Abstract
The ubiquity of DNA sequencing and the advent of medical imaging, electronic health records, and “omics” technologies have produced a deluge of data. Making meaning of those data—creating scientific knowledge and useful clinical information—will vastly exceed the capacity of even the largest institutions. Data must be shared to achieve the promises of genomic science and precision medicine.
National and international public–private partnerships, consortia, and other initiatives are being formed to collect and share data on a large scale. Many aspire to build, and some predict that the profusion of data will contribute to the creation of a medical information commons—a networked environment in which diverse sources of health, medical, and genomic data on large populations become widely shared resources. The ideal of an individual-centered information commons was a leitmotif of the 2011 National Academies report on precision medicine (National Research Council 2011). Many of those goals are embedded in large-scale efforts around the world, including the Precision Medicine Initiative and “cancer moonshot” in the United States, creating an ecosystem of medical information commons (Collins and Varmus 2015; Biden 2016). For example, the National Cancer Institute has established a Genomic Data Commons, and the NIH Commons is part of NIH's Big Data to Knowledge Initiative. The Wellcome Trust, international science funders, and many national funders have long supported data structures that constitute a nascent scientific commons, which is now broadening into clinical applications. Success of these initiatives depends on policies and practices that promote data sharing, are consistent with international standards, address barriers to participation, and attend to the ethical, legal, and social issues that arise when sharing medical data about individuals.
Genomics has strong precedents for broad data sharing and open science, most notably the Bermuda Principles of 1996, now two decades old. However, the very success of the Human Genome Project (HGP) has created more complex challenges—well beyond Bermuda—that will have to be addressed to achieve the aspiration of a global information commons. The design principles of data access and transparency can help achieve this goal by preserving the value of open science while protecting the rights and interests of individuals who contribute their data as well as institutions that generate, use, and share those data.
The Bermuda precedent
In February 1996, representatives from the major DNA sequencing centers in five nations convened in Bermuda and agreed upon daily release of DNA sequence information by laboratories participating in the HGP. Prepublication, rapid disclosure was intended to solve very practical problems as the HGP shifted from mapping to sequencing: how to allocate the work and how to monitor the accuracy of sequencing at different laboratories (Reardon et al. 2016). The Bermuda Principles were also a commitment to open science, in the spirit of producing data that would be available to laboratories of all sizes throughout the world (Contreras 2011; García-Sancho 2012). The Principles are thus rightly regarded as a touchstone for open science and a precedent for subsequent data-sharing policies (Arias et al. 2015).
The Bermuda Principles were primarily focused on enabling high-throughput DNA sequencing laboratories to assemble a human reference sequence—a global public good. Reaching consensus and ensuring compliance with the Bermuda Principles was no mean feat; indeed, the daily release rules initially conflicted with national policies to allow privileged data access to domestic companies in Germany and Japan and could have been interpreted to deviate from policies in the United States, France, and United Kingdom. However, the funders for over 90% of the initial sequencing under the HGP (the US National Institutes of Health and Department of Energy, and UK's Wellcome Trust) supported the daily data-release rule. When Germany and Japan did not formally adopt the Principles, US project leaders sent letters that indicated noncompliance with the Bermuda Principles would lead to exclusion from the global genome-sequencing consortium (Reardon et al. 2016; KM Jones, RA Ankeny, and R Cook-Deegan, unpubl.). The Principles held, even if compliance could not be closely monitored and was never fully enforced. It took a concerted effort, but rapid and extensive data sharing was built into the DNA of the HGP. This mattered, as data sharing was one of the main features that distinguished the publicly funded HGP from the corporate genome-sequencing efforts of Celera and other firms, and it served as a precedent for sharing of many other kinds of data in biomedical research, from clinical data to genome-wide association data and beyond. The Bermuda Principles became an icon of effective open science.
