Abstract
Fibroblast growth factors (FGF) and their tyrosine kinase receptors (FGFR) support cell proliferation, survival and migration during embryonic development, organogenesis and tissue maintenance and their deregulation is frequently observed in cancer development and progression. Consequently, increasing efforts are focusing on the development of strategies to target FGF/FGFR signaling for cancer therapy.
Among the FGFRs the family member FGFR4 is least well understood and differs from FGFRs1-3 in several aspects. Importantly, FGFR4 deletion does not lead to an embryonic lethal phenotype suggesting the possibility that its inhibition in cancer therapy might not cause grave adverse effects. In addition, the FGFR4 kinase domain differs sufficiently from those of FGFRs1-3 to permit development of highly specific inhibitors. The oncogenic impact of FGFR4, however, is not undisputed, as the FGFR4-mediated hormonal effects of several FGF ligands may also constitute a tissue-protective tumor suppressor activity especially in the liver.
Therefore it is the purpose of this review to summarize all relevant aspects of FGFR4 physiology and pathophysiology and discuss the options of targeting this receptor for cancer therapy.
Keywords: FGFR4, targeted therapy
1. Introduction
Several hallmarks of cancer including growth stimulation, cell death resistance, migration/invasion capability as well as immortality can be acquired or at least supported by deregulated activation of receptor tyrosine kinases (RTKs). Consequently, it is not surprising that RTKs are frequently activated during the process of malignant transformation and tumor progression by diverse genetic and epigenetic mechanisms. Many RTKs have been described as driving oncogenes in several tumor types so that they are in the center of multiple targeted anticancer therapy approaches. Additionally, representing enzymes with dynamic kinase domains and extracellular parts comprising ligand binding sites, RTKs are perfectly druggable.
Fibroblast growth factors (FGF) and their tyrosine kinase receptors (FGFR) regulate important cellular processes such as cell proliferation, survival and migration during embryonic development, organogenesis and tissue maintenance [1, 2]. Their expression and activity is stringently controlled in normal tissues and their deregulation is frequently observed in cancer development. FGF-dependent stimuli provide survival and migration signals for the cancer cells and also affect endothelial cells stimulating angiogenesis [3, 4]. Consequently, the topic of FGF/FGFR signaling and related questions of targeted therapy have recently been discussed in several reviews that mostly gave overviews of the complete FGFR family e.g. [2, 3, 5, 6], but none of them focused on FGFR4.
The 4 FGFR genes and the various receptor proteins expressed from differentially spliced FGFR-RNAs have moved into the focus as targets for therapy in several malignancies [7, 8]. While all FGFRs have common features in structure, physiological role, ligand-binding and down-stream signaling, FGFR4 differs from FGFRs 1-3 in several aspects. Firstly, there are no FGFR4 splice-variants that affect the immunglobulin-like (Ig-)loop III resulting in distinct ligand binding spectra [9]. Secondly, FGFR4 deletion does not lead to an embryonic lethal phenotype [10] suggesting the possibility that its inhibition might not cause grave adverse effects. Lastly, concerning inhibition by small molecule compounds IC50 concentrations for FGFR4 frequently differ by factors of 4 or higher from those for FGFRs 1-3 indicating structural differences in the kinase domain (see Table 4 [3, 11]). These characteristics could be exploited to develop more specific targeting agents than seems possible within other FGFR subtypes ([3] and see below). Therefore it is the purpose of this review to summarize all relevant aspects of FGFR4 physiology and pathophysiology and discuss the options of targeting this receptor for cancer therapy.
Table 4. Small Molecule TKIs Targeting FGFRs: Inhibitor Specificity (IC50 in nM)1 [11].
| Inhibitor [Citation] | Alias | Company | FGFR4 | FGFR1 | FGFR2 | FGFR3 | Other main target | Most developed (pre)clinical stage |
|---|---|---|---|---|---|---|---|---|
| AZD4547 [211] | AstraZenca | 165 | 0.2 | 2.5 | 1.8 | VEGFR2: 24 | Phase II | |
| BGJ398 [212] | NVP-BGJ398 | Novartis | 60 | 0.9 | 1.4 | 1 | VEGFR2: 180 | Phase I |
| BIBF 1120 [176, 213] | Nintedanib Vargatef | Böhringer Ingelheim | 451-937 | 47-69 | 37-63 | 108-122 | VEGFR2/3: 13 Lck:16 | Phase III |
| Brivanib [175–177] | BMS-582664 | Bristol-Myers Squibb | >1000 | 15-165 | 32-202 | 52-530 | VEGFR2: 4.2- | Phase III |
| Cediranib [176, 214] | Recentin, AZT2171 | AstraZeneca | 697 | 5-26 | 33 | 36 | VEGFR2: <1 VEGFR3: 3; KIT: 2 | Phase III |
| Dovitinib [177] | TKI-258 | Novartis | 470 | 13 | 21 | 18 | VEGFR2: 5.4 PDGFRß: 15 | Phase III |
| E-3810 [215] | - | EOS- | >1000 | 17.5 | 82.5 | 237.5 | VEGFR1: 7 CSF-1R: 5 | Phase I |
| JNJ-42756493 [177] | - | Astex/Jansen | 33 | 13 | 33 | 3 | VEGFR1:13 | Phase I |
| LY2874455 [174] | - | Lilly | 6 | 2.8 | 2.6 | 6.4 | VEGFR2: 7 | Phase I |
| Pazopanib [216] | Votrient | GlaxoSmithKline | ? | 80 | 350 | 138 | VEGFR3: 2 VEGFR1: 7; KIT (mut) 1 | Approved (STS, RCC) |
| Ponatinib [176, 217] | AP24534, Iclusig | Ariad | 8 | 2 | 2 | 18 | VEGFR2: 1.5 Abl: 0.37;PDGFR" : 1 Flt3: 13; KIT: 13 | Approved (TKI-resistant CML) |
| Sunitinib [216] | SU11248 Sutent | Pfizer | ? | 437 | 852 | 314 | KIT (mut): 0.45 VEGFR3: 3; Flt3: 4 | Approved for GIST, RCC, pNET |
| Sorafenib [216] | Nexavar | Bayer/Onyx | ? | 64 | 825 | 1019 | RET: 2 VEGFR3: 7; VEGFR1: 9 | Approved for HCC and RCC |
| XL228 [218] | - | Exelixis | ? | 8 | 2 | 3 | IGFR: 2 Aurora B: 0.5 | Phase I |
adapted from reference [11]. STS, soft tissue sarcoma; RCC, renal cell cancer; CML, chronic myeloid leukemia; GIST, gastrointestinal stromal tumor; pNET, pancreatic neuroendocrine tumor
2. Gene Structure and Regulation of FGFR4
a. Gene and Protein Structure
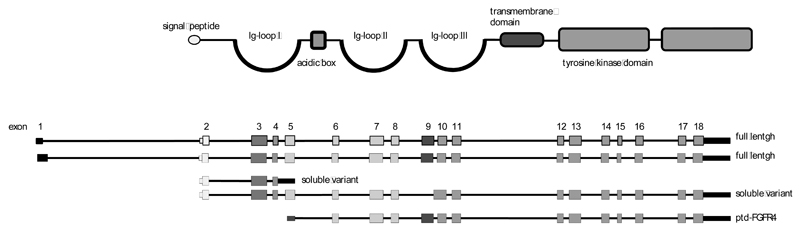
The vertebrate FGFR gene family is comprised of four members and originated by co-evolution with the FGF genes to permit increased ligand-receptor specificity and to enable functional diversity [12]. The four FGFR genes consist of up to 20 exons that together code for a highly conserved protein domain structure [13, 14]. The human FGFR4 gene is located on the long arm of chromosome 5 (5q 35.1) and spans more than 11kb. GenBank entries define 3 full length variants consisting of 17-18 exons and giving rise to mRNAs of about 3kb [15, 16]. Data mining at the UCSC sites indicates the existence of various additional transcript variants. Published isoforms encoding published protein variants are shown in (Fig. 1).
Fig. (1). The gene and protein structure of FGFR4.
The gene and protein structures of FGFR4 were extracted from the GenBank [219] and the nextProt [220] database and aligned to demonstrate the conserved exon protein domain relationships of the FGFR family. The figure depicts the two full-length transcripts NM002011 and NM213647, the soluble variant NM022963 that lacks exon 9 [41, 43], the soluble variant consisting of only exons 2-4 and the cytoplasmic ptd-FGFR4 variant (AF359246) [112]. Exons and protein domains are color coded to show which domain is derived from which exon.
The two full-length transcripts of the FGFR4 gene code for receptor proteins of 762 and 802 amino acids with an estimated molecular weight of 100-110kD. As for all FGFRs the FGFR4 protein domain structure reflects the exon structure (Fig. 1). The ATG start codon is located in exon 2 that mainly codes for the signal peptide. This is followed by the extracellular domain that contains 3 Ig-loops including autoinhibitory sequences in the Ig-loop I (exon 3), a heparin-binding site and the ligand-binding domain between Ig-loops II and III (exons 5-8) [3, 5]. The single transmembrane domain (exon 9) is about 25 amino acids long and the cytoplasmic part of the receptor features an interrupted kinase domain encoded in the remaining 9 exons (Fig. 1). While the Ig-loops III of FGFRs 1-3 are subject to alternative splicing that creates the IIIb and IIIc transcript variants coding for receptor isoforms with different ligand specificity, the FGFR4 gene lacks the respective alternative exon [17]. The resultant single ligand binding domain of FGFR4 enables high-affinity binding of a large number of FGFs, resembling the IIIc-splice variants of FGFRs 1-3 [3, 9] (Table 1). In addition to FGFs 1 and 2 FGFR4 ligands encompass members of the FGF4-, FGF8- and the hormonal FGF19 (hFGFs)- subfamilies [18, 19].
Table 1. Ligands with Affinity for FGFR4 According to [18,19].
| Mitogenic activity of FGFs | % biological response relative to FGF1 | |||||||
|---|---|---|---|---|---|---|---|---|
| FGFR4* | FGFR1IIIb | FGFR1IIIc | FGFR2IIIb | FGFR2IIIc | FGFR3IIIb | FGFR3IIIc | ||
| FGF1 subfamily | FGF1 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| FGF2 | 113.4 | 59.9 | 103.9 | 9 | 64 | 1.2 | 107.2 | |
| FGF4 subfamily | FGF4 | 108 | 15.6 | 102.3 | 14.9 | 94.3 | 1 | 69.1 |
| FGF5 | 7 | 3.8 | 59 | 5 | 25 | 1 | 11.8 | |
| FGF6 | 79.4 | 4.6 | 54.9 | 5.4 | 60.7 | 0.9 | 8.8 | |
| FGF7 subfamily | FGF3 | 5.8 | 34.4 | 0.3 | 44.6 | 4.2 | 1.5 | 0.6 |
| FGF7 | 17.8 | 8 | 14.2 | 168 | 10.4 | 6.7 | 3.3 | |
| FGF10 | 11.5 | 39.4 | 12.5 | 217 | 6.1 | 6 | 0.8 | |
| FGF22 | 12.5 | 40.3 | 10.2 | 232 | 7.1 | 5.3 | 1 | |
| FGF8 subfamily | FGF8 | 102 | 5.3 | 57.5 | 5.9 | 91.6 | 18.6 | 209 |
| FGF17 | 85.5 | 6 | 22.7 | 6.3 | 27.1 | 10.7 | 111 | |
| FGF18 | 52.8 | 6.3 | 4.7 | 7.8 | 28.9 | 12.5 | 77.7 | |
| FGF9 subfamily | FGF9 | 10.1 | 7.3 | 12.5 | 2.9 | 57.2 | 42.7 | 90.4 |
| FGF16 | 9.9 | 6.5 | 4.3 | 1.8 | 32.5 | 13 | 32.4 | |
| FGF20 | 26.6 | 7.3 | 28.1 | 12.3 | 68.4 | 44.3 | 89.5 | |
| FGF19 subfamily | FGF19 | 4.1 | 0.12 | 1.23 | 0.61 | 2.38 | 0.2 | 1.4 |
| FGF21 | 1.28 | 0.19 | 0.34 | 0.83 | 0.86 | 0.26 | 0.23 | |
| FGF23 | 2.22 | 0.11 | 0.23 | 0.66 | 0.89 | 0.21 | 0.43 | |
The relative biological response is calculated from publications by Ornitz et al. [18] and Zhang et al [19] using the 3H-thymidine uptake stimulated by 5nM of the respective FGF. For the FGF19 family members the activity was determined in the absence of klotho proteins.
Bold print indicates activity >50% or > than for any other FGFR variant
b. The FGFR4 Promoter
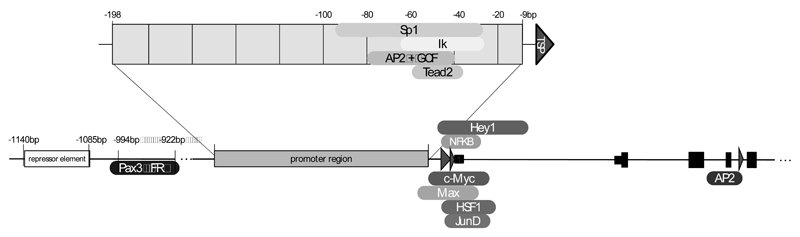
Systematic analysis of FGFR protein expression in normal human adult tissues representing the major organ systems resulted in the detection of FGFR4 expression in adult human adrenal, lung, kidney, intestine, pancreas, skeletal muscle, spleen, and liver [20]. The strict control of gene expression necessary for potent growth and survival factors and their receptors like FGFRs requires multiple regulatory elements in the promoter region. Promoter activity of the human FGFR4 gene was studied with reporter constructs up to - 1955 base pairs numbered relative to the major transcription start point (TSP) [21]. Our review considers regulatory elements defined within this region of human FGFR4 and downstream into introns 1 and 4 (Fig. 2).
Fig. (2). Promoter elements regulating FGFR4 gene expression.
Sequences from intron 4 of the FGFR4 gene to about 1500bp up-stream of the major TSP have been investigated in the ENCODE project. TSPs are marked by red arrow heads. Transcription factor binding sites are given as boxes at the appropriate site.
The human FGFR4 core promoter region reaches from position -198 to -9, is CG-rich and contains more than 1 TSP, but no TATA- or CCAAT-like elements [21]. This is a notable feature of many housekeeping genes, oncogenes, growth factors, and transcription factors [14, 22] and also seen in the promoters of FGFRs 1-3. Specifically, the human FGFR1 gene [23], the human FGFR2 gene [24], and the mouse and human FGFR3 gene [25, 26] display comparable characteristics. The FGFR4 promoter region harbors several binding motifs for the Sp1, AP2 and GCF transcription factors located ! 80 to ! 40 bp upstream of the TSPs as has been described for several TATA-less promoters [14, 22].
Tissue specific regulatory elements of FGFR4 promoters are mainly described for skeletal muscle and pituitary gland derived cells. For other tissues and cancers such elements have to be defined. Ets and Sp1 motifs and binding sites for the hematopoietic zinc finger-containing transcription factor Ikaros (Ik) were identified within the core promoter region of FGFR4 between sequence positions -65 to -26 and together regulate tissue specific FGFR4 expression in the pituitary gland [27]. Binding sites for Sp1 in the promoter region -95 to -56 are particularly important for FGFR4 expression in differentiating myotubes and its stimulating role in myogenesis and terminal skeletal muscle differentiation. Furthermore, the Sp1 transcription factor binding at sites within positions -95 to -56 and -65 to -26 controls FGFR4 transcription in sarcomas of skeletal muscle lineage [28].
Specifically, the mouse FGFR4 promoter region 49 bp upstream of the TSP binds the TEA domain transcriptional factor, Tead2, and regulates FGFR4 expression required for effective muscle regeneration in vivo [29]. Tead2 itself is induced by binding of MyoD, one of the main regulators of muscle differentiation, to the first intron of the Tead2 gene at day 3 during muscle regeneration. Recent work demonstrates that folate receptor alpha (FR") known as a glycosylphosphatidylinositol-anchored protein and a component of the caveolae fraction, is capable of translocating to the nucleus where it binds to cis-regulatory elements at the FGFR4 as well as other promoters [30]. In both mouse and human FGFR4 promoters, two Pax3 and one FR" binding regions are located at -994/-989, -980/-977 and -928/-922, respectively.
