Abstract
The innate immune system is a critical component of host defence against microbial pathogens, but effective responses require an ability to distinguish between infectious and non-infectious insult to prevent inappropriate inflammation. Using the important obligate intracellular human pathogen Chlamydia trachomatis; an organism that causes significant immunopathology, we sought to determine critical host and pathogen factors that contribute to the induction of inflammasome activation. We assayed inflammasome activation by immunoblotting and ELISA to detect IL-1β processing and LDH release to determine pyroptosis. Using primary murine bone marrow derived macrophages or human monocyte derived dendritic cells, infected with live or attenuated Chlamydia trachomatis we report that the live organism activates both canonical and non-canonical inflammasomes, but only canonical inflammasomes controlled IL-1β processing which preceded pyroptosis. NADPH oxidase deficient macrophages were permissive to Chlamydia trachomatis replication and displayed elevated type-1 interferon and inflammasome activation. Conversely, attenuated, non-replicating Chlamydia trachomatis, primed but did not activate inflammasomes and stimulated reduced type-1 interferon responses. This suggested bacterial replication or metabolism as important factors that determine interferon responses and inflammasome activation. We identified STING but not cGAS as a central mediator of interferon regulated inflammasome activation. Interestingly, exogenous delivery of a Chlamydia trachomatis metabolite and STING ligand—cyclic di-AMP, recovered inflammasome activation to attenuated bacteria in a STING dependent manner thus indicating that a bacterial metabolite is a key factor initiating inflammasome activation through STING, independent of cGAS. These data suggest a potential mechanism of how the innate immune system can distinguish between infectious and non-infectious insult and instigate appropriate immune responses that could be therapeutically targeted.
Author summary
Innate responses to bacterial infection such as Chlamydia trachomatis activate inflammasomes to enable the processing of IL-1β, IL-18 and the induction of an inflammatory form of cell death termed pyroptosis. Inflammasomes are crucial to host defence but require tight regulation in order to prevent inappropriate inflammation and immunopathology. Here, we demonstrate that the pro-inflammatory potential of an attenuated strain of Chlamydia trachomatis, that fails to activate the inflammasome, can be rescued by the addition of a bacterial metabolite. The requirement for this metabolite, highlights a novel mechanism of inflammasome regulation and reveals a crucial role for STING mediated interferon signalling independent of cGAS. These findings further our understanding of how the innate immune system can differentiate between potential infectious and non-infectious threats and mount appropriate immune responses.
Introduction
The obligate intracellular pathogen Chlamydia trachomatis is a major cause of infectious disease world-wide and can initiate inflammatory pathology such as pelvic inflammatory disease, reactive arthritis and infectious blindness (trachoma). Significantly, murine models of Chlamydia infection demonstrate that host inflammatory mediators, particularly the inflammatory cytokine IL-1β, type-1 interferons, caspase-1 and caspase-11 account for a significant proportion of infection associated pathology [1–3].
Inflammasomes are molecular scaffolds that facilitate the activation of inflammatory caspases resulting in the proteolytic processing of the cytokines IL-1β and IL-18 in addition to the induction of a form of programmed necrosis termed pyroptosis [4, 5]. Recently, a non-canonical inflammasome requiring caspase-11 (caspase-4/5 in humans) has been identified that responds specifically to LPS contamination of the cytosol, independent of TLR4 [6–9]. Uniquely, activation of the non-canonical inflammasome does not require an upstream sensor that is required for canonical caspase-1 activation, and occurs as a consequence of LPS being directly recognised by Caspase-11 (or Caspase-4/5) [10] and is critically dependent on the acylation status of the lipid-A moiety [9]. The non-canonical inflammasome is essential for pyroptosis and IL-1β maturation in response to infection with certain gram-negative bacterial pathogens, or the delivery of cytoplasmic LPS, and occurs as a consequence of the caspase-11 dependent cleavage of gasdermin-D (GSDMD) [11, 12] and pannexin-1 [13]. Activation of the non-canonical inflammasome in response to bacterial pathogens can occur as a direct result of cytosolic invasion by bacteria that occurs during infection with Burkolderia thailandensis, resulting in a rapid execution of caspase-11 mediated pyroptosis, essential to host defence [14]. Alternatively, non-canonical inflammasome activation can occur as a consequence of LPS being released from bacterial pathogen containing vacuoles (PV), via the action of Immune Related GTPases (IRG’s) and Guanylate Binding Proteins (GBP’s) [15].
Despite type-1 interferons being reported to inhibit canonical inflammasome activation [16], non-canonical inflammasomes require interferon signalling for up-regulation of caspase-11 [17], GBP and IRG expression [18].
Mechanisms controlling inflammasome activation in response to Chlamydia sp infection are still poorly understood, although NLRP3, reactive oxygen species, mitochondrial damage and oxidised mitochondrial DNA have been implicated in the process [19, 20] [21, 22]. In addition, recent work using interferon-primed macrophages has demonstrated that GBP’s, NLRP3 and AIM2 participate in IL-18 maturation in response to Chlamydia trachomatis infection, while cell death occurs via either caspase-1 or caspase-11 activity, suggesting engagement of the non-canonical inflammasome [23]. Importantly, Chlamydia muridarum infection of macrophages induces type-1 interferon expression via Stimulator of Interferon Gene (STING) activity, implying that Chlamydia sp infections of macrophages are ‘self-priming’ and exogenous interferon is not necessary [24]. However, mechanisms controlling this process are poorly understood.
Here, we report that inflammasome activation in un-primed bone marrow derived macrophages (BMDM) required replication or metabolic activity of Chlamydia trachomatis. We also provide evidence that production of a chlamydial metabolite, but not detection of host or microbial DNA by cGAS, activated STING initiating autocrine, type-1 interferon signalling leading to canonical and non-canonical inflammasome activation. We propose that intracellular Chlamydia trachomatis replication or metabolism, and the subsequent detection of cyclic di-AMP by STING, are key events initiating inflammasome activation in response to Chlamydia trachomatis infection.
Results
Cell death and IL-1β maturation in response to C. trachomatis infection occur asynchronously and are regulated through different inflammasomes
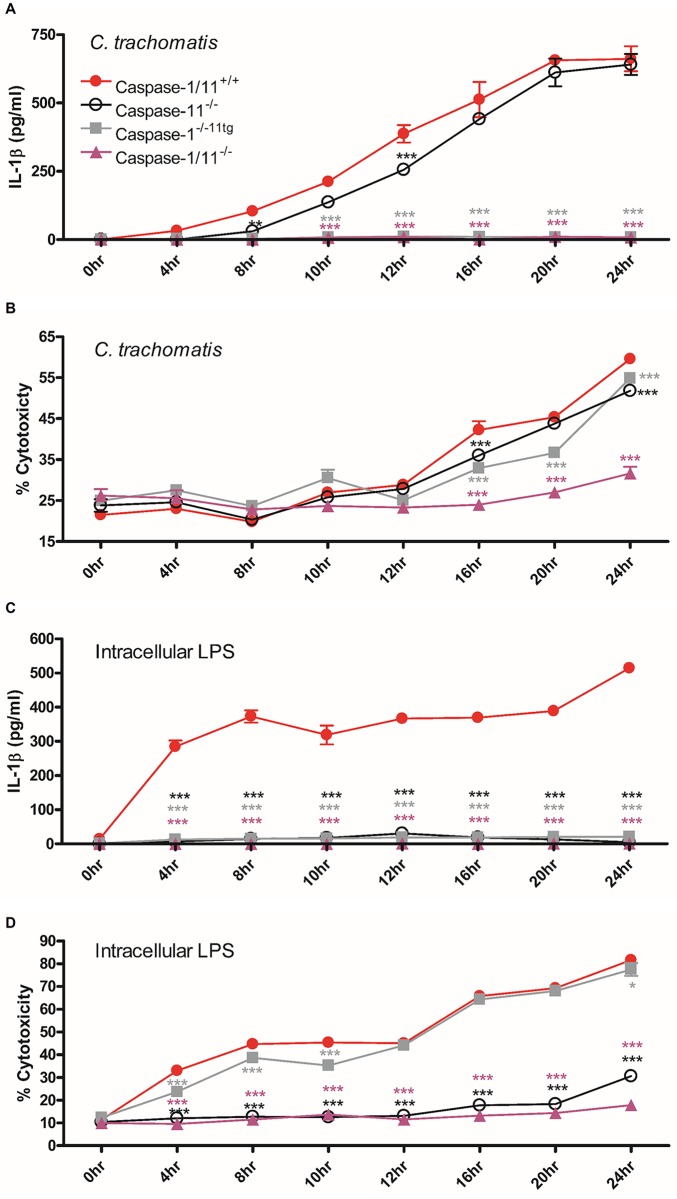
We sought to identify whether canonical and non-canonical inflammasomes regulate cytokine maturation and cell death in response to C. trachomatis infection of un-primed BMDM using cytosolic delivery of LPS as a control for non-canonical inflammasome activation. A time course infection of BMDM with C. trachomatis resulted in significant increases in IL-1β secretion as early as 8–10 hours post infection in wild-type and caspase-11 deficient but not caspase-1 deficient BMDM indicating that the non-canonical inflammasome was not required for IL-1β maturation in response to C. trachomatis infection (Fig 1A). We next analysed LDH release as an indicator of lytic cell death, associated with pyroptosis and necrosis over the same time course of C. trachomatis infection (Fig 1B). As expected C. trachomatis infection induced significant cytotoxicity in wild type BMDM but surprisingly, this was delayed compared to IL-1β release and individual deletion of either caspase-11 or caspase-1 did not confer resistance to cell death suggesting a potential non-pyroptotic cell death mechanism. However, deletion of both caspase-1 and caspase-11 resulted in a dramatic protection of cell death indicating that both inflammatory caspases contribute to the process and the form of cell death is therefore likely to be classical pyroptosis as both caspase-1 and caspase-11 have been demonstrated to cleave and activate the pore forming protein gasdermin D in order to initiate pyroptotic cell death [11, 12]. Intriguingly, despite the ability of both caspase-1 and caspase-11 to induce cell death in response to C. trachomatis infection, death appeared to be delayed in the caspase-1 ko / caspase-11 transgenic cells, particularly at the 20-hour time point. This suggests that caspase-1 may induce early cell death while caspase-11 contributes later. This is an attractive answer to the conundrum of why caspase-11 does not affect IL-1β but contributes to cell death. Late activity of caspase-11 may not be required for the NLRP3 dependent activation of caspase-1 that controls cytokine maturation, as this may have already occurred through canonical inflammasome activation. We confirmed the functional status of the caspase deficient cells by specific activation of the non-canonical inflammasome using cytosolic delivery of LPS. As expected, intracellular LPS induced IL-1β secretion was dependent on both capase-1 and caspase-11 (Fig 1C) while cell death was only dependent on caspase-11 (Fig 1D) as previously reported [6, 8, 9].
