Abstract
Litchi downy blight, caused by Peronophythora litchii, is one of the major diseases of litchi and has caused severe economic losses. P. litchii has the unique ability to produce downy mildew like sporangiophores under artificial culture. The pathogen had been placed in a new family Peronophytophthoraceae by some authors. In this study, the whole transcriptome of P. litchii from mycelia, sporangia, and zoospores was sequenced for the first time. A set of 23637 transcripts with an average length of 1284 bp was assembled. Using six open reading frame (ORF) predictors, 19267 representative ORFs were identified and were annotated by searching against several public databases. There were 4666 conserved gene families and various sets of lineage-specific genes among P. litchii and other four closely related oomycetes. In silico analyses revealed 490 pathogen-related proteins including 128 RXLR and 22 CRN effector candidates. Based on the phylogenetic analysis of 164 single copy orthologs from 22 species, it is validated that P. litchii is in the genus Phytophthora. Our work provides valuable data to elucidate the pathogenicity basis and ascertain the taxonomic status of P. litchii.
Introduction
Litchi (Litchi chinensis Sonn) is a delicious subtropical fruit in Southern China, Thailand, India, Vietnam, Australia, and South Africa. Litchi downy blight, caused by Peronophythora litchii Chen ex Ko et al., is one of the major diseases of litchi[1]. A previous study has reported that P. litchii exclusively infects litchi, but later research has indicated that it can also infect the fruits of longan, tomato, and pawpaw upon artificial inoculation[1–7]. The causal agent infects new shoots, young leaves, flowers, panicles, and fruits[6]. Diseased tissues turn brown and become covered with masses of white sporangia and sporangiophores. The pathogen further causes fruit rot and results in severe post-harvest losses (up to 60%)[8].
P. litchii is a hemi-biotrophic plant pathogen, which relies on living host cells at the early stage of infection and switches to a destructive necrotrophic lifestyle later. Similar to other oomycete pathogens, it often releases zoospores from sporangia under wet conditions[9]. Sporangia and zoospores of P. litchii attach to the hosts by swimming with motile flagella or dispersing through splash droplets. Upon docking at potential infection sites, zoospores stick onto host surfaces through adhesive glycoproteins and germinate[9]. The germinated zoospores penetrate the plant surface and invade host tissue by extending a germ tube. Host colonization then occurs and ultimately leads to cell death and tissue collapse[9]. At the later stage of pathogen infection, sporangia are formed to initiate the next infection cycle. The polycyclic nature of the pathogen results in the dispersal of inoculum over an extended time and wide area.
P. litchii is an oomycete, which is a fungus-like diploid eukaryote but evolutionarily related to brown algae. Peronosporomycetidae, a subclass of oomycete, include Peronosporales, Pythiales, and Albuginales. Many plant diseases are classified under Peronosporales, such as Phytophthora infestans (causes the potato late blight) and Hyaloperonospora arabidopsidis (causes downy mildew of Arabidopsis thaliana)[10–12]. P. litchii was first isolated by Chen and was described in a new genus[2,3]. The causal agent produces differentiated sporangiophores with dichotomous branchlets. When the growth of sporangiophores was terminated, sporangia on each tip of branchlets enlarged and matured simultaneously. The determinate sporangiophore is characteristic of downy mildews. P. litchii resembles downy mildews in terms of sporangiophore morphology, while characteristics of the mycelium and sexual reproduction, as well as the fact that the species can be easily cultured, renders it similar to Phytophthora and Pythium species. Before the advent of molecular phylogenetic studies in oomycetes, the former had been placed in the Peronosporaceae, while the latter were placed in the Pythiaceae. Consequently, the family Peronophythoraceae had been introduced[3]. Soon afterwards, the indeterminate sporangiophore was reported by Chi et al. It was suggested that P. litchii should be transferred to Phytophthora[4]. Further studies have indicated that the sporangiophores of P. litchii are basically determinate and occasionally indeterminate. However, the traditional taxonomic status of Peronophythora is mainly based on its morphological and physiological characteristics. In recent years, DNA sequences have been used in the phylogenetic studies of P. litchii. Analysis of ribosomal 28S and ITS sequences have revealed that P. litchii should be included in Phytophthora because of its closer phylogenetic relationship to Phytophthora than to downy mildews[13–15]. After considering both morphological and genetic characteristics, Zhang et al. insisted that the taxonomic status of P. litchii as a distinct transitional species should be retained[16]. Most recently, the sequenced genome of P. litchii has provided convincing evidence that the P. litchii belongs to Phytophthora[17]. But there is still a debate about the relationship between Phytophthora and downy mildews[18].
There is an evolutionary arms race between host plants and their pathogens[19]. Pathogens have developed weaponry to facilitate penetration, colonization, nutrient acquisition, and host-defense response. Conversely, plants have established a system to recognize and defend against invading pathogens. Pathogen-associated molecular patterns (PAMP)-triggered immunity (PTI) mechanisms have evolved to restrict infection and growth of pathogens in plants[19,20]. However, successful pathogens can overcome PTI by producing various effectors. In response, plants have developed an additional defense system that recognize pathogen effectors and lead to effector-triggered immunity (ETI)[19]. ETI is a robust response that can result in programmed cell death (hypersensitive response, HR), which can kill both pathogen and pathogen-infected cells. As the cycle of subversion and recognition evolves, hosts and pathogens are modulated by the interplay between pathogen effectors and their host counterparts[19,21].
Recent molecular studies on oomycete pathogens have focused on secreted effectors, which are ubiquitous in pathogens and manipulate the defense responses of host plants[22,23]. The apoplastic and cytosolic effectors are discerned dependent on the site of action[22,24]. Apoplastic effectors, which are located at the pathogen-host interface, fulfill functions outside the host cell to disturb defense[22]. Apoplastic effectors include several groups, such as elicitins and elictin-like, NPP1-like proteins (necrosis-inducing Phytophthora protein), CBEL (cellulose binding, elicitor, and lectin-like) proteins, and protease inhibitors. Elicitins and elictin-like proteins harbor a 98-amino-acid domain, which include a core of six conserved cysteines in the C-terminal. These are a type of PAMPs and can trigger the PTI of hosts[22]. NPP1-like proteins can promote programmed cell death in dicotyledonous plants with an NPP1 domain[25,26]. Cytoplasmic effectors, which translocate into plant cells and interfere with the host physiological process, include several effector groups such as RXLR and Crinkle (crinkling and necrosis, CRN)[22]. RXLR-type effectors are named for an N-terminal RXLR-dEER amino acid motif, which assists in the translocation of proteins into the host cytoplasm with a signal peptide[27,28]. Meanwhile, about half of RXLR effectors display a conserved core α-helical fold (termed “WY-domain”) on C-terminal regions, which execute the actual effector activity[29–31]. These effectors suppress PTI or activate ETI depending on the host genotype[22,27,28]. In addition to RXLR effectors, the CRN protein family produces leaf crinkling and necrosis phenotype with a conserved “LXLFLAK” motif[10,32].