Additional transcription factor binding sites downstream of the major TSP have been extracted from Chip-sequencing data of the Encode project [31]. Among others c-myc, max, junD, fos-like 2, hey1 and NF!B bind to the region around the untranslated exon 1 in tumor cell lines. Specifically, in pituitary tumors an alternative TSP within intron 4 can be activated by transcription factor AP-2 binding [32]. Further upstream the FGFR4 promoter region between -1140 and -1085 a potential repressor element is located, which down regulates transcriptional activity and might contribute to tissue specific expression [21].
c. Splice Variants of FGFR4
Within the FGFR-family multiple transcript variants are generated from the same gene by alternative initiation, alternative splicing, exon shuffling, and usage of variable polyadenylation sites [33]. These various transcripts contribute to the characteristic tissue-specific expression patterns observed for FGFRs. The FGFR4 gene lacks the alternative exon that creates the IIIb and IIIc variants of FGFRs1-3 with strong impact on ligand affinity [34] so that only one isoform was predicted from the genomic sequence [35]. The NCBI database lists two full-length transcripts, one variant transcript lacking exon 9, one transcript consisting of only exons 2-5 and one starting from an alternative TSP upstream of exon 5 (Fig. 1).
Alternative splicing may remove exon 9 which codes for the transmembrane domain or produces mRNAs containing premature stop codons and intronic polyadenylation sites as has been shown for e.g. the vascular endothelial growth factor receptor (VEGFR) [36] and for c-met [37]. Such receptor variants have been identified for many tyrosine kinase receptors including FGFR1 [38]. They act as dominant-negative decoy receptors and have a regulatory function in normal tissues [39]. Soluble receptor variants act as endogenous signaling inhibitors either by trapping respective ligands or by binding of Ig-loop I sequences to the ligand binding domain and are frequently down-regulated in malignant tumors [38]. They have therapeutic potential in situations of pathway over-activation, as has been shown for VEGFR [40]. Splice variants of FGFR4 expressed in breast cancer cells [41], normal pituitary gland [42] and the intestinal mucosa [43] code for soluble FGFR4 variants that lack a transmembrane domain. A soluble variant of FGFR4 has been shown to block FGF19-induced FGFR4 activation and prevent non-alcoholic fatty liver disease [44]. Genetic constructs coding for soluble FGFR variants or oligonucleotides forcing splice decisions towards soluble FGFR forms may therefore also have potential for cancer therapy.
3. Physiological Role of FGFR4
a. In Embryonic Development and Organogenesis
The FGFR4 expression pattern in fetal human tissue was recognized to be different from each of the other three FGFRs [15, 45]. FGFR4 together with FGFR3 is the highest-expressed FGFR in mouse pre-implantation blastocytes and on days E5 – E6 the receptor is found in primitive ectoderm [46]. Analysis of RNA isolated from whole embryos reveals an increase in FGFR4 RNA with time until day E14 – E15 after which expression declines again [47]. The spatial pattern of expression at days E8 – E9 shows FGFR4 signals in the gut endoderm and in the myotome of the somites. At day E14.5 expression is strong in the muscles at multiple sites of the mouse embryo but not in the heart muscle. It is widespread in cartilage, in the gut, the pancreatic ducts, the liver and the lung [47, 48]. Specific regions of the kidneys and the adrenal glands are positive, but FGFR4 is absent from both the brain and the spinal cord. After birth, expression remains high in the liver and the lung and somewhat lower in the kidneys [46, 48]. Specifically in the lung expression steadily increases after birth localizing to the vessels, the larger airway epithelium and the alveolar epithelium [49]. In the muscle FGFR4 becomes restricted to sites of muscle regeneration [50].
Despite the wide-spread FGFR4 expression during embryonic development deletion of the receptor gene does not cause developmental abnormalities, which indicates the presence of alternative receptors on target cells. Only the combined deletion of FGFR3 and FGFR4 blocks the formation of alveoli [10]. The FGFR4 knock-out mouse is born normally and remains viable. Symptoms due to lack of FGFR4 function in the liver only develop in later life [51].
b. In the Muscle
Tissue localization strongly suggests a role of FGFR4 in muscle differentiation. The fibroblast growth factor essential for this process was found to be FGF6 [52]. The growth factor is expressed mainly in the myogenic lineage, the developing muscle [53] and in myogenic precursor cells where it modulates expression of FGFRs 1 and 4 [54]. Deletion of FGF6 did not cause developmental defects in the myogenic lineage indicating some redundancy with other FGFs like FGF4 and FGF8. However, FGF6 knock-out mice were impaired in muscle regeneration (reviewed in [52]).
The FGFRs strongly stimulated by FGF6 are FGFR1-IIIc and FGFR4 [18]. Molecular analysis of muscle cell differentiation and muscle cell regeneration indicates that myogenesis is regulated by a controlled interplay between these two receptors [52]. FGFR1-IIIc dependent signaling blocks differentiation and stimulates migration of myogenic stem cells [55]. In regenerating muscle cells FGF6 shifts expression from FGFR1 to FGFR4 inducing dedifferentiation [56]. In chicken embryos myogenesis was blocked upon injection of a construct expressing a soluble FGFR4 variant that traps FGFR4 ligands, while a corresponding FGFR1 construct had no impact [57]. In fact, during muscle differentiation FGFR1 is down-regulated while FGFR4 expression transiently increases. This makes FGFR4 the major FGFR in newly formed muscle fibers and suggests that FGFR4 is necessary for muscle differentiation [50, 58]. In mature striated muscle the receptor has not been found [47, 50]. Regulation of FGFR4 expression was reported to occur by induction through MyoD [29, 59] but the FGFR4-promoter also contains Pax3 binding sites that permit direct control by this regulator of muscle stem cell behavior [60–62] (Fig. 2).
FGFR4 knock-out mice while having no defects in myogenic development [10] show abnormal muscle regeneration thus resembling FGF6 deficient animals. Their regenerating muscle fibers are smaller and more irregular with areas of calcification and fatty infiltration [29]. By contrast permanent up-regulation of FGFR4 signaling is a characteristic of rhabdomyosarcoma (RMS) [63], a childhood tumor that is supposed to derive from myogenic precursors blocked in differentiation [64].
c. Mediating Hormone-like FGF Effects
The hormone-like FGF subfamily (hFGF) consists of FGF19, its mouse orthologue FGF15, FGF21 and FGF23 that all have affinity for FGFR4 [19] (Table 1). Based on their protein domain structure the members of the hFGF family have lower affinity to both the FGFRs and heparansulfate (HS) than other FGFs [65] (Table 1). The latter characteristic is a prerequisite for the endocrine role of these FGFs, but it also hampers stimulation of FGF-FGFR binding by HS. The role of co-receptors boosting FGF-dependent signaling activity is played by the klotho proteins -klotho and #-klotho [66]. Klothos are membrane-bound, single-pass transmembrane domain proteins with an extracellular domain containing two internal repeats KL1 and KL2. FGF19 predominantly signals via #-klotho/FGFR4, although it can also activate -klotho/FGFR4 [67, 68], while FGF21 has a preference of #-klotho /FGFR1IIIc over #-klotho/FGFR4 [69], and FGF23 most efficiently activates - klotho/FGFR1IIIc [70]. It was suggested that presence of klotho-proteins directs FGFR signals towards the control of metabolic functions. In contrast to FGFR4 alone, signaling via the #-klotho/FGFR4 complex was shown to suppress activation of AKT and mTOR while still stimulating the mitogen activated protein kinase (MAPK) pathway, together resulting in apoptosis induction in hepatoma cells [71].
FGF19/15
Human FGF19 and its mouse orthologue FGF15 are the hFGF that most specifically signal through FGFR4. They can be found in fetal skin, retina and the small intestine as well as in adult gall bladder epithelium and intestinal mucosa [72, 73]. In addition, FGF15 was found to play an important role in embryonic heart [74] and brain development in the mouse [75, 76].
The liver is the organ of highest FGFR4 expression in adult vertebrates [46, 48] and the site of essential regulatory functions of the receptor. Deletion of FGFR4 does not cause any developmental defects in knock-out mice, but leads to alterations in cholesterol metabolism and elevated synthesis and excretion of bile acids [51]. The metabolic pathways are under control of hepatic FGFR4 and depend on the co-expression of ß-klotho and the FGFR4 stimulation by FGF19/FGF15 [65]. In the mouse intestine the expression and secretion of FGF15 [77] is stimulated postprandially by binding of bile acids to nuclear farnesyl-X-receptor. Via blood flow the factor reaches the liver and binds to the FGFR4 on the surface of hepatocytes, which causes activation of c-Jun N-terminal kinase (JNK) and repression of cholesterol 7alpha-hydroxylase (CYP7A1), the key enzyme of bile acid synthesis [77, 78]. The increased CYP7A1 levels in FGFR4 knock-out mice could not be reversed by adenoviral constructs expressing FGF15 demonstrating the dependency on FGFR4 signaling [77]. In humans FGF19 serum levels increase at a time when bile acids are reabsorbed in the distal colon, which could be evoked also by ingestion of chenodesoxycholic acid. This indicates that the same regulatory events occur as in the mouse model [79].
FGF15/19 also triggers the filling of the gallbladder. In FGF15 knock-out mice this function was absent but could be restored by FGF19 [80]. Both, #-klotho knock-out mice and FGFR4 knock-out mice have the depleted gallbladder phenotype. This underlines the importance of this signaling axis although the down-stream mediators of the effect remain to be identified [81].
As important mediator of homeostasis in the liver, absence of FGFR4 function may cause tissue damage in case of metabolic stress – a role that becomes even clearer in the model of carbon tetrachloride-induced liver damage. Lack of FGFR4 accelerated damage, caused increase in liver weight and delayed tissue repair during the acute toxic response. Accordingly, chronic carbon tetrachloride exposure caused liver fibrosis in FGFR4 knock-out but not in wild-type (wt) mice [82] clearly demonstrating the liver-protective effect of FGFR4.
In addition, FGF19 was shown to activate physiologically important, insulin-independent endocrine pathways that regulate hepatic protein and glycogen metabolism [83]. Accordingly, mice lacking FGF15 failed to properly maintain blood concentrations of glucose and normal postprandial amounts of liver glycogen, but showed normal insulin levels and normal insulin sensitivity [83, 84]. FGF19 treatment restored the loss of glycogen in diabetic animals lacking insulin. In humans, the physiological role of FGF19 in glucose homeostasis remains to be determined. Furthermore, over-expression of FGF19 in transgenic mice was shown to increase energy expenditure by elevated brown adipose tissue mass at the cost of fat mass through lower expression levels of coenzyme A carboxylase [85].
FGF21
Under fasting conditions FGF21 is expressed in adipose tissue [86] and the liver [87] to promote lipolysis and responses to fasting. Peroxisome proliferator-activated receptors PPAR# and PPAR" signaling mediates this process while the PI-3K/AKT signaling pathway is required in the thymus, in pancreatic islet beta-cells [88] and in the skeletal muscle [89]. Although FGF21 binds to all FGFRs its cognate receptor is FGFR1 together with the essential co-factor #-klotho, sometimes FGFR4/#-klotho also directs FGF21 towards metabolic activities [19, 90]. Although Tomiyama and colleagues could find FGF21 signaling in #-klotho knock-out mice [91], whole-body and adipose tissue-selective #-klotho knock-out mice failed to show any FGF21 effects on growth and metabolism [92].
FGF23
FGF23 is expressed in the bone [93] and regulates phosphate and vitamin D levels by inhibiting the secretion of parathyroid hormone, thereby decreasing phosphate uptake from the bone [94]. Lack of FGF23 action can lead to many diseases – among them hypophosphataemic rickets [95]. Binding affinities for all four receptors are present but FGF23 mainly acts through the FGFR1c in association with a klotho protein [19, 70, 96, 97]. A recent report also links the factor to FGFR3 and/or FGFR4 signaling, as FGFR3/4 double knock-out mice have lower serum phosphorus levels, lower renal phosphorus transporter expression and elevated Vitamin D3 serum levels as well as aberrant response to FGF23 [98].
4. FGF-Dependent Signaling
a. Canonical FGFR Signaling
Canonical signaling via FGFRs is dependent on receptor dimerization that leads to auto-phosphorylation of tyrosine residues in the cytoplasmic receptor domains. These act as binding sites for signaling proteins and docking proteins, which form a complex with an additional complement of signaling proteins [2, 4, 99]. Most of the studies performed on the details of receptor activation and downstream events have used FGFR1 and experimental validation for FGFR4 is more limited.
FGFR4 binds a specific subset of ligands of the FGF family (Table 1) [18, 19]. Like for other FGFRs binding is enhanced by the formation of quarternary complexes including heparin or heparan sulfate proteoglycans (HSPG) [100], which is instrumental for ligand-induced receptor activation. One study found that heparin alone may be sufficient to activate FGFR4 but not FGFR1 [101]. Also it was shown that cell surface HSPG may be involved in preferential binding of FGFR4 to FGF1 versus FGF2 in liver parenchymal cells [102].
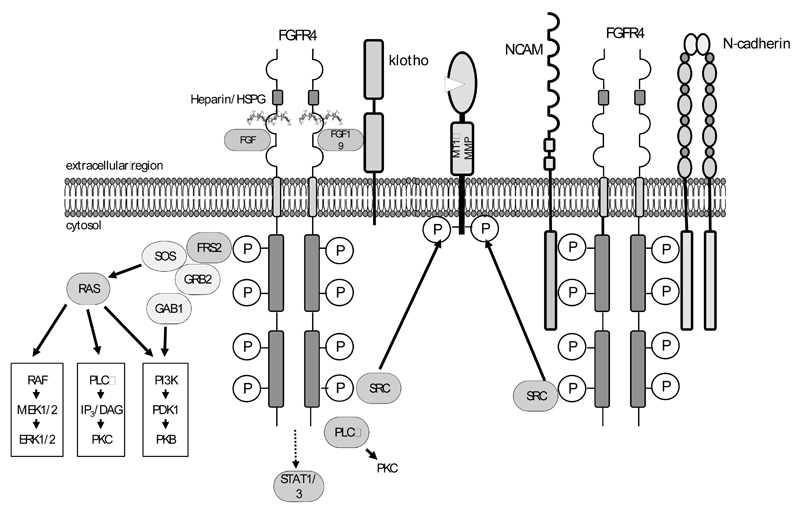
Like the other three members of the FGFR family, FGFR4 directly activates phospholipase C (PLC) and depends on FGFR substrate 2 (FRS2) or # for coupling to the MAPK and the phosphoinositide 3 kinase (PI3K) pathways, which are crucial for cellular proliferation and survival signals (Fig. 3). A high degree of overlap in the signaling effector proteins of different FGFRs including activation of Stat1 and Stat3 was suggested by experiments using constitutively active versions of FGFR1-4 [103]. Increased Stat3 signaling was demonstrated as a consequence of some of the activating mutations (K535, E550) of FGFR4 found in rhabdomyosarcoma [63]. Downstream of the MAPK and the PI3K pathway inhibition of FGFR4 together with ErbB2 cooperatively blocked S6 kinase 1 activity and cyclin D1 translation in MDA-MB-453 breast cancer cells [104]. Activation and phosphorylation of downstream signaling proteins by FGFR4 has been reported to be weaker than by FGFR1, probably due to specific differences in the kinase domain [34, 105–107].
Fig. (3). FGFR4 signaling options.
Left: Canonic down-stream signaling pathways adapted from [3].
Right: Non-canonical signaling options related to FGFR-CAM binding as described by [221]. In cancer cells non-canonical signaling is reported to involve c-src-mediated activation of MMPs [157].
b. Non-canonical Signaling Involving N-CAM and N-cadherin
In addition to the canonical signaling pathways, mounting evidence demonstrates a number of non-canonical signaling activities mainly involving different cell adhesion molecules. In the Rip1Tag2 transgenic mouse model of pancreatic beta cell carcinogenesis it was found that neural cell adhesion molecule (N-CAM) formed a complex with FGFR4 and a number of FGFR effector proteins including PLC, FRS2, Shc and c-src as well as with N-cadherin [108] (Fig. 3). In this model, ligand-independent, N-CAM-induced FGFR4 signaling was required for neurite outgrowth and cell-matrix attachment of the tumor cells. The dependency of cell-matrix attachment on N-CAM and FGFR4 signals provided an explanation for the observed metastasis formation upon N-CAM deletion. In contrast to these observations a study using recombinant proteins comprising the ligand-binding Ig-like domains 2 and 3 of FGFR1-4 found that all FGFRs except FGFR4 interacted with N-CAM [109].
In pituitary tumors a truncated FGFR4 isoform was identified and accordingly named pituitary-tumor derived (ptd) FGFR4 [110]. This isoform lacks a signal peptide and the first two extracellular Ig domains and consequently resides in the cytoplasm. Expression of ptd-FGFR4 in pituitary cells or mouse fibroblasts resulted in decreased cell adhesion to a collagen IV matrix [111]. Membrane-anchored wt-FGFR4 formed a proadhesive complex with N-CAM and N-cadherin which was sensitive to treatment with a beta1-integrin neutralizing antibody. In contrast, ptd-FGFR4 did not associate with N-CAM, and ptd-FGFR4 overexpressing cells formed invasive tumors showing loss of membrane localization of N-cadherin and beta-catenin. Treatment with the FGFR inhibitor PD173074 resulted in recovery of membranous N-cadherin localization and reduced tumor growth and invasiveness demonstrating dependency on FGFR kinase activity [112].
