Abstract
Kinetic analysis of 18F-fluorodeoxyglucose positron emission tomography data requires an accurate knowledge the arterial input function. The gold standard method to measure the arterial input function requires collection of arterial blood samples and is an invasive method. Measuring an image derived input function is a non-invasive alternative but is challenging due to partial volume effects caused by the limited spatial resolution of the positron emission tomography scanners. In this work, a practical image derived input function extraction method is presented, which only requires segmentation of the carotid arteries from MR images. The simulation study results showed that at least 92% of the true intensity could be recovered after the partial volume correction. Results from 19 subjects showed that the mean cerebral metabolic rate of glucose calculated using arterial samples and partial volume corrected image derived input function were 26.9 and 25.4 mg/min/100 g, respectively, for the grey matter and 7.2 and 6.7 mg/min/100 g for the white matter. No significant difference in the estimated cerebral metabolic rate of glucose values was observed between arterial samples and corrected image derived input function (p > 0.12 for grey matter and white matter). Hence, the presented image derived input function extraction method can be a practical alternative to noninvasively analyze dynamic 18F-fluorodeoxyglucose data without the need for blood sampling.
Keywords: Positron emission tomography, MRI, kinetic modelling, brain imaging, carotid artery
Introduction
Cerebral metabolic rate of glucose (cMRglc) in the brain can be quantitatively estimated using dynamic 18F-fluorodeoxyglucose (18F-FDG) positron emission tomography (PET) studies. To quantify this parameter, the dynamic behaviour of the 18F-FDG tracer needs to be described using a compartmental kinetic model1 or Patlak analysis.2,3 These kinetic analysis methods require an accurate knowledge of the available tracer concentration in plasma as a function of time, also known as the arterial input function (AIF). The AIF is conventionally measured by arterial cannulation and collection of blood samples, which is an invasive and uncomfortable procedure. Population-based input functions4,5 can be used as an alternative but these may introduce further errors due to the varying physiology across subjects and different injection protocols (i.e. injection rates) used during the introduction of the radiotracer.
Another noninvasive alternative to arterial blood samples is using image derived input functions (IDIFs), which are directly obtained from the reconstructed PET images. IDIFs can be derived by placing a volume of interest (VOI) over a suitable blood pool and creating a time activity curve (TAC) for whole blood signal. This method works successfully when vascular structures with large diameter such as aorta are in the field of view;6,7 however, it suffers from partial volume (PV) and spill-over effects in brain PET studies where carotid arteries are used. These artefacts are caused by the limited spatial resolution of the PET scanners, which may not be able to distinguish the exact source of the signal in small structures. Hence, the measured activity from carotid arteries will be affected by spill-out and spill-in effects and will not reflect the exact tracer concentration present in the blood. Variations in the blood to background ratio over time will result in changes in the shape of the derived IDIF.
There have been several studies trying to validate IDIF methods in brain imaging, aiming to avoid the need for blood samples. These methods included various approaches to delineate arterial voxels from PET images or coregistered anatomical images and to correct for PV effects.8 Some of these methods require one or more blood samples to scale the estimated IDIF,9,10 but there are also some fully blood-free methods, which have recently become available.11,12 Zanotti-Fregonara et al. compared eight of these methods13 and concluded that more accurate kinetic analysis results can be obtained with methods using blood samples to scale the AIF compared with the blood-free methods.
In this work, we present a non-invasive blood-free IDIF extraction method, which utilises magnetic resonance angiography (MRA) images to delineate the arterial voxels. A practical partial volume correction (PVC) technique, which only requires segmentation of the region of interest (i.e. carotid arteries), is applied to correct PV effects. IDIFs extracted using the proposed method was used to quantify cMRglc in human subjects and results were validated against the input function based on arterial samples.
Materials and methods
Theory
In this work, we use a recently proposed PVC method called the Single-Target Correction (STC) method.14 This method requires segmentation of one single VOI and is not dependent on any separately segmented background regions. Hence, it does not need complex segmentation of the background areas. The correction is performed on a voxel-by-voxel basis using an iterative procedure, and uniformity of activity concentration within the defined region is assumed for the spill-out correction. Individual voxel values were used for the spill-in correction. The new method is a modification of a previous method called Multi-Target Correction (MTC),15 which was an extension of the Müller–Gärtner method.16
The correction procedure starts with the blurred image. In each iteration, the algorithm first corrects for spill-over between the voxels inside and outside of the VOI. This is done by subtracting a background term, which is re-calculated in each iteration. Next, it corrects for spill-out by dividing by recovery coefficients, which depend only on the size and shape of the VOI and on the PSF of the system. This follows from the assumption of uniformity.
If is the true image, the image, , blurred by the PSF, can be described as follows:
| (1) |
where is the PSF of the system (which can be position-invariant, although this is not strictly necessary), and and are 3D spatial coordinates.
If a VOI is defined, called Ω, the STC method can be described as follows in pseudo-code:
where is the estimated image after k iterations, IΩ(·) is the indicator function for Ω, R(·) contains the recovery factors to correct for spill-out, gk(·) represents the spill-over term, estimated at each iteration and N is the number of iterations.
The main difference between STC and the Müller–Gärtner method16 is that STC does not require prior estimation of the mean value in the background region/regions for the spill-in correction. Instead, this correction term is estimated iteratively on a voxel-by-voxel basis. For this purpose, it is necessary to estimate the spill-over in both directions across the region boundary.
