Abstract
Model-based molecular phylogenetics plays an important role in comparisons of genomic data, and model selection is a key step in all such analyses. We present ModelFinder, a fast model-selection method that greatly improves the accuracy of phylogenetic estimates. The improvement is achieved by incorporating a model of rate-heterogeneity across sites not previously considered in this context, and by allowing concurrent searches of model-space and tree-space.
Model-based molecular phylogenetic analysis plays a key role in comparative genomics and evolutionary biology, allowing us to annotate genomes more accurately1, test our understanding of the evolution of species, genomes and genes2–6, and determine the likely origins and routes of dispersal of pathogens and pests7,8. Selecting an optimal model of sequence evolution (SE) is a critical step in all such analyses. Here we introduce ModelFinder, a model-selection method that combines substitution models used in other popular model-selection methods9,10 with a flexible rate-heterogeneity-across-sites (RHAS) model, and show that its use often leads to substantial improvements in the fit between tree, model and data.
Model selection is used to identify the best-fitting model of SE that led to the available data. Several methods for doing so are available for DNA9 and protein10. It is even possible to do so when different models are required for analysis of different sets of sites in an alignment11.
Finding an optimal model of SE for a given sequence alignment entails finding the best-fitting substitution model and the best-fitting model of RHAS. Usually, this means comparing three models of RHAS that assume: (i) all sites evolved at the same rate, (ii) some sites evolved at the same rate whilst the others were invariable (I), or (iii) RHAS follows a probability distribution, like the popular discrete Γ distribution12.
The discrete Γ distribution is parameterized using k rate categories, each comprising a rate (ri) and a weight (wi), where ri > 0, wi = 1/k, and Doing so imposes two constraints on the model: it is assumed RHAS can be modeled accurately by a Γ distribution, and that the probability that a site belongs to rate category i equals 1/k. These assumptions may be unrealistic and bias phylogenetic estimates.
One solution to this problem is to infer the weights from the data, as proposed by Yang13. The advantage offered by this probability-distribution-free (PDF) model of RHAS is that the distribution of rates-of-change across sites may take any shape, implying that estimates of rates and weights should be more accurate than those obtained under a Γ distribution. Until now, however, the PDF model was not available in the context of model selection.
To meet this need, we developed ModelFinder, a model-selection method for alignments of nucleotides, codons, amino acids, or other discrete data. ModelFinder is implemented in IQ-TREE14 and offers many features, including the choice of comparing models of SE inferred on the same tree (default) or on different trees (advanced). When the advanced option is used, ModelFinder searches tree space for every model of SE considered and, therefore, may find superior models of SE. ModelFinder incorporates 22 and 36 substitution models for DNA and protein, respectively, and 13 models of RHAS, including the PDF model with k = 2, … , kmax rate categories. By default, kmax = 10 but it can be increased if needed. Each PDF model, henceforth labelled Rk, is a family of RHAS models. The user can also specify the numbers and types of models to compare. In summary, ModelFinder considers models of RHAS that are more complex than those considered by other model-selection methods9–11.