In colon cancer cells a cross-talk between FGF19/FGFR4 and the beta-catenin pathway was found [113]. FGF19 treatment increased tyrosine phosphorylation of beta-catenin and caused loss of interaction between #-catenin and E-cadherin. Conversely, a FGF19 blocking antibody increased serine/threonine phosphorylation, and ubiquitination of #-catenin and reduced the expression of #-catenin target genes cyclinD1, CD44, c-jun, Cox-2 and uPAR.
Another form of interaction was demonstrated between FGFRs including FGFR4 and EphA4, a transmembrane tyrosine kinase of the ephrin receptor family [114]. Complex formation involved the juxtamembrane domain of FGFRs and the N-terminal part of the tyrosine kinase domain of EPhA4 and enabled trans-activation of FGFR by the Eph-receptor ligand ephrin-A1. A direct interaction was also detected between FGFR4 and inhibitor of NF$B Kinase beta, an essential component in the NF$B pathway. FGFR4 activation inhibited NF$B signaling and reduced the apoptotic effect of TNF treatment in prostate cancer cells [115].
5. FGFR4 and Cancer
a. Oncogenic mutations of FGFR4
FGFR4 has been found to be mutated [116] and thus constitutively activated in about 7% of embryonic rhabdomyosarcomas (RMS) and these mutations were correlated with advanced, aggressive tumors [63, 117]. Functional analysis demonstrated induction of both increased local growth and enhanced metastasis by mutated FGFR4 [63]. Kinase-activating FGFR4 mutations were also seen in a low percentage of adenocarcinomas of the lung [118–120] and glioblastomas [121]. Like for the other FGFRs, mutations in the extracellular domain (Ig-loops I and II) as well as the cytoplasmic kinase domain were observed (Table 2). However, only kinase-activating mutations have been well characterized functionally. In RMS strongly activating mutations affected amino acid positions 535 and 550 within the kinase domain. They caused increased FGFR4 phosphorylation as well as activated down-stream signaling through STAT3 in murine RMS cells, while phosphorylation of Akt and ERK was decreased [63]. Strong activating mutations in the extracellular and transmembrane domains that cause ligand-independent dimerization mediated by disulfide bonding through cystein residues have frequently been found in FGFR3 but were not among the FGFR4 mutations identified in RMS or lung cancers [5]. One such mutation was identified in the breast cancer cell line MDA-MB453 and was shown to result in constitutive FGFR4 activation and increased ERK activity [122].
Table 2. Kinase Activating Mutations [5].
b. Upregulation of FGFR4 Expression
FGFR4 was found upregulated in multiple tumor types and due to the broad ligand binding spectrum of FGFR4 [18, 19] (Table 1) its overexpression frequently resulted in autocrine stimulation through several candidate FGFs. Best characterized is the hyperactivation of FGFR4 signaling by FGF19 which has been observed in colorectal tumor cells [123] and in hepatocellular carcinomas (HCC) [124–126]. Several reports found FGFR4 to be a major driver of carcinogenesis in the liver [127]. FGFR4 expression correlated with worse prognosis and its inhibition reduced HCC aggressiveness [128]. Mice transgenic for FGF19 expression in the muscle developed HCC based on stimulation of hepatocyte proliferation via FGFR4 [126] and in human HCC FGF19 expression correlated with tumor progression [125, 128]. An amplicon on 11q13.3 present in about 15 % of the tumors and containing FGF19 together with the well-established proto-oncogene CCND1 was suggested as possible predictive marker for FGF19/FGFR4 inhibition therapy [129]. Accordingly, the serum level of FGF19 in HCC patients was significantly lower postoperatively than preoperatively [125]. Blocking FGF19 by a respective antibody abrogated liver cancer development in the transgenic mice and in xenografts of human carcinoma cells suggesting feasibility as a therapeutic target [123, 124].
Comparable to HCC, also in colon cancer xenograft models blockade of FGF19 or FGFR4 by an antibody or shRNA, respectively, reduced in vitro clonogenicity and in vivo tumor growth by attenuating wnt/ß-catenin signaling arguing for existence of an autocrine FGF19/FGFR4 oncogenic signal loop [113]. Additionally, our own group found that FGFR4 silencing inhibited colon cancer cell proliferation and migration again suggesting FGFR4 as therapeutic target in colon cancer [17].
Similarly, FGFR4 has been found overexpressed in pancreatic high-grade intra-epithelial neoplasia and pancreatic ductal carcinoma [130]. Stimulation by FGF19 increased cell adhesion suggesting a tumor suppressive function, however.
In RMS FGFR4 was also found over-expressed [63, 131] probably due to activation by Pax3 – a driving oncogene in RMS that can control FGFR4 expression [60]. Short hairpin (sh)RNA-mediated blockade of FGFR4 in embryonic RMS cells inhibited proliferation in vitro and tumorigenesis in vivo while in alveolar RMS models it induced cell death in line with the supposed driving role of FGFR4 in RMS [132].
In breast cancer upregulation was at least in part caused by gene amplification [7, 133, 134]. FGFR4 might represent a therapeutic target in this small patient subgroup [133]. However, the impact of molecular interventions in that respect needs to be established. In astrocytomas FGFR4 overexpression was correlated with advanced tumor stages and predictive for short survival [135]. Enhanced aggressiveness of high-grade serous ovarian cancer was also attributed to amplification of both the FGFR4 and FGF1 genes located in the identical amplicon at chromosome 5q. The oncogenic function was proven by exogenous overexpression of both these proteins forming an autocrine stimulation loop [136].
In prostate cancer several observations suggest the presence of autocrine growth-promoting loops of FGF ligands and FGFR4. FGFR4 expression was significantly upregulated in prostate cancer as compared to benign hyperplasia and expression level was correlated with higher Gleason score and unfavorable outcome [137]. Moreover, upregulation of the FGFR4 ligands FGF2 and FGF8 was also associated with advanced clinical stage [138]. Accordingly, shRNA-mediated knock-down of FGFR4 was recently demonstrated to reduce orthotopic primary tumor xenograft growth and lymph node metastasis [139]. Likewise targeting FGFR4 by RNA interference effectively blocked prostate cancer cell proliferation and invasion in response to exogenous stimuli [140].
Another cancer type with overexpression of FGFR4 especially within the more aggressive, less differentiated subgroup is thyroid cancer. Correspondingly, introduction of a dominant-negative, secretable FGFR4 protein attenuated cell proliferation specifically in FGFR4-positive, poorly differentiated cell lines with no appreciable effect in well-differentiated cells [141]. Accordingly, pharmacological inhibition of FGFR4 by PD173074 (see below) resulted in reduced xenograft growth in vivo [141]. Also in medullary thyroid carcinoma, a neoplasm of thyroid parafollicular C cells characterized by mutations in another RTK, the RET proto-oncogene, a synergistic effect of RET and FGFR4 inhibition by Imatinib and PD173074, respectively, was postulated [142].
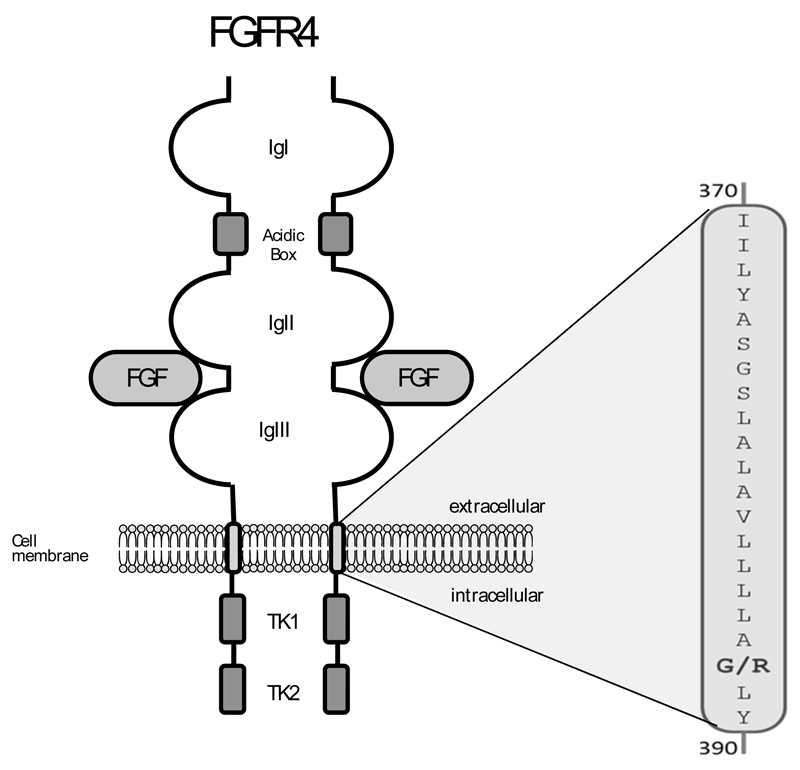
c. The FGFR4G388R Polymorphism (rs351855): Cancer Risk and Progression
Interest in FGFR4 was stimulated by the discovery of a polymorphic form of FGFR4 that causes an exchange of a glycine amino acid into an arginine in the transmembrane domain of the receptor by Axel Ulrich’s group [143] (Fig. 4). The polymorphic FGFR4R388 allele is found in about 55% of the human population (45% are heterozygous and about 10% are FGFR4G388 homozygous) while 45% have a FGFR4G388 homozygous genotype. The incidence is somewhat lower in african-americans than in caucasians [144]. Because FGFR4G388 is present in the majority of the human population it is referred to as the wild type allele [143].
Fig. (4). the FGFR4G388R SNP.
Among the many SNPs in the FGFR4 gene rs351855 has strong pathophysiological impact. It is located in the transmembrane domain of the protein.
Similar mutations in the transmembrane domain of other RTKs have previously been shown to activate kinase activity and/or cause prolonged signaling, especially when charged amino acids like arginine are introduced – most prominently in the erbB2 gene [145]. In the transmembrane domain of FGFR3 a mutation that exchanges a glycine to an arginine in codon 380 (homologous to the FGFR4G388R SNP) constitutes an activating mutation strongly associated with achondroplasia [146]. The same is observed for an A391E amino acid change in FGFR3 that also resides in the transmembrane domain [147]. These alterations lead to ligand independent activation and prolonged signaling due to slowed receptor internalization [146, 148–150].
For the FGFR4R388 substitution the impact is more subtle so that the consequences of the alteration for cell biology are more difficult to assess. For several tumor types controversial reports have been published during the past decade (Table 3). Moreover, the cancer types and parameters studied are heterogeneous. Three meta-analysis have hitherto been published and report sound evidence of pro-tumorigenic effects of FGFR4R388 for breast and prostate cancer while in other tumor types the evidence was found inconclusive [151, 152]. In spite of these difficulties the emerging picture shows a pro-tumorigenic and/or prometastatic impact of the FGFR4R388 allele resulting in poor outcome / survival in a wide spectrum of malignancies. Increased risk of developing a malignant tumor was only observed in the prostate while for HNSCC and soft tissue sarcomas it may even be decreased. Evidence for rapid tumor progression, early or increased metastasis and advanced stage in patients carrying the FGFR4R388 allele was found in breast, prostate, colon, and lung cancer at least by some authors. Some reports found reduced disease-free (DFS) or overall survival (OS), but even within one tumor type the data are contradictory in all cases. The impact on therapy response seems to be tissue specific (the respective literature is summarized in Table 3).
Table 3. Impact of FGFR4G388R on Cancer Risk and Progression.
| Tumor type | Cancer risk | Tumor aggressiveness | Survival | Therapy response | Reference |
|---|---|---|---|---|---|
| breast cancer | no impact | early node metastasis, advanced stage | reduced DFS | n.r. | [143] |
| n.r. | n.r. | no impact on OS | n.r. | [189] | |
| no impact | no impact | no impact on OS | n.r. | [190] | |
| n.r. | n.r. | reduced DFS in node-positive patients | poor | [183] | |
| no impact | increased node involvement, no impact on stage | n.r. | n.r. | [191] | |
| n.r. | increased node involvement | n.r. | improved | [185] | |
| n.r. | increased metastasis | shorter OS | n.r. | [192] | |
| prostate cancer | increased | increased node involvement | DSF decreased | n.r. | [144] |
| increased | increased metastasis | no impact on OS | n.r. | [193] | |
| no impact | no impact | little impact on OS | n.r. | [194] | |
| no impact | n.r. | n.r. | n.r. | [195] | |
| colon cancer | no impact | early node invovlement, advanced stage | n.r. | n.r. | [143] |
| no impact | no impact | n.r. | n.r. | [190] | |
| n.r. | n.r. | n.r. | therapy resistance | [196] | |
| no impact | higher stage | n.r. | n.r. | [17] | |
| HNSCC | n.r. | advanced stage in high FGFR4 tumors | reduced OS in high FGFR4 tumors | n.r. | [197] |
| n.r. | n.r. | reduced OS | n.r. | [198] | |
| reduced risk | n.r. | n.r. | increased cis-Pt sensitivity | [187] | |
| n.r. | n.r. | no impact | n.r. | [199] | |
| n.r. | increased node involvement | increased relapse and disease-related death | n.r. | [200] | |
| no impact | no impact on stage | increased disease-related death | n.r. | [201] | |
| lung cancer | early onset | increased node involvement, advanced stage | worse short term survival | n.r. | [202] |
| n.r. | no impact on stage | no impact | n.r. | [203] | |
| n.r. | n.r. | worse survival in node positive patients | no impact with platium treatment | [120] | |
| n.r. | advanced stage | decreased | n.r. | [161] | |
| melanoma | n.r. | thick tumors, nodular subtype | n.r. | n.r. | [204] |
| no impact | n.r. | n.r. | n.r. | [205] | |
| gastric cancer | n.r. | smaller tumors | decreased OS | n.r. | [206] |
| pancreatic neuroendocrine tumors | n.r. | increased metastasis and stage | n.r. | n.r. | [184] |
| Liver | no impact | portal venous tumor thrombosis | n.r. | n.r. | [207] |
| gliomas | n.r. | n.r. | no impact | n.r. | [208] |
| bladder cancer | no impact | no impact on stage | increased DFS | n.r. | [209] |
| bone sarcome | n.r. | n.r. | no impact | n.r. | [210] |
| soft-tissue sarcomas | n.r. | n.r. | improved | n.r. | |
| Ovarian cancer | no impact | portal venous tumor thrombosis | prolonged OS in R0 patients | increased platinum sensitivity | [186] |
The table lists studies describing the impact of FGFR4R388 on cancer risk, progression, treatment and prognosis. n.r. indicates that no information on the parameter has been reported
d. Differential Cell Biological Effects of FGFR4 Polymorphic Forms
Mechanistic studies have been performed using tumor cell models of the breast, the prostate and the colon [17, 143, 144, 153]. Transfected prostate epithelial cells and breast cancer cells grew in tightly attached colonies when they express FGFR4G388, whereas the FGFR4R388-cells had a more irregular morphology and grew in a scattered fashion [143, 144]. Moreover in transfected breast cancer cell lines [143], prostate cancer cell lines [153], and colorectal cancer cell lines [17] the FGFR4R388 allele induced stronger cell migration and invasion in vitro than the FGFR4G388 allele. By contrast, in metastatic breast cancer cells and in colon cancer cells expressing high levels of FGFR4R388 the FGFR4G388-allele even suppressed cell migration [17, 154].
Embryonic fibroblasts (MEFs) isolated from transgenic mice expressing the FGFR4R385 allele, which is the homologous mutation in the mouse, also displayed an increased migratory and invasive capacity in vitro compared to FGFR4G385 MEFs thus contributing to tumor progression [155]. In addition they were more easily transformed by EGFR or HER2 than MEFs expressing the FGFR4G385 indicating that the polymorphic allele in the mouse may not be an oncogene per se but may support other oncogenes like EGFR and HER2.
The main biochemical difference between the 2 polymorphic forms of FGFR4 is the higher stability of FGFR4R388 that results in prolonged signaling activity [144]. For FGFR1 prolonged signaling has been described after stimulation by N-CAM-derived peptides in Hela cells. This was related to activation of c-src and induction of cell migration [156]. In line with this report, src-upregulation was observed in FGFR4R388-transfected colon cancer cells that also had increased migration capacity [17]. However, the FGFR4G388 receptor was also shown to be stabilized in prostate cancer by interaction with an additional protein called Huntingtin-interacting protein 1 (HIP1) increasing the proliferative and anchorage-independent growth abilities in vitro [153]. The increased stability of both FGFR4 alleles led to an increased ERK phosphorylation which was even more sustained in FGFR4R388 transfected cells [139].