Simulation work
To evaluate the performance of the Single-Target PVC method, a simulation study based on real carotid arteries MR data was designed. Segmented carotid arteries from an MRI angiography image with voxel size of 0.39 × 0.39 × 1 mm3 were used to generate 3D emission and attenuation maps. An attenuation coefficient value of 0.096 cm−1 was assigned to soft tissue. To generate a digital 3D phantom, emission values were assigned to a uniform background region and inside carotid arteries on the segmented images. Six different phantoms with various background to artery intensity ratios were generated with ratios of 1/10, 1/5, 1/3, 1/1, 1.5/1 and 2/1. Each phantom was blurred using an isotropic Gaussian point spread function (PSF) with FWHM of 6.50 mm and forward projected using Software for Tomographic Reconstruction (STIR)17 to generate simulated 3D PET data. The Siemens Biograph mMR scanner geometry was simulated, taking account of attenuation and scatter. PET images were reconstructed using OSEM with three iterations and 21 subsets to a 3D matrix with a voxel size of 2.08 × 2.08 × 2.013 mm3.
The reconstructed PET images were resampled to MR angiography space (0.39 × 0.39 × 1 mm3) using tri-linear interpolation and the STC PVC method was applied with 15 iterations and the segmented carotid arteries as the mask image. To evaluate the effect of different PSFs in the correction, STC was applied to each of the reconstructed phantoms and the error between the recovered and true carotid activities were computed for different PSFs. The optimal PSF FWHM value was found by minimizing the mean error, averaged across the six phantoms.
An additional experiment was designed to measure the PSF from the reconstructed PET images. First, the centroids of the carotid arteries on axial PET and segmented MR images were found. The segmented images were blurred using a 3D Gaussian PSF. Line profiles were drawn through centroids of left and right carotid arteries in x and y directions on both PET and blurred segmented MR images. These line profiles were fitted with a Gaussian function and mean FWHMs of Gaussian fits were calculated for PET and blurred MRI data. This was repeated in an optimization routine to find the optimal PSF, which gave the lowest difference between the mean FWHM of fitted Gaussian functions on PET and blurred segmented MR line profiles.
Data acquisition
Dynamic PET and anatomical MRI images were acquired on 21 healthy male subjects (mean age: 28.3 years, range: 22–40 years). The protocol for this study was approved by the Danish National Committee on Health Research Ethics (h-4-2012-167) and was conducted in accordance to the Declaration of Helsinki. All subjects had given written informed consent.
Each subject received an intravenous bolus injection of 200 MBq of 18F-FDG over 20 s followed by 10 mL of saline flush and underwent a dynamic PET scan on a Siemens Biograph 64 mCT scanner (Siemens Healthcare, Erlangen, Germany). The dynamic scan lasted for 1 h and data were sorted in the following manner: 6 × 10 s, 2 × 30 s, 3 × 60 s, 2 × 150 s, 2 × 300 s and 3 × 600 s. The PET images were reconstructed using OSEM with four iterations and 24 subsets and a 4-mm Gaussian filter was applied. The data were reconstructed into a 128 × 128 matrix with a pixel size of 2.5 mm and 74 slices were acquired with a slice thickness of 3 mm. Attenuation correction was performed using information from the CT scan. The PET data were corrected for randoms, scatter, attenuation and radioactive decay. Arterial blood samples for measurement of radioactivity concentration in arterial plasma were drawn from the radial artery at 33 time points, with 10 s intervals in the first 2 min. Sampling was performed using vials with internal vacuum over 3 s at the beginning of each time interval. Immediately prior to each measurement, a similar vial was used to rinse residual activity out of the catheter. Plasma radioactivity concentrations were measured in a well-type gamma counter (COBRA 5003; Packard Instruments). Plasma glucose level and hematocrit were also measured for each subject.
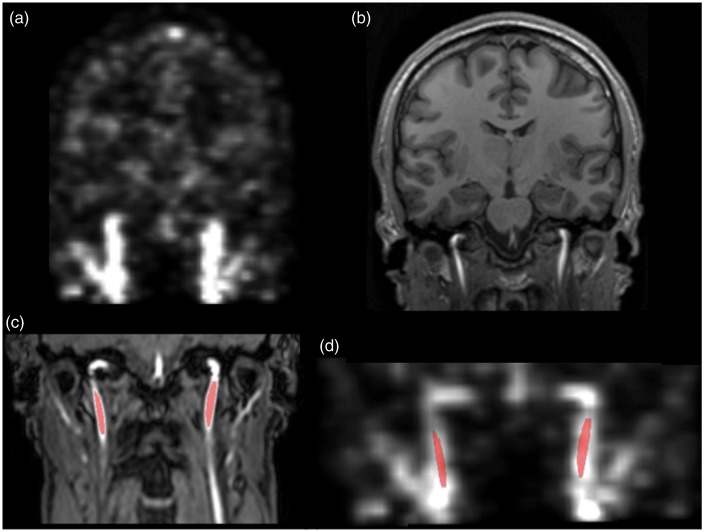
All MRI scans were performed on a 3T Achieva MRI scanner (Philips Medical Systems, Best, The Netherlands) using a 32-channel phased array head coil. Anatomical Magnetization Prepared Rapid Gradient Echo (MPRAGE) scans were obtained with a 3D T1-weighted turbo field echo sequence (150 slices, FOV = 241 ×180 × 165 mm3, TE = 2.78 ms, TR = 6.9 ms, flip angle = 9°) and were reconstructed into a 150 × 224 ×224 matrix with a voxel size of 1.1 × 1.1 × 1.1 mm3. High resolution time of flight angiography MR (MRA) images of the head and neck region were also acquired (100 slices, slice thickness = 1 mm, FOV = 200 × 200 cm2, TR = 23 ms, TE = 3.5 ms, flip angle = 18°) and were reconstructed with a voxel size of 0.39 × 0.39 × 1 mm3. Figure 1 shows examples of PET and MR images used in this work.