As we enter the third decade since the Principles were enunciated, the legacy of Bermuda is evident in the amount and diversity of data proliferating around the globe, with explicit attention to sharing data. The Principles have had to be modified somewhat. The Wellcome Trust convened a meeting at Fort Lauderdale that broadened the open science ideal to other organisms but also acknowledged a difference between “community resource projects” and other research, for which the same rules for prepublication data release might not apply, but also cited the importance of attribution and fairness (Wellcome Trust 2003). The HapMap Consortium was aggressive in fostering data sharing and discouraging patents (The International HapMap Consortium 2003), but the ENCODE Project, because it was focused on functional elements, acknowledged that some discoveries might be the proper subject of patents (National Human Genome Research Institute 2000; Contreras 2011). A Toronto meeting broadened the principles of data sharing further to include “omics” data beyond genomic variation (The ENCODE Project Consortium 2004; Kellis et al. 2014). Modern data-sharing policies provide heightened protection for individuals whose DNA fuels research and genomic medicine, while continuing to embody the spirit of broadly sharing sequence data (Wellcome Trust Sanger Institute 2014; National Institutes of Health 2015). Yet the task of building a medical information commons in coming decades will be vastly more complicated than sharing DNA sequence contributing to a single human reference genome and sequencing model organisms. Establishing the Bermuda Principles was hard; building a medical information commons will be much harder.
Toto, we are not in Bermuda anymore
The Bermuda Principles applied to a relatively small number of laboratories in five countries, later joined by China in 1999. It was possible to get 50 people in a room to develop the rules with representation from each group that would be contributing data. Those laboratories sent data to DNA sequence databases in the US, Japan, and Europe that had already been exchanging their curated data for over a decade. The sources of DNA for the reference sequence had given informed consent for that purpose. Sequencing was performed on a set of reference samples, and the data were sequence reads assembled into contiguous sequences. It was assumed the data could be sufficiently de-identified so as to avoid issues with individual privacy and security. The work was funded by government and nonprofit research organizations focused on a shared scientific objective: a human reference sequence.
Today, the situation is quite different (see Table 1). Genomic data are highly complex, generated by public and private laboratories, for both research and clinical purposes, and they are most useful if they are further linked to many other kinds of data. Even the starting elements—sequence-based data—come in diverse formats at different levels of organization: microarray SNPs, raw sequence reads, assembled sequence data, variant call files, and large-scale insertions and deletions. In addition, the data are generated by different platform technologies and instrument suites. Moreover, data on sequence variants must be linked to many other kinds of data such as health records, exposures, and genealogy in more diverse contexts and by varied groups that are not all governed by the same rules, regulations, or incentives. The value of linked data and the inherent identifiability of sequence data make it impossible to guarantee privacy and raises new ethical and legal concerns about people from whom the data are derived.
Table 1.
Comparison of Bermuda 1996 to current genomic data-sharing policy
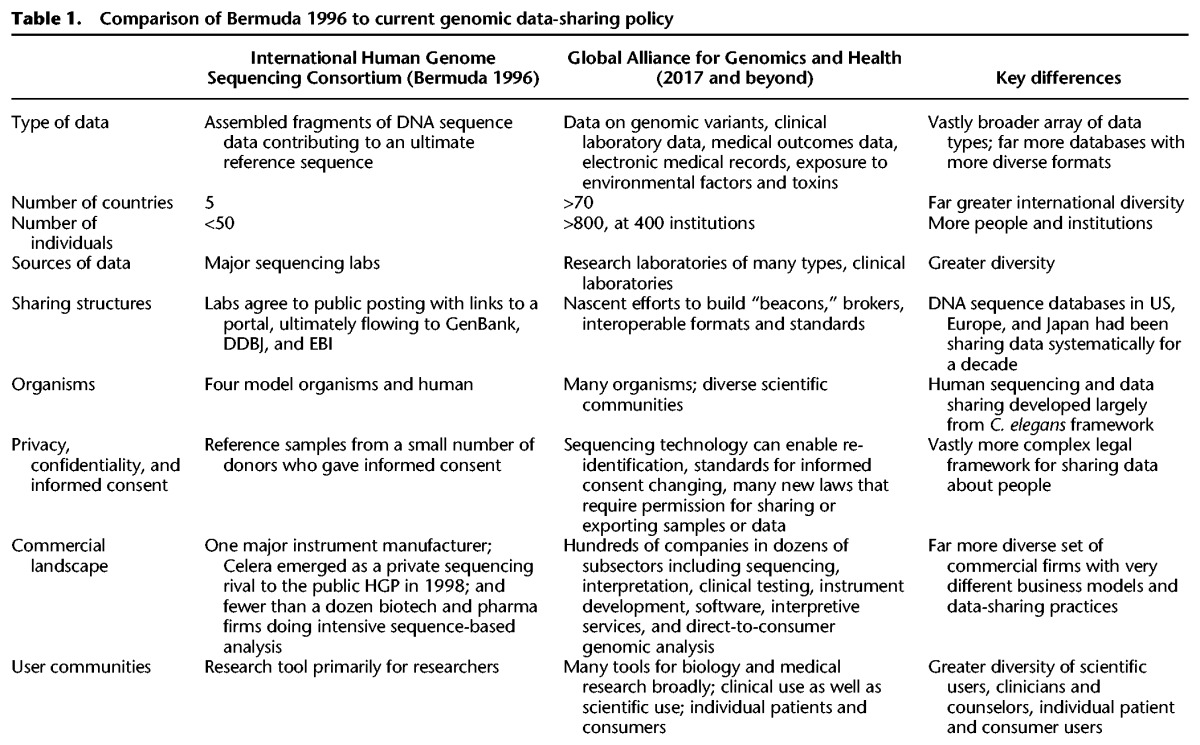
