Abstract
Large-scale, massively parallel sequencing of human cancer samples has revealed tremendous genetic heterogeneity within individual tumors. Indeed, tumors are composed of an admixture of diverse subpopulations—subclones—that vary in space and time. Here, we discuss a principal driver of clonal diversification in cancer known as chromosomal instability (CIN), which complements other modes of genetic diversification creating the multilayered genomic instability often seen in human cancer. Cancer cells have evolved to fine-tune chromosome missegregation rates to balance the acquisition of heterogeneity while preserving favorable genotypes, a dependence that can be exploited for a therapeutic benefit. We discuss how whole-genome doubling events accelerate clonal evolution in a subset of tumors by providing a viable path toward favorable near-triploid karyotypes and present evidence for CIN-induced clonal speciation that can overcome the dependence on truncal initiating events.
Tumors are composed of genetically diverse subclones. Chromosomal missegregation and instability play a powerful role in driving clonal diversification, enabling cells to acquire heterogeneity while preserving favorable genotypes.
Cancer originates from a single cell that has acquired a number of central genetic lesions allowing it to rescind normal multicellular development and expand, in an uncontrolled fashion at the expense of the host (Hanahan and Weinberg 2011). Genetic alterations that allow the initial expansion are typically defined as truncal and have been the focus of much investigation, as they represent attractive therapeutic targets harbored by the entirety of the malignant population (Barber et al. 2015). These studies have shed light on the number of mutations necessary for tumorigenesis, the order in which these mutations occur, and the specific types of alterations required to produce a malignant tumor (Bozic et al. 2010; Vogelstein et al. 2013; Tomasetti et al. 2014; Landau et al. 2015).
After acquiring truncal mutations, most tumors undergo a second phase of intratumoral diversification. Massively parallel sequencing of primary, metastatic, and relapsed tumors revealed frequent cancer subclones, which harbor genomic alterations that are not detected in the remainder of the tumor cell population. Notably, these subclones often show evidence supportive of positive selection in an active evolutionary process. In many tumors, subclonal mutations show marked enrichment of driver events, as well as convergent evolutionary processes whereby multiple subclones independently acquire mutations that activate similar oncogenic pathways (Gerlinger et al. 2012; Yates and Campbell 2012; Landau et al. 2013; 2015; Brastianos et al. 2015; Murugaesu et al. 2015; Stachler et al. 2015; Yates et al. 2015).
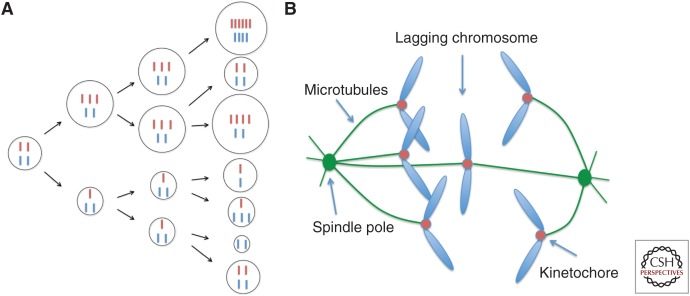
Fundamentally, subclonal heterogeneity stems from the ability of malignant cells to acquire novel mutations, a defining feature of many human tumors. Ongoing genomic diversification has been associated with poor prognosis and increased probability of tumor relapse after therapy (Landau et al. 2013, 2015; Andor et al. 2016; McGranahan et al. 2016). One of the central drivers of intratumoral diversification is copy number instability of particular loci, large chromosome segments, or entire chromosomes (Holland and Cleveland 2009; Navin et al. 2011; Wang et al. 2014). Indeed, cancer cells often show substantial diversity with respect to chromosomal content (karyotypes) (Gilgenkrantz and Rivera 2003). Aneuploidy, which represents a “state” of abnormal karyotype, has long been recognized as a hallmark of tumorigenesis (Boveri 1903). Karyotypic heterogeneity in tumor cells draws its origin from “chromosomal instability” (otherwise known as CIN), a hallmark that distinguishes most cancers from its normal tissue counterpart. CIN does not only generate abnormal aneuploid karyotypes, but continually expands phenotypic heterogeneity as tumor cell populations undergo consecutive cell divisions (Fig. 1A) (Lengauer et al. 1998, 1997).
Figure 1.
Chromosomal instability (CIN) in cancer. (A) Cancer cells experience frequent errors in the segregation of entire chromosomes, leading to karyotypic heterogeneity. Depending on the oncogenic and tumor suppressive effect of genes encoded by individual chromosomes, individual karyotypes will lead to distinct proliferative states and subclonal heterogeneity shaping the overall fitness of the population. Cell size depicts the relative cell fitness. Chromosomal instability (CIN) can enable rapid expansion of cells with variable fitness. (B) Lagging chromosomes during mitosis are a hallmark of CIN in cancer. They result from the erroneous attachments of chromosomes to spindle microtubules at the kinetochores. Most of the cellular defects that lead to CIN in cancer converge onto this process and produce lagging chromosomes.
Aneuploidy is present in ∼80% of human solid neoplasms (Ame et al. 2008) the majority of which often exhibit CIN (Storchova and Kuffer 2008). CIN can be present in two major forms: The first is numerical CIN whereby copy numbers of entire chromosomes vary within and among tumor cells. The second form is structural CIN, which is defined by copy number variations of subchromosomal regions (Roschke and Rozenblum 2014). Although both types of CIN arise through distinct cellular mechanisms and occur at different rates, they are nonetheless related and the presence of numerical CIN can induce, or accelerate, structural CIN and vice versa (Crasta et al. 2012; Bakhoum et al. 2014a). For the purpose of this review, we will focus primarily on the contribution of numerical CIN to intratumoral heterogeneity. However, we will also discuss how numerical and structural CIN are related and how the multilayered CIN phenotype can play different roles in tumor evolution under various selection pressures.
Numerical CIN is primarily defined by the “rate” at which chromosomes missegregate throughout consecutive cell divisions (Lengauer et al. 1998). The frequency of chromosome missegregation rates, directly measured in cancer-derived cell lines, are on the order of ∼1.3 × 10−3 per chromosome copy per cell division, which is ∼50–100-fold higher than in normal nontransformed cells (Thompson and Compton 2008; Bakhoum et al. 2009b; Laughney et al. 2015). This translates into a chromosome missegregation event every 3–10 cell divisions after accounting for cellular ploidy. In full-fledged malignancies, chromosome missegregation events may have a positive or negative effect on cellular fitness depending on the altered dosage of hundreds to thousands of genes carried by the missegregated chromosomes (Davoli et al. 2013; Laughney et al. 2015); making CIN a powerful tool to rapidly shape the phenotypic landscape of tumor cell populations.