Fig 1. IL-1β maturation is independent of caspase-11 and precedes pyroptosis which is regulated by both canonical and non-canonical inflammasomes in response to C. trachomatis infection.
Time course of IL-1β secretion analysed by ELISA or cell death measured by LDH release in supernatants from C. trachomatis infected (A+B) or LPS transfected (C+D) BMDM from wild-type (Caspase-1/11+/+), caspase-11 knock-out (Caspase-11-/-), caspase-1 knock-out caspase-11 transgenic (Caspase-1-/-11tg) or caspase-1/caspase-11 knock-out (Caspase-1/11-/-). Data represent the mean values from BMDM obtained from individual mice, performed in biological triplicate and are representative of 2-similar experiments. Error bars indicate ±SEM, *p = <0.05, **p = <0.01 and ***p = <0.001 vs Caspase-1/11+/+.
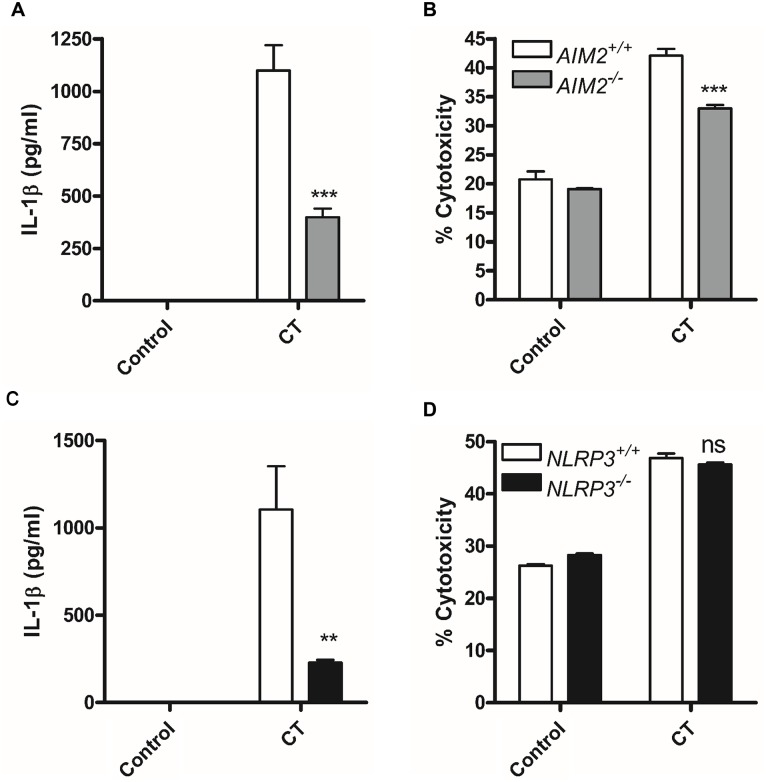
We next examined which canonical intracellular sensor activated caspase-1 in response to C. trachomatis infection. Both NLRP3 [19, 20] and AIM2 [23] are reported to be activated in response to C. trachomatis infection enabling caspase-1 activation and the processing of IL-1β. We therefore confirmed the requirement for both inflammasomes in our study (Fig 2A, 2B, 2C and 2D). Both NLRP3 and AIM2 deficient macrophages displayed reduced IL-1β responses to C. trachomatis infection as expected (Fig 2A and 2C). However, the absence of AIM2 but not NLRP3 had a small but significant effect on cell death in response to infection potentially accounting for the small reduction in cell death we observed with caspase-1 deficiency alone, but also reinforcing our findings that in contrast to IL-1β release, cell death was not solely dependent on caspase-1 activity (Fig 2B and 2D).
Fig 2. AIM2 and NLRP3 are required for canonical caspase-1 activation in response to C. trachomatis infection.
IL-1β release analysed by ELISA and cell death measured by LDH release following C. trachomatis infection of wild type (AIM2+/+ or NLRP3+/+) or AIM2 deficient (AIM2-/-) (A + B) or NLRP3 deficient (NLRP3-/-) (C + D) BMDM for 24 hours. Data represent the mean values from BMDM obtained from individual mice, performed in biological triplicate. Error bars indicate ±SEM, *p = <0.05, **p = <0.01 and ***p = <0.001, ns indicates no significant difference.
Mitochondrial dysfunction is known to contribute to inflammasome activation by sterile agonists [25] and Chlamydia sp infection [21, 22]. We therefore investigated whether canonical activation of caspase-1 resulting in IL-1β processing was inhibited by the presence of a mitochondrial anti-oxidant (Mito-Q) (S1A, S1B and S1C Fig). Mito-Q, in a pattern similar to NLRP3 deficiency, inhibited C. trachomatis induced IL-1β release, but had no effect on cell death, most likely due to the overriding effect of caspase-11 and further supporting our findings that C. trachomatis induced death could be regulated by both caspase-1 and caspase-11. Importantly, MitoQ did not block pro IL-1β upregulation by C. trachomatis indicating that MitoQ was blocking caspase-1 mediated IL-1β processing and not priming (S1B Fig). Infection of macrophages with Brucella abortus activates the NLRP3 inflammasome in a mechanism requiring IRE-1 mediated ER stress, mitochondrial damage and caspase-2 activity [26]. We have previously demonstrated that C. trachomatis infection of human dendritic cells results in IRE1α activation [27]. We therefore examined if IRE1α was relevant to C. trachomatis induced inflammasome activation by utilising IRE-1 deficient macrophages or the IRE-1 inhibitor; 4μ8c. IRE-1 deficient BMDM or BMDM pre-treated with 4μ8c demonstrated normal IL-1β release in response to C. trachomatis infection (S1D and S1E Fig) indicating that ER stress driven inflammasome activation was not relevant to C. trachomatis infection and the mitochondrial component differs from that of B. abortus. Furthermore, our data suggest that canonical activation of caspase-1 through release of DNA (either host or pathogen derived) in to the cytosol activating AIM2 is amplified by mitochondrial ROS to activate NLRP3 and induce maximal inflammasome responses to C. trachomatis. Both NLRP3 and AIM2 share the requirement for the adaptor ASC to provide a scaffold for the recruitment of caspase-1. The fact that deletion of either AIM2 or NLRP3 only partially attenuated inflammasome responses and that only one ASC speck is formed per cell suggests that maximal inflammasome response to C. trachomatis requires the co occupation of the ASC filament by both receptors. However, it remains unknown whether recruitment of both receptors occurs simultaneously to initiate the aggregation of the ASC complex resulting in an active inflammasome during infection.