Despite its economic importance, molecular study on P. litchii has lagged far behind other oomycete species. High-throughput sequencing techniques pave the way to collect genetic data rapidly and cost effectively for functional studies in non-model species. To ascertain the taxonomic status and gain more insight into the molecular pathogenicity mechanisms of P. litchii, we report the Illumina sequencing, de novo assembly, annotation and analysis of the P. litchii transcriptome herein. We investigated the potential effector arsenal (including RXLR effectors) and confirmed the phylogenetic status of P. litchii. Overall, our results serve as a crucial foundation for further study.
Materials and methods
Oomycete materials
P. litchii strain was isolated from diseased fruit of litchi in Hainan, cultured routinely on 10% V8 agar media at 25°C in the dark. Two-week-old plates were flooded with 10 mL of sterile water and kept at 4°C for 2 hours to promote zoospores release. Mycelia, sporangia and zoospores were collected by centrifugation at 2,000 × g for 10 min and immediately preserved in liquid nitrogen for RNA isolation.
RNA isolation, cDNA library preparation and Illumina sequencing
Total RNA was extracted using TRIzol reagent (Invitrogen, USA) according to the manufacturer’s instructions. The quality and quantity of RNA was checked by gel electrophoresis and spectrophotometry. Furthermore, Agilent 2100 Bioanalyzer (Agilent Technologies, Inc.) was applied to assess the integrity of the RNA sample. Prior to cDNA synthesis, RNA samples were treated with DNase I (Promega) to remove contaminating genomic DNA. The purified RNA was dissolved in RNase-free water and stored at -80℃ until used.
RNA-seq libraries were prepared following TruSeqTM RNA sample preparation Kit from Illumina (San Diego, CA), using 5 μg of total RNA. Shortly, mRNA was isolated with oligo (dT) magnetic beads and was fragmented into small pieces using fragmentation buffer. cDNA synthesis, end repair, A-base addition and ligation of the Illumina-indexed adaptors were performed according to Illumina’s protocol. The cDNA fragments of 300–500 bp on 2% Low Range Ultra Agarose were selected, then the products were enriched by PCR amplification using Phusion DNA polymerase (NEB) for 15 PCR cycles. After quantified by TBS380, cDNA library was sequenced using Illumina HiSeqTM 2000 (2 × 100 bp read length). All the experiments were carried out in the Majorbio corporations (Shanghai, China).
Transcriptome assembly and function annotation
The raw reads were cleaned by eliminating adapters, low-quality sequences and discarded reads with more than 10% of bases that had a q-value lower than 20. The filtered and trimmed reads were assembled de novo using Trinity program (v20140717) with the default settings[33]. To eliminate redundant sequences, transcripts were clustered based on sequence similarities, and the longest transcript in each cluster represented the final assembled genes that were subjected to function annotation. Open reading frames (ORFs) in the genes were predicted by TransDecoder (http://transdecoder.sourceforge.net/) with 180 bp set as the minimum ORF length. The longest ORF was extracted from each gene as the candidate protein sequence. The quality of the assembly was benchmarked against the core set of eukaryotic genes using the Core Eukaryotic Genes Mapping Approach (CEGMA) algorithm (with a cutoff e-value of 1e-5) and Benchmarking Universal Single-Copy Orthologs (BUSCO) referring to core eukaryote genes[34,35].
Assembled genes were compared to the following database: NCBI non-redundant protein sequences (Nr), SwissProt, Eukaryotic Orthologous Groups (KOG) and Kyoto Encyclopedia of Genes and Genomes (KEGG) using blastx with a cutoff e-value of 1e−5, NCBI non-redundant nucleotide sequences (Nt) using blastn with a cutoff e-value of 1e-5[36,37]. The candidate proteins were searched against Pfam database for conserved domains with hmmsearch[38,39]. Based on the results of the Nr and Pfam database annotation, Blast2GO was conducted to obtain Gene Ontology (GO) annotation[40].
Identification of putative secreted proteins and pathogenesis-related effectors
The genes were translated to all possible ORFs that encoded at least 60 amino acids (aa) from both strands. SignalP was used to identify an amino terminal signal peptide[41]. The secretome of P. litchii consisted of the proteins with a D-score more than 0.45 and without predicted transmembrane (TM) domains. RXLR and CRN effectors were initially identified based on the function annotation. In a second approach, RXLR and CRN effectors were identified with hmmsearch using Hidden Markov Model (HMM) profile[10,32]. For RXLR effectors identification, a regex method was used by script. String searches for the motif (^\w{10,40}\w[1,96]R\wLR\w{1,40}[ED][ED][KR]) within the secretome were conducted[10], which allow for a signal peptide between residues 10–40, followed by the RXLR motif within the next 100 residues, followed by the EER motif, allowing D and K replacements to E and R, respectively[10]. To search for the conserved WY-domain, HMMER was used to search the C-terminal sequences of the predicted RXLR effectors using the previously described HMM models[10]. Hits with a positive HMM score were considered as a putative effector protein.
Identification of orthologous genes and phylogenetic inference
Identification of single-copy orthologous genes are critical to comparative analysis. Here, we grouped the predicted proteins of P. litchii and other 21 oomycetes using blastp[36] (the sequences of Albugo laibachii, Hyaloperonospora arabidopsidis, Phaeodactylum tricornutum, Phytophthora infestans, Phytophthora kernoviae, Phytophthora lateralis, Phytophthora nicotianae, Phytophthora parasitica, Phytophthora ramorum, Phytophthora sojae, Plasmopara halstedii, Pythium aphanidermatum, Pythium arrhenomanes, Pythium irregulare, Pythium iwayamai, Pythium ultimum, Pythium vexans, Saprolegnia declina, Saprolegnia parasitica and Thalassiosira pseudonana obtained from the Ensembl Genomes database: ftp://ftp.ensemblgenomes.org/pub/protists/release-33/, and the sequences of Phytophthora capsici obtained from the DOE Joint Genome Institute (JGI) database: http://genome.jgi.doe.gov). The proteins of 22 oomycetes did self-blastp-self search with a cutoff e-value of 1e-10. The pairwise similarities were used to identify single-copy genes in each species with Markov cluster (MCL) algorithm at an inflation rate (I) of 2. The member of group with just one gene was identified as single-copy gene. On the other run, the predicted proteins of P. litchii were used as queries to search against other 21 species separately by reciprocal blast hit (RBH) approach. The best hits of each protein with a cutoff e-value of 1e–6 were retrieved. Secondly, the proteins of each 21 species were used as queries to search against P. litchii and the previous filters were carried out. The pair of genes was treated as orthologs if they met the “bidirectional best hit” law. Multiple orthologs-identify were conducted among the other 21 oomycetes. The orthologous group was ascertained if any two genes were orthologs among 22 species.
