Abstract
The impact of R-loops on the physiology and pathology of chromosomes has been demonstrated extensively by chromatin biology research. The progress in this field has been driven by technological advancement of R-loop mapping methods that largely relied on a single approach, DNA-RNA immunoprecipitation (DRIP). Most of the DRIP protocols use the experimental design that was developed by a few laboratories, without paying attention to the potential caveats that might affect the outcome of RNA-DNA hybrid mapping. To assess the accuracy and utility of this technology, we pursued an analytical approach to estimate inherent biases and errors in the DRIP protocol. By performing DRIP-sequencing, qPCR, and receiver operator characteristic (ROC) analysis, we tested the effect of formaldehyde fixation, cell lysis temperature, mode of genome fragmentation, and removal of free RNA on the efficacy of RNA-DNA hybrid detection and implemented workflows that were able to distinguish complex and weak DRIP signals in a noisy background with high confidence. We also show that some of the workflows perform poorly and generate random answers. Furthermore, we found that the most commonly used genome fragmentation method (restriction enzyme digestion) led to the overrepresentation of lengthy DRIP fragments over coding ORFs, and this bias was enhanced at the first exons. Biased genome sampling severely compromised mapping resolution and prevented the assignment of precise biological function to a significant fraction of R-loops. The revised workflow presented herein is established and optimized using objective ROC analyses and provides reproducible and highly specific RNA-DNA hybrid detection.
R-loops are three-stranded nucleic acid structures that are composed of an RNA-DNA hybrid and a displaced single-stranded DNA. Under physiological conditions, R-loops are prevalent along the chromosomes, constituting 5%–8% of the genome and impacting many cellular processes (Sanz et al. 2016; Wahba et al. 2016). For instance, R-loops (1) drive embryonic stem cell differentiation via modulating the chromosomal binding of chromatin-regulatory complexes (Chen et al. 2015), (2) ensure the optimal binding of transcriptional activators to the promoter of the human vimentin (VIM) gene (Boque-Sastre et al. 2015), (3) massively form on estrogen-responsive genes in human breast and other tissues upon estrogen-hormone stimulation (Stork et al. 2016), (4) induce heterochromatin formation in Schizosaccharomyces pombe (Nakama et al. 2012), and (5) inhibit the expression of an antisense noncoding RNA in Arabidopsis thaliana, associated with the flowering process (Sun et al. 2013). In a pathological context, perturbation or mutation of any of the following factors causes the chromosomal accumulation of RNA-DNA hybrids and consequent genomic instability: (1) mRNA splicing factors and RNA export factors (e.g., THO2, HPR1, MFT1, THP2, THOC1-7, SRSF1) (Huertas and Aguilera 2003; Li and Manley 2005; Domínguez-Sánchez et al. 2011; Gan et al. 2011); (2) RNA-DNA hybrid helicases (e.g., SETX/SEN1, AQR, PIF1) (Boulé and Zakian 2007; Mischo et al. 2011; Alzu et al. 2012; Sollier et al. 2014); (3) RNA-DNA ribonucleases (RNASEH1/RNH1, RNASEH2A-C/RNH201) (El Hage et al. 2010; Chon et al. 2013; Stuckey et al. 2015); (4) homologous recombination proteins (e.g., BRCA1, BRCA2, RTEL1, SRS2), (Bhatia et al. 2014; Hatchi et al. 2015); (5) Fanconi anemia proteins (FANCA, FANCB, FANCC) (García-Rubio et al. 2015; Schwab et al. 2015); and (6) topoisomerases (TOP1, TOP3B) (Wilson-Sali and Hsieh 2002; El Hage et al. 2010; Yang et al. 2014; Marinello et al. 2016).
The above examples clearly illustrate the massive progress in the field that has been driven by technological advancements of R-loop detection methods. These techniques involve, for instance, electrophoretic mobility shift assays (Yu et al. 2006), atomic force microscopy (Brown et al. 2008), transmission electron microscopy (Pohjoismäki et al. 2010), fluorescent microscopy (Székvölgyi et al. 2007), fluorescence in situ hybridization (Nadel et al. 2015), native bisulfite modification (Yu et al. 2003), immunoprecipitation (Skourti-Stathaki et al. 2011; Ginno et al. 2012), and computational prediction (Jenjaroenpun et al. 2015). The increasing numbers of R-loop mapping data relied on a single approach, DNA-RNA immunoprecipitation (DRIP) and its variations (RDIP, DRIPc, S1-DRIP, DRIP-RNA, DIP, ChIP). The DRIP method applies the S9.6 anti-RNA-DNA hybrid antibody (Hu et al. 2006) to capture RNA-DNA hybrids in their native chromosomal context, followed by mapping the enriched DNA fragments on a selected number of loci or across the whole genome, using quantitative PCR, microarray hybridization, or deep sequencing.
Having surveyed the published RNA-DNA hybrid mapping studies (Supplemental Table S1; El Hage et al. 2010, 2014; Mischo et al. 2011; Skourti-Stathaki et al. 2011; Alzu et al. 2012; Ginno et al. 2012; Castellano-Pozo et al. 2013; Sun et al. 2013; Wahba and Koshland 2013; Bhatia et al. 2014; Chan et al. 2014; Groh et al. 2014; Herrera-Moyano et al. 2014; Loomis et al. 2014; Rigby et al. 2014; Salvi et al. 2014; Yang et al. 2014, 2016; Zhang et al. 2014a, 2014b, 2015; Boque-Sastre et al. 2015; Chen et al. 2015; García-Rubio et al. 2015; Hatchi et al. 2015; Jenjaroenpun et al. 2015; Lim et al. 2015; Nadel et al. 2015; Pefanis et al. 2015; Cloutier et al. 2016; Marinello et al. 2016; Ohle et al. 2016; Romanello et al. 2016; Sanz et al. 2016; Stork et al. 2016; Wahba et al. 2016; Zeller et al. 2016), we found that most DRIP protocols used the experimental design that was developed by a few laboratories (Supplemental Fig. S1). The original protocols are still being used without paying attention to their potential caveats: several critical points have remained exceedingly heterogeneous among the DRIP studies (Supplemental Table S1) that might account for at least some of the contradictory results (Ginno et al. 2012; Chan et al. 2014; El Hage et al. 2014; Nadel et al. 2015; Wahba et al. 2016). One can reveal technical heterogeneities (1) in terms of the studied model organisms and cell types, (2) in whether the cells were fixed by formaldehyde (HCHO) or not, (3) in whether the immunoprecipitation was chromatin-based or DNA-based (ChIP vs. DIP), (4) in the cell lysis temperature (65°C, 55°C, 37°C), (5) in the mode of DNA fragmentation (restriction enzyme digestion vs. sonication), (6) in total nucleic acid extraction (solid-phase purification vs. organic extraction, or salting out extraction), and (7) in the application of ribonuclease A digestion to eliminate free RNA from the nucleic acid prep. Obviously, each of these variables can introduce substantial bias that might obscure the overall outcome of the experiment, but their consequence, alone or in combination, has remained unexplored.
In the current study, we aimed to assess possible confounding effects related to key experimental variables of the DRIP procedure. Combining DRIP-qPCR, DRIP-sequencing, and receiver operator characteristic (ROC) calculation, we devised an unbiased and systematic analytical pipeline in human T lymphoblastoid cells regarding the most important DRIP variables so that we suggest a reproducible and specific RNA-DNA hybrid detection, underlain by the objective criteria of ROC analysis.
Results
Introducing DRIP classifiers to assess true and false R-loop associations
Based on the available workflows of published DRIP protocols and considering the main technical variables that might contribute to the observed heterogeneities, we designed forty DRIP experimental schemes (binary classifiers) so that we assess how they rank different test loci according to their known RNA-DNA hybrid status (Fig. 1). The classifiers (“DRIP experiments” or “dependent variables”) were designed to systematically explore the main factors that might create experimental bias associated with the DRIP procedure.
