Abstract
Maintenance of chromatin homeostasis involves proper delivery of histone variants to the genome. The interplay between different chaperones regulating the supply of histone variants to distinct chromatin domains remains largely undeciphered. We report a role of promyelocytic leukemia (PML) protein in the routing of histone variant H3.3 to chromatin and in the organization of megabase-size heterochromatic PML-associated domains that we call PADs. Loss of PML alters the heterochromatic state of PADs by shifting the histone H3 methylation balance from K9me3 to K27me3. Loss of PML impairs deposition of H3.3 by ATRX and DAXX in PADs but preserves the H3.3 loading function of HIRA in these regions. Our results unveil an unappreciated role of PML in the large-scale organization of chromatin and demonstrate a PML-dependent role of ATRX/DAXX in the deposition of H3.3 in PADs. Our data suggest that H3.3 loading by HIRA and ATRX-dependent H3K27 trimethylation constitute mechanisms ensuring maintenance of heterochromatin when the integrity of these domains is compromised.
In the eukaryotic nucleus, chromatin is organized in domains distinguishable by their composition in post-translational histone modifications and histone variants. Proper routing and delivery of histone variants, including histone H3 variant H3.3, is essential for chromatin homeostasis (Chang et al. 2013; Ivanauskiene et al. 2014; Rapkin et al. 2015). Site-specific loading of H3.3 on chromatin is mediated by two major histone chaperones. Histone cell cycle regulator (HIRA) deposits H3.3 in transcribed regions (Goldberg et al. 2010; Pchelintsev et al. 2013), at sites of DNA damage (Adam et al. 2013), and in open chromatin areas (Ray-Gallet et al. 2011; Schneiderman et al. 2012). In contrast, the death domain–associated protein (DAXX) and alpha thalassemia/mental retardation syndrome X-linked (ATRX) complex loads H3.3 on telomeric and pericentric heterochromatin and in repeat regions (Drane et al. 2010; Goldberg et al. 2010; Wong et al. 2010; Eustermann et al. 2011; Chang et al. 2013).
The promyelocytic leukemia (PML) protein is a tumor suppressor initially found in acute promyelocytic leukemia as a fusion protein with retinoic acid receptor alpha (Lallemand-Breitenbach and de The 2010). PML is the main component of PML nuclear bodies, which is shown to play a key role in the interplay between nonnucleosomal H3.3 and H3.3 chaperones before the deposition of H3.3 in chromatin (Chang et al. 2013; Delbarre et al. 2013). PML bodies consist of a shell of PML protein that encloses up to more than 100 other proteins (Bernardi and Pandolfi 2007; de The et al. 2012), including newly synthesized H3.3 (Delbarre et al. 2013) and the H3.3 chaperones ATRX, DAXX (Ishov et al. 1999), HIRA (Delbarre et al. 2013), and DEK (Ivanauskiene et al. 2014). Heterogeneity in PML body composition may account for the variety of processes implicating PML, including tumor suppression, genome stability, transcription, and chromatin organization (Bernardi and Pandolfi 2007; Kumar et al. 2007; Hoemme et al. 2008; Torok et al. 2009; de The et al. 2012).
Although they seem to be devoid of DNA, PML bodies have been found to associate with transcribed regions (Bernardi and Pandolfi 2007), linking PML to gene activity. In contrast, PML bodies can also associate with transcriptional repressors such as histone deacetylases (Khan et al. 2001), CBX3 (Seeler et al. 1998; Ivanauskiene et al. 2014), and the H3K9 methyltransferase SETDB1 (Cho et al. 2011). This implicates PML not only in gene silencing but also as potentially a regulator of epigenetic states. Consistent with this notion, electron spectroscopic studies have identified PML bodies surrounded by decondensed and condensed chromatin (Torok et al. 2009), and fluorescence in situ hybridization (FISH) (Shiels et al. 2001), immuno-TRAP labeling (Ching et al. 2013), and chromatin immunoprecipitation (ChIP) (Kumar et al. 2007; Hoemme et al. 2008) reveal associations of PML with specific loci.
The ability of PML to associate with defined areas of the genome is supported by its interaction with telomeres in mouse embryonic stem (ES) cells, where it enables ATRX-mediated H3.3 deposition and maintenance of heterochromatin (Wong et al. 2009, 2010; Chang et al. 2013; Udugama et al. 2015). Cancer cells with an alternative lengthening of telomere (ALT) phenotype also anchor a subset of PML bodies at telomeres (Henson et al. 2005). In human somatic cells, a pool of H3.3 is recruited to PML bodies by DAXX before deposition into chromatin (Delbarre et al. 2013), including pericentric heterochromatin (Corpet et al. 2014). Thus, whether or not it is part of nuclear bodies, PML may organize chromatin domains containing H3.3. By using biochemical, imaging, and genomics approaches, we investigated whether PML also plays a role in chromatin organization on a broader scale.
Results
A pool of H3.3 relocalizes from a nuclease-sensitive to a nuclease-resistant chromatin compartment in the absence of PML
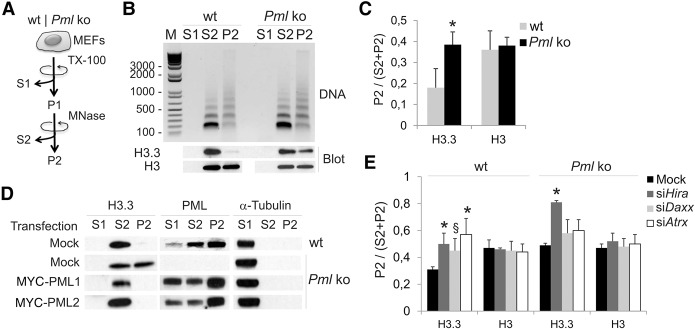
We investigated the role of PML in the deposition of H3.3 in chromatin using wild-type (Pml+/+; wt) and Pml knock-out (Pml−/−; ko) mouse embryonic fibroblasts (MEFs). We first established that levels of H3.3, HIRA, DAXX, and ATRX are similar in wt and Pml ko cells (Supplemental Fig. 1A) and verified the specificity of the anti-H3.3 antibody used here (Supplemental Fig. 1B). We then examined the distribution of endogenous H3.3 between chromatin compartments sensitive or refractory to micrococcal nuclease (MNase) digestion, which reflects their degree of compaction (Fig. 1A). Whereas in wt cells H3.3 is mainly found in an MNase-soluble fraction (S2), in Pml ko cells H3.3 is also detected in an MNase-resistant (P2) fraction (P < 0.05) (Fig. 1B,C). Since partitioning of total H3 between both fractions is not affected (Fig. 1B,C), we conclude that H3.3 is enriched in MNase-resistant chromatin in the absence of PML. This is confirmed by restoration of the H3.3 pool in S2 after introduction of the mouse PML isoforms PML-1 or PML-2 in ko cells (Fig. 1D). We also note that PML is found in the S2 fraction (Fig. 1D), indicating an association with chromatin. These results suggest that PML regulates the balance of H3.3 incorporation between loose and compact chromatin compartments.
Figure 1.