The single-copy orthologs were both single copy in each species and orthologs among all species. Each group of single-copy orthologs was aligned alone using MAFFT[42]. A concatenated alignment dataset was produced by putting each ortholog alignment in series by perl script. Phylogenetic analysis was carried out by using the RAxML program[43]. The PROTGAMMALGF model, which was the best model by using model-test program, was used to construct the phylogenetic tree with 1000 bootstrap trials. After consensus tree was constructed, the tree branch was estimated by proml program using phylip-3.696[44].
On the other hand, an all-against-all blastp (with cutoff e-value of 1e-10) was performed amongst predicted protein sequences of P. litchii and other four oomycete species: H. arabidopsidis, P. halstedii, P. infestans, and P. sojae[10,11,18,45]. The pairwise similarities served as input for clustering by the OrthoMCL approach with default settings[46]. The homologous proteins were grouped using Markov cluster (MCL) algorithm at an inflation rate (I) of 2.
Results
Illumina sequencing and sequence assembly
Illumina sequencing yielded about 52.5 million pair-end reads with an average length of 200 bp. After removing adaptor sequences, ambiguous reads, and low-quality reads, we obtained approximately 49.6 million cleaned high-quality reads (Table 1). Using the Trinity program, about 44.6 million cleaned reads were further assembled into 23637 transcripts (including isoforms) with an average length of 1284 bp and an N50 of 2176 bp (Table 1). The length of assembled transcripts ranged from 201 to 25551 bp. A set of 19267 unigenes (non-redundant transcripts) by selecting only the longest sequence among isoforms were used for subsequent analyses. More than half of the genes (10927, 56.7%) were longer than 500 bp, and 6538 (33.9%) genes were longer than 1 kb (S1 Fig). The GC content of assembly transcripts was about 51.83% (Table 1). Assembly completeness was evaluated using CEGMA and BUSCO pipeline. Totally, 241 of the 248 (97.2%) widely conserved core eukaryotic genes were mapped against the assembly of P. litchii transcriptome by CEGMA method. Among the 429 single-copy eukaryotic orthologous genes included in BUSCO analysis, there were 353 (82.3%) complete single-copy genes, 15 (3.5%) fragmented genes, and 61 (14.2%) genes missing in P. litchii.
Table 1. Summary of transcriptome sequencing and assembly for P. litchii.
| Category | Count |
|---|---|
| Raw reads | 52477156 |
| Clean reads | 49595837 |
| Assembled reads | 44613368 |
| Transcripts | 23637 |
| Maximum length of transcripts (bp) | 25511 |
| Minimum length of transcripts (bp) | 201 |
| Mean length of transcripts (bp) | 1284 |
| Transcripts size N50 (bp) | 2176 |
| Unigenes | 19267 |
| Maximum length of unigenes (bp) | 23217 |
| Minimum length of unigenes (bp) | 177 |
| Mean length of unigenes (bp) | 975 |
| Unigenes size N50 (bp) | 1533 |
| GC content of genes (%) | 51.83 |
Gene function annotation
To elucidate potential gene functions, the gene annotation was carried out against Nt, Nr, KOG, KEGG, SwissProt, and Pfam database. A total of 17647 genes (91.6%) were successfully annotated in at least one database (Table 2), whereas the remaining may represent additional genes that were not represented in the annotated protein databases or sequences that were too short to produce hits (Table 2). There is a large number of hits which matched the sequences of P. sojae (51.6%) and P. infestans (37.0%) (S2 Fig).
Table 2. Overview of functional annotation of the P. litchii transcriptome.
| Database | Number of annotated genes | Percentage (annotated/total number of genes, %) |
|---|---|---|
| Nr Annotation | 17245 | 89.50 |
| Nt Annotation | 6417 | 33.30 |
| SwissProt Annotation | 9121 | 47.34 |
| PFAM Annotation | 11080 | 57.50 |
| GO Annotation | 12933 | 67.12 |
| KOG Annotation | 6920 | 35.91 |
| KEGG Annotation | 4624 | 23.99 |
| Total annotated genes | 17647 | 91.59 |
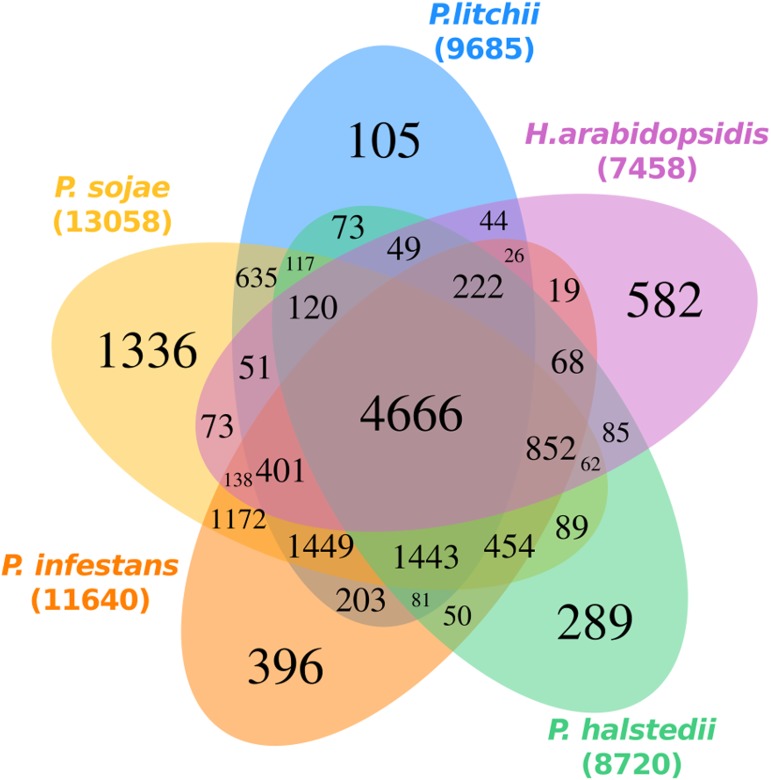
To better define conserved genes among P. litchii (19267), H. arabidopsidis (14321), P. halstedii (15469), P. infestans (17787), and P. sojae (26489), all protein from five species were clustered into 15350 groups (Fig 1). A total of 9685 orthologous groups for P. litchii, 7458 for H. arabidopsidis, 8720 for P. halstedii, 11640 for P. infestans, and 13058 for P. sojae were generated (Fig 1). Among them, 4666 orthologous groups were shared by all five species. A total of 635 gene families were shared only by P. litchii and P. sojae, whereas just 44 gene families were shared only by P. litchii and H. arabidopsidis (Fig 1). In addition, 105 gene families of P. litchii, including 257 proteins, had no orthologs in the other four oomycetes species (Fig 1).