THE ORIGINS OF CIN
A number of cellular mechanisms have been found to lead to CIN in cancer (reviewed in Thompson et al. 2010), including weakened sister chromatid cohesion (Zhang et al. 2008), deregulated centrosome duplication (Ganem et al. 2009; Silkworth et al. 2009), defects in microtubule attachments to chromosomes at the kinetochores (Cimini et al. 2001; Bakhoum et al. 2009a,b), premitotic replication stress (Burrell et al. 2013), and mitotic DNA damage (Crasta et al. 2012; Bakhoum et al. 2014a). Irrespective of the underlying cause, these mechanisms converge onto the process of chromosome segregation during mitosis, and lead to lagging chromosomes during anaphase, which is considered a hallmark of chromosomally unstable cells (Fig. 1B) (Cimini et al. 2001; Thompson and Compton 2011; Bakhoum et al. 2014b). Chromosomes lag as a result of their simultaneous attachments to microtubules emanating from opposite spindle poles (Cimini et al. 2003, 2002), which increases their potential for missegregation or encapsulation in micronuclei (Crasta et al. 2012).
The process of chromosome segregation is vulnerable in the face of many cellular defects that are known to occur in cancer. Activation of oncogenic pathways, loss of key tumor suppressor pathways, and a large number of therapeutic interventions can either facilitate or induce CIN (Orr and Compton 2013; Lee et al. 2016). This vulnerability stands in sharp contrast to the lack of aberrant karyotypes observed in normal human cells (Knouse et al. 2014). Although not yet fully understood, work in mice and cancer-derived cell lines revealed that the tumor suppressors p53 and p21 play a key role in safeguarding against aneuploidy. Mice without functional p53 generate tumors with aneuploid karyotypes (Shao et al. 2000). Furthermore, abrogation of p53 or p21 allows tolerance of aneuploidy and the propagation of instability (Thompson and Compton 2010). Accordingly, mutations in the p53 pathway are more prevalent in advanced tumors that typically contain significant chromosome copy number variations (Rivlin et al. 2011). More recently, the Hippo tumor suppressor signaling pathway was shown to be activated in response to cytokinesis failure, thereby acting as a selective safeguard against tetraploidy but not aneuploidy (Ganem et al. 2014).
THE CONSEQUENCES OF ANEUPLOIDY
Even in a potentially permissible cellular environment lacking p53, there is strong evidence to show that induction of aneuploidy in diploid cells is associated with reduced fitness and delays tumor formation (Rowald et al. 2016; Sheltzer et al. 2016). Work in yeast and human cells shows that aneuploidy induces metabolic and proteotoxic stress, which translate into lower proliferation rates (Torres et al. 2007, 2010; Williams et al. 2008; Sheltzer and Amon 2011; Oromendia et al. 2012; Sheltzer et al. 2012; Sheltzer 2013; Dürrbaum et al. 2014). Given the increased metabolic demand and oxidative stress imparted by aneuploidy, this has led some to postulate that aneuploidy may serve as one of the triggers for the Warburg effect often seen in cancer, whereby cells shift away from oxidative phosphorylation toward aerobic glycolysis (Vander Heiden et al. 2009; Siegel and Amon 2012). Inducing aneuploidy in vivo through depletion of Cenp-E—a mitotic centromere protein involved in chromosome segregation—reduces the incidence of hepatocellular carcinomas (Weaver et al. 2007) and computational evidence suggests that the percent reduction in normal cell fitness in response to aneuploidy could range anywhere from 6% to 30% (Gusev et al. 2001; Valind et al. 2013).
Although aneuploidy leads to a global reduction in cellular fitness, it also enhances the ability of malignant populations to sample the fitness landscape for superior evolutionary trajectories. There are many examples in which the induction of aneuploidy and CIN leads to tumorigenesis. For instance, induction of CIN through overexpression of mad2, a spindle assembly checkpoint protein, leads to spontaneous tumor formation (Schvartzman et al. 2011). Likewise, depletion of Cenp-E leads to the formation of lymphomas and lung tumors, and overexpression of hec1, a kinetochore protein involved in chromosome segregation, also leads to spontaneous tumor and adenoma formation (Díaz-Rodríguez et al. 2008). It is important to note, however, that in many of these cases, secondary alterations were required for tumorigenesis, such as the deletion of p53, p21, or p19Arf (Fujiwara et al. 2005; Weaver et al. 2007). Thus, aneuploidy can have both oncogenic and tumor suppressive phenotypes in a context-dependent manner. The determinants of this dichotomous phenotype are still poorly understood; some have postulated that the degree of aneuploidy is an important determinant of its effect on tumorigenesis (Silk et al. 2013); however, this concept requires additional experimental validation.
WHOLE-GENOME DOUBLING DURING TUMOR EVOLUTION
Chromosomally unstable clones have a wide array of karyotypes available for them to sample. For example, a cell containing up to six copies of any given chromosome has 623 unique karyotypic combinations. Our understanding of how CIN enables tumor subclones to sample this vast aneuploid fitness landscape is only beginning to emerge. Laughney et al. (2015) explored this by allowing clonal populations to sample the aneuploid fitness landscape in silico, and found that they strikingly converged onto a favorable near-triploid state that maximized the numbers of chromosomes that are more likely to harbor oncogenes and minimize the copies of chromosomes that are more likely to harbor tumor suppressor genes (Fig. 2). Importantly, both diploid-derived and tetraploid-derived clones achieved this optimal karyotype state, yet at a much lower fitness cost for the latter. This was due to the ability of tetraploid-derived clones to buffer chromosome loss and avoid lethal nullisomy. This suggests that a tetraploid intermediate arising from whole genome doubling is a desirable path toward the commonly observed favorable near-triploid state.
Figure 2.
Experimental and computational work reveals a favorable near-triploid karyotype, which favors clonal fitness. Diploid-derived and tetraploid-derived populations alike can achieve this karyotype, albeit at a much lower fitness cost for the latter, suggesting that whole-genome doubling provides a more cost-effective path toward a highly favorable state.