Figure 1.
Images used in this study: (a) PET frame showing the arteries, (b) MPRAGE image of the brain, (c) TOF MR angiography image of neck region and (d) PET frame co-registered to the MR angiography image. Marked voxels show the arterial voxels delineated from the MR angiography image.
Data analysis
Data from 2 of the 21 subjects were not included in the data analysis due to the movement of the head during the dynamic PET scan. Carotid arteries were segmented from MRA images using ITK-SNAP version 3.2,18 which includes an automatic active contour segmentation. Images were classified into two tissue classes using tissue classification and a region-growing algorithm was applied in the region with the arteries. Because of the different field of view of MRA images compared with the PET images, segmentation was limited to 3 cm below the petrous section of left and right internal carotids arteries (Figure 1(d)).
A linear image registration tool19 was used in the PET-MR image registration. Because of the different fields of view of the MRA and PET images, a two-step image registration method was applied where MPRAGE images were also utilized as they have the same field of view as the PET images. Within modality registration, MPRAGE to MRA was performed using translation (3 parameter) followed by a rigid registration (6 parameter) locally applied to masked carotid arteries. For the cross-modal registration, the sum of earliest 6 PET frames (0–60 s) was computed to maximize the intensity in the arteries and was registered to the MPRAGE image using a rigid registration (6 parameter). The combined matrix of these three transformations was used to resample PET frames onto MRA space.
PV effects in PET frames were corrected using STC where the segmented carotid arteries were used as the region of interest. A three-dimensional position-invariant isotropic Gaussian function with 6.8 mm FWHM was used to represent the scanner’s PSF. This PSF value was obtained by implementing the PSF measurement method by computing the FWHM of fitted Gaussian function to line profiles across the carotid centroids, as described above. This was performed for the two PET frames with highest arterial intensities (20 to 40 s) and the mean FWHM value between frames is calculated. This was done for each of the 19 subjects and the average calculated FWHM is used in the PVC of all subjects (6.80 ± 0.36 mm, range of 6.05 mm to 7.38 mm). Correction was applied to each PET frame individually. The PVC algorithm was found to converge in 10 iterations, after which the mean intensity within the carotid arteries stopped changing significantly.
After correcting for PV effects, whole blood TAC was measured by computing the mean intensity within the segmented carotid arteries. To obtain the IDIF, whole blood concentration was converted to plasma concentration using a rearranged version of equation (2), where HCT represents measured hematocrit and CBlood, CRBC and CPlasma represent the radioactivity concentration in whole blood, red blood cells and plasma, respectively. A population-based CRBC/CPlasma relationship was used in this conversion.20
| (2) |
As the arterial blood samples were taken from the radial artery, while the IDIFs were obtained from carotid arteries, there was a time difference and dispersion between these two input functions. To make a fair comparison against arterial blood samples, delay and dispersion were applied to the measured IDIFs. Delay was added to the IDIF by shifting the curve to later times to match the tracer arrival times of IDIF and arterial samples. Then it was convolved with a mono-exponential function to simulate dispersion. This was done in an optimization to estimate the delay and time constant of dispersion that gave the best match between the subject’s IDIF and arterial AIF peak shapes. This was performed for each subject independently and the averaged delay and dispersion values were used in the kinetic analysis. Finally, to see the effect of scaling, the measured IDIF curve with blood samples, the ratio between the mean of the last three arterial blood samples and the mean of the last three IDIF activities were calculated and the uncorrected and PV corrected IDIF curves were scaled using this ratio.
All PET kinetic analysis was done using PMOD (PMOD Technologies, Zurich, Switzerland). For each subject, two TACs were generated using average grey matter and white matter activity and these were fitted using the two-tissue compartment model20 with three rate constants (K1, k2 and k3). K1 and k2 represent the transfer from the vascular to the extra-vascular space and vice-versa, respectively, while k3 represents the transport across the cellular membrane and subsequent phosphorylation, leading to irreversible trapping of the tracer. The rate constant representing the transfer of tracer from metabolized state back to the unmetabolised state was assumed to be negligible (k4 = 0). Cerebral blood volume (Vb) was also estimated during the curve fitting. Equation (3) was used to compute net tracer uptake, Ki, and cerebral metabolic rate of glucose consumption, cMRglc, for each subject.
and
| (3) |
where LC is a lumped constant representing the ratio of 18F-FDG utilization to actual glucose utilization within the brain and Cglu is the cold glucose concentration. In this analysis, LC was set to be 0.8921 (irreversible FDG model) and Cglu was measured by taking a blood sample from each subject before the PET scan. The input functions derived before PVC (IDIFUncorrected), after PVC (IDIFPVC) and from plasma samples (AIFSamples) were fitted with Feng’s input function model, which consists of the sum of a gamma-variate function and two exponentials.4 Grey matter and white matter TACs were fitted using these three input functions and effects of PVC on estimated cMRglc values were evaluated. The performance of the proposed method was also tested by comparing the area under curve (AUC) of derived IDIFs and arterial samples. Paired Student’s t-test was used to evaluate the statistical difference between AUC and cMRglc values calculated using IDIFPVC and AIFSamples, where a significance level of 0.05 was adopted throughout.
