Abstract
Positron emission tomography (PET) with 15O-tracers is commonly used to measure brain hemodynamic parameters such as cerebral blood flow, cerebral blood volume, and cerebral metabolic rate of oxygen. Conventionally, the absolute quantification of these parameters requires an arterial input function that is obtained invasively by sampling blood from an artery. In this work, we developed and validated an image-derived arterial input function technique that avoids the unreliable and burdensome arterial sampling procedure for full quantitative 15O-PET imaging. We then compared hemodynamic PET imaging performed on a PET/MR hybrid scanner against a conventional PET only scanner. We demonstrated the proposed imaging-based technique was able to generate brain hemodynamic parameter measurements in strong agreement with the traditional arterial sampling based approach. We also demonstrated that quantitative 15O-PET imaging can be successfully implemented on a PET/MR hybrid scanner.
Keywords: Arterial input function, positron emission tomography, cerebral blood flow, cerebral metabolic rate of oxygen
Introduction
Functional brain imaging with positron emission tomography (PET) and magnetic resonance imaging (MRI) has been used extensively to map regional changes in brain metabolism and circulation. The use of 15O-tracer based PET imaging technique remains the gold standard technique for quantitative imaging of brain circulation and metabolism.1–3 In this technique, 15O-water scans are performed to measure the cerebral blood flow (CBF),2,4 15O-carbon monoxide scans are used to measure the cerebral blood volume (CBV),5 15O-oxygen scans are acquired in conjunction with the other two tracers to estimate oxygen extraction fraction (OEF) and cerebral metabolic rate of oxygen (CMRO2).1 The standard quantitative 15O-PET imaging protocol relies upon derivation of an arterial input function (AIF) through an invasive arterial blood sampling procedure, which is technically challenging, and inherently noisy6 and which poses as a hurdle for such studies due to both technical and ethical considerations. In one of our early studies, a complete quantitative analysis was not possible for 13 out of 81 patients due to the difficulties in quantifying the AIF through arterial sampling.7 A popular approach to avoid the arterial blood sampling procedure is the adoption of semi-quantitative methods that estimate brain metabolic parameters without the need of AIF.6–8 In these methods, a constant whole brain metabolic parameter is assumed and used to calibrate the regional parameters. On the other hand, evidence suggests that global CBF and brain metabolism may vary for many reasons.9–16 Therefore, it is important to obtain absolute quantification of the physiological parameters with estimated AIF. For that reason, we develop and validated an image-derived AIF (IDAIF) estimation technique to quantity brain hemodynamic parameters.
The IDAIF technique we are developing17,18 relies on co-registered MR images, i.e. anatomical T1-weighted MR and time-of-flight (TOF) MR angiogram (MRA), to identify the main arteries within the PET field of view. This technique is potentially sensitive to registration errors. A PET/MR hybrid scanner may resolve the co-registration problem since both images were acquired at the same time on the same scanner. On the other hand, whether this kind of scanner can give the same level of quantification as PET only scanners operating in 2D mode, especially for 15O-PET using inhaled radioactive gases, remains unknown because of the higher scatter fraction associated with the lack of septa. One study19 indicated the feasibility of performing quantitative 15O-PET imaging using a PET/CT scanner which only operate in 3D mode. Another recent study indicated that proper attention to the outside scatter compensation is needed for accurate quantification.20 Therefore, cross scanner comparisons remains important for 15O-PET imaging. In addition, arterial sampling is more difficult on a PET/MR scanner since measuring the radioactivity in the sampled blood is more challenging due to the strong magnetic field and requires specialized equipment. An image-based technique that avoids arterial blood sampling is ideal.
In recent years, a number of MR techniques for brain hemodynamics have been developed.21–24 However, the accuracy of these measurements remains controversial, since comparison studies to PET technique are either not done,24,25 or have not been very successful.26 One of the potential problems of such a validation study is the fact that these parameters can vary over time; and the conventional approach dictates that MR and PET data be acquired at different time. A PET/MR scanner will change this picture since MR and PET can be acquired simultaneously.
In our previous study, we validated an IDAIF technique in the context of 15O-water PET imaging and demonstrated that our proposed technique can be used successfully to quantify CBF.17 In this study, we further develop and validate the IDAIF technique for 15O-oxygen and 15O-carbon monoxide, we also perform cross scanner comparison between a Biograph mMR PET/MR hybrid scanner (mMR) (Siemens, Erlangen, Germany) and a conventional EXACT 962 HR + PET only scanner (HR+) (Siemens Medical Solutions USA, Inc., Malvern, PA, USA).
Materials and methods
Human subjects
Participants aged 18–35 years with no evidence of neurological disease were recruited from the surrounding community for this study. In the current analysis, 12 participants (age 23 ± 5, four females) were included. In addition, one female participant (age 46) with cerebral vascular disease was also included in this study to extend the dynamic range of observed hemodynamic parameters. All imaging procedures and assessments were approved by Washington University Human Research Protection Office, and written informed consent was obtained from all individuals in accordance with the ethical standards of the Helsinki Declaration of 1975 and its later amendments.
Imaging
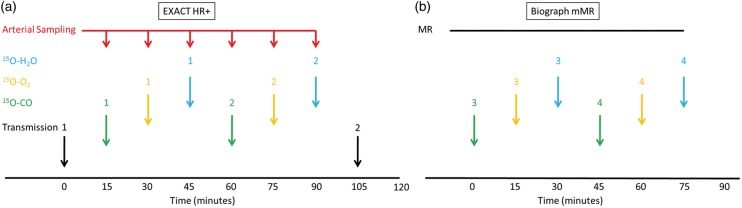
The experimental design for each subject was illustrated in Figure 1. All imaging sessions were performed in the resting state, without manipulation to the physiological state of the participants. Whenever possible, within each participant, two sets of 15O-PET imaging were performed on an HR + scanner, and another two sets of 15O-PET images were acquired on an mMR scanner, with simultaneous acquisition of MR data including Magnetization Prepared Rapid Gradient Echo (MPRAGE) T1-weighted imaging (TR = 2400 ms, TE = 2.13 ms, TI = 1000 ms, flip angle = 8°, and a voxel size of 1.0 × 1.0 × 1.0 mm3) and TOF-MRA (TR = 22 ms, TE = 3.6 ms, flip angle = 18°, and a voxel size of 0.26 × 0.26 × 0.5 mm3). The scans on the two scanners were performed immediately after one another. The mMR arm of the study was not performed on one of the participants (#4) due to claustrophobia concerns and his MR scans were acquired on a Siemens Tim Trio 3T scanner (Siemens Medical Solutions USA, Inc., Malvern, PA, USA) instead using identical MR sequences.