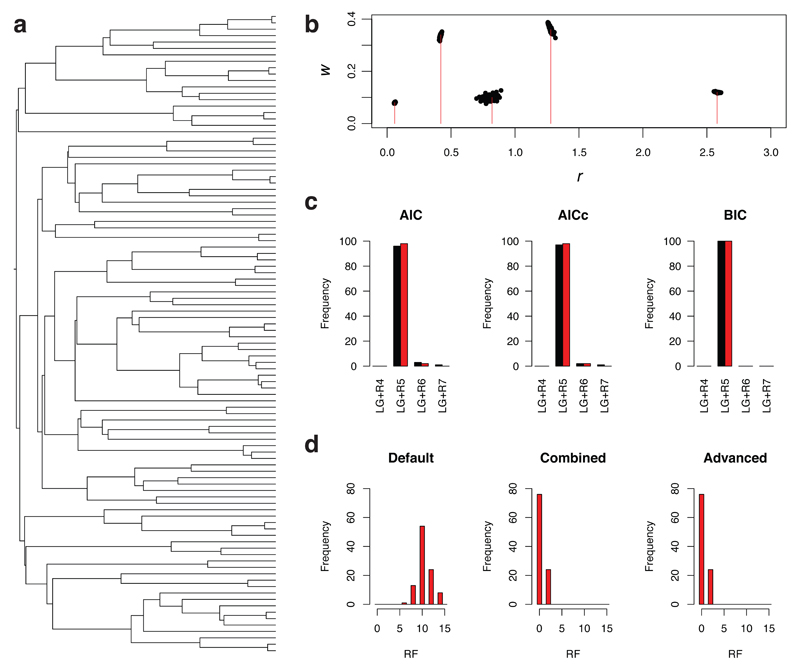
The PDF model is more parameter-rich than the discrete Γ model, so parameter estimation is a challenge. To tackle this challenge, ModelFinder uses the expectation-maximization (EM) algorithm15 to estimate the parameters for every Rk model, and an algorithm to identify the optimal value of k for the PDF model (Online Methods). The accuracy of ModelFinder was assessed by analysis of 100 amino-acid alignments generated on a 100-tipped tree (Fig. 1a). Alignments with 10,000 sites were generated using INDELible16 and the LG17+R5 model of SE. A bimodal distribution of RHAS was used. Figure 1b shows that ModelFinder estimated the model parameters accurately when the data were analyzed using the correct tree and model. Figure 1c shows that ModelFinder is accurate regardless of the optimality criterion (AIC, AICc, or BIC) and search option (default or advanced) used. When AIC or AICc were used, a 2-3% bias towards more parameter-rich RHAS models was found. The high success rate of BIC is noteworthy because the optimal model of SE was inferred even when the best tree found differed from the true tree. Figure 1d shows the distribution of Robinson-Foulds (RF) distances18 between the true tree and: (a) the parsimony tree (found using the default search option), (b) the tree inferred using the best model of SE found using the default search option, and (c) the tree found using the advanced search option. The RF distances ranged from 0 to 14, implying, in the best cases, that the trees were identical and, in the worst cases, that 7 of the 97 internal edges differed between the trees. In summary, ModelFinder is accurate and can identify models of SE that other model-selection methods are unable to detect.
Figure 1. Assessment of the accuracy of phylogenetic estimates obtained using ModelFinder.
(a) The rooted 100-tipped tree, with a root-to-tip distance of 0.5 substitutions/site, that was used to generate the simulated data. (b) Plot showing the true values of ri and wi (red lines; ri = (0.06, 0.42, 0.82, 1.28, 2.58) and wi = (0.08, 0.34, 0.10, 0.36, 0.12)) and the estimated values of (ri, wi) for the 100 simulated data sets (black dots). (c) Histograms showing the number of times different models of SE were identified under different criteria (AIC, AICc and BIC) using the default (black) and advanced (red) search options. (d) Graphs showing the distribution of Robinson-Foulds (RF) distances between the true tree and (a) the tree used during the default model search (Default), (b) the tree found, given the optimal model of SE found using the default model-search option (Combined), and (c) the tree found during the advanced model search (Advanced) (the BIC optimality criterion was used in this example).
The benefits of using ModelFinder are illustrated with an analysis of the alignment of amino acids that formed the basis for a genomic encyclopedia of Bacteria and Archaea19. The data were originally analyzed using the WAG+I+Γ5 model. The optimal model of SE was the same (LG+R14) for the two search options but the advanced option led to a better-parameterized model (BIC = 3,855,048) than the default option (BIC = 3,858,039) (when BIC scores differ by more than 10 (ΔBIC > 10) there is strong evidence against the model with the higher BIC score20). The large difference between these BIC scores (ΔBIC = 2,991) concurs with a large difference between the corresponding trees (RF = 138), implying that the default search option relied on a suboptimal tree. Doing so may lead to the selection of a suboptimal model of SE; that did not occur here, but it is a risk to consider when the default search option is used.
