congeners inhibit NADPH oxidase 2-dependent superoxide generation by interrupting the binding of regulatory subunits. Chem Biol. 2012;19:752–763. doi: 10.1016/j.chembiol.2012.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kuehne SA, et al. Importance of toxin A, toxin B, and CDT in virulence of an epidemic Clostridium difficile strain. J Infect Dis. 2014;209:83–86. doi: 10.1093/infdis/jit426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Carter GP, et al. Defining the roles of TcdA and TcdB in localized gastrointestinal disease, systemic organ damage, and the host response during Clostridium difficile infections. mBio. 2015;6:e00551. doi: 10.1128/mBio.00551-15. A histopathological analysis of the effects of TcdA and TcdB in a mouse model of CDI. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Drudy D, Fanning S, Kyne L. Toxin A-negative, toxin B-positive Clostridium difficile. Int J Infect Dis. 2007;11:5–10. doi: 10.1016/j.ijid.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 134.King AM, Mackin KE, Lyras D. Emergence of toxin A-negative, toxin B-positive Clostridium difficile strains: epidemiological and clinical considerations. Future Microbiol. 2015;10:1–4. doi: 10.2217/fmb.14.115. [DOI] [PubMed] [Google Scholar]
- 135.Gerding DN, Johnson S, Rupnik M, Aktories K. Clostridium difficile binary toxin CDT: mechanism, epidemiology, and potential clinical importance. Gut Microbes. 2014;5:15–27. doi: 10.4161/gmic.26854. A review of the different aspects of the binary toxin CDT. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Perelle S, Gibert M, Bourlioux P, Corthier G, Popoff MR. Production of a complete binary toxin (actin-specific ADP-ribosyltransferase) by Clostridium difficile CD196. Infect Immun. 1997;65:1402–1407. doi: 10.1128/iai.65.4.1402-1407.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Goncalves C, Decre D, Barbut F, Burghoffer B, Petit JC. Prevalence and characterization of a binary toxin (actin-specific ADP-ribosyltransferase) from Clostridium difficile. J Clin Microbiol. 2004;42:1933–1939. doi: 10.1128/JCM.42.5.1933-1939.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Stare BG, Delmee M, Rupnik M. Variant forms of the binary toxin CDT locus and tcdC gene in Clostridium difficile strains. J Med Microbiol. 2007;56:329–335. doi: 10.1099/jmm.0.46931-0. [DOI] [PubMed] [Google Scholar]
- 139.Carter GP, et al. Binary toxin production in Clostridium difficile is regulated by CdtR, a LytTR family response regulator. J Bacteriol. 2007;189:7290–7301. doi: 10.1128/JB.00731-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Metcalf DS, Weese JS. Binary toxin locus analysis in Clostridium difficile. J Med Microbiol. 2011;60:1137–1145. doi: 10.1099/jmm.0.028498-0. [DOI] [PubMed] [Google Scholar]
- 141.Sundriyal A, Roberts AK, Shone CC, Acharya KR. Structural basis for substrate recognition in the enzymatic component of ADP-ribosyltransferase toxin CDTa from Clostridium difficile. J Biol Chem. 2009;284:28713–28719. doi: 10.1074/jbc.M109.043018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Schwan C, et al. Cholesterol- and sphingolipid-rich microdomains are essential for microtubule-based membrane protrusions induced by Clostridium difficile transferase (CDT) J Biol Chem. 2011;286:29356–29365. doi: 10.1074/jbc.M111.261925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Androga GO, et al. Infection With toxin A-negative, toxin B-negative, binary toxin-positive Clostridium difficile in a young patient with ulcerative colitis. J Clin Microbiol. 2015;53:3702–3704. doi: 10.1128/JCM.01810-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Eckert C, et al. Prevalence and pathogenicity of binary toxin-positive Clostridium difficile strains that do not produce toxins A and B. New Microbes New Infect. 2015;3:12–17. doi: 10.1016/j.nmni.2014.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Hutton ML, Mackin KE, Chakravorty A, Lyras D. Small animal models for the study of Clostridium difficile disease pathogenesis. FEMS Microbiol Lett. 2014;352:140–149. doi: 10.1111/1574-6968.12367. [DOI] [PubMed] [Google Scholar]