Loss of PML partitions histone H3.3 into a compact micrococcal nuclease (MNase)–resistant chromatin compartment. (A) MEF fractionation into a cytosolic fraction (S1), a Triton X-100 (0.5%)-insoluble/MNase (0.5 U/µg DNA)-soluble fraction (S2), and a Triton X-100/MNase-insoluble fraction (P2). (B) Nucleosome laddering (DNA; top) and Western blot (bottom) of endogenous H3.3 and total H3 in each fraction, from wt and Pml ko MEFs. (C) Proportions of H3.3 and total H3 in P2; (*) P < 0.05 relative to wt; paired t-tests. (D) Distribution of H3.3 and PML in S1, S2, and P2 from mock-transfected wt and Pml ko MEFs and from ko MEFs expressing MYC-PML1 or MYC-PML2 (full-length PML); alpha-tubulin was used as loading control for S1. (E) Proportions of H3.3 and total H3 in P2 after knockdown of Hira, Daxx, or Atrx; mean ± SD, three experiments; (*) P = 0.03 (siHira wt); 0.04 (siAtrx wt); 1.4 × 10−5 (siHira ko); (§) P = 0.06 (siDaxx, wt); paired t-tests relative to mock control.
We next asked whether HIRA, DAXX, or ATRX would mediate this H3.3 redistribution. In wt MEFs, knock-down of Hira, Atrx, or Daxx by siRNA (Supplemental Fig. 1C) enhances accumulation of H3.3 (but not total H3) in P2, phenocopying the effect of PML loss (Fig. 1E). In Pml ko cells, none of the knock-downs reduce H3.3 partitioning in P2 (Fig. 1E). In fact, depletion of HIRA, but not ATRX or DAXX, further enhances H3.3 partitioning in P2 seen in the sole absence of PML (P = 1.4 × 10−5; paired t-tests). Thus, HIRA favors H3.3 incorporation in MNase-accessible chromatin independently of PML, whereas the involvement of the ATRX/DAXX complex in this process is PML dependent. This suggests that PML favors H3.3 loading in open chromatin regions or, alternatively, restricts H3.3 deposition in compact domains.
Absence of PML accelerates H3.3 deposition into chromatin
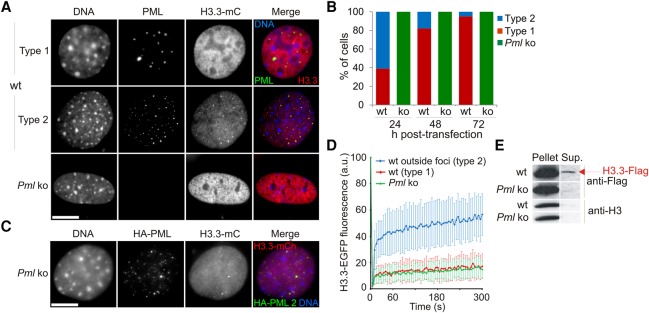
To address the mechanisms by which PML modulates H3.3 deposition, we expressed epitope-tagged forms of H3.3 in wt and Pml ko MEFs. Transient expression of fluorescently tagged H3.3 is well suited for imaging studies of H3.3 incorporation (Delbarre et al. 2013). Time-course imaging shows that as in human cells, H3.3-mCherry is detected at PML bodies (“type 2” cells) in wt MEFs and is gradually incorporated into chromatin (“type 1” cells) (Fig. 2A,B). At PML bodies, at least a fraction of H3.3 resists Triton X-100 solubilization but is extractable with DNase I, suggesting association with chromatin (Supplemental Fig. 2A). H3.3 recruitment to PML bodies is dynamic, as shown by fluorescence recovery after photobleaching (FRAP) (Supplemental Fig. 2B–D), and is DAXX dependent (Supplemental Fig. 2E).
Figure 2.
Loss of PML accelerates deposition of epitope-tagged H3.3 into chromatin. (A) Distribution of H3.3-mCherry (mC) in wt and Pml ko MEFs 24 h after H3.3-mC transfection. Cells were labeled using anti-PML antibodies and DNA stained with DAPI. Bar, 10 µm. (B) Proportions of cells of “type 1” and “type 2” over time after H3.3-mC transfection; no “type 2” Pml ko cells are detected at any time point. (C) Expression of HA-PML (full-length isoform PML2) in Pml ko MEFs restores H3.3-mC targeting to PML bodies. Bar, 10 µm. (D) FRAP analysis of H3.3-EGFP in wt and Pml ko cells; mean ± SD for 19–33 cells of each indicated type. See Supplemental Figure 2C for visualization of photobleached areas. (E) Western blot of H3.3-Flag and endogenous H3 in a 1% Triton X-100 soluble (Sup.) and insoluble (Pellet) fraction in wt and Pml ko MEFs.
In Pml ko MEFs, H3.3 is in contrast invariably detected in chromatin (Fig. 2A,B), and reintroduction of PML restores H3.3 detection at exogenous PML bodies (Fig. 2C). In addition, a SNAP-tagged H3.3 (Supplemental Fig. 3A,B) accumulates at PML bodies within 3 h after synthesis in wt cells, while it is already detectable in chromatin in Pml ko cells (Supplemental Fig. 3C). Thus, PML regulates H3.3 routing to chromatin by diverting a pool of H3.3 and restricting its deposition into chromatin.
To investigate the role of PML on global deposition of H3.3 in chromatin, we examined by FRAP the mobility of H3.3-EGFP outside PML foci (Fig. 2D; Supplemental Fig. 2C,D). In type 1 wt cells and in Pml ko cells, FRAP shows only ∼15% H3.3-EGFP fluorescence recovery (Fig. 2D; red and green lines), reflecting chromatin incorporation (Delbarre et al. 2013). In contrast, H3.3-EGFP in type 2 wt cells is significantly more mobile (P < 0.05; paired t-tests) (Fig. 2D; blue line). This rapid fluorescence recovery (>25% within 5 sec of photobleaching) reflects a diffusible nucleoplasmic pool of H3.3-EGFP. This pool is not detected by FRAP in Pml ko cells (Fig. 2D), and corroborating this observation, detergent extraction reveals a soluble fraction of epitope-tagged H3.3 only in wt cells (Fig. 2E). These results suggest an effect of PML on the deposition of H3.3, whether H3.3 is associated with PML bodies or not. We do not observe a difference by FRAP in the mobility of the canonical histone H3.1-EGFP in Pml ko versus wt cells (Supplemental Fig. 2F), indicating that only deposition of H3.3 is affected. Our biochemical and imaging data therefore suggest a regulatory role of PML in the deposition of H3.3 in chromatin by refraining incorporation into compact heterochromatic domains.
