Abstract
DNA methylation is an essential epigenetic modification, present in both unique DNA sequences and repetitive elements, but its exact function in repetitive elements remains obscure. Here, we describe the genome-wide comparative analysis of the 5mC, 5hmC, 5fC, and 5caC profiles of repetitive elements in mouse embryonic fibroblasts and mouse embryonic stem cells. We provide evidence for distinct and highly specific DNA methylation/oxidation patterns of the repetitive elements in both cell types, which mainly affect CA repeats and evolutionarily conserved mouse-specific transposable elements including IAP-LTRs, SINEs B1m/B2m, and L1Md-LINEs. DNA methylation controls the expression of these retroelements, which are clustered at specific locations in the mouse genome. We show that TDG is implicated in the regulation of their unique DNA methylation/oxidation signatures and their dynamics. Our data suggest the existence of a novel epigenetic code for the most recently acquired evolutionarily conserved repeats that could play a major role in cell differentiation.
DNA methylation is an epigenetic modification essential for mammalian development (Okano et al. 1999). In mammals, the cytosine bases at position 5 in CpG dinucleotides are modified genome-wide by dedicated methyl-transferases to produce 5-methylcytosine (5mC), which can be further oxidized by the Ten eleven translocation (TET1, 2, and 3) enzymes (Tahiliani et al. 2009; Ito et al. 2010, 2011; He et al. 2011) to 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC), and 5-carboxylcytosine (5caC). Recent data showed that both 5fC and 5caC can be excised and repaired to regenerate unmodified cytosines by the concerted action of thymine DNA glycosylase (TDG) and the base excision repair (BER) enzymes (Cortellino et al. 2011; He et al. 2011; Maiti and Drohat 2011). In mammalian genomes, the CpG dinucleotides are underrepresented. Unusually dense clusters of CpG dinucleotides, called CpG islands (CGIs), are present and overlap with the promoter regions of >70% of the genes (Illingworth et al. 2010; Deaton and Bird 2011). Despite their high CG content, CGIs are mainly unmethylated. The current view holds that the CpG dinucleotide remains the primary site for DNA methylation, but there is emerging evidence for non-CpG methylation in several mammalian cells and tissues, including embryonic stem cells (ESCs), induced pluripotent stem cells (iPSC), oocytes, and the brain (Lister et al. 2009, 2013; Laurent et al. 2010; Tomizawa et al. 2011; Xie et al. 2012; Ziller et al. 2013).
More than two-thirds of the mammalian genome consists of repetitive elements (de Koning et al. 2011), including long terminal repeats (LTR), long interspersed nuclear elements (LINE), and short interspersed nuclear elements (SINE), major satellites and simple repeats (Mouse Genome Sequencing Consortium 2002). Previous data suggest that at least some of these repeat elements are methylated in ESCs (Shen et al. 2013) and undergo an extensive demethylation during plant development (Gehring et al. 2009). No comprehensive data are, however, available for the DNA genome-wide methylation pattern of repeat elements in differentiated mammalian cells. The biological significance of repetitive element methylation/oxidation remains elusive. Whereas the hypomethylation of repetitive elements appears to be a hallmark of cancer cells (Howard et al. 2008; Ehrlich 2009; Baba et al. 2010), how this is related to cancer development is poorly understood.
In this study, by using DNA immunoprecipitation sequencing (DIP-seq), we have carried out a genome-wide comparative analysis of the DNA methylation/oxidation patterns of repetitive elements in both differentiated mouse embryonic fibroblasts (MEFs) and pluripotent ESCs. Our data decipher a dynamic combinatorial DNA methylation code at repetitive elements that could play a major role in cell differentiation and define novel DNA regulatory regions within the mouse genome.
Results
The available data showed that TDG could be implicated in the dynamics of genome-wide distribution of methylated cytosines (Cortellino et al. 2011; He et al. 2011; Maiti and Drohat 2011). To further analyze this in detail in differentiated cells, we performed, by using highly specific antibodies (Supplemental Fig. 1a), two independent 5mC, 5hmC, 5fC, and 5caC DIP-seq experiments in wild-type and Tdg-deficient MEFs (Supplemental Fig. 1b) and determined their genome-wide distribution patterns (Supplemental Fig. 1c). This approach allows a high coverage of repetitive elements and profiling of all CpNs within the genome (Down et al. 2008). We paid particular attention to the modified cytosine patterns of repeat elements, since no study has yet addressed this problem in mammalian cells at the genomic level. In parallel, RNA-seq experiments were carried out on the same samples. Correlation of the DIP-seq and the RNA-seq data allowed us to dissect the cross talk between the cytosine modifications and transcription.
Accumulation of cytosine modifications at repeats in MEFs
Analysis of genome-wide sequencing data identified 24%–42% of reads as uniquely mapped and 58–76% of reads with multiple hits (Fig. 1A). The unique reads can be mapped to specific genomic sites, whereas the multihit reads, which represent repetitive elements (Dreszer et al. 2012), can only be assigned to their specific repeat family. Since the majority of the reads mapped to repetitive elements, we first studied the DNA methylation/oxidation pattern of individual repeats by analyzing uniquely mapped reads (Fig. 1B) and then extended the study to include also “unmappable” multihit reads by assigning them to their unique repeat family (Fig. 1C; Methods). Importantly, this second analysis allowed extension of the results obtained with individual repeat elements to their corresponding family.
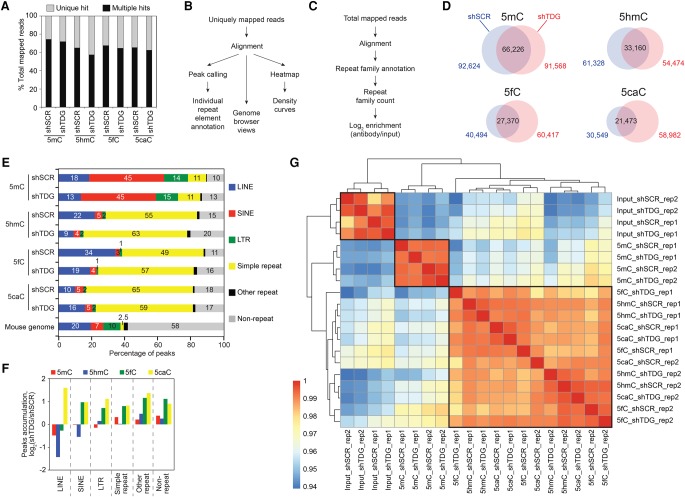
Figure 1.
Preferential accumulation of 5mC, 5hmC, 5fC, and 5caC at repetitive elements in MEFs. (A) Percentages of uniquely mapped and multihit reads in total mapped reads (average of two replicates). (B,C) Flowchart of computational analyses used in this study using uniquely mapped reads (B) and including multihit mapped reads (C). (D) Venn diagrams showing the overlap between 5mC, 5hmC, 5fC, and 5caC peaks in control (shSCR) and Tdg-deficient MEFs (shTDG). (E) Percentages of peaks overlapping with repetitive elements using the UCSC RepeatMasker database. (F) Bar graph representation of the peak accumulation (fold change, fc = log2-ratio shTDG/shSCR) at repetitive elements in response to Tdg knockdown in MEFs. (G) Heatmaps with hierarchical clustering showing Spearman's rank correlations between all pairwise comparisons. Spearman correlations were calculated using the raw read count across all types of repeats analyzed. Note that the 5hmC, 5fC and 5caC profiles were closely clustered.
Peak calling using uniquely mapped reads, which represent 24%–42% of tags, revealed that Tdg knockdown leads to a strong accumulation of 5fC and 5caC peaks and a slight decrease of 5mC and 5hmC peaks (Fig. 1D). Surprisingly, 80%–90% of these unique peaks overlap with the RepeatMasker database (RMSK), further supporting our finding that repetitive elements in MEF cells represent the vast majority of cytosine modification targets (Fig. 1E). In agreement with this, only 198 genes showed relatively marked expression change upon Tdg knockdown (Supplemental Fig. 1d). As expected, modified cytosines were strongly underrepresented at CGIs (Supplemental Fig. 1e). Forty-five percent of the 5mC called peaks were found at SINEs (Fig. 1E), which represent 7.5% of the mouse genome (Supplemental Fig. 1g). On the other hand, on average, >50% of the called 5hmC, 5fC, and 5caC peaks were found at simple repeats (Fig. 1E), which accounts for only 2.3% of the mouse genome (Supplemental Fig. 1g). Tdg knockdown leads to an accumulation of 5fC and 5caC peaks at all repetitive elements with the exception of LINEs, accumulating only 5caC peaks (Fig. 1F). In addition, a strong overlap between 5hmC, 5fC, and 5caC peaks was observed (Supplemental Fig. 1f). This result was further confirmed by the correlation matrix coefficient for the repetitive elements (calculated by using total mapped reads), which showed a close clustering between these three oxidized forms (Fig. 1G).
Of note, Tdg knockdown leads also to a twofold increase of 5fC and 5caC peaks at nonrepetitive elements, showing that the action of TDG is not restricted to repetitive elements. Taken collectively, these data revealed a marked and overall Tdg-deficient-dependent accumulation of 5fC and 5caC in MEFs.