- 146.Lawley TD, Young VB. Murine models to study Clostridium difficile infection and transmission. Anaerobe. 2013;24:94–97. doi: 10.1016/j.anaerobe.2013.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Best EL, Freeman J, Wilcox MH. Models for the study of Clostridium difficile infection. Gut Microbes. 2012;3:145–167. doi: 10.4161/gmic.19526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Collignon A. Methods for working with the mouse model. Methods Mol Biol. 2010;646:229–237. doi: 10.1007/978-1-60327-365-7_15. [DOI] [PubMed] [Google Scholar]
- 149.Lawley TD, et al. Targeted restoration of the intestinal microbiota with a simple, defined bacteriotherapy resolves relapsing Clostridium difficile disease in mice. PLoS Pathog. 2012;8:e1002995. doi: 10.1371/journal.ppat.1002995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Steele J, Feng H, Parry N, Tzipori S. Piglet models of acute or chronic Clostridium difficile illness. J Infect Dis. 2010;201:428–434. doi: 10.1086/649799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Baines SD, et al. Mixed infection by Clostridium difficile in an in vitro model of the human gut. J Antimicrob Chemother. 2013;68:1139–1143. doi: 10.1093/jac/dks529. [DOI] [PubMed] [Google Scholar]
- 152.Crowther GS, et al. Development and validation of a chemostat gut model to study both planktonic and biofilm modes of growth of Clostridium difficile and human microbiota. PLoS ONE. 2014;9:e88396. doi: 10.1371/journal.pone.0088396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Dobson G, Hickey C, Trinder J. Clostridium difficile colitis causing toxic megacolon, severe sepsis and multiple organ dysfunction syndrome. Intensive Care Med. 2003;29:1030. doi: 10.1007/s00134-003-1754-7. [DOI] [PubMed] [Google Scholar]
- 154.Cohen SH, et al. Clinical practice guidelines for Clostridium difficile infection in adults: 2010 update by the society for healthcare epidemiology of America (SHEA) and the infectious diseases society of America (IDSA) Infect Control Hosp Epidemiol. 2010;31:431–455. doi: 10.1086/651706. [DOI] [PubMed] [Google Scholar]
- 155.Debast SB, Bauer MP, Kuijper EJ. European Society of Clinical Microbiology and Infectious Diseases: update of the treatment guidance document for Clostridium difficile infection. Clin Microbiol Infect. 2014;20(Suppl 2):1–26. doi: 10.1111/1469-0691.12418. [DOI] [PubMed] [Google Scholar]
- 156.Yu H, et al. Identification of toxemia in patients with Clostridium difficile infection. PLoS ONE. 2015;10:e0124235. doi: 10.1371/journal.pone.0124235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Slimings C, Riley TV. Antibiotics and hospital-acquired Clostridium difficile infection: update of systematic review and meta-analysis. J Antimicrob Chemother. 2014;69:881–891. doi: 10.1093/jac/dkt477. [DOI] [PubMed] [Google Scholar]
- 158.Kwok CS, et al. Risk of Clostridium difficile infection with acid suppressing drugs and antibiotics: meta-analysis. Am J Gastroenterol. 2012;107:1011–1019. doi: 10.1038/ajg.2012.108. [DOI] [PubMed] [Google Scholar]
- 159.McDonald EG, Milligan J, Frenette C, Lee TC. Continuous proton pump inhibitor therapy and the associated risk of recurrent Clostridium difficile infection. JAMA Intern Med. 2015;175:784–791. doi: 10.1001/jamainternmed.2015.42. [DOI] [PubMed] [Google Scholar]
- 160.Novack L, et al. Acid suppression therapy does not predispose to Clostridium difficile infection: the case of the potential bias. PLoS ONE. 2014;9:e110790. doi: 10.1371/journal.pone.0110790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Tleyjeh IM, et al. Association between proton pump inhibitor therapy and Clostridium difficile infection: a contemporary systematic review and meta-analysis. PLoS ONE. 2012;7:e50836. doi: 10.1371/journal.pone.0050836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Bartlett JG, Gerding DN. Clinical recognition and diagnosis of Clostridium difficile infection. Clin Infect Dis. 2008;46(Suppl 1):S12–S18. doi: 10.1086/521863. [DOI] [PubMed] [Google Scholar]
- 163.Zollner-Schwetz I, et al. Role of Klebsiella oxytoca in antibiotic-associated diarrhea. Clin Infect Dis. 2008;47:e74–e78. doi: 10.1086/592074. [DOI] [PubMed] [Google Scholar]
- 164.Planche T, Wilcox MH. Diagnostic pitfalls in Clostridium difficile infection. Infect Dis Clin North Am. 2015;29:63–82. doi: 10.1016/j.idc.2014.11.008. [DOI] [PubMed] [Google Scholar]