PML associates with heterochromatic domains not necessarily associated with PML bodies
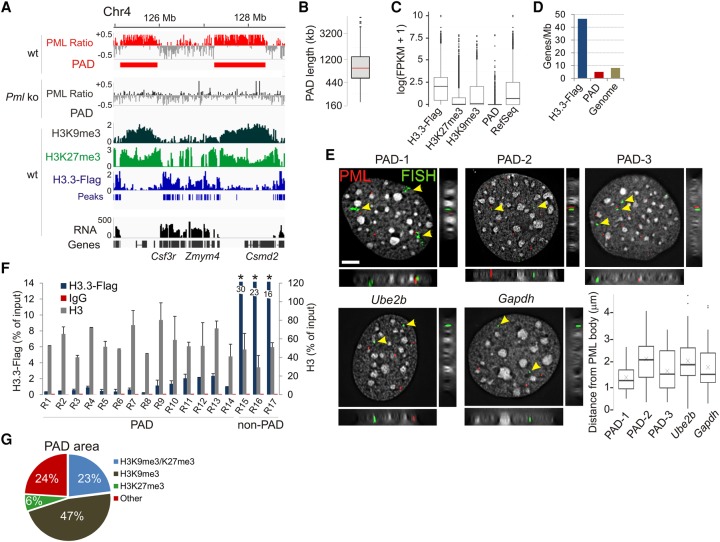
We next took a genomic approach to determine where in the genome PML would influence H3.3 deposition. By using anti-PML antibodies, we mapped by ChIP sequencing (ChIP-seq) the genome-wide association of endogenous PML with chromatin in MEFs. Detection of PML in a chromatin preparation for ChIP (Supplemental Fig. 4A) is consistent with PML fractionation in MNase-soluble chromatin (see Fig. 1D). This PML fraction is, however, not necessarily in the form of PML bodies, which are resistant to detergent and nuclease extraction (Supplemental Fig. 2A). In wt MEFs, ChIP-seq reveals megabase-sized regions associated with PML (Fig. 3A; Supplemental Fig. 4B). In contrast, both PML ChIP-seq and ChIP-qPCR data from Pml ko MEFs show no significant PML enrichment (Fig. 3A; Supplemental Fig. 4B,C). By using the Enriched Domain Detector algorithm (Lund et al. 2014), we identify in wt MEFs 484 domains enriched in PML, which we call PML-associated domains (PADs) (Fig. 3A; Supplemental Fig. 4B). We find that PADs encompass a total of 515 Mb, have a median length of 0.8 Mb, are gene-poor, and contain mostly inactive genes (Fig. 3B–D; Supplemental Tables 1, 2).
Figure 3.
PML associates with large heterochromatin domains. (A) ChIP-seq profiles of PML in wt and Pml ko MEFs and of H3K9me3, H3K27me3, and H3.3-Flag in wt MEFs; data are shown as log(ChIP/input) ratios. RNA-seq counts (FPKM) are shown. (B) PAD length (median, 0.8 Mb). (C) Expression levels of genes enriched in H3.3-Flag, H3K27me3, and H3K9me3; genes in PADs; and of all RefSeq genes in wt MEFs. (D) Gene density within H3.3-Flag peaks, in PADs, and in the whole genome. (E) Immuno-FISH analysis of PML (showing PML bodies; immunolabeling, red signal) and PADs (green signal; arrows; FISH probes). Ube2b and Gapdh are shown as genes outside PADs. Planar and orthogonal views are shown. Bar, 5 µm. Graph indicates distribution of FISH probe distances to nearest PML body (about 60 FISH signals analyzed). (F) ChIP-qPCR of H3.3-Flag and total H3 in indicated PAD and non-PAD regions in wt MEFs. (*) P < 0.01 relative to H3.3-Flag at all other sites; two-tailed t-tests; mean ± SD, three experiments. See Supplemental Table 4 for position of amplicons. (G) PAD area coverage by H3K9me3 and/or H3K27me3 peaks in wt cells.
To assess the relationship between PADs and PML bodies, we carried out FISH using probes designed in PADs, coupled to PML immunofluorescence. Immuno-FISH results show that FISH probes overlaying PADs do not necessarily localize in vicinity to PML bodies (Fig. 3E) and are not closer to PML bodies than probes covering genes not associated with PML (Ube2b, Gapdh; P > 0.02, Wilcoxon tests) (Fig. 3E). This confirms that immunoprecipitated PML in our ChIP experiments is predominantly not localized at PML bodies. Interaction of PML with chromatin detected here therefore mainly occurs from a pool of “nonbody” PML, providing an explanation as to how H3.3 dynamics and incorporation can be affected by PML loss, also outside regions interacting with PML bodies.
PADs are domains of low-level H3.3 incorporation
To assess the relationship between PADs and H3.3 deposition, we expressed H3.3-Flag for 24 h in wt MEFs and profiled its enrichment by ChIP-seq using anti-Flag antibodies. We identify 28,213 peaks of H3.3-Flag mainly in transcriptionally active gene-rich regions (Fig. 3A,C,D). In contrast, PADs strikingly display low levels of H3.3-Flag incorporation (Fig. 3A). Ruling out an epitope tag effect, ChIP-seq profiles of H3.3-Flag were validated by H3.3-Flag ChIP-qPCR (Fig. 3F), ChIP-seq of HA-tagged H3.3, and comparison with ChIP-seq profiles of endogenous H3.3 in mouse ES cells (Supplemental Fig. 4D,E; Elsasser et al. 2015).
The repressed state of PADs suggests a heterochromatic composition. Indeed, we find that PADs are enriched in H3K9me3 and H3K27me3 (Fig. 3A,G; Supplemental Fig. 5A), and the H3K9 methyltransferase SETDB1 is detected at PML bodies (Supplemental Fig. 5B; Cho et al. 2011). We then assessed PML association with DNA repeats (Day et al. 2010) that are maintained in a repressed state by H3K9me3 (Elsasser et al. 2015; Sadic et al. 2015) in a manner that involves H3.3 (Elsasser et al. 2015). We find that PML is detected in only 40 out of 952 mouse repeats (Supplemental Table 3); thus, PADs are not enriched in repeat elements in MEFs. We conclude that PML associates with heterochromatic regions of low-level H3.3 incorporation; this is consistent with the low turnover of H3.3 in heterochromatin (Huang and Zhu 2014).
In the absence of PML, H3.3 is targeted to PADs
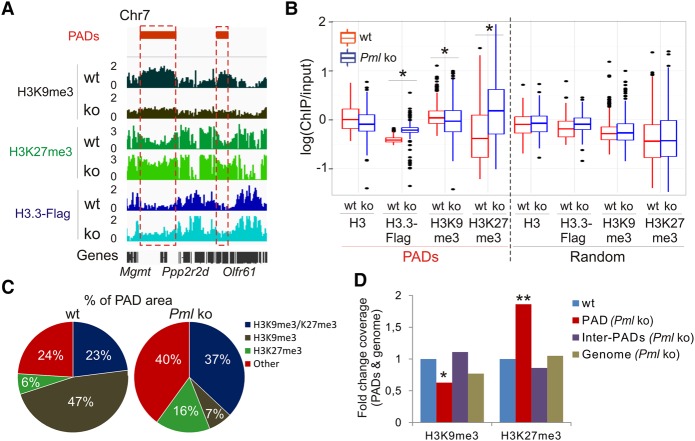
A view of PML withholding H3.3 deposition in PADs suggests that a loss of PML would enable H3.3 loading at these sites. First, we confirm H3.3 targeting to specific DNA repeat classes (Elsasser et al. 2015), albeit at similar levels in wt and Pml ko cells (Supplemental Table 3), suggesting that PML is not involved in loading H3.3 in these repeats. Further, we note that while H3.3-Flag is mostly detected in common regions in Pml ko and wt MEFs, it is also strikingly targeted to PADs in ko cells (P = 2.2 × 10−16, Wilcoxon test) (Fig. 4A,B; Supplemental Fig. 5C,D). This does not result from increased nucleosome density because total H3 levels are unaltered, and repeating the analysis on randomized PADs abolishes this H3.3 enrichment (Fig. 4B). These results strengthen the view that PML restricts but not fully abrogates H3.3 deposition in PADs.
Figure 4.