Specific DNA methylation profiles at recently integrated IAP LTRs in differentiated MEFs and pluripotent ESCs
In order to perform a comparative analysis of cytosine modification patterns between the repeat elements in differentiated MEFs and pluripotent ESCs, we compared our data on MEFs with reads overlapping with the RMSK database from previously published data sets for ESCs (Shen et al. 2013). In addition, we reanalyzed them independently. We first characterized the Tdg-deficient-dependent changes in DNA methylation at LTR retrotransposons (also known as endogenous retroviruses [ERVs]) in MEFs. For simplicity, we will collectively further refer to LTR retrotransposons as LTRs. The RMSK database distinguishes between elements corresponding to external domains (LTRext, containing the regulatory regions of the LTR) from those corresponding to internal domains (LTRint, containing the coding sequences of the proteins, necessary for the life cycle of the integrated viruses) (Fig. 2A) within the different LTR families. With this in mind, we carried out independent analyses for these two regions. Heatmaps were generated using uniquely mapped reads on internal domains (LTRint) with a 2-kb cutoff to eliminate truncated and degenerated LTRs (Fig. 2B). The corresponding normalized densities of 5mC, 5hmC, 5fC, and 5caC signals are presented in Figure 2C. Our data revealed a striking enrichment of 5mC exclusively at the external repeats of the Intracisternal A Particle (IAP) LTR family ERVK (Fig. 2C,D). These last results are intriguing, since IAP repeats represent 5.6% of the mouse genome (Supplemental Fig. 1g) and are the evolutionarily least truncated LTR subfamily (Fig. 2E) with the highest conservation score and CG content (Fig. 2B,F).
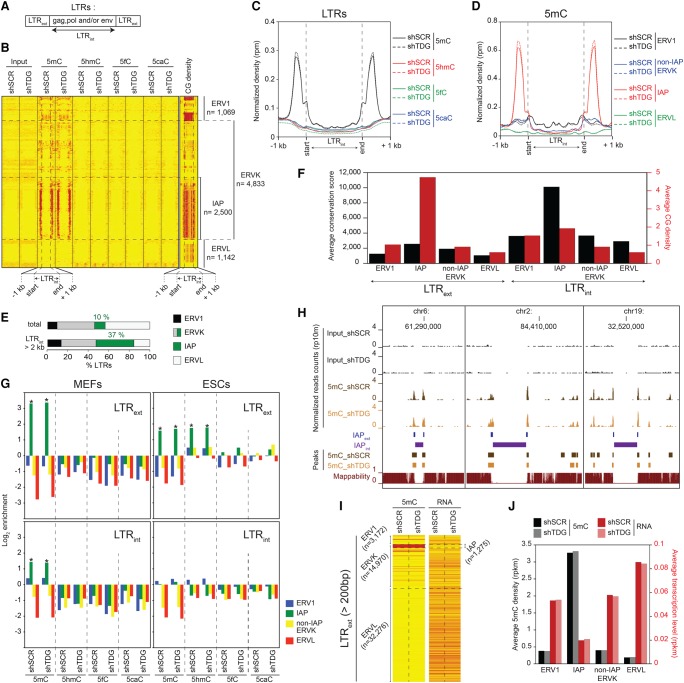
Figure 2.
Specific DNA methylation profiles at recently integrated IAP LTRs in MEFs and ESCs. (A) RepeatMasker database distinguishes within the LTR family elements corresponding to the external terminal repeats (LTRext) from those corresponding to the internal coding regions (LTRint). (B) Heatmaps of 5mC/5hmC/5fC/5caC/CG densities at full-length LTRs (LTRint > 2 kb) in control and Tdg-deficient MEFs. Tag densities were collected in 50-bp sliding windows spanning 1 kb (divided in 15 bins) of the length-normalized LTRint (divided in 30 bins). LTR retroelements were sorted by families. (C) Average 5mC/5hmC/5fC/5caC signals in control and Tdg-deficient MEFs at LTRs. (D) 5mC densities for the indicated LTR families in control and Tdg-deficient MEFs. (E) Distribution of LTR classes in the mouse genome (total or full-length retroelements). (F) Average conservation score (black columns) and CG density (number of CpG dinucleotides per 100 bases, red columns) of LTRext (left) and LTRint (right) elements. (G) Relative enrichment for each cytosine modification in MEFs (left) and ESCs (right) at the indicated LTR families (LTRext regions, upper; LTRint regions, lower). Note the significant enrichment of 5hmC at LTRext IAP in ESCs. (*) P-value <0.05. (H) Genome browser views showing the lack of mappability at IAPint elements. (I) Heatmaps of average 5mC densities and transcription levels at LTRext elements (length >200 bp) in control and Tdg-deficient MEFs. (J) Histogram showing the negative correlation between methylation density and transcription level of LTRext elements in control and Tdg-deficient MEFs.
Relative enrichment calculations including multihit reads (Fig. 2G, left; Supplemental Fig. 2a, left) further confirmed the 5mC enrichment of the IAP-LTR external regions (log2 > 3, ∼10-fold), but also revealed a moderate 5mC enrichment at their internal domain (log2 = 1.5, approximately threefold). This moderate methylation correlated with the intermediate CG level of the IAP-LTR internal regions (Fig. 2B,F), but was barely observed in the heatmap generated with uniquely mapped reads (Fig. 2B), due to their inability to be mapped by the existing approaches (Fig. 2H). Our data illustrate a direct correlation between the observed methylation level and the CG density of the different LTR retroelement regions. Overall, we observe 5mC enrichment at every LTR region whenever the CG density exceeds the average mouse genome density (0.83 CpG per 100 bp).
We next investigated how the methylcytosine density impacts LTR element transcription in MEF cells (Fig. 2I). The high levels of methylation of the IAP family suggest that their transcription is repressed. Accordingly, LTRext IAPs showed the highest 5mC density (10-fold higher than the rest) and the lowest transcription level (three- to fourfold lower than the rest) (Fig. 2J). In agreement with the absence of change in 5mC densities, the knockdown of Tdg had no effect on LTR transcription.
The methylation patterns of LTR retrotransposons in ESCs showed, however, striking differences when compared to these in MEFs. For example in ESCs, LTRext IAPs exhibited a threefold lower enrichment for 5mC than MEFs and a significant enrichment for 5hmC (Fig. 2G, upper right). Moreover, we detected an accumulation of 5fC and 5caC in response to Tdg knockdown at LTRext of several IAP subfamilies (Supplemental Fig. 2a, right). Similarly, the external domain of several mouse-specific non-IAP ERVK and ERV1 subfamilies showed an enrichment for oxidized methylated cytosines, with a specific accumulation of 5caC in the absence of TDG (Supplemental Fig. 2a, right). Importantly, ERVK and ERV1 internal domains (LTRint) tend to be depleted in 5mC (Fig. 2G, lower right; Supplemental Fig. 2b, right). Taken collectively, these data suggest that the evolutionarily conserved LTRs harboring a high CG content are dynamically regulated by TDG in ESCs, but stably methylated during cell differentiation.
DNA methylation dynamics at the evolutionarily youngest SINEs
SINEs are interspersed repeats that make up to 7.5% of the mouse genome (Supplemental Fig. 1g) and comprise two mouse-specific families, B1m and B2m, which correspond to the most recently integrated (Mouse Genome Sequencing Consortium 2002) and the most conserved elements (Supplemental Fig. 3a). Heatmaps of 5mC/5hmC/5fC/5caC/CG densities at SINEs ranked by families revealed a strong cytosine modification enrichment occurring at the mouse-specific CG-rich B1m and B2m families in MEFs (Fig. 3A). The CG density of B1m elements is on average twofold higher than that of B2m elements (Supplemental Fig. 3a). Accordingly, the 5mC density was approximately twofold higher at the B1m family than at the B2m family (Fig. 3B). Normalized density curves clearly showed that the mouse-specific SINEs are highly methylated and their oxidation is dynamically regulated by TDG (Fig. 3C; Supplemental Fig. 3b). The preceding results suggest that transcription of the most conserved SINEs is repressed by DNA methylation. To test this, individual mouse-specific B1m elements were ranked by conservation score, and heatmaps were generated. A strong correlation was observed between their phylogenetic conservation, CG density, and DNA modification density (Fig. 3D,E). Average transcription level revealed that the most conserved B1m SINEs are the most repressed and most methylated (Fig. 3D,E). Although a net conversion from 5hmC to 5caC was observed at B1m SINEs in the absence of TDG, their 5mC level was not affected. In line with this, knockdown of Tdg had no effect on B1m SINE transcription.