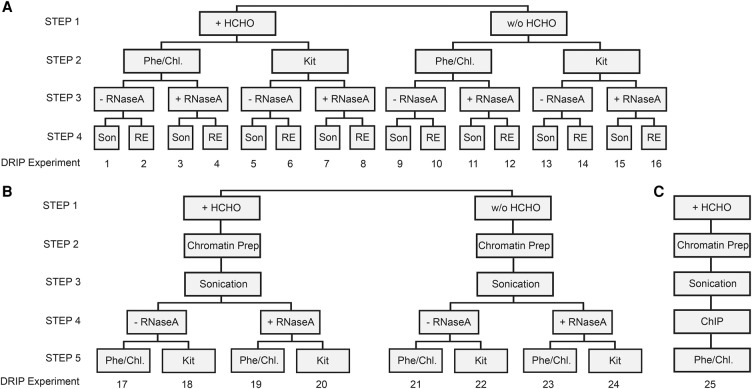
Figure 1.
Experimental design: constructing DRIP schemes. (A) Experiments 1–16 explore the effect of formaldehyde-fixation (Step 1), nucleic acid isolation (Step 2), removal of free RNA (Step 3), and nucleic acid fragmentation (Step 4) on the outcome of RNA-DNA hybrid detection. Each experiment was performed at two parallel cell lysis temperatures (65°C and 37°C), respectively. The temperature variable is not depicted in the cartoon, but it is referred in the main text. (B) Experiments 17–24 test the impact of acoustic sharing performed on a chromatin prep rather than on naked nucleic acid, similarly to the ChIP protocol. Each experiment was performed at 65°C cell lysis temperature. (C) Workflow of a ChIP experiment (shown only for comparison with the DRIP pipeline). (HCHO) Formaldehyde fixation, (Phe/Chl) phenol-chloroform extraction, (Kit) silica membrane-based nucleic acid purification, (RNase A) Ribonuclease A digestion performed at high (300 mM) NaCl concentration, (Son) sonication, (RE) restriction enzyme cocktail digestion (HindIII, EcoRI, BsrGI, XbaI, and SspI). As a negative control, RNase H digestion was applied in all DRIP experiments (not indicated in the cartoon).
Experiments 1–16 consider the effect of (1) formaldehyde (HCHO) fixation, (2) the method of nucleic acid isolation, (3) removal of free RNA, (4) the mode of nucleic acid fragmentation (Fig. 1A), and (5) cell lysis temperature (65°C as default vs. 37°C) (not shown in Fig. 1A, but referred to throughout the text as “37°C”).
Step 1: Formaldehyde fixation
The basic assumption behind HCHO–cross-linking is to maximize the DRIP yield while preserving biologically meaningful RNA-DNA hybrid interactions. However, formaldehyde has some well-known adverse effects: (1) the DNA accompanies a conformational change upon cross-linking, involving local denaturation or “breathing” of the double helix (McGhee and von Hippel 1977). This might create ectopic R-loop sites or abolish physiological R-loop contacts. (2) HCHO-treatment can reduce antigen accessibility or mask epitopes recognized by the antibody used for the immunoprecipitation. This might prevent a fraction of R-loops from being detected. (3) HCHO-fixation elicits spurious localization of irrelevant proteins at highly expressed genes (Baranello et al. 2016) and induces massive poly(ADP)ribose polymer formation in live cells (Beneke et al. 2012). These examples warrant deeper investigation of the usage of HCHO-fixation in RNA-DNA hybrid mapping; therefore, we classified our DRIP samples as HCHO-treated and nontreated categories (Fig. 1A,B).
Step 2: Nucleic acid purification
Two common methods were compared: organic (phenol/chloroform) extraction versus solid-phase (silica membrane) purification of total nucleic acids (Fig. 1A,B).
Step 3: Ribonucleolytic treatment (RNase A, RNase H, and sodium hydroxide)
Most DRIP protocols do not treat the isolated nucleic acid with ribonucleases to remove free RNA; however, the S9.6 antibody can recognize RNA duplexes with an approximately fivefold reduced affinity compared to RNA-DNA hybrids (Phillips et al. 2013). At this point, four kinds of ribonucleoleolytic digestion were incorporated into our DRIP pipelines: (1) RNase H1 digestion that removes RNA-DNA hybrids (negative control #1); (2) alkaline hydrolysis by sodium hydroxide that degrades free RNA and RNA-DNA hybrids (negative control #2); (3) RNase A digestion at high (300 mM) NaCl concentration that removes free RNA; and (4) RNase A digestion at low (25 mM) NaCl concentration that removes free RNA and RNA-DNA hybrids.
RNase H1 treatment is an accepted negative control of the DRIP procedure since it degrades the RNA strand in the hybrids, preventing their recognition by the S9.6 antibody. Half of the nucleic acid prep was digested by RNase H1 before the DNA fragmentation step that let us estimate the bulk level of RNA-DNA hybrids (dot blot setting; Supplemental Fig. S2A). The other half was digested just before the S9.6 immunoprecipitation step that let us obtain crucial information about the specificity of the IP signal (see DRIP-qPCR). As expected, RNA-DNA hybrids were sensitive to RNase H1 digestion in vitro. Similarly to RNase H1, alkaline hydrolysis by 50 mM NaOH also efficiently eliminated the RNA-DNA hybrid signal (Supplemental Fig. S2A). Less is known about the salt-dependent RNase H-like activity of RNase A that is supposed to digest RNA-DNA hybrids as an efficient hybridase at low ionic strength (https://www.thermofisher.com/order/catalog/product/EN0531). As shown in Supplemental Figure S2B, the hybrids were indeed resistant to RNase A digestion at high ionic strength, but they became highly sensitive to RNase A as a function of decreasing monovalent concentration. The RNase H-like activity of RNase A at low salt condition was confirmed by an independent method (Supplemental Fig. S2C,D) applying fluorescent microscopic detection. Based on these experiences, RNase A digestion at high salt concentration (300 mM NaCl) was integrated into our DRIP protocol to test if removal of competing free RNA improves the specificity of the RNA-DNA hybrid signal. Also, RNase H1 digestion of the fragmented nucleic acid was kept as an obligatory negative control of the immunoprecipitation.
Step 4: Nucleic acid fragmentation
The choice of restriction enzymes defines the cleavage pattern of DNA that is critical to achieve optimal fragment length distribution and mapping resolution. Based on the original DRIP protocol (Ginno et al. 2012), we combined five enzymes (HindIII, EcoRI, BsrGI, XbaI, and SspI) for in silico digestion, resulting in a median restriction fragment length of 314 bp (Supplemental Fig. S3A). In contrast to the theoretical fragment size distribution, we observed a broad DNA size range in a real digestion reaction (between 100–10,000 bp) (Supplemental Fig. S4A). As a control, we repeated the restriction enzyme cleavage in varying reaction conditions without detecting any improvement in the digestion efficacy (Supplemental Fig. S3B). When a budding yeast genomic DNA was digested in a parallel experiment, we managed to obtain the expected (in silico) fragment size distribution (Supplemental Fig. S3C). These observations necessitate the proper control of DNA fragment length distribution in DRIP samples that derive from restriction enzyme-fragmented nucleic acid.
As opposed to restriction enzyme digestion, sonication creates random DNA fragments with a typical size of 150–500 bp that dictate the spatial resolution of the DRIP assay (Supplemental Fig. S4B). However, excessive sonication can introduce strand breaks in the DNA or simply shake off a subset of R-loops from the chromosomes, potentially compromising their detectability by qPCR. Because of the above, the mode of DNA fragmentation (restriction enzymes and sonication) was introduced as an important parameter in our DRIP pipeline (Fig. 1A).
