Abstract
In many animal embryos, a specific gene expression pattern is established along the animal-vegetal axis soon after zygotic transcription begins. In the embryo of the ascidian Ciona intestinalis, soon after the division that separates animal and vegetal hemispheres into distinct blastomeres, maternal Gata.a and β-catenin activate specific genes in the animal and vegetal blastomeres, respectively. On the basis of these initial distinct gene expression patterns, gene regulatory networks promote animal cells to become ectodermal tissues and vegetal cells to become endomesodermal tissues and a part of the nerve cord. In the vegetal hemisphere, β-catenin directly activates Foxd, an essential transcription factor gene for specifying endomesodermal fates. In the present study, we found that Foxd also represses the expression of genes that are activated specifically in the animal hemisphere, including Dmrt1, Prdm1-r.a (Bz1), Prdm1-r.b (Bz2), and Otx. A reporter assay showed that Dmrt1 expression was directly repressed by Foxd, and a chromatin immunoprecipitation assay showed that Foxd was bound to the upstream regions of Dmrt1, Prdm1-r.a, Prdm1-r.b, and Otx. Thus, Foxd has a dual function of activating specific gene expression in the vegetal hemisphere and of repressing the expression of genes that are normally expressed in the animal hemisphere. This dual function stabilizes the initial patterning along the animal-vegetal axis by β-catenin and Gata.a.
Author summary
In embryogenesis of most animals, a specific gene expression pattern is established along the animal-vegetal axis first. In the embryo of the ascidian Ciona intestinalis, the activity of the maternal factor Gata.a is suppressed by β-catenin, which is active only in the vegetal hemisphere, and thereby these two factors activate specific genes in the animal and vegetal blastomeres, respectively. We found that a gene encoding a transcription factor, Foxd, which is a direct target of β-catenin, works as a promoter for endomesodermal fate and an inhibitor for ectodermal fate. In the ascidian embryo, the animal-vegetal axis initially established by the maternal factors is not stable enough for subsequent developmental processes, and needs to be maintained by Foxd. Thus, the animal hemisphere fate is suppressed first by the maternal factor β-catenin, and then by Foxd, which is activated by β-catenin. The primary embryonic axis is not stable initially, and stabilized by a transcription factor, which is expressed differentially along the axis.
Introduction
In many animal embryos, localized maternal factors create differential gene expression patterns along the animal-vegetal axis [1–3], and the subsequent developmental program proceeds on the basis of this initial patterning. In ascidian unfertilized eggs, several identified and unidentified maternal factors are unequally distributed along the animal-vegetal axis [4]. At the 8-cell stage, the animal and vegetal hemispheres become separated into distinct blastomeres, and the difference along the animal-vegetal axis is clearly established; when blastomeres are experimentally isolated at the 8-cell stage, endomesodermal cells are differentiated from vegetal hemisphere cells [5–8], and epidermal cells are differentiated from animal cells [9]. In 16-cell embryos of the ascidian Ciona intestinalis, the maternal transcription factor Gata.a activates Ephrina.d and Tfap2-r.b specifically in the animal hemisphere, and a complex of β-catenin and Tcf7 activates Foxd and Fgf9/16/20 in the vegetal hemisphere [10–15]. In the vegetal hemisphere, β-catenin/Tcf7 weakens the Gata.a-binding activity for target sites through a physical interaction, and thereby the animal hemisphere genes are not expressed in the vegetal hemisphere at the 16-cell stage [15]. In this manner, the initial difference between the animal and vegetal hemispheres is set up.
Foxd and Fgf9/16/20, which are activated by β-catenin/Tcf7, encode a transcription factor and a signaling molecule, respectively. These molecules are required for expression of endodermal and mesodermal genes including Lhx3/4, Zic-r.b (ZicL), and Brachyury in the vegetal hemisphere [16–18]. In addition, Fgf9/16/20 signaling also induces expression of neural genes including Dmrt1, Otx, Prdm1-r.a and Prdm1-r.b in the neural lineage of the animal hemisphere [11, 19–22]. Animal hemisphere cells that are not induced by Fgf9/16/20 signaling give rise to epidermal cells under the control of Tfap2-r.b, which encodes a transcription factor [23]. Thus, the difference between the animal and vegetal hemispheres are critically important for subsequent developmental programs.
However, the initial difference between the animal and vegetal hemispheres, which is established by Gata.a and β-catenin/Tcf7, may not be sufficient for explaining differential gene expression patterns between them at the 32-cell stage and thereafter, because two animal hemisphere genes Dmrt1 and Dlx.b are expressed ectopically in the vegetal hemisphere of Foxd morphants at the early gastrula stage [19]. Dmrt1 is important for anterior neural and palp (a placode-like structure) fate specification [19, 24], and Dlx.b is important for neural and epidermal fate specification [23]. In the present study, we examined how the animal-vegetal axis is maintained at the 32-cell stage and thereafter, and showed that Foxd acts as a robust binary switch to stabilize the initial patterning along the animal-vegetal axis by Gata.a and β-catenin/Tcf7.
Results
Candidate genes under the control of Foxd in early embryos
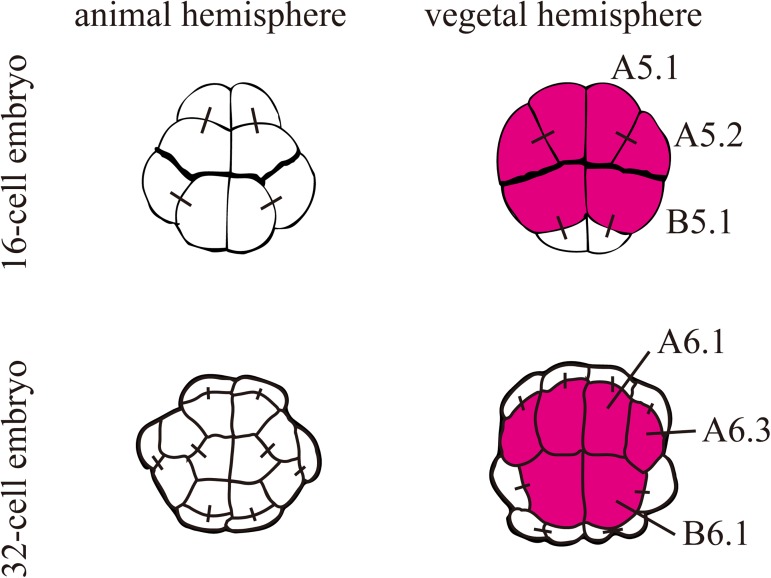
Foxd is expressed under the direct control of β-catenin/Tcf7 in three vegetal cell pairs (A5.1, A5.2, and B5.1) of 16-cell embryos (Fig 1). After the next division, among their daughter cells, cells with endodermal fate continue to express Foxd (A6.1, A6.3, and B6.1), and the expression becomes undetectable at the 64-cell stage. To identify genes regulated by Foxd in early embryos, we performed RNA-seq analysis at the 32-cell, 64-cell, and 112-cell stages to compare transcriptomes between unperturbed and Foxd knocked-down embryos. For Foxd knockdown, we used a morpholino oligonucleotide (MO) against Foxd. We picked up genes encoding transcription factors and signaling molecules that are known to be expressed zygotically between the 32-cell and 112-cell stages [25], and compared expression levels between unperturbed and Foxd morphant embryos (Fig 2). We did not utilize biological replicates because we used these data for screening purposes and because we performed this analysis at three successive time points. Fourteen genes were identified to be differentially expressed at one or more stages by a computer program called NOISeq [26] (> 80%, probability of differential expression by NOIseq-sim, which simulates technical replicates). Among them, nine genes were previously known to be regulated by Foxd: Zic-r.b (ZicL), Brachyury, Fgf8/17/18, Fgf9/16/20, Foxb, Lhx3/4, and Mnx were known to be positively regulated by Foxd, and Dmrt1 and Foxd itself are known to be negatively regulated [16, 19, 27]. These observations indicate that the RNA-seq experiments successfully identified genes under the control of Foxd.