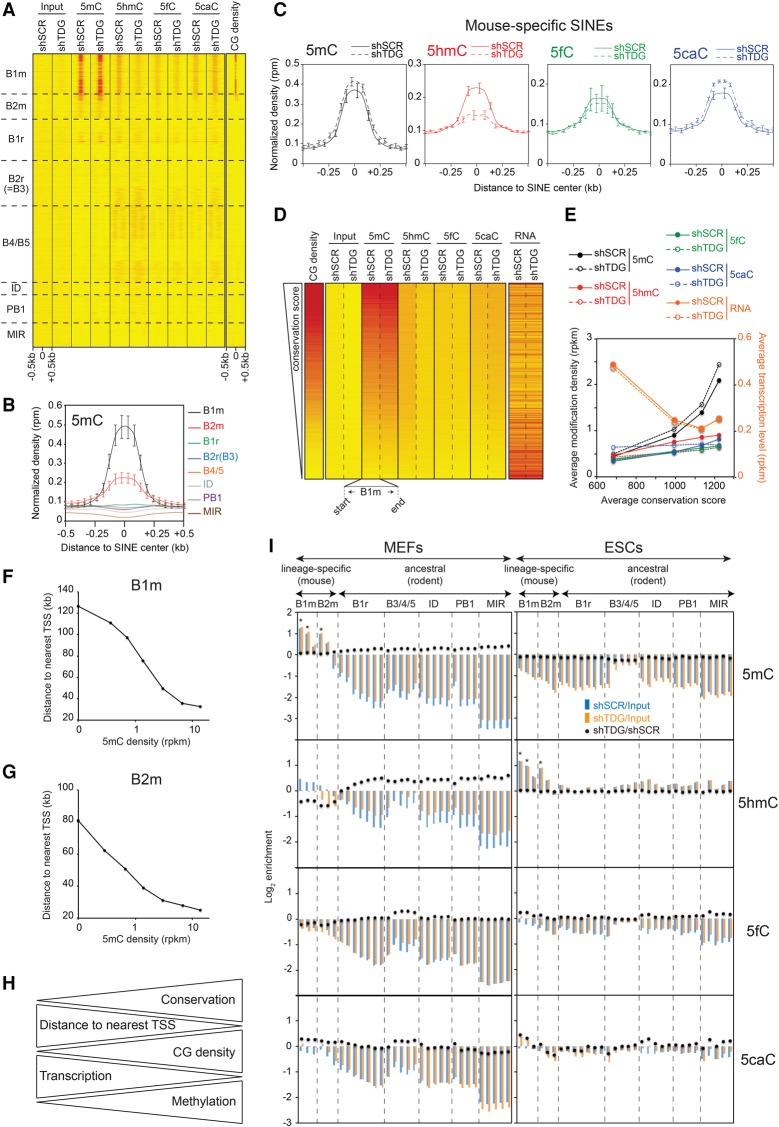
Figure 3.
Specific TDG-dependent DNA methylation dynamics at the evolutionarily youngest SINEs. (A) Heatmaps of 5mC/5hmC/5fC/5caC/CG densities at SINEs (n = 1,511,580), sorted by family. Tags were counted within 500 bp around the SINE center. (B) 5mC densities in control MEFs for the indicated SINE families. (C) Average 5mC/5hmC/5fC/5caC signals in control and Tdg-deficient MEFs at mouse-specific SINEs. (B,C) Values represent means of two biological replicates. Error bars represent the range of the duplicate values. (D) Heatmaps of average 5mC/5hmC/5fC/5caC/CG densities and transcription levels in control and Tdg-deficient MEFs at B1m retroelements ranked by conservation score (n = 185,667). (E) Curves representing cytosine modification densities and transcription levels of B1m SINEs as a function of their conservation. B1m retroelements were sorted into quartiles based on their conservation score. Note the negative correlation between the methylation density and the transcription level of B1m SINEs. (F,G) Average distance to TSS of B1m (F) and B2m (G) elements as a function of their 5mC density. (H) Diagram illustrating the relationship between DNA methylation, CG density, transcription level, and distance to TSS for the mouse-specific B1m and B2m families. (I) Relative enrichment for each cytosine modification at each SINE subfamily in control and Tdg-deficient MEFs (left) and ESCs (right). SINE subfamilies were sorted in two groups relative to their appearance in the rodent lineage, the mouse-specific group and the ancestral group (common in rodents). Note that mouse-specific SINEs are specifically hypermethylated in MEFs but hydroxymethylated in ESCs. (*) P-value <0.05.
The available data indicated that SINEs are implicated in chromatin domain anchoring and gene regulation (Lunyak et al. 2007; Ichiyanagi 2013). If this was the case, these species-specific SINEs should be located at distinct functional loci within the mouse genome and should show a distinct methylation pattern. To test this, we first analyzed the distance to a transcription start site (TSS) distribution of mouse-specific SINEs according to their 5mC level. A highly statistically significant correlation between 5mC density and proximity to the TSS was observed (Supplemental Fig. 3c). Genome browser visualization revealed that the methylated mouse-specific SINEs are clustered around CGIs (Supplemental Fig. 3d, upper), close to the TSS (Supplemental Fig. 3d, lower). This result was further validated by measuring the average distance to the TSS of B1m and B2m elements as a function of their 5mC level (Fig. 3F,G). Altogether, our data revealed that the transcription of species-specific SINEs integrated around TSSs are highly regulated by DNA methylation (Fig. 3H).
These data were further supported by the calculated relative enrichment of all cytosine modifications using total mapped reads. The results confirmed the global hypermethylation status of mouse-specific SINEs over ancestral SINEs and their TDG-dependent regulation in MEFs (Fig. 3I, left). Comparative analysis revealed a specific hydroxymethylation of these mouse-specific SINEs in ESCs (Fig. 3I, right).
DNA methylation patterns of LINEs
LINEs are autonomous retrotransposons making up ∼20% of the mouse genome (Supplemental Fig. 1g; Mouse Genome Sequencing Consortium 2002). Most of LINEs are defective due to truncation or accumulation of mutations over time (for review, see Edgell et al. 1987; Sookdeo et al. 2013). Indeed, only 1% of annotated LINEs are intact (length > 5 kb) and potentially active (Castro-Diaz et al. 2014). The majority of full-length LINEs are represented by the strongly conserved mouse-specific L1Md family, which covers 2.8% of the mouse genome (Supplemental Fig. 1g) and exhibits the highest CG density (Supplemental Fig. 4a,b). This family contains a 5′ UTR functioning as a promoter, two open reading frames, ORF1 and ORF2, and a 3′ UTR containing a poly(A) signal. To analyze the 5mC/5hmC/5fC/5caC distribution at intact LINEs in MEFs, we generated heatmaps of 5mC/5hmC/5fC/5caC/CG densities and transcription levels at L1Md ranked by their appearance in the mouse genome (Castro-Diaz et al. 2014). Two distinct clusters were identified: cluster 1, containing the youngest subfamilies L1Md_T, L1Md_A, and L1Mf_Gf (0.5–1.5 million years old) with a CG-rich 5′ UTR region; and cluster 2, containing the oldest subfamilies L1Md_F, L1Md_F2, and L1Md_F3 (3.5–4.5 million years old) with a lower CG density at their 5′ UTR region (Fig. 4A). Average 5mC/5hmC/5fC/5caC signals at L1Md in control and Tdg-deficient MEFs show two distinct profiles: (1) a hypermethylated 5′ UTR region of the evolutionarily recent L1Md subfamilies; and (2) TDG-dependent dynamics of the 5mC oxidation derivatives along their coding sequence in the oldest subfamilies (Fig. 4B). In addition, three individual L1Md elements representative of the two described clusters are visualized in Supplemental Fig. 4c.
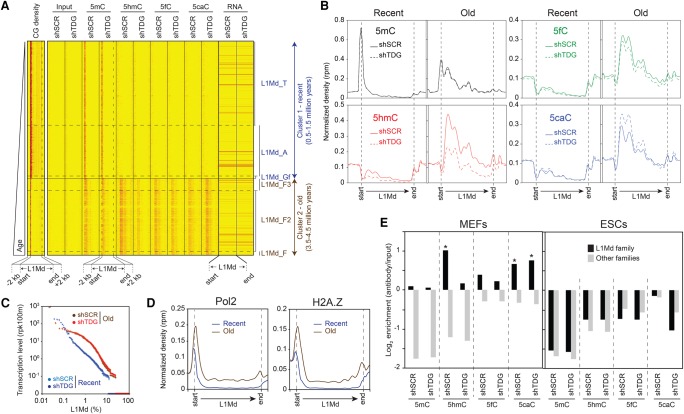
Figure 4.
Distinct TDG-dependent DNA methylation patterns at full-length L1Md. (A) Heatmaps of 5mC/5hmC/5fC/5caC/CG densities and transcription levels in control and Tdg-deficient MEFs at full-length L1Md elements (length > 5 kb, n = 12,916), sorted relative to their appearance in the mouse genome. Tag densities were collected in 50-bp sliding windows spanning 2 kb (divided in 10 bins) of the length-normalized L1Md (divided in 40 bins). Two distinct clusters are identified: cluster 1, containing the youngest subfamilies L1Md_T, L1Md_A, and L1Mf_Gf (0.5–1.5 million years old); and cluster 2, containing the oldest subfamilies L1Md_F, L1Md_F2, and L1Md_F3 (3.5–4.5 million years old). (B) Average 5mC/5hmC/5fC/5caC signals at L1Md in control and Tdg-deficient MEFs reveal two distinct profiles. The evolutionarily recent L1Md subfamilies contain a hypermethylated 5′ UTR region, whereas the oldest subfamilies show a TDG-dependent dynamic of 5mC oxidation derivatives along their coding sequence. (C) Transcription levels of each individual element of the recent and old L1Md subfamilies in control and Tdg-deficient MEFs. (D) Normalized densities of Pol2 (left), H2A.Z (right) at recent (T/A/Gf), and old (F/F2/F3) L1Md subfamilies. (E) Relative enrichment for each cytosine modification at indicated LINE families in control and Tdg-deficient MEFs (left) and ESCs (right). (*) P-value <0.05.