Attenuated C. trachomatis induces inflammasome priming but not activation
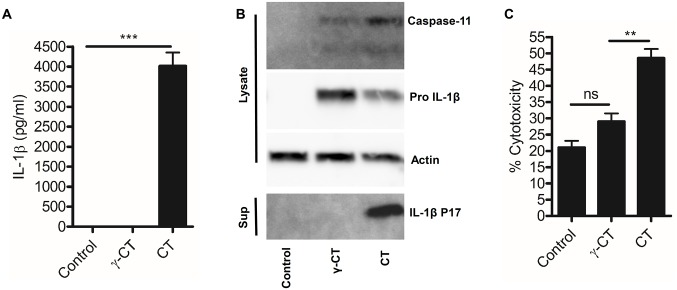
We next investigated whether inflammasome responses to C. trachomatis infection required factors produced by the live-replicating organism. We attenuated C. trachomatis using gamma-irradiation or used a Chlamydia Protease-like Activity Factor (CPAF) deficient mutant strain that exhibited delayed growth kinetics in Hela cells [28]. Attenuated C. trachomatis (both gamma-irradiated and the CPAF mutant) failed to induce IL-1β secretion and cell death (Fig 3A and 3C and S2 Fig) but still induced pro IL-1β expression indicating that the attenuated C. trachomatis particles were not biologically inert and pathogen recognition receptor (PRR) and NFκB responses were intact, but an inflammasome activatory signal was absent (Fig 3B). Importantly, we confirmed that lack of inflammasome activation was not due to reduced internalisation of attenuated C. trachomatis particles as intracellular staining demonstrated equivalent levels to live C. trachomatis (S3 Fig) to the attenuated forms. We also noted that although pro IL-1β priming was normal in response to attenuated C. trachomatis, expression of caspase-11 was markedly reduced in cells stimulated with irradiated but not CPAF deficient C. trachomatis. This indicated that replication or metabolism of C. trachomatis intracellularly, provided additional, essential signals required for inflammasome activation and caspase-11 expression in addition to CPAF function.
Fig 3. Attenuated C. trachomatis can prime but not activate inflammasomes.
(A) IL-1β secretion analysed by ELISA of supernatants from wild-type BMDM stimulated with attenuated, gamma-irradiated C. trachomatis particles (γ-CT) or infection with live C. trachomatis (CT) for 24hrs. Data represent the mean from BMDM obtained from three-individual mice, error bars indicate ±SEM, ***p = <0.001. (B) Caspase-11 expression, IL-1β maturation and inflammasome priming, analysed by western blot of cell lysates and supernatants from wild-type BMDM stimulated with attenuated, gamma-irradiated C. trachomatis particles (γ-CT) or infection with live C. trachomatis (CT) for 24hrs. (C) Cell death analysed by LDH release in wild-type BMDM in response to stimulation with attenuated, gamma-irradiated C. trachomatis particles (γ-CT) or infection with live C. trachomatis (CT) for 24hrs. Data represent the mean from BMDM obtained from three-individual mice, error bars indicate ±SEM. **p = <0.01, ***p = <0.001 ns indicates no significant statistical difference.
NADPH oxidase regulates C. trachomatis replication within BMDM and human mDC
Our experiments utilising attenuated C. trachomatis indicated that intracellular replication could be contributing to signals that activate the inflammasome. We therefore wished to identify macrophages that were permissive to C. trachomatis replication to investigate this hypothesis further.
The NADPH oxidase system is a key component of host defence against microbial pathogens via the generation of anti-microbial reactive oxygen species (ROS). Individuals with inherited, disabling mutations of genes that contribute to the NADPH oxidase system, develop a primary immunodeficiency termed chronic granulomatous disease (CGD) that is characterised by recurrent bacterial and fungal infections [29]. Generation of ROS by NADPH oxidase contributes to the anti-microbial action of macrophages in response to infection with bacteria such as Salmonella [30]. Previous studies using chemical inhibition of NADPH oxidase in Hela cells suggested that ROS amplify C. trachomatis replication through a caspase-1 mediated mechanism [31]. However, the role of NADPH oxidase in the control of C. trachomatis replication within primary macrophages is unknown.
We therefore investigated Chlamydia trachomatis growth in murine macrophages or human monocyte derived dendritic cells obtained from CGD patients deficient in gp91 Phox (Cybb); a critical component of the NADPH oxidase system in phagocytic cells.
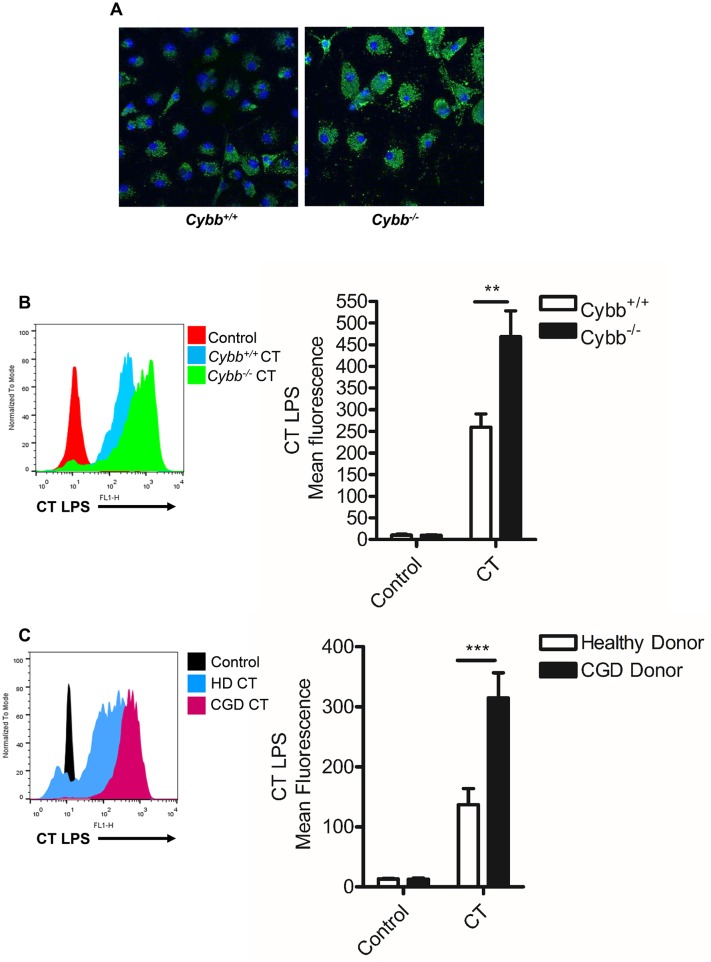
Intracellular staining of C. trachomatis LPS and analysis of Chlamydia 16s RNA expression was employed to determine the bacterial burden within infected cells Significantly, infection of macrophages deficient in gp91 phox exhibited a marked increase in the burden of C. trachomatis compared to wild-type controls (Fig 4A). The intracellular staining of Chlamydia LPS in murine macrophages (Fig 4B) or human dendritic cells (Fig 4C) was also quantified using FACS and further confirmed by analysis of Chlamydia 16s RNA in BMDM (S4 Fig), a technique employed by others to analyse Chlamydia replication [31]. Both murine and human cells deficient in gp91 phox displayed a significant increase in intracellular C. trachomatis burden compared to control (gp91 phox sufficient) cells, indicating that NADPH oxidase activity was a key regulator of C. trachomatis replication and survival within myeloid cells in both mice and humans.