The heterochromatic state of PADs is remodeled in the absence of PML. (A) Profiles of H3K9me3, H3K27me3, and H3.3-Flag inside and outside PADs in wt and Pml ko MEFs. (B) Median enrichment level of indicated marks in PADs and in randomized PADs. (*) P < 2.2 × 10−16; Wilcoxon tests. (C) Proportions of PAD area enriched in H3K9me3 and H3K27me3. (D) Fold change of H3K9me3 and H3K27me3 coverage in PADs (red bars), between PADs (purple bars), and in the whole genome (brown bars) in Pml ko versus wt cells. Reference (wt) coverage is set to one (blue bars). (*) P < 0.01, (**) P < 0.001 relative to wt; one-sample t-tests.
Loss of PML switches the heterochromatic states of PADs
Deposition of H3.3 in PADs in Pml ko cells raises the possibility that the heterochromatic state of PADs may be affected. Indeed, we find that in Pml ko cells, both H3K9me3 level (P < 5.3 × 10−8, Wilcoxon test) (Fig. 4A,B) and peak coverage (P < 10−4, one sample t-test) (Fig. 4C,D) are reduced within PADs but not between PADs. In contrast, H3K27me3 level (P < 2.2 × 10−16) (Fig. 4A,B) and coverage (P < 10−4) (Fig. 4C,D) are increased in PADs. These findings were validated by ChIP-qPCR (Supplemental Fig. 5A) and by PAD randomization (Fig. 4B). Consistent with these changes, most genes in PADs remain repressed in ko cells (Supplemental Fig. 5E,F). Thus, in the absence of PML, the heterochromatic landscape is rearranged in regions no longer associated with PML. Moreover, the shift in H3 methylation from K9 to K27 suggests the importance of PML in maintaining a repressed heterochromatic state in these domains.
In the absence of PML, HIRA loads H3.3 in PAD regions but is not responsible for the increase in H3K27me3
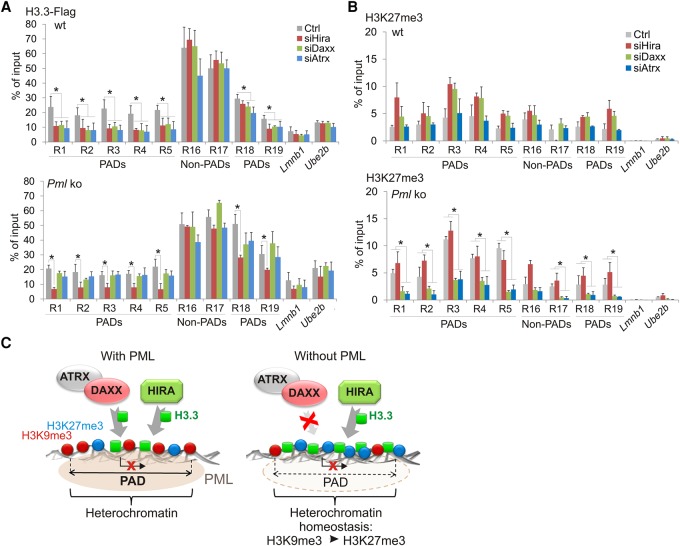
We next determined, using knock-down and ChIP approaches, which H3.3 chaperone might be responsible for H3.3 deposition in PAD regions in the absence of PML. Figure 5A reveals in wt MEFs a significant impairment of H3.3-Flag levels in PADs after knock-down of Hira, Atrx, or Daxx (R1-5, R18-19), while no effect is seen in non-PAD regions (R16-17). Lower H3.3 deposition in PADs after knock-down of Atrx/Daxx is consistent with the H3K9me3 enrichment of PADs and with the H3K9me3-binding property of ATRX (Eustermann et al. 2011; Iwase et al. 2011), while the effect of HIRA depletion may be explained by the deposition of H3.3 at genes in PADs (see Fig. 4A). Thus, both the ATRX/DAXX and HIRA complexes are capable of H3.3 deposition in PADs in wt cells, in agreement with the genic and intergenic content of these domains. In Pml ko cells, however, the effect of ATRX or DAXX depletion on H3.3-Flag levels in PAD regions is strikingly abrogated (Fig. 5A), This suggests that the H3.3 loading function of the ATRX/DAXX complex in PADs requires PML. Nevertheless, HIRA depletion still hampers H3.3-Flag deposition in these regions (Fig. 5A), indicating that HIRA retains its H3.3 loading function in the absence of PML.
Figure 5.
Absence of PML promotes H3.3 loading by HIRA in PADs. (A) ChIP-qPCR of H3.3-Flag in PAD and non-PAD sites in wt and Pml ko MEFs after Hira, Daxx, or Atrx knock-down. (B) ChIP-qPCR of H3K27me3 under the same conditions as in A. All ChIPs: mean ± SD of three to five experiments. (*) P < 0.02; Fisher's exact tests. Position of amplicons is given in Supplemental Table 4. (C) PML organizes heterochromatin domains and modulates H3.3 loading. (Left) In wt cells, PML associates with heterochromatic H3K9me3-rich domains (PADs). Deposition of H3.3 in PADs is low relative to inter-PAD regions, and can be mediated by HIRA, DAXX, or ATRX. (Right) In the absence of PML, H3K9me3 levels are reduced in PADs. The ATRX/DAXX complex loses is ability to load H3.3 in these regions. In contrast, HIRA retains its ability to deposit H3.3 in PADs, supporting the idea of a gap-filling mechanism (Ray-Gallet et al. 2011) established to preserve chromatin integrity. A heterochromatic state is maintained in PADs by an increase in H3K27me3 in a manner implicating the ATRX/DAXX complex, perhaps through the ability of ATRX to recruit the PRC2 complex to chromatin (Sarma et al. 2014). Our results suggest an interplay between the HIRA-H3.3 and ATRX/DAXX-Polycomb pathways to maintain heterochromatin homeostasis in domains where H3K9me3 is compromised in the absence of PML, with the purpose of keeping these regions transcriptionally silent.
To determine whether the increase in H3K27me3 reported above in PADs in Pml ko cells (see Fig. 4B) could be linked to the rise in H3.3 in these domains, we assessed the dependency of H3K27me3 levels on HIRA, which deposits H3.3 in PADs in the absence of PML. We find that HIRA depletion does not affect H3K27me3 levels in PADs in ko cells (Fig. 5B). Thus, the H3K27me3 increase in PADs is unlikely to result from a higher level of H3.3. In contrast, depletion of ATRX or DAXX reduces H3K27me3 levels within and outside PADs (P < 0.02) (Fig. 5B). Again, this is likely unrelated to H3.3 deposition, because knock-down of ATRX or DAXX in Pml ko cells does not affect H3.3 deposition (Fig. 5A). Rather, ATRX/DAXX may modulate H3K27me3 by a mechanism distinct from its chaperone activity. This finding notably agrees with evidence of ATRX being essential for targeting of the H3K27 methylating Polycomb repressor complex 2 (PRC2) to chromatin (Sarma et al. 2014).