- 165.Polage CR, et al. Overdiagnosis of Clostridium difficile infection in the molecular test era. JAMA Intern Med. 2015;175:1792–1801. doi: 10.1001/jamainternmed.2015.4114. Together with reference 34, this report confirms that reliance on molecular tests alone for diagnosing CDI will probably lead to overdiagnosis, overtreatment and increased health-care costs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Crobach MJ, et al. European Society of Clinical Microbiology and Infectious Diseases: update of the diagnostic guidance document for Clostridium difficile infection. Clin Microbiol Infect. doi: 10.1016/j.cmi.2016.03.010. in the press. [DOI] [PubMed] [Google Scholar]
- 167.Louie TJ, et al. Fidaxomicin preserves the intestinal microbiome during and after treatment of Clostridium difficile infection (CDI) and reduces both toxin reexpression and recurrence of CDI. Clin Infect Dis. 2012;55(Suppl 2):S132–S142. doi: 10.1093/cid/cis338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Eastwood K, Else P, Charlett A, Wilcox M. Comparison of nine commercially available Clostridium difficile toxin detection assays, a real-time PCR assay for C difficile tcdB, and a glutamate dehydrogenase detection assay to cytotoxin testing and cytotoxigenic culture methods. J Clin Microbiol. 2009;47:3211–3217. doi: 10.1128/JCM.01082-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Ota KV, McGowan KL. Clostridium difficile testing algorithms using glutamate dehydrogenase antigen and C difficile toxin enzyme immunoassays with C difficile nucleic acid amplification testing increase diagnostic yield in a tertiary pediatric population. J Clin Microbiol. 2012;50:1185–1188. doi: 10.1128/JCM.05620-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Reigadas E, et al. Missed diagnosis of Clostridium difficile infection; a prospective evaluation of unselected stool samples. J Infect. 2015;70:264–272. doi: 10.1016/j.jinf.2014.10.013. [DOI] [PubMed] [Google Scholar]
- 171.Faust SN, et al. Lack of evidence for an unmet need to treat Clostridium difficile infection in infants aged <2 years: expert recommendations on how to address this issue. Clin Infect Dis. 2015;60:912–918. doi: 10.1093/cid/ciu936. [DOI] [PubMed] [Google Scholar]
- 172.Schutze GE, Willoughby RE. Clostridium difficile infection in infants and children. Pediatrics. 2013;131:196–200. doi: 10.1542/peds.2012-2992. [DOI] [PubMed] [Google Scholar]
- 173.Spina A, et al. Spectrum of enteropathogens detected by the FilmArray GI Panel in a multicentre study of community-acquired gastroenteritis. Clin Microbiol Infect. 2015;21:719–728. doi: 10.1016/j.cmi.2015.04.007. [DOI] [PubMed] [Google Scholar]
- 174.Bruijnesteijn van Coppenraet LE, et al. Case–control comparison of bacterial and protozoan microorganisms associated with gastroenteritis: application of molecular detection. Clin Microbiol Infect. 2015;21:592. doi: 10.1016/j.cmi.2015.02.007. [DOI] [PubMed] [Google Scholar]
- 175.Curry SR, et al. Use of multilocus variable number of tandem repeats analysis genotyping to determine the role of asymptomatic carriers in Clostridium difficile transmission. Clin Infect Dis. 2013;57:1094–1102. doi: 10.1093/cid/cit475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Kyne L, Warny M, Qamar A, Kelly CP. Asymptomatic carriage of Clostridium difficile and serum levels of IgG antibody against toxin A. N Engl J Med. 2000;342:390–397. doi: 10.1056/NEJM200002103420604. [DOI] [PubMed] [Google Scholar]
- 177.Donskey CJ, Kundrapu S, Deshpande A. Colonization versus carriage of Clostridium difficile. Infect Dis Clin North Am. 2015;29:13–28. doi: 10.1016/j.idc.2014.11.001. [DOI] [PubMed] [Google Scholar]
- 178.Ziakas PD, et al. Asymptomatic carriers of toxigenic C difficile in long-term care facilities: a meta-analysis of prevalence and risk factors. PLoS ONE. 2015;10:e0117195. doi: 10.1371/journal.pone.0117195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Riggs MM, et al. Asymptomatic carriers are a potential source for transmission of epidemic and nonepidemic Clostridium difficile strains among long-term care facility residents. Clin Infect Dis. 2007;45:992–998. doi: 10.1086/521854. [DOI] [PubMed] [Google Scholar]
- 180.Loo VG, et al. Host and pathogen factors for Clostridium difficile infection and colonization. N Engl J Med. 2011;365:1693–1703. doi: 10.1056/NEJMoa1012413. [DOI] [PubMed] [Google Scholar]
- 181.Samore MH, Venkataraman L, DeGirolami PC, Arbeit RD, Karchmer AW. Clinical and molecular epidemiology of sporadic and clustered cases of nosocomial Clostridium difficile diarrhea. Am J Med. 1996;100:32–40. doi: 10.1016/s0002-9343(96)90008-x. [DOI] [PubMed] [Google Scholar]