Fragmenting chromatin rather than purified genomic DNA (experiments 17–24)
In comparison to the original DRIP protocol, classical chromatin immunoprecipitation (ChIP) involves the capture of RNA-DNA hybrids by immunoprecipitation from cross-linked and sonicated chromatin (rather than naked DNA) followed by phenol/chloroform purification (Fig. 1C). Since sonication, performed on purified genomic DNA, led to loss of ∼80% of the DRIP signal in yeast (Wahba et al. 2016), we tested if acoustic shearing performed on a chromatin prep rather than on naked nucleic acid (Supplemental Fig. S4C) could improve the signal-to-noise ratio of the DRIP measurement (Fig. 1B).
Varying the cell lysis temperature
Published DRIP protocols apply various cell lysis temperatures, ranging from 37°C to 65°C and lasting from a couple of hours to overnight. To test the effect of temperature on the specificity of RNA-DNA hybrid detection, we lysed the samples at 65°C for 7 h or at 37°C overnight. Experiments 1–16 were processed in parallel at both temperatures, while exp. 17–24 were omitted from the temperature analysis since cross-link reversal typically occurs at 65°C.
Taken together, the above experimental variables resulted in 40 (16 × 2 + 8) autonomous DRIP classifiers (schemes) for which RNA-DNA hybrid enrichment scores were determined at several test loci. This allowed us to assess whether the S9.6 signal represented true or false R-loop associations within the applied condition.
Making a reference R-loop set for benchmarking the DRIP classifiers
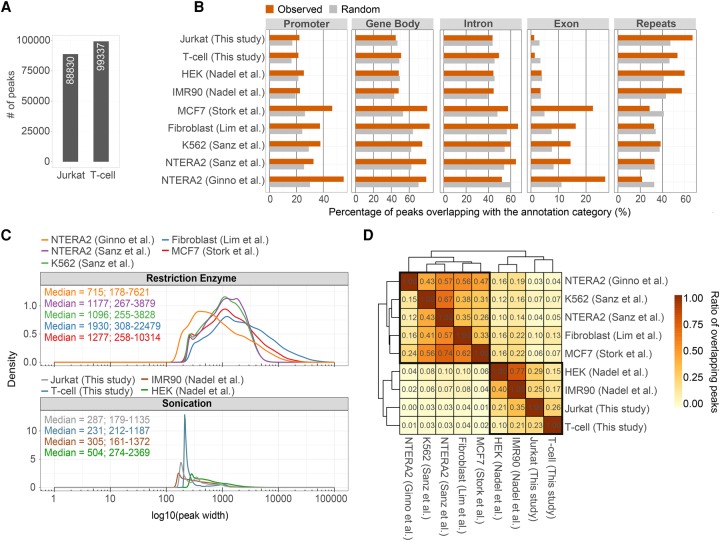
To derive the parameters of the DRIP classifiers, known positive and negative examples (genomic sites) could be chosen from the scientific literature based on their known R-loop profiles; however, the heterogeneity of the available DRIP-qPCR and DRIP-seq data sets (see Introduction) prompted us to establish our independent R-loop training set. We performed DNA-RNA hybrid mapping (DRIP-seq) in two closely related human cell types (Jurkat T cell leukemia cell line and naive CD4+ T lymphocytes) and identified 88,830 and 99,337 R-loop enriched regions, respectively (Fig. 2A). A high-confidence R-loop peak set was generated from the identified binding sites, and their chromosomal distribution was characterized. The peaks were significantly enriched at gene promoters and repetitive elements (Fig. 2B), consistent with previously published DRIP-seq results (Ginno et al. 2012; Nadel et al. 2015). R-loop sites were underrepresented at protein coding exons, similarly to earlier DRIP experiments performed with sonicated nucleic acid; however, restriction enzyme-fragmented DRIP samples were positively biased toward exons. Sonicated and restriction enzyme-digested samples were strikingly different in their R-loop length distributions (narrow: 179–2369 bp vs. wide: 178–22,479 bp) (Fig. 2C), and the identified R-loop binding sites significantly overlapped within each group but sharply stood apart between the two groups (Fig. 2D). We attribute these differences to the extensive variation of R-loop lengths and heterogeneities of the studied cell types. Biological implications of having too wide peak sizes will be discussed later. With the observed variances in mind, our consensus R-loop set was regarded as an amenable reference to benchmark the DRIP classifiers.
Figure 2.
Summary of available human DRIP-seq experiments. (A) Bar chart showing the number of identified R-loop peaks in human Jurkat cells and naive T cells (this study). (B) Annotation of R-loop binding sites over functional genomic elements. DRIP-seq peaks were determined in Jurkat cells and naive T cells, and in other published cell types (NTERA2, K562, Fibroblast, MCF7, IMR90, HEK293T). The upper four rows represent DRIP experiments fragmenting the nucleic acid by sonication, while the lower five rows highlight restriction enzyme-digested DRIP samples. The difference between the two groups is especially noticeable over exons (associated to 14%–27% and 1%–3.5% of R-loops, respectively) and repeat elements (SINEs, LINEs, LTRs, simple and low complexity repeats) that involve 22%–38% and 54%–67% of the R-loop peaks, respectively. At other annotation categories (gene body, introns, and promoters), the difference was not significant between the two groups. (C) Density plots showing the distribution of R-loop peak sizes, classified by fragmentation method (restriction enzyme vs. sonication). Median peak length and 2.5%–97.5% quantiles are indicated. Peak length distributions differ significantly between the two fragmentation methods. (D) Heat map showing the overlap of R-loop binding sites between independent DRIP-seq experiments. Values and cell colors represent pairwise and unique overlap ratios between each peak set. The difference between the two nucleic acid fragmentation methods is clearly apparent, as peak sets from the same fragmentation process better resemble each other (highlighted in black).
Measuring RNA-DNA hybrid enrichment over the DRIP classifiers
Positive and negative test regions were selected from the identified R-loop set (Supplemental Fig. S5) and were systematically probed for RNA-DNA hybrid enrichment across the DRIP classifiers (Supplemental Fig. S6). Five test regions were frequently used as positive and negative controls in various published DRIP studies (SNRPN, ZNF554, MYADM, FMR1, APOE) (Ginno et al. 2012; Bhatia et al. 2014; Groh et al. 2014; Herrera-Moyano et al. 2014; Loomis et al. 2014; Yang et al. 2014; Boque-Sastre et al. 2015; García-Rubio et al. 2015; Marinello et al. 2016), while the remaining sites were picked at random from the consensus R-loop set (PRR5L, LOC440704, NOP58, VIM, ING3). The reference DRIP-seq signal (benchmarking the classifiers) is shown over selected test regions along with DRIP-seq patterns taken from published studies (Supplemental Fig. S5). DRIP-qPCR yields were measured in control and RNase H-treated samples for 40 (16 × 2 + 8) DRIP classifiers, at 10 test regions, in five independent experiments. The resulting 4000 (40 × 2 × 10 × 5) DRIP enrichment scores were then readily used as an input parameter of receiver operator characteristics calculation.