Fig 1. Five-way-Venn-diagram of unique and shared gene families among oomycete species.
Homologous proteins in P. litchii, H. arabidopsidis, P. halstedii, P. infestans, and P. sojae were clustered into gene families using OrthoMCL. Numbers in each section refer to number of gene families (not genes). Overlapping regions denote groups with at least one protein of all species that are part of the intersection. The number under the organism name refer to the total number of clusters.
Based on GO annotation, 12933 (67.12%) genes were assigned to 50 subcategories belonging to three major functional categories (biological process, cellular component, and molecular function) (S3 Fig). Among the categories of biological process, a significant proportion of clusters was assigned to “cellular process” (8384, 64.8%) and “metabolic process” (7772, 60.1%). In the categories of cellular component, the great majority of genes were related to the terms “cell” (6907, 53.4%) and “cell part” (6907, 53.4%). In the categories of molecular function, the most represented terms were “binding” (8369, 64.7%) and “catalytic activity” (6755, 52.2%) (S3 Fig).
To classify orthologous gene products, 6920 (35.9%) genes were functionally classified into 25 KOG categories (Table 2, S4 Fig). Among these groups, the cluster of “general function prediction only” (1078, 15.6%) represented the largest group, followed by “signal transduction mechanisms” (930, 13.4%), “posttranslational modification, protein turnover, chaperones” (844, 12.2%) and “translation, ribosomal structure and biogenesis” (473, 6.8%). The cluster of “cell motility” (12, 0.2%) and “extracellular structures” (21, 0.3%) represented the smallest groups (S4 Fig).
To identify the biological functions and interactions of genes, the genes were mapped to the reference canonical pathways in KEGG. A total of 4,624 (24.0%) genes were assigned to 32 groups, including 260 specific pathways (Table 2, S1 Table, S5 Fig). Major KEGG pathways can be divided into five categories: (i) cellular processes, (ii) environmental information processing, (iii) genetic information processing, (iv) metabolism, and (v) organismal systems (S5 Fig). Abundant genes (540, 11.7%) were involved in signal transduction (S1 Table, S5 Fig). Among them, some important pathways such as “MAPK signaling pathway” (126, 2.7%) and “calcium signaling pathway” (60, 1.3%) were identified. “T cell receptor signaling pathway” (61, 1.3%), “p53 signaling pathway” (25, 0.5%), “plant hormone signal transduction” (19, 0.4%), and “notch signaling pathway” (16, 0.3%) potentially involved in pathogenesis, were also identified (S1 Table, S5 Fig).
In silico-predicted P. litchii pathogenesis-related proteins
Like other plant oomycete pathogens, P. litchii presumably secretes a battery of virulence proteins to promote infection. A total of 978 secrete proteins were identified in P. litchii using Signal[41]. Pathogenicity-related proteins (except for RXLR and CRN) were identified by comparison of P. litchii genes against public databases. A total of 490 pathogenicity-related genes with known or putative roles in virulence were identified (Table 3). Compared with two other oomycete species (P. sojae and H. arabidopsidis), the families of glycosyl hydrolases were significantly reduced in P. litchii (Table 3), whereas the families encoding ABC transporters were expanded (Table 3). The P. litchii families encoding host target, degradative enzymes (polygalacturonases, pectin methylesterases, pectinesterases, and extracellular proteases) and elicitin-like proteins were about half of these in P. sojae and more than two-fold of H. arabidopsidis (Table 3). Results further revealed that P. litchii contained relatively fewer genes involved in hydrolytic enzymes than P. sojae. Protease inhibitors and NPP1-like proteins, which have been implicated in the transition from biotrophy to necrotrophy were abundant in P. litchii and P. sojae (Table 3). Other apoplastic effectors groups including CBEL-like proteins, crinklers, and RXLR proteins had a similar level in P. litchii and H. arabidopsidis (Table 3).
Table 3. Gene families of P. litchii potentially implicated in plant pathogenesis.
| Gene families | H. arabidopsidisa | P. litchii | P. sojaea |
|---|---|---|---|
| ABC transporters, all | 55 | 165 | 134 |
| Glycosyl hydrolases | >60 | 27 | 125 |
| Polygalacturonases | 3 | 12 | 25 |
| Pectin methylesterases | 3 | 7 | 19 |
| Pectinesterases | 3 | 10 | 19 |
| Extracellular proteasesb | 18 | 30 | 47 |
| Elicitins and Elicitin-like | 15 | 35 | 57 |
| CBEL and CBEL-like | 2 | 4 | 13 |
| NPP1-like | 10 | 29 | 29 |
| Protease inhibitors, all | 3 | 21 | 22 |
| Crinklers | 20 | 22 | 40 |
| RXLR | 134 | 128 | 396 |
a Data from other oomycete species are from Tyler et al. for P. sojae and Baxter et al. for H. arabidopsidis. Counts of annotated pseudogenes are omitted.
b Extracellular proteases refer to proteases with signal peptide within N-terminate.