- 182.Enoch DA, Butler MJ, Pai S, Aliyu SH, Karas JA. Clostridium difficile in children: colonisation and disease. J Infect. 2011;63:105–113. doi: 10.1016/j.jinf.2011.05.016. [DOI] [PubMed] [Google Scholar]
- 183.Furuichi M, et al. Characteristics of Clostridium difficile colonization in Japanese children. J Infect Chemother. 2014;20:307–311. doi: 10.1016/j.jiac.2014.01.009. [DOI] [PubMed] [Google Scholar]
- 184.Leibowitz J, Soma VL, Rosen L, Ginocchio CC, Rubin LG. Similar proportions of stool specimens from hospitalized children with and without diarrhea test positive for Clostridium difficile. Pediatr Infect Dis J. 2015;34:261–266. doi: 10.1097/INF.0000000000000556. [DOI] [PubMed] [Google Scholar]
- 185.Bergstrom A, et al. Establishment of intestinal microbiota during early life: a longitudinal, explorative study of a large cohort of Danish infants. Appl Environ Microbiol. 2014;80:2889–2900. doi: 10.1128/AEM.00342-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Shim JK, Johnson S, Samore MH, Bliss DZ, Gerding DN. Primary symptomless colonisation by Clostridium difficile and decreased risk of subsequent diarrhoea. Lancet. 1998;351:633–636. doi: 10.1016/S0140-6736(97)08062-8. [DOI] [PubMed] [Google Scholar]
- 187.Gerding DN, et al. Administration of spores of nontoxigenic Clostridium difficile strain M3 for prevention of recurrent C difficile infection: a randomized clinical trial. JAMA. 2015;313:1719–1727. doi: 10.1001/jama.2015.3725. [DOI] [PubMed] [Google Scholar]
- 188.Brouwer MS, et al. Horizontal gene transfer converts non-toxigenic Clostridium difficile strains into toxin producers. Nat Commun. 2013;4:2601. doi: 10.1038/ncomms3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Clabots CR, Johnson S, Olson MM, Peterson LR, Gerding DN. Acquisition of Clostridium difficile by hospitalized patients: evidence for colonized new admissions as a source of infection. J Infect Dis. 1992;166:561–567. doi: 10.1093/infdis/166.3.561. [DOI] [PubMed] [Google Scholar]
- 190.Lanzas C, Dubberke ER, Lu Z, Reske KA, Grohn YT. Epidemiological model for Clostridium difficile transmission in healthcare settings. Infect Control Hosp Epidemiol. 2011;32:553–561. doi: 10.1086/660013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Vonberg RP, et al. Infection control measures to limit the spread of Clostridium difficile. Clin Microbiol Infect. 2008;14(Suppl 5):2–20. doi: 10.1111/j.1469-0691.2008.01992.x. [DOI] [PubMed] [Google Scholar]
- 192.Dubberke ER, et al. Strategies to prevent Clostridium difficile infections in acute care hospitals: 2014 update. Infect Control Hosp Epidemiol. 2014;35(Suppl 2):S48–S65. doi: 10.1017/s0899823x00193857. [DOI] [PubMed] [Google Scholar]
- 193.Surawicz CM, et al. Guidelines for diagnosis, treatment, and prevention of Clostridium difficile infections. Am J Gastroenterol. 2013;108:478–498. doi: 10.1038/ajg.2013.4. [DOI] [PubMed] [Google Scholar]
- 194.Dancer SJ. Controlling hospital-acquired infection: focus on the role of the environment and new technologies for decontamination. Clin Microbiol Rev. 2014;27:665–690. doi: 10.1128/CMR.00020-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Feazel LM, et al. Effect of antibiotic stewardship programmes on Clostridium difficile incidence: a systematic review and meta-analysis. J Antimicrob Chemother. 2014;69:1748–1754. doi: 10.1093/jac/dku046. [DOI] [PubMed] [Google Scholar]
- 196.Barbut F. How to eradicate Clostridium difficile from the environment. J Hosp Infect. 2015;89:287–295. doi: 10.1016/j.jhin.2014.12.007. [DOI] [PubMed] [Google Scholar]
- 197.Johnson S, et al. Vancomycin, metronidazole, or tolevamer for Clostridium difficile infection: results from two multinational, randomized, controlled trials. Clin Infect Dis. 2014;59:345–354. doi: 10.1093/cid/ciu313. This paper describes two large randomized studies and was the first paper to show the significantly improved outcome following vancomycin versus metronidazole therapy for CDI on an intent-to-treat basis. [DOI] [PubMed] [Google Scholar]
- 198.Cornely OA, Miller MA, Louie TJ, Crook DW, Gorbach SL. Treatment of first recurrence of Clostridium difficile infection: fidaxomicin versus vancomycin. Clin Infect Dis. 2012;55(Suppl 2):S154–S161. doi: 10.1093/cid/cis462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Neal MD, Alverdy JC, Hall DE, Simmons RL, Zuckerbraun BS. Diverting loop ileostomy and colonic lavage: an alternative to total abdominal colectomy for the treatment of severe, complicated Clostridium difficile associated disease. Ann Surg. 2011;254:423–427. doi: 10.1097/SLA.0b013e31822ade48. [DOI] [PubMed] [Google Scholar]