Fig 1. Expression of Foxd at the 16- and 32-cell stages.
Schematics of the animal and vegetal hemispheres of the bilaterally symmetrical 16-cell and 32-cell embryos. Cells expressing Foxd are colored in magenta. Their blastomere names are indicated in the right halves. Note that Foxd is expressed only at the 16-cell and 32-cell stages in early embryos. Black bars connecting two cells indicate their sister cell relationship.
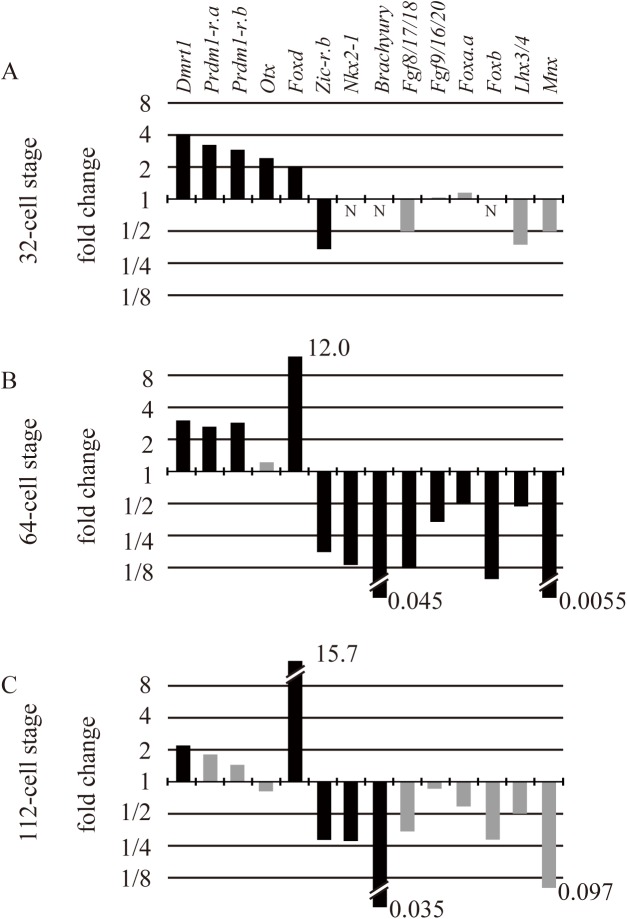
Fig 2. Differential gene expression upon Foxd knockdown revealed by RNA-seq analysis.
(A–C) Among regulatory genes that are expressed zygotically in early embryos, genes greatly upregulated or downregulated in morphant embryos of Foxd are shown (black bars, q-values in NOIseq-sim > 0.80; gray bars, q-values in NOIseq-sim < 0.80). No bars are shown for genes in which no sequence tags were detected (N). The analyses were performed at the 32-cell (A), 64-cell (B), and 112-cell stages (C).
In addition to these nine previously characterized genes, there were five differentially expressed regulatory genes identified: Foxa.a was downregulated at the 64-cell stage, Nkx2-1 (Ttf1) was downregulated at the 64-cell and 112-cell stages, Otx was upregulated at the 32-cell stage, and Prdm1-r.a (Bz1) and Prdm1-r.b (Bz2) were upregulated at the 32- and 64-cell stages in Foxd morphants. These genes were candidates for Foxd targets that had not yet been identified.
Genes positively regulated by Foxd were expressed in the vegetal hemisphere
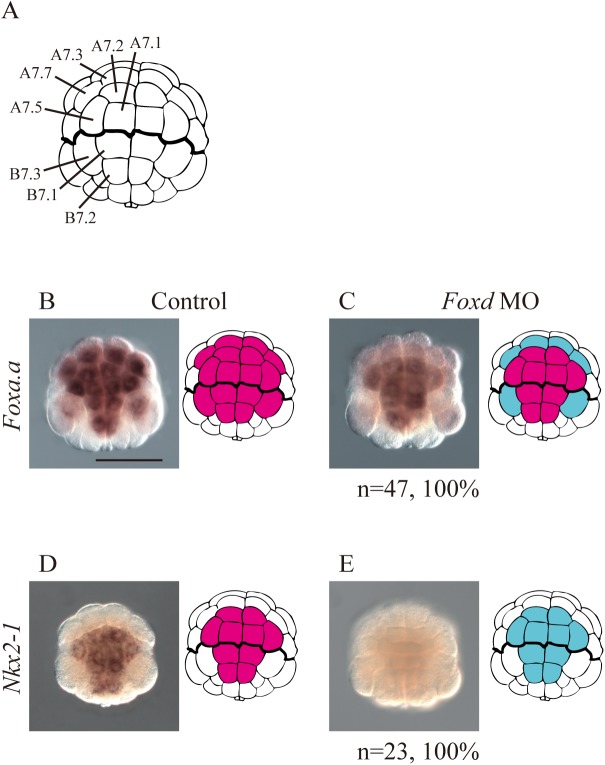
To confirm downregulation of Foxa.a and Nkx2-1 in Foxd morphant embryos, we performed in situ hybridization. Foxa.a was normally expressed strongly in the vegetal blastomeres designated A7.1, A7.2, A7.3, A7.5, A7.7, B7.1, and B7.2, and weakly in B7.3 at the 64-cell stage (Fig 3A and 3B). Foxa.a expression was lost only in A7.3, A7.7, and B7.3 in Ciona Foxd morphants (Fig 3C). Foxa.a expression begins at the 8-cell stage, and our data did not indicate downregulation of Foxa.a at the 32-cell stage (Fig 2A), which is consistent with a recent study [16]. Nkx2-1 was normally expressed in the vegetal blastomeres designated A7.1, A7.2, A7.5, B7.1, and B7.2 at the 64-cell stage (Fig 3D), whereas it was not expressed in Foxd morphants (Fig 3E), as recently shown at the early gastrula stage [16]. Thus, Foxd positively regulated Foxa.a and Nkx2-1.
Fig 3. In situ hybridization analysis to determine if Foxa.a and Nkx2-1 are controlled by Foxd at the 64-cell stage.
(A) Illustrations of the vegetal hemisphere of the 64-cell embryo. Blastomere names are indicated in the left half of the bilaterally symmetrical embryo. (B–E) The expression of (B, C) Foxa.a, and (D, E) Nkx2-1 in (B, D) control unperturbed embryos and (C, E) Foxd morphant embryos. Illustrations on the right indicate the expression patterns. Blastomeres with expression are filled in magenta. Blastomeres that lost expression in Foxd morphants are shown in cyan. The number of morphant embryos examined and the proportion of embryos that each panel represents are shown below the panels. Scale bar, 100 μm.
In addition to Foxa.a and Nkx2-1, the genes Brachyury, Fgf8/17/18, Fgf9/16/20, Foxb, Mnx, and Zic-r.b, which were found to be positively regulated by Foxd (Fig 2), are all expressed in the vegetal hemisphere [17, 25, 27, 28]. Namely, genes that were identified to be positively regulated by Foxd in early embryos were all expressed in the vegetal hemisphere.