We hypothesized that the LINEs, characterized by methylcytosine oxidation dynamics along their ORFs are more prone to activation compared with LINEs hypermethylated on their 5′ UTR. Accordingly, RNA-seq analysis revealed that the oldest L1Md elements are globally more transcribed than the youngest (Fig. 4C). In addition, a stronger enrichment of H2A.Z and Pol2 at promoters of the oldest L1Md elements was found. We conclude that the CG-rich promoters of the recently integrated LINEs are silenced by DNA methylation, whereas the oldest ones are more transcribed, oxidized along their coding sequences, which is actively removed by TDG. Of note, a net conversion from 5hmC to 5caC observed in the absence of TDG had no effect on their transcription states (Fig. 4C).
We next investigated the L1Md LINEs distribution throughout the genome. We observed a strong enrichment of full-length LINEs in genes involved in brain development (Supplemental Fig. 4e). The specific DNA methylation pattern found at L1Md family in MEF cells was further validated by analyses of total mapped reads (Fig. 4E, left; Supplemental Fig. 4d, left). Surprisingly, L1Md were depleted in cytosine modifications in ESCs (Fig. 4E, right; Supplemental Fig. 4d, right), suggesting that the regulation of LINE transcription by methylation takes place during differentiation.
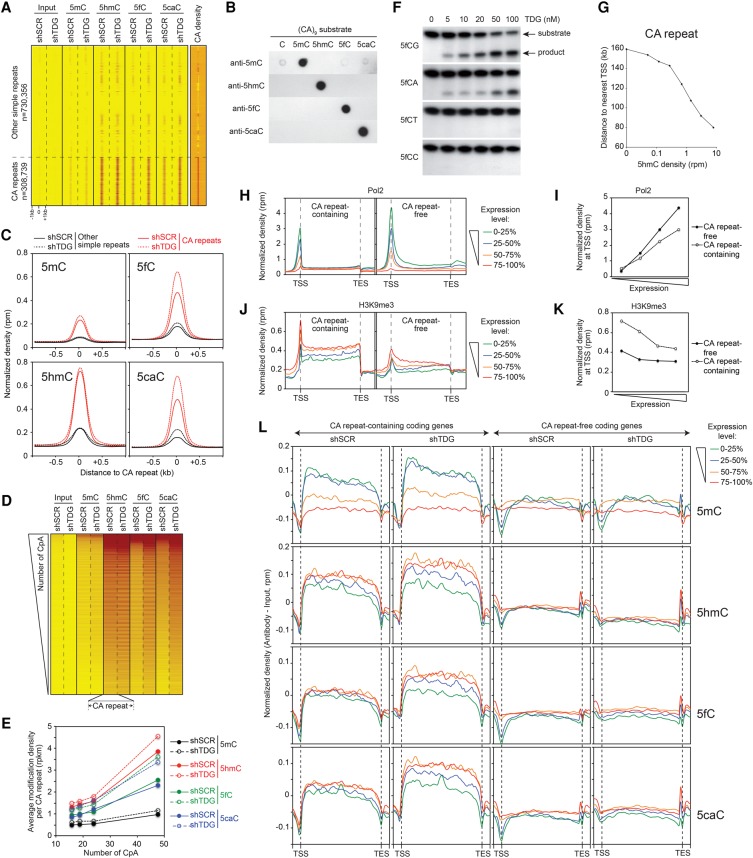
DNA methylation dynamics at CA repeats
Simple repeats are made up of variable numbers of successive repeating units with various lengths (for review, see Ellegren 2004). Heatmaps of 5mC/5hmC/5fC/5caC/CA densities for simple repeats revealed a clear enrichment of all 5mC oxidation derivatives specifically at CA repeats in MEFs (Fig. 5A). Note that the antibodies we have used are highly specific for the individual 5mC oxidized forms of the CA repeats, thus ruling out the possibility of nonspecific association of the antibodies with repeats (Fig. 5B). Normalized density curves (Fig. 5C) and genome browser views (Supplemental Fig. 5b,c) confirmed that CpA dinucleotides are mainly oxidized and regulated by TDG. Indeed, Tdg knockdown leads to a strong density accumulation of both 5fC and 5caC (∼50% increase) (Fig. 5C, right). Of note, the depletion of TDG weakly affects the density of both 5mC and 5hmC (Fig. 5C, left). Since the 5hmC, 5fC, and 5caC patterns track closely the CpA content of simple repeats (Fig. 5A), and the CA repeats exhibit a 50-fold higher CA than CG density (Supplemental Fig. 5a), the preceding results suggest that the cytosine within the CpA dinucleotides could also be methylated. To analyze this, CA repeats were ranked by CpA dinucleotide repetition number, and heatmaps were generated (Fig. 5D). Interestingly, a strong correlation is seen between CA density and 5hmC, 5caC, and 5fC densities. Average modification density calculations further confirmed the close correlation between the number of CpA repetitions and their modification enrichments (Fig. 5E). The occurrence of 5mC oxidation at CA repeats prompted us to study the ability of TDG to excise an oxidized cytosine in sequences that do not contain CpG. In vitro assays showed that recombinant TDG efficiently excised a formylcytosine in both CpG and CpA contexts, but not in synthesized sequences containing either CpC or a CpT (Fig. 5F). Likewise, the dioxygenase activity of the Naegleria Tet-like protein has a strong preference for 5mCpG and 5mCpA (Hashimoto et al. 2014). These results further validate the occurrence of DNA methylation/oxidation at CA repeats in MEFs.
Figure 5.
DNA methylation dynamics at CA repeats. (A) Heatmaps of 5mC/5hmC/5fC/5caC/CA densities in control and Tdg-deficient MEFs at simple repeats sorted by family. Note the specific enrichment of 5mC oxidation derivatives at CA repeats. Tags were counted within 1 kb around the simple repeat center. (B) Anti-5mC, anti-5hmC, anti-5fC, and anti-5caC antibodies do not recognize CA repeats nonspecifically. Dot blot assays showing that 5mC, 5hmC, 5fC, and 5caC antibodies specifically recognize 5mC, 5hmC, 5fC, and 5caC-containing substrates, respectively in (CA)9 repeat contexts. (C) Average 5mC/5hmC/5fC/5caC signals at CA repeats reveal a specific accumulation of 5fC and 5caC in the absence of TDG. (D) Heatmaps of 5mC/5hmC/5fC/5caC levels in control and Tdg-deficient MEFs at CA repeats ranked in descending order based on their number of CpA dinucleotides. (E) Curves showing the positive correlation between cytosine modification densities and CA densities at CA repeats. CA repeats were sorted in quartiles based on their number of CpA dinucleotides. (F) In vitro glycosylase assays revealing that the recombinant protein TDG excises formylcytosine exclusively in a CpG or CpA context. (G) Average distance to TSS of CA repeats as a function of their 5hmC level. (H–K) Normalized densities of Pol2 (H,I) and H3K9me3 (J,K) within gene bodies (H,J) or at TSSs (I,K) expressed at different levels. Genes were sorted into two groups according to the presence or the absence of CA repeats (length >100 bp) within their gene body (CA repeat-containing and CA repeat-free, respectively). Tag densities were collected in 100-bp sliding windows spanning 2 kb (divided in 10 bins) of the length-normalized gene bodies (divided in 40 bins). Within both groups, genes were then sorted in quartiles according to their expression level. (L) Average 5mC/5hmC/5fC/5caC signals in control and Tdg-deficient MEFs within CA repeat-containing and CA repeat-free gene bodies expressed at different levels. Tag densities were collected in 100-bp sliding windows spanning 2 kb (divided in five bins) of the length-normalized gene bodies (divided in 50 bins).
Since CA repeats have been implicated in transcription and splicing regulation (Naylor and Clark 1990; Gebhardt et al. 1999; Pravica et al. 1999; Shimajiri et al. 1999; Gabellini 2001; Hui et al. 2003), we sought to determine whether CA repeat modifications occurred randomly or at specific locations within the mouse genome. Annotation of CA repeats using HOMER software revealed that >40% of CA repeats are associated with introns, and their 5hmC density increases with their proximity to a TSS (Fig. 5G; Supplemental Fig. 5d). Altogether, these data revealed that the densest CA repeats are the most modified and the closest to TSSs, which point to their potential role in transcription regulation. Bearing this in mind, we next studied the relationship between cytosine modifications, Pol2, and H3K9me3 distributions and the expression level of genes containing or not CA repeats (with a cutoff of [CA]n > 100 bp) (Supplemental Fig. 5e). Using coding genes from the Ensembl 67 database, we were able to identify 8,595 (CA)n-containing genes and 10,456 (CA)n-free genes. Interestingly, the two classes of genes showed a very distinct Pol2 and H3K9me3 profiles (Fig. 5H–K). Although the density of Pol2 followed the transcription level, we observed a much stronger accumulation of Pol2 on (CA)n-free genes than (CA)n-containing genes at equivalent transcription level (Fig. 5H,I). As expected, we found a strong accumulation of H3K9me3 at silenced genes with a much stronger association with (CA)n-containing genes (Fig. 5J,K).
We then analyzed the methylation/oxidation profiles of (CA)n-free and (CA)n-containing genes. Surprisingly, we observed a positive correlation between gene body methylation and transcription level specific for (CA)n-containing genes (Fig. 5L, first row of panels). In the absence of TDG, an accumulation of 5mC density specifically for highly transcribed (CA)n-containing genes (Fig. 5L, first row of panels) was observed. Since 90% of 5mC peaks were detected within repetitive elements (Fig. 1E), we conclude that the methylation of repeats within gene bodies positively correlate with their transcription. This correlation is only seen for (CA)n-containing genes, which suggest that this class of genes contains more repetitive elements and hence could correspond to longer genes. In agreement, (CA)n-containing genes were found eight times longer than (CA)n-free genes (96 versus 12 kb) (Supplemental Fig. 5f). In contrast to 5mC distribution, the oxidation patterns of (CA)n-containing genes negatively correlate with their transcription (Fig. 5L, second to fourth row of panels). In support, Tdg knockdown leads to a stronger accumulation of 5fC and 5caC at repressed (CA)n-containing genes (Fig. 5L, third and fourth row of panels).