Figure 1.
Imaging protocol on the Siemens EXACT HR + scanner (a) and the Siemens Biograph mMR scanner (b). The MR session include MPRAGE, time-of-flight (TOF) MRA.
On the HR + scanner, each set of 15O-PET imaging included a 5-min static scan beginning 2 min after brief inhalation of 40–75 mCi of 15O-carbon monoxide in room air; a 2-min dynamic emission scan (60 × 2 s) after brief inhalation of 40–75 mCi of 15O-oxygen in room air; and a 2-min dynamic emission scan (60 × 2 s) after rapid intravenous injection of 25–50 mCi of 15O-water in saline. The minimum interval between two 15O-tracer administrations was 12 min to allow decay of the previous tracer. The image acquisition on the HR + scanner was performed in 2D mode (septa extended). Two sets of 15O-PET imaging with a total of six tracer administration were performed during each HR + imaging session whenever possible. Some scans were skipped due to cyclotron failure or other technical issues. A transmission scan was performed before the first tracer administration, and a second transmission scan was performed at the end of the imaging session. A filtered back-projection algorithm was used to reconstruct the emission data with random, attenuation, scatter, and decay correction. During these HR + imaging studies, whenever possible, arterial blood was withdrawn at 5 mL/min from the radial artery through narrow bore tubing to a lead shielded scintillation detector that measured positron emissions with 1 s temporal resolution.27 Blood sampling started simultaneously with tracer administration.
On the mMR scanner, another two sets of 15O-PET scans were performed with lower dose to reduce the radiation exposure and allow optimal counting statistics (15–37 mCi for oxygen and carbon monoxide, and 15–35 mCi for water). The emission data were acquired in list mode immediately after tracer administration to allow custom reconstruction post acquisition. By default, the mMR scanner used an MR derived attenuation map for attenuation and scatter correction.28 However, it has been shown that MR-based attenuation correction led to significant signal reduction while the amount of reduction was location and individual dependent,29,30 which may be problematic for quantification. To avoid this problem, in this study, we used e7tools (Siemens, Knoxville, Tennessee, US) to perform custom reconstruction of the emission data acquired on the mMR scanner using coregistered attenuation map derived from the transmission scans performed on the HR + scanner. The transmission scan obtained on the HR + scanner was coregistered with the MPRAGE scan,31 which is in turn registered to the non-attenuation corrected emission scan obtained on the mMR, so that the attenuation map to emission scan transformation matrix could be obtained. An ordered-subset expectation maximization algorithm32 with three iterations and 21 subsets was used for image reconstruction with standard normalization, dead time, random, and scatter correction33 in a fashion similar to our previous work.30 Similar to our previous works,1,34 the radioactive gases used in this study was pumped into a shielded rubber air bag in an MR compatible ionization chamber at the participant’s side. When the activity reaches the intended range (15–37 mCi), the participant is asked to inhale the gas through a plastic ventilator hose (approximately 1.6 m in length).
Image processing
FreeSurfer v5.1 (Martinos Center for Biomedical Imaging, Charlestown, Massachusetts, USA) was used to segment the MPRAGE image to enable regional analysis. For each participant, within modality image registration, e.g. 15O-water to 15O-water, was performed for data acquired on the same scanner before the PET to MR registration was performed independently for each tracer. PET to MR and TOF-MRA to MR image registration were performed using a vector gradient method.31 Within modality registration was achieved using standard techniques implemented with an in-house software.35 Atlas registration (12-parameter affine) was performed via the MPRAGE against an atlas template. Similar to our earlier works,17,18 a modified adaptive segmentation algorithm36 was used to automatically segment the MRA images to identify arteries. To reduce inter-scanner difference in the spatial resolution of PET images, the PET data were smoothed to a common resolution of 8 mm full-width-half-max (FWHM) using an established protocol37 before further analysis. Regional time-activity curves (TACs) were extracted for FreeSurfer ROIs and arterial ROI. Based on the arterial ROI and the PET resolution (8-mm FWHM), the recovery coefficient for the arterial ROI (ra) is also determined. This parameter varies from one participant to another and depends on the exact definition of the ROI. The value for this parameter is approximately 0.12 in this study.
Models
To facilitate discussion, a summary of the variables and acronyms used in this article is summarized in Table 1. For 15O-water imaging, IDAIF is derived using previously described method17 with joint estimation of blood flow for whole brain (rCBFWB), cortical gray matter (rCBFCG), deep white matter (rCBFDW), and the arterial ROI background tissue blood flow (rCBFa) by minimizing the cost function (Q) defined as equation (1):
| (1) |
Table 1.
Variables and acronyms used in this article.