Genes negatively regulated by Foxd were expressed in the animal hemisphere
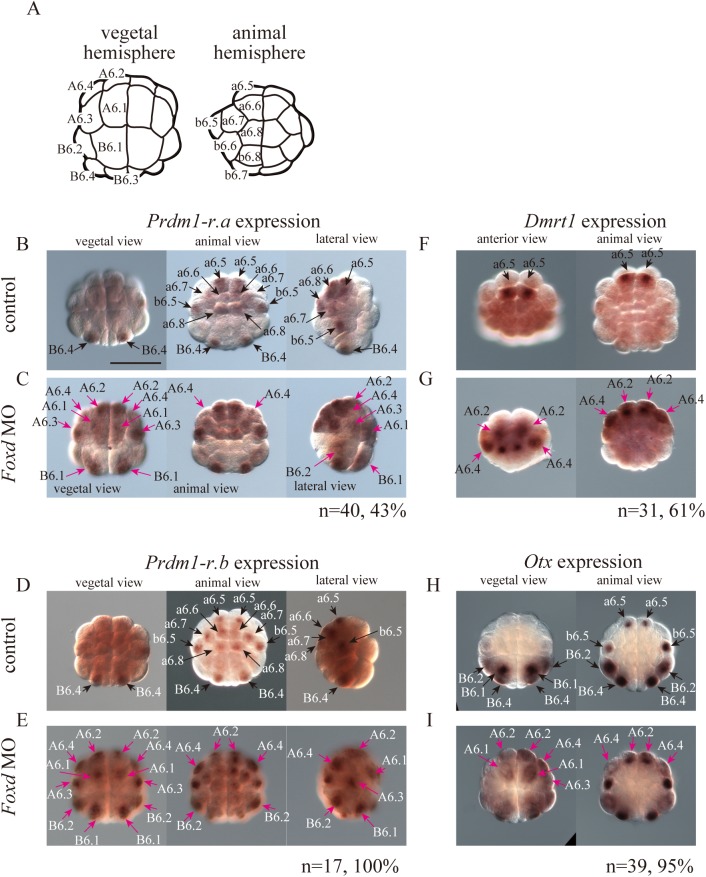
Prdm1-r.a, Prdm1-r.b, Foxd, Dmrt1, and Otx were found to be negatively regulated by Foxd (Fig 2). While Prdm1-r.a and Prdm1-r.b are normally expressed in five pairs of animal cells (a6.5 to a6.8 and b6.5) and a pair of vegetal cells (B6.4) at the 32-cell stage [29], these two genes were ectopically expressed in vegetal cells of Foxd morphants (A6.1 to A6.4, B6.1, and B6.2) (Fig 4A–4E).
Fig 4. Ectopic expression of animal hemisphere genes in the vegetal hemisphere of Foxd morphant embryos at the 32-cell stage.
(A) Illustrations of the 32-cell embryo. Blastomere names are indicated in the left half of the bilaterally symmetrical embryo. (B–I) The expression of (B, C) Prdm1-r.a (Bz1), (D, E) Prdm1-r.b (Bz2) (F, G) Dmrt1, and (H, I) Otx in (B, D, F, H) control unperturbed embryos, and (C, E, G, I) Foxd morphant embryos. Black arrows in (B, D, F, H) indicate expression in normal embryos, while magenta arrows in (C, E, G, I) indicate ectopic expression in Foxd morphant embryos. The number of morphant embryos examined and the proportion of embryos that each panel represents are shown below the panels. Scale bar, 100 μm. Note that Fgf9/16/20 is activated independently of Foxd at the 16-cell and 32-cell stages (S1D and S1E Fig; Fig 2A) [14–17], although it is later activated by Foxd (Fig 2B) [19]. Therefore, it is not strange that Otx expression in the animal hemisphere, which is under control of Fgf9/16/20 [11, 20, 21, 45, 46], was not affected at the 32-cell stage.
Foxd expression was examined in Foxd morphants (S1A and S1B Fig). Foxd mRNA was detected in Foxd morphants at the 64-cell stage, while it was rarely detected in normal 64-cell embryos. This might suggest that Foxd negatively regulates itself, or alternatively, that Foxd mRNA was stabilized by binding the MO. To discriminate between these possibilities, we injected synthetic Foxd mRNA into Ciona eggs. Because the synthetic mRNA lacked the endogenous 3’-UTR, we were able to measure the amount of the endogenous Foxd mRNA by RT-qPCR with primers designed to its 3’-UTR. While levels of the maternal control mRNA Pou2 were unchanged, Foxd mRNA levels were greatly reduced by injection of synthetic Foxd mRNA (S1C Fig). Therefore, Foxd indeed regulates itself negatively.
We previously showed that Dmrt1 is expressed at the 64- and 112-cell stages in the anterior neural lineage of the animal hemisphere [25], and the RNA-seq result of the present study suggested that this gene was expressed in 32-cell embryos under the control of Foxd. Indeed, upon careful re-examination, we detected a weak signal in the anterior animal cells (a6.5) at the 32-cell stage of normal embryos. This expression pattern was expanded to the anterior vegetal cells (A6.2 and A6.4) of Foxd morphants (Fig 4F and 4G). Consistently, injection of Foxd mRNA reduced Dmrt1 expression (S1C Fig).
Otx is expressed in three pairs of vegetal cells (B6.1, B6.2, and B6.4) and two pairs of animal cells (a6.5 and b6.5) at the 32-cell stage in normal embryos [20]. This gene was expressed ectopically in the anterior vegetal cells (A6.1 to A6.4) of Foxd morphants (Fig 4H and 4I).
Otx and Dmrt1 are activated by Fgf signaling [19, 20], and Fgf9/16/20 is downregulated at later stages in Foxd morphants [19], which was consistent with the RNA-seq result at the 64-cell stage (Fig 2B). On the other hand, Fgf9/16/20 is not downregulated at the 32-cell stage [16, 17], which was also consistent with the RNA-seq result at the 32-cell stage (Fig 2A). Indeed, Fgf9/16/20 was not downregulated at the 16-cell stage in Foxd morphants (S1D and S1E Fig). Therefore, it is likely that the earliest expression of Fgf9/16/20, which is controlled by maternal β-catenin [15] but not by Foxd, induced Otx and Dmrt1 expression, even in Foxd morphants.
Because Tfap2-r.b is regulated directly by a maternal factor [15] and expressed in the animal hemisphere at the 16-cell stage [25], and because expression of Tfap2-r.b was not significantly changed in our RNA-seq experiment (~1.7 fold-increase), we examined the expression of this gene as a negative control. We confirmed by in situ hybridization that the expression of this gene was not affected in Foxd morphants (S1F and S1G Fig).
Our results showed that Foxd represses Prdm1-r.a, Prdm1-r.b, Dmrt1, and Otx expression in vegetal cells at the 32-cell stage, although Otx is expressed in the posterior vegetal cells of normal embryos and Foxd morphants. In addition, Dlx.b, which is expressed in the entire animal hemisphere, is known to be regulated negatively by Foxd [19], although this gene was not identified to be downstream of Foxd in our RNA-seq experiment (Fig 2); this is probably because the number of cells with ectopic Dlx.b expression is much smaller than the number of animal hemisphere cells with Dlx.b expression. Because Prdm1-r.a, Prdm1-r.b, Dmrt1, Otx, and Dlx.b play essential roles in the specification of epidermal and neural fates [11, 19, 20, 22–24, 29], Foxd is likely to suppress ectodermal fates in the vegetal hemisphere.
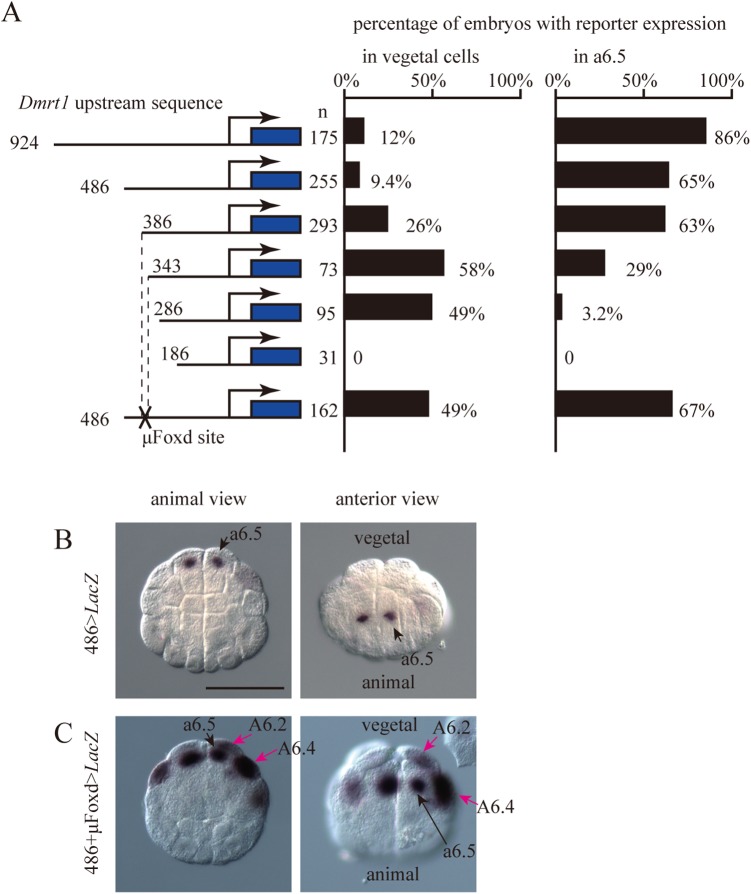
A putative Foxd binding site within the Dmrt1 upstream sequence was important for suppressing ectopic expression in the vegetal hemisphere
To understand the mechanism by which Foxd negatively regulates ectodermal fates, we analyzed the upstream regulatory sequence of Dmrt1 by introducing lacZ reporter constructs using electroporation. Experimental embryos were fixed at the 32-cell stage, and reporter expression was examined by in situ hybridization.