Discussion
We identify a role of PML in the organization of broad heterochromatic domains that we call PML-associated domains, or PADs. We used a ChIP-seq approach, which is distinct from the immuno-TRAP technique developed to map genomic regions close to PML bodies, and differences between the two have been discussed (Ching et al. 2013). ChIP identifies regions associated with the PML protein, whether it is found in “bodies” or not. In contrast, the immuno-TRAP study focused on PML bodies; yet, the genomic regions identified in this assay may not necessarily interact with PML and would not be detected by ChIP. In addition, our immuno-FISH data indicate that PADs identified here are not necessarily associated with PML bodies. Interestingly, PML can also be detected in dense chromatin regions outside PML bodies (de The et al. 2012), consistent with an association with heterochromatin. These regions could correspond to PML covering megabase-size portions of the genome or to PML dynamically scanning broad genome areas. Both alternatives are plausible: By analogy, lamin-associated domains have been reported to encompass >25% of the genome, but this does not imply that lamins associate with such large genome fractions in single cells (Kind et al. 2015; Paulsen et al. 2017).
Our results lead to a model of organization of chromatin domains involving a PML-ATRX-H3.3 axis (Fig. 5C). In wt cells, PML interacts with H3K9me3-rich domains where H3.3 can be incorporated, yet at low level, by HIRA, DAXX, or ATRX. FRAP and ChIP analyses suggest that PML restricts access of H3.3 to regions of low transcriptional activity, perhaps because of low nucleosome turnover (Huang and Zhu 2014). In the absence of PML, deposition of H3.3 in PADs may reflect higher nucleosome turnover. The ATRX/DAXX complex loses its H3.3 loading ability (Fig. 5C). This may be because ATRX/DAXX needs to associate with PML in PADs to function as an H3.3 chaperone or, alternatively, ATRX association with chromatin is decreased due to lower H3K9me3 levels in these regions. In contrast, HIRA retains its H3.3 loading capacity in PADs in the absence of PML, lending additional support to the idea of a gap-filling process delivering H3.3 to regions of altered chromatin integrity (Ray-Gallet et al. 2011; Adam et al. 2013; Ivanauskiene et al. 2014).
The loss of PML results in a remodeling of the chromatin state of PADs through a shift from H3K9 toward H3K27me3, maybe as a way to preserve heterochromatin integrity (Fig. 5C). A compensatory mechanism maintaining heterochromatin by a shift in the H3K9me3/K27me3 balance is not unlike that reported in instances of PRC2 recruitment to pericentric heterochromatin to counteract deficiencies in the H3K9 methyltransferase SUV39H1 (Saksouk et al. 2014). In ES cells, H3.3 is implicated in maintaining integrity of heterochromatin in repeat elements and at telomeres (Elsasser et al. 2015; Udugama et al. 2015) and is required for H3K27me3 and interaction of PRC2 with HIRA (Banaszynski et al. 2013). We show here that while H3.3 deposition in PADs in Pml ko cells is HIRA dependent, H3K27 trimethylation in these domains relies on the ATRX/DAXX complex. This argues against a contribution of neo-deposited H3.3 to H3K27 methylation at these sites.
Rather, ATRX/DAXX may be involved in recruiting PRC2 to PADs. Supporting this possibility, ATRX has been shown to direct PRC2 loading on chromatin on a broad scale (Sarma et al. 2014). The underlying mechanism remains to be elucidated but may entail an RNA mediator, since ATRX binds a wide range of RNAs. Alternatively, ATRX, while displaying weak chromatin remodeling activity, harbors ATPase activity that may remodel a cofactor for PRC2 to a conformation enabling PRC2 binding. These possibilities remain to be examined.
Clustering of functionally related genes in PADs may provide the right spatial conformation to coregulate expression. PADs notably include olfactory receptor gene clusters (including Olfr, Vmn1r, and Vmn2r genes) and nonclassical MHC class I genes that cooperate with the former in specific neuron subtypes in which they are expressed (Dulac and Torello 2003). This suggests that PML is involved in clustering functionally related genes. Interestingly, the SATB1-PML complex has been implicated in a chromatin loop conformation proposed to regulate expression of MHC class I genes (Kumar et al. 2007), and immuno-TRAP findings are compatible with a view of chromatin loops tethering loci near PML (Ching et al. 2013). These observations raise the hypothesis that PML may be involved in the topological organization of chromatin.
We ascribe a novel role of PML in organizing chromatin domains through two mechanisms. One involves the regulation of a pool of H3.3 available to its various chaperones prior to deposition into chromatin (Delbarre et al. 2013). The other, evidenced here, involves the interaction of PML with the genome to regulate site-specific H3.3 deposition and maintain heterochromatin integrity.
Methods
Cells and transfection
MEFs were derived from wt and Pml−/− mice. 129Sv Pml−/− mice (Wang et al. 1998) from the National Cancer Institute Repository were backcrossed to C57BL/6 mice for eight generations. wt controls were C57BL/6. MEFs were cultured in DMEM/F12/10% fetal calf serum. For ChIP and chromatin fractionation, cells were from confluent cultures. Cells were transfected by electroporation (Nucleofector, Lonza) using 5 µg plasmid or 200 pmol relevant siRNA per 3 × 106 cells. For expression of epitope-tagged H3.3 in siRNA-treated cells, a first siRNA transfection was followed 96 h later by a second transfection with both siRNA and the relevant H3.3 plasmid (Delbarre et al. 2013).
Antibodies
Antibodies to ATRX (sc-10078, immunofluorescence; sc-15408, blots), SETDB1 (sc-66884), and DAXX (sc-7152) were from Santa Cruz Biotechnology; PML (05-718), H3.3 (09-838), and H3K9me3 (05-1250, blots) from Millipore; H3 (ab1791) and HA (ab9110) from Abcam; H3K27me3 (pAb-069-050) and H3K9me3 (pAb-056-050) from Diagenode; and Flag (F1804) and alpha-tubulin (T5168) from Sigma. DyLight 549- (711-505-152), Cy3-, Cy2-, Dylight 488-, and HRP-conjugated antibodies were from Jackson ImmunoResearch Laboratories. Antibodies to SNAP-tag were from New England Biolabs. Antibodies to lamin A/C were from Brigitte Buendia. Antibodies to HIRA were from Peter Adams. For immunolabeling, primary antibodies were diluted 1:100 (DAXX, ATRX), 1:200 (PML, SNAP), and 1:1,000 (HA, lamin A/C); secondary antibodies were diluted 1:200 except DyLight 549 (1:800). For immunoblotting, antibodies were diluted as follows: ATRX, DAXX, HIRA, and H3.3 1:1000; H3 1:7000; alpha-tubulin 1:20,000; and HRP-conjugated 1:7000.
Plasmids and siRNAs
Plasmids encoding H3.3-mCherry and H3.3-EFGP were as previously described (Delbarre et al. 2010). Plasmids encoding HA-PML2, MYC-PML1, and MYC-PML2 were from Shu-Yong Lin (Xiamen University, China). The pH3.3-Flag 3× plasmid was made in two steps. H3.3 cDNA was amplified from pH3.3-EGFP (Delbarre et al. 2010). Primers contained an EcoRI site (5′ end of sense primer 5′-GCGAATTCATGGCACGTACCAAGCAAACAGC-3′) and a BglII site (5′ end of antisense primer 5′-ATAGATCTAAGGCCCGCTCGCCACGGATGCGT-3′). Products were digested with EcoRI and BglII and ligated into the EcoRI and BglII cloning sites of pFlag-CMV5 (Sigma) to make pH3.3-Flag. Primers containing two Flag cDNAs in tandem with a BglII site (5′ end of sense primer (5′-GCAGATCTATCCATAGCAGATTACAAGGACGACGATGACAAGTCGATCTCGGACTATAAGGATGACGACGACAAAGTCGACCG-3′) and a SalI site (5′ end of antisense primer 5′-CGGTCGACTTTGTCGTCGTCATCCTTATAGTCCGAGATCGACTTGTCATCGTCGTCCTTGTAATCTGCTATGGATAGATCTGC-3′) were annealed. The double-stranded DNA was digested with BglII and SalI and ligated into the BglII and SalI cloning sites of pH3.3-Flag.
siRNA oligonucleotides were from Invitrogen (MSS212902 (Atrx siRNA), MSS273927 (Daxx siRNA), and MSS205135 (Hira siRNA).