A unitary set of Bermuda-like Principles to share sequence reads will simply not suffice. The prospect of building a medical information commons faces far more daunting challenges, which can only be addressed by understanding the major differences in how data are collected, used, and shared now as compared to 20 years ago (see Box 1).
BOX 1. Medical information commons, 2017, compared to DNA sequence, Bermuda 1996.
More countries are involved. Data-intensive genomic initiatives are cropping up all over the globe. BGI in China has sequencing capacity comparable to Europe and North America. Major cohort studies are being established all over the globe. Yet, many nations have laws that constrain the export of genetic resources, sometimes requiring approval for the export of data. Countries differ in their informed consent standards and in the oversight authority of ethics review boards. Two special issues of the Journal of Law, Medicine and Ethics have analyses of 20 countries by 40 authors, a massive effort to assess the legal complexities (Rothstein and Knoppers 2015). Yet, this is still only a slice of the pie. The need for a global infrastructure, standards, norms, and practices was highlighted by a white paper in June 2013 that gave rise to the Global Alliance for Genomics and Health (Global Alliance for Genomics and Health 2013), which has proposed a global framework to enable data sharing to build a medical information commons (Global Alliance for Genomics and Health 2014).
Privacy, identifiability, and informed consent. Genomic data about people come with very real concerns about privacy, confidentiality, and need for consent for use of their data. It is no longer possible to completely “de-identify” sequence data (McGuire and Gibbs 2006), and there is tremendous value in being able to recontact individuals to collect longitudinal data and communicate clinically important findings. One central insight of the 2011 National Academies Precision Medicine report is that data should be organized so that the link to the person can be retained (National Research Council 2011), and this is recognized in the framework of the Global Alliance for Genomics and Health (Global Alliance for Genomics and Health 2014). Strong international protections against unauthorized re-identification and misuse of data, as well as innovative solutions to facilitate longitudinal bidirectional engagement of participants in the medical information commons are essential.
Data are clinical, not just scientific. Most new genomic variants are already being uncovered through clinical laboratories doing tests for medical decision making rather than for academic research. These data must comply with more stringent standards of analytic validity and are most valuable when linked to other health-related information. The medical information commons will draw on such data. Thousands of laboratories around the globe, of many different kinds, are generating data relevant to the exploration of human genomic variation. The key is to capture such data and make them available to improve inferences about health outcomes and risks, and for other practical uses. Most data generated by clinical laboratories, however, fail to feed back into a learning health care system that uses data to build evidence to guide medical decisions, with more data improving outcomes over time. The vast majority of data are used to make decisions about the individual; it is not wasted, but restricted data flow impedes the improvement of medical decision making. The American College of Medical Genetics and Genomics has recognized the importance of data sharing to foster clinical interpretation (American College of Medical Genetics and Genomics, Board of Directors 2017).