Recent studies have shown a vast repertoire of cytoplasmic effector proteins, including RXLR and CRN families. The P. litchii gene dataset was searched against known oomycete effectors. As a result, 74 and 70 potential RXLR effectors were identified by blastx and hmmsearch, respectively. String searches for the motif within secretome yielded six additional potential RXLR effectors by Regex method. Finally, a total of 128 genes were identified as potential RXLR effectors (S2 Table). Furthermore, 22 genes were ascertained as RXLR effectors by all three methods (S2 Table). Among the 128 potential effectors, 87 candidates contained RXLR and/or dEER motif (S2 Table). The remaining 31 RXLR effector candidates contain either RXLR or dEER motifs (S2 Table), which may due to short sequences. Fifty genes both with signal peptide and RXLR motif were identified in potential RXLR effectors by using SignalP (S2 Table). WY-domain was another structure unit conserved and found in tandem repeats in many effectors. A total of 48 out of 128 (37.5%) potential RXLR effectors contained WY-domain, and 36 candidate effectors harbor more than one WY-domains (S2 Table). Furthermore, 14 out of 22 (63.6%) ascertained RXLR had WY-domains. Therefore, many RXLR genes of P. litchii comprised WY-domains (S2 Table). We performed similar searches to identify CRN effectors in P. litchii. In total, 22 potential CRN proteins with “LXLFLAK” motif were identified (Table 3).
Phylogenetic analysis of P. litchii
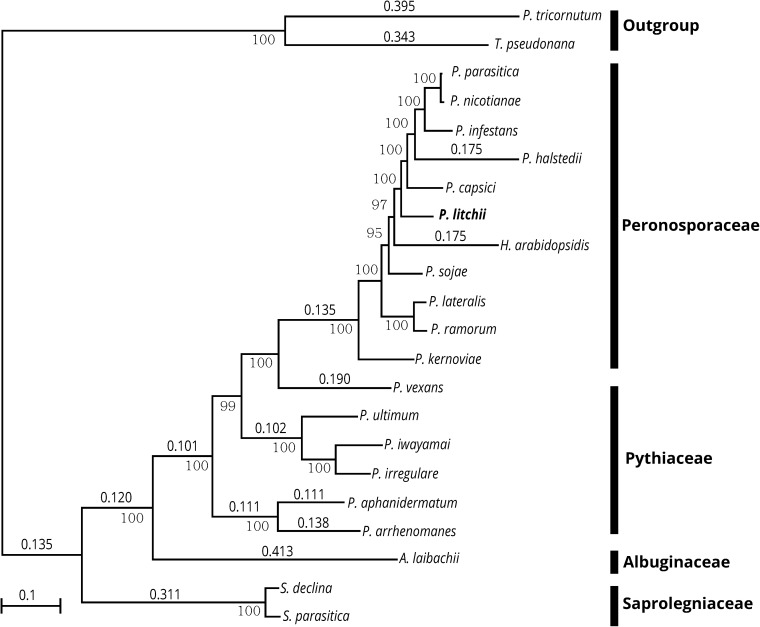
The accuracy of species-tree estimates highly depends on the quality of initial phylogenetic data, usually gene trees, and multiple alignments in selected cases. Due to rapid gene evolution, gene recombination, and horizontal gene transfer, the trees constructed from different genes often were conflicting and resulted in an incorrect species tree. To reconstruct a reliable species phylogeny, the multiple families of single-copy orthologs were identified among 22 species (Fig 2). We concatenated alignments of 164 single-copy orthologs (included 143458 amino acid positions) and inferred the species tree using a maximum likelihood approach[43]. The obtained species phylogeny was highly supported with bootstrap values more than 90% for all nodes. With P. tricornutum and T. pseudonana set as outgroup, the branch of Saprolegniaceae (S. declina and S. parasitica) was off firstly and followed by the branch of Albuginaceae. After the six Pythium species were off in the evolutionary tree, the eleven members of Peronosporaceae clustered into a group (Fig 2). P. litchii was the neighbor of P. capsica and H. arabidopsidis(Fig 2). Two species of downy mildews (H. arabidopsidis and P. halstedii) were nested within Phytophthora clade with high bootstrap support and were separated by P. litchii and P. capsici. The branch length of P. halstedii and H. arabidopsidis both represented 0.175 genetic distance and were much longer than other nine members of Peronosporaceae (Fig 2).
Fig 2. Phylogenetic tree of 22 oomycete species.
Phylogenetic tree infered from the 164 single-copy proteins dataset by RAxML under the GAMMALGF model. ML bootstrap support (lower value) and genetic distance (upper value) were estimated under the uniform model.
Discussion
Despite the severe economic pre- and post-harvest losses caused by P. litchii, little is known about the genetic information and molecular basis of its pathogenicity. The availability of genetic sequences from other pathogens has greatly advanced the understanding of this pathogen. A lag still exists in developing these resources for P. litchii. High-throughput sequencing is a powerful and economical technology to uncover new genes and their involved biochemical pathways in non-model species[47–50]. Transcriptome analysis provide abundant sequence information and shed light on the molecular mechanism of pathogenicity. In the present work, 23637 transcripts of P. litchii were assembled. The average length (1284 bp) of transcripts was longer than that (1116 bp) of Phytophthora cactorum obtained using a similar approach[49]. The gene number of P. litchii transcriptome was comparable to that of P. infestans[10], but less than that of P. sojae[45] and more than that of H. arabidopsidis[11] and P. halstedii[18]. There were 13155 gene models in the genome assembly of P. litchii [17]. The total number of P. litchii genes was probably underestimated in genome project. It could be more accurately predicted if the transcriptome was incorporated into genome annotation. In addition, 97.2% of conserved core eukaryotic genes were mapped against the P. litchii transcriptome. The high mapped rate was comparable to that (93.6%) in A. laibachii and (95.0%) in H. arabidopsidis genome assemblies[11,51]. The similar result was displayed by BUSCO analysis. The high rate of identified CEGMA and BUSCO genes suggested a good assembly of P. litchii. Our work provided a high-quality transcriptome, which could be advantageous for further gene function validation and functional genomics approaches.
The conserved genes globally were examined by OrthoMCL across other four oomycete species. A total of 4666 gene families shared by all five species could be defined as the core gene set of Peronosporomycetidae (Fig 1). On the contrary, these genes lacked homologs in other species represented potentially P. litchii-specific members. Based on gene annotations and pathway analyses, the unigenes of P. litchii were predominantly involved in cellular and metabolic processes, and signal transduction. In addition, some important pathways such as “MAPK signaling pathway”, “calcium signaling pathway”, and “plant hormone signal transduction”, were identified and potentially involved in pathogenesis. The predicted pathways together with the gene annotations can be used to further investigate gene function in the future.