| AIF | Arterial input function |
| ART | Arterial ROI |
| BG | Background |
| Ca | Arterial input function |
| 15O-water component of the arterial input function | |
| 15O-oxygen component of the arterial input function | |
| CBF | Cerebral blood flow |
| CBV | Cerebral blood volume |
| CMRO2 | Cerebral metabolic rate of oxygen |
| TAC of an ROI calculated from a model | |
| TAC of an ROI measured by PET imaging | |
| CROI | ROI TAC |
| C(t) | ROI TAC |
| HR+ | Siemens EXACT962 HR + PET only scanner |
| ICC | Intraclass correlation coefficient |
| IDAIF | Image-derived arterial input function |
| k | Decay constant in the systematic oxygen metabolism model |
| λ | Partition coefficient of water of the brain |
| mMR | Siemens Biograph mMR PET/MR hybrid scanner |
| MPRAGE | Magnetization Prepared Rapid Gradient Echo |
| MRA | MR angiogram |
| MRI | Magnetic resonance imaging |
| PET | Positron emission tomography |
| OEF | Oxygen extraction fraction |
| Q | Cost function in the 15O-water IDAIF model |
| r | Pearson correlation coefficient |
| ra | Recovery coefficients of the arterial ROI |
| rCBF | Regional blood flow |
| rCBFa | Background tissue blood flow |
| rCBFWB | Whole brain mean blood flow |
| rCBFCG | Cortical gray matter blood flow |
| rCBFDW | Deep white matter blood flow |
| rCBV | Regional blood volume |
| rCBVWB | Whole brain mean blood volume |
| rCMRO2 | Regional metabolic rate of oxygen |
| rCMRO2WB | Whole brain mean metabolic rate of oxygen |
| rOEF | Regional oxygen extraction fraction |
| ROI | Region of interest |
| t | time |
| TAC | Time-activity curve |
| TOF | Time-of-flight |
| τ | Time |
| Δt | Delay constant in the systematic oxygen metabolism model |
| w1,w2,w3 | Weighting factors in the 15O-water IDAIF model |
An optimization procedure searches for the optimal set of regional blood flow values that minimizes the differences between observed ROI TAC (i.e. ) and model-based ROI TAC (i.e.). The model ROI TAC can be calculated from AIF and rCBF using standard CBF model.2 For any given estimation of whole brain blood flow (rCBFWB), the IDAIF (Ca(t)) can be derived according to equation (2) and the measured whole brain TAC:
| (2) |
where λ is the water partition coefficient between blood and brain tissue. For more details of the IDAIF estimation and CBF estimation, please refer to our previous work.17
For 15O-carbon monoxide and 15O-oxygen imaging, the IDAIF (Ca(t)) is derived according to the following equation:
| (3) |
where is the PET measured arterial ROI TAC; is the measured background ROI TAC; and ra is the recovery coefficients of the arterial ROI. Regional CBV is calculates as rCBV = CROI/Ca. Regional cerebral oxygen extraction fraction (rOEF) is calculated using a linearized version38 of the original OEF model:1
| (4) |
where and represent the radioactivity in the arterial blood that are contributed by 15O-oxygen and 15O-water, respectively; and C(t) represents the regional TAC for a particular ROI. Since we do not directly measure the respective contribution of 15O-oxygen and 15O-water to the overall blood radioactivity in the 15O-oxygen PET study, a modeling based approach39 is used instead according to the following equations:
| (5) |
| (6) |
With rCBF and rOEF estimated from the PET data, rCMRO2 can then be calculated as the product of rCBF, rOEF and blood oxygen content.1 For k and Δt in equation (5), we used the reported value,39 i.e. k = 0.0722 min−1; Δt = 20 s.
Analysis
For each participant, CBF, CBV and CMRO2 were estimated using both IDAIF and arterial sampling based AIF whenever possible. The quantification with arterial sampling data was performed following the original 15O-PET imaging quantification models.1,2,34,40 To validate the proposed IDAIF technique, estimated whole brain hemodynamic parameters were compared between the two approaches based on data acquired on the HR + scanner. Both Pearson correlation (r) and intraclass correlation coefficient (ICC) were calculated to assess the agreement between the two sets of parameters. For scanner comparisons, hemodynamic parameters estimated using the IDAIF technique were assessed for agreement. Agreement between quantification methods and across scanners were assessed using r and ICC based on the first measurement obtained, to account for the fact that not all dataset had repeated measurements on the same scanner. To assess the variability of the hemodynamic parameter measurements, inter-subject standard deviation (SDi), within-scanner intra-subject standard deviation (SDw), and between-scanner intra-subject standard deviation (SDb) were estimated. To allow comparison among these variability measures, only the set of participants that had repeated measurements on both scanners were included for these assessments. In addition, spatial correlation between parametric maps of CBF, CBV, and CMRO2 were evaluated at individual level between the two scanners.
Results
IDAIF validation
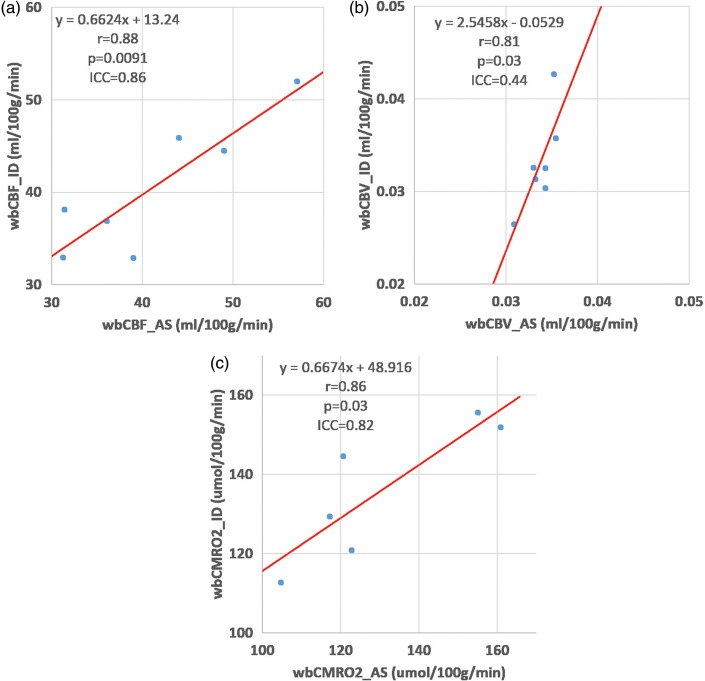
Arterial blood sampling procedure was fully successful for six participants and partially successful for another participant. Arterial data were not available on the cerebral vascular disease patient. Measured rCBFWB (r = 0.88, p = 0.0091, ICC = 0.86) and rCMRO2WB (r = 0.86, p = 0.03, ICC = 0.82) were in strong agreement between IDAIF based approach and arterial sampling based approach (Figure 2(a) and (c) and Table 2). The agreement of measured rCBVWB (r = 0.81, p = 0.03, ICC = 0.44) between the two approach was less strong (Figure 2(b) and Table 2), possibly due to the narrow range of this parameter in healthy young participants and the relatively high level of noise in this measurement. Based on paired Student’s t-test, the two sets of measurements were not different statistically (p > 0.05). Inter subject variability as well as within subject reproducibility data were also reported in Table 2.
Figure 2.
Comparison of arterial sampling and IDAIF based measurements of cerebral blood flow (a), cerebral blood volume (b), and cerebral metabolic rate of oxygen (c). These data were obtained on the HR + scanner.
Table 2.