- 200.Bauer MP, et al. Renal failure and leukocytosis are predictors of a complicated course of Clostridium difficile infection if measured on day of diagnosis. Clin Infect Dis. 2012;55(Suppl 2):S149–S153. doi: 10.1093/cid/cis340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Kelly CP. Can we identify patients at high risk of recurrent Clostridium difficile infection? Clin Microbiol Infect. 2012;18(Suppl 6):21–27. doi: 10.1111/1469-0691.12046. [DOI] [PubMed] [Google Scholar]
- 202.Cornely OA, et al. Fidaxomicin versus vancomycin for infection with Clostridium difficile in Europe, Canada, and the USA: a double-blind, non-inferiority, randomised controlled trial. Lancet Infect Dis. 2012;12:281–289. doi: 10.1016/S1473-3099(11)70374-7. [DOI] [PubMed] [Google Scholar]
- 203.Louie TJ, et al. Fidaxomicin versus vancomycin for Clostridium difficile infection. N Engl J Med. 2011;364:422–431. doi: 10.1056/NEJMoa0910812. Together with reference 202, this study formed the basis for the addition of fidaxomicin as a therapeutic for the treatment of recurrent CDI. [DOI] [PubMed] [Google Scholar]
- 204.D’Agostino RB, Sr, Collins SH, Pencina KM, Kean Y, Gorbach S. Risk estimation for recurrent Clostridium difficile infection based on clinical factors. Clin Infect Dis. 2014;58:1386–1393. doi: 10.1093/cid/ciu107. [DOI] [PubMed] [Google Scholar]
- 205.Goldenberg JZ, et al. Probiotics for the prevention of Clostridium difficile-associated diarrhea in adults and children. Cochrane Database Syst Rev. 2013;5:CD006095. doi: 10.1002/14651858.CD006095.pub3. [DOI] [PubMed] [Google Scholar]
- 206.van Nood E, et al. Duodenal infusion of donor feces for recurrent Clostridium difficile. N Engl J Med. 2013;368:407–415. doi: 10.1056/NEJMoa1205037. The first randomized controlled trial demonstrating the superiority of FMT over vancomycin for the treatment of patients with multiple recurrences of CDI. [DOI] [PubMed] [Google Scholar]
- 207.Sokol H, et al. Faecal microbiota transplantation in recurrent Clostridium difficile infection: recommendations from the French Group of Faecal microbiota Transplantation. Dig Liver Dis. 2015;48:242–247. doi: 10.1016/j.dld.2015.08.017. [DOI] [PubMed] [Google Scholar]
- 208.Varier RU, et al. Cost-effectiveness analysis of fecal microbiota transplantation for recurrent Clostridium difficile infection. Infect Control Hosp Epidemiol. 2015;36:438–444. doi: 10.1017/ice.2014.80. [DOI] [PubMed] [Google Scholar]
- 209.Kump PK, et al. Recommendations for the use of faecal microbiota transplantation ‘stool transplantation’: consensus of the Austrian Society of Gastroenterology and Hepatology (OGGH) in cooperation with the Austrian Society of Infectious Diseases and Tropical Medicine. Z Gastroenterol. 2014;52:1485–1492. doi: 10.1055/s-0034-1385562. (in German) [DOI] [PubMed] [Google Scholar]
- 210.Youngster I, et al. Oral, capsulized, frozen fecal microbiota transplantation for relapsing Clostridium difficile infection. JAMA. 2014;312:1772–1778. doi: 10.1001/jama.2014.13875. [DOI] [PubMed] [Google Scholar]
- 211.Petrof EO, et al. Stool substitute transplant therapy for the eradication of Clostridium difficile infection: ‘RePOOPulating’ the gut. Microbiome. 2013;1:3. doi: 10.1186/2049-2618-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Tvede M, Tinggaard M, Helms M. Rectal bacteriotherapy for recurrent Clostridium difficile-associated diarrhoea: results from a case series of 55 patients in Denmark 2000–2012. Clin Microbiol Infect. 2015;21:48–53. doi: 10.1016/j.cmi.2014.07.003. [DOI] [PubMed] [Google Scholar]
- 213.Khanna S, et al. A novel microbiome therapeutic increases gut microbial diversity and prevents recurrent Clostridium difficile infection. J Infect Dis. 2016 doi: 10.1093/infdis/jiv766. http://dx.doi.org/10.1093/infdis/jiv766. [DOI] [PubMed]
- 214.Wilcox M, et al. Bezlotoxumab alone and with actoxumab for prevention of recurrant C. difficile infection in patients on standard of care antibiotics: integrated results of 2 Phase 3 studies (MODIFY I and MODIFY II) Open Forum Infect Dis. 2015 [online], http://ofid.oxfordjournals.org/content/2/suppl_1/S3.1.full.