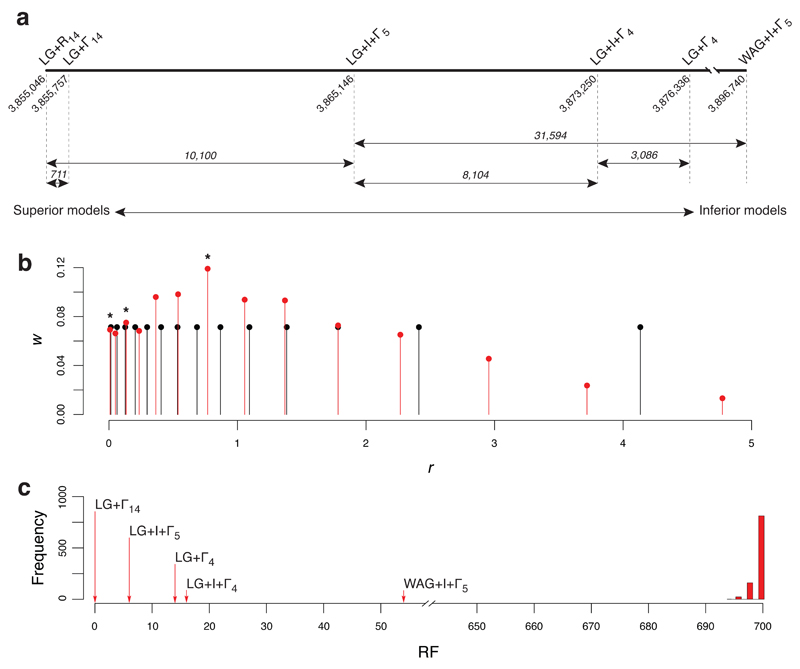
We then did a phylogenetic analysis to compare the estimates for selected models. Figure 2a confirms that the LG+R14 model is the best. Factors contributing to its superior fit include changes in substitution model (WAG+I+Γ5→LG+I+Γ5:ΔBIC = 31,954) and the RHAS model (LG+I+Γ5→LG+R14:ΔBIC = 10,100). Other models considered reveal the effects of the I model of RHAS (LG+Γ4→LG+I+Γ4:ΔBIC = 3,086) and the number of rate categories used to model the Γ distribution (LG+I+Γ4→LG+I+Γ5:ΔBIC = 8,104). Given this last result, we wondered whether the LG+Γ14 model might fit the data better than the LG+R14 model, but this was not the case (ΔBIC = 711). Figure 2b shows the estimates of ri and wi for the R14 and Γ14 models. Unlike the Γ14 model, the R14 model is trimodal and has a larger maximum/minimum rate ratio (rmax/rmin = 575 for R14 and 274 for Γ14). In summary, for these data, RHAS is best modeled by the R14 model.
Figure 2. Illustration of the advantages provided by ModelFinder.
(a) One-dimensional plot showing the BIC scores of selected models of SE, given the alignment of amino acids used by Wu et al.19 The models are listed above the line. Numbers drawn at a 45° angle are the BIC scores and those shown in italics are the ΔBIC scores. The relative position of each model of SE is shown on the axis, with the worst model on the right and the best model on the left. (b) Plot showing the values of ri and wi obtained under the R14 model of RHAS (red lines and balls) and the Γ14 model of RHAS (black lines and balls) for the alignment analyzed by Wu et al.19 Stars (*) indicate local peaks in the R14 model of RHAS. (c) Plot showing the RF distances between the most likely tree inferred under the LG+R14 model of SE and the most likely trees inferred under the LG+Γ14, LG+Γ4, LG+I+Γ4, LG+I+Γ5 and WAG+I+Γ5 models of SE. For comparison, a histogram with the distribution of 1,000 RF distances is included; each of these distances was obtained by comparing the most likely tree inferred under the LG+R14 model of SE to a randomly-generated tree with the same number of leaves.
Finally, we wanted to see whether the optimal tree for these data was model-dependent. Figure 2c shows the RF distances between the most likely tree inferred under the LG+R14 model and those inferred under the other models. The RF distances ranged from 0 to 54, so the optimal tree for these data is clearly model-dependent. Interestingly, although the trees inferred under the other models differ from that inferred under the LG+R14 model, they are still significantly more like the tree inferred under the LG+R14 model than random trees are, so the other models are not too misleading. That said, the best explanation for these data is provided by the tree inferred under the LG+R14 model.
Similar results emerged from analyses of other phylogenetic data (Table 1). In each of these cases, the best model of SE involved the PDF model of RHAS, and the best tree inferred using this model often differed from that found using the best model identified using other model-selection methods. Clearly, using ModelFinder can lead to a significant improvement in the fit between tree, model, and data irrespective of the source and type of data. A survey of 130 other data sets from TreeBASE21 reinforces this conclusion (Supplementary Table 1): in 122 of the cases, the fit between tree, model, and data improved (in 111 cases significantly), and in 118 of the cases, the tree topology changed. When the default and advanced search options were compared, a better fit between tree, model, and data was found using the advanced search option in 75 of the 130 cases. In 46 of these 75 cases, the models of SE differed, and in every one of these 46 cases the optimal trees differed; hence, the advanced search option provides a significant advantage over the default search option.