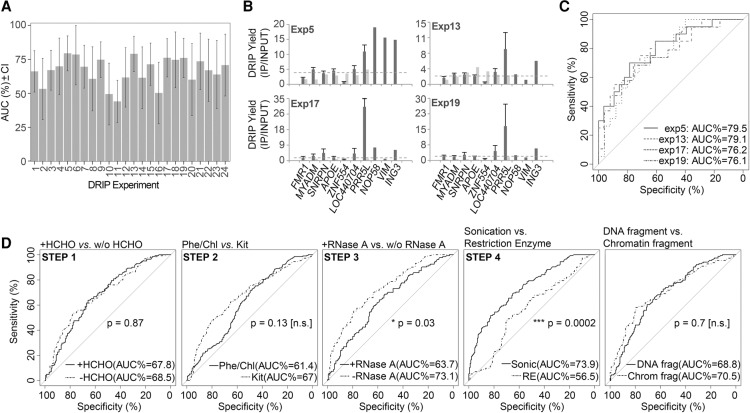
Determining the sensitivity and specificity of RNA-DNA hybrid detection: ROC analysis
We quantitated the relative trade-offs between true positive hits and experimental errors (false R-loop associations) by performing ROC analysis (Robin et al. 2011) on the DRIP-qPCR screen characterizing the classifiers (Supplemental Figs. S6–S10). The sensitivity, specificity, and the area under the curve (AUC) values were extracted from the ROC plots (Supplemental Table S2) and used as an objective measure of the robustness of the 40 experiments. High (>0.7) AUC values were obtained for 10 DRIP classifiers (exp. 5, 6, 13, 15, 17, 18, 19, 21, and 24), implying that those experiments could predict the presence or absence of an RNA-DNA hybrid with high efficacy (Fig. 3A). AUC values close to 0.5 were obtained in four experiments (exp. 2, 10, 11, and 16), implying that the classifiers gave random answers without any predictive power as to the presence of an R-loop. Based on these considerations, the top four DRIP classifiers were: exp. 5, 13, 17, and 19 (Fig. 3B,C), with a sensitivity of 68.5%–75% and specificity of 68%–79%. Similar (or even higher) ROC parameters were obtained in a repeated experiment using a B lymphoblastoid cell line (Supplemental Fig. S8), demonstrating the reliability of the tested DRIP protocols in other cell types.
Figure 3.
Good DRIP practice. (A) Bar charts showing the distribution of AUC (area under the curve) values of ROC plots for 24 DRIP classifiers. Error bars represent the confidence interval of AUCs. High (>0.7) AUC values were obtained for 10 DRIP classifiers (exp. 5, 6, 13, 15, 17, 18, 19, 21, and 24). Low (∼0.5) AUC values were obtained in four DRIP experiments (exp. 2, 10, 11, and 16). We highlight these groups as “preferred” and “not preferred,” respectively. (B,C) The top four DRIP experiments ranked by AUCs (exp. 5, 13, 17, and 19). (B) DRIP-qPCR enrichment scores are displayed over the test regions. Horizontal dotted lines represent the cutoff value (calculated from the ROC curves) separating the true R-loop signal from background. (C) ROC curves of the top four experiments. (D) Paired-ROC plots, comparing the main variables (steps) of the DRIP experiments. The level of statistical significance was 0.05.
Pairwise comparison of the main experimental variables (Fig. 3D) revealed no significant difference between (1) formaldehyde-fixed vs. unfixed samples, (2) phenol-chloroform extracted vs. silica membrane-purified nucleic acid samples, and (3) DNA-fragmented (exp. 1–16) vs. chromatin-fragmented DRIP samples (exp. 17–24). Cell lysis temperature (65°C vs. 37°C) did not change the specificity and sensitivity of the DRIP assay (Supplemental Figs. S9, S10). A statistically significant difference was obtained for RNase A-treated vs. untreated samples (P = 0.03), suggesting that addition of RNase A does not improve the efficacy of RNA-DNA hybrid detection (Step 3, Fig. 3D). We explain the adverse effect of RNase A by its reported DNA binding activity (Benore-Parsons and Ayoub 1997; Dona and Houseley 2014) that selectively eliminates a vast amount (micrograms) of melted DNA regions upon nucleic acid purification (Dona and Houseley 2014). We confirmed the strong DNA binding of RNase A as migration defects on DNA gels, when a plasmid DNA was incubated with the enzyme (Supplemental Fig. S11). The observed electrophoretic mobility shift was prevalent on supercoiled, nicked-circular, and linearized DNA templates.
Finally, by comparing sonicated and restriction enzyme fragmented DRIP samples (Step 4, Fig. 3D), we found a statistically significant difference (P = 0.0002) in the ROC parameters, suggesting that sonication is more efficient in discriminating true positive signals from false positives, at least within the tested conditions.
Good DRIP practice: impact on the annotation and basic biological function of R-loops
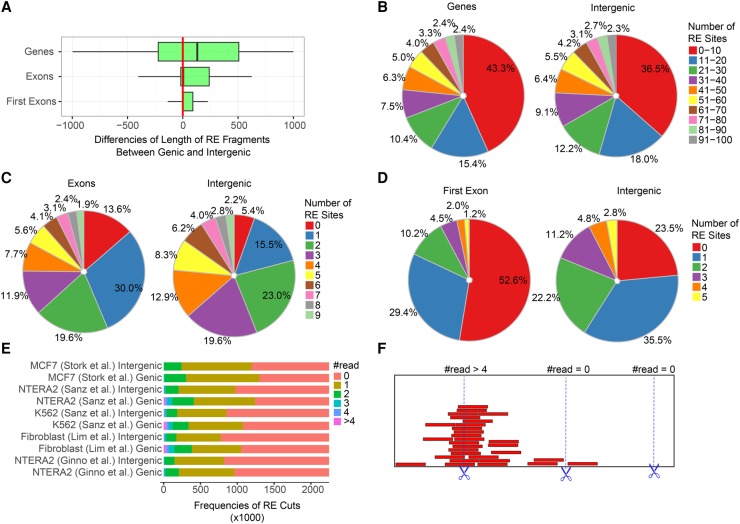
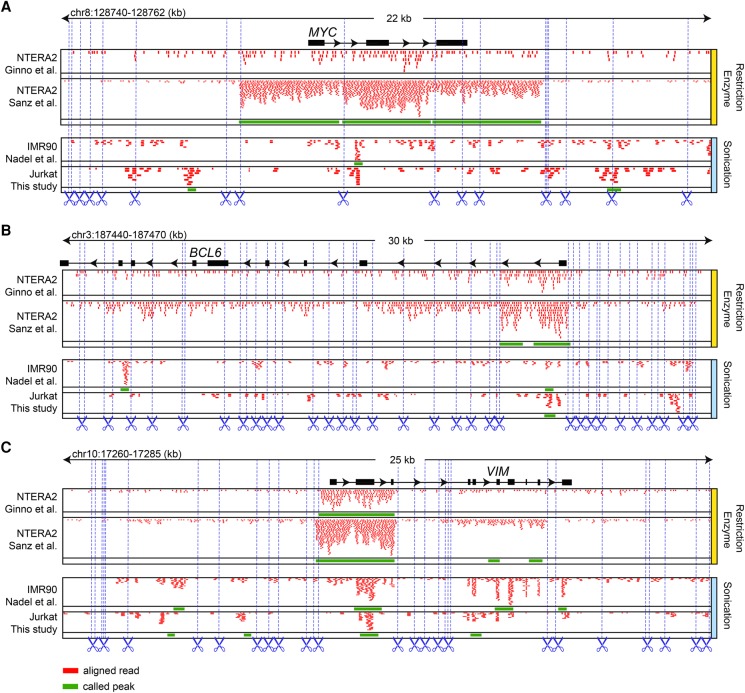
Suboptimal DRIP conditions might prevent the assignment of precise biological function to a significant fraction of R-loops. Although the average DNA fragment size resulting from restriction enzyme digestion fits the requirements of the DRIP assay, we found that the frequency of cutting sites was significantly higher within intergenic regions, producing lengthy restriction fragments over protein coding ORFs (Fig. 4). Biased genome sampling, related to the nonrandom distribution of restriction enzyme recognition sequences, was even more pronounced over exons (Fig. 4C), especially over the first exons (Fig. 4D). In 82% of first exons, there were only 0–1 suitable restriction sites, compared to intergenic regions (59%). We estimated the digestion efficiency of restriction enzyme cutting sites as ∼50% over intergenic regions (based on the proportion of zero reads over restriction enzyme cutting sequences, representing cleaved sites), which was significantly reduced over gene coding regions (Fig. 4E,F). Consequently, genic regions void of suitable restriction sites appear as long DRIP fragments that potentially compromise mapping resolution. The MYC, BCL6, and VIM genes are shown as representative examples for large, restriction fragment-sized DRIP peaks (Fig. 5). Precise genomic position of R-loops could be resolved by sonication.
Figure 4.