FGFR4R388–induced c-scr-activation has been shown to increase the secretion of MMP2 [157, 158] which was activated mainly by membrane type 1 matrix metalloproteinase (MT1-MMP, MMP14) [159]. FGFR4G388 or any other FGFR was inactive in this regard. It seems that the FGFR4R388 receptor stabilized MT1-MMP by reducing its lysosomal degradation and induced its phosphorylation but was also activated in reverse. In contrast FGFR4G388 and MT1-MMP suppressed and down-regulated each other. Interaction of FGFR4R388 with MT1-MMP enhanced the proteolytic activity and stimulated invasive characteristics [157]. In transfected prostate cancer cell lines FGFR4R388 silencing decreased MT1-MMP protein expression, N-cadherin expression and other metastatic parameters whereas E-cadherin increased, resulting in an inhibition in tumor growth as well as vascular and lymph node invasion in vivo. Silencing FGFR4G388 resulted in the opposite effect and even increased MT1-MMP protein expression and invasive properties of the cells in vivo. This effect of the FGFR4R388 was not triggered by mitogenic signaling because silencing did not impair ERK1/2 and PI3K/AKT pathways [160]. This suggests that the FGFR4G388 wt-allele has an anti-metastatic effect that is lost through the G388R amino acid substitution. Upregulation of c-src was observed in SW480 colon carcinoma cells overexpressing FGFR4R388 compared to control cells or cells overexpressing the G388 variant [17]. Specific phosphorylation of Src at tyrosine 418 and of FRS2, PLC, ERK, GSK and S6 was similar between the FGFR4G388 and FGFR4R388 overexpressing cells, although the G388-variants showed a higher fraction of FRS2 and membrane-associated PLC.
Other reports suggested that the aggressive effect of FGFR4R388 might be caused by interaction with uPAR (cell surface receptor for urokinase-type plasminogen activator) [144] and Notch signaling [161]. In pituitary tumor cells FGFR4G388 and FGFR4R388 differentially regulated growth hormone production via src-activation and increased mitochondrial serine phosphorylation but decreased tyrosine phosphorylation of Stat3 [162].
Overall these observations support a pro-metastatic effect of the FGFR4R388 allele, but more tumor types, specifically those for which a protective impact of the FGFR4R388 receptors has been reported – need to be investigated to completely understand the role of the polymorphism in tumor progression.
e. Tumor Suppressor or Oncogene
Based on the published data the question whether FGFR4 acts as oncogene or tumor-suppressor cannot be unequivocally answered and it may well be that the impact is dependent on the exact tissue context of tumor development. The controversy is most obvious in the liver. While there is strong evidence for a tumor-driving oncogenic role of FGFR4 in HCC (see above [124–127]), a tumor suppressive function has also been described. It is related to the tissue protective effect of FGFR4 and is consistent with a caretaker function capable of preventing tumor development [82, 163]. This role of FGFR4 is restricted to models of chemically induced hepatocarcinogenesis associated with severe tissue damage. Accordingly, diethylnitrosamine (DEN)-induced liver tumor formation was significantly accelerated in FGFR4-deficient mice, while spontaneous tumor formation remained unaltered [163]. It has been suggested that the tumorsuppressor function of FGFR4 in the liver may depend on coexpression with #-klotho which turns FGF19 signaling to metabolic and growth-suppressing functions [71, 163, 164]. These data suggest that a tumor-suppressive function of FGFR4 probably depends on the lineage-specific expression of klotho proteins, which are known anticancer and antiaging proteins [66]. One might hypothesize that the anticancer function of FGFR4/klotho competes with the pro-oncogenic RTK signaling function of FGFR4 in tumors probably based on the lack of klotho.
However, in marked contrast to this hypothesis Guagano et al. recently reported that sensitivity to the pan-FGFR inhibitor NVP-BGJ398 (see below) in FGFR4-driven liver cancer cell lines was strictly dependent on the overexpression of both FGF19 and #-klotho supporting FGF19/FGFR4 interaction [165]. Additionally, it was shown that FGF19-induced hepatocarcinogenesis was strongly enhanced by DEN but completely blocked in FGFR4 knock-out mice and by both FGF19- and FGFR4-blocking antibodies [124]. A similar effect could be seen when treating FGF19-over-expressing/FGFR4wt animals with antibodies blocking FGF19 or the dimerization of FGFR4 [123, 124]. It remains to be elucidated whether the protumorigenic effects of FGFR4 occur at supraphysiological concentrations of FGF19 only and whether the unique metabolic functions of this FGFR4 ligand contribute to this effect. Nevertheless, these contradictory data call for caution when targeting FGFR4-mediated signals as anticancer strategies and stress the urgent need for well-established models and specific inhibitors for FGFR4.
Another case for FGFR4 as a tumor suppressor comes from the frequent but often undiagnosed pituitary tumors. The oncogenic ptd-FGFR4 is thought to interrupt the pro-adhesive and potentially tumor-suppressive interaction of FGFR4 with N-CAM and N-cadherin and to diminish cell adhesion inducing a transformed phenotype [111], [112]. Pharmacological inhibition by a small-molecule tyrosine kinase inhibitor (TKI) PD173074 (see below) was only moderately effective in cells overexpressing ptd-FGFR4 but not the wt-receptor [112]. Based on a recently established FGFR4R388 crosstalk with c-src and STAT3, src inhibitors have been shown to block FGFR4R388-positive pituitary tumor cells [162]. However, the reason why in that case upstream FGFR4 inhibition is ineffective remains to be explained.
While breast cancer is one of the tumor types for which FGFR4 overexpression was observed already in the 1990’s (see above [133, 134]), loss of heterozygosity was found in 50% of the heterozygous cases suggesting a tumor-suppressing role of FGFR4. In this study DNA methylation of the FGFR4 gene affected both gene variants, but was twice as likely to silence the FGFR4G388 versus the FGFR4R388 allele arguing for a stronger tumor suppressor activity of FGFR4G388 [166].
With regard to the FGFR4G388R polymorphism the hypothesis has been discussed that FGFR4G388 may be a tumor suppressor loosing its function through the G to R amino acid substitution. This is supported by reports of FGFR4G388 inhibiting cell migration and metastasis [154] even though this was restricted to a high FGFR4R388 expression background in colon cancer cells [3]. At the same time contradicting observations show increased tumor growth from FGFR4G388 transfected cells [17]. The data certainly support an anti-metastatic impact of FGFR4G388 that is also supported by signaling and matrix degradation results [157, 160]. However, they do not unequivocally show a tumor suppressor function.
Knock-in mice carrying a FGFR4R385 that have been constructed to resolve this question initially do not show an obvious phenotype. However, in a chemically induced breast tumor model they display a higher tumor number, earlier tumor onset, more aggressive behavior and earlier metastasis onset compared to FGFR4G385 mice [155]. The observation is consistent with both a tumor/metastasis suppressing function of FGFR4G388 and an oncogenic function of FGFR4R388. Our own observations in colon cancer xenografts show increased local tumor growth induced by FGFR4G388 and earlier metastasis from FGFR4R388 tumors – which are both oncogenic effects [17].
Even though the case for an oncogenic activation of FGFR4 is strong in several tumor types the possibility of an additional tumor suppressive function has to be kept in mind when designing strategies of FGFR4-targeting therapy.
6. Targeting FGFR4
a. Is FGFR4 a Suitable Target for Cancer Therapy?
The number of studies investigating specifically FGFR4 as a target for anticancer therapy is comparably limited so far [3, 167]. Reasons might be that the FGFR4 activating mutations and amplifications are restricted to rather small patient subgroups as compared to successfully developed therapeutic targets like EGFR in lung adenocarcinoma, Her2 in case of breast cancer and FGFR1 in case of squamous cell carcinomas of lung. In addition the cellular mechanisms underlying FGFR4 pathophysiology are still insufficiently understood so that predictive markers for therapy response are widely lacking.
Additionally, within the FGFR tyrosine kinase family, FGFR4 shows the lowest homology to the other members suggesting that this gene underwent the strongest alterations during evolution. This might reflect the need for FGFR4 to take over functions other than growth factor-mediated proliferation and survival signaling including cell adhesion and migration/invasion as well as important metabolic functions (see chapter 3). Such it is not surprising that most of the small FGFR inhibitors described so far (and partly already tested in clinical studies) are significantly less active against FGFR4 as compared to the other FGFR family members. In fact none of the known inhibitors has specificity for FGFR4. Even a somewhat higher inhibition constant against FGFR4 compared to FGFR1, 2 and/or 3 has not yet been observed (see Table 4) [167, 168]. Consequently, development of specific FGFR4 tyrosine kinase inhibitors (TKIs) is of utmost importance to clarify the feasibility of this therapeutic strategy in respective cancer patients harboring tumors with alterations in FGFR4. A respective strategy based on in silico docking scores has been very recently reported resulting in relatively active FGFR4 TKIs which show activity against FGFR4-overexpressing cancer models in vitro [169].
Nevertheless, when directly targeting FGFR4 in cancer therapy, the impact on the important metabolic functions of FGFR4 outlined above, has to be considered and thoroughly tested to avoid strong unwanted adverse effects based on metabolic deregulation. As mice devoid of FGFR4 show elevated cholesterol metabolism and bile acid synthesis [51] and enhanced vulnerability to chemically-induced liver damage [82] (see above), unwanted adverse effects in the liver need to be closely monitored.
Additionally, it has to be considered that in some studies especially in breast cancer, hepatoma and pituitary tumors also convincing evidence for a tumor-suppressive function has been assigned to wt-FGFR4 [154, 163, 166]. Although these studies are controversially discussed, their results have to be kept in mind when developing FGFR4 as target for cancer therapy.
Despite these caveats several studies based on genetic and/or biochemical as well as pharmacologic interventions clearly demonstrated that inhibition of FGFR4-mediated functions/signals might induce anticancer activity especially in tumors with enhanced FGFR4 expression. The respective methodological approaches include siRNA/shRNA, expression of dominant-negative FGFR constructs (dnFGFR), FGF ligand- or receptor-blocking antibodies and FGFR4-specific ligand trap proteins containing the extracellular domain of FGFR4 and thus depleting the FGF ligand pool of the endogenous receptor. These effects were especially prominent in those cell types with known oncogenic alteration in the FGFR4 gene (for literature see chapter 5a and b).
While predictive markers have not yet been defined, FGFR4 mutations or overexpression of autocrine ligands seem to be a pre-requisite for a response to FGFR4 blockade based on the reports discussed in chapter 5. For HCC also #-klotho expression may be a suitable predictive marker [165]. Tumor-specific FGFR4-dependent down-stream signaling involves STAT3 in RMS suggesting the possibility that STAT3 levels may also be a suitable marker for drug response [63]. With regard to the G388R polymorphic forms of FGFR4 there are several reports regarding significant effects on the response to other oncological therapies suggesting quality as a predictive marker (Table 3). However, any impact of the polymorphic forms on response to FGFR4-silencing still remains to be determined.
b. Small Molecule FGFR Inhibitors
Small molecule TKIs targeting the ATP-binding pocket are of central interest in personalized cancer therapy and some of them have even been already approved for clinical use in defined patient subgroups. Accordingly, several companies have developed TKI targeting members of the FGFR family (Table 4). Although no declared FGFR inhibitor has been approved for anticancer therapy so far, several of these drugs are in clinical development [3, 4, 167, 170]. Nevertheless, it needs to be mentioned that several of the known antiangiogenic VEGFR or platelet-derived growth factor receptor (PDGFR) inhibitors (sorafenib, sunitinib, dovitinib), based on the strong homology of the kinase domains, also exert some inhibitory activity against FGFRs and the contribution to the clinical efficacy needs to be determined. However, for many of these substances the activity against FGFR4 is not published.
Unfortunately in multiple preclinical studies aiming on pharmacological inhibition of FGFR4, inhibitors with weaker potency against FGFR4 as compared to the other FGFRs were used due to lack of appropriate substances. Thus the TKI PD173074 [171], a synthetic compound of the pyrido-[2,3-d] pyrimidine class, characterized in some studies or by suppliers as FGFR1- and -3 specific was widely used in preclinical studies and shown to be able to inhibit (mutant) FGFR4-driven (transgenic) activities [63, 112, 141, 164, 172, 173]. However, if endogenous FGFR4-mediated effects are investigated the application of such unspecific inhibitors is problematic considering the high activity against other FGFR family members.
Generally, as mentioned above only a few FGFR TKIs published so far show activity against FGFR4 comparable to the other FGFR family members and for many substances data for FGFR4 have not been reported (Table 4). One TKI having equally high activity against all four FGFRs is (R)-(E)-2-(4-(2-(5-(1-(3,5-dichloropyridin-4-yl)ethoxy)-1H-indazol-3yl)vinyl)-1H-pyrazol-1-yl)ethanol, developed by Lilly and designated as LY2874455 [174]. This substance was shown to be highly active against FGFR-driven cancer xenografts but an FGFR4-driven model was not assessed. Promising activities have been determined also for ponatinib, clinically mainly developed for Imatinib-refractory chronic myeloid leukemia based on a potent abl kinase inhibition [175], but also highly active against the entire FGFR family [176]. Accordingly, ponatinib was shown to inhibit FGFR4 transgenic cell models with almost equal potency as compared to the other FGFR molecules [176]. However, despite investigating multiple FGFR-driven cell models and despite a stronger anticancer activity of ponatinib as compared to several other FGFR inhibitors like BIBF1120, brivanib, cediranib and dovitinib, no single model was mentioned in this study to be selected for FGFR4 dependence. Also Astex Pharmaceuticals recently reported on the development of pan-FGFR inhibitors with comparable activities against all four FGFRs and one of them (JNJ-42756493) being already evaluated as orally available drug in a clinical phase I study [177]. However, data on FGFR4-driven cancers are widely missing so far. Intersection of genome-wide gene expression and genomic alteration data of a large cancer cell line panel with sensitivity against the pan-FGFR inhibitor BGJ398 suggested that responsiveness is closely related to FGF/FGFR deregulation [165]. Although no FGFR4 mutant or amplified cell model was included, hepatoma cell models with FGF19 amplification and #-klotho overexpression responded to BGJ398 in an FGFR4-dependend manner. These data suggest that biomarkers might be feasible for identification of patient subgroups likely to respond to FGFR4 inhibition.
c. Antibodies and Ligand Traps
The lack of specificity for FGFR4, evident for all small molecule FGFR TKIs, might be solved by the development of targeting antibodies against an FGFR4-specific epitope at the extracellular domain of FGFR4. Following this strategy, researchers at Genentech have developed the monoclonal anti-FGFR4 antibody LD1 with exclusive specificity for FGFR4 [178] and tested for activity against the FGF19/FGFR4 driven hepatocellular carcinoma model in mice described above [124]. The antibody was able to inhibit ligand binding to FGFR4 as well as downstream signals and blocked tumor formation in the FGF19-transgenic mouse model even after induction by DEN. The group of Axel Ullrich has described the generation of another mouse monoclonal anti-FGFR4 blocking antibody (10F10) raised against the purified extracellular domain of FGFR4 and now developed by U3 Pharma [179]. Interestingly, this antibody effectively blocked signaling by wt FGFR4 but was less efficient against the Y367C mutant, the dominant oncogene in MDA-MB453 breast cancer cells [122]. Additionally, antibodies against the FGFR4 specific ligand FGF19 were able to block preclinical colorectal and liver cancer models [113, 123]. An alternative approach is the use of extracellular ligand traps. Thus a FGFR4 fusion trap protein FTP-091 (FGFR4mut:Fc:R4-Trap; FGFR4:Fc) was generated by Five Prime Therapeutics, representing a chimeric fusion protein consisting of the three Ig-like extracellular domains of FGFR4 with the acid box linker between Ig-loops I and II replaced by the corresponding acid box region of FGFR1. The resulting molecule exhibited the highest affinities for FGF17 and FGF18 followed by FGF1 and FGF2 [136]. Although this agent has been shown to inhibit FGF1/FGFR4-mediated growth signals, it cannot be considered as a FGFR4-specific in view of the promiscuous binding of the latter ligands to FGFRs. For none of the FGFR4-targeting antibodies initiation of a clinical study has been reported so far. This holds also true for the FGFR4 FTP-091 ligand trap in contrast to the FGFR1 extracellular domain-containing FP-1039[180].
d. Impact of FGFR4 on Sensitivity Against Chemotherapy and Targeted Agents
FGF and FGFR mediated signals in cancer cells have been repeatedly associated with chemotherapy failure well in agreement with a survival promoting function of this signal cascade [170]. Interestingly, also FGFR4 hyperactivation or presence of the oncogenic FGFR4R388 variant has been associated with altered anticancer therapy response. Roidl et al. [179] observed distinct upregulation of FGFR4 expression in breast cancer cell clones surviving doxorubicin treatment. Accordingly, overexpression and knock-down experiments confirmed that FGFR4 is able to mediate chemoresistance conferred by upregulation of anti-apoptotic bcl-xl via MAPK signal transduction. This resistance could be reversed by a specific FGFR4 blocking antibody 10F10. In addition to chemotherapy response, FGFR4 was also associated with the efficacy of endocrine therapy and of novel targeted anticancer agents. Thus, FGFR4 expression predicts failure on tamoxifen therapy in patients with recurrent breast cancer [181] and FGFR4 expression correlated with erlotinib resistance in glioblastoma cell lines suggesting that it might substitute for oncogenic EGFR signals [182].