The 924-base pair (bp) upstream sequence of Dmrt1, which was slightly longer than the sequence used in previous studies [29, 30], drove reporter expression specifically in anterior neural cells (a6.5) at the 32-cell stage (Fig 5A; S2 Fig). A construct containing the 486-bp upstream sequence showed almost the same activity (Fig 5A and 5B). Ectopic expression in vegetal cells was increased in constructs containing 386-, 343-, and 286-bp upstream regions, while expression in the a6.5 neural lineage was decreased in the constructs containing 343-, and 286-bp upstream regions. The construct containing the 186-bp upstream sequence did not drive reporter expression. This observation indicated that cis-elements important for expression in the animal hemisphere are present between bases -286 and -386, and that cis-elements important for repression in the vegetal hemisphere are present between bases -343 and -386.
Fig 5. Reporter assay to determine the importance of the Foxd binding site upstream of Dmrt1.
(A) Analysis of the upstream regulatory region of Dmrt1. Illustrations on the left depict the constructs. Blue boxes indicate the LacZ gene and SV40 polyadenylation signal. The numbers indicate the relative nucleotide positions from the transcription start site. Mutated Foxd-binding sites are indicated by the letter X. Graphs show the percentage of embryos expressing the reporter in vegetal cells and a6.5 blastomeres. Note that not all cells or embryos could express the reporter because of mosaic incorporation of the electroporated plasmid. (B, C) LacZ mRNA expression in embryos electroporated with the constructs containing the 486-bp long upstream region of Dmrt1 with an intact (B) or mutant (C) Fox binding site. Black arrows indicate normal expression in a6.5, while magenta arrows indicate ectopic expression in Foxd morphant embryos. Scale bar, 100 μm.
We searched candidate Foxd binding sites using the Patser program [31] and a position weight matrix for human FOXD2 [32], which identified one putative Foxd binding site between -343 and -386 (S3 Fig). This site was conserved in the genome of the closely related species Ciona savignyi (S3 Fig). Therefore, we mutated this putative binding site. The mutant upstream sequence drove reporter expression in the vegetal hemisphere (Fig 5A and 5C), suggesting that Foxd directly represses Dmrt1 expression via this site.
Foxd directly bound to the upstream region of Dmrt1 and other animal hemisphere-specific genes
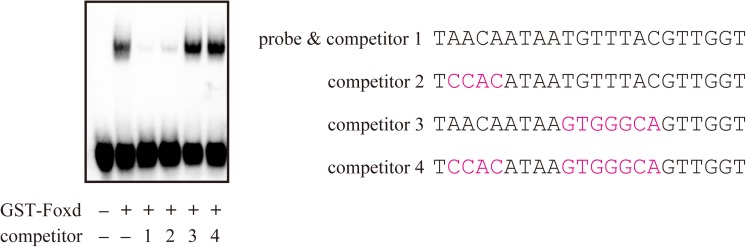
To confirm if the identified site could bind Foxd, we performed gel-shift assays (Fig 6). Foxd binding was observed as a shifted band that disappeared upon incubation with a specific competitor (competitor 1) but did not disappear upon incubation with competitors containing a mutation in the putative Fox binding site (competitors 3 and 4). Because the gel-shift probe contained an additional sequence similar to the Fox binding site (AACA), we tested whether this sequence also bound Foxd. The competitor containing a mutation in this second site (competitor 2) did not compete, suggesting that this site does not bind Foxd efficiently.
Fig 6. Gel-shift assay to determine Foxd binding to the putative Fox binding site upstream of Dmrt1.
Foxd.b-GST fusion protein produced in E. coli were incubated with a probe containing a putative Fox binding site (TGTTTAC). The shifted band disappeared by co-incubation with competitors 1 and 2, in which the Fox binding site was intact, but not by co-incubation with competitors 3 and 4, in which the Fox binding site was mutated. Sequences of the probe and competitors are shown on the right. Mutated nucleotides are shown in magenta. In competitors 2 and 4, a secondary site (AACA) similar to the primary site was mutated.
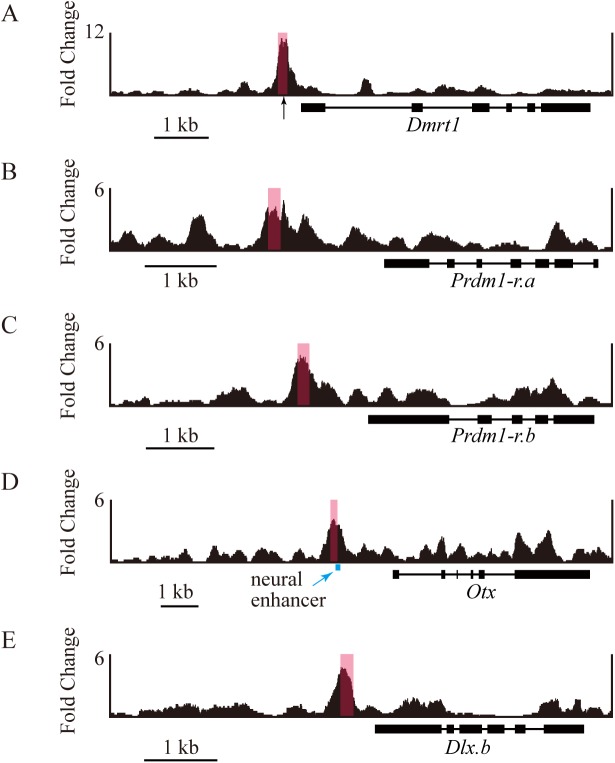
Finally, we performed a chromatin-immunoprecipitation assay followed by high-throughput DNA sequencing (ChIP-seq) to confirm that Foxd bound to the regions containing the above putative Foxd binding site in vivo at the 32-cell stage (Fig 7). We electroporated an expression construct encoding a Foxd-Gfp fusion protein under the control of the Foxd upstream regulatory sequence, and performed a ChIP assay using 32-cell embryos with an anti-Gfp antibody. Two different computer programs identified 114 and 799 peaks, respectively (false discovery rate < 0.1%), of which 63 peaks were common and considered in the subsequent analysis.
Fig 7. A chromatin-immunoprecipitation assay to determine Foxd binding to the upstream regions of genes that are negatively regulated by Foxd.
A construct driving expression of a gene encoding a Foxd-Gfp fusion protein was introduced by electroporation, and anti-Gfp antibodies were used for chromatin immunoprecipitation. Precipitated DNA fragments were analyzed by deep-sequencing. Chromosomal regions containing the upstream regions and exons of (A) Dmrt1, (B) Prdm1-r.a, (C) Prdm1-r.b, (D) Otx, and (E) Dlx.b are shown. Peaks were called separately for two biological duplicates, and the graphs include data of two biological duplicates. Pink rectangles indicate peak regions commonly identified by the two different peak caller programs; these peak regions were identified in both of the duplicates. A black arrow in (A) indicates the positions of the Foxd binding site identified in the reporter analysis. A cyan line in (D) indicates the location of the neural enhancer revealed by previous studies [11, 35].