H3.3-SNAP labeling and immunofluorescence
MEFs were transfected with H3.3-SNAP and treated 24 h later as described in Supplemental Figure 3 (Ray-Gallet et al. 2011). After treatment, cells were processed for immunostaining with anti-PML or anti-SNAP antibodies and analyzed using the same acquisition parameters. Immunostaining was done after cell fixation with 3% paraformaldehyde and permeabilization with 0.1% Triton X-100/0.01% Tween 20/2% BSA. Images were captured at 100×/1.4 NA on a DeltaVision personalDV (Applied Precision) and analyzed using ImageJ 1.48v (National Institutes of Health).
FRAP
FRAP was done as previously described (Delbarre et al. 2013) 24 h after plasmid transfection. Bleaching was done within a ∼2-µm diameter area with a 405-nm laser, and fluorescence followed using a 488-nm laser. Images were taken every 5 sec. Fluorescence recovery was analyzed with corrections for photobleaching using ImageJ and data normalized as a percentage of prebleach fluorescence intensity. Statistics were done using unpaired Student's t-tests.
Cell fractionation
Cells were lysed for 15 min on ice with 1% Triton X-100 in 10 mM HEPES (pH 7.9), 10 mM KCl, 1.5 mM MgCl2, 1 mM DTT, 0.1 mM PMSF and protease inhibitors and sedimented at 20,000g (10 min, 4°C), and the insoluble fraction was dissolved in 1× Laemmli buffer at a 10,000 cell equivalent per microliter. Proteins of the soluble fraction were precipitated with methanol/chloroform and solubilized in 1× Laemmli buffer at a 10,000 cell-equivalent per microliter. For MNase digestion, cells were lysed (20 min, 4°C) on a shaker with 0.5% Triton X-100 in digestion buffer (250 mM sucrose, 50 mM Tris-HCl at pH 7.5, 5 mM MgCl2, 1 mM CaCl2, 5 mM Na-butyrate, 1 mM DTT, 0.1 mM PMSF, protease inhibitors). The lysate was centrifuged at 20,000g (10 min, 4°C) and the supernatant collected (S1). The pellet was washed, suspended in digestion buffer, and chromatin digested for 10 min at 37°C with MNase at 0.5 U/µg DNA. After centrifugation at 20,000g (10 min, 4°C), the supernatant (S2) was collected, and the pellet washed and dissolved in digestion buffer (P2). Twenty microliters was taken from S1, S2, and P2 for DNA isolation. Proteins were purified and solubilized in 1× Laemmli buffer at a 10,000 cell-equivalent per microliter for electrophoresis.
Western blotting
Proteins were resolved in 4%–20% or 10% SDS-PAGE gels and immunoblotted and detected as previously described (Delbarre et al. 2013).
Immuno-FISH
MEFs grown on glass coverslips were fixed with 3% paraformaldehyde for 15 min, washed 3 × 5 min in PBS, blocked, and permeabilized in PBS with 0.01% Tween-20, 2% BSA, and 0.1% Triton X-100 for 30 min. Cells were incubated for 45 min each with primary and secondary antibodies diluted in PBS with 0.01% Tween-20 and 2% BSA. Antibodies were anti-mouse PML (1:100; Millipore Mab3738) and donkey anti-mouse Alexa 594 (2.3 µg/mL; Invitrogen). Slides were washed 3 × 5 min in PBS with 0.01% Tween-20 and 2% BSA between and after the incubations. Immunostaining was fixed with 3% PFA for 10 min. Slides were washed 3× in PBS, 2 × 2 min in 2× SSC and 3 min in 2× SSC at 80°C. Slides were denatured for 20 min in 80°C in 70% deionized formamide (Ambion) in 2× SSC (pH 7.5). BAC FISH probe DNA (BacPac Resource Center) was labeled using a nick translation kit (Roche) and Biotin-16-dUTP (Roche).
Probes used were RP24-355G21 (Chr14:4997796–5168126; PAD-1), RP24-95N11 (Chr5:126000257–126123539; PAD-2), RP23-101019 (Chr7:22580017–22580829; PAD-3), RP24-147C4 (Chr6:125120297–125253818; Gapdh), and RPA24-340021 (Chr11:51908305–52087687; Ube2b). Per slide, 200 ng labeled probe was mixed with 8 µg mouse Cot-1 DNA (Invitrogen) and 30 µg salmon sperm DNA (Invitrogen) and precipitated. DNA pellet was dissolved in 11 µL hybridization mix (50% deionized formamide [Ambion], 2× SSC, 1% Tween 20, 10% dextran sulphate) for 20 min at 42°C and preannealed for 1 h at 37°C. Probes were denatured for 5 min at 80°C and preannealed for 15 min at 37°C. Ten microliters of probe was applied onto coverslips (22 × 22 mm), which were then mounted on a slide. Slides were hybridized overnight at 37°C. Slides were washed in 2× SSC (45°C for 2 min and then 3 × 5 min) and in 0.1× SSC (60°C for 4 × 4 min). Slides were blocked in 5% skim milk in 4× SSC for 15 min at 37°C and incubated for 30–60 min at 37°C with Avidin Alexa Fluor 488 conjugate (Invitrogen) at 1.7 µg/mL in blocking buffer. Slides were washed in 4× SSC with 0.1% Tween 20 for 3 × 5 min and incubated with biotinylated anti-Avidin D conjugate (goat; vector; 1.0 µg/mL in blocking buffer) for 30 min at 37°C. Slides were washed as above and incubated again with Avidin Alexa Fluor 488 conjugate as above. Slides were mounted with DAPI (0.2 µg/ml) in Dako fluorescent mounting medium and examined on a wide-field Olympus microscope. Images were processed with ImageJ (National Institutes of Health).
ChIP
ChIP of PML, H3.3-Flag, H3.3-HA, and modified histones was done as previously described (Lund et al. 2014) using chromatin from 3 × 106 cells (107 cells for ChIP-seq). Immunoprecipitation was done at 4°C using 2.5 µg anti-Flag, HA, H3K9me3, or H3K27me3 antibody (20 µg for ChIP-seq); ChIP DNA was purified and libraries prepared and sequenced on an Illumina HiSeq 2500. ChIP DNA was also used for qPCR (Supplemental Table 4) using SYBR green (BioRad), with 3 min at 95°C and 40 cycles of 30 sec at 95°C, 30 sec at 60°C, and 30 sec at 72°C.