- 215.Wilcox MH, Cunniffe JG, Trundle C, Redpath C. Financial burden of hospital-acquired Clostridium difficile infection. J Hosp Infect. 1996;34:23–30. doi: 10.1016/s0195-6701(96)90122-x. [DOI] [PubMed] [Google Scholar]
- 216.Vonberg RP, et al. Costs of nosocomial Clostridium difficile-associated diarrhoea. J Hosp Infect. 2008;70:15–20. doi: 10.1016/j.jhin.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 217.Wiegand PN, et al. Clinical and economic burden of Clostridium difficile infection in Europe: a systematic review of healthcare-facility-acquired infection. J Hosp Infect. 2012;81:1–14. doi: 10.1016/j.jhin.2012.02.004. A systematic review of European data on CDI-related mortality, recurrence, length of hospital stay and cost. [DOI] [PubMed] [Google Scholar]
- 218.Mitchell BG, Gardner A. Prolongation of length of stay and Clostridium difficile infection: a review of the methods used to examine length of stay due to healthcare associated infections. Antimicrob Resist Infect Control. 2012;1:14. doi: 10.1186/2047-2994-1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Ghantoji SS, Sail K, Lairson DR, DuPont HL, Garey KW. Economic healthcare costs of Clostridium difficile infection: a systematic review. J Hosp Infect. 2010;74:309–318. doi: 10.1016/j.jhin.2009.10.016. [DOI] [PubMed] [Google Scholar]
- 220.Nanwa N, et al. The economic impact of Clostridium difficile infection: a systematic review. Am J Gastroenterol. 2015;110:511–519. doi: 10.1038/ajg.2015.48. [DOI] [PubMed] [Google Scholar]
- 221.Zimlichman E, et al. Health care-associated infections: a meta-analysis of costs and financial impact on the US health care system. JAMA Intern Med. 2013;173:2039–2046. doi: 10.1001/jamainternmed.2013.9763. [DOI] [PubMed] [Google Scholar]
- 222.Levy AR, et al. Incidence and costs of Clostridium difficile infections in Canada. Open Forum Infect Dis. 2015;2:ofv076. doi: 10.1093/ofid/ofv076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Kwon JH, Olsen MA, Dubberke ER. The morbidity, mortality, and costs associated with Clostridium difficile infection. Infect Dis Clin North Am. 2015;29:123–134. doi: 10.1016/j.idc.2014.11.003. [DOI] [PubMed] [Google Scholar]
- 224.Vuong NN, et al. Use of PROMIS network to evaluate patient-reported health status associated with Clostridium difficile infection. Am Soc Microbiol Abstr. 2015 [online], http://www.abstractsonline.com/Plan/ViewAbstract.aspx?sKey=568b5770-3a64-49e2-9209-0c8c05a52334&cKey=72a0bada-33bf-4ef9-9436-0ce89e73ac95&mKey=%7b7A574A80-EAB1-4B50-B343-4695DF14907E%7d.
- 225.Fimlaid KA, Shen A. Diverse mechanisms regulate sporulation sigma factor activity in the Firmicutes. Curr Opin Microbiol. 2015;24:88–95. doi: 10.1016/j.mib.2015.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Battistuzzi FU, Feijao A, Hedges SB. A genomic timescale of prokaryote evolution: insights into the origin of methanogenesis, phototrophy, and the colonization of land. BMC Evol Biol. 2004;4:44. doi: 10.1186/1471-2148-4-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Merrigan M, et al. Human hypervirulent Clostridium difficile strains exhibit increased sporulation as well as robust toxin production. J Bacteriol. 2010;192:4904–4911. doi: 10.1128/JB.00445-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Burns DA, Heap JT, Minton NP. The diverse sporulation characteristics of Clostridium difficile clinical isolates are not associated with type. Anaerobe. 2010;16:618–622. doi: 10.1016/j.anaerobe.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 229.Borgmann S, et al. Increased number of Clostridium difficile infections and prevalence of Clostridium difficile PCR ribotype 001 in southern Germany. Euro Surveill. 2008;13:5. [PubMed] [Google Scholar]
- 230.Knetsch CW, et al. Genetic markers for Clostridium difficile lineages linked to hypervirulence. Microbiology. 2011;157:3113–3123. doi: 10.1099/mic.0.051953-0. [DOI] [PubMed] [Google Scholar]