Because Foxd-Gfp might be overexpressed above physiological levels, it is possible that the above binding interactions were stronger than interactions that would normally occur in normal embryos. However, among 52,518 of ‘GTAAACA’ sequences found in the genome, only 9 sites were included in the 63 peaks identified by the ChIP-seq assay, suggesting that Foxd-Gfp does not bind non-specifically to all potential binding sites.
As shown in Fig 7A, the upstream region of Dmrt1 around the Fox binding site identified above bound Foxd, suggesting direct regulation of Dmrt1 by Foxd. In addition, we found peak regions in the upstream sequences of Prdm1-r.a, Prdm1-r.b, Otx, and Dlx.b (Fig 7B–7E). All these peak regions contained Foxd binding motifs that were identifiable by the Patser program [31] and a position weight matrix for human FOXD2 [32], although their scores were less than the scores of Dmrt1 (S4 Fig). Meanwhile, the 63 significant peaks were not found in the upstream regulatory region of Tfap2-r.b, which is not regulated by Foxd as described above, although a weak, insignificant peak was observed (S5 Fig). Therefore, it is conceivable that Dmrt1, Prdm1-r.a, Prdm1-r.b, Otx, and Dlx.b are direct targets of Foxd.
Discussion
Foxd maintains differential gene expression along the animal-vegetal axis
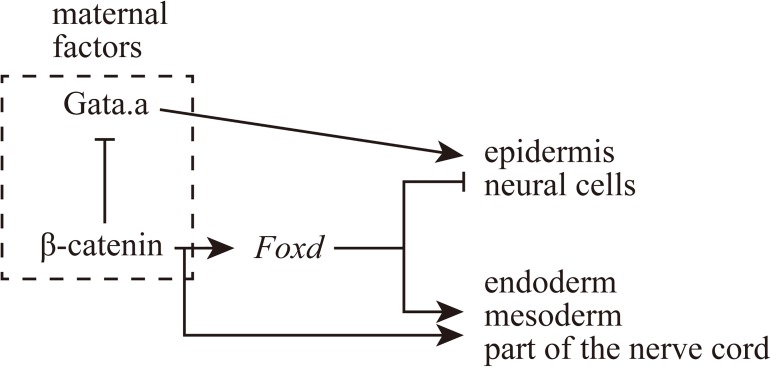
In ascidian embryos, maternal factors establish differential gene expression patterns between the animal and vegetal hemispheres, which largely correspond to the ectodermal and endomesodermal lineages (with the exception of part of the nerve cord, which is derived from the vegetal hemisphere). Gata.a and β-catenin/Tcf7 activate specific gene expression in these two domains at the 16-cell stage. However, our present results indicated that this segregation between the animal and vegetal hemisphere lineages was not robust enough to maintain this segregation alone. We found that Foxd activity commits vegetal cells to the endomesoderm fate by repressing ectoderm genes including Prdm1-r.a, Prdm1-r.b, Dmrt1, Otx, and Dlx.b (Fig 8), although Foxd may not necessarily repress all genes that are expressed in the animal hemisphere. In other words, maternal factors generated a transient regulatory stage, which was maintained by Foxd activity. Thus, animal hemisphere gene expression is suppressed in the vegetal hemisphere continuously during early embryogenesis. First, Gata.a activity is suppressed by β-catenin/Tcf7 in the vegetal hemisphere of the 16-cell embryo [15], and then Foxd, which is activated by β-catenin/Tcf7, directly represses animal hemisphere genes in the vegetal hemisphere at the 32-cell stage and thereafter.
Fig 8. Schematic representation of a gene regulatory circuit essential for determining endomesodermal fate in the vegetal hemisphere.
Two domains with different gene expression patterns along the animal-vegetal axis are established initially by maternal β-catenin and Gata.a, and then stabilized by Foxd. These two domains mostly correspond to the ectoderm and endomesoderm.
In Foxd morphants, Prdm1-r.a and Prdm1-r.b were activated ectopically in both the anterior and posterior vegetal cells, while Dmrt1 and Otx were activated ectopically only in the anterior cells. Activators for Dmrt1 might not be present in the posterior vegetal cells. In normal embryos, Dmrt1 is activated only in the anterior neural cells, because Foxa.a, which encodes an activator for Dmrt1, is not expressed in the posterior neural cells [19, 25, 33]. Foxa.a expression indeed begins in the anterior half of the 8-cell embryo, although it is expressed in posterior vegetal cells at the 16-cell stage and thereafter [33, 34]. Meanwhile, in normal embryos, Otx is expressed in the posterior vegetal cells except the most posterior cells, in addition to the animal neural cells [20]. Different enhancers are responsible for expression in these two regions [11, 35]. Therefore, even if the neural enhancer of Otx is ectopically activated in these posterior vegetal cells, this ectopic activation cannot be detected by in situ hybridization. Indeed, one of the peak regions in the Otx upstream region partly overlaps the neural enhancer identified in previous studies [11, 35] (Fig 7D). Activation of Otx in the vegetal hemisphere by different enhancers may explain why we detected differential expression of Otx only at the 32-cell stage by the RNA-seq experiments.
In addition to the repressive function shown above, Foxd functions as an activator; it activates Zic-r.b and Lhx3/4 cooperatively with Foxa.a and Fgf9/16/20 [16, 19, 27]. A ChIP assay showed that Foxd binds to upstream regions of Foxd-regulated genes at the 64-cell stage [36]. Reporter assays also showed that two Fox-binding sites within the upstream sequence of Zic-r.b, which is activated by Foxd, are essential for its expression [37]. Lhx3/4 is also likely to be a direct target of Foxd, because Lhx3/4 is expressed at the 32-cell stage under the control of Foxd [16], and because Foxd is bound to the upstream region of Lhx3/4 at the 64-cell stage [36]. Thus, Foxd is a dual-functional protein; it simultaneously promotes endomesodermal fates and inhibits ectodermal fates.
It has been proposed that there are sub-circuits responsible for locking down regulatory states [38]. In Ciona early embryos, Foxd maintains the regulatory state of the vegetal hemisphere, and therefore this dual-functional protein may alone work like such a sub-circuit to lock down dynamic states.
Foxd works as a transcriptional activator and repressor
In Xenopus, FoxD4L1.1 has a dual role as a transcriptional activator and repressor in the neural ectoderm; it activates genes that keep cells in a proliferative state and represses genes that promote differentiation [39]. Xenopus FoxD4L1.1 is also involved in repressing BMP signaling, thereby suppressing epidermal fate. The activating function is mediated in Xenopus by an acidic domain near the N-terminus and the repressing function at least partly depends on an Engrailed homology region-1 (Eh-1) located in the C-terminal region. Ciona Foxd also contains a putative acidic domain near the N-terminus and an Eh-1 motif in the C-terminal region (S6 Fig). In both Ciona and Xenopus, Foxd acts as a robust binary switch that promotes one fate and suppresses the other fate. This might be an evolutionarily conserved function of Foxd.
A previous study identified two critical Fox-binding sites to which Foxd might bind in the upstream region of Zic-r.b [37]. The sequences of these sites are slightly different from the sequence of the Foxd-binding site for Dmrt1 and those found in the peak regions in the upstream regions of Prdm1-r.a, Prdm1-r.b, Otx, and Dlx.b. In the ChIP-seq assay of the present study, we did not find clear binding peaks upstream of Zic-r.b, although our previous ChIP-chip assay using slightly older embryos exhibited peaks [36]. Therefore, the binding sites in the upstream regions of Prdm1-r.a, Prdm1-r.b, Otx, and Dlx.b might be stronger than the binding sites upstream of Zic-r.b. Indeed, at least one Foxd binding motif in each of the peak regions in the upstream regions of Prdm1-r.a, Prdm1-r.b, Otx, and Dlx.b gave a higher score than the Fox binding sites found in Zic-r.b (S4B Fig). Such a qualitative difference might be important for Foxd to work as an activator or a repressor. In the ascidian embryo, Sox1/2/3, and Gata.a are important for specification of ectodermal fate [11, 12, 15, 23]. Because there are clear Sox and Gata binding motifs in the peak regions of Dmrt1, Prdm1-r.a, Prdm1-r.b, Otx, and Dlx.b, it is possible that Sox1/2/3 and Gata.a help Foxd to act as a repressor.