Comparison of whole brain mean hemodynamic parameters measured using an arterial sampling approach and the IDAIF approach.
| CBF (ml/100 g/min) |
CBV (ml/g) |
CMRO2 (µmol/100 g/min) |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PID | AS |
ID |
AS |
ID |
AS |
ID |
||||||
| 1 | 2 | 1 | 2 | 1 | 2 | 1 | 2 | 1 | 2 | 1 | 2 | |
| 2 | 57.1 | 44.1 | 52.0 | 50.8 | 0.035 | 0.034 | 0.036 | 0.035 | 117.3 | 168.5 | 129.3 | 127.9 |
| 3 | 31.4 | 31.4 | 38.1 | 38.8 | 0.033 | 0.033 | 0.034 | 160.8 | 151.0 | 151.8 | 170.7 | |
| 4 | 44.0 | 46.0 | 45.9 | 43.2 | 0.034 | 0.034 | 0.033 | 0.034 | 122.8 | 119.9 | 120.9 | 117.1 |
| 5 | 36.1 | 32.9 | 36.9 | 40.9 | 0.033 | 0.034 | 0.031 | 0.031 | 104.7 | 139.0 | 112.7 | 109.5 |
| 7 | 49.0 | 53.8 | 44.5 | 42.6 | 0.035 | 0.035 | 0.043 | 0.041 | 155.0 | 142.7 | 155.6 | 169.1 |
| 9 | 39.0 | 34.5 | 32.9 | 31.6 | 0.034 | 0.033 | 0.030 | 0.027 | 120.6 | 144.5 | 113.5 | |
| 10 | 31.3 | 33.8 | 33.0 | 33.5 | 0.031 | 0.031 | 0.026 | 0.025 | ||||
| Mean | 41.1 | 39.5 | 40.5 | 40.2 | 0.034 | 0.034 | 0.033 | 0.032 | 132.1 | 144.2 | 134.1 | 138.9 |
| SDi | 9.5 | 8.5 | 7.2 | 6.4 | 0.002 | 0.001 | 0.005 | 0.005 | 24.5 | 17.7 | 18.9 | 29.1 |
| SDw | 4.2 | 1.5 | 0.001 | 0.001 | 21.2 | 7.9 | ||||||
For each hemodynamic parameter, Mean, SDi, and SDw, were only estimated for the set of subjects that had within scanner test–retest data for both methods.
AS: arterial sampling based measurements; ID: IDAIF based measurements; SDi: inter-subject standard deviation; SDw: within-scanner, intra-subject standard deviation.
mMR to HR + comparison
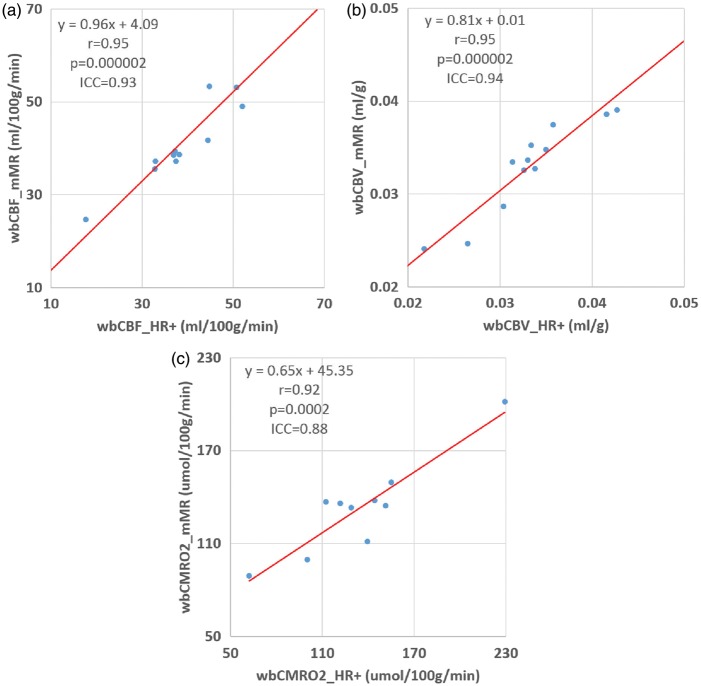
15O-oxygen scan failed in two participants on the mMR scanner due to technical reasons. Strong agreement was observed between hemodynamic parameters measured on the two scanners. The Pearson correlation coefficients and ICC were r = 0.95, p = 0.000002, ICC = 0.93 for rCBFWB; r = 0.95, p = 0.000002, ICC = 0.94 for rCBVWB; and r = 0.92, p = 0.0002, ICC = 0.88 for rCMRO2WB (Figure 3 and Table 3). Hemodynamic parameters obtained on the two scanners did not differ from each other based on paired Student’s t-test (p > 0.05). Inter subject variability as well as within subject reproducibility data were also reported in Table 3. Example parametric images for a healthy young participant (#7) and the CVD patient (#11) obtained from both scanners were shown in Figure 4 along with MR images. The voxel-wise spatial correlation within a whole brain mask was strong at single subject level for all three physiological parameters between the two scanners: rCBF, 0.92 (0.83–0.94); rCBV, 0.97 (0.95–0.0.98); and rCMRO2, 0.89 (0.87–0.92). The voxel-wise correlation was assessed with the PET data smoothed to 12 mm FWHM as in our previous studies.6,41 Strong spatial correlation was also observed in our regional analysis as summarized in the Supplementary Material.
Figure 3.
Comparison of measurements of cerebral blood flow (a), cerebral blood volume (b), and cerebral metabolic rate of oxygen (c) obtained on the HR + and mMR scanners.
Table 3.
Comparison of whole brain mean hemodynamic parameters measured on two different PET scanners (i.e. HR+ and mMR).