Relative enrichment calculations including multihit reads for each cytosine modification at different simple repeat families confirmed that CA repeats are specifically enriched in 5mC oxidized forms (Supplemental Fig. 5g, left; Supplemental Fig. 5h, left). Surprisingly, these repeats were strongly enriched in 5mC in ESCs (Supplemental Fig. 5g, right). Clustered heatmap densities of cytosine modification levels at CA repeats in ESCs and MEFs revealed that the highly methylated CA repeats in ESCs correspond to those harboring the strongest 5mC oxidized forms in MEFs (Supplemental Fig. 5i). Together our data show that the densest CA repeats are preferentially methylated in ESCs, oxidized, and dynamically regulated by TDG during differentiation in a transcription-dependent manner. Since CA methylation was shown by bisulfite sequencing to occur in Drosophila and mammals mainly in the CAC trinucleotide context (Laurent et al. 2010; Lister et al. 2013; Guo et al. 2014; Takayama et al. 2014), we investigated whether this motif is also preferentially modified at simple repeats. Our analysis identified the CAC trinucleotide as the main motif targeted by 5mC oxidation at simple repeats in MEFs (Supplemental Fig. 5h, right). Collectively, these results highlight the TDG-dependent DNA methylation dynamics at conserved CA repeats that are located closer to TSSs compared to degenerate CA repeats. The distinct methylation/oxidation patterns found in MEFs and ESCs may reflect an active role of these modifications in shaping the transcriptional reprogramming taking place during differentiation.
We finally analyzed the occurrence of cytosine modifications at major satellites and DNA transposons. Major satellites showed a unique cytosine modification pattern conserved between MEFs and ESCs, characterized by a specific 5mC and 5fC enrichment (Supplemental Fig. 6a). In contrast, DNA transposons did not show any enrichment in cytosine modifications (Supplemental Fig. 6b).
Discussion
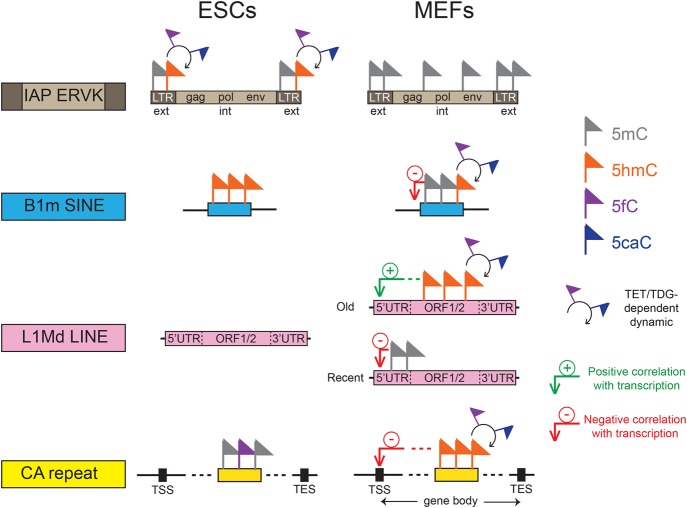
Here, we present a genome-wide comparative analysis of DNA methylation/oxidation profiles of repetitive elements in both MEFs and ESCs. We found major differences in the DNA methylation/oxidation patterns of repetitive elements in these cells. The majority of DNA methylation/oxidation patterns are dynamically regulated by TDG and occur mainly at both the CA repeats and the most recently acquired transposable elements corresponding to mouse-specific repeats with high CG content. We show that these elements are not distributed randomly throughout the mouse genome, but are clustered with respect to the TSS and hence, may act as novel cis-acting regulatory elements (Fig. 6).
Figure 6.
Combinatorial DNA methylation code at repetitive elements. Diagram summarizing the TDG-dependent DNA methylation dynamics at repetitive elements in MEFs and ESCs, which affect essentially both the CA repeats and the evolutionarily youngest mouse-specific transposable elements including IAP LTRs, B1m SINEs, and L1Md LINEs.
Overall, we observed an enrichment of DNA methylation in repetitive elements whenever the CG density exceeded 0.83 CpG per 100 bp, which is the average mouse genome density. For example, the IAP retroviruses, which have the highest CG density, showed the highest methylation enrichment. In ESCs, this subfamily was partially methylated and enriched in 5hmC, but was fully methylated in MEFs, which suggests that their permanent inactivation during differentiation is important to prevent insertional mutagenesis. Accordingly, IAP transcription is constrained by methylation (Walsh et al. 1998), and LTR elements were found excluded from gene-rich regions (Medstrand et al. 2002), likely because of their potential to alter transcription. LTR families harboring an intermediate CG density (such as ERV1 and non-IAP ERVK) showed DNA methylation/oxidation dynamics specific to ESCs accumulating the more oxidized forms in the absence of TDG, whereas the evolutionarily oldest CG-poor ERVL family escaped methylation. Collectively, our data revealed that methylation level of CG-rich LTRs is highly dynamic during differentiation.
DNA methylation-regulated, mouse-specific SINEs are concentrated around CGIs. This peculiar localization could have profound consequences on neighboring gene expression. Human B1 SINEs have been shown to influence the activity of downstream gene promoters, with acquisition of DNA methylation and loss of active histone marks (Estecio et al. 2012). Mouse B1m and B2m SINEs might act as boundary elements that protect CGIs against pervasive methylation; hence, they could be used by ESCs (where the SINEs are not methylated) to maintain the undifferentiated state. Consequently, SINE hydroxymethylation could regulate the transcriptional circuit that sustains the pluripotent state before subsequent methylation silencing during differentiation. Nevertheless, we cannot rule out the possibility that the higher concentration of recently acquired SINEs close to a TSS reflects their higher probability to insert into highly transcribed, accessible chromatin structure. In agreement with this, genes bound by insulator proteins, known to function as barriers to heterochromatin spreading, show a higher frequency of SINEs than unbound genes (Estecio et al. 2012).
The lineage-specific LINE families L1Md showed TDG-dependent cytosine modification dynamics in MEFs according to their evolutionary age: the youngest subfamilies present mainly a hypermethylated 5′ UTR, whereas the oldest subfamilies show a TDG-dependent methylation dynamic throughout their ORFs. In agreement, the oldest L1Md elements are more active, and their highly dynamic 5hmC/5fC/5caC profiles follow transcriptional directionality, peaking at the beginning of the first ORF and diminishing toward the 3′ UTR. These L1Mds are, as expected, characterized by high Pol2 and H2A.Z association levels. These intact LINEs also exhibited a nonrandom genomic distribution, being concentrated in genes involved in synapse and neuron function. The biological significance of this genomic distribution could reflect a specific role in gene regulation and genome organization in brain given that LINEs have been implicated in several fundamental processes such as differentiation and development (Speek 2001; Nigumann et al. 2002; Matlik et al. 2006; Slotkin and Martienssen 2007; Faulkner et al. 2009; Muotri et al. 2010). Consistent with this, DNA methylation dynamics at L1Md were not observed in ESCs suggesting a specific function in differentiated cells.
Another important aspect of this study is the identification of CA methylation enrichment at simple repeats. Our data show that simple repeats with the highest CA density are preferentially methylated in ESCs, but hydroxymethylated and formyl/carboxylated in MEFs, accumulating the more oxidized forms in the absence of TDG. The biological significance of the switch from methylation to oxidation during differentiation remains unclear for the moment, but the occurrence of 5mC/5hmC on CA repeats at close distances to the TSS suggests an important role of these elements in the control of gene expression (Naylor and Clark 1990; Gebhardt et al. 1999; Pravica et al. 1999; Shimajiri et al. 1999; Gabellini 2001; Hui et al. 2003). In agreement, 5hmC and 5fC/5caC accumulation in the absence of TDG target preferentially silenced (CA)n-containing genes. The ability of recombinant TDG protein to excise formylcytosine exclusively in CpG and CpA contexts further validates the implication of CA repeats in genome regulation/organization. Our data support previous observations, obtained by bisulfite sequencing or 5hmC-specific restriction enzyme PvuRts1l approaches, describing non-CG methylation in brain, oocytes, ESCs, iPSC, and flies (Laurent et al. 2010; Tomizawa et al. 2011; Xie et al. 2012; Lister et al. 2013; Ziller et al. 2013; Sun et al. 2015) and provide straightforward evidence for its occurrence in the CAC motif at simple repeats. CA repeat methylation/oxidation dynamics could also play an important role in brain development. Recent work reported their hypomethylation is associated with autism (Papale et al. 2015), and MECP2 has been shown to repress gene expression in brain tissues by binding to methylated CA sites within long genes (Gabel et al. 2015).
We hypothesize that the TDG-dependent cytosine DNA methylation/oxidation dynamics, specific to both CA repeats and the youngest lineage-specific transposable elements, may constitute a novel epigenetic code with an as yet unknown role in genome organization and function (Fig. 6). Indeed, retroelement insertions are major drivers of evolutionary changes within species (Cordaux and Batzer 2009; Burns and Boeke 2012; Mita and Boeke 2016), and the observed retroelement methylation dynamics could be strongly implicated in evolution. Alterations of this code could be associated with disease development. This may be particularly true for tumorigenesis, since strong hypomethylation of repeats is observed in cancer cells (Howard et al. 2008; Ehrlich 2009; Baba et al. 2010).