Materials and methods
Animals, whole-mount in situ hybridization, and gene identifiers
Ciona intestinalis (type A; this type is also called Ciona robusta) adults were obtained from the National Bio-Resource Project for Ciona. cDNA clones were obtained from our EST clone collection [40]. Whole-mount in situ hybridization was performed as described previously [25]. Identifiers for genes examined in the present study are shown in S1 Table, according to the nomenclature rule proposed in a recent paper [41].
Gene knockdown, overexpression and reporter assays
A morpholino oligonucleotide (MO; Gene Tools, LLC) for Foxd knock-down is designed to block translation of two paralogous Foxd genes, Foxd.a and Foxd.b (5′-GCACACAACACTGCACTGTCATCAT-3′). This MO has been used previously, and its specificity has been evaluated [19, 25]. The MO was introduced by microinjection under a microscope.
The coding sequence of Foxd.b was cloned into pBluscript RN3 [42], and Foxd mRNA was transcribed using the mMESSAGE mMACHINE T3 Transcription Kit (Life technologies).
Reporter constructs were introduced into fertilized eggs by electroporation. Chromosomal positions of the upstream sequences for reporter constructs and the mutated sequence are indicated in S2 Fig. We randomly chose embryos introduced with reporter constructs to examine reporter construct expression by in situ hybridization.
We performed all gene knockdown experiments and reporter gene assays at least twice with different batches of embryos.
Gel-shift assay
Recombinant Foxd.b protein was produced as a fusion protein of the Foxd DNA-binding domain and glutathione S-transferase in Escherichia coli BL21 star DE3 strain (Thermo Fisher Scientific), and the protein was purified under a native condition using glutathione Sepharose 4B (GE Healthcare). After annealing two complementary oligonucleotides (5’-AAATAACAATAATGTTTACGTTGGT-3’ and 5’-AAAACCAACGTAAACATTATTGTTA-3’), both protruding ends of the double-stranded oligonucleotides were filled with biotin-11-dUTP, and this biotin-labelled oligonucleotide was used as a probe. Proteins and the biotin-labeled probe were mixed in 10 mM Tris (pH 7.5), 50 mM KCl, 1 mM DTT, 1 mM EDTA, 50 ng/μL poly(dIdC), 2.5% glycerol, and 0.05% NP40 with or without competitor double-stranded DNAs (a 100 fold molar excess) shown in Fig 6. Proteins amounts were empirically determined. Protein–DNA complexes were detected using an AP-conjugated anti-biotin antibody (Roche) and CDP-star substrate (Roche).
RNA sequencing (RNA-seq)
For RNA-seq experiments, 50 unperturbed and Foxd-morphant embryos were collected at the 32-, 64-, and 112-cell stages. RNA was extracted using a Dynabeads mRNA DIRECT Purification Kit (Thermo Fischer Scientific) and libraries were made with an Ion Total RNA-Seq kit ver 2 (Thermo Fischer Scientific). The libraries were sequenced with an Ion PGM instrument (Thermo Fischer Scientific) (SRA accession number: DRA005206). We did not utilize duplicates because we used this experiment for screening purposes, and the obtained results were confirmed using other methods, as explained in the Results section. NOISeq [26] was used to identify differentially expressed genes.
Chromatin immunoprecipitation
We used a DNA construct encoding GFP-tagged Foxd under the control of the Foxd promoter [36]. Embryos were fixed at the 32-cell stage. The embryos were subjected to ChIP analysis using anti-GFP antibodies, and the immunoprecipitated DNA was amplified by ligation-mediated PCR [36]. Whole cell extract DNA was used as a control. Then, high-throughput DNA sequencing was performed with the Ion PGM instrument (SRA accession number: DRA005285). To identify peak regions, we used two different programs called Homer [43] with options “-style factor -F 4 -P 0.01 -L 4 -localSize 3000” and MACS2 [44] with an option “--nomodel -q 0.001".
RT-qPCR
For RT-qPCR, we extracted RNA from wild-type embryos and embryos injected with Foxd mRNA. The RNA was converted to cDNA by the Cells-to-Ct kit (Thermo Fisher Scientific). The obtained cDNA samples were then analyzed by quantitative PCR with the SYBR green method. For each qPCR, the amount of cDNA used was equivalent to two-thirds of an embryo. Primers used were: Dmrt1, 5’-CGCTGAACGACAACGAGTCAT-3’ and 5’-TTCGTTTTCCTCTTGTGCTTGTT-3’; Foxd.a, 5’-AGTTTCTTCCCCACAGTTCCAA-3’ and 5’-GGTTTGTTGTATCCGGGATGTT-3’; Foxd.b, 5’-GCAGTACGCATTCCGCAAT-3’ and 5’-CGGAACAAAAACACAAAAGTCAAA-3’; Pou2, 5’- AAGATGGTTGCTGGATGCTAATAAT-3’ and 5’-TTGGATTGGAGTGGGAATAACAA-3’.
Ethics statement
Ciona intestinalis is excluded from legislation regulating scientific research on animals in Japan. Although there is no scientific evidence that Ciona intestinalis can experience pain, discomfort or stress, we made our best efforts to minimize potential harm that Ciona individuals might experience when we obtained eggs and sperm from them.
Supporting information
(A, B) The expression of Foxd revealed by in situ hybridization in (A) control and (B) Foxd morphants at the 64-cell stage. (C) The amount of endogenous Foxd mRNA was measured by RT-qPCR in uninjected control embryos and embryos injected with 2.3 pg of Foxd mRNA. The relative amount of mRNA in the experimental embryos compared with control embryos is shown. A maternal mRNA, Pou2, was used as an endogenous control. Error bars indicate mean±s.d. between two technical duplicates. The results of two independent experiments are shown in different colors. (D–G) The expression of (D, E) Fgf9/16/20, and (F, G) Tfap2-r.b revealed by in situ hybridization in (D, F) control unperturbed embryos and (E, G) Foxd morphant embryos at the 16-cell (D, E) and 32-cell (F, G) stages. Note that Foxd expression was not downregulated in the vegetal hemisphere of Foxd morphants (A, B) and that the expression of Fgf9/16/20 and Tfap2-r.b was not changed (D–G), although Fgf9/16/20 expression is downregulated in later embryos (Fig 2) [19]. The number of morphant embryos examined and the proportion of embryos that each panel represents are shown within the panels. Scale bar, 100 μm.
(TIF)
The numbers indicate the relative nucleotide positions from the transcriptional start site. The sequence of the fragment used for the gel-shift assay in Fig 6 is underlined, and the putative Foxd site is indicated in cyan. The mutation introduced in the Foxd site is shown in magenta.
(TIF)
(A) Asterisks indicate conserved nucleotides. T-Coffee [47] was used for generating this alignment. Putative Foxd binding sites, which were identified by Patser [31], are shown in cyan with scores. (B) A consensus sequence for human FOXD2 [32], which was used for identifying the putative Foxd binding sites, is shown as a sequence logo [48].
(TIF)
(A) Nucleotide sequences of the Foxd-binding regions in the upstream regions of Dmrt1, Prdm1-r.a, Prdm1-r.b, Otx, and Dlx.b, which were identified by the chromatin-immunoprecipitation and are shown in pink boxes in Fig 7. Putative Foxd-binding sites are shown in magenta. (B) An alignment of the putative Foxd binding sites found in (A) and those in the Zic-r.b (ZicL) upstream region identified previously [37]. Scores on the right were calculated by the Patser program and a position weight matrix for human FOXD2 binding sites [32], which is represented in S3B Fig.
(TIF)
Because Tfap2-r.b was not regulated by Foxd (S1F and S1G Fig), the upstream region of Tfap2-r.b is shown as a negative control for genes shown in Fig 7. Significant peaks were not identified by the computer programs in this genomic region. The graphs include data of two biological duplicates.