Commercial laboratories are highly diverse and some business models entail hoarding data. Genomic technology has bred many kinds of companies of varying sizes, carrying out many different functions (from making instruments to developing software, test kits, or supply reagents for genomic analysis; from providing genetic testing services to interpreting or storing data to performing genetic counseling) (Curnutte et al. 2014). Firms differ not only in what they do and how they do it, but also in how they expect to make money. Some firms are committed to open science and data sharing. For example, some genetic testing services that spun out of nonprofit or government laboratories to do rigorous genetic testing, such as GeneDx, have a strong norm of data sharing and tolerate but do not relish gene patents. InVitae, Ambry, ARUP, Counsyl, and Pathway Genomics all contribute data to ClinVar, the freely available public genomic variant database maintained by the National Library of Medicine (Rehm et al. 2015). PrecisionFDA contains data from many commercial laboratories. The largest US laboratory testing companies, LabCorp and Quest, are collaborating with the Universal Mutation Database in Paris (Ray 2015), and the Global Alliance for Genomics and Health is sponsoring the BRCA Challenge and BRCA Exchange, an open science flagship project to foster interpretation of genetic variants associated with cancer risk (BRCA Challenge 2015). Other companies, however, are pursuing strategies that foresee permanent, proprietary databases. The salient example is Myriad Genetics, which premises its strategy on keeping the results of its genetic tests proprietary and binds users of its portal to maintain variant data as trade secrets. The value of a proprietary database depends in part on how good public databases become. Any firm that uses public databases but does not contribute to them is, by definition, a free rider and will have a proprietary advantage. This was true for Celera relative to the HGP, but it is true in spades for clinical data for which the pathologies of free-riding are not restricted to a research database but affect medical decisions. As public data sources “catch up” to proprietary ones, however, proprietary advantage dissipates, and eventually the cost of maintaining a private database will exceed its marginal value. This sets up a classic multiplayer prisoners’ dilemma scenario for policy (Ostrom 1990). The system as a whole will be more efficient and effective with a robust commons, but individual firms relying on proprietary databases will have incentives at the margin to contribute little to the commons and even to disparage it. A robust commons actually discourages the formation of proprietary data siloes, but absence of a commons invites a proliferation of siloes.
Genomic data must link to many other kinds of data. Genomic data will be linked to imaging and clinical data, medical records, and laboratory results beyond genomics, as well as family history and genealogy. Where there was one set of collaborating DNA sequence databases when the Bermuda meetings took place 1996–1998, the data in the medical information commons will be in many kinds of databases. Some will be public, some private, and some hybrids.
A commons is not a free-for-all
A medical information commons will have stakeholders of at least three types: (1) individuals whose data populate the commons; (2) researchers, clinical laboratories, and other for-profit and not-for-profit institutions that contribute data; and (3) many different kinds of users. Forming a commons means finding rules to which stakeholders—donors, data generators, and users—can agree. The envisioned medical information commons is unlikely to be a single central database, but rather a linked network of resources. As the President's Council of Advisors on Science and Technology notes, interoperablility will be key (President's Council of Advisors on Science and Technology 2014).
A commons has rules, means of monitoring compliance, and sanctions for violating them (Ostrom 2000). Experience with genomic commons suggests that developing those capacities will take time and effort. The furor that greeted Longo and Drazen's New England Journal of Medicine editorial, which characterized clinical researchers’ concerns about “research parasites” feeding on open science, suggests that clinical researchers have work to do convincing bioinformatics users and others who do not wish to impede the benefits of unforeseen uses and users to strengthen incentives for contributors (Longo and Drazen 2016). Conflict is common in building a commons; it can be messy and contentious. Open debate is part of the process and should be welcomed. Commons take work to build, and that work is starting, through organizations like the Global Alliance for Genomics and Health.
Data access and transparent analysis as design principles
The challenges of creating a medical information commons are daunting but not a counsel for despair. Design principles can guide policy decisions for those funding science and paying for medical goods and services that can foster a learning system that builds a robust medical information commons. Three National Academies reports all point toward the importance of having access to data and the ability to reproduce the interpretation of the data, either for making scientific inferences (scientific replication) or for making clinical decisions (evidence-based medicine) (Cech et al. 2003; National Research Council 2011; Micheel et al. 2012). Two general principles stand out: data access and transparent analysis (DA-TA).