On the contrary under certain circumstances even blockade of FGFR4-mediated signals might induce therapy resistance. Thus blockade of both #-klotho or FGFR4 via siRNA technology and PD173074 selected for resistant cell clones overexpressing the stem cell markers CD133 and CD44 implicating survival of therapy refractory slow growing tumor subclones [164].
With regard to isoforms, the poor survival of FGFR4R388 carriers with node-positive breast cancer was at least partly attributed to poor response to adjuvant therapy mainly with cyclophosphamide/methotrexate/fluorouracil-based regimens [183]. In a model for pancreatic neuroendocrine tumors, expression of FGFR4R388 but not FGFR4G388 diminished responsiveness to the mTOR inhibitor everolimus correlating with reduced clinical response in patients harboring this genotype [184]. While in a recent study on neoadjuvant chemotherapy for breast cancer the poor prognosis of FGFR4R388 carriers was confirmed, this genotype surprisingly indicated enhanced responsiveness and complete pathological response to anthracyclines/taxane-based regimens [185]. Accordingly, the same research group found hypersensitivity towards platinum-based chemotherapy associated with FGFRR388 in advanced ovarian cancer resulting in prolonged patient survival [186]. Analysis of preclinical HNSCC models suggest the same relationship between FGFR4R388 and cis-platin response in this tumor type [187]. These apparently conflicting data might be based on the different therapy approaches and agents, their dependence on tumor cell proliferation, and specific interactions with tumor cell subpopulations like cancer stem cell compartments. Nevertheless, all these studies point towards an involvement of FGFR4-mediated mechanisms in anticancer therapy responses and call for precise elucidation of the underlying mechanisms to extract potential predictive information for diverse therapeutic interventions.
7. Conclusion
The collected results indicate that FGFR4 can be a suitable target for therapy of several cancer types and thus further development of specific therapeutic agents is well worth the effort. Markers predicting response to FGFR4-targeting therapy, however, are still lacking. In addition, the tissue protective impact of FGFR4 in the liver and its possible tumor suppressive function has to be kept in mind when planning systemic FGFR4 signaling blockade.
Acknowledgements
Grant support: Work on FGFR4 was supported by the Austrian National Bank (Projects 12684, 14211), the Austrian Science Foundation (P19920, P23693).
Abbreviations
- CYP7A1
cholesterol 7alpha-hydroxylase
- DEN
diethylnitrosamine
- DFS
disease-free survival
- FGF(R)
fibroblast growth factor (receptor)
- Fr
folate receptor alpha
- FRS
FGFR substrate alpha
- HCC
hepatocellular carcinoma
- HS
heparansulfate
- hFGF
hormonal FGF
- HNSCC
head and neck squamous cell carcinoma
- HS
heparan sulfate
- Ig
immunoglobuline-like
- JNK
c-Jun N-terminal kinase
- MAPK
mitogen activated protein kinase
- MMP
matrix metalloproteinase
- MT1-MMP, MMP14
membrane type 1 matrix metalloproteinase
- N-CAM
neural cell adhesion molecule
- OS
overall survival
- PDGFR
platelet-derived growth factor receptor
- PI3K
phosphoinositide 3 kinase
- PLC
phospholipase C gamma
- ptd-FGFR4
pituitary-tumor derived FGFR4
- RMS
rhabdomyosarcomas
- RTK
receptor tyrosine kinase
- TKI
tyrosine kinase inhibitor
- TSP
transcription start point
- VEGFR
vascular endothelial growth factor receptor
- wt
wildtype
Footnotes
Conflict of Interest
The authors confirm that this article content has no conflicts of interest.
Disclosures
No conflicting interests to disclose
References
- [1].Powers CJ, McLeskey SW, Wellstein A. Fibroblast growth factors, their receptors and signaling. Endocr Relat Cancer. 2000;7:165–97. doi: 10.1677/erc.0.0070165. [DOI] [PubMed] [Google Scholar]
- [2].Beenken A, Mohammadi M. The FGF family: biology, pathophysiology and therapy. Nat Rev Drug Discov. 2009;8:235–53. doi: 10.1038/nrd2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Heinzle C, Sutterluty H, Grusch M, et al. Targeting fibroblast-growth-factor-receptor-dependent signaling for cancer therapy. Expert Opin Ther Targets. 2011;15(7):829–46. doi: 10.1517/14728222.2011.566217. [DOI] [PubMed] [Google Scholar]
- [4].Turner N, Grose R. Fibroblast growth factor signalling: from development to cancer. Nat Rev Cancer. 2010;10:116–29. doi: 10.1038/nrc2780. [DOI] [PubMed] [Google Scholar]
- [5].Haugsten EM, Wiedlocha A, Olsnes S, Wesche J. Roles of Fibroblast Growth Factor Receptors in Carcinogenesis. Mol Cancer Res. 2010;8(11):1439–52. doi: 10.1158/1541-7786.MCR-10-0168. [DOI] [PubMed] [Google Scholar]
- [6].Knights V, Cook SJ. De-regulated FGF receptors as therapeutic targets in cancer. Pharmacol Ther. 2010;125:105–17. doi: 10.1016/j.pharmthera.2009.10.001. [DOI] [PubMed] [Google Scholar]
- [7].Tenhagen M, van Diest PJ, Ivanova IA, van der Wall E, van der Groep P. Fibroblast growth factor receptors in breast cancer: expression, downstream effects, and possible drug targets. Endocr-Relat Cancer. 2012;19:R115–R129. doi: 10.1530/ERC-12-0060. [DOI] [PubMed] [Google Scholar]
- [8].Hadari Y, Schlessinger J. FGFR3-targeted mAb therapy for bladder cancer and multiple myeloma. J Clin Invest. 2009;119:1077–9. doi: 10.1172/JCI38948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Holzmann K, Grunt T, Heinzle C, et al. Alternative Splicing of Fibroblast Growth Factor Receptor IgIII Loops in Cancer. J Nucleic Acids. 2012;2012:950508. doi: 10.1155/2012/950508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Weinstein M, Xu X, Ohyama K, Deng CX. FGFR-3 and FGFR-4 function cooperatively to direct alveogenesis in the murine lung. Development. 1998;125:3615–23. doi: 10.1242/dev.125.18.3615. [DOI] [PubMed] [Google Scholar]
- [11].Dy G. Emerging systemic therapeutic approaches for personalized medicine in squamous cell carcinoma of the lung. J Cancer Sci Ther. 2012;S11:1–7. [Google Scholar]
- [12].Itoh N, Ornitz DM. Evolution of the Fgf and Fgfr gene families. Trends Genet. 2004;20:563–9. doi: 10.1016/j.tig.2004.08.007. [DOI] [PubMed] [Google Scholar]
- [13].Rebscher N, Deichmann C, Sudhop S, et al. Conserved intron positions in FGFR genes reflect the modular structure of FGFR and reveal stepwise addition of domains to an already complex ancestral FGFR. Dev Genes Evol. 2009;219:455–68. doi: 10.1007/s00427-009-0309-5. [DOI] [PubMed] [Google Scholar]
- [14].Zhang Y, Gorry MC, Post JC, Ehrlich GD. Genomic organization of the human fibroblast growth factor receptor 2 (FGFR2) gene and comparative analysis of the human FGFR gene family. Gene. 1999;230:69–79. doi: 10.1016/s0378-1119(99)00047-5. [DOI] [PubMed] [Google Scholar]
- [15].Partanen J, Makela TP, Eerola E, et al. FGFR-4, a novel acidic fibroblast growth factor receptor with a distinct expression pattern. EMBO J. 1991;10:1347–54. doi: 10.1002/j.1460-2075.1991.tb07654.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Ron D, Reich R, Chedid M, et al. Fibroblast growth factor receptor 4 is a high affinity receptor for both acidic and basic fibroblast growth factor but not for keratinocyte growth factor. J Biol Chem. 1993;268:5388–94. [PubMed] [Google Scholar]
- [17].Heinzle C, Gsur A, Hunjadi M, et al. Differential effects of polymorphic alleles of FGF receptor 4 on colon cancer growth and metastasis. Cancer Res. 2012;72:5767–77. doi: 10.1158/0008-5472.CAN-11-3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Ornitz DM, Xu J, Colvin JS, et al. Receptor Specificity of the Fibroblast Growth Factor Family. J Biol Chem. 1996;271:15292–15297. doi: 10.1074/jbc.271.25.15292. [DOI] [PubMed] [Google Scholar]
- [19].Zhang X, Ibrahimi OA, Olsen SK, et al. Receptor Specificity of the Fibroblast Growth Factor Family: the complete mammalian FGF family. J Biol Chem. 2006;281:15694–15700. doi: 10.1074/jbc.M601252200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Hughes SE. Differential expression of the fibroblast growth factor receptor (FGFR) multigene family in normal human adult tissues. J Histochem Cytochem. 1997;45:1005–19. doi: 10.1177/002215549704500710. [DOI] [PubMed] [Google Scholar]
- [21].Becker M, Brauninger A, Wolf G, Kaufmann M, Strebhardt K. Identification and functional characterization of the human and murine fibroblast growth factor receptor 4 promoters. Biochem Biophys Res Commun. 2000;276:493–501. doi: 10.1006/bbrc.2000.3483. [DOI] [PubMed] [Google Scholar]
- [22].Pugh BF, Tjian R. Transcription from a TATA-less promoter requires a multisubunit TFIID complex. Genes Dev. 1991;5:1935–45. doi: 10.1101/gad.5.11.1935. [DOI] [PubMed] [Google Scholar]
- [23].Saito H, Kouhara H, Kasayama S, Kishimoto T, Sato B. Characterization of the promoter region of the murine fibroblast growth factor receptor 1 gene. Biochem Biophys Res Commun. 1992;183:688–93. doi: 10.1016/0006-291x(92)90537-u. [DOI] [PubMed] [Google Scholar]
- [24].Avivi A, Skorecki K, Yayon A, Givol D. Promoter region of the murine fibroblast growth factor receptor 2 (bek/KGFR) gene. Oncogene. 1992;7:1957–62. [PubMed] [Google Scholar]
- [25].McEwen DG, Ornitz DM. Regulation of the fibroblast growth factor receptor 3 promoter and intron I enhancer by Sp1 family transcription factors. J Biol Chem. 1998;273:5349–57. doi: 10.1074/jbc.273.9.5349. [DOI] [PubMed] [Google Scholar]
- [26].Perez-Castro AV, Wilson J, Altherr MR. Genomic organization of the human fibroblast growth factor receptor 3 (FGFR3) gene and comparative sequence analysis with the mouse Fgfr3 gene. Genomics. 1997;41:10–6. doi: 10.1006/geno.1997.4616. [DOI] [PubMed] [Google Scholar]
- [27].Yu S, Asa SL, Ezzat S. Fibroblast growth factor receptor 4 is a target for the zinc-finger transcription factor Ikaros in the pituitary. Mol Endocrinol. 2002;16:1069–78. doi: 10.1210/mend.16.5.0832. [DOI] [PubMed] [Google Scholar]
- [28].Yu SJ, Zheng L, Ladanyi M, Asa SL, Ezzat S. Sp1-mediated transcriptional control of fibroblast growth factor receptor 4 in sarcomas of skeletal muscle lineage. Clin Cancer Res. 2004;10:6750–8. doi: 10.1158/1078-0432.CCR-04-0223. [DOI] [PubMed] [Google Scholar]
- [29].Zhao P, Caretti G, Mitchell S, et al. Fgfr4 is required for effective muscle regeneration in vivo. Delineation of a MyoD-Tead2-Fgfr4 transcriptional pathway. J Biol Chem. 2006;281:429–38. doi: 10.1074/jbc.M507440200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Boshnjaku V, Shim KW, Tsurubuchi T, et al. Nuclear localization of folate receptor alpha: a new role as a transcription factor. Sci Rep. 2012;2:980. doi: 10.1038/srep00980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].encode. 2012.