(TIF)
Conserved and similar amino acids are shown by black and gray boxes, respectively. The forkhead domains, Eh-1 domains, and putative acidic domains are enclosed by black lines, and acidic amino acids in the putative acidic region of the N-terminal half are shown in magenta.
(TIF)
(DOCX)
Acknowledgments
We thank the National Bio-resource project (MEXT, Japan) for providing experimental animals.
Data Availability
All RNA sequencing and ChIP-seq data files are available from the SRA database (accession numbers DRA005206 and DRA005285).
Funding Statement
This study was supported by a CREST program of Japan Science and Technology Agency (http://www.jst.go.jp/EN/index.html) to YS. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.White JA, Heasman J. Maternal control of pattern formation in Xenopus laevis. J Exp Zool Part B. 2008;310b(1):73–84. [DOI] [PubMed] [Google Scholar]
- 2.Ma J, He F, Xie G, Deng WM. Maternal AP determinants in the Drosophila oocyte and embryo. Wiley Interdiscip Rev Dev Biol. 2016;5(5):562–581. doi: 10.1002/wdev.235 [DOI] [PubMed] [Google Scholar]
- 3.Stein DS, Stevens LM. Maternal control of the Drosophila dorsal-ventral body axis. Wires Dev Biol. 2014;3(5):301–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yamada A, Nishida H. Distribution of cytoplasmic determinants in unfertilized eggs of the ascidian Halocynthia roretzi. Development Genes and Evolution. 1996;206(5):297–304. doi: 10.1007/s004270050056 [DOI] [PubMed] [Google Scholar]
- 5.Whittaker JR, Ortolani G, Farinella-Ferruzza N. Autonomy of acetylcholinesterase differentiation in muscle lineage cells of ascidian embryos. Dev Biol. 1977;55(1):196–200. [DOI] [PubMed] [Google Scholar]
- 6.Whittaker JR. Determination of Alkaline-Phosphatase Expression in Endodermal Cell Lineages of an Ascidian Embryo. Biol Bull. 1990;178(3):222–230. [DOI] [PubMed] [Google Scholar]
- 7.Makabe KW, Fujiwara S, Nishida H, Satoh N. Failure of Muscle Myosin Heavy-Chain Gene-Expression in Quarter Ascidian Embryos Developed from the Secondary Muscle Lineage Cells. Zool Sci. 1992;9(3):569–573. [Google Scholar]
- 8.Nishikata T, Satoh N. Specification of notochord cells in the ascidian embryo analysed with a specific monoclonal antibody. Cell Differ Dev. 1990;30(1):43–53. [DOI] [PubMed] [Google Scholar]
- 9.Nishikata T, Mita-Miyazawa I, Deno T, Takamura K, Satoh N. Expression of epidermis-specific antigens during embryogenesis of the ascidian, Halocynthia roretzi. Dev Biol. 1987;121(2):408–416. [DOI] [PubMed] [Google Scholar]
- 10.Imai K, Takada N, Satoh N, Satou Y. β-catenin mediates the specification of endoderm cells in ascidian embryos. Development. 2000;127(14):3009–3020. [DOI] [PubMed] [Google Scholar]
- 11.Bertrand V, Hudson C, Caillol D, Popovici C, Lemaire P. Neural tissue in ascidian embryos is induced by FGF9/16/20, acting via a combination of maternal GATA and Ets transcription factors. Cell. 2003;115(5):615–627. [DOI] [PubMed] [Google Scholar]
- 12.Rothbächer U, Bertrand V, Lamy C, Lemaire P. A combinatorial code of maternal GATA, Ets and β-catenin-TCF transcription factors specifies and patterns the early ascidian ectoderm. Development. 2007;134(22):4023–4032. doi: 10.1242/dev.010850 [DOI] [PubMed] [Google Scholar]
- 13.Horikawa Y, Matsumoto H, Yamaguchi F, Ishida S, Fujiwara S. Transcriptional regulation in the early ectodermal lineage of ascidian embryos. Dev Growth Differ. 2013;55(9):776–785. doi: 10.1111/dgd.12100 [DOI] [PubMed] [Google Scholar]
- 14.Imai KS, Satoh N, Satou Y. An essential role of a FoxD gene in notochord induction in Ciona embryos. Development. 2002;129(14):3441–3453. [DOI] [PubMed] [Google Scholar]
- 15.Oda-Ishii I, Kubo A, Kari W, Suzuki N, Rothbacher U, Satou Y. A Maternal System Initiating the Zygotic Developmental Program through Combinatorial Repression in the Ascidian Embryo. PLoS genetics. 2016;12(5):e1006045 doi: 10.1371/journal.pgen.1006045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hudson C, Sirour C, Yasuo H. Co-expression of Foxa.a, Foxd and Fgf9/16/20 defines a transient mesendoderm regulatory state in ascidian embryos. Elife. 2016;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Imai K, Satoh N, Satou Y. Early embryonic expression of FGF4/6/9 gene and its role in the induction of mesenchyme and notochord in Ciona savignyi embryos. Development. 2002;129(7):1729–1738. [DOI] [PubMed] [Google Scholar]
- 18.Satou Y, Imai KS, Satoh N. Early embryonic expression of a LIM-homeobox gene Cs-lhx3 is downstream of beta-catenin and responsible for the endoderm differentiation in Ciona savignyi embryos. Development. 2001;128(18):3559–3570. [DOI] [PubMed] [Google Scholar]
- 19.Imai KS, Levine M, Satoh N, Satou Y. Regulatory blueprint for a chordate embryo. Science. 2006;312(5777):1183–1187. doi: 10.1126/science.1123404 [DOI] [PubMed] [Google Scholar]
- 20.Hudson C, Lemaire P. Induction of anterior neural fates in the ascidian Ciona intestinalis. Mech Dev. 2001;100(2):189–203. [DOI] [PubMed] [Google Scholar]
- 21.Ohta N, Satou Y. Multiple signaling pathways coordinate to induce a threshold response in a chordate embryo. PLoS genetics. 2013;9(10):e1003818 doi: 10.1371/journal.pgen.1003818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ikeda T, Satou Y. Differential temporal control of Foxa.a and Zic-r.b specifies brain versus notochord fate in the ascidian embryo. Development. 2017;144(1):38–43. doi: 10.1242/dev.142174 [DOI] [PubMed] [Google Scholar]
- 23.Imai KS, Hikawa H, Kobayashi K, Satou Y. Tfap2 and Sox1/2/3 cooperatively specify ectodermal fates in ascidian embryos. Development. 2017;144(1):33–37. doi: 10.1242/dev.142109 [DOI] [PubMed] [Google Scholar]
- 24.Tresser J, Chiba S, Veeman M, El-Nachef D, Newman-Smith E, Horie T, et al. doublesex/mab3 related-1 (dmrt1) is essential for development of anterior neural plate derivatives in Ciona. Development. 2010;137(13):2197–2203. doi: 10.1242/dev.045302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Imai KS, Hino K, Yagi K, Satoh N, Satou Y. Gene expression profiles of transcription factors and signaling molecules in the ascidian embryo: towards a comprehensive understanding of gene networks. Development. 2004;131(16):4047–4058. doi: 10.1242/dev.01270 [DOI] [PubMed] [Google Scholar]
- 26.Tarazona S, Garcia-Alcalde F, Dopazo J, Ferrer A, Conesa A. Differential expression in RNA-seq: A matter of depth. Genome Res. 2011;21(12):2213–2223. doi: 10.1101/gr.124321.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Imai KS, Satou Y, Satoh N. Multiple functions of a Zic-like gene in the differentiation of notochord, central nervous system and muscle in Ciona savignyi embryos. Development. 2002;129(11):2723–2732. [DOI] [PubMed] [Google Scholar]
- 28.Yasuo H, Satoh N. Function of vertebrate T gene. Nature. 1993;364(6438):582–583. [DOI] [PubMed] [Google Scholar]
- 29.Ikeda T, Matsuoka T, Satou Y. A time delay gene circuit is required for palp formation in the ascidian embryo. Development. 2013;140(23):4703–4708. doi: 10.1242/dev.100339 [DOI] [PubMed] [Google Scholar]