Table 1.
Results from analyses of five other data sets. For each data set is shown: the numbers of sequences in the alignment, the number of sites in the alignment, the optimal models of SE identified using ModelFinder and IQ-TREE’s implementations of jModelTest9 and ProtTest10 (Other Methods), and the differences in terms of the ∆BIC score and RF distance between phylogenetic estimates inferred using these optimal models of SE.
| Data type, source & origin | Sequences | Sites | ModelFinder | BIC | Other Methods | BIC | ∆BIC | RF |
|---|---|---|---|---|---|---|---|---|
| DNA, Lassa virus7 | 179 | 3,186 | SYM+R5 | 131,325 | SYM+I+Γ4 | 131,540 | 215 | 16 |
| DNA, mitochondrial, mammals3 | 274 | 7,370 | GTR+R8 | 681,837 | GTR+I+Γ4 | 684,469 | 2,632 | 16 |
| DNA, nuclear, birds4 | 200 | 394,684 | GTR+R8 | 18,891,706 | GTR+I+Γ4 | 18,969,054 | 77,348 | 4 |
| Protein, plastids, green plants5 | 360 | 19,449 | JTT+F+R10 | 2,830,471 | JTT+F+I+Γ4 | 2,838,957 | 8,486 | 4 |
| Protein, nuclear, yeast6 | 23 | 634,530 | LG+F+R7 | 25,629,204 | LG+F+I+Γ4 | 25,638,043 | 8,839 | 0 |
ModelFinder is fast and more flexible than other model-selection methods9–11 and can detect models of SE that the other methods are unable to detect (e.g., multi-modal distributions of RHAS). Based on surveys of simulated and real data, ModelFinder proved accurate (Fig. 1) and often outperformed other model-selection methods in terms of the fit between tree, model and data (Table 1, Supplementary Table 1). Fears of over-parameterization have traditionally led users of model-based phylogenetic methods to avoid parameter-rich models of SE, but the use of the BIC, AIC and AICc criteria should alleviate this concern. Although the accuracy and benefits of ModelFinder were demonstrated using proteins generated under time-reversible conditions, the method is also suitable to other data that have evolved under such conditions. If, however, the data have evolved under more non-time-reversible conditions, then ModelFinder is not suitable for model selection. When data have evolved under non-time-reversible conditions, model selection is a challenge because different edges in the tree may require different models of SE. In practical terms, the HAL-HAS model22 addresses this need for nucleotides but a similar solution for other data is not yet available.
Software
ModelFinder is implemented in IQ-TREE version 1.5.4 (http://www.iqtree.org).
Data
Data and scripts used in this study are available from http://www.iqtree.org/ModelFinder/.
Online Methods
ModelFinder is included in IQ-TREE version 1.5.4. and available from http://www.iqtree.org. ModelFinder complements other methods for identifying the optimal model of SE9–11,23–30 for data comprising alignments of nucleotides or amino acids, but it differs from most of these other methods in three important ways:
ModelFinder considers alignments of nucleotides, codons, amino acids, and other discrete data (e.g., binary and morphological data). Like the methods cited above, but not PartitionFinder11, ModelFinder defines the alignment as a single partition of sites;
ModelFinder includes the PDF model of RHAS proposed by Yang13, thus increasing the variety of models of RHAS that are considered during model selection. The PDF model has since been used elsewhere31, but its suitability is not yet widely recognized;
ModelFinder allows the tree topology to vary during the search for an optimal model of SE, thus reducing the chance of entrapment in local optima during model selection. This search strategy has been used previously28, but its suitability is under-recognized.
ModelFinder uses three algorithms to search model space. Algorithm 1 (default search option), uses the following steps:
-
0.
Given an alignment of characters (D);
-
1.
Find a reasonable tree T (inferred using parsimony);
-
2.
Obtain L(D|T, Si, Hi) over i and j, where Si is a list of substitution models and Hj is a list of RHAS models;
-
3.
Identify (Sopt,Hopt) using AIC, AICc or BIC (default).
where L(D|T, Si, Hj) denotes the likelihood of the data, given a tree, T, the i-th substitution model and the j-th model of RHAS, Sopt denotes the optimal substitution model, and Hopt denotes the optimal RHAS model. Algorithm 2 (advanced search option), uses the following steps:
-
0.