| CBF (ml/100 g/min) |
CBV (ml/g) |
CMRO2 (µmol/100 g/min) |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PID | HR+ |
mMR |
HR+ |
mMR |
HR+ |
mMR |
||||||
| 1 | 2 | 1 | 2 | 1 | 2 | 1 | 2 | 1 | 2 | 1 | 2 | |
| 1 | 37.3 | 46.5 | 39.4 | 40.6 | 0.035 | 0.036 | 0.035 | 121.8 | 120.2 | 136.1 | 122.1 | |
| 2 | 52.0 | 50.8 | 49.0 | 0.036 | 0.035 | 0.037 | 128.9 | 127.5 | 133.3 | |||
| 3 | 38.1 | 38.8 | 38.6 | 41.8 | 0.033 | 0.034 | 0.033 | 0.032 | 151.3 | 170.1 | 134.6 | 128.2 |
| 5 | 36.9 | 40.9 | 38.6 | 37.1 | 0.031 | 0.031 | 0.033 | 0.032 | 112.3 | 109.2 | 137.0 | 123.0 |
| 6 | 37.5 | 37.2 | 37.3 | 38.8 | 0.033 | 0.036 | 0.035 | 0.036 | 100.1 | 103.9 | 99.6 | 105.6 |
| 7 | 44.5 | 42.6 | 41.7 | 41.2 | 0.043 | 0.041 | 0.039 | 0.039 | 155.1 | 168.5 | 149.6 | 152.1 |
| 8a | 44.8 | 41.4 | 53.3 | 59.1 | 0.033 | 0.031 | 0.034 | |||||
| 9 | 32.9 | 31.6 | 35.6 | 36.4 | 0.030 | 0.027 | 0.029 | 0.030 | 144.0 | 113.2 | 137.9 | 151.2 |
| 10 | 33.0 | 33.5 | 37.2 | 0.026 | 0.025 | 0.025 | 139.4 | 123.3 | 111.2 | |||
| 12 | 63.4 | 67.8 | 70.4 | 67.6 | 0.042 | 0.040 | 0.039 | 0.039 | 229.2 | 201.7 | ||
| 16a | 50.7 | 51.1 | 53.2 | 0.034 | 0.033 | |||||||
| 11b | 17.7 | 24.7 | 0.022 | 0.024 | 62.3 | 89.1 | ||||||
| Mean | 41.9 | 43.4 | 44.4 | 45.3 | 0.035 | 0.035 | 0.035 | 0.034 | 130.8 | 130.8 | 132.4 | 130.4 |
| SDi | 9.5 | 10.8 | 11.8 | 11.5 | 0.005 | 0.005 | 0.004 | 0.004 | 22.6 | 30.3 | 17.0 | 18.1 |
| SDw | 3.0 | 2.0 | 0.002 | 0.001 | 11.7 | 7.7 | ||||||
| SDb | 3.1 | 0.002 | 10.3 | |||||||||
15O-oxygen scan failed on the mMR scanner due to technical reasons.
Cerebral vascular disease patient.
Mean and SD are assessed excluding the patient. For each hemodynamic parameter, Mean, SDi, SDw, and SDb were only estimated for the set of subjects that had within scanner test–retest data on both scanners.
SDi: inter-subject standard deviation; SDw: within-scanner, intra-subject standard deviation; SDb: between-scanner, intra-subject standard deviation.
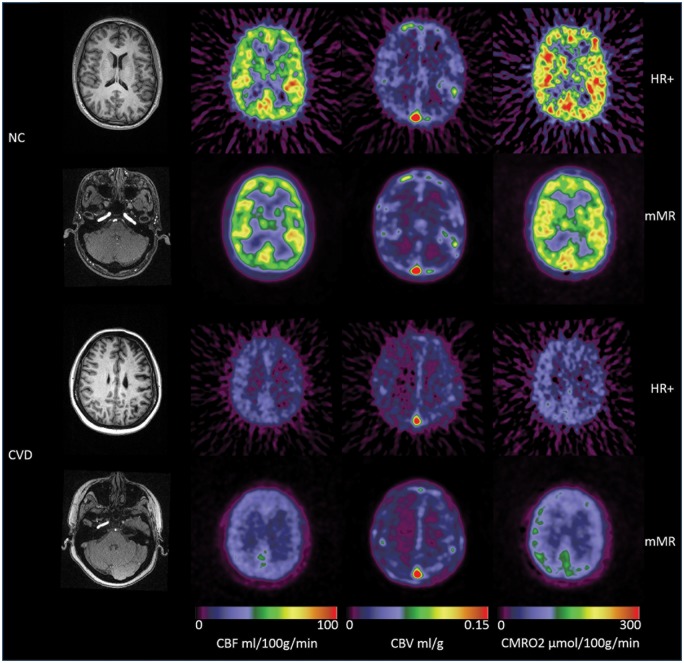
Figure 4.
Example hemodynamic parametric images from a normal control (NC, #7) participants (top two rows) and a cerebral vascular disease patient (CVD, #11) (bottom two rows). Also included were the T1-weighted MR images and the TOF-MRA data at the level of internal carotid artery. It can be observed that the left internal carotid artery was occluded for the CVD patient, and in addition to the overall reduction in CBF and CMRO2, asymmetry can be observed in the CVD patient, whose left hemisphere had lower CBF and CMRO2, and higher CBV. The parametric images obtained on the mMR scanner had better signal to noise ratio.
Discussion
PET imaging using 15O-tracers, primarily 15O-water, was the primary tool in the early days of human brain mapping although it is less used nowadays with the development of functional MRI.42 Nevertheless, along with 15O-oxygen, 15O-carbon monoxide and 18F-fluorodeoxyglucose (FDG), 15O-water provides the only truly quantitative approach to measure human brain circulation and metabolism in health and disease.43–45 With the recent association of aerobic glycolysis with Alzheimer disease and other neurological disorders,8,41,44 there is a renewed interest in this technique, which is an essential component in assessing glycolysis in conjunction with quantitative FDG imaging. Therefore, it is our goal to modernize this technique to avoid arterial sampling and to use PET/MR hybrid scanners. The very fact that we were only able to obtain the complete set of arterial sampling data in 6 out of the 13 participants studied here underscored the weakness of the traditional approach, although it also limited our sample size. Nevertheless, we demonstrated that the IDAIF approach successfully generated hemodynamic parameters in strong agreement with the traditional arterial sampling technique. It is worthwhile to point out that although the arterial sampling based approach was used as the gold standard here, it could be compromised by the fact that the arterial blood is commonly collected from a radial artery, which does not directly supply the brain as we have elaborated before.17
Traditionally, 15O-PET studies are performed on PET scanners that were able to operate in 2D mode,1,2 i.e. with septa extended. However, in the interest of better sensitivity and lower radiation dose, modern PET scanners are no longer equipped with septa and only operate in 3D mode, which results in substantially higher scatter fraction.46 Because of this, the quantitative accuracy of 3D PET scanners47 especially in the context of 15O-PET remains a concern.19 To address this concern, Ibaraki et al. demonstrated that with proper scatter correction, 3D PET was able to perform quantitative brain 15O-PET study with the same degree of accuracy as that in traditional 2D PET scanners.19 In this work, we confirmed the previous work and demonstrated that quantitative hemodynamic and metabolic parameters estimated using the mMR scanner, which operated only in 3D mode, were in strong agreement with those obtained using HR + scanner operating in 2D mode. We further demonstrated that there was a strong voxel-wise correlation between parametric images generated from the two scanners. It is worthwhile pointing out that, although PET/MR hybrid scanner is the targeting scanner of this study, the proposed IDAIF methodology is applicable to PET/CT scanners and older PET only scanners provided that the necessary anatomical imaging data are acquired.