Data access
The philosophy and sociology of science do not always agree on details, but they do agree on some features that make scientific knowledge reliable, including the ability to replicate scientific work (Merton 1942; Popper 1959; Ziman 1978). If data are important for making scientific inference, they need to be shared so others can verify the findings, and those findings need to be subjected to “organized skepticism,” in the phrase of Robert Merton (Merton 1942). Another, equally important rationale for data sharing is that some findings are simply not possible without expanding beyond the scale of individual institutions, or even countries. An analogous principle of data sharing to enable clinical validation underlies evidence-based medicine (Cochrane 1972; Eddy 2011), for example, when formulating clinical practice guidelines (Field and Lohr 1992).
Access to data is not just for science and medicine, however; it is also emerging as an individual right (Lunshof et al. 2014). Access to data about oneself is becoming a norm, both for those participating in research and in being able to get laboratory data and access to medical records from clinical care. Access to underlying data is distinct from access to “research results” interpreted by researchers, which may come with great uncertainty about their clinical or other significance. Access to data does not necessarily imply ownership, because the data may also be retained by the laboratory or institution that generated it. Rather, it is a right to get the underlying data and to transfer those data to others, rights to “get” and “send.” Policies to codify such rights are beginning to be implemented. Until October 2014, for example, US laboratories certified under the Clinical Laboratory Improvement Amendments of 1988 (CLIA) were exempt from the requirement to share medical information with individuals under the Health Insurance Portability and Accountability Act (HIPAA). However, that laboratory exemption was removed, and individuals may now request the relevant record set of a laboratory test. Laboratories are obligated to provide the complete designated record set in the format specified by the requestor at a reasonable cost (DHHS Office of Civil Rights, http://www.hhs.gov/hipaa/for-professionals/privacy/guidance/access/index.html).
Data access is a core principle for both moral and practical reasons, but it has not yet become a design principle for genomic and other databases and resources. Access is essential to respect the rights and interests of individuals, to comply with norms of scientific evidence, and to lay the foundation for evidence-based medicine, but it is not built into the system and will not be until and unless policy incentives are in place. A commons needs rules and ways to enforce them; this will fall to organizations like the Global Alliance or other forms of governance that may yet form.
Transparent analysis
Access to data is necessary but not sufficient. The canons of science also require that the methods used to interpret data as well as other forms of metadata needed to make scientific inferences be shared. Assessment of clinical validity rests on a bedrock of being able to verify and validate the interpretation of clinical data. Both scientific and clinical practice thus entail sharing data in sufficient detail to enable independent replication and verification.
The DA-TA principles have real consequences. They can guide policies to ensure that science and precision medicine rest on firm ground. They can be translated into policies for funding science. NIH, for example, has required data-sharing policies to be explicit in grants over $500,000 for over a decade. NIH broadened its data-sharing policy in 2014 (National Institutes of Health 2014). The International Committee of Medical Journal Editors has long since agreed on the need for data access and transparent analysis (International Committee of Medical Journal Editors 2013), but the principles also apply to medical goods and services that are not published. The American Medical Association Resolution 460.971 “encourages laboratories to place all clinical variants and the data that were used to assess the clinical significance of these results, into the public domain which would allow appropriate interpretation and surveillance for these variations that can impact the public's health” (American Medical Association 2013). The January 2017 statement of the American College of Medical Genetics and Genomics observes that “no single provider, laboratory, medical center, state, or even individual country will typically possess sufficient knowledge to deliver the best care for patients in need of care” (American College of Medical Genetics, Board of Directors 2017). Criteria for certification of laboratories by the College of American Pathologists, or under CLIA, should ensure compliance with this principle. Insurers and health plans that pay for laboratory tests should ensure that clinical inferences can be independently verified, which, in the case of genomic variants, necessarily implies deposit of data into public databases and sharing interpretive algorithms. Recent guidelines from the US Food and Drug Administration are addressed to “publicly accessible databases of human genetic variants,” and the procedures for curation and proficiency testing presume sharing of data (Food and Drug Administration 2016).
Failure to implement policies that embody the DA-TA principles invites a “race to the bottom” of hoarding data for proprietary advantage in commercial medical services and in highly competitive academic research. Sharing costs money, takes time, and involves players outside the laboratory. It will always be cheaper to hoard than share. However, if all laboratories play by the same DA-TA rules, they can compete on other grounds, and the system will incrementally grow a medical information commons that all can draw upon to improve science and the practice of medicine.
Acknowledgments
Research was supported in part by grants from the National Human Genome Research Institute (R01 HG008918; P50 HG003391). R.C.-D. is a senior fellow at FasterCures, a Center of the Milken Institute.