ChIP-seq data processing
Scripts were written in Perl (Stajich et al. 2002) or R (R Core Team 2015), and graphs were plotted using ggplot2 in R (Supplemental Scripts 1–4; R Core Team 2015). ChIP-seq reads were mapped to mm10 using Bowtie v1.0.0 (Langmead et al. 2009). Mapped PML ChIP-seq reads were used to call peaks (PADs) using Enhanced Domain Detector (Supplemental Scripts 2, 3a,b; Lund et al. 2014). Significance testing was done by Monte Carlo simulation and P-values adjusted for multiple testing with FDR 0.01. H3.3-Flag, and H3.3-HA, and histone peaks were called using RSEG (Song and Smith 2011). Intersects and randomizations were computed using BEDTools v2.21.0 (Quinlan and Hall 2010). “Random PADs” were nonoverlapping regions of the same sizes as PADs distributed in a genome subtracted of PADs (Supplemental Scripts 4a,b). Browser files were generated from ChIP/input ratios for each of 1-kb bins with input normalized to ratio of total ChIP reads/total input reads (Supplemental Script 1).
Repeat analysis
ChIP-seq reads for ChIP and input were assigned to Repbase repeat classes and families using the Repeat Enrichment Estimator algorithm provided with the software (Day et al. 2010) after extraction from Repbase tracks of the UCSC Genome Browser. The application computes the maximum likelihood estimate fold enrichment in ChIP versus input for each repeat type. Maximum likelihood enrichment estimates for H3.3-Flag, H3, H3K9me3, and H3K27me3 in wt and Pml ko were computed using the R function “heatmap” (R Core Team 2015).
RNA sequencing
Total RNA was isolated using the Ambion TRIzol reagent RNA extraction kit from triplicate cell cultures. Libraries were prepared and paired-end sequenced on an Illumina NextSeq500. RNA-seq reads were processed using Tuxedo (Trapnell et al. 2010). TopHat (Trapnell et al. 2012) was used to align reads with no mismatch against mm10 applying Bowtie 2 (Langmead and Salzberg 2012) preset “-very sensitive”. Cufflinks and cuffdiff were run using default settings and bias correction in R (https://bioconductor.org/packages/release/bioc/html/cummeRbund.html). Genes with log(fold change) > 2 and α < 0.05 were considered differentially expressed. Gene ontology analysis was done using GOSlim (Ashburner et al. 2000).
Data viewing
Browser views are shown using Integrated Genomics Viewer (Robinson et al. 2011). Genes are from Illumina iGenomes with UCSC source for mm10. Plots were generated using ggplot2 or LSD libraries in R (R Core Team 2015).
Data access
Sequence data from this study have been submitted to the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE66364.
Supplementary Material
Acknowledgments
We thank Andrew Reiner for repeat analysis, Anita Sørensen and Núria Peñuelas Peñarroya for laboratory assistance, Brigitte Buendia (Institut Jacques Monod, Paris) for lamin A/C antibodies, Peter Adams (CR-UK Beatson Institute, Glasgow) for HIRA antibodies, and Shu-Yong Lin (Xiamen University, Fujian, China) for PML plasmids. This work was supported by The Research Council of Norway, Czech-Norwegian program CZ09 grant 7F14369, the Norwegian Cancer Society, South East Health Norway, and the University of Oslo.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.215830.116.
References
- Adam S, Polo SE, Almouzni G. 2013. Transcription recovery after DNA damage requires chromatin priming by the H3.3 histone chaperone HIRA. Cell 155: 94–106. [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. 2000. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25: 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banaszynski LA, Wen D, Dewell S, Whitcomb SJ, Lin M, Diaz N, Elsasser SJ, Chapgier A, Goldberg AD, Canaani E, et al. 2013. Hira-dependent histone H3.3 deposition facilitates PRC2 recruitment at developmental loci in ES cells. Cell 155: 107–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardi R, Pandolfi PP. 2007. Structure, dynamics and functions of promyelocytic leukaemia nuclear bodies. Nat Rev Mol Cell Biol 8: 1006–1016. [DOI] [PubMed] [Google Scholar]
- Chang FT, McGhie JD, Chan FL, Tang MC, Anderson MA, Mann JR, Andy Choo KH, Wong LH. 2013. PML bodies provide an important platform for the maintenance of telomeric chromatin integrity in embryonic stem cells. Nucleic Acids Res 41: 4447–4458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ching RW, Ahmed K, Boutros PC, Penn LZ, Bazett-Jones DP. 2013. Identifying gene locus associations with promyelocytic leukemia nuclear bodies using immuno-TRAP. J Cell Biol 201: 325–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S, Park JS, Kang YK. 2011. Dual functions of histone-lysine N-methyltransferase Setdb1 protein at promyelocytic leukemia-nuclear body (PML-NB): maintaining PML-NB structure and regulating the expression of its associated genes. J Biol Chem 286: 41115–41124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpet A, Olbrich T, Gwerder M, Fink D, Stucki M. 2014. Dynamics of histone H3.3 deposition in proliferating and senescent cells reveals a DAXX-dependent targeting to PML-NBs important for pericentromeric heterochromatin organization. Cell Cycle 13: 249–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day DS, Luquette LJ, Park PJ, Kharchenko PV. 2010. Estimating enrichment of repetitive elements from high-throughput sequence data. Genome Biol 11: R69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de The H, Le Bras M, Lallemand-Breitenbach V. 2012. The cell biology of disease: acute promyelocytic leukemia, arsenic, and PML bodies. J Cell Biol 198: 11–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delbarre E, Jacobsen BM, Reiner AH, Sorensen AL, Kuntziger T, Collas P. 2010. Chromatin environment of histone variant H3.3 revealed by quantitative imaging and genome-scale chromatin and DNA immunoprecipitation. Mol Biol Cell 21: 1872–1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delbarre E, Ivanauskiene K, Kuntziger T, Collas P. 2013. DAXX-dependent supply of soluble (H3.3-H4) dimers to PML bodies pending deposition into chromatin. Genome Res 23: 440–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drane P, Ouararhni K, Depaux A, Shuaib M, Hamiche A. 2010. The death-associated protein DAXX is a novel histone chaperone involved in the replication-independent deposition of H3.3. Genes Dev 24: 1253–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulac C, Torello AT. 2003. Molecular detection of pheromone signals in mammals: from genes to behaviour. Nat Rev Neurosci 4: 551–562. [DOI] [PubMed] [Google Scholar]
- Elsasser SJ, Noh KM, Diaz N, Allis CD, Banaszynski LA. 2015. Histone H3.3 is required for endogenous retroviral element silencing in embryonic stem cells. Nature 522: 240–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eustermann S, Yang JC, Law MJ, Amos R, Chapman LM, Jelinska C, Garrick D, Clynes D, Gibbons RJ, Rhodes D, et al. 2011. Combinatorial readout of histone H3 modifications specifies localization of ATRX to heterochromatin. Nat Struct Mol Biol 18: 777–782. [DOI] [PubMed] [Google Scholar]