In our estimation of rCMRO2 using IDAIF, we used an empirically determined exponential model39 as well as population average values for the required parameters to separate water and oxygen components in the IDAIF. It should be pointed out that the original model and the associated parameters was derived assuming a slow inhalation of the labelled oxygen, and a relatively long scan duration.39 Adopting the original model and parameters in our work may lead to biases and noises in the estimated rCMRO2. A change of imaging protocol using the slow inhalation procedure may reduce this problem.
As discussed in our previous work,17 the IDAIF technique is sensitive to registration errors between PET and structural MR and the assumed scanner resolution. A translational error of 1 mm or a 1°rotational error could lead to ∼10% underestimation of the AIF because of underestimation of arterial ROI signal and hence causing over estimation of the hemodynamic parameters. An underestimation of the FWHM of scanner point spread function by 0.5 mm will lead to ∼10% overestimation of the recovery coefficient (ra) for the arterial ROI, ∼10% underestimation of the AIF, and ∼10% overestimation of the hemodynamic parameter. Another potential source of variability for IDAIF estimation is the variability in TOF-MRA imaging. In a separate study where we acquired multiple TOF-MRA scans during the same imaging session, the estimated arterial ROI recovery coefficient (ra) is quite consistent with coefficient of variation on the order of 2%. This observation suggests the TOF-MRA scan is reproducible. In this study, in general, the observed within scanner reproducibility is better when the mMR scanner is used. This is consistent with our expectation due to the reduced registration errors and higher signal-to-noise ratio for a more modern PET scanner. The within scanner reproducibility also appears to be better when IDAIF approach is used in general. This may reflect the uncertainty in the arterial sampling approach.
It is noted that the inter-subject variation in whole brain rCBV was greater using the IDAIF approach while the same trend was not observed for rCBF and rCMRO2. We attribute this observation as a consequence of the less reliable registration between 15O-CO images to the anatomical MR due to the lack of spatial contrast other than the vasculatures. As we mentioned earlier, registration uncertainty will lead to variability in AIF estimation and the associated physiological parameters. On the other hand, within scanner reproducibility remains good for rCBV due to the fact that within modality registration is robust for 15O-CO images, and in our implementation, within modality registration is performed before each modality is aligned to the anatomical reference, i.e. the MPRAGE data. In this study, substantial inter-subject variability is observed for rCBF and rCMRO2, while the test–retest reproducibility is high. It should be noted however the observed inter-subject variability is in line with previous studies by our group44 as well as by others.19 The contrast of large inter-individual variability versus small between scan variations may be attributable to natural variability of these parameters among individuals and the fact that these parameters may be stable over a short period of time but could vary over longer intervals. By study design, we did not record a comprehensive battery of physiological parameters as some previous studies19 did and partly due to the difficulty we encountered in obtaining arterial access. Therefore, we could not test the hypothesis that the inter-subject variability in hemodynamic parameters is a consequence of the variability in physiological states of the participants.
Our choice of a PET/MR hybrid scanner as the target platform for 15O-PET imaging was motivated by several factors. In addition to the better sensitivity and signal to noise ratio provided by a modern PET system, the simultaneous PET and MR acquisition capability allows substantial reduction in total study time for complex imaging protocols involving both modalities. The simultaneous acquisition also allows improved spatial alignment between the structural information provided by MR and the physiological information derived from the PET. This is especially important since our IDAIF technique relies on the coherent analysis of PET and MR data and is sensitive to any mismatches between the two modalities as we have discussed before.17 As a new generation of PET imaging technology, however, one of the major concerns for using PET/MR in quantitative studies is the attenuation correction, especially for neuroimaging. In this aspect, we30 and others48,49 have demonstrated that current MR-based attenuation correction can lead to spatially varying biases of as much as 20% in reconstructed PET images. Because of this concern, instead of using MR-based attenuation correction provided by the scanner, the measured attenuation map acquired in the HR + arm of the study was used in an offline reconstruction procedure. Lack of bone information in the MR-based attenuation map is a particular issue for our IDAIF technique, because we derive AIF from the internal carotid arteries, part of which goes through bones. Not accounting for bone attenuation, the AIF can be under estimated and hence leads to over estimation of physiological parameters. A suboptimal attenuation map may also affect the quantitative accuracy through its impact to scatter correction. In several ongoing studies, we are acquiring a separate CT scan for attenuation purposes to address this issue. A recent study demonstrated that using CT-based attenuation map, emission data acquired on a PET/MR system can be reconstructed to minimize the quantitative difference from scans performed natively on a PET/CT scanner.50 It is worth noting that, in this study, the benefit of using a correct attenuation map might be negatively impacted by the potential registration error between the separately acquired attenuation map and emission data. More advanced techniques that derive bone information from MR are also under investigation and hold great potential to resolve this issue.51,52
In summary, we developed and validated an image-derived AIF technique in the context of quantitative 15O-PET imaging, and demonstrated the proposed technique was able to generate brain hemodynamic parameter measurements in strong agreement with the traditional arterial sampling based approach. We further demonstrated quantitative 15O-PET imaging can be successfully implemented on a PET/MR hybrid scanner. In fact, although technical challenges remains, PET/MR may be an ideal platform to realize the full quantitative potential of PET imaging.
Supplementary Material
Acknowledgments
We thank Washington University cyclotron facility and their staff for their assistance in PET tracer production; we thank Washington University Center for Clinical Imaging Research and Neurological/Neurosurgical Intensive Care Unit PET facility and their staff for assistance in data acquisition; we also thank the technical assistance from Drs. Michael Casey, David Faul, Judson Jones, and Agus Priatna from Siemens Medical Solutions.