Footnotes
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.216911.116.
References
- American College of Medical Genetics and Genomics, Board of Directors. 2017. Laboratory and clinical genomic data sharing is crucial to improving genetic health care: a position statement of the American College of Medical Genetics and Genomics. Genet Med 10.1038/gim.2016.196. [DOI] [PubMed] [Google Scholar]
- American Medical Association. 2013. D-460.971 Genome analysis and variant identification policy statement. https://searchpf.ama-assn.org/SearchML/searchDetails.action?uri=%2FAMADoc%2Fdirectives.xml-0-1601.xml. [Google Scholar]
- Arias JJ, Pham-Kanter G, Campbell EG. 2015. The growth and gaps of genetic data sharing policies in the United States. J Law Biosci 2: 56–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biden J. 2016. Beginning cancer ‘moonshot’ at Penn Medicine. Penn Medicine Almanac 62: 1. [Google Scholar]
- BRCA Challenge. 2015. Global Alliance for Genomics and Health. https://genomicsandhealth.org/work-products-demonstration-projects/brca-challenge-0. [Google Scholar]
- Cech TR, Eddy SR, Eisenberg D, Hersey K, Holtzman SH, Poste GH, Raikhel NV, Scheller RH, Singer DB, Waltham MC, et al. 2003. Sharing publication-related data and materials: responsibilities of authorship in the life sciences. Plant Physiol 132: 19–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochrane AL. 1972. Effectiveness and efficiency: random reflections on health services. Nuffield Provincial Hospitals Trust, London, UK. [Google Scholar]
- Collins FS, Varmus H. 2015. A new initiative on precision medicine. N Engl J Med 372: 793–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras JL. 2011. Bermuda's legacy: policy, patents, and the design of the genome commons. Minn J Law Sci Technol 12: 61–125. [Google Scholar]
- Curnutte MA, Frumovitz KL, Bollinger JM, McGuire AL, Kaufman DJ. 2014. Development of the clinical next-generation sequencing industry in a shifting policy climate. Nat Biotechnol 32: 980–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy DM. 2011. The origins of evidence-based medicine—a personal perspective. Virtual Mentor 13: 55–60. [DOI] [PubMed] [Google Scholar]
- The ENCODE Project Consortium. 2004. The ENCODE (ENCyclopedia Of DNA Elements) Project. Science 306: 636–640. [DOI] [PubMed] [Google Scholar]
- Field MJ, Lohr KN. 1992. Guidelines for clinical practice: from development to use. National Academies Press, Washington, DC. [PubMed] [Google Scholar]
- Food and Drug Administration, US Department of Health and Human Services, Center for Devices and Radiological Health, Office of In Vitro Diagnostics and Radiological Health, and Center for Biologics Evaluation and Research. 2016. Use of public human genetic variant databases to support clinical validity for next generation sequencing (NGS)-based in vitro diagnostics: draft guidance for stakeholders and Food and Drug Administration staff. http://www.fda.gov/downloads/MedicalDevices/DeviceRegulationandGuidance/GuidanceDocuments/UCM509837.pdf. [Google Scholar]
- García-Sancho M. 2012. Biology, computing, and the history of molecular sequencing: from proteins to DNA. Palgrave Macmillan UK, Basingstoke, UK. [Google Scholar]
- Global Alliance for Genomics and Health. 2013. White paper: creating a global alliance to enable responsible sharing of genomic and clinical data. http://genomicsandhealth.org/about-the-global-alliance/key-documents/white-paper-creating-global-alliance-enable-responsible-shar. [Google Scholar]
- Global Alliance for Genomics and Health. 2014. Framework for responsible sharing of genomic and health-related data. http://genomicsandhealth.org/about-the-global-alliance/key-documents/framework-responsible-sharing-genomic-and-health-related-data. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Committee of Medical Journal Editors. 2013. Recommendations for the conduct, reporting, editing, and publication of scholarly work in medical journals. ICMJE: 1–17. [PubMed] [Google Scholar]
- The International HapMap Consortium. 2003. The International HapMap Project. Nature 426: 789–796. [DOI] [PubMed] [Google Scholar]