Given an alignment of characters (D);
-
1.
Obtain L(D|Th, Si, Hj) over h, i, and j, where Th is a list of trees (generated by IQ-TREE), Si is a list of substitution models and Hj is a list of RHAS models;
-
2.
Identify (Sopt,Hopt) using AIC, AICc or BIC.
Algorithm 3 identifies the optimal PDF model of RHAS and is a key component of Algorithm 1 and Algorithm 2 (it is used whenever the PDF model of RHAS is considered). In the example given below, the BIC optimality criterion is used (but the AIC and AICc optimality criteria can be used if the user chooses to do so):
-
0.
Given an alignment of characters (D), a tree (T), and a substitution model (S);
-
1.
Set k = 2;
-
2.
Obtain L(D|T, S, Rk) and L(D|T, S, Rk+1);
-
3.
If BIC(L(D|T, S, Rk)) > BIC(L(D|T, S, Rk+1)),
-
4.
Increment k by one unit, and go to 2;
-
5.
Else stop, and report Rk as the optimal PDF model.
In practice, Algorithm 1 is invoked with this command (given here for an alignment of amino acids):
iqtree -s data.fst -st AA -m MF
while Algorithm 2 is invoked using:
iqtree -s data.fst -st AA -m MF -mtree
IQ-TREE includes several other options (Supplementary Table 2) that will cause ModelFinder to conduct the search under different constraints. For example, the -m TEST and -m TESTONLY options cause ModelFinder to operate like jModelTest9 and ProtTest10 while the -m TESTMERGE and -m TESTMERGEONLY options cause it to operate like PartitionFinder11. However, none of these options consider the PDF model of RHAS. To do so, it is necessary to use the -m MF and -m MFP options.
When the PDF model is used, it is often necessary to optimize more than two parameters (the I+Γ4 model is parameterized using two parameters). To ensure that these parameters are estimated as accurate as possible, we initially compared parameter estimates obtained using two parameter optimization procedures: the expectation-maximization (EM) algorithm15 (see subsection below) and the quasi-Newton BFGS algorithm32. We found the EM algorithm to be most accurate (results not shown).
ModelFinder is fast. For example, when benchmarking time required by the standard model-selection procedure of ModelFinder, we saw a 39- to 289-fold speedup when compared with jModelTest9 (based on 70 alignments of DNA) and a 16- to 52-fold speedup when compared to ProtTest10 (based on 45 alignments of amino acids).
Model selection for the alignment used by Wu et al.19 (i.e., 6,597 sites and 353 species) was done using two commands:
iqtree -s data.fst -st AA -m MF -msub nuclear -cmax 20
iqtree -s data.fst -st AA -m MF -msub nuclear -cmax 20 -mtree
Having found the optimal model of SE for the data, phylogenetic analyses were done under six models of SE using the following commands:
iqtree -s data.fst -st AA -m WAG+I+G5
iqtree -s data.fst -st AA -m LG+I+G5
iqtree -s data.fst -st AA -m LG+I+G4
iqtree -s data.fst -st AA -m LG+G4
iqtree -s data.fst -st AA -m LG+R14
iqtree -s data.fst -st AA -m LG+G14
Each of these analyses was repeated 100 times to reduce the likelihood of being caught in local optima. The fact that the fit between tree, model and data varied across the 100 results for each of these models of SE shows that this problem is an issue to consider, as done here.
Model selection for the alignments considered in Table 1 was done using commands like those above, albeit with some variations to accommodate, for example, the type of data.
Model selection for the data considered in Supplementary Table 1 was done using two commands:
iqtree -s data.fst -m MF -mtree
iqtree -s data.fst -m TEST
The first command causes IQ-TREE to run the advanced version of ModelFinder; the second command causes IQ-TREE to run its implementation of jModelTest9 or ProtTest10, followed by a phylogenetic analysis under the optimal model of SE.
The PDF model is available in three other phylogenetic programs (i.e., PhyML33, PhyTime34, and BEAST35), so users of ModelFinder are not limited to using IQ-TREE to solve their phylogenetic questions.