Funding
The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: The Knight Alzheimer’s Disease Research Center (http://knightadrc.wustl.edu/) pilot award; Washington University Institute of Clinical and Translational Sciences (http://icts.wustl.edu/) Pilot Grant supported by the Clinical and Translational Science Award (CTSA) program (https://www.ctsacentral.org/) of National Institute of Health (http://www.nih.gov/): UL1TR000448; MIR Research and Development fund; McDonnell Center for System Neuroscience New Resource Proposal; National Institute of Neurological Disorders and Stroke P30NS048056, P01NS080675; National Institute of Ageing: P01AG026276, U19AG032438, P50AG005681, P01AG003991.
Declaration of conflicting interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Authors’ contributions
YS, TLSB, AZS, CPD, and MER conceived and designed the experiment and interpreted the data; YS, AGV, and CPD carried out the experiments; YS and LEC analyzed the data; YS drafted the manuscript; all authors helped with manuscript revision and approved the final version.
Supplementary material
Supplementary material for this paper can be found at http://jcbfm.sagepub.com/content/by/supplemental-data
References
- 1.Mintun MA, Raichle ME, Martin WR, et al. Brain oxygen utilization measured with O-15 radiotracers and positron emission tomography. J Nucl Med 1984; 25: 177–187. [PubMed] [Google Scholar]
- 2.Raichle ME, Martin WR, Herscovitch P, et al. Brain blood flow measured with intravenous H2(15)O. II. Implementation and validation. J Nucl Med 1983; 24: 790–798. [PubMed] [Google Scholar]
- 3.Wintermark M, Sesay M, Barbier E, et al. Comparative overview of brain perfusion imaging techniques. Stroke 2005; 36: e83–e99. [DOI] [PubMed] [Google Scholar]
- 4.Herscovitch P, Markham J, Raichle ME. Brain blood flow measured with intravenous H2(15)O. I. Theory and error analysis. J Nucl Med 1983; 24: 782–789. [PubMed] [Google Scholar]
- 5.Grubb RL, Jr, Raichle ME, Higgins CS, et al. Measurement of regional cerebral blood volume by emission tomography. Ann Neurol 1978; 4: 322–328. [DOI] [PubMed] [Google Scholar]
- 6.Vaishnavi SN, Vlassenko AG, Rundle MM, et al. Regional aerobic glycolysis in the human brain. Proc Natl Acad Sci USA 2010; 107: 17757–17762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Derdeyn CP, Videen TO, Simmons NR, et al. Count-based PET method for predicting ischemic stroke in patients with symptomatic carotid arterial occlusion1. Radiology 1999; 212: 499–506. [DOI] [PubMed] [Google Scholar]
- 8.Vlassenko AG, Vaishnavi SN, Couture L, et al. Spatial correlation between brain aerobic glycolysis and amyloid-{beta} (A{beta}) deposition. Proc Natl Acad Sci U S A 2010; 107: 17763–17767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Braun AR, Balkin TJ, Wesenten NJ, et al. Regional cerebral blood flow throughout the sleep-wake cycle. An H2(15)O PET study. Brain 1997; 120(Pt 7): 1173–1197. [DOI] [PubMed] [Google Scholar]
- 10.Dai W, Lopez OL, Carmichael OT, et al. Mild cognitive impairment and alzheimer disease: patterns of altered cerebral blood flow at MR imaging. Radiology 2009; 250: 856–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meltzer CC, Cantwell MN, Greer PJ, et al. Does cerebral blood flow decline in healthy aging? A PET study with partial-volume correction. J Nucl Med 2000; 41: 1842–1848. [PubMed] [Google Scholar]
- 12.Martin AJ, Friston KJ, Colebatch JG, et al. Decreases in regional cerebral blood flow with normal aging. J Cereb Blood Flow Metab 1991; 11: 684–689. [DOI] [PubMed] [Google Scholar]
- 13.Ances BM, Sisti D, Vaida F, et al. Resting cerebral blood flow: a potential biomarker of the effects of HIV in the brain. Neurology 2009; 73: 702–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kennan RP, Takahashi K, Pan C, et al. Human cerebral blood flow and metabolism in acute insulin-induced hypoglycemia. J Cereb Blood Flow Metab 2005; 25: 527–534. [DOI] [PubMed] [Google Scholar]
- 15.Boyle PJ, Scott JC, Krentz AJ, et al. Diminished brain glucose metabolism is a significant determinant for falling rates of systemic glucose utilization during sleep in normal humans. J Clin Invest 1994; 93: 529–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goyal MS, Hawrylycz M, Miller JA, et al. Aerobic glycolysis in the human brain is associated with development and neotenous gene expression. Cell Metab 2014; 19: 49–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Su Y, Arbelaez AM, Benzinger TL, et al. Noninvasive estimation of the arterial input function in positron emission tomography imaging of cerebral blood flow. J Cereb Blood Flow Metab 2013; 33: 115–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Su Y, Blazey TM, Snyder AZ, et al. Quantitative amyloid imaging using image-derived arterial input function. PLoS One 2015; 10: e0122920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ibaraki M, Miura S, Shimosegawa E, et al. Quantification of cerebral blood flow and oxygen metabolism with 3-dimensional PET and 15O: validation by comparison with 2-dimensional PET. J Nucl Med 2008; 49: 50–59. [DOI] [PubMed] [Google Scholar]
- 20.Hori Y, Hirano Y, Koshino K, et al. Validity of using a 3-dimensional PET scanner during inhalation of 15O-labeled oxygen for quantitative assessment of regional metabolic rate of oxygen in man. Phys Med Biol 2014; 59: 5593–5609. [DOI] [PubMed] [Google Scholar]
- 21.Ances BM, McGarvey ML, Abrahams JM, et al. Continuous arterial spin labeled perfusion magnetic resonance imaging in patients before and after carotid endarterectomy. J Neuroimaging 2004; 14: 133–138. [PubMed] [Google Scholar]
- 22.Wang J, Aguirre GK, Kimberg DY, et al. Arterial spin labeling perfusion fMRI with very low task frequency. Magn Reson Med 2003; 49: 796–802. [DOI] [PubMed] [Google Scholar]