Consistent with this framework, copy number analysis across cancer revealed a propensity for a near-triploid state (Storchova and Kuffer 2008; Laughney et al. 2015). Interestingly, Storchova and Kuffer found that intratumor karyotypic heterogeneity was maximal in tumors with near-triploid karyotypes, which may signify that higher rates of CIN allow a more rapid convergence onto this favorable state, or that the near-triploid state allows more facile clonal diversification (Storchova and Kuffer 2008). Second, genomic analysis also supports the presence of a tetraploid intermediate occurring early during tumor evolution followed by gradual chromosome loss in at least a third of tumor samples examined (Carter et al. 2012; Zack et al. 2013). Accordingly, experimental work in yeast, cancer cell lines, and xenograft mouse models shows that inducing whole-genome doubling promotes tumorigenesis and propagates further genomic instability (Fujiwara et al. 2005; Dewhurst et al. 2014; Selmecki et al. 2015).
Given the facilitating impact of genome duplication on cancer, it is not surprising that many factors safeguard against the proliferation of tetraploid cells either before transformation or in the early stages of tumorigenesis (Ganem and Pellman 2007). Just like aneuploidy induction, polyploidization induces a p53-mediated cell-cycle arrest (Carter 1967; Fujiwara et al. 2005). This arrest is likely instigated by DNA damage and other osmotic and proteotoxic stressors that are mediated by p38/MAP kinase (Castedo et al. 2006; Srsen et al. 2006). More recently, Ganem et al. (2014) identified the Hippo tumor suppressor pathway as a tetraploid-specific checkpoint that responds to cytokinesis failure. Activation of this pathway is mediated in part by the presence of extra centrosomes that alter small G-protein signaling, which activate LATS2 kinase. LATS2 in turn inhibits the transcriptional regulators YAP and TAZ through a p53-dependent mechanism. Interestingly, this pathway was selectively activated in response to whole-genome doubling and not in response to whole-chromosome missegregation, highlighting the specificity of this pathway toward tetraploidy.
Thus, whole-genome doubling is a key step in the “evolution” of the multicellular genome to the unicellular-like cancer genome. Consistently, powerful mechanisms have evolved to prevent whole-genome doubling (Ganem and Pellman 2007; Mable 2013); however, once cells are able to overcome these safeguards whole-genome doubling generates a more plastic genome that empowers the evolutionary process and confers higher adaptive potential to the malignant population. The exact contribution of whole-genome doubling during early and late tumor evolution remains to be established. Although tetraploidization may make it harder for cells to fully inactivate key tumor suppressor genes, it is likely that the ratio of oncogenes-to-tumor suppressor genes is the major determinant of tumor fitness. Moreover, the presence of multiple copies of each chromosome allows the cells to shuffle their chromosome numbers with a lower risk of lethal nullisomy, where all copies of an essential gene are lost. In summary, the recurrent patterns of chromosome copy number changes seen across human cancer, as well as the observation of convergent evolution toward near triploid karyotypes, show that chromosome missegregation is not a mere bystander during tumor evolution, but actively contributes to malignant speciation.
LINKING NUMERICAL CIN TO STRUCTURAL CIN AND CHROMOTHRIPSIS
Numerical CIN does not exist in isolation. Numerical chromosomal aberrations are often coinherited with complex structural damage that alters the juxtaposition of genomic loci (e.g., translocations, inversions, duplications, etc.). Although they occur through distinct mechanisms and at different rates, structural and numerical CIN are interrelated (Sheltzer et al. 2011; Crasta et al. 2012; Burrell et al. 2013; Bakhoum et al. 2014a). Replication stress and structural chromosomal damage can lead to defective mitotic chromosome segregation as both acentric chromosomes and dicentric chromosomes cannot form proper bi-oriented attachments to the mitotic spindle (Murnane 2012; Burrell et al. 2013). Additionally, activation of the DNA damage response during mitosis, either through extrinsic factors or uncorrected premitotic defects, perturbs microtubule attachments to chromosomes and leads to whole-chromosome segregation errors and CIN (Bakhoum et al. 2014a; Orthwein et al. 2014).
Conversely, chromosome missegregation during mitosis can also lead to abrupt structural chromosomal aberrations. Lagging chromosomes are repeatedly encapsulated in micronuclei, which are defective in DNA replication and repair and have dysfunctional micronuclear envelopes (Crasta et al. 2012). Such micronuclei undergo catastrophic micronuclear envelope collapse associated with invasion of the endoplasmic reticulum tubules into their enclosed chromatin (Hatch et al. 2013). Through a yet unidentified mechanism, exposure of this double-stranded DNA to the cytosol leads to dramatic chromosome pulverization (Fig. 3) (Crasta et al. 2012). By performing single-cell sequencing, Zhang et al. (2015) were able to show that pulverized chromosomes can be repaired through nonhomologous end joining, leading to massive chromosome rearrangements, a process known as “chromothripsis” (Stephens et al. 2011). This mitotic pathway offers the means to dramatically alter the genome in the span of a single cell division. Chromothripsis can also occur as a result of erroneous segregation of dicentric chromosomes resulting from telomere fusion (Maciejowski et al. 2015). It is likely that most chromothripsis events lead to unviability—as cells lose chromosome fragments containing essential genes (Zhang et al. 2015)—although rarely chromothripsis leads to amplification or fusion events that significantly enhance cellular fitness. Indeed, in their original report of chromothripsis, Stephens et al. (2011) found that this process is associated with the formation of oncogene-harboring double-minute chromosomes that existed in hundreds of copies per cell.
Figure 3.
Diagram depicting the link between numerical and structural chromosomal instability (CIN). In addition to aneuploid karyotypes, chromosome segregation during mitosis also leads to the formation of chromosome-containing micronuclei. This in turn leads to pulverization of their enclosed chromosomes and persistence of DNA damage into the subsequent mitosis. Activation of a partial mitotic DNA damage response in turn leads to whole chromosome segregation forming a cycle linking numerical and structural CIN.
The rapid, punctuated genomic changes that result from such genomic “catastrophes,” stands in stark contrast to the progressive genomic alteration caused by numerical changes in chromosome copy numbers. The interconnectedness between structural and numerical CIN offers tumor cell populations the ability to alternate between these two rates of genomic transformation during the process of cancer evolution.
THE DYNAMICS OF CIN IN TUMOR CLONAL EVOLUTION AND THERAPEUTIC RESPONSE
How distinct chromosome copy numbers alter tumor cell fitness remains a poorly understood process. Chromosome (or arm)-level amplifications and deletions alter transcript and protein levels of hundreds to thousands of genes (Torres et al. 2007; Williams et al. 2008) and have often been associated with altered prognosis and different tumor behavior.