The transcriptome of P. litchii contained mass of genes encoding proteins that were potentially involved in pathogenicity. ABC transporters, which are found in all species and transport substrates across cellular membranes, is essential in cell viability, virulence, and pathogenicity in oomycetes. Compared to P. sojae and H.arabidopsidis, the expansion of ABC transporters was observed in P. litchii. It was implied that the transport activity between host and pathogen was more frequent than others. The families of glycosyl hydrolases were significantly reduced. Meanwhile, the families encoding degradative enzymes, polygalacturonases, pectin methylesterases, pectinesterases, and extracellular proteases, were less than those in P. sojae and more than those in H. arabidopsidis. In general, P. litchii contained a relatively fewer genes involved hydrolytic enzymes than P. sojae, in accordance with the fact that the fruit of litchi has a gap-peel structure and is penetrated through the crack of pericarp by P. litchii. Thus, it is not necessary to maintain mass hydrolases for cell wall collapse.
The species of Phytophthora and downy mildews secrete a vast repertoire of effector proteins that modulate host defenses and enable pathogenicity[10,19,29]. In the present study, we examined the similarity to known oomycete effectors. NPP1-like proteins have been implicated in the transition from biotrophy to necrotrophy. Compared to 30 NPP1-like proteins in assembly of genome, there were 29 members identified in transcriptome of P. litchii. These genes were abundant in P. sojae and P. litchii. The NPP1-like proteins may have contributed to necrotrophy life style of P. litchii. CBEL-like proteins, another apoplastic effectors, were also detected but had a similar level in P. litchii and H. arabidopsidis (Table 3). In addition to apoplastic effectors, there were also a large repertoire of candidate CRN effectors that elicit crinkling and necrosis in planta. The transcriptome of P. litchii annotated 22 candidate CRN effectors, which were little more than 14 members in assembly of genome. The number discrepancies between the assembly of genome and transcriptome were most likely the result of incomplete assembly of genome or chimeric assembly of transcriptome. We identified 128 potential RXLR effectors in the P. litchii transcriptome. The C-terminal regions of RXLR effectors fulfill the effector activity. About half of them share a conserved WY-domain, which is an adaptive structure[31]. Using HMM-based method, WY-domains were also detected in our potential RXLR effectors, 37.5% of which contained the WY-domain. These candidate RXLR effectors contained sequence features and motifs characteristic of the canonical RXLR effectors. There were 245 candidate RXLR effectors in genome annotation[17]. The transcripts of P. litchii represented these genes expressed during culture but did not represent all genes of genome. The effector data set could be expanded by including additional life cycle and infection stages of P. litchii. Our data indicated that P. litchii exhibited the same kind of pathogenicity-related proteins found in other oomycetes.
The relationship of Phytophthora, Peronophythora, and downy mildews is still under debate. Phytophthora and Peronophythora can grow on artificial media, whereas downy mildews cannot be culture under in vitro conditions. Meanwhile, Peronophythora and downy mildews are determinate and produce differentiated sporangiophores. Peronophythora is a genus with characteristics intermediate between downy mildews and Phytophthora[2,3]. Phytophthora and downy mildews share an intimate relationship. These are both morphologically and phylogenetically connected by “bridging taxa” of the graminicolous downy mildews such as Viennotia, Poakatesthia, and Sclerophthora[52]. The traditional classification has the limitation of the taxonomy-based-morphology. These methods quickly lose power when too many species are included or when dealing with specimens whose closest phylogenetic relatives are unknown. Molecular data, especially DNA sequence, can be used for the rapid identification and delimitation of species. Through species classification analysis, P. litchii and P. sojae shared the highest level of similarity across all NCBI Nr databases (S2 Fig). Meanwhile, P. litchii shared more specific gene families with P. sojae than H. Arabidopsis. These results implied that P. litchii was closer to Phytophthora than downy mildews. Recently studies have suggested that the three genera are included in one monophyly, in which Phytophthora is paraphyletic and downy mildews is nested within clade 4 of Phytophthora[15]. P. litchii is also included into clade 4 of Phytophthora[15]. Previous studies on P. litchii phylogenies have focused primarily on 28S and ITS sequences. In phylogenetic analyses, different single-gene data sets have resulted in controversial phylogenies in certain cases even within the same species group. But large numbers of characters and independent evidence from many genetic loci often result in well-resolved and highly supported phylogenetic hypotheses. For these reasons, the analysis of multi-gene sequence data is becoming increasingly common. 164 single-copy orthologs were used to determine the taxonomic position of P. litchii and the relationship of Phytophthora and downy mildews. The obtained species phylogeny was supported with high bootstrap values (>94) for all nodes (Fig 2). Phylogenetic analysis revealed a closer relationship between P. litchii and P. capsici (Fig 2). P. capsici falls within clade 2 of Phytophthora, which is neighbor to clade 4[15]. It is confirmed that P. litchii is a member of Phytophthora. However, two downy mildews, P. halstedii and H. arabidopsidis, were separated by P. litchii and P. capsica. The results refuted monophyly of the downy mildews and were consistent with the phylogenies analysis in recently studies[18]. The longer branch length of two downy mildews was obvious. It indicated that the two of downy mildews had more divergent mutation rates than nine Phytophthora species (Fig 2). It maybe is a result of biotrophic life in downy mildews. It should be noted that P. litchii was adjacent to H. arabidopsidis in genetic levels. Considering the character of sporanigia, P. litchii can be classified as bridging species between Phytophthora and downy mildews. In summary, the molecular evidence presented in this work confirms that P. litchii belong to Phytophthora and is a bridging species between Phytophthora and downy mildews.
Supporting information
(XLSX)
(XLSX)
(TIFF)
(TIFF)
The results are summarized in three main categories: biological process, cellular component and molecular function. A set of 17647 genes were assigned to GO term based on blastx matches to known proteins.
(TIFF)
In total, 6920 of the 19627 P. litchii genes with Nr hits were grouped into 25 KOG classification.
(TIFF)
Pathway assignment was summarized for five main categories: Cellular Processes (C), Environmental Information Processing (E), Genetic Information Processing (G), Metabolism (M) and Organismal Systems (O). A total of 4624 genes were assigned to 32 groups, and abundant genes were involved signal transduction.
(TIFF)
Acknowledgments
This study was supported by the National Natural Science Foundation (31301766) and the Special Funds of the Modern Agricultural Industry Technology System of China (CARS-33-02).
Data Availability
All relevant transcriptome data are available from GenBank (SRA accession: SRP079825).
Funding Statement
Funded by the National Natural Science Foundation (31301766) JHS the Special Funds of the Modern Agricultural Industry Technology System of China (CARS-33-02) JBW.