- 231.Eyre DW, et al. Emergence and spread of predominantly community-onset Clostridium difficile PCR ribotype 244 infection in Australia, 2010 to 2012. Euro Surveill. 2015;20:21059. doi: 10.2807/1560-7917.es2015.20.10.21059. [DOI] [PubMed] [Google Scholar]
- 232.Rea MC, et al. Thuricin CD, a posttranslationally modified bacteriocin with a narrow spectrum of activity against Clostridium difficile. Proc Natl Acad Sci USA. 2010;107:9352–9357. doi: 10.1073/pnas.0913554107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Hopkins MJ, Macfarlane GT. Changes in predominant bacterial populations in human faeces with age and with Clostridium difficile infection. J Med Microbiol. 2002;51:448–454. doi: 10.1099/0022-1317-51-5-448. [DOI] [PubMed] [Google Scholar]
- 234.Bingley PJ, Harding GM. Clostridium difficile colitis following treatment with metronidazole and vancomycin. Postgrad Med J. 1987;63:993–994. doi: 10.1136/pgmj.63.745.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Gebhart D, et al. A modified R-type bacteriocin specifically targeting Clostridium difficile prevents colonization of mice without affecting gut microbiota diversity. mBio. 2015;6:e02368–14. doi: 10.1128/mBio.02368-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Hargreaves KR, Clokie MR. Clostridium difficile phages: still difficult? Front Microbiol. 2014;5:184. doi: 10.3389/fmicb.2014.00184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Rasko DA, Sperandio V. Anti-virulence strategies to combat bacteria-mediated disease. Nat Rev Drug Discov. 2010;9:117–128. doi: 10.1038/nrd3013. [DOI] [PubMed] [Google Scholar]
- 238.Lowy I, et al. Treatment with monoclonal antibodies against Clostridium difficile toxins. N Engl J Med. 2010;362:197–205. doi: 10.1056/NEJMoa0907635. [DOI] [PubMed] [Google Scholar]
- 239.Martin CE, et al. Immunological evaluation of a synthetic Clostridium difficile oligosaccharide conjugate vaccine candidate and identification of a minimal epitope. J Am Chem Soc. 2013;135:9713–9722. doi: 10.1021/ja401410y. [DOI] [PubMed] [Google Scholar]
- 240.Kandalaft H, et al. Targeting surface-layer proteins with single-domain antibodies: a potential therapeutic approach against Clostridium difficile-associated disease. Appl Microbiol Biotechnol. 2015;99:8549–8562. doi: 10.1007/s00253-015-6594-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Abou Chakra CN, Pepin J, Valiquette L. Prediction tools for unfavourable outcomes in Clostridium difficile infection: a systematic review. PLoS ONE. 2012;7:e30258. doi: 10.1371/journal.pone.0030258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Na X, et al. A multi-center prospective derivation and validation of a clinical prediction tool for severe Clostridium difficile infection. PLoS ONE. 2015;10:e0123405. doi: 10.1371/journal.pone.0123405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Hensgens MP, Dekkers OM, Goorhuis A, LeCessie S, Kuijper EJ. Predicting a complicated course of Clostridium difficile infection at the bedside. Clin Microbiol Infect. 2014;20:O301–O308. doi: 10.1111/1469-0691.12391. [DOI] [PubMed] [Google Scholar]
- 244.Zilberberg MD, Reske K, Olsen M, Yan Y, Dubberke ER. Development and validation of a recurrent Clostridium difficile risk-prediction model. J Hosp Med. 2014;9:418–423. doi: 10.1002/jhm.2189. [DOI] [PubMed] [Google Scholar]
- 245.European Commission. The 2015 Ageing Report. Underlying Assumptions and Projection Methodologies. European Economy. 2014 [online], http://ec.europa.eu/economy_finance/publications/european_economy/2014/pdf/ee8_en.pdf.
- 246.Ortman JM, Velkoff VA, Howard H. An Aging Nation: The Older Population in the United States. US Census Bureau. 2014 [online], https://www.census.gov/prod/2014pubs/p25-1140.pdf.