The identification of genes on these chromosome segments responsible for altered prognosis has been challenging. A recent attempt by Roy et al. (2016) used a multitiered genomic dissection strategy and patient data to identify a handful of genes responsible for poor prognosis in low-grade gliomas harboring loss of the 9p chromosome arm. The investigators found that reductions in messenger RNA levels in response to 9p loss was due to haploinsufficiency in subset of genetic loci. In another subset, there was evidence for epigenetic inactivation on the remaining copies of the 9p chromosome arms leading to significant down-regulation of transcript levels. Interestingly, many genes on this chromosome arm were not significantly affected suggesting a dosage compensation mechanism.
In a separate study, Davoli et al. (2013) mapped tumor suppressor genes and oncogenes on human chromosomes inferred from the mutational patterns of these genes in human tumors. By analyzing the potency and density of oncogenes and tumor suppressor genes, they found that human chromosomes are not equivalent with respect to the net cumulative oncogenic and tumor suppressive effect of the genes they encode, suggesting that cumulative haploinsufficiency and triplosensitivity of pro- and antiproliferative genes shape tumor karyotypes and in part explains chromosome-level amplifications and deletions in cancer. This work provides a framework to explain how chromosome copy number changes that affect hundreds of genes may still output a coherent fitness effect, and therefore subjected to evolutionary selection.
To quantitatively model the evolutionary impact of CIN, Laughney et al. (2015) simulated the process of clonal evolution under chromosomally unstable conditions. Cell viability was determined by the net dosage of pro- and antiproliferative genes they contained, dictated by the respective chromosome copy numbers (Fig. 1A). Interestingly, the dependence of clonal fitness on chromosome missegregation rates was nonlinear and parabolic—fitness rapidly declined when missegregation rates were either too high or too low. At very low rates, populations lacked the ability to generate phenotypic heterogeneity induced by karyotypic diversity. As such, clonal populations were less capable of sampling fitter karyotypic states. On the other hand, excessively high rates of chromosome missegregation prevented populations from preserving highly fit karyotypic states, and also resulted in frequent nullisomy—the lethal loss of all copies of any given chromosome. Thus, tumor populations thrive when they balance the ability to diversify with the ability to preserve favorable genotypes. Strikingly, When missegregation frequencies were experimentally measured in cancer-derived cell lines they largely clustered around the range predicted to be optimal for population fitness (∼1.9 × 10−3 chromosome per copy per cell division), suggesting a strong evolutionary pressure to optimize CIN rates in human cancer (Fig. 4) (Laughney et al. 2015).
Figure 4.
Relative tumor clonal fitness as a function of chromosomal instability (CIN). Excessively low CIN would suppress genomic heterogeneity leading to reduction in tumor adaptability, whereas excessively high CIN would lead to genomic collapse, DNA damage, and frequent nullisomy events incompatible with viability.
The concept of an optimal range of chromosome missegregation rates has important therapeutic implications (Silk et al. 2013; Laughney et al. 2015). It predicts that tumors with moderate CIN (within the optimal range) would show the highest likelihood of stable plasticity promoting therapeutic resistance. On the other hand, tumors would be vulnerable to either a decrease or an increase of CIN rates outside of the optimal range that supports clonal evolution. This is indeed supported by a number of clinical observations and experimental evidence (Bakhoum et al. 2011; Birkbak et al. 2011; Roylance et al. 2011; Jamal-Hanjani et al. 2015; Andor et al. 2016). Among colorectal cancer-derived cell lines, chromosomally unstable cells are more resistant to targeted kinase inhibition as compared with their chromosomally stable counterparts (Lee et al. 2011). In patients with breast cancer, the presence of moderate CIN is associated with inferior prognosis and resistance to taxanes whereas extreme CIN is associated with improved survival and reduced tumor relapse after primary therapy (Swanton et al. 2009; Roylance et al. 2011; Jamal-Hanjani et al. 2015). Furthermore, in a pancancer genomic analysis of intratumor heterogeneity, patients that had tumors in the middle quartiles of chromosome copy number variations were at a significantly greater risk for death compared with those with tumors in the lowest or highest quartiles of chromosome copy number variations (Andor et al. 2016).
This notion offers a largely untapped opportunity to target chromosomally unstable tumors, by increasing CIN beyond the tolerable threshold. Support for this strategy stems from observations that CIN is a predictor of favorable response to therapies that induce chromosome missegregation, particularly conventional DNA-damaging cytotoxic therapies. For instance, CIN correlates with sensitivity to platinum-based therapies in ovarian cancer (Swanton et al. 2009), and in those with locally advanced rectal cancer, considerably elevated rates of chromosome missegregation are associated with a higher likelihood of pathologic response after treatment with radiation therapy and concurrent 5-fluorouracil (Zaki et al. 2014). Consistent with this notion, in the pancancer analysis performed by Andor et al. (2016), the negative impact of CIN on survival was abolished in the subset of patients who received DNA-damaging therapies (Fig. 4).
Thus, elevating chromosome missegregation rates in inherently chromosomally unstable cancers has been proposed as a potential antineoplastic strategy. Experimental evidence shows that cells are rendered more sensitive to agents that increase chromosome missegregation when they already experience preexisting CIN (Janssen et al. 2009; Bakhoum et al. 2015). The recent development of specific inhibitors that are capable of inducing massive chromosome missegregation is encouraging and has shown substantial preclinical promise (Mason et al. 2014). These agents can be rationally combined with traditional therapies, such as DNA-damaging agents, to achieve greater therapeutic benefits. Collectively, a better understanding of the pretreatment CIN status as well as the reciprocal effect of therapy on chromosome stability will enable us to design strategies that aim at displacing cells from their acquired favorable copy number states.
CIN AND THE EMERGENCE OF SUBCLONAL THERAPEUTIC RESISTANCE
The ability for CIN to enable phenotypic diversity among tumor cells can accelerate the emergence of subclones with specific genotypes that confer therapeutic resistance. For instance, treatment with EGFR inhibitors rapidly selects for glioblastoma-derived cells with amplification of the number of copies of chromosome 7, which harbors the EGFR gene (Chen et al. 2015).