Methods
Isolation of primary MEFs
Embryonic fibroblasts were isolated from mouse embryos at embryonic day 10.5 (genetic background C57BL/6) as previously described (Obri et al. 2014). MEFs were kept in culture for no more than 1 mo.
Lentiviral knockdown of Tdg
shRNA targeting Tdg (shTDG-1, 5′-CCAGCAGGATTTAATGGTATT-3′ and shTDG-2, 5′-GCCACGAATAGCGGTGTTTAA-3′) or the control shRNA (5′-CCTAAGGTTAAGTCGCCCTCG-3′) were cloned into pLKO.1-blast vector (Addgene). To generate lentiviruses, the transducing vectors were cotransfected into 293T cells using Effectene Transfection Reagent (Qiagen). The supernatant was harvested at 48 h after transfection. To generate control and Tdg-knockdown cells, MEFs were infected with lentivirus in a six-well plate. Blasticidine (10 µg/mL) was added to the medium (DMEM containing 10% FBS) 24 h after infection to select infected cells. Cells were selected by blasticidin for 10 d and were subcultured when necessary until being harvested.
RT-qPCR analysis
Total RNAs were purified from subconfluent MEFs using standard methods and subjected to reverse transcription using random primers (Promega) and the SuperScript II reverse transcriptase (Invitrogen). Real-time quantitative PCR was done with the LightCycler 480 SYBR Green I Master kit (Roche) and the Mastercycler Realplex apparatus (Eppendorf). PCRs were performed with the oligonucleotide pairs 5′-GCCAGATGTGCTCAGTTTCC-3′ and 5′-CTGCCTCATAGCCTGGATCA-3′ for Tdg and 5′-GGCTGTATTCCCCTCCATCG-3′ and 5′-CCAGTTGGTAACAATGCCATGT-3′ for Actb. Results were normalized to Actb.
RNA-seq
After isolation of total cellular RNA from subconfluent MEFs, libraries of template molecules suitable for strand-specific high-throughput DNA sequencing were created using the TruSeq Stranded Total RNA with Ribo-Zero Gold Prep Kit (# RS-122-2301, Illumina). Briefly, starting with 300 ng total RNA, the first step involved the removal of cytoplasmic and mitochondrial ribosomal RNA (rRNA) using biotinylated, target-specific oligos combined with Ribo-Zero rRNA removal beads. Following purification, the RNA was fragmented into small pieces using divalent cations under elevated temperature. The cleaved RNA fragments were copied into first-strand cDNA using reverse transcriptase and random primers, followed by second-strand cDNA synthesis using DNA Polymerase I and RNase H. The double-stranded cDNA fragments were blunted using T4 DNA polymerase, Klenow DNA polymerase and T4 PNK. A single “A” nucleotide was added to the 3′ ends of the blunt DNA fragments using a Klenow fragment (3′ to 5′ exo minus) enzyme. The cDNA fragments were ligated to double-stranded adapters using T4 DNA Ligase. The ligated products were enriched by PCR amplification (30 sec at 98°C; [10 sec at 98°C, 30 sec at 60°C, 30 sec at 72°C] × 12 cycles; 5 min at 72°C). Then surplus PCR primers were removed by purification using AMPure XP beads (Agencourt Biosciences). Final cDNA libraries were checked for quality and quantified using the 2100 Bioanalyzer (Agilent). The libraries were loaded in the flow cell at 7 pM concentration, and clusters were generated in the Cbot and sequenced in the Illumina HiSeq 2500 as single-end 50 base reads following Illumina's instructions. Image analysis and base calling were performed using RTA 1.17.20 and CASAVA 1.8.2. Reads were mapped onto the mm9 assembly of the mouse genome by using TopHat (Trapnell et al. 2009) and the Bowtie aligner (Langmead et al. 2009). Quantification of gene expression was performed using HTSeq-0.5.4p3 (http://www-huber.embl.de/users/anders/HTSeq) and gene annotations from Ensembl release 67. Read counts have been normalized across libraries with the statistical method proposed by Anders and Huber (Anders and Huber 2010) and implemented in the DESeq Bioconductor library.
5mC/5hmC/5fC/5caC DNA immunoprecipitation assays
DNA immunoprecipitation assays were done as previously described (Shen et al. 2013) with some modifications. Briefly, 10 µg DNA was used as input, then 2 µL 5mC antibody (Active Motif, 39649), 4 µL 5hmC antibody (Active Motif, 39791), 1 µL 5fC anti-serum or 0.5 µL 5caC anti-serum (Yi Zhang) were used to immunoprecipitate modified DNA. DNA and antibodies were incubated at 4°C overnight in a final volume of 500 µL DIP buffer (10 mM sodium phosphate pH 7.0, 140 mM NaCl, 0.05% Triton X-100). The bound material was recovered after incubation with 30 µL blocked protein G Dynabeads (beads washed three times with 1 mL DIP buffer and incubated for 4 h minimum with BSA 1 mg mL−1 and yeast tRNA 0.5 mg mL−1). The beads were washed three times with 1 mL DIP buffer, then treated overnight with RNase A at 65°C in the presence of 300 mM NaCl and then treated 4 h with proteinase K at 55°C. Immunoprecipitated DNA was purified by phenol-chloroform extraction followed by ethanol precipitation. Four independent DNA immunoprecipitations per replicate were pooled for each condition before sequencing analysis. For each independent replicate, 5mC, 5hmC, 5fC, and 5caC DIP assays were performed using the same batches of genomic DNA.
ChIP assay
H3K9me3 and Pol2 ChIP experiments were performed as previously described (Obri et al. 2014). Briefly, 50 µg sonicated chromatin isolated from subconfluent MEFs was immunoprecipitated using 1 µL antibody anti-H3K9me3 (Abcam, ab8898) or 5 µg antibody anti-Pol2 (Santa Cruz, sc-9001 X). Five independent chromatin immunoprecipitations were pooled for each antibody before sequencing analysis.
ChIP-seq, DIP-seq, and computational analyses
ChIP-seq and DIP-seq were performed on an Illumina HiSeq 2500 as single-end 50 base reads following Illumina's instructions. Image analysis and base calling were performed using RTA 1.17.20 and CASAVA 1.8.2. Reads were mapped to the mouse genome (mm9) using Bowtie (Langmead et al. 2009) using the following arguments “-m 1 --strata --best -y -S -l 40 -p 2.” Peak detection was performed running MACS (Zhang et al. 2008) (http://liulab.dfci.harvard.edu/MACS/) using data sets normalized to 10 million uniquely mapped reads under settings in which the input fraction was used as negative control (effective genome size: 1.87×109; tag size: 50; b width: 300; P-value cutoff for peak detection: 1×10−5). Detected peak summits were annotated using HOMER (http://homer.ucsd.edu/homer/ngs). Heatmaps, global clustering, and quantitative comparisons of the ChIP-seq/DIP-seq data were performed running seqMINER (Ye et al. 2011) (http://bips.u-strasbg.fr/seqminer/), using data sets representing the average of replicates normalized to 10 million uniquely mapped reads. As reference coordinates, we used the annotated RepeatMasker (RMSK) database (for repetitive elements) or the Ensembl 67 database (for coding genes) of the mouse genome (mm9). The replicate average densities were normalized in reads per million mapped reads (rpm). In order to characterize the relationship between cytosine modification density and transcription level in repetitive elements, we computed average normalized densities (for DIP-seq data sets) and average transcription level (for RNA-seq data sets) per repetitive element (reads per kilobase per million reads [rpkm] or reads per kilobase per hundred million reads [rpk100m], as indicated). When clustered, the collected values were subjected to k-means clustering coupled to linear-based normalization. The conservation score assigned to each individual repeat element corresponds to the Smith Waterman alignment score provided by the RMSK database.
Generation of WIG tracks for peak visualization
To visualize peaks in the genome browser, WIG track files were generated using WigMaker for each data set. Scores represent tag numbers (average of two replicates) per 10 million uniquely mapped reads (reads per 10 million reads [rp10m]).
Repeat analysis
For repeat analyses, reads were aligned to repetitive elements in two passes. In the first pass, reads were aligned to the nonmasked mouse reference genome (NCBI37/mm9) using BWA v.0.6.2 (Li and Durbin 2009). Positions of the reads mapped uniquely to the mouse genome were cross-compared with the positions of the repeats extracted from UCSC (RMSK table in the UCSC database for mouse genome mm9), and reads overlapping a repeat sequence were annotated with the repeat family. In the second pass, reads not mapped or multimapped to the mouse genome in the previous pass were aligned to RepBase v.18.07 (Jurka et al. 2005) repeat sequences for rodent. Reads mapped to a unique repeat family were annotated with their corresponding family name. Finally, we summed up the read counts per repeat family of the two annotation steps. Data were normalized based upon library size. For enrichment analysis, normalized read counts of DIP samples per repeat family were divided by normalized read counts of matched input samples and expressed as log2 fold enrichment (average of the two replicates). Significance of the difference of repeat read counts between DIP and input samples was assessed using the Bionconductor package DESeq. To avoid over- or underestimating fold enrichments due to low sequence representation, repeat families with less than 100 mapped reads per DIP sample were excluded from further analysis.