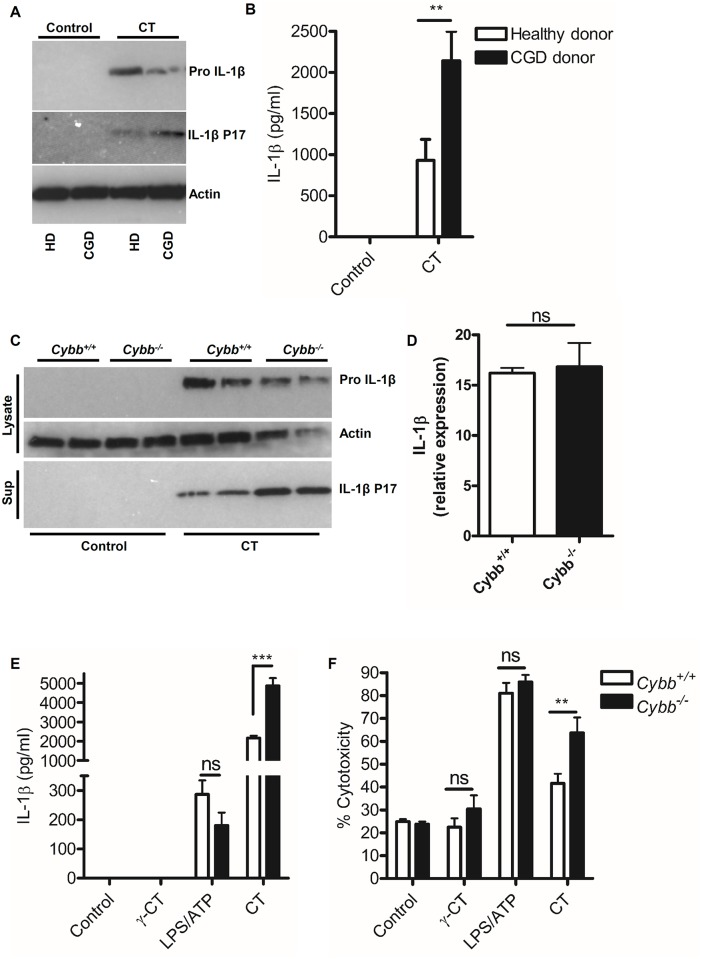
Fig 4. NADPH oxidase controls C. trachomatis intracellular replication in BMDM and mDC.
(A) Assessment of intracellular C. trachomatis burden at 24hrs post-infection in wild-type (Cybb+/+) and NADPH oxidase deficient (Cybb-/-) BMDM by confocal fluorescent microscopy of Chlamydia LPS (green) and nuclei counter-stain with DAPI (blue). (B) Representative FACS histogram plot of C. trachomatis LPS staining 24hrs post-infection in wild-type (Cybb+/+) or NADPH oxidase deficient (Cybb-/-) BMDM (left panel) and Quantification of geo-mean fluorescence of Chlamydia LPS staining in wild-type (Cybb+/+) or NADPH oxidase deficient (Cybb-/-) BMDM infected with C. trachomatis for 24hrs (right panel). Data represent the mean from BMDM obtained from three-individual mice, error bars indicate ±SEM **p = <0.01. (C) Representative FACS histogram plot of C. trachomatis LPS staining 24hrs post-infection in healthy-donor or NADPH oxidase deficient CGD patient mDC (left panel) and quantification of geo-mean fluorescence of Chlamydia LPS staining in healthy or CGD mDC infected with C. trachomatis for 24hrs (right panel). Data represent the mean from mDC obtained from four-individual donors from each group, error bars indicate ±SEM, ***p = <0.001.
NADPH oxidase deficient cells exhibit elevated inflammasome activation in response to live C. trachomatis infection but not sterile agonists
Given that we had observed an apparent requirement for C. trachomatis replication or metabolism in the induction of inflammasome activation, we tested the hypothesis that increased C. trachomatis replication observed in NADPH oxidase deficient cells, would correspond with increased inflammasome activation. C. trachomatis infection of dendritic cells from CGD patients (Fig 5A and 5B) or gp91 phox deficient murine macrophages (Cybb-/-) (Fig 5C, 5E and 5F) resulted in increased IL-1β maturation and pyroptosis indicating increased inflammasome activation. Furthermore, expression of IL-1β mRNA was equivalent in wild-type and gp91 phox deficient cells in response to infection indicating that the increased bacterial burden was having a direct effect on inflammasome activation, and was not just due to increased priming (Fig 5D). Crucially, inflammasome responses to sterile agonists such as LPS/ATP or gamma attenuated C. trachomatis, were not elevated in the absence of NADPH oxidase activity, indicating that the increased bacterial burden was the critical factor leading to inflammasome activation and that this was not simply a consequence of elevated background inflammasome activation in the absence of NADPH oxidase. These data also reinforced our hypothesis that aspects of C. trachomatis metabolism or replication were factors influencing inflammasome activation.
Fig 5. Increased C. trachomatis replication is associated with elevated inflammasome activation.
(A) IL-1β maturation in response to C. trachomatis infection, analysed by western blot of cell lysates in mDC obtained from healthy donors (HD) or CGD donors (CGD). Data representative of three-independent donors in each patient group. (B) IL-1β secretion analysed by ELISA from mDC obtained from healthy donors or CGD patient donors infected with C. trachomatis (CT) for 24hrs. Data represents the mean from mDC obtained from seven-donors from each group, error bars indicate ±SEM **p = <0.01. (C) IL-1β maturation in response to C. trachomatis infection, analysed by western blot of cell lysates and supernatants from wild-type (Cybb+/+) or NADPH oxidase deficient (Cybb-/-) BMDM. Data represents the response from BMDM obtained from two-individual mice. (D) IL-1β mRNA expression relative to HPRT analysed by qRT-PCR of RNA obtained from wild-type (Cybb+/+) or NADPH oxidase deficient (Cybb-/-) BMDM following infection with C. trachomatis for 6-hours. Data represent the mean from three-individual mice, error bars indicate ±SEM, ns indicates no significant difference. (E) IL-1β secretion analysed by ELISA or (F) Cell death analysed by LDH release from BMDM obtained from wild-type (Cybb+/+) or NADPH oxidase deficient (Cybb-/-) mice following stimulation with gamma-attenuated C. trachomatis (γ-CT), live C. trachomatis (CT) or LPS/ATP for 24hrs. Data represent the mean from 12-individual mice (CT) or six-individual mice (control, γ-CT and LPS/ATP), error bars indicate ±SEM. **p = <0.01, ***p = <0.001, ns indicates no significant difference.
Chlamydia trachomatis replication and metabolism contribute to type-1 interferon responses in macrophages
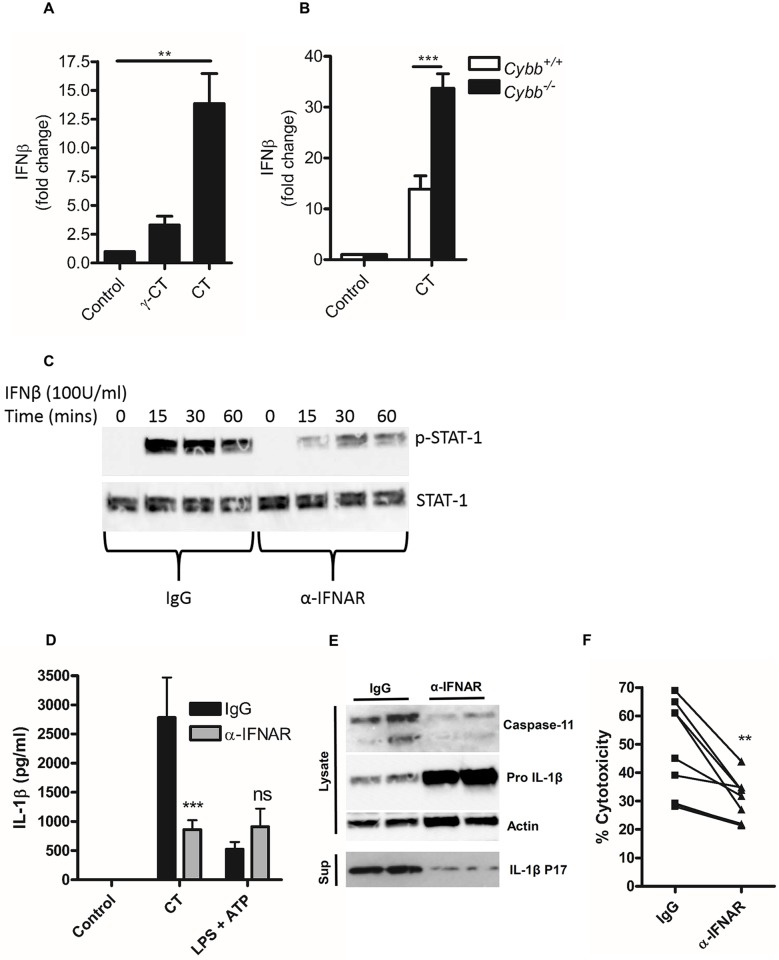
Recently, type-1 interferon signalling has been demonstrated to play an important role in host defence and inflammasome activation to bacterial pathogens [32] and is crucial for the expression of caspase-11 in response to a range of gram-negative bacteria [17]. We therefore investigated whether intracellular metabolism or replication of C. trachomatis within macrophages contributed to type-1 interferon expression. Significantly, gamma-irradiated attenuated C. trachomatis or the CPAF deficient mutant (S5 Fig) failed to induce significant interferon-β expression compared to non-attenuated bacteria (Fig 6A). Furthermore, gp91 phox deficient macrophages, harbouring increased bacterial burdens also displayed elevated interferon-β expression compared to wild-type controls (Fig 6B). These data indicated that type-1 interferon responses could be critical mediators of inflammasome activation in macrophages during C. trachomatis infection. We therefore tested the hypothesis that autocrine, type-1 interferon signalling through the type-1 interferon receptor, IFNAR, contributed to inflammasome activation during C. trachomatis infection. To do this, we infected macrophages with C. trachomatis in the presence of an IFNAR blocking antibody or isotype control. We confirmed the effectiveness of blocking IFNAR by examining STAT-1 phosphorylation in response to interferon-β stimulation of wild-type BMDM (Fig 6C). Blocking IFNAR during C. trachomatis infection resulted in reduced IL-1β secretion (Fig 6D), IL-1β maturation, caspase-11 expression (Fig 6E) and cell death (Fig 6F) but, as expected, did not significantly affect canonical inflammasome activation with LPS/ATP. Furthermore, the effect on IL-1β secretion was of reduced proteolytic processing and not priming of pro IL-1β, as blocking IFNAR during C. trachomatis infection actually resulted in increased pro IL-1β, presumably due to reduced maturation by caspase-1 and accumulation of the un-processed substrate and the inhibitory effect of interferon signalling on pro IL-1β synthesis [16] (Fig 6E). These data suggested that C. trachomatis replication or metabolism induced a type-1 interferon signature that controlled both canonical and non-canonical inflammasome activation.