Analysis of restriction sites over genic and intergenic regions. (A) Restriction fragment lengths over genic regions (gene bodies, exons, first exons) are significantly larger compared to intergenic regions. The plot shows the difference of genic (observed) and intergenic (expected) fragment sizes in base pairs. The following enzymes were applied in combination: HindIII, EcoRI, BsrGI, XbaI, and SspI. (B–D) The number of restriction sites over genic regions is significantly lower compared to intergenic regions. Colors indicate the proportion of cutting sites in each category. Red and blue slices, marking the rarest restriction site frequencies, are prevalent over genic elements in each pie chart. (E) Cutting efficiency of restriction enzymes applied in the indicated DRIP-seq experiments. Zero read: the restriction site was cut. Greater equal than one read: the restriction site was uncut in a fraction of cells. There were uncut reads (sites) over half of the theoretical restriction sites. The proportion of uncut reads was even higher within gene coding regions compared to intergenic regions. See the model of cutting efficiency in panel F.
Figure 5.
Large restriction fragments over gene bodies cause uncertainty in the precise localization of R-loops, potentially impeding their functional annotation. (A–C) Genome browser tracks showing three representative examples (MYC, BCL6, and VIM). Upper two tracks: restriction fragment-sized R-loops are prevalent over the 5′ end of genes, vastly exceeding the gene borders in the case of MYC. Lower two tracks: the precise genomic position of R-loops was resolved in the sonicated group of samples. Green boxes represent R-loop enriched regions predicted by the peak callers. Blue dashed lines represent cutting sites for restriction enzymes (HindIII, EcoRI, BsrGI, XbaI, and SspI).
Discussion
The increasing recognition of RNA-DNA hybrid structures in the physiology and pathology of chromosomes has prompted us to develop an analytical approach to estimate the inherent biases and errors of existing DRIP protocols and to assess the power of the technology. The determined ROC parameters (AUC, sensitivity, specificity, threshold) served as an objective measure for the efficacy of predicting the presence or absence of RNA-DNA hybrids. In the tested experimental conditions, we managed to find and verify DRIP workflows that were able to distinguish complex or weak DRIP-qPCR signals from a noisy background with high confidence across a number of genomic regions (exp. 5, 13, 17, and 19). On the contrary, some DRIP workflows performed unreliably and generated random answers (exp. 2, 10, 11, and 16). Under our experimental conditions, we highlight these groups as “preferred” and “not preferred.” By testing the main parameters of the DRIP experimental scheme—involving formaldehyde fixation, cell lysis temperature, nucleic acid isolation, free RNA removal, and DNA fragmentation—we found that fragmenting the nucleic acid by sonication and omitting RNase A digestion could improve the precision and specificity of RNA-DNA hybrid detection (Fig. 3D). At this point, we emphasize the lack of correlation between the DRIP scores (IP/input ratios) and AUC values, as these quantities are not related to each other. The former highlights the yield of immunoprecipitation, while the latter is a quantitative measure of true and false R-loop associations. For instance, the worst and best DRIP schemes (exp. 2 and exp. 5) had a qPCR yield of 10%–95% and 1%–18% over the studied regions, respectively (Supplemental Figs. S6, S8). Consequently, high DRIP enrichment is not necessarily accompanied by increased accuracy, and vice versa.
We also showed that genome fragmentation by restriction enzymes led to the overrepresentation of long DRIP fragments over ORFs, which was especially enhanced over the first exons of protein coding genes (Figs. 4–5). Biased genome sampling severely compromised mapping resolution and, as a consequence, the assignment of clear biological function to a fraction of R-loops. For instance, correct estimation of evolutionary conservation between R-loop binding sites, relying on sequence homologies of exons that are associated with R-loops (Sanz et al. 2016), becomes uncertain.
Based on the above experiences, we suggest the following refinements of DRIP workflows to obtain accurate estimates of RNA-DNA hybrid occupancies: (1) Omission of HCHO-fixation and RNase A treatment, isolation of nucleic acid by silica membrane (kit) purification, nucleic acid fragmentation by sonication, followed by immunoprecipitation with the S9.6 antibody (see Methods). (2) If formaldehyde-fixation is applied, we recommend preparing soluble chromatin and fragmenting the prep by sonication (similarly to the ChIP protocol), followed by organic extraction and immunoprecipitation with the S9.6 antibody. (3) If restriction enzyme fragmentation needs to be applied (e.g., in some cases, sonication might be too harsh to capture transient or very weak RNA-DNA hybrid interactions), we advise the careful control of DNA fragment size distribution before immunoprecipitation.
An important premise is that our recommendations apply to the experimental conditions investigated by this study. Generalization should be avoided since altering critical parameters in the experiment (e.g., incorporating S1 nuclease [S1-DRIP] [Wahba et al. 2016] or lambda exonuclease digestion [DRIP-exo] [Ohle et al. 2016], or changing the model organism) might significantly affect the outcome of RNA-DNA hybrid detection.
In conclusion, the DRIP method remains a gold-standard for identifying bona fide R-loop binding sites across individual chromosomes, but a continued effort is needed to find alternatives and test complementary protocols. We hope that this aim has been achieved, at least in part, by this study, and will help recognize real R-loop binding events and enable a better interpretation of DRIP-seq mapping data.
Methods
Detection of RNA-DNA hybrids by DNA-RNA immunoprecipitation
DRIP classifiers 1–16
Cross-linking (Step 1)
Cross-linking of Jurkat cells (experiments 1–8) was done with 1% paraformaldehyde (UP) for 10 min, then quenched with 2.5 M glycine (pH 6, final concentration: 500 mM) for 5 min at room temperature. Cross-linking was omitted from experiments 9–16.
Cell lysis
Cells were lysed in 1 mL lysis buffer composed of 500 µL 2× lysis buffer (1% SDS, 20 mM Tris-HCl pH 7.5, 40 mM EDTA pH 8, 100 mM NaCl, ddH2O) plus 500 µL TE buffer (100 mM Tris-HCl pH 8, 10 mM EDTA pH 8) per 5 million cells. Cell lysis was performed at two different temperatures: either at 65°C for 7 h, or at 37°C overnight, as indicated in the text.
Phenol-chloroform extraction of total nucleic acid (Step 2)
In experiments 1–4 and 9–12, total nucleic acid was prepared by phenol-chloroform extraction. Before the phenol-chloroform extraction step, the nucleic acid preps were treated with 10 µL of Proteinase K (20 mg/mL; Thermo Fisher Scientific) at 65°C for 7 h, or at 37°C overnight, to remove the proteins. The extracted DNA was precipitated with 1/10 volume 3 M Na-acetate (pH 5.2) plus 1 volume of isopropanol. The DNA pellet was dissolved in 200 µL of 10 mM Tris-HCl pH 8.
Silica membrane-based (kit) extraction of total nucleic acid (Step 2)
In experiments 5–8 and 13–16, total nucleic acid was isolated by the NucleoSpin Tissue kit (Macherey-Nagel) according to the manufacturer's protocol, except the cell lysis step was performed either at 65°C for 7 h (according to the kit protocol), or at 37°C overnight, where indicated in the text. Nucleic acids were eluted in 500 µL of elution buffer (5 mM Tris-HCl pH 8.5).
Removal of free RNA by RNase A treatment (Step 3)
In experiments 3–4, 7–8, 11–12, and 15–16, the DNA purification step was directly followed by the RNase A digestion of free ribonucleic acids. The purified DNA preps (from Step 2) were supplemented with 18 µL of 5 M NaCl and 2 µL of RNase A (10 mg/mL; UD-GenoMed Ltd.) in a buffer containing 10 mM Tris-HCl (pH 8) and 300 mM NaCl (V = 300 µL) at 37°C for 1 h. RNase A-treated samples were repurified either by phenol-chloroform extraction (experiments 4, 12) or by the NucleoSpin Gel and PCR Clean-up kit (Macherey-Nagel) (experiments 8, 16). Phenol-chloroform–extracted DNA was dissolved in 100 µL of 5 mM Tris-HCl pH 8.5. The DNA purified with the kit was eluted in 5 mM Tris-HCl pH 8.5.