Dinucleotide composition analyses
CG and CA heatmaps were generated using custom perl scripts wrapping BEDTools v.2.26.0 utilities (Quinlan and Hall 2010) and developed with custom functions. The scripts generated matrices of dinucleotide counts per bin in regions of interest. TreeView v.3.0 (Saldanha 2004) was then used to plot heatmaps from generated matrices.
Dot blot assay
Ten nanograms denatured oligos (CG-containing oligos: 5′-GCATCGTACGGAATCGCTTCTAGCCGGACATTAGCGATXGATCGATCAGGCTCGTAGGTACTCGACGGCAATCGTTAG-3′ or (CA)9-containing oligos: 5′-CTAACGATTGCCGTCGCACACACAXACACACACAGATCGCTAATGTCCGC-3′; X = C, 5mC, 5hmC, 5fC, or 5caC) were spotted onto a positively charged membrane (Amersham Hybond-XL). The membrane was then baked at 80°C and blocked for 1 h with 5% nonfat milk in TBST buffer (10 mM Tris-HCl pH 7.6, 150 mM NaCl, 0.1% Tween-20). Membranes were then incubated overnight with a 1:500 dilution of 5mC, 5hmC, 5fC, or 5caC antibodies. After three rounds of washing with the blocking solution, membranes were incubated with a 1:20,000 dilution of HRP-conjugated anti-mouse (for 5mC) or anti-rabbit (for 5hmC, 5fC, and 5caC) IgG secondary antibody. The membranes were then washed with TBST and treated with ECL.
TDG purification
The mouse His-tagged Tdg cDNA was cloned in a pET28b vector and expressed in the BL21-CodonPlus-RIL-pLysS (Stratagene) strain. A 1-L culture was grown in LB medium at 37°C until OD600 of 0.5 was reached before induction with 1 mM IPTG for 3 h at 25°C. Cells were lysed in 15 mL of buffer containing 20 mM Tris-HCl pH 7.65, 500 mM NaCl, 10% glycerol, 0.01% NP40, 20 mM imidazole, and 0.2 mM PMSF, and protease inhibitor cocktail tablets (Roche) in the presence of lysozyme at 1 mg/mL and sonicated on ice. The clarified supernatant was applied to cOmplete His-Tag Purification Resin (Roche), washed with 50 mM imidazole, and eluted with 300 mM imidazole using buffer containing 10 mM Tris-HCl pH 7.65, 150 mM NaCl, 10% glycerol, and 0.01% NP40. The eluate fraction was diluted two times with sodium phosphate buffer (50 mM sodium phosphate pH 7, 1 mM DTT, 1 mM EDTA), incubated with SP sepharose fast flow bead (GE Healthcare), extensively washed with sodium phosphate buffer containing 100 mM NaCl, and eluted with sodium phosphate buffer containing 500 mM NaCl. The eluate fraction was desalted with PD-10 Sephadex G-25 columns (GE Healthcare) previously equilibrated with TGEN buffer (20 mM Tris-HCl pH 7.65, 10% glycerol, 3 mM MgCl2, 0.1 mM EDTA, 0.01% NP40).
Glycosylase assay
The DNA substrates for enzymatic activity assays were prepared by annealing equimolar amounts of the following oligonucleotides (top_78mer, 5′-CTAACGATTGCCGTCGAGTACCTACGAGCCTGATCGATCXATCGCTAATGTCCGGCTAGAAGCGATTCCGTACGATGC-3′ ; X= G, A, T, or C; and bottom_78mer, 5′-GCATCGTACGGAATCGCTTCTAGCCGGACATTAGCGAT5fCYATCGATCAGGCTCGTAGGTACTCGACGGCAATCGTTAG-3′; Y=G, A, T, or C) in a buffer containing 10 mM Tris-HCl (pH 7.5), 1 mM EDTA, and 100 mM NaCl. DNA substrates were 5′-end labeled on the bottom strand (bottom_78mer) with [γ-32P]ATP and T4 polynucleotide kinase. Reaction mixtures (10 µL) containing 20 mM Tris-HCl pH 7.65, 50 mM NaCl, 5% glycerol, 1 mM DTT, 0.1 µg/µL BSA, and 2 nM of end-labeled substrates was incubated 15 min at 37°C with the indicated concentration of TDG (from 5 to 100 nM). The reaction was stopped by adding 1 µL 1 M NaOH and 10 µL formamide buffer (90% formamide, 10 mM EDTA, 0.1% blue bromophenol). The mixture was heated 5 min at 95°C before loading on a 12% denaturing polyacrylamide gel.
Published data sets
To calculate normalized density of H2A.Z at L1Md (Fig. 4D), we used our previously published data sets obtained in MEFs and deposited in GEO under accession number GSE51579 (Obri et al. 2014). To determine the relative enrichment for each cytosine modification at repeat families in control and Tdg-deficient ESCs, we downloaded data sets deposited in GEO under accession number GSE42250 (Shen et al. 2013) and performed an independent analysis of reads as described in the “repeat analysis” section.
Data access
All raw and processed data obtained in MEFs (RNA-seq, ChIP-seq, and DIP-seq) have been submitted to the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE66438.
Supplementary Material
Acknowledgments
We thank Irwin Davidson for critical reading of the manuscript and Yi Zhang and Shen Li for kindly providing us with anti-5fC and anti-5caC antibodies. This work was supported by institutional funds from CNRS, INSERM, Université de Strasbourg (UDS), Université de Grenoble Alpes, and by grants from La Ligue Nationale contre le Cancer Equipe labellisée (A.H.), ITMO Cancer AVIESAN (Alliance Nationale pour les Sciences de la Vie et de la Santé, National Alliance for Life Sciences and Health) within the framework of the Cancer Plan EPIG201409, INCA (INCa_4496 and INCa_4454), and ANR (VariZome, contract number ANR-12-BSV8-0018-01). Sequencing was performed by the IGBMC Microarray and Sequencing platform, a member of the France Génomique consortium (ANR-10-INBS-0009). C.P. thanks the Fondation pour la Recherche Médicale for financial support. A.I. thanks the Libyan Ministry of Higher Education and Scientific Research for financial support.
Author contributions: C.P. and A.I. performed the experiments. C.P., S.L.G., and A.V. conducted bioinformatics analyses. I.S. and H.M. performed protein purification. B.J. provided support for the high-throughput experiments. C.B. helped design the studies. C.P., S.D., and A.H. designed experiments, analyzed the data, and wrote the paper. A.H. conceived and supervised the project.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.213983.116.
References
- Anders S, Huber W. 2010. Differential expression analysis for sequence count data. Genome Biol 11: R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba Y, Huttenhower C, Nosho K, Tanaka N, Shima K, Hazra A, Schernhammer ES, Hunter DJ, Giovannucci EL, Fuchs CS, et al. 2010. Epigenomic diversity of colorectal cancer indicated by LINE-1 methylation in a database of 869 tumors. Mol Cancer 9: 125–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns KH, Boeke JD. 2012. Human transposon tectonics. Cell 149: 740–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro-Diaz N, Ecco G, Coluccio A, Kapopoulou A, Yazdanpanah B, Friedli M, Duc J, Jang SM, Turelli P, Trono D. 2014. Evolutionally dynamic L1 regulation in embryonic stem cells. Genes Dev 28: 1397–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordaux R, Batzer MA. 2009. The impact of retrotransposons on human genome evolution. Nat Rev Genet 10: 691–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortellino S, Xu J, Sannai M, Moore R, Caretti E, Cigliano A, Le Coz M, Devarajan K, Wessels A, Soprano D, et al. 2011. Thymine DNA glycosylase is essential for active DNA demethylation by linked deamination-base excision repair. Cell 146: 67–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Koning AP, Gu W, Castoe TA, Batzer MA, Pollock DD. 2011. Repetitive elements may comprise over two-thirds of the human genome. PLoS Genet 7: e1002384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deaton AM, Bird A. 2011. CpG islands and the regulation of transcription. Genes Dev 25: 1010–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Down TA, Rakyan VK, Turner DJ, Flicek P, Li H, Kulesha E, Graf S, Johnson N, Herrero J, Tomazou EM, et al. 2008. A Bayesian deconvolution strategy for immunoprecipitation-based DNA methylome analysis. Nat Biotechnol 26: 779–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreszer TR, Karolchik D, Zweig AS, Hinrichs AS, Raney BJ, Kuhn RM, Meyer LR, Wong M, Sloan CA, Rosenbloom KR, et al. 2012. The UCSC Genome Browser database: extensions and updates 2011. Nucleic Acids Res 40: D918–D923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgell MH, Hardies SC, Loeb DD, Shehee WR, Padgett RW, Burton FH, Comer MB, Casavant NC, Funk FD, Hutchison CA III. 1987. The L1 family in mice. Prog Clin Biol Res 251: 107–129. [PubMed] [Google Scholar]
- Ehrlich M. 2009. DNA hypomethylation in cancer cells. Epigenomics 1: 239–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellegren H. 2004. Microsatellites: simple sequences with complex evolution. Nat Rev Genet 5: 435–445. [DOI] [PubMed] [Google Scholar]
- Estecio MR, Gallegos J, Dekmezian M, Lu Y, Liang S, Issa JP. 2012. SINE retrotransposons cause epigenetic reprogramming of adjacent gene promoters. Mol Cancer Res 10: 1332–1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulkner GJ, Kimura Y, Daub CO, Wani S, Plessy C, Irvine KM, Schroder K, Cloonan N, Steptoe AL, Lassmann T, et al. 2009. The regulated retrotransposon transcriptome of mammalian cells. Nat Genet 41: 563–571. [DOI] [PubMed] [Google Scholar]
- Gabel HW, Kinde B, Stroud H, Gilbert CS, Harmin DA, Kastan NR, Hemberg M, Ebert DH, Greenberg ME. 2015. Disruption of DNA-methylation-dependent long gene repression in Rett syndrome. Nature 522: 89–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabellini N. 2001. A polymorphic GT repeat from the human cardiac Na+Ca2+ exchanger intron 2 activates splicing. Eur J Biochem 268: 1076–1083. [DOI] [PubMed] [Google Scholar]