Fig 6. Type-1 interferon signalling regulates C. trachomatis inflammasome activation.
(A) Induction of IFNβ mRNA expression in wild-type BMDM analysed by quantitative RT-PCR stimulated with gamma attenuated C. trachomatis (γ-CT) or live C. trachomatis (CT) for 8hrs. (B) Induction of IFNβ mRNA expression in wild-type BMDM (Cybb+/+) or NADPH oxidase deficient BMDM (Cybb-/-) in response to infection with live C. trachomatis for 8hrs. Data represent the mean from BMDM obtained from three-individual mice in each group, error bars indicate SEM **p = <0.01, ***p = <0.001. (C) STAT-1 phosphorylation analysed by western blot from BMDM stimulated with 100U/ml IFNβ for indicated times in the presence of isotype (IgG) or IFNAR blocking antibody (α-IFNAR). (D) IL-1β secretion analysed by ELISA of supernatants from wild-type BMDM infected with C. trachomatis (CT) or stimulated with LPS/ATP in the presence of an IFNAR blocking antibody (αIFNAR) or isotype control (isotype IgG) for 24hrs. Data represent the mean from BMDM obtained from nine-individual mice, error bars indicate ±SEM ***p = <0.001. (E) Caspase-11 expression and IL-1β maturation analysed by western blot of lysates and supernatants from wild-type BMDM in response to C. trachomatis infection in the presence of an IFNAR blocking antibody (αIFNAR) or isotype control (isotype IgG) for 24hrs. (F) Cell death analysed by LDH release from wild-type BMDM in response to C. trachomatis infection (CT) in the presence of an IFNAR blocking antibody (αIFNAR) or isotype control (isotype IgG) for 24hrs. Data represent the mean from BMDM obtained from nine-individual mice, error bars indicate ±SEM **p = <0.01.
STING regulates type-1 interferon expression and inflammasome activation in response to C. trachomatis infection
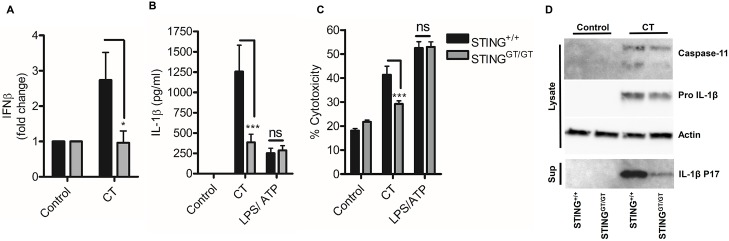
Stimulator of interferon gene (STING) is a critical mediator of type-1 interferon expression in response to Chlamydia sp infection [24, 33, 34]. Given that we have demonstrated a crucial role for autocrine type-1 interferon signalling in C. trachomatis induced inflammasome activation, we tested the hypothesis that STING was a central regulator of inflammasome activation. Infection of macrophages from STING deficient mice with C. trachomatis failed to induce interferon-β expression (Fig 7A) and exhibited reduced IL-1β secretion (Fig 7B), cell death (Fig 7C) and reduced IL-1β processing and caspase-11 expression (Fig 7D) but had equivalent responses to LPS/ATP stimulation. Thus, STING is a critical mediator of interferon dependent inflammasome activation in response to C. trachomatis infection of macrophages.
Fig 7. STING regulates interferon induced inflammasome activation in response to C. trachomatis infection.
(A) Induction of IFNβ mRNA in wild-type (STING+/+) or STING deficient (STINGGT/GT) BMDM infected with C. trachomatis (CT) for 8hrs. Data represent the mean of BMDM obtained from six-individual mice, error bars indicate SEM *p = <0.05. (B) IL-1β secretion analysed by ELISA of supernatants from wild-type (STING+/+) or STING deficient (STINGGT/GT) BMDM infected with C. trachomatis (CT) or stimulated with LPS /ATP for 24hrs. Data represent the mean of BMDM obtained from twelve-(CT) or six-(LPS/ATP) individual mice, error bars indicate SEM *p = <0.05, **p = <0.01, ***p = <0.001. (C) Cell death analysed by LDH release in wild-type (STING+/+) or STING deficient (STINGGT/GT) BMDM. Data represent the mean from BMDM obtained from six-(CT) or three-(LPS/ATP) individual mice, error bars indicate SEM *p = <0.05, **p = <0.01, ***p = <0.001. (D) Capase-11 expression and IL-1β maturation analysed by western blot of lysates and supernatants from wild-type (STING+/+) or STING deficient (STINGGT/GT) BMDM infected with C. trachomatis.
STING dependent canonical inflammasome responses to C. trachomatis infection are independent of cGAS
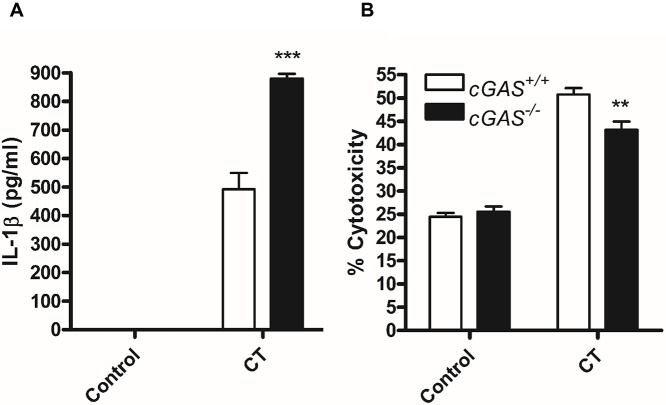
Activation of STING to induce type-1 interferon responses can occur through two distinct pathways: 1. the conversion of cytosolic DNA to cGAMP catalysed by cGAS which is then recognised by STING or 2. the direct recognition of cyclic di-nucleotides (cyclic di-AMP/GMP) produced by certain bacteria. Recently, cGAS was shown to be crucial for STING dependent inflammasome responses to Francisella tularensis infection of macrophages [35, 36]. We therefore analysed whether cGAS was required for STING mediated inflammasome activation in response to C. trachomatis infection using cGAS deficient BMDM. In contrast to inflammasome responses to F. tularensis, cGAS deficient BMDM produced elevated IL-1β in response to C. trachomatis infection indicating that STING dependent canonical caspase-1 activation through NLRP3 and AIM2 activation was independent of cGAS conversion of host or microbial DNA to cGAMP (Fig 8A). Surprisingly however, there was a small but significant protection from C. trachomatis induced pyroptosis in cGAS deficient cells (Fig 8B). Given that we demonstrate cell death responses are delayed compared to IL-1β release and death is governed by both canonical activation of caspase-1 and non-canonical caspase-11 activation, suggests that later release of microbial DNA to the cytosol may amplify STING responses that are not required for canonical inflammasome activation.
Fig 8. cGAS is not required for canonical inflammasome responses to C. trachomatis infection.
(A) IL-1β release measured by ELISA or (B) cell death measured by LDH release in supernatants from wild-type (cGAS+/+) or cGAS deficient (cGAS-/-) BMDM infected with C. trachomatis (CT) for 24hrs. Data represent the mean from BMDM obtained from three individual mice, error bars indicate ±SEM **p = <0.01, ***p = <0.001.