References
- 1.Kao CW, Leu LS (1980) Sporangium germination of Peronophythora litchii, the causal of litchi downy blight. Mycologia 72: 737–748. [Google Scholar]
- 2.Chen CC (1961) A species of Peronophythora gen. nov. parasitic on litchi fruit in Taiwan. Special Publication of College of Agriculture, National Taiwan University 10: 1–37. [Google Scholar]
- 3.Ko WH, Chang HS, Su HJ, Chen CC, Leu LS (1978) Peronophythoraceae, a new family of Peronosporales. Mycologia 70: 380–384. [Google Scholar]
- 4.Chi PK, Pang SP, Liu R (1982) Studies on Phytophthora disease of litchi. Ⅰ. Identification of causal agent. In Proceedings of the Annual Meeting of the Chinese Phytopathological Society: 72–73.
- 5.Huang H, Wang CP, Xu DY (1983) On Peronophythora litchii. Acta Mycologica Sinica 2: 201–206. [Google Scholar]
- 6.Ann PJ, Ko WH (1984) Blossom Blight of Litchi in Taiwan Caused by Peronohythora litchi. Plant Disease 68: 826. [Google Scholar]
- 7.Chi PK, Pang SP, Liu R (1984) On downy blight of Litchi chinensis Sonn. Ⅰ. The pathogen and its infection process. Acta Phytopatholgica Sinica 14: 113–119. [Google Scholar]
- 8.Wang HC, Sun HY, Stammler G, Ma JX, Zhou MG (2009) Baseline and differential sensitivity of Peronophythora litchii (lychee downy blight) to three carboxylic acid amide fungicides. Plant Pathology 58: 571–576. [Google Scholar]
- 9.Hardham AR (2007) Cell biology of plant-oomycete interactions. Cellular Microbiology 9: 31–39. doi: 10.1111/j.1462-5822.2006.00833.x [DOI] [PubMed] [Google Scholar]
- 10.Haas BJ, Kamoun S, Zody MC, Jiang RH, Handsaker RE, et al. (2009) Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans. Nature 461: 393–398. doi: 10.1038/nature08358 [DOI] [PubMed] [Google Scholar]
- 11.Baxter L, Tripathy S, Ishaque N, Boot N, Cabral A, et al. (2010) Signatures of Adaptation to Obligate Biotrophy in the Hyaloperonospora arabidopsidis Genome. Science 330: 1549–1551. doi: 10.1126/science.1195203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thines M (2014) Phylogeny and evolution of plant pathogenic oomycetes—a global overview. European Journal of Plant Pathology 138: 431–447. [Google Scholar]
- 13.Riethmuller A, Voglmayr H, Goker M, Weiss M, Oberwinkler F (2002) Phylogenetic relationships of the downy mildews (Peronosporales) and related groups based on nuclear large subunit ribosomal DNA sequences. Mycologia 94: 834–849. [DOI] [PubMed] [Google Scholar]
- 14.Voglmayr H (2003) Phylogenetic relationships of Peronospora and related genera based on nuclear ribosomal ITS sequences. Mycol Res 107: 1132–1142. [DOI] [PubMed] [Google Scholar]
- 15.Goker M, Voglmayr H, Riethmuller A, Oberwinkler F (2007) How do obligate parasites evolve? A multi-gene phylogenetic analysis of downy mildews. Fungal Genetics and Biology 44: 105–122. doi: 10.1016/j.fgb.2006.07.005 [DOI] [PubMed] [Google Scholar]
- 16.Zhang ZG, Zheng XB, Wang YC, Ko WH (2007) Evaluation of the rearrangement of taxonomic position of Peronophythora litchii based on partial DNA sequences. Botanical Studies 48: 79–89. [Google Scholar]
- 17.Ye W, Wang Y, Shen D, Li D, Pu T, et al. (2016) Sequencing of the Litchi Downy Blight Pathogen Reveals It Is a Phytophthora Species With Downy Mildew-Like Characteristics. Mol Plant Microbe Interact 29: 573–583. doi: 10.1094/MPMI-03-16-0056-R [DOI] [PubMed] [Google Scholar]
- 18.Sharma R, Xia X, Cano LM, Evangelisti E, Kemen E, et al. (2015) Genome analyses of the sunflower pathogen Plasmopara halstedii provide insights into effector evolution in downy mildews and Phytophthora. BMC Genomics 16: 741 doi: 10.1186/s12864-015-1904-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jones JDG, Dangl JL (2006) The plant immune system. Nature 444: 323–329. doi: 10.1038/nature05286 [DOI] [PubMed] [Google Scholar]
- 20.Ausubel FM (2005) Are innate immune signaling pathways in plants and animals conserved? Nature Immunology 6: 973 doi: 10.1038/ni1253 [DOI] [PubMed] [Google Scholar]
- 21.Tsuda K, Katagiri F (2010) Comparing signaling mechanisms engaged in pattern-triggered and effector-triggered immunity. Current Opinion in Plant Biology 13: 459–465. doi: 10.1016/j.pbi.2010.04.006 [DOI] [PubMed] [Google Scholar]
- 22.Kamoun S (2006) A catalogue of the effector secretome of plant pathogenic oomycetes. Annual Review of Phytopathology 44: 41–60. doi: 10.1146/annurev.phyto.44.070505.143436 [DOI] [PubMed] [Google Scholar]
- 23.Wawra S, Belmonte R, Löbach L, Saraiva M, Willems A, et al. (2012) Secretion, delivery and function of oomycete effector proteins. Current Opinion in Microbiology 15: 685–691. doi: 10.1016/j.mib.2012.10.008 [DOI] [PubMed] [Google Scholar]
- 24.Stassen JH, Ackerveken GVd (2011) how do oomycete effectors interfere with plant life? Current Opinion in Plant Biology 14: 407–414. doi: 10.1016/j.pbi.2011.05.002 [DOI] [PubMed] [Google Scholar]
- 25.Gijzen M, Nuernberger T (2006) Nep1‐Like Proteins from Plant Pathogens: Recruitment and Diversification of the NPP1 Domain Across Taxa. Phytochemistry 37. [DOI] [PubMed] [Google Scholar]
- 26.Ottmann C, Luberacki B, Kufner I, Koch W, Brunner F, et al. (2009) A common toxin fold mediates microbial attack and plant defense. Proceedings of the National Academy of Sciences of the United States of America 106: 10359–10364. doi: 10.1073/pnas.0902362106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rehmany AP, Gordon A, Rose LE, Allen RL, Armstrong M, et al. (2005) Differential Recognition of Highly Divergent Downy Mildew Avirulence Gene Alleles by RPP1 Resistance Genes from Two Arabidopsis Lines. The Plant Cell 17: 1839–1850. doi: 10.1105/tpc.105.031807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Birch PRJ, Rehmany AP, Pritchard L, Kamoun S, Beynon JL (2006) Trafficking arms: oomycete effectors enter host plant cells. Trends in Microbiology 14: 8–11. doi: 10.1016/j.tim.2005.11.007 [DOI] [PubMed] [Google Scholar]