Results
Simulation results
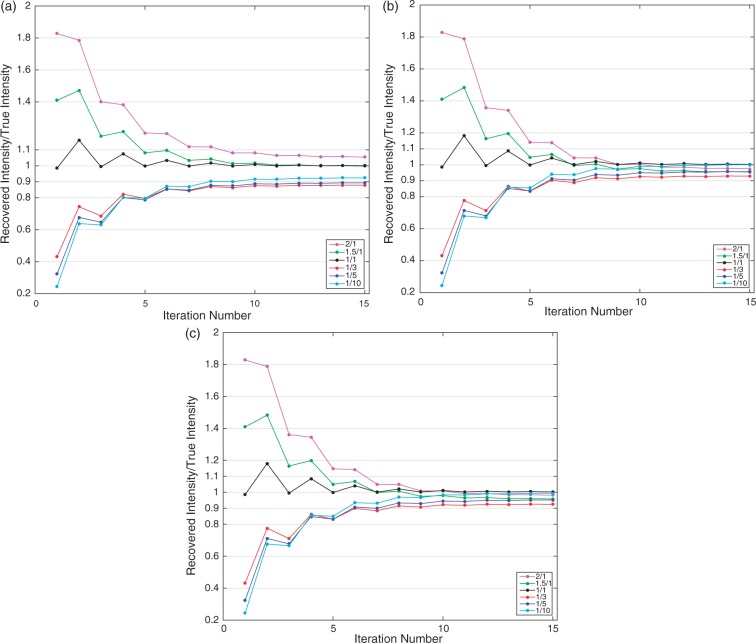
Figure 2 illustrates the ratio of recovered intensity after PVC to the true intensity within the carotid arteries for each iteration. The first value (iteration 1) represents the intensities before the PVC is applied and it can be seen that the PV effect increases when the difference between the carotid artery intensity and background intensity is increased. This is caused by the reduced relative spill-in effect from background tissues to the arteries as the intensity within the arteries is increased. When the PSF with 6.50 mm FWHM was used, which is identical to the PSF used to blur the mask image in the phantom generation, the correction converged to a solution after 10 iterations. As can be seen in Figure 2(a), at least 87.4% of the true intensity was recovered for all of the six phantoms. PSF with 6.74 mm FWHM was found to give the smallest difference between the recovered and the true intensities across the all datasets. Figure 2(b) shows the results when this optimal PSF was used and it can be seen that a better recovery was reached with this PSF where 92.9% of true intensity was recovered at minimum. A similar PSF FWHM value, 6.72 mm, was obtained when it was measured by drawing line profiles to carotid centroids of the reconstructed PET and MR images and results with this PSF is illustrated in Figure 2(c). At minimum, 92.4% of the carotid artery intensity could be recovered with this PSF.
Figure 2.
Recovered intensity to true intensity ratios for each iteration of partial volume correction. Results are shown for six different background to carotid intensity ratios. (a) The result when PSF FWHM is 6.5 mm, (b) PSF is 6.74 mm and (c) 6.72 mm.
Clinical data results
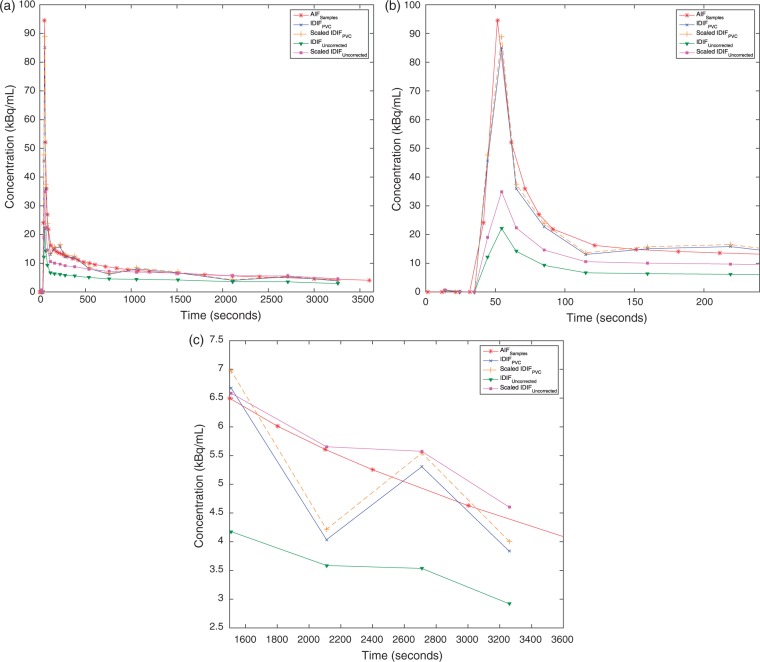
The mean value for prescan blood glucose was 5.07 mmol/L (4.3–5.7 mmol/L) and average hematocrit was 41% (36–45%). Figure 3 shows the IDIFPVC derived from one subject plotted together with IDIFUncorrected and AIFSamples as well as IDIFPVC and IDIFUncorrected scaled with blood samples. It can be seen that application of the PVC increases the intensities both at early and late parts of the input function. The tail of the IDIFPVC (after 3 min) was well matched with AIFSamples, which shows that the PVC method worked successfully to recover the accurate intensity values within the carotid arteries. The first frames of the IDIFUncorrected and IDIFPVC after the delivery of the tracer showed a much higher difference where the PVC increased the peak of the input function by 300% in average. After PVC, a close match between AIFSamples and IDIFPVC peak shapes was observed (Figure 3(b)). Across the 19 subjects, an average time shift of 9.49 s was observed between the peaks of these two input functions. Similarly, the average dispersion time constant was computed as 4.70 s between the carotid artery and radial artery TACs.
Figure 3.
(a) IDIFUncorrected, IDIFPVC, IDIFPVC scaled with blood samples, IDIFUncorrected scaled with blood samples and AIFSamples plotted together for one subject. (b) Input function curves plotted for first 4 min only to show the peaks. Dispersion was added to IDIFPVC and IDIFUncorrected curves. (c) Input function curves plotted for last 30 min.