- [32].Yu S, Asa SL, Weigel RJ, Ezzat S. Pituitary tumor AP-2alpha recognizes a cryptic promoter in intron 4 of fibroblast growth factor receptor 4. J Biol Chem. 2003;278:19597–602. doi: 10.1074/jbc.M212432200. [DOI] [PubMed] [Google Scholar]
- [33].Yan G, Wang F, Fukabori Y, et al. Expression and transforming activity of a variant of the heparin-binding fibroblast growth factor receptor (flg) gene resulting from splicing of the alpha exon at an alternate 3'-acceptor site. Biochem Biophys Res Commun. 1992;183:423–30. doi: 10.1016/0006-291x(92)90498-a. [DOI] [PubMed] [Google Scholar]
- [34].Vainikka S, Partanen J, Bellosta P, et al. Fibroblast growth factor receptor-4 shows novel features in genomic structure, ligand binding and signal transduction. EMBO J. 1992;11:4273–80. doi: 10.1002/j.1460-2075.1992.tb05526.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Kostrzewa M, Muller U. Genomic structure and complete sequence of the human FGFR4 gene. Mamm Genome. 1998;9:131–5. doi: 10.1007/s003359900703. [DOI] [PubMed] [Google Scholar]
- [36].He Y, Smith SK, Day KA, et al. Alternative splicing of vascular endothelial growth factor (VEGF)-R1 (FLT-1) pre-mRNA is important for the regulation of VEGF activity. Mol Endocrinol. 1999;13:537–45. doi: 10.1210/mend.13.4.0265. [DOI] [PubMed] [Google Scholar]
- [37].Tiran Z, Oren A, Hermesh C, et al. A novel recombinant soluble splice variant of Met is a potent antagonist of the hepatocyte growth factor/scatter factor-Met pathway. Clin Cancer Res. 2008;14:4612–21. doi: 10.1158/1078-0432.CCR-08-0108. [DOI] [PubMed] [Google Scholar]
- [38].Vorlová S, Rocco G, LeFave Clare V, et al. Induction of Antagonistic Soluble Decoy Receptor Tyrosine Kinases by Intronic PolyA Activation. Mol Cell. 2011;43:927–939. doi: 10.1016/j.molcel.2011.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Lemmon MA, Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2010;141:1117–34. doi: 10.1016/j.cell.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Uehara H, Cho Y, Simonis J, et al. Dual suppression of hemangiogenesis and lymphangiogenesis by splice-shifting morpholinos targeting vascular endothelial growth factor receptor 2 (KDR) FASEB J. 2013;27:76–85. doi: 10.1096/fj.12-213835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Ezzat S, Zheng L, Yu S, Asa SL. A soluble dominant negative fibroblast growth factor receptor 4 isoform in human MCF-7 breast cancer cells. Biochem Biophys Res Commun. 2001;287:60–5. doi: 10.1006/bbrc.2001.5546. [DOI] [PubMed] [Google Scholar]
- [42].Abbass SAA, Asa SL, Ezzat S. Altered Expression of Fibroblast Growth Factor Receptors in Human Pituitary Adenomas. J Clin Endocrinol Metabol. 1997;82:1160–1166. doi: 10.1210/jcem.82.4.3896. [DOI] [PubMed] [Google Scholar]
- [43].Takaishi S, Sawada M, Morita Y, et al. Identification of a novel alternative splicing of human FGF receptor 4: soluble-form splice variant expressed in human gastrointestinal epithelial cells. Biochem Biophys Res Commun. 2000;267:658–62. doi: 10.1006/bbrc.1999.2010. [DOI] [PubMed] [Google Scholar]
- [44].Chen Q, Jiang Y, An Y, et al. Soluble FGFR4 extracellular domain inhibits FGF19-induced activation of FGFR4 signaling and prevents nonalcoholic fatty liver disease. Biochem Biophys Res Commun. 2011;409:651–656. doi: 10.1016/j.bbrc.2011.05.059. [DOI] [PubMed] [Google Scholar]
- [45].Keegan K, Johnson DE, Williams LT, Hayman MJ. Isolation of an additional member of the fibroblast growth factor receptor family, FGFR-3. Proc Natl Acad Sci USA. 1991;88:1095–9. doi: 10.1073/pnas.88.4.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Rappolee DA, Patel Y, Jacobson K. Expression of fibroblast growth factor receptors in peri-implantation mouse embryos. Mol Reprod Dev. 1998;51:254–264. doi: 10.1002/(SICI)1098-2795(199811)51:3<254::AID-MRD4>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- [47].Korhonen J, Partanen J, Alitalo K. Expression of FGFR-4 mRNA in developing mouse tissues. Int J Dev Biol. 1992;36:323–9. [PubMed] [Google Scholar]
- [48].Stark KL, McMahon JA, McMahon AP. FGFR-4, a new member of the fibroblast growth factor receptor family, expressed in the definitive endoderm and skeletal muscle lineages of the mouse. Development. 1991;113:641–51. doi: 10.1242/dev.113.2.641. [DOI] [PubMed] [Google Scholar]
- [49].Powell PP, Wang C-C, Horinouchi H, et al. Differential Expression of Fibroblast Growth Factor Receptors 1 to 4 and Ligand Genes in Late Fetal and Early Postnatal Rat Lung. Am J Respir Cell Mol Biol. 1998;19:563–572. doi: 10.1165/ajrcmb.19.4.2994. [DOI] [PubMed] [Google Scholar]
- [50].Zhao P, Hoffman EP. Embryonic myogenesis pathways in muscle regeneration. Dev Dyn. 2004;229:380–92. doi: 10.1002/dvdy.10457. [DOI] [PubMed] [Google Scholar]
- [51].Yu C, Wang F, Kan M, et al. Elevated cholesterol metabolism and bile acid synthesis in mice lacking membrane tyrosine kinase receptor FGFR4. J Biol Chem. 2000;275:15482–9. doi: 10.1074/jbc.275.20.15482. [DOI] [PubMed] [Google Scholar]
- [52].Armand A-S, Laziz I, Chanoine C. FGF6 in myogenesis. Biochim Biophys Acta. 2006;1763:773–778. doi: 10.1016/j.bbamcr.2006.06.005. [DOI] [PubMed] [Google Scholar]
- [53].Han JK, Martin GR. Embryonic expression of Fgf-6 is restricted to the skeletal muscle lineage. Dev Biol. 1993;158:549–54. doi: 10.1006/dbio.1993.1212. [DOI] [PubMed] [Google Scholar]
- [54].Pizette S, Coulier F, Birnbaum D, DeLapeyriere O. FGF6 modulates the expression of fibroblast growth factor receptors and myogenic genes in muscle cells. Exp Cell Res. 1996;224:143–51. doi: 10.1006/excr.1996.0122. [DOI] [PubMed] [Google Scholar]
- [55].Itoh N, Mima T, Mikawa T. Loss of fibroblast growth factor receptors is necessary for terminal differentiation of embryonic limb muscle. Development. 1996;122:291–300. doi: 10.1242/dev.122.1.291. [DOI] [PubMed] [Google Scholar]
- [56].Armand AS, Launay T, Pariset C, et al. Injection of FGF6 accelerates regeneration of the soleus muscle in adult mice. Biochim Biophys Acta. 2003;1642:97–105. doi: 10.1016/s0167-4889(03)00103-4. [DOI] [PubMed] [Google Scholar]
- [57].Marics I, Padilla F, Guillemot J-F, Scaal M, Marcelle C. FGFR4 signaling is a necessary step in limb muscle differentiation. Development. 2002;129:4559–4569. doi: 10.1242/dev.129.19.4559. [DOI] [PubMed] [Google Scholar]
- [58].Cool SM, Sayer RE, van Heumen WR, Pickles JO, Nurcombe V. Temporal and spatial expression of fibroblast growth factor receptor 4 isoforms in murine tissues. Histochem J. 2002;34:291–7. doi: 10.1023/a:1023326524562. [DOI] [PubMed] [Google Scholar]
- [59].Delfini MC, Duprez D. Ectopic Myf5 or MyoD prevents the neuronal differentiation program in addition to inducing skeletal muscle differentiation, in the chick neural tube. Development. 2004;131:713–23. doi: 10.1242/dev.00967. [DOI] [PubMed] [Google Scholar]
- [60].Cao L, Yu Y, Bilke S, et al. Genome-wide identification of PAX3-FKHR binding sites in rhabdomyosarcoma reveals candidate target genes important for development and cancer. Cancer Res. 2010;70:6497–508. doi: 10.1158/0008-5472.CAN-10-0582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Lagha M, Kormish JD, Rocancourt D, et al. Pax3 regulation of FGF signaling affects the progression of embryonic progenitor cells into the myogenic program. Genes Dev. 2008;22:1828–37. doi: 10.1101/gad.477908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Lagha M, Sato T, Bajard L, et al. Regulation of skeletal muscle stem cell behavior by Pax3 and Pax7. Cold Spring Harb Symp Quant Biol. 2008;73:307–15. doi: 10.1101/sqb.2008.73.006. [DOI] [PubMed] [Google Scholar]
- [63].Taylor JGt, Cheuk AT, Tsang PS, et al. Identification of FGFR4-activating mutations in human rhabdomyosarcomas that promote metastasis in xenotransplanted models. J Clin Invest. 2009;119:3395–407. doi: 10.1172/JCI39703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Tapscott SJ, Thayer MJ, Weintraub H. Deficiency in rhabdomyosarcomas of a factor required for MyoD activity and myogenesis. Science. 1993;259:1450–3. doi: 10.1126/science.8383879. [DOI] [PubMed] [Google Scholar]
- [65].Beenken A, Mohammadi M. The structural biology of the FGF19 subfamily. Adv Exp Med Biol. 2012;728:1–24. doi: 10.1007/978-1-4614-0887-1_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Kuro-o M. Klotho and betaKlotho. Adv Exp Med Biol. 2012;728:25–40. doi: 10.1007/978-1-4614-0887-1_2. [DOI] [PubMed] [Google Scholar]
- [67].Lin BC, Wang M, Blackmore C, Desnoyers LR. Liver-specific activities of FGF19 require Klotho beta. J Biol Chem. 2007;282:27277–84. doi: 10.1074/jbc.M704244200. [DOI] [PubMed] [Google Scholar]
- [68].Wu X, Ge H, Gupte J, et al. Co-receptor requirements for fibroblast growth factor-19 signaling. J Biol Chem. 2007;282:29069–72. doi: 10.1074/jbc.C700130200. [DOI] [PubMed] [Google Scholar]
- [69].Yang C, Jin C, Li X, et al. Differential Specificity of Endocrine FGF19 and FGF21 to FGFR1 and FGFR4 in Complex with KLB. PLoS ONE. 2012;7:e33870. doi: 10.1371/journal.pone.0033870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Urakawa I, Yamazaki Y, Shimada T, et al. Klotho converts canonical FGF receptor into a specific receptor for FGF23. Nature. 2006;444:770–4. doi: 10.1038/nature05315. [DOI] [PubMed] [Google Scholar]
- [71].Luo Y, Yang C, Lu W, et al. Metabolic Regulator %Klotho Interacts with Fibroblast Growth Factor Receptor 4 (FGFR4) to Induce Apoptosis and Inhibit Tumor Cell Proliferation. J Biol Chem. 2010;285:30069–30078. doi: 10.1074/jbc.M110.148288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Xie MH, Holcomb I, Deuel B, et al. FGF-19, a novel fibroblast growth factor with unique specificity for FGFR4. Cytokine. 1999;11:729–35. doi: 10.1006/cyto.1999.0485. [DOI] [PubMed] [Google Scholar]
- [73].Miyake A, Itoh N. Rat fibroblast growth factor receptor-4 mRNA in the brain is preferentially expressed in cholinergic neurons in the medial habenular nucleus. Neurosci Lett. 1996;203:101–4. doi: 10.1016/0304-3940(95)12272-9. [DOI] [PubMed] [Google Scholar]
- [74].Vincentz JW, McWhirter JR, Murre C, Baldini A, Furuta Y. Fgf15 is required for proper morphogenesis of the mouse cardiac outflow tract. Genesis. 2005;41:192–201. doi: 10.1002/gene.20114. [DOI] [PubMed] [Google Scholar]
- [75].Gimeno L, Brulet P, Martinez S. Study of Fgf15 gene expression in developing mouse brain. Gene Expr Patterns. 2003;3:473–81. doi: 10.1016/s1567-133x(03)00059-0. [DOI] [PubMed] [Google Scholar]
- [76].Nakayama Y, Miyake A, Nakagawa Y, et al. Fgf19 is required for zebrafish lens and retina development. Dev Biol. 2008;313:752–66. doi: 10.1016/j.ydbio.2007.11.013. [DOI] [PubMed] [Google Scholar]
- [77].Inagaki T, Choi M, Moschetta A, et al. Fibroblast growth factor 15 functions as an enterohepatic signal to regulate bile acid homeostasis. Cell Metab. 2005;2:217–25. doi: 10.1016/j.cmet.2005.09.001. [DOI] [PubMed] [Google Scholar]
- [78].Holt JA, Luo G, Billin AN, et al. Definition of a novel growth factor-dependent signal cascade for the suppression of bile acid biosynthesis. Genes Dev. 2003;17:1581–91. doi: 10.1101/gad.1083503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Lundasen T, Galman C, Angelin B, Rudling M. Circulating intestinal fibroblast growth factor 19 has a pronounced diurnal variation and modulates hepatic bile acid synthesis in man. J Intern Med. 2006;260:530–6. doi: 10.1111/j.1365-2796.2006.01731.x. [DOI] [PubMed] [Google Scholar]
- [80].Choi M, Moschetta A, Bookout AL, et al. Identification of a hormonal basis for gallbladder filling. Nat Med. 2006;12:1253–5. doi: 10.1038/nm1501. [DOI] [PubMed] [Google Scholar]
- [81].Portincasa P, Di Ciaula A, Wang HH, et al. Coordinate regulation of gallbladder motor function in the gut-liver axis. Hepatology. 2008;47:2112–26. doi: 10.1002/hep.22204. [DOI] [PubMed] [Google Scholar]
- [82].Yu C, Wang F, Jin C, et al. Increased carbon tetrachloride-induced liver injury and fibrosis in FGFR4-deficient mice. Am J Pathol. 2002;161:2003–10. doi: 10.1016/S0002-9440(10)64478-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Kir S, Beddow SA, Samuel VT, et al. FGF19 as a postprandial, insulin-independent activator of hepatic protein and glycogen synthesis. Science. 2011;331:1621–4. doi: 10.1126/science.1198363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Schaap FG. Role of fibroblast growth factor 19 in the control of glucose homeostasis. Curr Opin Clin Nutr Metab Care. 2012;15:386–91. doi: 10.1097/MCO.0b013e3283547171. [DOI] [PubMed] [Google Scholar]
- [85].Tomlinson E, Fu L, John L, et al. Transgenic mice expressing human fibroblast growth factor-19 display increased metabolic rate and decreased adiposity. Endocrinology. 2002;143:1741–7. doi: 10.1210/endo.143.5.8850. [DOI] [PubMed] [Google Scholar]
- [86].Zhang X, Yeung DC, Karpisek M, et al. Serum FGF21 levels are increased in obesity and are independently associated with the metabolic syndrome in humans. Diabetes. 2008;57:1246–53. doi: 10.2337/db07-1476. [DOI] [PubMed] [Google Scholar]
- [87].Nishimura T, Nakatake Y, Konishi M, Itoh N. Identification of a novel FGF, FGF-21, preferentially expressed in the liver. Biochim Biophys Acta. 2000;1492:203–6. doi: 10.1016/s0167-4781(00)00067-1. [DOI] [PubMed] [Google Scholar]
- [88].Wente W, Efanov AM, Brenner M, et al. Fibroblast growth factor-21 improves pancreatic beta-cell function and survival by activation of extracellular signal-regulated kinase 1/2 and Akt signaling pathways. Diabetes. 2006;55:2470–8. doi: 10.2337/db05-1435. [DOI] [PubMed] [Google Scholar]
- [89].Izumiya Y, Bina HA, Ouchi N, et al. FGF21 is an Akt-regulated myokine. FEBS Lett. 2008;582:3805–10. doi: 10.1016/j.febslet.2008.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Itoh N. Hormone-like (endocrine) Fgfs: their evolutionary history and roles in development, metabolism, and disease. Cell Tissue Res. 2010;342:1–11. doi: 10.1007/s00441-010-1024-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Tomiyama K, Maeda R, Urakawa I, et al. Relevant use of Klotho in FGF19 subfamily signaling system in vivo. Proc Natl Acad Sci USA. 2010;107:1666–71. doi: 10.1073/pnas.0913986107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Ding X, Boney-Montoya J, Owen BM, et al. betaKlotho is required for fibroblast growth factor 21 effects on growth and metabolism. Cell Metab. 2012;16:387–93. doi: 10.1016/j.cmet.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Riminucci M, Collins MT, Fedarko NS, et al. FGF-23 in fibrous dysplasia of bone and its relationship to renal phosphate wasting. J Clin Invest. 2003;112:683–92. doi: 10.1172/JCI18399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Razzaque MS. The FGF23-Klotho axis: endocrine regulation of phosphate homeostasis. Nat Rev Endocrinol. 2009;5:611–9. doi: 10.1038/nrendo.2009.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Consortium. A. Autosomal dominant hypophosphataemic rickets is associated with mutations in FGF23. Nat Genet. 2000;26:345–8. doi: 10.1038/81664. [DOI] [PubMed] [Google Scholar]
- [96].Kurosu H, Kuro OM. The Klotho gene family as a regulator of endocrine fibroblast growth factors. Mol Cell Endocrinol. 2009;299:72–8. doi: 10.1016/j.mce.2008.10.052. [DOI] [PubMed] [Google Scholar]