- 247.Tenover FC, et al. Comparison of strain typing results for Clostridium difficile isolates from North America. J Clin Microbiol. 2011;49:1831–1837. doi: 10.1128/JCM.02446-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Fawley WN, et al. Development and validation of an internationally-standardized, high-resolution capillary gel-based electrophoresis PCR-ribotyping protocol for Clostridium difficile. PLoS ONE. 2015;10:e0118150. doi: 10.1371/journal.pone.0118150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Baines SD, O’Connor R, Saxton K, Freeman J, Wilcox MH. Activity of vancomycin against epidemic Clostridium difficile strains in a human gut model. J Antimicrob Chemother. 2009;63:520–525. doi: 10.1093/jac/dkn502. [DOI] [PubMed] [Google Scholar]
- 250.Paredes-Sabja D, Cofre-Araneda G, Brito-Silva C, Pizarro-Guajardo M, Sarker MR. Clostridium difficile spore-macrophage interactions: spore survival. PLoS ONE. 2012;7:e43635. doi: 10.1371/journal.pone.0043635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Spigaglia P, Barbanti F, Mastrantonio P. Multidrug resistance in European Clostridium difficile clinical isolates. J Antimicrob Chemother. 2011;66:2227–2234. doi: 10.1093/jac/dkr292. [DOI] [PubMed] [Google Scholar]
- 252.Peltier J, et al. Genomic and expression analysis of the vanG-like gene cluster of Clostridium difficile. Microbiology. 2013;159:1510–1520. doi: 10.1099/mic.0.065060-0. [DOI] [PubMed] [Google Scholar]
- 253.Ammam F, et al. The functional vanGCd cluster of Clostridium difficile does not confer vancomycin resistance. Mol Microbiol. 2013;89:612–625. doi: 10.1111/mmi.12299. [DOI] [PubMed] [Google Scholar]
- 254.Amy J, Johanesen P, Lyras D. Extrachromosomal and integrated genetic elements in Clostridium difficile. Plasmid. 2015;80:97–110. doi: 10.1016/j.plasmid.2015.04.006. [DOI] [PubMed] [Google Scholar]
- 255.Hansen LH, Vester B. A cfr-like gene from Clostridium difficile confers multiple antibiotic resistance by the same mechanism as the cfr gene. Antimicrob Agents Chemother. 2015;59:5841–5843. doi: 10.1128/AAC.01274-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Roberts AP, Johanesen PA, Lyras D, Mullany P, Rood JI. Comparison of Tn5397 from Clostridium difficile, Tn916 from Enterococcus faecalis and the CW459tet(M) element from Clostridium perfringens shows that they have similar conjugation regions but different insertion and excision modules. Microbiology. 2001;147:1243–1251. doi: 10.1099/00221287-147-5-1243. [DOI] [PubMed] [Google Scholar]
- 257.Chong PM, et al. Proteomic analysis of a NAP1 Clostridium difficile clinical isolate resistant to metronidazole. PLoS ONE. 2014;9:e82622. doi: 10.1371/journal.pone.0082622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Lynch T, et al. Characterization of a stable, metronidazole-resistant Clostridium difficile clinical isolate. PLoS ONE. 2013;8:e53757. doi: 10.1371/journal.pone.0053757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Leeds JA, Sachdeva M, Mullin S, Barnes SW, Ruzin A. In vitro selection, via serial passage, of Clostridium difficile mutants with reduced susceptibility to fidaxomicin or vancomycin. J Antimicrob Chemother. 2014;69:41–44. doi: 10.1093/jac/dkt302. [DOI] [PubMed] [Google Scholar]
- 260.Freeman J, et al. Pan-European longitudinal surveillance of antibiotic resistance among prevalent Clostridium difficile ribotypes. Clin Microbiol Infect. 2015;21:248.e9–248.e16. doi: 10.1016/j.cmi.2014.09.017. [DOI] [PubMed] [Google Scholar]
- 261.Centers for Disease Control and Prevention. CDC; 2013. Threat Report 2013. [online], http://www.cdc.gov/drugresistance/threat-report-2013/ A report from the CDC that qualifies C. difficile as an urgent antibiotic-resistance threat. [Google Scholar]
- 262.Spigaglia P, et al. Fluoroquinolone resistance in Clostridium difficile isolates from a prospective study of C difficile infections in Europe. J Med Microbiol. 2008;57:784–789. doi: 10.1099/jmm.0.47738-0. [DOI] [PubMed] [Google Scholar]
- 263.Papatheodorou P, et al. Lipolysis-stimulated lipoprotein receptor (LSR) is the host receptor for the binary toxin Clostridium difficile transferase (CDT) Proc Natl Acad Sci USA. 2011;108:16422–16427. doi: 10.1073/pnas.1109772108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Martin J, Monaghan T, Wilcox MH. Clostridium difficile infection: advances in epidemiology, diagnosis and understanding of transmission. Nat Rev Gastroenterol Hepatol. 2016 doi: 10.1038/nrgastro.2016.25. http://dx.doi.org/10.1038/nrgastro.2016.25. [DOI] [PubMed]
- 265.Le MA, et al. Hospital cost of Clostridium difficile infection including the contribution of recurrences in French acute-care hospitals. J Hosp Infect. 2015;91:117–122. doi: 10.1016/j.jhin.2015.06.017. [DOI] [PubMed] [Google Scholar]
- 266.Gabriel L, Beriot-Mathiot A. Hospitalization stay and costs attributable to Clostridium difficile infection: a critical review. J Hosp Infect. 2014;88:12–21. doi: 10.1016/j.jhin.2014.04.011. [DOI] [PubMed] [Google Scholar]