- 30.Wagner E, Levine M. FGF signaling establishes the anterior border of the Ciona neural tube. Development. 2012;139(13):2351–2359. doi: 10.1242/dev.078485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hertz GZ, Stormo GD. Identifying DNA and protein patterns with statistically significant alignments of multiple sequences. Bioinformatics. 1999;15(7–8):563–577. [DOI] [PubMed] [Google Scholar]
- 32.Jolma A, Yan J, Whitington T, Toivonen J, Nitta KR, Rastas P, et al. DNA-Binding Specificities of Human Transcription Factors. Cell. 2013;152(1–2):327–339. doi: 10.1016/j.cell.2012.12.009 [DOI] [PubMed] [Google Scholar]
- 33.Lamy C, Rothbacher U, Caillol D, Lemaire P. Ci-FoxA-a is the earliest zygotic determinant of the ascidian anterior ectoderm and directly activates Ci-sFRP1/5. Development. 2006;133(15):2835–2844. doi: 10.1242/dev.02448 [DOI] [PubMed] [Google Scholar]
- 34.Shimauchi Y, Chiba S, Satoh N. Synergistic action of HNF-3 and Brachyury in the notochord differentiation of ascidian embryos. Int J Dev Biol. 2001;45(4):643–652. [PubMed] [Google Scholar]
- 35.Oda-Ishii I, Bertrand V, Matsuo I, Lemaire P, Saiga H. Making very similar embryos with divergent genomes: conservation of regulatory mechanisms of Otx between the ascidians Halocynthia roretzi and Ciona intestinalis. Development. 2005;132(7):1663–1674. doi: 10.1242/dev.01707 [DOI] [PubMed] [Google Scholar]
- 36.Kubo A, Suzuki N, Yuan X, Nakai K, Satoh N, Imai KS, et al. Genomic cis-regulatory networks in the early Ciona intestinalis embryo. Development. 2010;137(10):1613–1623. doi: 10.1242/dev.046789 [DOI] [PubMed] [Google Scholar]
- 37.Anno C, Satou A, Fujiwara S. Transcriptional regulation of ZicL in the Ciona intestinalis embryo. Dev Genes Evol. 2006;216(10):597–605. doi: 10.1007/s00427-006-0080-9 [DOI] [PubMed] [Google Scholar]
- 38.Davidson EH. Emerging properties of animal gene regulatory networks. Nature. 2010;468(7326):911–920. doi: 10.1038/nature09645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Neilson KM, Klein SL, Mhaske P, Mood K, Daar IO, Moody SA. Specific domains of FoxD4/5 activate and repress neural transcription factor genes to control the progression of immature neural ectoderm to differentiating neural plate. Dev Biol. 2012;365(2):363–375. doi: 10.1016/j.ydbio.2012.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Satou Y, Kawashima T, Shoguchi E, Nakayama A, Satoh N. An integrated database of the ascidian, Ciona intestinalis: Towards functional genomics. Zool Sci. 2005;22(8):837–843. doi: 10.2108/zsj.22.837 [DOI] [PubMed] [Google Scholar]
- 41.Stolfi A, Sasakura Y, Chalopin D, Satou Y, Christiaen L, Dantec C, et al. Guidelines for the nomenclature of genetic elements in tunicate genomes. Genesis. 2015;53(1):1–14. doi: 10.1002/dvg.22822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lemaire P, Garrett N, Gurdon JB. Expression cloning of Siamois, a Xenopus homeobox gene expressed in dorsal-vegetal cells of blastulae and able to induce a complete secondary axis. Cell. 1995;81(1):85–94. [DOI] [PubMed] [Google Scholar]
- 43.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38(4):576–589. doi: 10.1016/j.molcel.2010.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, et al. Model-based Analysis of ChIP-Seq (MACS). Genome Biology. 2008;9(9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hudson C, Darras S, Caillol D, Yasuo H, Lemaire P. A conserved role for the MEK signalling pathway in neural tissue specification and posteriorisation in the invertebrate chordate, the ascidian Ciona intestinalis. Development. 2003;130(1):147–159. [DOI] [PubMed] [Google Scholar]
- 46.Ohta N, Waki K, Mochizuki A, Satou Y. A Boolean Function for Neural Induction Reveals a Critical Role of Direct Intercellular Interactions in Patterning the Ectoderm of the Ascidian Embryo. PLoS Comput Biol. 2015;11(12):e1004687 doi: 10.1371/journal.pcbi.1004687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Notredame C, Higgins DG, Heringa J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J Mol Biol. 2000;302(1):205–217. doi: 10.1006/jmbi.2000.4042 [DOI] [PubMed] [Google Scholar]
- 48.Schneider TD, Stephens RM. Sequence Logos—a New Way to Display Consensus Sequences. Nucleic Acids Research. 1990;18(20):6097–6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A, B) The expression of Foxd revealed by in situ hybridization in (A) control and (B) Foxd morphants at the 64-cell stage. (C) The amount of endogenous Foxd mRNA was measured by RT-qPCR in uninjected control embryos and embryos injected with 2.3 pg of Foxd mRNA. The relative amount of mRNA in the experimental embryos compared with control embryos is shown. A maternal mRNA, Pou2, was used as an endogenous control. Error bars indicate mean±s.d. between two technical duplicates. The results of two independent experiments are shown in different colors. (D–G) The expression of (D, E) Fgf9/16/20, and (F, G) Tfap2-r.b revealed by in situ hybridization in (D, F) control unperturbed embryos and (E, G) Foxd morphant embryos at the 16-cell (D, E) and 32-cell (F, G) stages. Note that Foxd expression was not downregulated in the vegetal hemisphere of Foxd morphants (A, B) and that the expression of Fgf9/16/20 and Tfap2-r.b was not changed (D–G), although Fgf9/16/20 expression is downregulated in later embryos (Fig 2) [19]. The number of morphant embryos examined and the proportion of embryos that each panel represents are shown within the panels. Scale bar, 100 μm.
(TIF)
The numbers indicate the relative nucleotide positions from the transcriptional start site. The sequence of the fragment used for the gel-shift assay in Fig 6 is underlined, and the putative Foxd site is indicated in cyan. The mutation introduced in the Foxd site is shown in magenta.
(TIF)
(A) Asterisks indicate conserved nucleotides. T-Coffee [47] was used for generating this alignment. Putative Foxd binding sites, which were identified by Patser [31], are shown in cyan with scores. (B) A consensus sequence for human FOXD2 [32], which was used for identifying the putative Foxd binding sites, is shown as a sequence logo [48].
(TIF)
(A) Nucleotide sequences of the Foxd-binding regions in the upstream regions of Dmrt1, Prdm1-r.a, Prdm1-r.b, Otx, and Dlx.b, which were identified by the chromatin-immunoprecipitation and are shown in pink boxes in Fig 7. Putative Foxd-binding sites are shown in magenta. (B) An alignment of the putative Foxd binding sites found in (A) and those in the Zic-r.b (ZicL) upstream region identified previously [37]. Scores on the right were calculated by the Patser program and a position weight matrix for human FOXD2 binding sites [32], which is represented in S3B Fig.
(TIF)
Because Tfap2-r.b was not regulated by Foxd (S1F and S1G Fig), the upstream region of Tfap2-r.b is shown as a negative control for genes shown in Fig 7. Significant peaks were not identified by the computer programs in this genomic region. The graphs include data of two biological duplicates.
(TIF)
Conserved and similar amino acids are shown by black and gray boxes, respectively. The forkhead domains, Eh-1 domains, and putative acidic domains are enclosed by black lines, and acidic amino acids in the putative acidic region of the N-terminal half are shown in magenta.
(TIF)
(DOCX)
Data Availability Statement
All RNA sequencing and ChIP-seq data files are available from the SRA database (accession numbers DRA005206 and DRA005285).