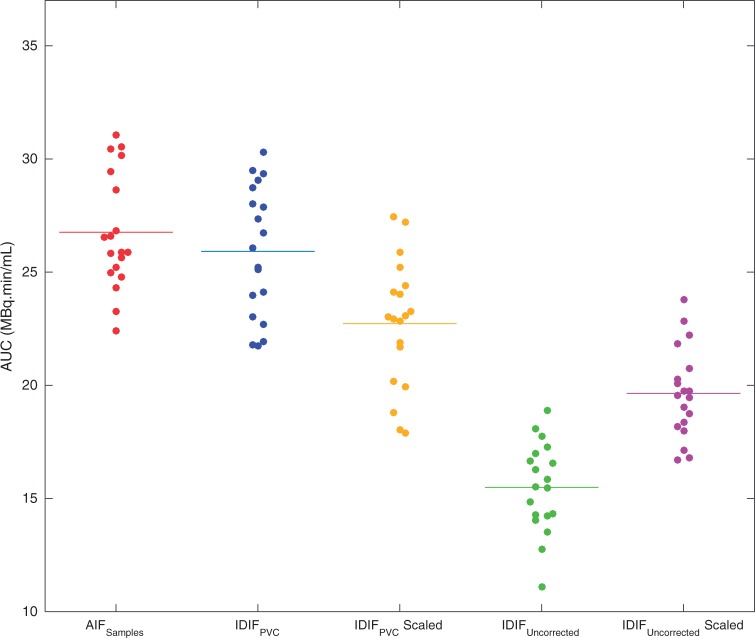
Comparison of the computed AUC values for each of the three input function curves was illustrated in Figure 4. The mean and standard deviation of AUC values in MBq.min/mL was 15.5 ± 2.0 for IDIFUncorrected, 19.7 ± 2.0 for IDIFUncorrected scaled with blood samples, 26.0 ± 2.9 for IDIFPVC, 22.7 ± 2.8 for IDIFPVC scaled with blood samples and 26.8 ± 2.6 for AIFSamples. In all of the 19 subjects, IDIFUncorrected had significantly lower AUC values (paired t-test, p < 0.0001), which confirms the underestimation of the input function due to PV effects. It was seen that applying the PVC brought the area under the IDIF curve to a good agreement with the AIFSamples. Comparing the area under the IDIFPVC and AIFSamples for each subject, there was no statistically significant difference between AUC of these curves with a p value of 0.16. AUC of IDIFPVC scaled with late blood samples was also found to be significantly different than AIFSamples (p < 0.001).
Figure 4.
Area under curves (AUCs) of AIFSamples, IDIFPVC, IDIFPVC scaled with blood samples, IDIFUncorrected and IDIFUncorrected scaled with blood samples. Dots represent individual AUCs and lines represent the mean AUC across the group.
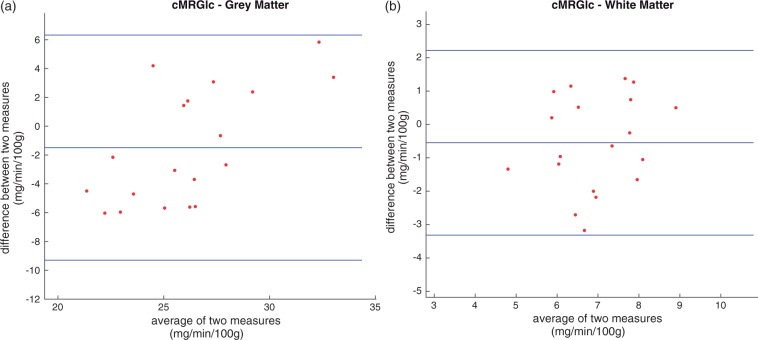
Tables 1 and 2 list the summary of the kinetic parameter and cMRglc results estimated for grey matter and white matter using IDIFUncorrected, IDIFPVC and AIFSamples. When IDIFUncorrected was used, cMRglc in grey matter was overestimated by 50.9% in average compared with the estimates with AIFSamples. Similarly, cMRglc in white matter was overestimated by 45.4% when IDIFUncorrected was used. Using the IDIFPVC largely improved the estimates, bringing the average absolute error to 5.6% and 7.5%, respectively. Performing a paired t-test on the results obtained with IDIFPVC and AIFSamples showed no statistically significant difference for both regions (p = 0.12). Bland–Altman plots of individual cMRglc estimates for white and grey matter are shown in Figure 5, illustrating that there was no consistent bias in the produced white matter results. However, there is a statistically significant positive trend in the grey matter cMRglc results (Pearson correlation, p < 0.005). At present, this effect remains unexplained.
Table 1.
Grey matter results.
| Grey matter |
|||||||
|---|---|---|---|---|---|---|---|
| Vb | K 1 | k 2 | k 3 | K1/k2 | Ki | cMRglc | |
| AIFSamples | 0.059 ± 0.019 | 0.123 ± 0.014 | 0.121 ± 0.045 | 0.079 ± 0.023 | 1.175 ± 0.581 | 0.049 ± 0.004 | 26.88 ± 2.36 |
| IDIFPVC | 0.050 ± 0.020 | 0.149 ± 0.022* | 0.124 ± 0.044 | 0.056 ± 0.025* | 1.323 ± 0.490 | 0.046 ± 0.008 | 25.38 ± 4.66 |
| Scaled IDIFPVC | 0.050 ± 0.020 | 0.166 ± 0.038* | 0.118 ± 0.044 | 0.053 ± 0.024* | 1.592 ± 0.645* | 0.049 ± 0.005 | 26.95 ± 2.92 |
| IDIFUncorrected | 0.076 ± 0.015* | 0.427 ± 0.098* | 0.219 ± 0.063* | 0.046 ± 0.007* | 2.017 ± 0.429* | 0.074 ± 0.009* | 40.56 ± 4.82* |
| Scaled IDIFUncorrected | 0.101 ± 0.022* | 0.325 ± 0.070* | 0.220 ± 0.070* | 0.047 ± 0.008* | 1.531 ± 0.293* | 0.057 ± 0.007* | 31.35 ± 3.91* |
Mean value and standard deviation of kinetic parameters and cMRglc estimates for grey matter calculated using arterial samples, PV corrected IDIF, PV corrected IDIF scaled with blood samples, uncorrected IDIF and uncorrected IDIF scaled with blood samples for 19 subjects.