In addition to directly altering the copy number of the target gene, CIN has the potential to induce resistance by circumventing driver alterations and relieving tumors from their dependence on oncogenic events that underlie their genesis. This was experimentally tested by Sotillo et al. (2010) where they induced CIN in Kras-driven lung tumors through transient overexpression of Mad2. Subsequent withdrawal of Kras led to tumor regression; however, transient Mad2 overexpression promoted tumor relapse after Kras withdrawal. These Kras-independent tumors were characterized by markedly elevated rates of aneuploidy and CIN. In these tumors, the investigators found up-regulation of proproliferative pathways as evidenced by elevated levels of phosphorylated forms of ERK, Stat3, and AKT. Other studies revealed that CIN can circumvent the loss-of-function of otherwise essential genes in yeast (Rancati et al. 2008; Liu et al. 2015). Collectively, these findings support the notion that CIN represents an adaptive cellular tool to overcome inhibition of essential genes or truncal genetic alterations that occur early during tumor evolution.
CONCLUDING REMARKS
Understanding the mechanisms and dynamics of tumor genomic diversification is critical to our ability to overcome the challenge of drug resistance. To target CIN therapeutically, we require a deeper understanding of several key aspects of CIN in cancer. These include identifying more comprehensively the cellular mechanisms that underlie CIN and enable tolerance for a chromosomally unstable state. Second, because CIN is defined by a rate of chromosome missegregation, methods interrogating this process in cancer should be refined to not only measure the extent of chromosome-level copy number alterations in cancer but also the rate at which chromosome number changes throughout consecutive cell divisions. Widely used methods such as comparative genomic hybridization and DNA sequencing of bulk tumor samples significantly mask the extent of karyotypic cell-to-cell variation, which may now be extensively probed through single-cell analyses (Laughney et al. 2015; Bakker et al. 2016). Third, our efforts to understand chromosome copy number changes would need to be coupled with quantitative models that aim to predict the behavior of tumor cell populations derived from our knowledge of chromosome segregation at the single-cell level. Ultimately, these models hold the promise of providing robust predictive tools enabling precision therapeutic manipulation of the rapidly evolving cancer genome.
ACKNOWLEDGMENTS
The authors thank Ashley Laughney Bakhoum, Luciano Martelotto (both at Memorial Sloan Kettering Cancer Center), and Jason Sheltzer (Cold Spring Harbor Laboratory) for comments and feedback. D.A.L. is supported by the Burroughs Wellcome Fund Career Award for Medical Scientists, Stand Up to Cancer Innovative Research Grant, the Kimmel Scholar’s Award, the ASH Scholar Award, Leukemia Lymphoma Society translational research program, and by the National Institutes of Health (NIH) Big Data to Knowledge initiative (BD2K, 1K01ES025431-01). S.F.B. is supported by The Department of Defense Breast Cancer Research Breakthrough Award No. W81XWH-16-1-0315, the Elsa U. Pardee Foundation, and the NCI MSKCC Cancer Center Core Grant P30 CA008748.
Footnotes
Editors: Charles Swanton, Alberto Bardelli, Kornelia Polyak, Sohrab Shah, and Trevor A. Graham
Additional Perspectives on Cancer Evolution available at www.perspectivesinmedicine.org
REFERENCES
- Ame JC, Cimini D, Fouquerel E, Gauthier LR, Biard D, Boussin FD, Dantzer F, de Murcia G, Schreiber V. 2008. Merotelic kinetochore orientation, aneuploidy, and cancer. Biochim Biophys Acta 1786: 32–40. [DOI] [PubMed] [Google Scholar]
- Andor N, Graham TA, Jansen M, Xia LC, Aktipis CA, Petritsch C, Ji HP, Maley CC. 2016. Pan-cancer analysis of the extent and consequences of intratumor heterogeneity. Nat Med 22: 105–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakhoum SF, Genovese G, Compton DA. 2009a. Deviant kinetochore microtubule dynamics underlie chromosomal instability. Curr Biol 19: 1937–1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakhoum SF, Thompson SL, Manning AL, Compton DA. 2009b. Genome stability is ensured by temporal control of kinetochore-microtubule dynamics. Nat Cell Biol 11: 27–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakhoum SF, Danilova OV, Kaur P, Levy NB, Compton DA. 2011. Chromosomal instability substantiates poor prognosis in patients with diffuse large B-cell lymphoma. Clin Cancer Res 17: 7704–7711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakhoum SF, Kabeche L, Murnane JP, Zaki BI, Compton DA. 2014a. DNA-damage response during mitosis induces whole-chromosome missegregation. Cancer Discov 4: 1281–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakhoum SF, Silkworth WT, Nardi IK, Nicholson JM, Compton DA, Cimini D. 2014b. The mitotic origin of chromosomal instability. Curr Biol 24: R148–R149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakhoum SF, Kabeche L, Wood MD, Laucius CD, Qu D, Laughney AM, Reynolds GE, Louie RJ, Phillips J, Chan DA, et al. 2015. Numerical chromosomal instability mediates susceptibility to radiation treatment. Nat Commun 6: 5990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakker B, Taudt A, Belderbos ME, Porubsky D, Spierings DCJ, de Jong TV, Halsema N, Kazemier HG, Hoekstra-Wakker K, Bradley A, et al. 2016. Single-cell sequencing reveals karyotype heterogeneity in murine and human malignancies. Genome Biol 17: 115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber LJ, Davies MN, Gerlinger M. 2015. Dissecting cancer evolution at the macro-heterogeneity and micro-heterogeneity scale. Curr Opin Genet Dev 30: 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkbak NJ, Eklund AC, Li Q, McClelland SE, Endesfelder D, Tan P, Tan IB, Richardson AL, Szallasi Z, Swanton C. 2011. Paradoxical relationship between chromosomal instability and survival outcome in cancer. Cancer Res 71: 3447–3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boveri T. 1903. Über mehrpolige mitosen als mittel zur analyse des zellkerns [On multipolar mitosis as a means of analysis of the cell nucleus]. Verh Phys Med Ges Wurzburg 35: 67–90. [Google Scholar]