Nucleic acid fragmentation by sonication (Step 4)
In experiments 1, 3, 5, 7, 9, 11, 13, 15, the purified nucleic acid preps were sonicated in a buffer of 10 mM Tris-HCl pH 8.5 supplemented with 300 mM NaCl (V = 300 µL) for 2 × 5 min (30 sec ON, 30 sec OFF, LOW; Bioruptor, Diagenode) to yield an average DNA fragment size of ∼300 bp.
Nucleic acid fragmentation by restriction enzyme digestion (Step 4)
In exp. 2, 4, 6, 8, 10, 12, 14, and 16, purified DNA samples (∼25 µg each) were fragmented using a restriction enzyme cocktail of 1 µL HindIII (20 U/µL), 1 µL EcoRI (20 U/µL), 2 µL BsrGI (10 U/µL), 1 µL XbaI (20 U/µL), and 4 µL SspI (5 U/µL) in NEB Buffer 2 (NEB) (V = 300 µL) at 37°C for 4 h.
The fragmented DNA samples were repurified either by phenol-chloroform extraction (experiments 1–4, 9–12) or by the NucleoSpin Gel and PCR Clean-up kit (Macherey-Nagel) (experiments 5–8, 13–16). The DNA was dissolved in 100 µL of 5 mM Tris-HCl pH 8.5.
Two percent (V/V%) of the DNA preps were kept as input DNA for the DRIP-qPCR measurement. Half of samples were treated with 8 µL of RNase H (5000 U/mL; NEB) in a total volume of 80 µL, at 37°C overnight.
DRIP classifiers 17–24
Cross-linking (Step 1)
Cross-linking of Jurkat cells (experiments 17–20) was done with 1% paraformaldehyde (UP) for 10 min, then quenched with 2.5 M glycine (pH 6, final concentration: 500 mM) for 5 min at room temperature. Cross-linking was omitted from experiments 21–24.
Chromatin preparation (Step 2): cell lysis
Cells were lysed in 750 µL of ChIP lysis buffer (50 mM HEPES-KOH at pH 7.5, 140 mM NaCl, 1 mM EDTA at pH 8, 1% Triton X-100, 0.1% Na-Deoxycholate, 1% SDS) per 10 million cells and homogenized using Fast Prep-24 5G (MP Biomedicals, speed: 6 m/sec; time: 40 sec; 2 cycles; pause time: 120 sec; A lysing matrix).
Chromatin fragmentation by sonication (Step 3)
Three hundred microliters of chromatin preps were sonicated for 2 × 5 min (30 sec ON, 30 sec OFF, LOW, Bioruptor) to yield an average DNA fragment size of ∼300 bp.
Removal of free RNA by RNase A treatment (Step 4)
In experiments 19, 20, 23, and 24, the sonication step was directly followed by the RNase A digestion of free ribonucleic acids. The fragmented chromatin was supplemented with 270 µL of 5 M NaCl (300 mM) and 10 µL of RNase A (10 mg/mL; UD-GenoMed Ltd.) in 4500 µL of TE buffer (10 mM Tris-HCl pH 8, 10 mM EDTA pH 8) at 37°C for 1 h.
Before Step 5, the chromatin preps were treated with 30 µL of Proteinase K (20 mg/mL; Thermo Fisher Scientific) at 65°C overnight to remove the proteins and reverse the cross-links.
Phenol-chloroform extraction of total nucleic acid (Step 5)
In experiments 17, 19, 21, and 23, total nucleic acid was prepared by phenol-chloroform extraction. The extracted DNA was precipitated with 1/10 volume 3 M Na-acetate (pH 5.2) plus 1 volume of isopropanol. The DNA pellet was dissolved in 100 µL of 5 mM Tris-HCl pH 8.5.
Silica membrane-based (kit) extraction of total nucleic acid (Step 5)
In experiments 18, 20, 22, and 24, total nucleic acids were isolated by the NucleoSpin Gel and PCR Clean-up kit (Macherey-Nagel) according to the manufacturer's protocol. Nucleic acids were eluted in 100 µL of elution buffer (5 mM Tris-HCl pH 8.5).
Two percent (V/V%) of the DNA preps were kept as input DNA for the DRIP-qPCR measurement. Half of the samples were treated with 8 µL of RNase H (5000 U/mL; NEB) in a total volume of 80 µL at 37°C overnight.
RNA-DNA hybrid immunoprecipitation with the S9.6 antibody
Dynabeads Protein A magnetic beads (Thermo Fisher Scientific) were pre-blocked with PBS/EDTA containing 0.5% BSA. To immobilize the S9.6 antibody, 50 µL pre-blocked Dynabeads Protein A were incubated with 10 µg of S9.6 antibody in IP buffer (50 mM Hepes/KOH at pH 7.5; 0.14 M NaCl; 5 mM EDTA; 1% Triton X-100; 0.1% Na-Deoxycholate, ddH2O) at 4°C for 4 h with rotation. Six micrograms of digested genomic DNA were added to the mixture and gently rotated at 4°C overnight. Beads were recovered and washed successively with 1 mL lysis buffer (low salt, 50 mM Hepes/KOH pH 7.5, 0.14 M NaCl, 5 mM EDTA pH 8, 1% Triton X-100, 0.1% Na-Deoxycholate), 1 mL lysis buffer (high salt, 50 mM Hepes/KOH pH 7.5, 0.5 M NaCl, 5 mM EDTA pH 8, 1% Triton X-100, 0.1% Na-Deoxycholate), 1 mL wash buffer (10 mM Tris-HCl pH 8, 0.25 M LiCl, 0.5% NP-40, 0.5% Na-Deoxycholate, 1 mM EDTA pH 8), and 1 mL TE (100 mM Tris-HCl pH 8, 10 mM EDTA pH 8) at 4°C, two times. Elution was performed in 100 µL of elution buffer (50 mM Tris-HCl pH 8, 10 mM EDTA, 1% SDS) for 15 min at 65°C. After purification with the NucleoSpin Gel and PCR Clean-up kit (Macherey-Nagel), nucleic acids were eluted in 55 µL of elution buffer (5 mM Tris-HCl pH 8.5). The recovered DNA was then analyzed by quantitative real-time PCR (qPCR). qPCR was performed with a LightCycler 480 SYBR Green I Master (Roche) and analyzed on a QuantStudio 12K Flex Real-Time PCR System (Thermo Fisher Scientific). Primer sequences are listed in Supplemental Table S3. qPCR results were analyzed using the comparative CT method. The RNA-DNA hybrid enrichment was calculated based on the IP/Input ratio.
DRIP-sequencing
DRIP-sequencing was performed in human Jurkat cells and naive T CD4+ lymphocytes. A full description of the DRIP-seq experiment and bioinformatics analysis can be found in the Supplemental Material.
Receiver operating characteristic analysis
ROC curves were obtained for each DRIP variable (DRIP experiments) by ranking the studied genomic loci having known RNA-DNA hybrid states (based on the training set) according to their DRIP-qPCR profile, starting from the lowest to the highest estimated DRIP scores and then calculating sensitivity and specificity. The ROC curves plotted the sensitivity or true-positive rate (TPR) against the false-positive rate (FPR) or 1-specificity, estimated as follows: TPR = P(positive DRIP-qPCR result|R-loop present), FPR = P(positive DRIP-qPCR result|R-loop absent), where P means conditional probability. The AUC values were then calculated from the observed DRIP-qPCR (IP/input) yields using the pROC algorithm.
Data access
DRIP Sequencing data from this study have been submitted to the NCBI Sequence Read Archive (SRA; https://www.ncbi.nlm.nih.gov/sra) under accession number SRP095885.