- 29.Bos JIB, Kanneganti T, Young C, Cakir C, Huitema E, et al. (2006) The C‐terminal half of Phytophthora infestans RXLR effector AVR3a is sufficient to trigger R3a‐mediated hypersensitivity and suppress INF1‐induced cell death in Nicotiana benthamiana. Plant Journal 48: 165–176. doi: 10.1111/j.1365-313X.2006.02866.x [DOI] [PubMed] [Google Scholar]
- 30.Dou D, Kale SD, Wang X, Jiang RHY, Bruce NA, et al. (2008) RXLR-Mediated Entry of Phytophthora sojae Effector Avr1b into Soybean Cells Does Not Require Pathogen-Encoded Machinery. The Plant Cell 20: 1930–1947. doi: 10.1105/tpc.107.056093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van Poppel PMJA Jiang RHY, Śliwka J, Govers F (2009) Recognition of Phytophthora infestans Avr4 by potato R4 is triggered by C‐terminal domains comprising W motifs. Molecular Plant Pathology 10: 611–620. doi: 10.1111/j.1364-3703.2009.00556.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Torto TA, Li S, Styer A, Huitema E, Testa A, et al. (2003) EST mining and functional expression assays identify extracellular effector proteins from the plant pathogen Phytophthora. Genome Research 13: 1675–1685. doi: 10.1101/gr.910003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grabherr M, Haas BJ, Yassour M, Levin JZ, Thompson DA, et al. (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nature Biotechnology 29: 644 doi: 10.1038/nbt.1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parra G, Bradnam K, Korf I (2007) CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics 23: 1061–1067. doi: 10.1093/bioinformatics/btm071 [DOI] [PubMed] [Google Scholar]
- 35.Simao FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM (2015) BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31: 3210–3212. doi: 10.1093/bioinformatics/btv351 [DOI] [PubMed] [Google Scholar]
- 36.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, et al. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M (2007) KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Research 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eddy SR (1998) Profile hidden Markov models. Bioinformatics 14: 755–763. [DOI] [PubMed] [Google Scholar]
- 39.Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, et al. (2016) The Pfam protein families database: towards a more sustainable future. Nucleic Acids Research 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Conesa A, Gotz S, Garciagomez JM, Terol J, Talon M, et al. (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21: 3674–3676. doi: 10.1093/bioinformatics/bti610 [DOI] [PubMed] [Google Scholar]
- 41.Petersen TN, Brunak S, Von Heijne G, Nielsen HB (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nature Methods 8: 785 doi: 10.1038/nmeth.1701 [DOI] [PubMed] [Google Scholar]
- 42.Katoh K, Standley DM (2016) A simple method to control over-alignment in the MAFFT multiple sequence alignment program. Bioinformatics 32: 1933–1942. doi: 10.1093/bioinformatics/btw108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stamatakis A (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. doi: 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Retief JD (2000) Phylogenetic analysis using PHYLIP. Methods Mol Biol 132: 243–258. [DOI] [PubMed] [Google Scholar]
- 45.Tyler BM, Tripathy S, Zhang X, Dehal P, Jiang RHY, et al. (2006) Phytophthora Genome Sequences Uncover Evolutionary Origins and Mechanisms of Pathogenesis. Science 313: 1261–1266. doi: 10.1126/science.1128796 [DOI] [PubMed] [Google Scholar]
- 46.Li L, Stoeckert CJ, Roos DS (2003) OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Research 13: 2178–2189. doi: 10.1101/gr.1224503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kunjeti SG, Evans TA, Marsh AG, Gregory NF, Kunjeti S, et al. (2012) RNA‐Seq reveals infection‐related global gene changes in Phytophthora phaseoli, the causal agent of lima bean downy mildew. Molecular Plant Pathology 13: 454–466. doi: 10.1111/j.1364-3703.2011.00761.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen XR, Xing YP, Li YP, Tong YH, Xu JY (2013) RNA-Seq reveals infection-related gene expression changes in Phytophthora capsici. PLOS ONE 8: e74588–e74588. doi: 10.1371/journal.pone.0074588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen X, Zhang B, Xing Y, Li Q, Li Y, et al. (2014) Transcriptomic analysis of the phytopathogenic oomycete Phytophthora cactorum provides insights into infection-related effectors. BMC Genomics 15: 980 doi: 10.1186/1471-2164-15-980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Krajaejun T, Lerksuthirat T, Garg G, Lowhnoo T, Yingyong W, et al. (2014) Transcriptome analysis reveals pathogenicity and evolutionary history of the pathogenic oomycete Pythium insidiosum. Fungal Biology 118: 640–653. doi: 10.1016/j.funbio.2014.01.009 [DOI] [PubMed] [Google Scholar]
- 51.Kemen E, Gardiner A, Schultzlarsen T, Kemen A, Balmuth AL, et al. (2011) Gene gain and loss during evolution of obligate parasitism in the white rust pathogen of Arabidopsis thaliana. PLOS Biology 9: e1001094 doi: 10.1371/journal.pbio.1001094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thines M (2009) Bridging the Gulf: Phytophthora and Downy Mildews Are Connected by Rare Grass Parasites. PLOS ONE 4(3): e4790 doi: 10.1371/journal.pone.0004790 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
(XLSX)
(TIFF)
(TIFF)
The results are summarized in three main categories: biological process, cellular component and molecular function. A set of 17647 genes were assigned to GO term based on blastx matches to known proteins.
(TIFF)
In total, 6920 of the 19627 P. litchii genes with Nr hits were grouped into 25 KOG classification.
(TIFF)
Pathway assignment was summarized for five main categories: Cellular Processes (C), Environmental Information Processing (E), Genetic Information Processing (G), Metabolism (M) and Organismal Systems (O). A total of 4624 genes were assigned to 32 groups, and abundant genes were involved signal transduction.
(TIFF)
Data Availability Statement
All relevant transcriptome data are available from GenBank (SRA accession: SRP079825).