- Bozic I, Antal T, Ohtsuki H, Carter H, Kim D, Chen S, Karchin R, Kinzler KW, Vogelstein B, Nowak MA. 2010. Accumulation of driver and passenger mutations during tumor progression. Proc Natl Acad Sci 107: 18545–18550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brastianos PK, Carter SL, Santagata S, Cahill DP, Taylor-Weiner A, Jones RT, Van Allen EM, Lawrence MS, Horowitz PM, Cibulskis K, et al. 2015. Genomic characterization of brain metastases reveals branched evolution and potential therapeutic targets. Cancer Discov 5: 1164–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrell RA, McClelland SE, Endesfelder D, Groth P, Weller M-C, Shaikh N, Domingo E, Kanu N, Dewhurst SM, Gronroos E, et al. 2013. Replication stress links structural and numerical cancer chromosomal instability. Nature 494: 492–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter SB. 1967. Effects of cytochalasins on mammalian cells. Nature 213: 261–264. [DOI] [PubMed] [Google Scholar]
- Carter SLS, Cibulskis KK, Helman EE, McKenna AA, Shen HH, Zack TT, Laird PWP, Onofrio RCR, Winckler WW, Weir BAB, et al. 2012. Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol 30: 413–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castedo M, Coquelle A, Vivet S, Vitale I, Kauffmann A, Dessen P, Pequignot MO, Casares N, Valent A, Mouhamad S, et al. 2006. Apoptosis regulation in tetraploid cancer cells. EMBO J 25: 2584–2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Mulla WA, Kucharavy A, Tsai H-J, Rubinstein B, Conkright J, McCroskey S, Bradford WD, Weems L, Haug JS, et al. 2015. Targeting the adaptability of heterogeneous aneuploids. Cell 160: 771–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimini D, Howell B, Maddox P, Khodjakov A, Degrassi F, Salmon ED. 2001. Merotelic kinetochore orientation is a major mechanism of aneuploidy in mitotic mammalian tissue cells. J Cell Biol 153: 517–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimini D, Fioravanti D, Salmon ED, Degrassi F. 2002. Merotelic kinetochore orientation versus chromosome mono-orientation in the origin of lagging chromosomes in human primary cells. J Cell Sci 115: 507–515. [DOI] [PubMed] [Google Scholar]
- Cimini DD, Ben B Moree, Canman JCJ, Salmon EDE. 2003. Merotelic kinetochore orientation occurs frequently during early mitosis in mammalian tissue cells and error correction is achieved by two different mechanisms. J Cell Sci 116: 4213–4225. [DOI] [PubMed] [Google Scholar]
- Crasta KK, Ganem NJ, Dagher RR, Lantermann AB, Ivanova EVE, Pan YY, Nezi LL, Protopopov AA, Chowdhury DD, Pellman DD. 2012. DNA breaks and chromosome pulverization from errors in mitosis. Nature 482: 53–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davoli T, Xu AW, Mengwasser KE, Sack LM, Yoon JC, Park PJ, Elledge SJ. 2013. Cumulative haploinsufficiency and triplosensitivity drive aneuploidy patterns and shape the cancer genome. Cell 155: 948–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewhurst SM, McGranahan N, Burrell RA, Rowan AJ, Gronroos E, Endesfelder D, Joshi T, Mouradov D, Gibbs P, Ward RL, et al. 2014. Tolerance of whole-genome doubling propagates chromosomal instability and accelerates cancer genome evolution. Cancer Discov 4: 175–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz-Rodríguez E, Sotillo R, Schvartzman JM, Benezra R. 2008. Hec1 overexpression hyperactivates the mitotic checkpoint and induces tumor formation in vivo. Proc Natl Acad Sci 105: 16719–16724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dürrbaum M, Kuznetsova AY, Passerini V, Stingele S, Stoehr G, Storchova Z. 2014. Unique features of the transcriptional response to model aneuploidy in human cells. BMC Genomics 15: 139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara T, Bandi M, Nitta M, Ivanova EV, Bronson RT, Pellman D. 2005. Cytokinesis failure generating tetraploids promotes tumorigenesis in p53-null cells. Nature 437: 1043–1047. [DOI] [PubMed] [Google Scholar]
- Ganem NJ, Pellman D. 2007. Limiting the proliferation of polyploid cells. Cell 131: 437–440. [DOI] [PubMed] [Google Scholar]
- Ganem NJ, Godinho SA, Pellman D. 2009. A mechanism linking extra centrosomes to chromosomal instability. Nature 460: 278–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem NJ, Cornils H, Chiu SY, O’Rourke KP, Arnaud J, Yimlamai D, Théry M, Camargo FD, Pellman D. 2014. Cytokinesis failure triggers Hippo tumor suppressor pathway activation. Cell 158: 833–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, Martinez P, Matthews N, Stewart A, Tarpey P, et al. 2012. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med 366: 883–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilgenkrantz S, Rivera EM. 2003. The history of cytogenetics. Portraits of some pioneers. Ann Genet 46: 433–442. [DOI] [PubMed] [Google Scholar]
- Gusev Y, Kagansky V, Dooley WC. 2001. Long-term dynamics of chromosomal instability in cancer: A transition probability model. Math Comput Model 33: 1253–1273. [Google Scholar]
- Hanahan D, Weinberg RA. 2011. Hallmarks of cancer: The next generation. Cell 144: 646–674. [DOI] [PubMed] [Google Scholar]
- Hatch EM, Fischer AH, Deerinck TJ, Hetzer MW. 2013. Catastrophic nuclear envelope collapse in cancer cell micronuclei. Cell 154: 47–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland AJ, Cleveland DW. 2009. Boveri revisited: Chromosomal instability, aneuploidy and tumorigenesis. Nat Rev Mol Cell Biol 10: 478–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamal-Hanjani M, A’Hern R, Birkbak NJ, Gorman P, Gronroos E, Ngang S, Nicola P, Rahman L, Thanopoulou E, Kelly G, et al. 2015. Extreme chromosomal instability forecasts improved outcome in ER-negative breast cancer: A prospective validation cohort study from the TACT trial. Ann Oncol 26: 1340–1346. [DOI] [PubMed] [Google Scholar]
- Janssen A, Kops GJPL, Medema RH. 2009. Elevating the frequency of chromosome mis-segregation as a strategy to kill tumor cells. Proc Natl Acad Sci 106: 19108–19113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knouse KA, Wu J, Whittaker CA, Amon A. 2014. Single cell sequencing reveals low levels of aneuploidy across mammalian tissues. Proc Natl Acad Sci 111: 13409–13414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landau DA, Carter SL, Stojanov P, McKenna A, Stevenson K, Lawrence MS, Sougnez C, Stewart C, Sivachenko A, Wang L, et al. 2013. Evolution and impact of subclonal mutations in chronic lymphocytic leukemia. Cell 152: 714–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landau DA, Tausch E, Taylor-Weiner AN, Stewart C, Reiter JG, Bahlo J, Kluth S, Bozic I, Lawrence M, Böttcher S, et al. 2015. Mutations driving CLL and their evolution in progression and relapse. Nature 526: 525–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laughney AM, Elizalde S, Genovese G, Bakhoum SF. 2015. Dynamics of tumor heterogeneity derived from clonal karyotypic evolution. Cell Rep 12: 809–820. [DOI] [PubMed] [Google Scholar]