Parameters with significant difference to arterial samples (paired t-test, p < 0.05).
Table 2.
White matter results.
| White matter |
|||||||
|---|---|---|---|---|---|---|---|
| Vb | K 1 | k 2 | k 3 | K1/k2 | Ki | cMRglc | |
| AIFSamples | 0.029 ± 0.008 | 0.044 ± 0.009 | 0.092 ± 0.030 | 0.040 ± 0.013 | 0.512 ± 0.125 | 0.013 ± 0.002 | 7.22 ± 1.10 |
| IDIFPVC | 0.027 ± 0.010 | 0.054 ± 0.012* | 0.101 ± 0.032 | 0.030 ± 0.009* | 0.573 ± 0.136 | 0.012 ± 0.003 | 6.68 ± 1.35 |
| Scaled IDIFPVC | 0.027 ± 0.008 | 0.061 ± 0.014* | 0.096 ± 0.024 | 0.027 ± 0.009* | 0.655 ± 0.157* | 0.013 ± 0.002 | 7.26 ± 1.07 |
| IDIFUncorrected | 0.038 ± 0.008* | 0.151 ± 0.037* | 0.181 ± 0.040* | 0.027 ± 0.006* | 0.850 ± 0.180* | 0.019 ± 0.004* | 10.50 ± 1.89* |
| Scaled IDIFUncorrected | 0.048 ± 0.010* | 0.114 ± 0.028* | 0.179 ± 0.041* | 0.027 ± 0.006* | 0.652 ± 0.134* | 0.015 ± 0.003* | 8.05 ± 1.16* |
Mean value and standard deviation of kinetic parameters and cMRglc estimates for white matter calculated using arterial samples, PV corrected IDIF and uncorrected IDIF for 19 subjects.
Parameters with significant difference to arterial samples (paired t-test, p < 0.05).
Figure 5.
Bland–Altman plots of cMRglc estimates with arterial blood samples and PV corrected IDIF for (a) grey matter and (b) white matter.
For individual kinetic parameter estimates, using the IDIFUncorrected overestimated the K1 parameter by 247.0% on average for grey matter and white matter, respectively. This error was reduced to 21.0% and 22.7% after the PVC correction. There was no statistically significant difference on the k2 parameter values produced with IDIFPVC and AIFSamples (p = 0.98). On the other hand, using the IDIFUncorrected underestimated k3 values but IDIFPVC reduced this difference by a factor of 2, similar to the improvement seen for k2 results. IDIFPVC was able to return Vb, k2 and cMRglc values with no significant differences.
Using three arterial blood samples to scale the IDIFPVC slightly improved the mean of cMRglc estimates, with an average absolute percent difference of 0.3% and 1.5% and no statistically significant difference (p = 0.93 and p = 0.72) against AIFSamples for grey and white matters, respectively. Scaling increased the error on the K1 parameter, with 35.0% and 38.6% error against the AIFSamples. Similar to IDIFPVC results, scaled IDIFPVC was able to return Vb, k2 and cMRglc with no significant difference to AIFSamples. Scaling the IDIFUncorrected with blood caused an improvement on the cMRglc estimates. However, there was a significant difference between scaled IDIFUncorrected and AIFSamples cMRglc estimates (p < 0.01). Furthermore, it yielded large errors on the individual kinetic parameter estimates.
Discussions
Simulated data results showed that STC can be accurately applied to regions with different region-to-background activity ratios and can correct for spill-in and spill-out effects accurately. Using a PSF value with a larger FWHM than the original PSF used to blur the phantoms resulted in more accurate PVC. This indicate that further blurring was caused by the forward-projection, reconstruction and re-sampling of the data, mainly due to interpolation effects. A PSF FWHM value closer to the optimal value could be accurately measured from the reconstructed PET images and using this PSF resulted in similar recovery performance for all of the datasets. This indicates that a reliable PSF can be measured from reconstructed PET images if the true shape and size of the region of interest is known.
Because of the small diameter of carotid arteries, IDIFs directly estimated from reconstructed PET frames are severely affected by the PV effects. Results from this study show that such IDIFs have significantly lower tracer concentration even when the arterial VOIs are delineated from high-resolution MR anatomical images. Errors caused by PV effects are especially observed in the early frames, where the peak of the input function is highly underestimated. Performing a PVC greatly improved the recovery of signal intensities within the carotids and a good visual agreement was observed against input function curves acquired from serial arterial sampling. Using the PV corrected IDIFs also yielded a good agreement between the estimated cMRglc values with the arterial samples. Scaling the PV corrected IDIF with late blood samples resulted in smaller AUC values compared with the arterial samples. This can be due to the variation of the error in the PVC at different time points as the early frames are more affected from spill-out effects than later time points. Therefore, a single scaling constant measured at the latest part of the curve may not be sufficient to calibrate the whole input function.