- Gebhardt F, Zanker KS, Brändt B. 1999. Modulation of epidermal growth factor receptor gene transcription by a polymorphic dinucleotide repeat in intron 1. J Biol Chem 274: 13176–13180. [DOI] [PubMed] [Google Scholar]
- Gehring M, Bubb KL, Henikoff S. 2009. Extensive demethylation of repetitive elements during seed development underlies gene imprinting. Science 324: 1447–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JU, Su Y, Shin JH, Shin J, Li H, Xie B, Zhong C, Hu S, Le T, Fan G, et al. 2014. Distribution, recognition and regulation of non-CpG methylation in the adult mammalian brain. Nat Neurosci 17: 215–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto H, Pais JE, Zhang X, Saleh L, Fu ZQ, Dai N, Correa IR Jr, Zheng Y, Cheng X. 2014. Structure of a Naegleria Tet-like dioxygenase in complex with 5-methylcytosine DNA. Nature 506: 391–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, Ding J, Jia Y, Chen Z, Li L, et al. 2011. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science 333: 1303–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard G, Eiges R, Gaudet F, Jaenisch R, Eden A. 2008. Activation and transposition of endogenous retroviral elements in hypomethylation induced tumors in mice. Oncogene 27: 404–408. [DOI] [PubMed] [Google Scholar]
- Hui J, Stangl K, Lane WS, Bindereif A. 2003. HnRNP L stimulates splicing of the eNOS gene by binding to variable-length CA repeats. Nat Struct Biol 10: 33–37. [DOI] [PubMed] [Google Scholar]
- Ichiyanagi K. 2013. Epigenetic regulation of transcription and possible functions of mammalian short interspersed elements, SINEs. Genes Genet Syst 88: 19–29. [DOI] [PubMed] [Google Scholar]
- Illingworth RS, Gruenewald-Schneider U, Webb S, Kerr AR, James KD, Turner DJ, Smith C, Harrison DJ, Andrews R, Bird AP. 2010. Orphan CpG islands identify numerous conserved promoters in the mammalian genome. PLoS Genet 6: e1001134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, D'Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. 2010. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature 466: 1129–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y. 2011. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 333: 1300–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurka J, Kapitonov VV, Pavlicek A, Klonowski P, Kohany O, Walichiewicz J. 2005. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet Genome Res 110: 462–467. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent L, Wong E, Li G, Huynh T, Tsirigos A, Ong CT, Low HM, Kin Sung KW, Rigoutsos I, Loring J, et al. 2010. Dynamic changes in the human methylome during differentiation. Genome Res 20: 320–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, et al. 2009. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462: 315–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Mukamel EA, Nery JR, Urich M, Puddifoot CA, Johnson ND, Lucero J, Huang Y, Dwork AJ, Schultz MD, et al. 2013. Global epigenomic reconfiguration during mammalian brain development. Science 341: 1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunyak VV, Prefontaine GG, Nuñez E, Cramer T, Ju BG, Ohgi KA, Hutt K, Roy R, García-Díaz A, Zhu X, et al. 2007. Developmentally regulated activation of a SINE B2 repeat as a domain boundary in organogenesis. Science 317: 248–251. [DOI] [PubMed] [Google Scholar]
- Maiti A, Drohat AC. 2011. Thymine DNA glycosylase can rapidly excise 5-formylcytosine and 5-carboxylcytosine: potential implications for active demethylation of CpG sites. J Biol Chem 286: 35334–35338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matlik K, Redik K, Speek M. 2006. L1 antisense promoter drives tissue-specific transcription of human genes. J Biomed Biotechnol 2006: 71753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medstrand P, van de Lagemaat LN, Mager DL. 2002. Retroelement distributions in the human genome: variations associated with age and proximity to genes. Genome Res 12: 1483–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mita P, Boeke JD. 2016. How retrotransposons shape genome regulation. Curr Opin Genet Dev 37: 90–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouse Genome Sequencing Consortium. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520–562. [DOI] [PubMed] [Google Scholar]
- Muotri AR, Marchetto MC, Coufal NG, Oefner R, Yeo G, Nakashima K, Gage FH. 2010. L1 retrotransposition in neurons is modulated by MeCP2. Nature 468: 443–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naylor LH, Clark EM. 1990. d(TG)n·d(CA)n sequences upstream of the rat prolactin gene form Z-DNA and inhibit gene transcription. Nucleic Acids Res 18: 1595–1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigumann P, Redik K, Matlik K, Speek M. 2002. Many human genes are transcribed from the antisense promoter of L1 retrotransposon. Genomics 79: 628–634. [DOI] [PubMed] [Google Scholar]
- Obri A, Ouararhni K, Papin C, Diebold ML, Padmanabhan K, Marek M, Stoll I, Roy L, Reilly PT, Mak TW, et al. 2014. ANP32E is a histone chaperone that removes H2A.Z from chromatin. Nature 505: 648–653. [DOI] [PubMed] [Google Scholar]
- Okano M, Bell DW, Haber DA, Li E. 1999. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99: 247–257. [DOI] [PubMed] [Google Scholar]
- Papale LA, Zhang Q, Li S, Chen K, Keles S, Alisch RS. 2015. Genome-wide disruption of 5-hydroxymethylcytosine in a mouse model of autism. Hum Mol Genet 24: 7121–7131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pravica V, Asderakis A, Perrey C, Hajeer A, Sinnott PJ, Hutchinson IV. 1999. In vitro production of IFN-gamma correlates with CA repeat polymorphism in the human IFN-γ gene. Eur J Immunogenet 26: 1–3. [DOI] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldanha AJ. 2004. Java Treeview—extensible visualization of microarray data. Bioinformatics 20: 3246–3248. [DOI] [PubMed] [Google Scholar]
- Shen L, Wu H, Diep D, Yamaguchi S, D'Alessio AC, Fung HL, Zhang K, Zhang Y. 2013. Genome-wide analysis reveals TET- and TDG-dependent 5-methylcytosine oxidation dynamics. Cell 153: 692–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimajiri S, Arima N, Tanimoto A, Murata Y, Hamada T, Wang KY, Sasaguri Y. 1999. Shortened microsatellite d(CA)21 sequence down-regulates promoter activity of matrix metalloproteinase 9 gene. FEBS Lett 455: 70–74. [DOI] [PubMed] [Google Scholar]
- Slotkin RK, Martienssen R. 2007. Transposable elements and the epigenetic regulation of the genome. Nat Rev Genet 8: 272–285. [DOI] [PubMed] [Google Scholar]
- Sookdeo A, Hepp CM, McClure MA, Boissinot S. 2013. Revisiting the evolution of mouse LINE-1 in the genomic era. Mob DNA 4: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speek M. 2001. Antisense promoter of human L1 retrotransposon drives transcription of adjacent cellular genes. Mol Cell Biol 21: 1973–1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Dai N, Borgaro JG, Quimby A, Sun D, Correa IR Jr, Zheng Y, Zhu Z, Guan S. 2015. A sensitive approach to map genome-wide 5-hydroxymethylcytosine and 5-formylcytosine at single-base resolution. Mol Cell 57: 750–761. [DOI] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, et al. 2009. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324: 930–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama S, Dhahbi J, Roberts A, Mao G, Heo SJ, Pachter L, Martin DI, Boffelli D. 2014. Genome methylation in D. melanogaster is found at specific short motifs and is independent of DNMT2 activity. Genome Res 24: 821–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomizawa S, Kobayashi H, Watanabe T, Andrews S, Hata K, Kelsey G, Sasaki H. 2011. Dynamic stage-specific changes in imprinted differentially methylated regions during early mammalian development and prevalence of non-CpG methylation in oocytes. Development 138: 811–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh CP, Chaillet JR, Bestor TH. 1998. Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nat Genet 20: 116–117. [DOI] [PubMed] [Google Scholar]
- Xie W, Barr CL, Kim A, Yue F, Lee AY, Eubanks J, Dempster EL, Ren B. 2012. Base-resolution analyses of sequence and parent-of-origin dependent DNA methylation in the mouse genome. Cell 148: 816–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye T, Krebs AR, Choukrallah MA, Keime C, Plewniak F, Davidson I, Tora L. 2011. seqMINER: an integrated ChIP-seq data interpretation platform. Nucleic Acids Res 39: e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. 2008. Model-based analysis of ChIP-Seq (MACS). Genome Biol 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziller MJ, Gu H, Müller F, Donaghey J, Tsai LT, Kohlbacher O, De Jager PL, Rosen ED, Bennett DA, Bernstein BE, et al. 2013. Charting a dynamic DNA methylation landscape of the human genome. Nature 500: 477–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.