- Goldberg AD, Banaszynski LA, Noh KM, Lewis PW, Elsaesser SJ, Stadler S, Dewell S, Law M, Guo X, Li X, et al. 2010. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell 140: 678–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henson JD, Hannay JA, McCarthy SW, Royds JA, Yeager TR, Robinson RA, Wharton SB, Jellinek DA, Arbuckle SM, Yoo J, et al. 2005. A robust assay for alternative lengthening of telomeres in tumors shows the significance of alternative lengthening of telomeres in sarcomas and astrocytomas. Clin Cancer Res 11: 217–225. [PubMed] [Google Scholar]
- Hoemme C, Peerzada A, Behre G, Wang Y, McClelland M, Nieselt K, Zschunke M, Disselhoff C, Agrawal S, Isken F, et al. 2008. Chromatin modifications induced by PML-RARα repress critical targets in leukemogenesis as analyzed by ChIP-Chip. Blood 111: 2887–2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Zhu B. 2014. H3.3 turnover: a mechanism to poise chromatin for transcription, or a response to open chromatin? BioEssays 36: 579–584. [DOI] [PubMed] [Google Scholar]
- Ishov AM, Sotnikov AG, Negorev D, Vladimirova OV, Neff N, Kamitani T, Yeh ET, Strauss JF III, Maul GG. 1999. PML is critical for ND10 formation and recruits the PML-interacting protein daxx to this nuclear structure when modified by SUMO-1. J Cell Biol 147: 221–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanauskiene K, Delbarre E, McGhie JD, Kuntziger T, Wong LH, Collas P. 2014. The PML-associated protein DEK regulates the balance of H3.3 loading on chromatin and is important for telomere integrity. Genome Res 24: 1584–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwase S, Xiang B, Ghosh S, Ren T, Lewis PW, Cochrane JC, Allis CD, Picketts DJ, Patel DJ, Li H, et al. 2011. ATRX ADD domain links an atypical histone methylation recognition mechanism to human mental-retardation syndrome. Nat Struct Mol Biol 18: 769–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan MM, Nomura T, Kim H, Kaul SC, Wadhwa R, Shinagawa T, Ichikawa-Iwata E, Zhong S, Pandolfi PP, Ishii S. 2001. Role of PML and PML-RARα in Mad-mediated transcriptional repression. Mol Cell 7: 1233–1243. [DOI] [PubMed] [Google Scholar]
- Kind J, Pagie L, de Vries SS, Nahidiazar L, Dey SS, Bienko M, Zhan Y, Lajoie B, de Graaf CA, Amendola M, et al. 2015. Genome-wide maps of nuclear lamina interactions in single human cells. Cell 163: 134–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar PP, Bischof O, Purbey PK, Notani D, Urlaub H, Dejean A, Galande S. 2007. Functional interaction between PML and SATB1 regulates chromatin-loop architecture and transcription of the MHC class I locus. Nat Cell Biol 9: 45–56. [DOI] [PubMed] [Google Scholar]
- Lallemand-Breitenbach V, de The H. 2010. PML nuclear bodies. Cold Spring Harb Perspect Biol 2: a000661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund EG, Oldenburg AR, Collas P. 2014. Enriched Domain Detector: a program for detection of wide genomic enrichment domains robust against local variations. Nucleic Acids Res 42: e92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen J, Sekelja M, Oldenburg AR, Barateau A, Briand N, Delbarre E, Shah A, Sørensen AL, Vigouroux C, Buendia B, et al. 2017. Chrom3D: three-dimensional genome modeling from Hi-C and lamin-genome contacts. Genome Biol 18: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pchelintsev NA, McBryan T, Rai TS, van TJ, Ray-Gallet D, Almouzni G, Adams PD. 2013. Placing the HIRA histone chaperone complex in the chromatin landscape. Cell Rep 13: 1012–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2015. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: https:/www.R-project.org/. [Google Scholar]
- Rapkin LM, Ahmed K, Dulev S, Li R, Kimura H, Ishov AM, Bazett-Jones DP. 2015. The histone chaperone DAXX maintains the structural organization of heterochromatin domains. Epigenetics Chromatin 8: 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray-Gallet D, Woolfe A, Vassias I, Pellentz C, Lacoste N, Puri A, Schultz DC, Pchelintsev NA, Adams PD, Jansen LE, et al. 2011. Dynamics of histone H3 deposition in vivo reveal a nucleosome gap-filling mechanism for H3.3 to maintain chromatin integrity. Mol Cell 44: 928–941. [DOI] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. 2011. Integrative genomics viewer. Nat Biotechnol 29: 24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadic D, Schmidt K, Groh S, Kondofersky I, Ellwart J, Fuchs C, Theis FJ, Schotta G. 2015. Atrx promotes heterochromatin formation at retrotransposons. EMBO Rep 16: 836–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saksouk N, Barth TK, Ziegler-Birling C, Olova N, Nowak A, Rey E, Mateos-Langerak J, Urbach S, Reik W, Torres-Padilla ME, et al. 2014. Redundant mechanisms to form silent chromatin at pericentromeric regions rely on BEND3 and DNA methylation. Mol Cell 56: 580–594. [DOI] [PubMed] [Google Scholar]
- Sarma K, Cifuentes-Rojas C, Ergun A, Del Rosario A, Jeon Y, White F, Sadreyev R, Lee JT. 2014. ATRX directs binding of PRC2 to Xist RNA and Polycomb targets. Cell 159: 869–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneiderman JI, Orsi GA, Hughes KT, Loppin B, Ahmad K. 2012. Nucleosome-depleted chromatin gaps recruit assembly factors for the H3.3 histone variant. Proc Natl Acad Sci 109: 19721–19726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeler JS, Marchio A, Sitterlin D, Transy C, Dejean A. 1998. Interaction of SP100 with HP1 proteins: a link between the promyelocytic leukemia-associated nuclear bodies and the chromatin compartment. Proc Natl Acad Sci 95: 7316–7321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiels C, Islam SA, Vatcheva R, Sasieni P, Sternberg MJ, Freemont PS, Sheer D. 2001. PML bodies associate specifically with the MHC gene cluster in interphase nuclei. J Cell Sci 114: 3705–3716. [DOI] [PubMed] [Google Scholar]
- Song Q, Smith AD. 2011. Identifying dispersed epigenomic domains from ChIP-Seq data. Bioinformatics 27: 870–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stajich JE, Block D, Boulez K, Brenner SE, Chervitz SA, Dagdigian C, Fuellen G, Gilbert JG, Korf I, Lapp H, et al. 2002. The Bioperl toolkit: Perl modules for the life sciences. Genome Res 12: 1611–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torok D, Ching RW, Bazett-Jones DP. 2009. PML nuclear bodies as sites of epigenetic regulation. Front Biosci 14: 1325–1336. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28: 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7: 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udugama M, Chang FT, Chan FL, Tang MC, Pickett HA, McGhie JD, Mayne L, Collas P, Mann JR, Wong LH. 2015. Histone variant H3.3 provides the heterochromatic H3 lysine 9 tri-methylation mark at telomeres. Nucleic Acids Res 43: 10227–10237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZG, Delva L, Gaboli M, Rivi R, Giorgio M, Cordon-Cardo C, Grosveld F, Pandolfi PP. 1998. Role of PML in cell growth and the retinoic acid pathway. Science 279: 1547–1551. [DOI] [PubMed] [Google Scholar]
- Wong LH, Ren H, Williams E, McGhie J, Ahn S, Sim M, Tam A, Earle E, Anderson MA, Mann J, et al. 2009. Histone H3.3 incorporation provides a unique and functionally essential telomeric chromatin in embryonic stem cells. Genome Res 19: 404–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong LH, McGhie JD, Sim M, Anderson MA, Ahn S, Hannan RD, George AJ, Morgan KA, Mann JR, Choo KH. 2010. ATRX interacts with H3.3 in maintaining telomere structural integrity in pluripotent embryonic stem cells. Genome Res 20: 351–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.