In this work, we have used a novel image-based PVC method in the IDIF estimation. This method only requires an accurate segmentation of the region of interest, which is the carotid artery in this application. This eliminates the need for complete parcellation of the image into separate regions, which is a necessary step for most image-based PVC methods to operate,22,23 or even definition of representative background regions.13 Therefore, here we have implemented a more practical PVC method for the purpose of IDIF measurements.
The performance of the presented method highly depends on the accuracy of segmentation, image registration and PSF estimation. As presented here, time of flight MR angiography images can be easily used to segment carotid arteries, as they clearly distinguish signal coming from inflowing blood compared with neighbouring voxels. Typical T1-weighted MPRAGE images have larger voxel size, which can affect the accuracy of the segmentation of internal carotid arteries with small diameter. Furthermore, background regions neighbouring the carotid arteries have higher intensity on MPRAGE images and can interfere with the region-growing algorithm. We attempted to apply the same segmentation method on the MPRAGE images but observed discontinuities in the segmented arteries and leakage to background tissues. MR angiography is now routinely performed in many clinical and research brain imaging studies and a high resolution 3D MR angiography image of the carotid region can be easily acquired.
Mismatches in the PET/MR registration can lead to errors in the measured IDIF curves, as they may cause inclusion of signal from non-arterial voxels in the measurement. To minimize the errors caused by misregistration, we developed a two-step registration method and alignment of carotid arteries was visually inspected for each dataset. However, it is likely that small mismatch errors were present in these data since the PET and MRI images were acquired on separate scanners and patients may have had different head positioning across scans, making the registration step more problematic. Using a simultaneous PET/MRI system is likely to improve the performance of IDIF extraction, as it could reduce co-registration errors.
One limitation of this study was the arterial blood sampling protocol used to obtain the AIF. AIF was sampled for every 10 s for the first 2 min, which might cause errors in the definition of the AIF peak. The sampling thereby closely matches the framing initially. However, the measured AIFs and IDIFs were fitted with an analytical input function model in the kinetic analysis, which could reduce the negative effects caused by the sampling protocol. During the AIF fitting, we took into account the fact that each blood samples was taken over a 3-s period, while the IDIF represented an average over each PET frame duration. The accuracy of the IDIFs would depend on the accuracy of segmentation, registration and PSF measurement methods, while the AIF could be affected by issues related to the blood sampling. These can be possible explanations for the significantly different K1 and k3 parameters.
A small time difference between the peaks of the IDIFPVC and AIFSamples was observed, which can be caused by different PET tracer arrival times to the carotid arteries and the radial artery, tubing used in arterial sampling and variation in blood velocity in different arteries. This time difference in AIF peaks may also introduce errors in the estimated kinetic parameter values. There are several methods present in the literature to correct arterial samples for these time delay and dispersion effects,24,25 but in this case, we have chosen not to modify the gold standard arterial blood sample curve. Therefore, to make a fair comparison, time delay and dispersion were applied to the measured IDIFs instead. It could be argued that due to its proximity to brain tissues, an IDIF measured from carotid arteries, corrected for PV effects, might be a more accurate representation of the input function in brain kinetic analysis than arterial samples obtained from a radial artery.
There was a good agreement between IDIFPVC and AIFSamples in terms of the macro-parameter Ki. However, the micro-parameters K1 and k3 were significantly different. This may suggest that, when using IDIFPVC, the outcome values should be restricted to Ki, in which case it would be possible to use a simple Patlak analysis,2,3 obviating the need for full kinetic modelling. On the other hand, it is not entirely clear which set of estimated micro-parameters are closer to the true values. The K1, k2, k3 and cMRglc values obtained with our method are well within the range of previously published values for grey matter tissues in healthy subjects (K1: 0.068–0.161, k2: 0.071–0.301, k3: 0.03–0.10, cMRglc: 28.60 ± 4.73).18
Conclusions
In conclusion, we have described an IDIF estimation technique, utilizing STC to correct for contamination from neighbouring tissues. This method only requires segmentation of the carotid arteries, which was performed using coregistered MR angiography images with excellent soft tissue contrast. The proposed method does not require any arterial or venous blood samples to be used in PVC and curve scaling. Results were validated against AIFs determined from serial arterial blood samples and no significant difference was seen in the area under input function curves and estimated cMRglc values. Only two of four microparameters could be retrieved with no significant differences.
Funding
The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was taken at UCLH/UCL which receives a proportion of funding from the Department of Health’s NIHR Biomedical Research Centres funding scheme. HS was supported by an IMPACT Studentship funded jointly by Siemens and the UCL Faculty of Engineering Sciences. KE was supported by a grant from EPSRC (EP/K005278/1).
Declaration of conflicting interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Authors’ contributions
HS contributed with analysis and interpretation of data, drafting and revising the manuscript, and final approval of manuscript. KE contributed with interpretation of data, revising and approving the manuscript. IL contributed with acquisition of data, revising and approving the manuscript. HBL contributed with acquisition of data, revising and approving the manuscript. SO contributed with revising and approving the manuscript. SA contributed with revising and approving the manuscript. DA contributed with revising and approving the manuscript. BFH contributed with interpretation of data, revising and approving the manuscript.