- [97].Kurosu H, Ogawa Y, Miyoshi M, et al. Regulation of fibroblast growth factor-23 signaling by klotho. J Biol Chem. 2006;281:6120–3. doi: 10.1074/jbc.C500457200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Gattineni J, Twombley K, Goetz R, Mohammadi M, Baum M. Regulation of serum 1,25(OH)2 vitamin D3 levels by fibroblast growth factor 23 is mediated by FGF receptors 3 and 4. Am J Physiol Renal Physiol. 2011;301:F371–7. doi: 10.1152/ajprenal.00740.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Eswarakumar VP, Lax I, Schlessinger J. Cellular signaling by fibroblast growth factor receptors. Cytokine Growth Factor Rev. 2005;16:139–49. doi: 10.1016/j.cytogfr.2005.01.001. [DOI] [PubMed] [Google Scholar]
- [100].Loo BM, Kreuger J, Jalkanen M, Lindahl U, Salmivirta M. Binding of heparin/heparan sulfate to fibroblast growth factor receptor 4. J Biol Chem. 2001;276:16868–76. doi: 10.1074/jbc.M011226200. [DOI] [PubMed] [Google Scholar]
- [101].Gao G, Goldfarb M. Heparin can activate a receptor tyrosine kinase. Embo J. 1995;14:2183–90. doi: 10.1002/j.1460-2075.1995.tb07212.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Kan M, Wu X, Wang F, McKeehan WL. Specificity for fibroblast growth factors determined by heparan sulfate in a binary complex with the receptor kinase. J Biol Chem. 1999;274:15947–52. doi: 10.1074/jbc.274.22.15947. [DOI] [PubMed] [Google Scholar]
- [103].Hart KC, Robertson SC, Kanemitsu MY, et al. Transformation and Stat activation by derivatives of FGFR1, FGFR3, and FGFR4. Oncogene. 2000;19:3309–20. doi: 10.1038/sj.onc.1203650. [DOI] [PubMed] [Google Scholar]
- [104].Koziczak M, Hynes NE. Cooperation between fibroblast growth factor receptor-4 and ErbB2 in regulation of cyclin D1 translation. J Biol Chem. 2004;279:50004–11. doi: 10.1074/jbc.M404252200. [DOI] [PubMed] [Google Scholar]
- [105].Shaoul E, Reich-Slotky R, Berman B, Ron D. Fibroblast growth factor receptors display both common and distinct signaling pathways. Oncogene. 1995;10:1553–61. [PubMed] [Google Scholar]
- [106].Wang JK, Gao G, Goldfarb M. Fibroblast growth factor receptors have different signaling and mitogenic potentials. Mol Cell Biol. 1994;14:181–188. doi: 10.1128/mcb.14.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Wang JK, Goldfarb M. Amino acid residues which distinguish the mitogenic potentials of two FGF receptors. Oncogene. 1997;14:1767–78. doi: 10.1038/sj.onc.1201021. [DOI] [PubMed] [Google Scholar]
- [108].Cavallaro U, Niedermeyer J, Fuxa M, Christofori G. N-CAM modulates tumour-cell adhesion to matrix by inducing FGF-receptor signalling. Nat Cell Biol. 2001;3:650–7. doi: 10.1038/35083041. [DOI] [PubMed] [Google Scholar]
- [109].Christensen C, Berezin V, Bock E. Neural cell adhesion molecule differentially interacts with isoforms of the fibroblast growth factor receptor. Neuroreport. 2011;22:727–32. doi: 10.1097/WNR.0b013e3283491682. [DOI] [PubMed] [Google Scholar]
- [110].Ezzat S, Zheng L, Zhu XF, Wu GE, Asa SL. Targeted expression of a human pituitary tumor-derived isoform of FGF receptor-4 recapitulates pituitary tumorigenesis. J Clin Invest. 2002;109:69–78. doi: 10.1172/JCI14036. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [111].Ezzat S, Zheng L, Asa SL. Pituitary tumor-derived fibroblast growth factor receptor 4 isoform disrupts neural cell-adhesion molecule/N-cadherin signaling to diminish cell adhesiveness: a mechanism underlying pituitary neoplasia. Mol Endocrinol. 2004;18:2543–52. doi: 10.1210/me.2004-0182. [DOI] [PubMed] [Google Scholar]
- [112].Ezzat S, Zheng L, Winer D, Asa SL. Targeting N-Cadherin through Fibroblast Growth Factor Receptor-4: Distinct Pathogenetic and Therapeutic Implications. Mol Endocrinol. 2006;20:2965–2975. doi: 10.1210/me.2006-0223. [DOI] [PubMed] [Google Scholar]
- [113].Pai R, Dunlap D, Qing J, et al. Inhibition of Fibroblast Growth Factor 19 Reduces Tumor Growth by Modulating {beta}-Catenin Signaling. Cancer Res. 2008;68:5086–5095. doi: 10.1158/0008-5472.CAN-07-2325. [DOI] [PubMed] [Google Scholar]
- [114].Yokote H, Fujita K, Jing X, et al. Trans-activation of EphA4 and FGF receptors mediated by direct interactions between their cytoplasmic domains. Proc Natl Acad Sci USA. 2005;102:18866–71. doi: 10.1073/pnas.0509741102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Drafahl KA, McAndrew CW, Meyer AN, Haas M, Donoghue DJ. The Receptor Tyrosine Kinase FGFR4 Negatively Regulates NF-kappaB Signaling. PLoS ONE. 2010;5:e14412. doi: 10.1371/journal.pone.0014412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Greenman C, Stephens P, Smith R, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–8. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Shukla N, Ameur N, Yilmaz I, et al. Oncogene mutation profiling of pediatric solid tumors reveals significant subsets of embryonal rhabdomyosarcoma and neuroblastoma with mutated genes in growth signaling pathways. Clin Cancer Res. 2012;18:748–57. doi: 10.1158/1078-0432.CCR-11-2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Ding L, Getz G, Wheeler DA, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–75. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Marks JL, McLellan MD, Zakowski MF, et al. Mutational Analysis of EGFR and Related Signaling Pathway Genes in Lung Adenocarcinomas Identifies a Novel Somatic Kinase Domain Mutation in FGFR4. PLoS ONE. 2007;2:e426. doi: 10.1371/journal.pone.0000426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Sasaki H, Okuda K, Kawano O, et al. Fibroblast growth factor receptor 4 mutation and polymorphism in Japanese lung cancer. Oncol Rep. 2008;20:1125–30. [PubMed] [Google Scholar]
- [121].Masica DL, Karchin R. Correlation of somatic mutation and expression identifies genes important in human glioblastoma progression and survival. Cancer Res. 2011;71:4550–61. doi: 10.1158/0008-5472.CAN-11-0180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Roidl A, Foo P, Wong W, et al. The FGFR4 Y367C mutant is a dominant oncogene in MDA-MB453 breast cancer cells. Oncogene. 2010;29:1543–1552. doi: 10.1038/onc.2009.432. [DOI] [PubMed] [Google Scholar]
- [123].Desnoyers LR, Pai R, Ferrando RE, et al. Targeting FGF19 inhibits tumor growth in colon cancer xenograft and FGF19 transgenic hepatocellular carcinoma models. Oncogene. 2008;27:85–97. doi: 10.1038/sj.onc.1210623. [DOI] [PubMed] [Google Scholar]
- [124].French DM, Lin BC, Wang M, et al. Targeting FGFR4 inhibits hepatocellular carcinoma in preclinical mouse models. PLoS ONE. 2012;7:e36713. doi: 10.1371/journal.pone.0036713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Miura S, Mitsuhashi N, Shimizu H, et al. Fibroblast growth factor 19 expression correlates with tumor progression and poorer prognosis of hepatocellular carcinoma. BMC Cancer. 2012;12:56. doi: 10.1186/1471-2407-12-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Nicholes K, Guillet S, Tomlinson E, et al. A mouse model of hepatocellular carcinoma: ectopic expression of fibroblast growth factor 19 in skeletal muscle of transgenic mice. Am J Pathol. 2002;160:2295–307. doi: 10.1016/S0002-9440(10)61177-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Cheng AL, Shen YC, Zhu AX. Targeting fibroblast growth factor receptor signaling in hepatocellular carcinoma. Oncology. 2011;81:372–80. doi: 10.1159/000335472. [DOI] [PubMed] [Google Scholar]
- [128].Ho HK, Pok S, Streit S, et al. Fibroblast growth factor receptor 4 regulates proliferation, anti-apoptosis and alpha-fetoprotein secretion during hepatocellular carcinoma progression and represents a potential target for therapeutic intervention. J Hepatol. 2009;50:118–27. doi: 10.1016/j.jhep.2008.08.015. [DOI] [PubMed] [Google Scholar]
- [129].Sawey ET, Chanrion M, Cai C, et al. Identification of a therapeutic strategy targeting amplified FGF19 in liver cancer by Oncogenomic screening. Cancer Cell. 2011;19:347–58. doi: 10.1016/j.ccr.2011.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Motoda N, Matsuda Y, Onda M, et al. Overexpression of fibroblast growth factor receptor 4 in high-grade pancreatic intraepithelial neoplasia and pancreatic ductal adenocarcinoma. Int J Oncol. 2011;38:133–43. [PubMed] [Google Scholar]
- [131].Paulson V, Chandler G, Rakheja D, et al. High-resolution array CGH identifies common mechanisms that drive embryonal rhabdomyosarcoma pathogenesis. Genes Chromosomes Cancer. 2011;50:397–408. doi: 10.1002/gcc.20864. [DOI] [PubMed] [Google Scholar]
- [132].Crose LE, Etheridge KT, Chen C, et al. FGFR4 blockade exerts distinct antitumorigenic effects in human embryonal versus alveolar rhabdomyosarcoma. Clin Cancer Res. 2012;18:3780–90. doi: 10.1158/1078-0432.CCR-10-3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Jaakkola S, Salmikangas P, Nylund S, et al. Amplification of fgfr4 gene in human breast and gynecological cancers. Int J Cancer. 1993;54:378–82. doi: 10.1002/ijc.2910540305. [DOI] [PubMed] [Google Scholar]
- [134].Penault-Llorca F, Bertucci F, Adelaide J, et al. Expression of FGF and FGF receptor genes in human breast cancer. Int J Cancer. 1995;61:170–6. doi: 10.1002/ijc.2910610205. [DOI] [PubMed] [Google Scholar]
- [135].Yamada SM, Yamada S, Hayashi Y, et al. Fibroblast growth factor receptor (FGFR) 4 correlated with the malignancy of human astrocytomas. Neurol Res. 2002;24:244–8. doi: 10.1179/016164102101199864. [DOI] [PubMed] [Google Scholar]
- [136].Zaid T, Yeung TL, Thompson MS, et al. Identification of FGFR4 as a Potential Therapeutic Target for Advanced-Stage High-grade Serous Ovarian Cancer. Clin Cancer Res. 2013;19(4):809–20. doi: 10.1158/1078-0432.CCR-12-2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Gowardhan B, Douglas DA, Mathers ME, et al. Evaluation of the fibroblast growth factor system as a potential target for therapy in human prostate cancer. Br J Cancer. 2005;92:320–7. doi: 10.1038/sj.bjc.6602274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Murphy T, Darby S, Mathers ME, Gnanapragasam VJ. Evidence for distinct alterations in the FGF axis in prostate cancer progression to an aggressive clinical phenotype. J Pathol. 2010;220:452–60. doi: 10.1002/path.2657. [DOI] [PubMed] [Google Scholar]
- [139].Yu W, Feng S, Dakhova O, et al. FGFR-4 Arg(3)(8)(8) enhances prostate cancer progression via extracellular signal-related kinase and serum response factor signaling. Clin Cancer Res. 2011;17:4355–66. doi: 10.1158/1078-0432.CCR-10-2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Sahadevan K, Darby S, Leung HY, et al. Selective over-expression of fibroblast growth factor receptors 1 and 4 in clinical prostate cancer. J Pathol. 2007;213:82–90. doi: 10.1002/path.2205. [DOI] [PubMed] [Google Scholar]
- [141].Bernard R, Zheng L, Liu W, et al. Fibroblast growth factor receptors as molecular targets in thyroid carcinoma. Endocrinology. 2005;146:1145–53. doi: 10.1210/en.2004-1134. [DOI] [PubMed] [Google Scholar]
- [142].Ezzat S, Huang P, Dackiw A, Asa SL. Dual Inhibition of RET and FGFR4 Restrains Medullary Thyroid Cancer Cell Growth. Clin Cancer Res. 2005;11:1336–41. [PubMed] [Google Scholar]
- [143].Bange J, Prechtl D, Cheburkin Y, et al. Cancer progression and tumor cell motility are associated with the FGFR4 Arg(388) allele. Cancer Res. 2002;62:840–7. [PubMed] [Google Scholar]
- [144].Wang J, Stockton DW, Ittmann M. The fibroblast growth factor receptor-4 Arg388 allele is associated with prostate cancer initiation and progression. Clin Cancer Res. 2004;10:6169–78. doi: 10.1158/1078-0432.CCR-04-0408. [DOI] [PubMed] [Google Scholar]
- [145].Bargmann CI, Hung MC, Weinberg RA. Multiple independent activations of the neu oncogene by a point mutation altering the transmembrane domain of p185. Cell. 1986;45:649–57. doi: 10.1016/0092-8674(86)90779-8. [DOI] [PubMed] [Google Scholar]
- [146].Rousseau F, Bonaventure J, Legeai-Mallet L, et al. Mutations in the gene encoding fibroblast growth factor receptor-3 in achondroplasia. Nature. 1994;371:252–4. doi: 10.1038/371252a0. [DOI] [PubMed] [Google Scholar]
- [147].Meyers GA, Orlow SJ, Munro IR, Przylepa KA, Jabs EW. Fibroblast growth factor receptor 3 (FGFR3) transmembrane mutation in Crouzon syndrome with acanthosis nigricans. Nat Genet. 1995;11:462–4. doi: 10.1038/ng1295-462. [DOI] [PubMed] [Google Scholar]
- [148].Bellus GA, Spector EB, Speiser PW, et al. Distinct missense mutations of the FGFR3 lys650 codon modulate receptor kinase activation and the severity of the skeletal dysplasia phenotype. Am J Hum Genet. 2000;67:1411–21. doi: 10.1086/316892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Parafioriti A, del Bianco S, Barisani D, et al. Increased p21 expression in chondrocytes of achondroplasic children independently from the presence of the G380R FGFR3 mutation. J Orthop Sci. 2009;14:623–30. doi: 10.1007/s00776-009-1355-6. [DOI] [PubMed] [Google Scholar]
- [150].Webster MK, Donoghue DJ. FGFR activation in skeletal disorders: too much of a good thing. Trends Genet. 1997;13:178–82. doi: 10.1016/s0168-9525(97)01131-1. [DOI] [PubMed] [Google Scholar]
- [151].Xu W, Li Y, Wang X, et al. FGFR4 transmembrane domain polymorphism and cancer risk: A meta-analysis including 8555 subjects. Eur J Cancer. 2010;46:3332–8. doi: 10.1016/j.ejca.2010.06.017. [DOI] [PubMed] [Google Scholar]
- [152].Frullanti E, Berking C, Harbeck N, et al. Meta and pooled analyses of FGFR4 Gly388Arg polymorphism as a cancer prognostic factor. Eur J Cancer Prev. 2011;20(4):340–7. doi: 10.1097/CEJ.0b013e3283457274. [DOI] [PubMed] [Google Scholar]
- [153].Wang J, Yu W, Cai Y, Ren C, Ittmann MM. Altered fibroblast growth factor receptor 4 stability promotes prostate cancer progression. Neoplasia. 2008;10:847–56. doi: 10.1593/neo.08450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Stadler CR, Knyazev P, Bange J, Ullrich A. FGFR4 GLY388 isotype suppresses motility of MDA-MB-231 breast cancer cells by EDG-2 gene repression. Cell Signal. 2006;18:783–94. doi: 10.1016/j.cellsig.2005.07.002. [DOI] [PubMed] [Google Scholar]
- [155].Seitzer N, Mayr T, Streit S, Ullrich A. A single nucleotide change in the mouse genome accelerates breast cancer progression. Cancer Res. 2010;70:802–12. doi: 10.1158/0008-5472.CAN-09-3239. [DOI] [PubMed] [Google Scholar]
- [156].Francavilla C, Cattaneo P, Berezin V, et al. The binding of NCAM to FGFR1 induces a specific cellular response mediated by receptor trafficking. J Cell Biol. 2009;187:1101–1116. doi: 10.1083/jcb.200903030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Sugiyama N, Varjosalo M, Meller P, et al. FGF receptor-4 (FGFR4) polymorphism acts as an activity switch of a membrane type 1 matrix metalloproteinase–FGFR4 complex. Proc Natl Acad Sci. 2010;107:15786–15791. doi: 10.1073/pnas.0914459107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Nyalendo C, Michaud M, Beaulieu E, et al. Src-dependent phosphorylation of membrane type I matrix metalloproteinase on cytoplasmic tyrosine 573: role in endothelial and tumor cell migration. J Biol Chem. 2007;282:15690–9. doi: 10.1074/jbc.M608045200. [DOI] [PubMed] [Google Scholar]
- [159].Ueda J, Kajita M, Suenaga N, Fujii K, Seiki M. Sequence-specific silencing of MT1-MMP expression suppresses tumor cell migration and invasion: importance of MT1-MMP as a therapeutic target for invasive tumors. Oncogene. 2003;22:8716–22. doi: 10.1038/sj.onc.1206962. [DOI] [PubMed] [Google Scholar]
- [160].Sugiyama N, Varjosalo M, Meller P, et al. Fibroblast growth factor receptor 4 regulates tumor invasion by coupling fibroblast growth factor signaling to extracellular matrix degradation. Cancer Res. 2010;70:7851–61. doi: 10.1158/0008-5472.CAN-10-1223. [DOI] [PubMed] [Google Scholar]