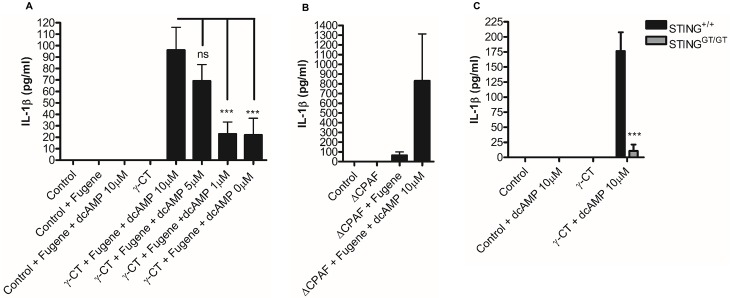
The bacterial metabolite cyclic di-AMP recovers STING dependent inflammasome responses to attenuated C. trachomatis
STING induced interferon responses during C. trachomatis infection, have been demonstrated to occur as a consequence of recognition of a metabolite; cyclic di-AMP, that is produced by metabolically active C. trachomatis [33]. Given that we identified STING as a critical regulator of canonical and non-canonical inflammasome responses to C. trachomatis infection and that cGAS was not required for canonical caspase-1 activation to induce IL-1β release in addition to finding a requirement for C. trachomatis metabolism or replication in the induction of interferon-β expression and subsequent inflammasome activation, we investigated whether cyclic di-AMP was a critical factor in STING mediated activation of the inflammasome. To test this hypothesis, we stimulated wild-type BMDM with attenuated C. trachomatis that failed to induce inflammasome activation and transfected the metabolite cyclic di-AMP in to the cells. Crucially, transfection of titrated amounts of cyclic di-AMP in to cells stimulated with attenuated gamma irradiated (Fig 9A) or CPAF deficient (Fig 9B) C. trachomatis resulted in partial recovery of IL-1β secretion that was dependent on STING (Fig 9C). These data provide an attractive explanation as to how STING mediated inflammasome responses could be regulated in the absence of a requirement for cGAS conversion of DNA to cGAMP during infection with C. trachomatis that requires a replicating or metabolically active organism.
Fig 9. Exogenous cyclic di-AMP recovers inflammasome activation in response to attenuated C. trachomatis.
(A) IL-1β secretion analysed by ELISA in wild type BMDM in response to stimulation with gamma attenuated C. trachomatis (γ-CT) transfected with cyclic di-AMP (dcAMP) at indicated concentrations. Data represent the mean of BMDM obtained from six to nine individual mice, error bars indicate ±SEM. (B) IL-1β secretion analysed by ELISA of supernatants from wild-type BMDM following infection with CPAF deficient C. trachomatis (ΔCPAF) transfected with 10μM cyclic di-AMP (dcAMP) for 24hrs. Data represented as the mean of one experiment performed on cells from three individual mice, error bars indicate ±SEM. (C) IL-1β secretion analysed by ELISA in wild-type (STING+/+) or STING deficient (STINGGT/GT) in response to stimulation with gamma attenuated C. trachomatis (γ-CT) transfected with cyclic di-AMP (dcAMP) (10μM). Data represent the mean of BMDM from three-individual mice, error bars indicate ±SEM *p = <0.05, **p = <0.01, ***p = <0.001.
Discussion
Innate host defence relies upon a repertoire of pathogen recognition systems that detect a diverse array of conserved microbial components in order to initiate inflammatory responses. Here, we provide evidence that inflammasome activation in response to Chlamydia trachomatis infection is dependent on detection of a microbial metabolite produced by the live organism. The inflammasome is an important component of innate host defence against bacterial pathogens including Chlamydia sp, but excessive or inappropriate activation contributes to infection associated immunopathology. Therefore, inflammasome activation must be tightly regulated. Our data demonstrate a novel mechanism of inflammasome regulation that allows the host to differentiate between live (a potential infectious threat) or attenuated Chlamydia trachomatis and mount appropriate responses. We suggest that this distinction is critical for effective host defence, but also regulates inappropriate or excessive responses by only responding to live organisms. Conversely, these data also suggest a potential mechanism of immune evasion that could be adopted by C. trachomatis by modulating production of cyclic di-AMP. Bacterial pathogens are known to evade non-canonical inflammasome activation by modulating the acylation status of their lipid-A moieties [9], and we suggest that modulating production of inflammasome activating metabolites could also be a strategy utilised by pathogenic bacteria to evade this response.
We have shown that STING, independent of cGAS, is a critical component of interferon mediated inflammasome activation in response to C. trachomatis infection, and differs from recent studies utilising F. tularensis that required conversion of bacterial DNA to cGAMP, rather than detection of a microbial metabolite to induce STING mediated AIM2 inflammasome activation [35, 36]. These previous studies raise an important paradoxical question because AIM2 activation also requires the release of DNA from the pathogen in an interferon dependent process, yet the interferon signal also requires conversion of this same DNA by cGAS to induce the STING mediated interferon signal. This would suggest that either cGAS is a more sensitive sensor of DNA than AIM2 and responds to low levels within the cytosol that would not initiate AIM2 activation or that something else is providing the initiating interferon signal leading to loss of compartmentilisation and amplification of responses. We envisage that in our model—microbial metabolism and the production of cyclic di-AMP precedes any involvement of cGAS that is only partially required for cell death responses. Instead, cyclic di-AMP induction of STING activation results in small, but significant increases in type-1 interferon expression that facilitates the up-regulation of IRG’s and GBP’s that are known to be recruited to Chlamydial containing vacuoles [37–39]. Damage to the Chlamydia containing vacuole, mediated by IRG’s and GBP’s, could then result in the further release of vacuole contents including microbial DNA that further enhances STING activation through cGAS [34] and also activates the AIM2 inflammasome as we and others have reported recently [23]. In this regard, we would not discount cGAS from C. trachomatis induced inflammasome activation, but would suggest it enhances cell death responses, rather than initiates, the response. Furthermore, our experiments using attenuated C. trachomatis support this hypothesis, as attenuated organisms did not activate the inflammasome despite containing a full complement of unaltered DNA and LPS that would be immunostimulatory if detected by cytosolic receptors. This suggests that a loss of compartmentalisation of the Chlamydia containing vacuole is a key event that delivers inflammasome activatory ligands to the cytosol. Thus release of cyclic di-AMP activates STING which in turn mediates type-1 interferon signalling.
We also provide evidence that cell death observed during C. trachomatis is true pyroptosis. Deletion of both caspase-1 and caspase-11 prevented cell death and this observation would rule out other forms of lytic cell death such as necroptosis. It is unusual that both caspases could induce pyroptosis, but this is not without precedent [40], and has also been reported for Chlamydia infection by others independently of our studies [23]. We think it is also important that cell death was delayed compared to IL-1β release and suggests IL-1β release is controlled via an active process rather than accidental release as a consequence of cell death, although we cannot rule out that sensitivity of the assays employed could also contribute to these findings. However, in support of our findings, Recent studies have identified that cleavage of GSDMD by inflammatory caspases is a critical mediator of pyroptosis [11, 12, 41] and IL-1β release [12, 41]. It is also hypothesised that the number of gasdermin pores formed with the plasma membrane of the cell could control the balance between release of mature cytokine and commitment to cell death so that fewer pores allow release of IL-1β without cell death [42]. This would be an attractive explanation of the asynchronous behaviour of IL-1β release and cell death responses observed during C. trachomatis infection. Given that C. trachomatis induced pyroptosis could utilise both caspases, the role of GSDMD would be an interesting line of enquiry and will form part of our future studies. Furthermore, although inflammatory caspases are known to play a role in Chlamydia induced immunopathology [3], the role of pyroptosis has yet to be defined.
Finally, we have demonstrated a striking role for the control of intracellular growth of C. trachomatis by the NADPH oxidase system in both human and murine cells. Increased intracellular growth correlated with elevated inflammasome activation reinforcing our hypothesis that C. trachomatis replication or metabolism is a critical factor inducing inflammasome activation. There are very few published data investigating the role of NADPH oxidase during Chlamydia infection. Previous studies have utilised chemical inhibition to study ROS function during Chlamydia infection of Hela cells which suggested that ROS were important for C. trachomatis replication within these cells [31]. However, Hela cells provide a wholly different cellular environment to primary macrophages and C. trachomatis exists within a specially constructed pathogen containing vacuole that may well protect the organism from the toxic effects of ROS generation within non-phagocytic cells. It is not well defined which compartment C. trachomatis occupies within macrophages. We speculate from our work that this compartment is likely to be formed as a consequence of phagocytic activity as uptake of the bacteria was independent of high speed centrifugation often employed to achieve infection of non-phagocytic cells such as Hela. We would therefore suggest that Chlamydia trachomatis is likely to be existing within a different compartment in macrophages compared to Hela cells and therefore may be more susceptible to the toxic effects of ROS generated through NADPH oxidase activity which could account for the intriguing differences between our findings and others. Intriguingly, other members of the Chlamydiaceae family have been shown to express functional catalase activity [43] suggesting that ROS is an important host defence mechanism against Chlamydia infection and may limit the replicative ability of C. trachomatis within phagocytic cells such as macrophages. Importantly, a recent report has linked macrophage pyroptosis and neutrophil oxidative killing of bacteria as an important axis in innate host defence against bacterial infection [44]. Neutrophils from CGD patients display reduced bactericidal effects on Chlamydia trachomatis [45]. We would therefore predict that CGD patients may be at risk from more severe immunopathology during C. trachomatis infection due to elevated inflammasome responses of macrophages and reduced bacterial killing by neutrophils. This would also be in agreement with the consensus that CGD patients have a pro-inflammatory phenotype [46].