References
- 1.Gunn RN, Gunn SR, Turkheimer FE, et al. Positron emission tomography compartmental models: a basis pursuit strategy for kinetic modelling. J Cereb Blood Flow Metab 2002; 22: 1425–1439. [DOI] [PubMed] [Google Scholar]
- 2.Patlak CS, Blasberg RG, Fenstermacher JD. Graphical evaluation of blood-to-brain transfer constants from multiple-time uptake data. J Cereb Blood Flow Metab 1983; 3: 1–7. [DOI] [PubMed] [Google Scholar]
- 3.Gjedde A. Calculation of cerebral glucose phosphorylation from brain uptake of glucose analogs in vivo: a re-examination. Brain Res Rev 1982; 4: 237–274. [DOI] [PubMed] [Google Scholar]
- 4.Feng D, Huang SC, Wang X. Models for computer simulation studies of input functions for tracer kinetic modeling with positron emission tomography. Int J Biomed Comput 1993; 32: 95–110. [DOI] [PubMed] [Google Scholar]
- 5.Eberl S, Anayat AR, Fulton RR, et al. Evaluation of two population-based input functions for quantitative neurological FDG PET studies. Eur J Nucl Med 1997; 24: 299–304. [DOI] [PubMed] [Google Scholar]
- 6.De Geus-Oei LF, Visser EP, Krabbe PF, et al. Comparison of image-derived and arterial input functions for estimating the rate of glucose metabolism in therapy-monitoring 18F-FDG PET studies. J Nucl Med 2006; 47: 945–949. [PubMed] [Google Scholar]
- 7.Van der Weerdt AP, Klein LJ, Boullaard R, et al. Image-derived input functions for determination of MRGlu in cardiac (18)F-FDG PET scans. J Nucl Med 2001; 42: 1622–1629. [PubMed] [Google Scholar]
- 8.Zanotti-Fregonara P, Chen K, Liow JS, et al. Image-derived input function for brain PET studies: many challenges and few opportunities. J Cereb Blood Flow Metab 2011; 31: 1986–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berradja K, Boughanmi N and Bentourkia M. Kinetic modeling of brain FDG data with input function derived from images by independent component analysis. In: IEEE Nucl Sci Symp Conf Rec, Orlando, FL, 2009, pp. 2920–2923.
- 10.Chen K, Bandy D, Reiman E, et al. Noninvasive quantification of the cerebral metabolic rate for glucose using positron emission tomography, 18F-fluoro-2-deoxyglucose, the Patlak method, and an image-derived input function. J Cereb Blood Flow Metab 1998; 18: 716–723. [DOI] [PubMed] [Google Scholar]
- 11.Carson R, Planeta-Wilson B, Mulnix T, et al. Image-based input functions from the carotid arteries with the HRRT. J Nucl Med Meeting Abstr 2006; 47: 57. [Google Scholar]
- 12.Su Y, Arbelaez AM, Benzinger TL, et al. Noninvasive estimation of the arterial input function in positron emission tomography imaging of cerebral blood flow. J Cereb Blood Flow Metab 2013; 33: 115–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zanotti-Fregonara P, Fadaili el M, Maroy R, et al. Comparison of eight methods for the estimation of the image-derived input function in dynamic [(18)F]-FDG PET human brain studies. J Cereb Blood Flow Metab 2009; 29: 1825–1835. [DOI] [PubMed] [Google Scholar]
- 14.Erlanddsson K, Hutton BF. A novel voxel-based partial volume correction method for single regions of interest. J Nucl Med Meeting Abstr 2014; 55: 2023. [Google Scholar]
- 15.Erlandsson K, Wong AT, Van Heertum R, et al. An improved method for voxel-based partial volume correction in PET and SPECT. Neuroimage 2006; 31(Supplement 2): T84. [Google Scholar]
- 16.Muller-Gartner HW, Links JM, Prince JL, et al. Measurement of radiotracer concentration in brain gray matter using positron emission tomography: MRI-based correction for partial volume effects. J Cereb Blood Flow Metab 1992; 12: 571–583. [DOI] [PubMed] [Google Scholar]
- 17.Thielemans K, Mustafovic S, Tsoumpas C. STIR: Software for tomographic image reconstruction release 2. IEEE Nucl Sci Symp Conf Rec 2007; 4: 2174–2176. [DOI] [PubMed] [Google Scholar]
- 18.Yushkevich PA, Piven J, Hazlett HC, et al. User-guided 3D active contour segmentation of anatomical structures: Significantly improved efficiency and reliability. Neuroimage 2006; 31: 1116–1128. [DOI] [PubMed] [Google Scholar]
- 19.Jenkinson M, Smith S. A global optimisation method for robust affine registration of brain images. Med Image Anal 2001; 5: 143–156. [DOI] [PubMed] [Google Scholar]
- 20.Phelps ME, Huang SC, Hoffman EJ, et al. Tomographic measurement of local cerebral glucose metabolic rate in humans with (F-18)2-fluoro-2-deoxy-D-glucose: validation of method. Ann Neurol 1979; 6: 371–388. [DOI] [PubMed] [Google Scholar]
- 21.Graham MM, Muzi M, Spence AM, et al. The FDG lumped constant in normal human brain. J Nucl Med 2002; 43: 1157–1166. [PubMed] [Google Scholar]
- 22.Rousset OG, Ma Y, Evans AC. Correction for partial volume effects in PET: principle and validation. J Nucl Med 1998; 39: 904–911. [PubMed] [Google Scholar]
- 23.Erlandsson K, Buvat I, Pretorius PH, et al. A review of partial volume correction techniques for emission tomography and their applications in neurology, cardiology and oncology. Phys Med Biol 2012; 57: R119–R159. [DOI] [PubMed] [Google Scholar]
- 24.Meyer E. Simultaneous correction for tracer arrival delay and dispersion in CBF measurements by the H215O autoradiographic method and dynamic PET. J Nucl Med 1989; 30: 1069–1078. [PubMed] [Google Scholar]
- 25.Iida H, Higano S, Tomura N, et al. Evaluation of regional differences of tracer appearance time in cerebral tissues using [15O]water and dynamic positron emission tomography. J Cereb Blood Flow Metab 1988; 8: 285–288. [DOI] [PubMed] [Google Scholar]