- [161].Falvella FS, Frullanti E, Galvan A, et al. FGFR4 Gly388Arg polymorphism may affect the clinical stage of patients with lung cancer by modulating the transcriptional profile of normal lung. Int J Cancer. 2009;124:2880–5. doi: 10.1002/ijc.24302. [DOI] [PubMed] [Google Scholar]
- [162].Tateno T, Asa SL, Zheng L, et al. The FGFR4-G388R polymorphism promotes mitochondrial STAT3 serine phosphorylation to facilitate pituitary growth hormone cell tumorigenesis. PLoS Genet. 2011;7:e1002400. doi: 10.1371/journal.pgen.1002400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Huang X, Yang C, Jin C, et al. Resident hepatocyte fibroblast growth factor receptor 4 limits hepatocarcinogenesis. Mol Carcinog. 2009;48:553–562. doi: 10.1002/mc.20494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Poh W, Wong W, Ong H, et al. Klotho-beta overexpression as a novel target for suppressing proliferation and fibroblast growth factor receptor-4 signaling in hepatocellular carcinoma. Mol Cancer. 2012;11:14. doi: 10.1186/1476-4598-11-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165].Guagnano V, Kauffmann A, Wohrle S, et al. FGFR genetic alterations predict for sensitivity to NVP-BGJ398, a selective pan-FGFR inhibitor. Cancer Discov. 2012;2:1118–33. doi: 10.1158/2159-8290.CD-12-0210. [DOI] [PubMed] [Google Scholar]
- [166].Zhu X, Zheng L, Asa SL, Ezzat S. Loss of heterozygosity and DNA methylation affect germline fibroblast growth factor receptor 4 polymorphism to direct allelic selection in breast cancer. Am J Pathol. 2010;177:2860–9. doi: 10.2353/ajpath.2010.100509. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [167].Brooks AN, Kilgour E, Smith PD. Molecular Pathways: Fibroblast Growth Factor Signaling: A New Therapeutic Opportunity in Cancer. Clin Cancer Res. 2012;18:1855–1862. doi: 10.1158/1078-0432.CCR-11-0699. [DOI] [PubMed] [Google Scholar]
- [168].Kumar SB, Narasu L, Gundla R, Dayam R, J ARPS. Fibroblast growth factor receptor inhibitors. Curr Pharm Des. 2013;19:687–701. [PubMed] [Google Scholar]
- [169].Ho HK, Nemeth G, Ng YR, et al. Developing FGFR4 inhibitors as potential anti-cancer agents via in silico design, supported by in vitro and cell-based testing. Curr Med Chem. 2013;20:1203–17. doi: 10.2174/0929867311320100001. [DOI] [PubMed] [Google Scholar]
- [170].Korc M, Friesel RE. The role of fibroblast growth factors in tumor growth. Curr Cancer Drug Targets. 2009;9:639–51. doi: 10.2174/156800909789057006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Mohammadi M, Froum S, Hamby James M, et al. Crystal structure of an angiogenesis inhibitor bound to the FGF receptor tyrosine kinase domain. EMBO J. 1998;17:5896–5904. doi: 10.1093/emboj/17.20.5896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].Koziczak M, Holbro T, Hynes NE. Blocking of FGFR signaling inhibits breast cancer cell proliferation through downregulation of D-type cyclins. Oncogene. 2004;23:3501–8. doi: 10.1038/sj.onc.1207331. [DOI] [PubMed] [Google Scholar]
- [173].Loilome W, Joshi A, ap Rhys C, et al. Glioblastoma cell growth is suppressed by disruption of fibroblast growth factor pathway signaling. J Neuro-Oncol. 2009;94:359–366. doi: 10.1007/s11060-009-9885-5. [DOI] [PubMed] [Google Scholar]
- [174].Zhao G, Li WY, Chen D, et al. A novel, selective inhibitor of fibroblast growth factor receptors that shows a potent broad spectrum of antitumor activity in several tumor xenograft models. Mol Cancer Ther. 2011;10:2200–10. doi: 10.1158/1535-7163.MCT-11-0306. [DOI] [PubMed] [Google Scholar]
- [175].Huang WS, Metcalf CA, Sundaramoorthi R, et al. Discovery of 3-[2-(imidazo[1,2-b]pyridazin-3-yl)ethynyl]-4-methyl-N-{4-[(4-methylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl} benzamide (AP24534), a potent, orally active pan-inhibitor of breakpoint cluster region-abelson (BCR-ABL) kinase including the T315I gatekeeper mutant. J Med Chem. 2010;53:4701–19. doi: 10.1021/jm100395q. [DOI] [PubMed] [Google Scholar]
- [176].Gozgit JM, Wong MJ, Moran L, et al. Ponatinib (AP24534), a multitargeted pan-FGFR inhibitor with activity in multiple FGFR-amplified or mutated cancer models. Mol Cancer Ther. 2012;11:690–9. doi: 10.1158/1535-7163.MCT-11-0450. [DOI] [PubMed] [Google Scholar]
- [177].Squires M, Ward G, Saxty G, et al. Potent, Selective Inhibitors of Fibroblast Growth Factor Receptor Define Fibroblast Growth Factor Dependence in Preclinical Cancer Models. Mol Cancer Ther. 2011;10:1542–1552. doi: 10.1158/1535-7163.MCT-11-0426. [DOI] [PubMed] [Google Scholar]
- [178].Bumbaca D, Wong A, Drake E, et al. Highly specific off-target binding identified and eliminated during the humanization of an antibody against FGF receptor 4. MAbs. 2011;3:376–86. doi: 10.4161/mabs.3.4.15786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [179].Roidl A, Berger HJ, Kumar S, et al. Resistance to chemotherapy is associated with fibroblast growth factor receptor 4 up-regulation. Clin Cancer Res. 2009;15:2058–66. doi: 10.1158/1078-0432.CCR-08-0890. [DOI] [PubMed] [Google Scholar]
- [180].Marshall ME, Hinz TK, Kono SA, et al. Fibroblast growth factor receptors are components of autocrine signaling networks in head and neck squamous cell carcinoma cells. Clin Cancer Res. 2011;17:5016–25. doi: 10.1158/1078-0432.CCR-11-0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [181].Meijer D, Sieuwerts AM, Look MP, et al. Fibroblast growth factor receptor 4 predicts failure on tamoxifen therapy in patients with recurrent breast cancer. Endocr Relat Cancer. 2008;15:101–11. doi: 10.1677/ERC-07-0080. [DOI] [PubMed] [Google Scholar]
- [182].Halatsch ME, Low S, Mursch K, et al. Candidate genes for sensitivity and resistance of human glioblastoma multiforme cell lines to erlotinib. Laboratory investigation. J Neurosurg. 2009;111:211–8. doi: 10.3171/2008.9.JNS08551. [DOI] [PubMed] [Google Scholar]
- [183].Thussbas C, Nahrig J, Streit S, et al. FGFR4 Arg388 allele is associated with resistance to adjuvant therapy in primary breast cancer. J Clin Oncol. 2006;24:3747–55. doi: 10.1200/JCO.2005.04.8587. [DOI] [PubMed] [Google Scholar]
- [184].Serra S, Zheng L, Hassan M, et al. The FGFR4-G388R Single-Nucleotide Polymorphism Alters Pancreatic Neuroendocrine Tumor Progression and Response to mTOR Inhibition Therapy. Cancer Res. 2012;72:5683–91. doi: 10.1158/0008-5472.CAN-12-2102. [DOI] [PubMed] [Google Scholar]
- [185].Marme F, Werft W, Benner A, et al. FGFR4 Arg388 genotype is associated with pathological complete response to neoadjuvant chemotherapy for primary breast cancer. Ann Oncol. 2010;21:1636–42. doi: 10.1093/annonc/mdq017. [DOI] [PubMed] [Google Scholar]
- [186].Marme F, Hielscher T, Hug S, et al. Fibroblast growth factor receptor 4 gene (FGFR4) 388Arg allele predicts prolonged survival and platinum sensitivity in advanced ovarian cancer. Int J Cancer. 2012;131:E586–91. doi: 10.1002/ijc.27329. [DOI] [PubMed] [Google Scholar]
- [187].Ansell A, Farnebo L, Grenman R, Roberg K, Thunell LK. Polymorphism of FGFR4 in cancer development and sensitivity to cisplatin and radiation in head and neck cancer. Oral Oncol. 2009;45:23–9. doi: 10.1016/j.oraloncology.2008.03.007. [DOI] [PubMed] [Google Scholar]
- [188].Davies H, Hunter C, Smith R, et al. Somatic mutations of the protein kinase gene family in human lung cancer. Cancer Res. 2005;65:7591–5. doi: 10.1158/0008-5472.CAN-05-1855. [DOI] [PubMed] [Google Scholar]
- [189].Jezequel P, Campion L, Joalland MP, et al. G388R mutation of the FGFR4 gene is not relevant to breast cancer prognosis. Br J Cancer. 2004;90:189–93. doi: 10.1038/sj.bjc.6601450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [190].Spinola M, Leoni VP, Tanuma J, et al. FGFR4 Gly388Arg polymorphism and prognosis of breast and colorectal cancer. Oncol Rep. 2005;14:415–9. [PubMed] [Google Scholar]
- [191].Naidu R, Har YC, Taib NA. Polymorphism of FGFR4 Gly388Arg does not confer an increased risk to breast cancer development. Oncol Res. 2009;18:65–71. doi: 10.3727/096504009789954609. [DOI] [PubMed] [Google Scholar]
- [192].Batschauer AP, Cruz NG, Oliveira VC, et al. HFE, MTHFR, and FGFR4 genes polymorphisms and breast cancer in Brazilian women. Mol Cell Biochem. 2011;357:247–53. doi: 10.1007/s11010-011-0895-1. [DOI] [PubMed] [Google Scholar]
- [193].Ma Z, Tsuchiya N, Yuasa T, et al. Polymorphisms of fibroblast growth factor receptor 4 have association with the development of prostate cancer and benign prostatic hyperplasia and the progression of prostate cancer in a Japanese population. Int J Cancer. 2008;123:2574–9. doi: 10.1002/ijc.23578. [DOI] [PubMed] [Google Scholar]
- [194].FitzGerald LM, Karlins E, Karyadi DM, et al. Association of FGFR4 genetic polymorphisms with prostate cancer risk and prognosis. Prostate Cancer Prostatic Dis. 2009;12:192–7. doi: 10.1038/pcan.2008.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [195].Ho CK, Anwar S, Nanda J, Habib FK. FGFR4 Gly388Arg polymorphism and prostate cancer risk in Scottish men. Prostate Cancer Prostatic Dis. 2010;13:94–6. doi: 10.1038/pcan.2009.49. [DOI] [PubMed] [Google Scholar]
- [196].Gordon MA, Gil J, Lu B, et al. Genomic profiling associated with recurrence in patients with rectal cancer treated with chemoradiation. Pharmacogenomics. 2006;7:67–88. doi: 10.2217/14622416.7.1.67. [DOI] [PubMed] [Google Scholar]
- [197].Streit S, Bange J, Fichtner A, et al. Involvement of the FGFR4 Arg388 allele in head and neck squamous cell carcinoma. Int J Cancer. 2004;111:213–7. doi: 10.1002/ijc.20204. [DOI] [PubMed] [Google Scholar]
- [198].da Costa Andrade VC, Parise O, Jr, Hors CP, et al. The fibroblast growth factor receptor 4 (FGFR4) Arg388 allele correlates with survival in head and neck squamous cell carcinoma. Exp Mol Pathol. 2007;82:53–7. doi: 10.1016/j.yexmp.2006.05.003. [DOI] [PubMed] [Google Scholar]
- [199].Azad AK, Bairati I, Samson E, et al. Validation of genetic sequence variants as prognostic factors in early-stage head and neck squamous cell cancer survival. Clin Cancer Res. 2012;18:196–206. doi: 10.1158/1078-0432.CCR-11-1759. [DOI] [PubMed] [Google Scholar]
- [200].Dutra RL, de Carvalho MB, Dos Santos M, et al. FGFR4 Profile as a Prognostic Marker in Squamous Cell Carcinoma of the Mouth and Oropharynx. PLoS ONE. 2012;7:e50747. doi: 10.1371/journal.pone.0050747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [201].Tanuma J, Izumo T, Hirano M, et al. FGFR4 polymorphism, TP53 mutation, and their combinations are prognostic factors for oral squamous cell carcinoma. Oncol Rep. 2010;23:739–44. [PubMed] [Google Scholar]
- [202].Spinola M, Leoni V, Pignatiello C, et al. Functional FGFR4 Gly388Arg polymorphism predicts prognosis in lung adenocarcinoma patients. J Clin Oncol. 2005;23:7307–11. doi: 10.1200/JCO.2005.17.350. [DOI] [PubMed] [Google Scholar]
- [203].Matakidou A, El Galta R, Rudd MF, et al. Further observations on the relationship between the FGFR4 Gly388Arg polymorphism and lung cancer prognosis. Br J Cancer. 2007;96:1904–7. doi: 10.1038/sj.bjc.6603816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [204].Streit S, Mestel DS, Schmidt M, Ullrich A, Berking C. FGFR4 Arg388 allele correlates with tumour thickness and FGFR4 protein expression with survival of melanoma patients. Br J Cancer. 2006;94:1879–86. doi: 10.1038/sj.bjc.6603181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [205].Nan H, Qureshi AA, Hunter DJ, Han J. Genetic variants in FGFR2 and FGFR4 genes and skin cancer risk in the Nurses' Health Study. BMC Cancer. 2009;9:172. doi: 10.1186/1471-2407-9-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [206].Ye Y, Shi Y, Zhou Y, et al. The fibroblast growth factor receptor-4 Arg388 allele is associated with gastric cancer progression. Ann Surg Oncol. 2010;17:3354–61. doi: 10.1245/s10434-010-1323-6. [DOI] [PubMed] [Google Scholar]
- [207].Yang Y, Zhou Y, Lu M, et al. Association between fibroblast growth factor receptor 4 polymorphisms and risk of hepatocellular carcinoma. Mol Carcinog. 2012;51:515–21. doi: 10.1002/mc.20805. [DOI] [PubMed] [Google Scholar]
- [208].Mawrin C, Kirches E, Diete S, et al. Analysis of a single nucleotide polymorphism in codon 388 of the FGFR4 gene in malignant gliomas. Cancer Lett. 2006;239:239–45. doi: 10.1016/j.canlet.2005.08.013. [DOI] [PubMed] [Google Scholar]
- [209].Yang YC, Lu ML, Rao JY, et al. Joint association of polymorphism of the FGFR4 gene and mutation TP53 gene with bladder cancer prognosis. Br J Cancer. 2006;95:1455–8. doi: 10.1038/sj.bjc.6603456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [210].Morimoto Y, Ozaki T, Ouchida M, et al. Single nucleotide polymorphism in fibroblast growth factor receptor 4 at codon 388 is associated with prognosis in high-grade soft tissue sarcoma. Cancer. 2003;98:2245–50. doi: 10.1002/cncr.11778. [DOI] [PubMed] [Google Scholar]
- [211].Gavine PR, Mooney L, Kilgour E, et al. AZD4547: an orally bioavailable, potent, and selective inhibitor of the fibroblast growth factor receptor tyrosine kinase family. Cancer Res. 2012;72:2045–56. doi: 10.1158/0008-5472.CAN-11-3034. [DOI] [PubMed] [Google Scholar]
- [212].Guagnano V, Furet P, Spanka C, et al. Discovery of 3-(2,6-dichloro-3,5-dimethoxy-phenyl)-1-{6-[4-(4-ethyl-piperazin-1-yl)-phenylamin o]-pyrimidin-4-yl}-1-methyl-urea (NVP-BGJ398), a potent and selective inhibitor of the fibroblast growth factor receptor family of receptor tyrosine kinase. J Med Chem. 2011;54:7066–83. doi: 10.1021/jm2006222. [DOI] [PubMed] [Google Scholar]
- [213].Hilberg F, Roth GJ, Krssak M, et al. BIBF 1120: triple angiokinase inhibitor with sustained receptor blockade and good antitumor efficacy. Cancer Res. 2008;68:4774–82. doi: 10.1158/0008-5472.CAN-07-6307. [DOI] [PubMed] [Google Scholar]
- [214].Wedge SR, Kendrew J, Hennequin LF, et al. AZD2171: a highly potent, orally bioavailable, vascular endothelial growth factor receptor-2 tyrosine kinase inhibitor for the treatment of cancer. Cancer Res. 2005;65:4389–400. doi: 10.1158/0008-5472.CAN-04-4409. [DOI] [PubMed] [Google Scholar]
- [215].Bello E, Colella G, Scarlato V, et al. E-3810 is a potent dual inhibitor of VEGFR and FGFR that exerts antitumor activity in multiple preclinical models. Cancer Res. 2011;71:1396–405. doi: 10.1158/0008-5472.CAN-10-2700. [DOI] [PubMed] [Google Scholar]
- [216].Kumar R, Crouthamel MC, Rominger DH, et al. Myelosuppression and kinase selectivity of multikinase angiogenesis inhibitors. Br J Cancer. 2009;101:1717–23. doi: 10.1038/sj.bjc.6605366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [217].Gozgit JM, Wong MJ, Wardwell S, et al. Potent activity of ponatinib (AP24534) in models of FLT3-driven acute myeloid leukemia and other hematologic malignancies. Mol Cancer Ther. 2011;10:1028–35. doi: 10.1158/1535-7163.MCT-10-1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [218].Dy G. Emerging systemic therapeutic approaches for personalized medicine in squamous cell carcinoma of the lung. J Cancer Sci Ther. 2012;S11:1–7. [Google Scholar]
- [219].NCBI. [access 2012/12/15]; Available from: http://www.ncbi.nlm.nih.gov/gene/
- [220].nextProt. [access 2012/12/15]; Available from: http://www.nextprot.org/
- [221].Cavallaro U, Christofori G. Cell adhesion and signalling by cadherins and Ig-CAMS in cancer. Nat Rev Cancer. 2004;4:118–132. doi: 10.1038/nrc1276. [DOI] [PubMed] [Google Scholar]