In summary, we propose that intracellular replication of Chlamydia trachomatis and production of the metabolite cyclic di-AMP is a key pathogen associated molecular pattern detected by STING that is crucial for activation of both canonical and non-canonical inflammasomes. This requires type-1 interferon and allows the host to initiate appropriate immune responses. We also suggest that modulation of cyclic di-AMP production by Chlamydia trachomatis could provide a mechanism of immune evasion and contribute to mechanisms of infection latency. Finally, targeting of the STING/interferon pathway may provide useful vaccine adjuvant and therapeutic targets to aid the treatment of Chlamydia trachomatis infection and its associated inflammatory pathology.
Materials and methods
Ethics statement
All studies involving human subjects were performed in accordance with the Declaration of Helsinki, with approval of the Cambridge Regional Ethical Committee (01/363). All donors gave written informed consent.
All animal related work was conducted by trained and appropriately licensed staff, under the authority and conditions of a Home Office project licence. This licence is issued under the UK Animal (Scientific Procedures) Act 1986 (ASPA), following local ethical approval. Home Office Inspectors provide governmental supervision of the work. Local ethical approval and supervision of standards of work and animal husbandry is carried out by the University of Cambridge Animal Welfare Ethical Review Body (AWERB) and its delegated representatives. Animals used for tissue were culled by fully trained personnel using approved humane methods under Schedule 1 of ASPA.
Cell culture and infection
Bone marrow derived macrophages (BMDM) were prepared from femurs of littermate wild type and genetically deficient animals by culturing bone marrow isolates for 7-days in RPMI containing 10% v/v HIFCS, L-glutamine, 5% v/v L929 conditioned medium with gentamicin. Of note, we became aware that macrophages produced from mice housed in different facilities displayed extremely variable responses to stimulation. It was therefore imperative that comparisons between wild-type and knock-out animals were made using littermates or, as a minimum, using age and sex matched mice housed in the same facility, and not between mice housed at different facilities. Unprimed macrophages were infected with live Chlamydia trachomatis at a multiplicity of infection (MOI) of 20. Attenuated Chlamydia trachomatis was achieved by γ-irradiating live C. trachomatis elementary bodies purified by high speed density centrifugation for 6-hours in a Gammacell-1000 irradiator (Atomic Energy of Canada Ltd) or using a Chlamydia Protease Activity like Factor (CPAF) deficient C. trachomatis mutant (ΔCPAF) described previously [28]. Attenuated C. trachomatis were used at a multiplicity of infection of 20 and attenuation was determined by the absence of replication in Hela cells. Intracellular LPS stimulation of macrophages was achieved by priming BMDM with 100ng/ml LPS for 4-hours in L929 conditioned RPMI. After priming, the supernatants were removed and replaced with serum free optiMEM (Gibco) containing 5μg/ml LPS with 5μl/ml Fugene (Promega). Cells were incubated for 24-hours before supernatants were harvested for ELISA and LDH assays. Recovery of inflammasome activation with attenuated C. trachomatis was achieved by stimulation of BMDM for 4-hours with γ-attenuated or CPAF deficient C. trachomatis mutant (ΔCPAF) in L929 conditioned RPMI as per intracellular LPS stimulation. The supernatant and non-internalised C. trachomatis were then removed and the media replaced with optiMEM containing cyclic-di-AMP (Invivogen, France) at indicated concentrations with Fugene. BMDM were then incubated for 24hrs before supernatants were harvested for ELISA.
Human monocyte derived dendritic cells (mDC) were obtained by IL-4/GMCSF differentiation [27, 47] of peripheral blood monocytes from age and sex matched chronic granulomatous disease (CGD) patients (recruited at the Royal Free Hospital, London) or healthy donors, (recruited at the University of Cambridge Department of Medicine, Cambridge) conforming to ethical guidelines of each institution.
Intracellular staining of Chlamydia for fluorescent microscopy and FACS analysis
Assessment of relative amounts of intracellular C. trachomatis was determined by fluorescent staining of Chlamydia LPS. BMDM or mDC were plated on coverslips (microscopy) or without coverslips (FACS) in 24-well plates at 2.5x105 cells/well and infected as described above for 24-hours. Intracellular staining of LPS was achieved by washing cells three times in PBS before fixation and permeabilisation using CellFix (BD Bioscience, USA) and incubating cells at room temperature with 1μg/ml FITC conjugated monoclonal mouse anti-chlamydia LPS (SourceBioscience U.K.). Cells were washed a further three times and mounted on slides using DAPI counterstain for fluorescent microscopy or scraped and assayed by FACS (FacsCalibur, BD Bioscience, USA). Uninfected cells were stained as described and acted as a negative control.
ELISA and LDH assay
Detection of mature IL-1β in cell culture supernatants was achieved by ELISA following the manufacturer’s instructions (Mouse or human Ready-SET-Go IL-1β ELISA, Ebioscience, USA). Macrophages were seeded in 96-plates at 1x105 cells/well and cultured for 24hrs in the presence of cell stimulations. For assessment of cell death, a lactate dehydrogenase (LDH) release assay was employed (promega U.K.) following the manufacturer’s instructions. The percentage cytotoxicity was calculated from the absorbance of the test well divided by the absorbance of the corresponding 100% lysis control wells.
SDS PAGE and western blotting
For analysis of protein expression cytosolic protein extracts and cell culture supernatants were assayed by SDS PAGE and western blotting as described previously [27]. Briefly, equal amounts of cytosolic protein, determined by Bradford Assay (Thermo U.K.) was mixed with an appropriate volume of 6x reducing buffer and boiled for 10 minutes before loading on to 4–20% gradient SDS pre-cast gels (BioRad U.K.). Separated protein was then transferred to PVDF membrane (BioRad U.K.) using a Transblot Turbo (BioRad U.K.). Membranes were blocked for 1-hour at RT in 5% w/v milk protein before incubation with appropriate primary antibody overnight at 4°C. Membranes were washed 3 times in TBS-TWEEN (0.05%v/v) before incubating with appropriate secondary HRP conjugated antibodies for 1-hour at RT. Membranes were developed via ECL (Lightning super signal, Perkin Elmer, USA) and visualised using a G-box (Syngene U.K.) or chemiluminescence film (Amersham Hyperfilm GE Healthcare U.K). Membranes were stripped using low pH stripping buffer for 30-minutes at RT followed by blocking and incubation with antibody. The following primary antibodies were used in this study: Monoclonal rat anti-mouse Caspase-11 (clone 17D9) (Sigma U.K.), Goat anti-mouse IL-1β (R&D Systems U.K.), Goat anti-human IL-1β (R&D Systems U.K.) and monoclonal mouse anti-actin (Abcam U.K.).
RNA extraction and qRT-PCR
Macrophages were cultured at 0.5x106 cells per well of 24-well plate and were infected with an MOI of 20 live C. trachomatis, γ-irradiated C. trachomatis or a CPAF deficient C. trachomatis for 8hrs. Cells were lysed via addition of RNA lysis buffer (Norgen, Canada) directly to the well. RNA was purified using the Norgen RNA kit according to manufacturer’s instructions. For interferon-β and IL-1β expression qRT-PCR was employed using commercial probe/primer sets (LifeTechnologies U.K.) and analysed using the Taqman ‘one-step’ system (LifeTechnologies U.K.). Analysis of C. trachomatis 16s expression was determined by quantitative PCR of cDNA prepared from total RNA extracted as above.
Statistical analysis
All numeric data were analysed using Graphpad Prism (USA). Analysis of multiple data groups with single variables was analysed using 1-way ANOVA with Dunnet’s post-test while multiple data groups with two variables was analysed using a 2-way ANOVA with Bonferonni’s post-test. Comparison of data between two-paired data sets was analysed using a paired Student’s t-test. All data are represented as ±SEM of the mean performed on BMDM obtained from at least three individual mice or mDC obtained from at least three individual human donors unless stated otherwise. A p value <0.05 was deemed significant.