- 23.He X, Yablonskiy DA. Quantitative BOLD: Mapping of human cerebral deoxygenated blood volume and oxygen extraction fraction: Default state. Magn Reson Med 2007; 57: 115–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.An H, Liu Q, Chen Y, et al. Evaluation of MR-derived cerebral oxygen metabolic index in experimental hyperoxic hypercapnia, hypoxia, and ischemia. Stroke 2009; 40: 2165–2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He X, Zhu M, Yablonskiy DA. Validation of oxygen extraction fraction measurement by qBOLD technique. Magn Reson Med 2008; 60: 882–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Donahue MJ, Lu H, Jones CK, et al. An account of the discrepancy between MRI and PET cerebral blood flow measures. A high-field MRI investigation. NMR Biomed 2006; 19: 1043–1054. [DOI] [PubMed] [Google Scholar]
- 27.Quarles RP, Mintun MA, Larson KB, et al. Measurement of regional cerebral blood flow with positron emission tomography: a comparison of [15O]water to [11C]butanol with distributed-parameter and compartmental models. J Cereb Blood Flow Metab 1993; 13: 733–747. [DOI] [PubMed] [Google Scholar]
- 28.Delso G, Furst S, Jakoby B, et al. Performance measurements of the Siemens mMR integrated whole-body PET/MR scanner. J Nucl Med 2011; 52: 1914–1922. [DOI] [PubMed] [Google Scholar]
- 29.Samarin A, Burger C, Wollenweber SD, et al. PET/MR imaging of bone lesions–implications for PET quantification from imperfect attenuation correction. Eur J Nucl Med Mol Imaging 2012; 39: 1154–1160. [DOI] [PubMed] [Google Scholar]
- 30.Su Y, Rubin BB, McConathy J, et al. Impact of MR based attenuation correction on neurological PET studies. J Nucl Med 2016; 57: 913–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rowland DJ, Garbow JR, Laforest R, et al. Registration of [18F]FDG microPET and small-animal MRI. Nucl Med Biol 2005; 32: 567–572. [DOI] [PubMed] [Google Scholar]
- 32.Comtat C, Bataille F, Michel C, et al. OSEM-3D reconstruction strategies for the ECAT HRRT. IEEE Nucl Sci Symp Conf Record 2004; 6: 3492–3496. [Google Scholar]
- 33.Watson CC. New, faster, image-based scatter correction for 3D PET. IEEE Trans Nucl Sci 2000; 47: 1587–1594. [Google Scholar]
- 34.Martin WR, Powers WJ, Raichle ME. Cerebral blood volume measured with inhaled C15O and positron emission tomography. J Cereb Blood Flow Metab 1987; 7: 421–426. [DOI] [PubMed] [Google Scholar]
- 35.Hajnal JV, Saeed N, Soar EJ, et al. A registration and interpolation procedure for subvoxel matching of serially acquired MR images. J Comput Assist Tomogr 1995; 19: 289–296. [DOI] [PubMed] [Google Scholar]
- 36.Wilson DL, Noble JA. An adaptive segmentation algorithm for time-of-flight MRA data. IEEE Trans Med Imaging 1999; 18: 938–945. [DOI] [PubMed] [Google Scholar]
- 37.Joshi A, Koeppe RA, Fessler JA. Reducing between scanner differences in multi-center PET studies. Neuroimage 2009; 46: 154–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Su Y, Couture L, Vlassenko A, et al. Linearized models for quantitative analysis of cerebral blood flow and oxygen extraction. J Nucl Med 2010; 51(S2): 245P. [Google Scholar]
- 39.Iida H, Jones T, Miura S. Modeling approach to eliminate the need to separate arterial plasma in oxygen-15 inhalation positron emission tomography. J Nucl Med 1993; 34: 1333–1340. [PubMed] [Google Scholar]
- 40.Videen TO, Perlmutter JS, Herscovitch P, et al. Brain blood volume, flow, and oxygen utilization measured with 15O radiotracers and positron emission tomography: revised metabolic computations. J Cereb Blood Flow Metab 1987; 7: 513–516. [DOI] [PubMed] [Google Scholar]
- 41.Vlassenko AG, McConathy J, Couture LE, et al. Aerobic glycolysis as a marker of tumor aggressiveness: preliminary data in high grade human brain tumors. Dis Markers 2015; 2015: 874–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Raichle ME. A brief history of human brain mapping. Trends Neurosci 2009; 32: 118–126. [DOI] [PubMed] [Google Scholar]
- 43.Shannon BJ, Dosenbach RA, Su Y, et al. Morning-evening variation in human brain metabolism and memory circuits. J Neurophysiol 2013; 109: 1444–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Powers WJ, Videen TO, Markham J, et al. Selective defect of in vivo glycolysis in early Huntington's disease striatum. Proc Natl Acad Sci USA 2007; 104: 2945–2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marshall RS, Festa JR, Cheung YK, et al. Randomized evaluation of carotid occlusion and neurocognition (RECON) trial: main results. Neurology 2014; 82: 744–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Adam LE, Zaers J, Ostertag H, et al. Performance evaluation of the whole-body PET scanner ECAT EXACT HR+ following the IEC standard. IEEE Trans Nucl Sci 1997; 44: 1172–1179. [Google Scholar]
- 47.Sossi V, Oakes TR, Chan GL, et al. Quantitative comparison of three- and two-dimensional PET with human brain studies. J Nucl Med 1998; 39: 1714–1719. [PubMed] [Google Scholar]
- 48.Akbarzadeh A, Ay MR, Ahmadian A, et al. MRI-guided attenuation correction in whole-body PET/MR: assessment of the effect of bone attenuation. Ann Nucl Med 2013; 27: 152–162. [DOI] [PubMed] [Google Scholar]
- 49.Andersen FL, Ladefoged CN, Beyer T, et al. Combined PET/MR imaging in neurology: MR-based attenuation correction implies a strong spatial bias when ignoring bone. Neuroimage 2014; 84C: 206–216. [DOI] [PubMed] [Google Scholar]
- 50.Teuho J, Johansson J, Liden J, et al. Effect of attenuation correction to regional quantification between PET/MR and PET/CT: a multi-centre study using a three-dimensional brain phantom. J Nucl Med 2016; 57: 818–824. [DOI] [PubMed] [Google Scholar]
- 51.Juttukonda MR, Mersereau BG, Chen Y, et al. MR-based attenuation correction for PET/MRI neurological studies with continuous-valued attenuation coefficients for bone through a conversion from R2* to CT-Hounsfield units. Neuroimage 2015; 112: 160–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen Y, Juttukonda M, Su Y, et al. Probabilistic air segmentation and sparse regression estimated pseudo CT for PET/MR attenuation correction. Radiology 2014; 275: 562–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.