- Lee AJX, Endesfelder D, Rowan AJ, Walther A, Birkbak NJ, Futreal PA, Downward J, Szallasi Z, Tomlinson IPM, Howell M, et al. 2011. Chromosomal instability confers intrinsic multidrug resistance. Cancer Res 71: 1858–1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H-S, Lee NCO, Kouprina N, Kim J-H, Kagansky A, Bates S, Trepel JB, Pommier Y, Sackett D, Larionov V. 2016. Effects of anticancer drugs on chromosome instability and new clinical implications for tumor-suppressing therapies. Cancer Res 76: 902–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengauer C, Kinzler KW, Vogelstein B. 1997. Genetic instability in colorectal cancers. Nature 386: 623–627. [DOI] [PubMed] [Google Scholar]
- Lengauer C, Kinzler KW, Vogelstein B. 1998. Genetic instabilities in human cancers. Nature 396: 643–649. [DOI] [PubMed] [Google Scholar]
- Liu G, Yong MYJ, Yurieva M, Srinivasan KG, Liu J, Lim JSY, Poidinger M, Wright GD, Zolezzi F, Choi H, et al. 2015. Gene essentiality is a quantitative property linked to cellular evolvability. Cell 163: 1388–1399. [DOI] [PubMed] [Google Scholar]
- Mable BK. 2013. Polyploids and hybrids in changing environments: Winners or losers in the struggle for adaptation? Heredity (Edinb) 110: 95–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciejowski J, Li Y, Bosco N, Campbell PJ, de Lange T. 2015. Chromothripsis and kataegis induced by telomere crisis. Cell 163: 1641–1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason JM, Lin DC-C, Wei X, Che Y, Yao Y, Kiarash R, Cescon DW, Fletcher GC, Awrey DE, Bray MR, et al. 2014. Functional characterization of CFI-400945, a Polo-like kinase 4 inhibitor, as a potential anticancer agent. Cancer Cell 26: 163–176. [DOI] [PubMed] [Google Scholar]
- McGranahan N, Furness AJS, Rosenthal R, Ramskov S, Lyngaa R, Saini SK, Jamal-Hanjani M, Wilson GA, Birkbak NJ, Hiley CT, et al. 2016. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science 351: 1463–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murnane JP. 2012. Telomere dysfunction and chromosome instability. Mutat Res 730: 28–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murugaesu N, Wilson GA, Birkbak NJ, Watkins TBK, McGranahan N, Kumar S, Abbassi-Ghadi N, Salm M, Mitter R, Horswell S, et al. 2015. Tracking the genomic evolution of esophageal adenocarcinoma through neoadjuvant chemotherapy. Cancer Discov 5: 821–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navin N, Kendall J, Troge J, Andrews P, Rodgers L, McIndoo J, Cook K, Stepansky A, Levy D, Esposito D, et al. 2011. Tumour evolution inferred by single-cell sequencing. Nature 472: 90–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oromendia AB, Dodgson SE, Amon A. 2012. Aneuploidy causes proteotoxic stress in yeast. Genes Dev 26: 2696–2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr B, Compton DA. 2013. A double-edged sword: How oncogenes and tumor suppressor genes can contribute to chromosomal instability. Front Oncol 3: 164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orthwein A, Fradet-Turcotte A, Noordermeer SM, Canny MD, Brun CM, Strecker J, Escribano-Diaz C, Durocher D. 2014. Mitosis inhibits DNA double-strand break repair to guard against telomere fusions. Science 344: 189–193. [DOI] [PubMed] [Google Scholar]
- Rancati G, Pavelka N, Fleharty B, Noll A, Trimble R, Walton K, Perera A, Staehling-Hampton K, Seidel CW, Li R. 2008. Aneuploidy underlies rapid adaptive evolution of yeast cells deprived of a conserved cytokinesis motor. Cell 135: 879–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivlin N, Brosh R, Oren M, Rotter V. 2011. Mutations in the p53 tumor suppressor gene: Important milestones at the various steps of tumorigenesis. Genes Cancer 2: 466–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roschke AV, Rozenblum E. 2014. Multi-layered cancer chromosomal instability phenotype. Front Oncol 3: 302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowald K, Mantovan M, Passos J, Buccitelli C, Mardin BR, Korbel JO, Jechlinger M, Sotillo R. 2016. Negative selection and chromosome instability induced by Mad2 overexpression delay breast cancer but facilitate oncogene-independent outgrowth. Cell Rep 15: 2679–2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy DM, Walsh LA, Desrichard A, Huse JT, Wu W, Gao J, Bose P, Lee W, Chan TA. 2016. Integrated genomics for pinpointing survival loci within arm-level somatic copy number alterations. Cancer Cell 29: 737–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roylance R, Endesfelder D, Gorman P, Burrell RA, Sander J, Tomlinson I, Hanby AM, Speirs V, Richardson AL, Birkbak NJ, et al. 2011. Relationship of extreme chromosomal instability with long-term survival in a retrospective analysis of primary breast cancer. Cancer Epidemiol Biomarkers Prev 20: 2183–2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schvartzman J-M, Duijf PHG, Sotillo R, Coker C, Benezra R. 2011. Mad2 is a critical mediator of the chromosome instability observed upon Rb and p53 pathway inhibition. Cancer Cell 19: 701–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selmecki AM, Maruvka YE, Richmond PA, Guillet M, Shoresh N, Sorenson AL, De S, Kishony R, Michor F, Dowell R, et al. 2015. Polyploidy can drive rapid adaptation in yeast. Nature 519: 349–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao C, Deng L, Henegariu O, Liang L, Stambrook PJ, Tischfield JA. 2000. Chromosome instability contributes to loss of heterozygosity in mice lacking p53 Proc Natl Acad Sci 97: 7405–7410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheltzer JM. 2013. A transcriptional and metabolic signature of primary aneuploidy is present in chromosomally unstable cancer cells and informs clinical prognosis. Cancer Res 73: 6401–6412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheltzer JM, Amon A. 2011. The aneuploidy paradox: Costs and benefits of an incorrect karyotype. Trends Genet 27: 446–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheltzer JM, Blank HM, Pfau SJ, Tange Y, George BM, Humpton TJ, Brito IL, Hiraoka Y, Niwa O, Amon A. 2011. Aneuploidy drives genomic instability in yeast. Science 333: 1026–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheltzer JM, Torres EM, Dunham MJ, Amon A. 2012. Transcriptional consequences of aneuploidy. Proc Natl Acad Sci 109: 12644–12649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheltzer JM, Ko JH, Burgos N, Chung ES. 2016. Single-chromosome aneuploidy commonly functions as a tumor suppressor. bioRxiv 040162; 10.1101/040162. [DOI] [Google Scholar]