Practical considerations
When using ModelFinder, it is important to remember that it optimizes the likelihood of the tree and model, given the data, whenever it searches for the optimal values of parameters considered. Therefore, it is possible that the search algorithms may become trapped in local optima. To reduce the chance of this occurring, we strongly recommend model selection be repeated many times for each data set, as noted above. Doing so may entail using much more computing time, especially when long, species-rich alignments are considered or the advanced search option of ModelFinder is used. Therefore, when the alignment is very long, we recommend the following set of strategies to reduce the amount of time used on model selection:
If the computational resources allow distributed computing, invoke the –nt x option to spread the processes over x threads;
If the data are characters encoded by a specific type of genome (e.g., mitochondrial), invoke the –msub source option to limit the search to this specific type of data;
If the optimal model turns out to include the R10 model of RHAS, we recommend the analysis be rerun with both the –cmin x and –cmax y options invoked (e.g., –cmin 8, –cmax 20). Doing so will ensure that PDF models with k = 8, 9, … , 20 are considered (i.e., lower values of k are ignored). The program will stop when the optimal value of k has been found, even if this value turns out to be 10.
Use the default search option to find the optimal model of SE. Having identified this model, use the advanced search option with the optimal substitution model selected (e.g., –mset LG) to search for the optimal model of RHAS. While there is no guarantee that this approach will identify the optimal model of SE, our experience suggests that the choice of RHAS model is highly influenced by the topology of the tree while that of the substitution model is not.
The EM algorithm to estimate PDF model parameters
Let = {W1, …, Wk, r1, …, rk} be the weights and rates of the PDF model Rk that we want to estimate. First, we initialize using a discrete Γk model12 (i.e., the initial values of and are derived from the discrete Γ distribution with k categories and a shape parameter α = 1). This becomes the current estimate . The EM algorithm iteratively performs an expectation (E-) step and a maximization (M-) step to update the current estimate until a (local) maximum in likelihood is reached.
E-step:
For the i-th site in the alignment Di and the j-th category compute the posterior probability of Di belonging to category j based on the current estimate :
where is the likelihood of the tree T, substitution model S, and relative rate for the alignment site Di.
M-step:
For each category j the log-likelihood function:
is maximized to obtain the next where N is the number of sites in the alignment. This can be done with standard numerical optimization such as Brent’s method36. The weights are updated using:
that is, the new weight for category j is the mean posterior probability of each alignment site belonging to class j. This completes the proposal of the new estimate . If the likelihood of is higher than that of , then is replaced by and the E- and M-steps will be repeated. Otherwise, the EM algorithm stops and reports as the maximum-likelihood estimates of the PDF model Rk.
This EM algorithm allows estimation of the parameters of the Rk model, given a fixed tree T and a substitution model S. ModelFinder then iteratively estimates branch lengths of T, model parameters of S, and Rk until the likelihood converges.
Supplementary Material
Acknowledgements
We thank D.Y. Wu, J.A. Eisen, P. Donoghue, and A. Rokas for access to their data, E. Susko for discussions about the EM algorithm, and V. Jayaswal for constructive feedback. B.Q.M. and A.v.H. were supported by the Austrian Science Fund (FWF I-2805-B29).
Footnotes
Author Contributions
S.K., T.K.F.W. and L.S.J. conceived the method and executed a pilot study to assess the likely impact. B.Q.M. and T.K.F.W. implemented the method in IQ-TREE, with contributions from S.K., L.S.J. and A.v.H. S.K., T.K.F.W. L.S.J. and B.Q.M. assessed the performance and accuracy of the method. S.K., T.K.F.W. and L.S.J. carried out the analyses of simulated and real data. L.S.J., S.K., T.K.F.W., B.Q.M., and A.v.H. wrote the paper.
Competing Financial Interests
The authors declare not competing financial interests.