Supplementary Material
Acknowledgments
L.S. received funding from the Hungarian Academy of Sciences (Lendület programme, Magyar Tudományos Akadémia, LP2015-9/2015), from the International Center for Genetic Engineering and Biotechnology (CRP-ICGEB), Italy (CRP/HUN13-01), from the European Union, Seventh Framework Programme (FP7/Marie Curie Actions/CIG_#292259), from IMéRA/Inserm and the Aix-Marseille University (France), from NKFIH_ERC_HU_#117670, and from H2020/NKFIH_GINOP-2.3.2-15-2016-00024 and -00043 (G.H.). We thank Ibolya Fürtös for the excellent technical assistance and for the Genomic Medicine and Bioinformatics Core Facility (University of Debrecen) for the NGS service. We thank Dr. György Fenyőfalvi for critical discussions on the topic and for his idea about the RNase H-like activity of RNase A. We thank Dr. Gábor Szabó for providing us with the S9.6 antibody.
Author contributions: B.B.O., T.K.R., É.S., É.N., and Á.M.L. conceived and performed the experiments; A.M., É.R., G.H., and Z.K. provided reagents, expertise, and feedback; L.H., Z.K., and L.S. analyzed the data; and L.S. wrote the manuscript and secured funding.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.219394.116.
References
- Alzu A, Bermejo R, Begnis M, Lucca C, Piccini D, Carotenuto W, Saponaro M, Brambati A, Cocito A, Foiani M, et al. 2012. Senataxin associates with replication forks to protect fork integrity across RNA-polymerase-II-transcribed genes. Cell 151: 835–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranello L, Kouzine F, Sanford S, Levens D. 2016. ChIP bias as a function of cross-linking time. Chromosom Res 24: 175–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beneke S, Meyer K, Holtz A, Hüttner K, Bürkle A. 2012. Chromatin composition is changed by poly(ADP-ribosyl)ation during chromatin immunoprecipitation. PLoS One 7: e32914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benore-Parsons M, Ayoub MA. 1997. Presence of RNase A causes aberrant DNA band shifts. Biotechniques 23: 128–131. [DOI] [PubMed] [Google Scholar]
- Bhatia V, Barroso SI, García-Rubio ML, Tumini E, Herrera-Moyano E, Aguilera A. 2014. BRCA2 prevents R-loop accumulation and associates with TREX-2 mRNA export factor PCID2. Nature 511: 362–365. [DOI] [PubMed] [Google Scholar]
- Boque-Sastre R, Soler M, Oliveira-Mateos C, Portela A, Moutinho C, Sayols S, Villanueva A, Esteller M, Guil S. 2015. Head-to-head antisense transcription and R-loop formation promotes transcriptional activation. Proc Natl Acad Sci 112: 5785–5790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulé J-B, Zakian VA. 2007. The yeast Pif1p DNA helicase preferentially unwinds RNA DNA substrates. Nucleic Acids Res 35: 5809–5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown TA, Tkachuk AN, Clayton DA. 2008. Native R-loops persist throughout the mouse mitochondrial DNA genome. J Biol Chem 283: 36743–36751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellano-Pozo M, Santos-Pereira JM, Rondón AG, Barroso S, Andújar E, Pérez-Alegre M, García-Muse T, Aguilera A. 2013. R loops are linked to histone H3 S10 phosphorylation and chromatin condensation. Mol Cell 52: 1–8. [DOI] [PubMed] [Google Scholar]
- Chan YA, Aristizabal MJ, Lu PYT, Luo Z, Hamza A, Kobor MS, Stirling PC, Hieter P. 2014. Genome-wide profiling of yeast DNA:RNA hybrid prone sites with DRIP-chip. PLoS Genet 10: e1004288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen PB, Chen HV, Acharya D, Rando OJ, Fazzio TG. 2015. R loops regulate promoter-proximal chromatin architecture and cellular differentiation. Nat Struct Mol Biol 22: 999–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chon H, Sparks JL, Rychlik M, Nowotny M, Burgers PM, Crouch RJ, Cerritelli SM. 2013. RNase H2 roles in genome integrity revealed by unlinking its activities. Nucleic Acids Res 41: 3130–3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloutier SC, Wang S, Ma WK, Al Husini N, Dhoondia Z, Ansari A, Pascuzzi PE, Tran EJ. 2016. Regulated formation of lncRNA-DNA hybrids enables faster transcriptional induction and environmental adaptation. Mol Cell 61: 393–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domínguez-Sánchez MS, Barroso S, Gómez-González B, Luna R, Aguilera A. 2011. Genome instability and transcription elongation impairment in human cells depleted of THO/TREX. PLoS Genet 7: e1002386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dona F, Houseley J. 2014. Unexpected DNA loss mediated by the DNA binding activity of ribonuclease A. PLoS One 9: e115008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Hage A, French SL, Beyer AL, Tollervey D. 2010. Loss of Topoisomerase I leads to R-loop-mediated transcriptional blocks during ribosomal RNA synthesis. Genes Dev 24: 1546–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Hage A, Webb S, Kerr A, Tollervey D. 2014. Genome-wide distribution of RNA-DNA hybrids identifies RNase H targets in tRNA genes, retrotransposons and mitochondria. PLoS Genet 10: e1004716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan W, Guan Z, Liu J, Gui T, Shen K, Manley JL, Li X. 2011. R-loop-mediated genomic instability is caused by impairment of replication fork progression. Genes Dev 25: 2041–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Rubio ML, Pérez-Calero C, Barroso SI, Tumini E, Herrera-Moyano E, Rosado IV, Aguilera A. 2015. The Fanconi anemia pathway protects genome integrity from R-loops. PLoS Genet 11: e1005674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginno PA, Lott PL, Christensen HC, Korf I, Chédin F. 2012. R-loop formation is a distinctive characteristic of unmethylated human CpG island promoters. Mol Cell 45: 814–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groh M, Lufino MMP, Wade-Martins R, Gromak N. 2014. R-loops associated with triplet repeat expansions promote gene silencing in Friedreich ataxia and fragile X syndrome. PLoS Genet 10: e1004318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatchi E, Skourti-Stathaki K, Ventz S, Pinello L, Yen A, Kamieniarz-Gdula K, Dimitrov S, Pathania S, McKinney KM, Eaton ML, et al. 2015. BRCA1 recruitment to transcriptional pause sites is required for R-loop-driven DNA damage repair. Mol Cell 57: 636–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera-Moyano E, Mergui X, García-Rubio ML, Barroso S, Aguilera A. 2014. The yeast and human FACT chromatin-reorganizing complexes solve R-loop-mediated transcription-replication conflicts. Genes Dev 1: 735–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Zhang A, Storz G, Gottesman S, Leppla SH. 2006. An antibody-based microarray assay for small RNA detection. Nucleic Acids Res 34: e52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huertas P, Aguilera A. 2003. Cotranscriptionally formed DNA:RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Mol Cell 12: 711–721. [DOI] [PubMed] [Google Scholar]
- Jenjaroenpun P, Wongsurawat T, Yenamandra SP, Kuznetsov VA. 2015. QmRLFS-finder: amodel, web server and stand-alone tool for prediction and analysis of R-loop forming sequences. Nucleic Acids Res 43: W527–W534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Manley JL. 2005. Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell 122: 365–378. [DOI] [PubMed] [Google Scholar]