- Siegel JJ, Amon A. 2012. New insights into the troubles of aneuploidy. Annu Rev Cell Dev Biol 28: 189–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silk AD, Zasadil LM, Holland AJ, Vitre B, Cleveland DW, Weaver BA. 2013. Chromosome missegregation rate predicts whether aneuploidy will promote or suppress tumors. Proc Natl Acad Sci 110: E4134–E4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silkworth WT, Nardi IK, Scholl LM, Cimini D. 2009. Multipolar spindle pole coalescence is a major source of kinetochore mis-attachment and chromosome mis-segregation in cancer cells. PLoS ONE 4: e6564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotillo R, Schvartzman J-M, Socci ND, Benezra R. 2010. Mad2-induced chromosome instability leads to lung tumour relapse after oncogene withdrawal. Nature 464: 436–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srsen V, Gnadt N, Dammermann A, Merdes A. 2006. Inhibition of centrosome protein assembly leads to p53-dependent exit from the cell cycle. J Cell Biol 174: 625–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stachler MD, Taylor-Weiner A, Peng S, McKenna A, Agoston AT, Odze RD, Davison JM, Nason KS, Loda M, Leshchiner I, et al. 2015. Paired exome analysis of Barrett’s esophagus and adenocarcinoma. Nat Genet 47: 1047–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens PJ, Greenman CD, Fu B, Yang F, Bignell GR, Mudie LJ, Pleasance ED, Lau KW, Beare D, Stebbings LA, et al. 2011. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell 144: 27–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storchova Z, Kuffer C. 2008. The consequences of tetraploidy and aneuploidy. J Cell Sci 121: 3859–3866. [DOI] [PubMed] [Google Scholar]
- Swanton C, Nicke B, Schuett M, Eklund AC, Ng C, Li Q, Hardcastle T, Lee A, Roy R, East P, et al. 2009. Chromosomal instability determines taxane response. Proc Natl Acad Sci 106: 8671–8676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson SL, Compton DA. 2008. Examining the link between chromosomal instability and aneuploidy in human cells. J Cell Biol 180: 665–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson SL, Compton DA. 2010. Proliferation of aneuploid human cells is limited by a p53-dependent mechanism. J Cell Biol 188: 369–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson SL, Compton DA. 2011. Chromosome missegregation in human cells arises through specific types of kinetochore–microtubule attachment errors. Proc Natl Acad Sci 108: 17974–17978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson SL, Bakhoum SF, Compton DA. 2010. Mechanisms of chromosomal instability. Curr Biol 20: R285–R295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasetti C, Marchionni L, Nowak MA, Parmigiani G, Vogelstein B. 2014. Only three driver gene mutations are required for the development of lung and colorectal cancers. Proc Natl Acad Sci 112: 118–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres EM, Sokolsky T, Tucker CM, Chan LY, Boselli M, Dunham MJ, Amon A. 2007. Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science 317: 916–924. [DOI] [PubMed] [Google Scholar]
- Torres EM, Dephoure N, Panneerselvam A, Tucker CM, Whittaker CA, Gygi SP, Dunham MJ, Amon A. 2010. Identification of aneuploidy-tolerating mutations. Cell 143: 71–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valind A, Jin Y, Gisselsson D. 2013. Elevated tolerance to aneuploidy in cancer cells: Estimating the fitness effects of chromosome number alterations by in silico modelling of somatic genome evolution. PLoS ONE 8: e70445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB. 2009. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science 324: 1029–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Kinzler KW. 2013. Cancer genome landscapes. Science 339: 1546–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Waters J, Leung ML, Unruh A, Roh W, Shi X, Chen K, Scheet P, Vattathil S, Liang H, et al. 2014. Clonal evolution in breast cancer revealed by single nucleus genome sequencing. Nature 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver BAA, Silk AD, Montagna C, Verdier-Pinard P, Cleveland DW. 2007. Aneuploidy acts both oncogenically and as a tumor suppressor. Cancer Cell 11: 25–36. [DOI] [PubMed] [Google Scholar]
- Williams BR, Prabhu VR, Hunter KE, Glazier CM, Whittaker CA, Housman DE, Amon A. 2008. Aneuploidy affects proliferation and spontaneous immortalization in mammalian cells. Science 322: 703–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates LR, Campbell PJ. 2012. Evolution of the cancer genome. Nat Rev Genet 13: 795–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates LR, Gerstung M, Knappskog S, Desmedt C, Gundem G, Van Loo P, Aas T, Alexandrov LB, Larsimont D, Davies H, et al. 2015. Subclonal diversification of primary breast cancer revealed by multiregion sequencing. Nat Med 21: 751–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zack TI, Schumacher SE, Carter SL, Cherniack AD, Saksena G, Tabak B, Lawrence MS, Zhang C-Z, Wala J, Mermel CH, et al. 2013. Pan-cancer patterns of somatic copy number alteration. Nat Genet 45: 1134–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki BI, Suriawinata AA, Eastman AR, Garner KM, Bakhoum SF. 2014. Chromosomal instability portends superior response of rectal adenocarcinoma to chemoradiation therapy. Cancer 120: 1733–1742. [DOI] [PubMed] [Google Scholar]
- Zhang N, Ge G, Meyer R, Sethi S, Basu D, Pradhan S, Zhao YJ, Li XN, Cai WW, El-Naggar AK, et al. 2008. Overexpression of Separase induces aneuploidy and mammary tumorigenesis. Proc Natl Acad Sci 105: 13033–13038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang CZ, Spektor A, Cornils H, Francis JM, Jackson EK, Liu S, Meyerson M, Pellman D. 2015. Chromothripsis from DNA damage in micronuclei. Nature 522: 179–184. [DOI] [PMC free article] [PubMed] [Google Scholar]