References
- 1.Eisen JA. Genome Res. 1998;8:163–167. doi: 10.1101/gr.8.3.163. [DOI] [PubMed] [Google Scholar]
- 2.Hardy MP, Owczarek CM, Jermiin LS, Ejdebäck M, Hertzog PJ. Genomics. 2004;84:331–345. doi: 10.1016/j.ygeno.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 3.dos Reis M, et al. Proc R Soc B. 2012;279:3491–3500. doi: 10.1098/rspb.2012.0683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prum RO, et al. Nature. 2015;526:569–U247. doi: 10.1038/nature15697. [DOI] [PubMed] [Google Scholar]
- 5.Ruhfel BR, Gitzendanner MA, Soltis PS, Soltis DE, Burleigh JG. BMC Evol Biol. 2014;14:26. doi: 10.1186/1471-2148-14-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Salichos L, Rokas A. Nature. 2013;497:327–331. doi: 10.1038/nature12130. [DOI] [PubMed] [Google Scholar]
- 7.Andersen KG, et al. Cell. 2015;162:738–750. doi: 10.1016/j.cell.2015.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tay WT, et al. Sci Rep. 2017;7:45302. doi: 10.1038/srep45302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Darriba D, Taboada GL, Doallo R, Posada D. Nature Meth. 2012;9:772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Darriba D, Taboada GL, Doallo R, Posada D. Bioinformatics. 2011;27:1164–1165. doi: 10.1093/bioinformatics/btr088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lanfear R, Calcott B, Ho SYW, Guindon S. Mol Biol Evol. 2012;29:1695–1701. doi: 10.1093/molbev/mss020. [DOI] [PubMed] [Google Scholar]
- 12.Yang Z. J Mol Evol. 1994;39:306–314. doi: 10.1007/BF00160154. [DOI] [PubMed] [Google Scholar]
- 13.Yang Z. Genetics. 1995;139:993–1005. doi: 10.1093/genetics/139.2.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nguyen L-T, Schmidt HA, Von Haeseler A, Minh BQ. Mol Biol Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dempster AP, Laird NM, Rubin DB. J R Stat Soc Ser B. 1977;39:1–38. [Google Scholar]
- 16.Fletcher W, Yang ZH. Mol Biol Evol. 2009;26:1879–1888. doi: 10.1093/molbev/msp098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Le SQ, Gascuel O. Mol Biol Evol. 2008;25:1307–1320. doi: 10.1093/molbev/msn067. [DOI] [PubMed] [Google Scholar]
- 18.Robinson DF, Foulds LR. Math Biosci. 1981;53:131–147. [Google Scholar]
- 19.Wu DY, et al. Nature. 2009;462:1056–1060. doi: 10.1038/nature08656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kass RE, Raftery AE. J Am Stat Assoc. 1995;90:773–795. [Google Scholar]
- 21.Sanderson MJ, Donoghue MJ, Piel W, Eriksson T. Am J Bot. 1994;81:183. [Google Scholar]
- 22.Jayaswal V, Wong TKF, Robinson J, Poladian L, Jermiin LS. Syst Biol. 2014;63:726–742. doi: 10.1093/sysbio/syu036. [DOI] [PubMed] [Google Scholar]
- 23.Posada D, Crandall KA. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 24.Chiotis M, Jermiin LS, Crozier RH. Mol Phylogenet Evol. 2000;17:108–116. doi: 10.1006/mpev.2000.0821. [DOI] [PubMed] [Google Scholar]
- 25.Abascal F, Zardoya R, Posada D. Bioinformatics. 2005;21:2104–2105. doi: 10.1093/bioinformatics/bti263. [DOI] [PubMed] [Google Scholar]
- 26.Keane TM, Creevey CJ, Pentony MM, Naughton TJ, McInerney JO. BMC Evol Biol. 2006;6:29. doi: 10.1186/1471-2148-6-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Posada D. Nucl Acid Res. 2006;34:W700–W703. doi: 10.1093/nar/gkl042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Posada D. Mol Biol Evol. 2008;25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- 29.Santorum JM, Darriba D, Taboada GL, Posada D. Bioinformatics. 2014;30:1310–1311. doi: 10.1093/bioinformatics/btu032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Whelan S, Allen JE, Blackburne BP, Talavera D. Syst Biol. 2015;64:42–55. doi: 10.1093/sysbio/syu062. [DOI] [PubMed] [Google Scholar]
- 31.Soubrier J, et al. Mol Biol Evol. 2012;29:3345–3358. doi: 10.1093/molbev/mss140. [DOI] [PubMed] [Google Scholar]
- 32.Fletcher R. Practical Methods of Optimization Second Edition. John Wiley & Sons; 2000. [Google Scholar]
- 33.Guindon S, et al. Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 34.Guindon S. Syst Biol. 2013;62:22034. doi: 10.1093/sysbio/sys063. [DOI] [PubMed] [Google Scholar]
- 35.Bouckaert R, et al. PLoS Comp Biol. 2014;10:6. [Google Scholar]
- 36.Brent RP. Algorithms for minimization without derivatives. Prentice Hall; 1973. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.