- Lim YW, Sanz LA, Xu X, Hartono SR, Chédin F. 2015. Genome-wide DNA hypomethylation and RNA:DNA hybrid accumulation in Aicardi-Goutières syndrome. eLife 4: e08007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loomis EW, Sanz LA, Chédin F, Hagerman PJ. 2014. Transcription-associated R-loop formation across the human FMR1 CGG-repeat region. PLoS Genet 10: e1004294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinello J, Bertoncini S, Aloisi I, Cristini A, Tagliazucchi GM, Forcato M, Sordet O, Capranico G. 2016. Dynamic effects of topoisomerase i inhibition on R-loops and short transcripts at active promoters. PLoS One 11: e0147053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGhee JD, von Hippel PH. 1977. Formaldehyde as a probe of DNA structure. 4. Mechanism of the initial reaction of formaldehyde with DNA. Biochemistry 16: 3276–3293. [DOI] [PubMed] [Google Scholar]
- Mischo HE, Gómez-González B, Grzechnik P, Rondón AG, Wei W, Steinmetz L, Aguilera A, Proudfoot NJ. 2011. Yeast Sen1 helicase protects the genome from transcription-associated instability. Mol Cell 41: 21–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadel J, Athanasiadou R, Lemetre C, Wijetunga NA, Broin PÓ, Sato H, Zhang Z, Jeddeloh J, Montagna C, Golden A, et al. 2015. RNA:DNA hybrids in the human genome have distinctive nucleotide characteristics, chromatin composition, and transcriptional relationships. Epigenetics Chromatin 8: 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakama M, Kawakami K, Kajitani T, Urano T, Murakami Y. 2012. DNA-RNA hybrid formation mediates RNAi-directed heterochromatin formation. Genes Cells 17: 218–233. [DOI] [PubMed] [Google Scholar]
- Ohle C, Tesorero R, Schermann G, Dobrev N, Sinning I, Fischer T. 2016. Transient RNA-DNA hybrids are required for efficient double-strand break repair. Cell 167: 1001–1013. [DOI] [PubMed] [Google Scholar]
- Pefanis E, Wang J, Rothschild G, Lim J, Kazadi D, Sun J, Federation A, Chao J, Elliott O, Liu Z-P, et al. 2015. RNA exosome-regulated long non-coding RNA transcription controls super-enhancer activity. Cell 161: 774–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips DD, Garboczi DN, Singh K, Hu Z, Leppla SH, Leysath CE. 2013. The sub-nanomolar binding of DNA-RNA hybrids by the single-chain Fv fragment of antibody S9.6. J Mol Recognit 26: 376–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohjoismäki JLO, Holmes JB, Wood SR, Yang M-Y, Yasukawa T, Reyes A, Bailey LJ, Cluett TJ, Goffart S, Willcox S, et al. 2010. Mammalian mitochondrial DNA replication intermediates are essentially duplex but contain extensive tracts of RNA/DNA hybrid. J Mol Biol 397: 1144–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigby RE, Webb LM, Mackenzie KJ, Li Y, Leitch A, Reijns MAM, Lundie RJ, Revuelta A, Davidson DJ, Diebold S, et al. 2014. RNA:DNA hybrids are a novel molecular pattern sensed by TLR9. EMBO J 33: 542–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robin X, Turck N, Hainard A, Tiberti N, Lisacek F, Sanchez J-C, Müller M. 2011. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics 12: 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanello M, Schiavone D, Frey A, Sale JE. 2016. Histone H3.3 promotes IgV gene diversification by enhancing formation of AID-accessible single-stranded DNA. EMBO J 35: 1452–1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvi JS, Chan JNY, Szafranski K, Liu TT, Wu JD, Olsen JB, Khanam N, Poon BPK, Emili A, Mekhail K. 2014. Roles for Pbp1 and caloric restriction in genome and lifespan maintenance via suppression of RNA-DNA hybrids. Dev Cell 30: 177–191. [DOI] [PubMed] [Google Scholar]
- Sanz LA, Hartono SR, Lim YW, Ginno PA, Sanz LA, Hartono SR, Lim YW, Steyaert S, Rajpurkar A, Ginno PA, et al. 2016. Prevalent, dynamic, and conserved R-loop structures associate with specific epigenomic signatures in mammals. Mol Cell 63: 167–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab RA, Nieminuszczy J, Shah F, Langton J, Lopez Martinez D, Liang CC, Cohn MA, Gibbons RJ, Deans AJ, Niedzwiedz W. 2015. The Fanconi anemia pathway maintains genome stability by coordinating replication and transcription. Mol Cell 60: 351–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skourti-Stathaki K, Proudfoot NJ, Gromak N. 2011. Human senataxin resolves RNA/DNA hybrids formed at transcriptional pause sites to promote Xrn2-dependent termination. Mol Cell 42: 794–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollier J, Stork CT, García-Rubio ML, Paulsen RD, Aguilera A, Cimprich KA. 2014. Transcription-coupled nucleotide excision repair factors promote R-loop-induced genome instability. Mol Cell 56: 777–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stork CT, Bocek M, Crossley MP, Sollier J, Sanz LA, Chédin F, Swigut T, Cimprich KA. 2016. Co-transcriptional R-loops are the main cause of estrogen-induced DNA damage. eLife 5: e17548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuckey R, García-Rodríguez N, Aguilera A, Wellinger RE. 2015. Role for RNA:DNA hybrids in origin-independent replication priming in a eukaryotic system. Proc Natl Acad Sci 112: 5779–5784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Csorba T, Skourti-Stathaki K, Proudfoot NJ, Dean C. 2013. R-loop stabilization represses antisense transcription at the Arabidopsis FLC locus. Science 340: 619–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Székvölgyi L, Rákosy Z, Bálint BL, Kókai E, Imre L, Vereb G, Bacsó Z, Goda K, Varga S, Balázs M, et al. 2007. Ribonucleoprotein-masked nicks at 50-kbp intervals in the eukaryotic genomic DNA. Proc Natl Acad Sci 104: 14964–14969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahba L, Koshland D. 2013. The Rs of biology: R-loops and the regulation of regulators. Mol Cell 50: 611–612. [DOI] [PubMed] [Google Scholar]
- Wahba L, Costantino L, Tan FJ, Zimmer A, Koshland D. 2016. S1-DRIP-seq identifies high expression and polyA tracts as major contributors to R-loop formation. Genes Dev 30: 1327–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson-Sali T, Hsieh T-S. 2002. Preferential cleavage of plasmid-based R-loops and D-loops by Drosophila topoisomerase IIIβ. Proc Natl Acad Sci 99: 7974–7979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, McBride KM, Hensley S, Lu Y, Chedin F, Bedford MT. 2014. Arginine methylation facilitates the recruitment of TOP3B to chromatin to prevent R loop accumulation. Mol Cell 53: 484–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, La H, Tang K, Miki D, Yang L, Wang B, Duan C-G, Nie W, Wang X, Wang S, et al. 2016. SAC3B, a central component of the mRNA export complex TREX-2, is required for prevention of epigenetic gene silencing in Arabidopsis. Nucleic Acids Res 45: 181–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu K, Chedin F, Hsieh C-L, Wilson TE, Lieber MR. 2003. R-loops at immunoglobulin class switch regions in the chromosomes of stimulated B cells. Nat Immunol 4: 442–451. [DOI] [PubMed] [Google Scholar]
- Yu K, Roy D, Huang F-T, Lieber MR. 2006. Detection and structural analysis of R-loops. Methods Enzymol 409: 316–329. [DOI] [PubMed] [Google Scholar]
- Zeller P, Padeken J, van Schendel R, Kalck V, Tijsterman M, Gasser SM. 2016. Histone H3K9 methylation is dispensable for Caenorhabditis elegans development but suppresses RNA:DNA hybrid-associated repeat instability. Nat Genet 48: 1385–1395. [DOI] [PubMed] [Google Scholar]
- Zhang ZZ, Pannunzio NR, Han L, Hsieh C-L, Yu K, Lieber MR. 2014a. The strength of an Ig switch region is determined by its ability to drive R loop formation and its number of WGCW sites. Cell Rep 8: 557–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ZZ, Pannunzio NR, Hsieh C-L, Yu K, Lieber MR. 2014b. The role of G-density in switch region repeats for immunoglobulin class switch recombination. Nucleic Acids Res 42: 13186–13193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ZZ, Pannunzio NR, Hsieh C-L, Yu K, Lieber MR. 2015. Complexities due to single-stranded RNA during antibody detection of genomic RNA:DNA hybrids. BMC Res Notes 8: 127. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.