- Kellis M, Wold B, Snyder MP, Bernstein BE, Kundaje A, Marinov GK, Ward LD, Birney E, Crawford GE, Dekker J, et al. 2014. Defining functional elements in the human genome. Proc Natl Acad Sci 111: 6131–6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longo DL, Drazen JM. 2016. Data sharing. N Engl J Med 374: 276–277. [DOI] [PubMed] [Google Scholar]
- Lunshof JE, Church GM, Prainsack B. 2014. Raw personal data: providing access. Science 343: 373–374. [DOI] [PubMed] [Google Scholar]
- McGuire AM, Gibbs RA. 2006. No longer de-identified. Science 312: 370–371. [DOI] [PubMed] [Google Scholar]
- Merton RK. 1942. The normative structure of science. In The sociology of science: theoretical and empirical investigations, pp. 267–278. University of Chicago Press, Chicago, IL. [Google Scholar]
- Micheel CM, Nass SJ, Omenn GS. 2012. Evolution of translational omics: lessons learned and the path forward. The National Academies Press, Washington, DC. [PubMed] [Google Scholar]
- National Human Genome Research Institute. 2000. ENCODE consortia data release, data use, and publication policies. https://www.genome.gov/10000910/policy-on-release-of-human-genomic-sequence-data-2000. [Google Scholar]
- National Institutes of Health, Department of Health and Human Services. 2014. Final NIH genomic data sharing policy. Federal Register 79: 51345–51354. [Google Scholar]
- National Institutes of Health. 2015. NIH security best practices for controlled-access data subject to the NIH Genomic Data Sharing (GDS) policy. http://www.ncbi.nlm.nih.gov/projects/gap/pdf/dbgap_2b_security_procedures.pdf. [Google Scholar]
- National Research Council. 2011. Toward precision medicine: building a knowledge network for biomedical research and a new taxonomy of disease. The National Academies Press, Washington, DC. [PubMed] [Google Scholar]
- Ostrom E. 1990. Governing the commons: the evolution of institutions for collective action. Cambridge University Press, New York. [Google Scholar]
- Ostrom E. 2000. Collective action and the evolution of social norms. J Econ Perspect 14: 137–158. [Google Scholar]
- Popper KR. 1959. The logic of scientific discovery. Genomics 83: 349–360. [Google Scholar]
- President's Council of Advisors on Science and Technology. 2014. Better health care and lower costs: accelerating improvement through systems engineering. https://obamawhitehouse.archives.gov/sites/default/files/microsites/ostp/systems_engineering_and_healthcare.pdf. [Google Scholar]
- Ray T. 2015. With BRCA Share, Quest hopes to provide field with variant database to support clinical decisions. GenomeWeb. https://www.genomeweb.com/business-news/brca-share-quest-hopes-provide-field-variant-database-support-clinical-decisions. [Google Scholar]
- Reardon J, Ankeny RA, Bangham J, Darling KW, Hilgartner S, Jones KM, Shapiro B, Stevens H. 2000. The genomic open workshop group. Bermuda 2.0: reflections from Santa Cruz. https://academic.oup.com/gigascience/article/doi/10.1093/gigascience/giw003/2756884/Bermuda-2-0-reflections-from-Santa-Cruz. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehm HR, Berg JS, Brooks LD, Bustamante CD, Evans JP, Landrum MJ, Ledbetter DH, Maglott DR, Martin CL, Nussbaum RL, et al. 2015. ClinGen—the Clinical Genome Resource. N Engl J Med 372: 2235–2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein MA, Knoppers BM. 2015. INTRODUCTION: harmonizing privacy laws to enable international biobank research. J Law Med Ethics 43: 673–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellcome Trust. 2003. Sharing data from large-scale biological research projects: a system of tripartite responsibility. Report of a meeting organized by the Wellcome Trust, January 14–15, 2003, Fort Lauderdale, FL http://www.sanger.ac.uk/datasharing/docs/fortlauderdalereport.pdf [Google Scholar]
- Wellcome Trust Sanger Institute. 2014. Data sharing policy. http://www.sanger.ac.uk/legal/assets/policy-and-guidelines.pdf. [Google Scholar]
- Ziman JM. 1978. Reliable knowledge: an exploration of the grounds for belief in science. Cambridge University Press, Cambridge, UK. [Google Scholar]