Supporting information
(A) IL-1β secretion analysed by ELISA of supernatants wild-type BMDM infected with C. trachomatis (CT) presence of the mitochondrial anti-oxidant; MitoQ (1μM) for 24hrs (B) Pro IL-1β expression analysed by western blotting of lysates from unstimulated or C. trachomatis infected BMDM in the presence (+) or absence (-) of 1μM MitoQ for 24hrs. (C) Cell death analysed by LDH release from wild-type BMDM infected with C. trachomatis (CT) presence of the mitochondrial anti-oxidant; MitoQ (1μM) for 24hrs. (D) IL-1β secretion analysed by ELISA of supernatants from C. trachomatis (CT) infected wild-type (IRE-1+/+) or IRE1 knock-out (IRE-1-/-) BMDM for 24hrs. (E) IL-1β secretion analysed by ELISA of supernatants from wild-type BMDM infected with C. trachomatis (CT) in the presence of the IRE1 inhibitor 4μ8c (30μM) for 24hrs. Data represented as the mean of one experiment performed on cells from three individual mice, error bars indicate ±SEM. *p = <0.05, **p = <0.01 and ***p = <0.001. ns indicates no statistical significance between samples.
(TIF)
(A) IL-1β secretion analysed by ELISA of supernatants from wild-type BMDM infected with CPAF deficient (ΔCPAF) or CPAF sufficient control (CPAF WT) C. trachomatis. (B) IL-1β maturation and priming analysed by western blotting of cell lysates and supernatants from wild-type BMDM infected with CPAF deficient (ΔCPAF) or CPAF sufficient control (CPAF WT) C. trachomatis for 24hrs. (C) cell death analysed by LDH release from wild-type BMDM infected with CPAF deficient (ΔCPAF) or CPAF sufficient control (CPAF WT) C. trachomatis for 24hrs. Data represented as the mean of one experiment performed on cells from three individual mice, error bars indicate ±SEM. (D) Caspase-11 expression analysed by western blotting of lysates from BMDM infected with deficient (ΔCPAF) or CPAF sufficient control (CPAF WT) C. trachomatis for 24hrs.
(TIF)
(A) Intracellular staining of C. trachomatis LPS in BMDM analysed by FACS following infection with irradiated C. trachomatis (γ-CT) or non-attenuated C. trachomatis (CT). (B) Intracellular staining of C. trachomatis LPS in BMDM analysed by FACS following infection with CPAF deficient C. trachomatis (ΔCPAF) or wild-type C. trachomatis (WT CT).
(TIF)
C. trachomatis replication in wild type (Cybb+/+) or Cybb deficient (Cybb-/-) BMDM analysed by qRT-PCR of C. trachomatis 16s RNA expression following C. trachomatis infection for 6-hours. Data represented as the mean of one experiment performed on BMDM from three individual mice, error bars indicate ±SEM *p = <0.05.
(TIF)
Induction of IFNβ mRNA expression in wild type BMDM analysed by quantitative RT-PCR following infection with CPAF deficient (ΔCPAF) or CPAF sufficient control (CPAF WT) C. trachomatis for 8hrs. Data represented as the mean of one experiment performed on cells from three individual mice, error bars indicate ±SEM. *p = <0.05, **p = <0.01.
(TIF)
Acknowledgments
We are extremely grateful to Dr Vishva Dixit at Genentech, California for providing the caspase-1 knock-out caspase-11 transgenic mice, Dr Jan Rehwinkel and Chiara Cursi, University of Oxford, Weatherall Institute of Molecular Medicine, for provision of bone marrow tissue from wildtype and cGas deficient mice and Dr Thomas Henry, CIRI-Centre International de Recherche en Infectiologie for providing tissue from AIM2 deficient and wildtype mice. We are also extremely grateful to Dr Raphael Valdivia at Duke University, North Carolina for providing the CPAF deficient C. trachomatis. We thank Dr Mike Murphy at the MRC Mitochondrial Biology Unit Cambridge, U.K. for providing the Mito-Q. We also thank Sarita Workman at the Royal Free Hospital London, for her assistance in providing the CGD patient samples and to staff of the Cambridge University Phenotyping Hub for assistance with Flow Cytometry and fluorescent microscopy. We thank Professor Ziad Mallat at the University of Cambridge U.K. for helpful discussion of the data.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by a Medical Research Council grant (RG61445) to JCG and HJSG http://www.mrc.ac.uk/, an Arthritis Research UK Senior Fellowship (RG61798) grant to JCG http://www.arthritisresearchuk.org/, the National Institute for Health Research http://www.nihr.ac.uk/ Cambridge Biomedical Research Centre http://www.cambridge-brc.org.uk/ and Cambridge Arthritis Research Endeavour (CARE) http://www.cambridge-arthritis.org.uk/. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Nagarajan UM, Prantner D, Sikes JD, Andrews CW Jr., Goodwin AM, Nagarajan S, et al. Type I interferon signaling exacerbates Chlamydia muridarum genital infection in a murine model. Infect Immun. 2008;76(10):4642–8. Epub 2008/07/30. 10.1128/IAI.00629-08 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prantner D, Darville T, Sikes JD, Andrews CW Jr., Brade H, Rank RG, et al. Critical role for interleukin-1beta (IL-1beta) during Chlamydia muridarum genital infection and bacterial replication-independent secretion of IL-1beta in mouse macrophages. Infect Immun. 2009;77(12):5334–46. Epub 2009/10/07. 10.1128/IAI.00883-09 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheng W, Shivshankar P, Li Z, Chen L, Yeh IT, Zhong G. Caspase-1 contributes to Chlamydia trachomatis-induced upper urogenital tract inflammatory pathologies without affecting the course of infection. Infect Immun. 2008;76(2):515–22. Epub 2007/11/21. 10.1128/IAI.01064-07 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lamkanfi M, Dixit VM. Mechanisms and functions of inflammasomes. Cell. 2014;157(5):1013–22. Epub 2014/05/27. 10.1016/j.cell.2014.04.007 . [DOI] [PubMed] [Google Scholar]
- 5.Wallach D, Kang TB, Dillon CP, Green DR. Programmed necrosis in inflammation: Toward identification of the effector molecules. Science. 2016;352(6281):aaf2154. Epub 2016/04/02. 352/6281/aaf2154 [pii] 10.1126/science.aaf2154 . [DOI] [PubMed] [Google Scholar]
- 6.Kayagaki N, Warming S, Lamkanfi M, Vande Walle L, Louie S, Dong J, et al. Non-canonical inflammasome activation targets caspase-11. Nature. 2011;479(7371):117–21. Epub 2011/10/18. 10.1038/nature10558 . [DOI] [PubMed] [Google Scholar]
- 7.Casson CN, Yu J, Reyes VM, Taschuk FO, Yadav A, Copenhaver AM, et al. Human caspase-4 mediates noncanonical inflammasome activation against gram-negative bacterial pathogens. Proc Natl Acad Sci U S A. 2015;112(21):6688–93. Epub 2015/05/13. 10.1073/pnas.1421699112 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kayagaki N, Wong MT, Stowe IB, Ramani SR, Gonzalez LC, Akashi-Takamura S, et al. Noncanonical inflammasome activation by intracellular LPS independent of TLR4. Science. 2013;341(6151):1246–9. Epub 2013/07/28. 10.1126/science.1240248 . [DOI] [PubMed] [Google Scholar]
- 9.Hagar JA, Powell DA, Aachoui Y, Ernst RK, Miao EA. Cytoplasmic LPS activates caspase-11: implications in TLR4-independent endotoxic shock. Science. 2013;341(6151):1250–3. Epub 2013/09/14. 10.1126/science.1240988 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shi J, Zhao Y, Wang Y, Gao W, Ding J, Li P, et al. Inflammatory caspases are innate immune receptors for intracellular LPS. Nature. 2014;514(7521):187–92. Epub 2014/08/15. 10.1038/nature13683 . [DOI] [PubMed] [Google Scholar]
- 11.Kayagaki N, Stowe IB, Lee BL, O'Rourke K, Anderson K, Warming S, et al. Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling. Nature. 2015;526(7575):666–71. Epub 2015/09/17. 10.1038/nature15541 . [DOI] [PubMed] [Google Scholar]
- 12.Shi J, Zhao Y, Wang K, Shi X, Wang Y, Huang H, et al. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature. 2015;526(7575):660–5. Epub 2015/09/17. 10.1038/nature15514 . [DOI] [PubMed] [Google Scholar]
- 13.Yang D, He Y, Munoz-Planillo R, Liu Q, Nunez G. Caspase-11 Requires the Pannexin-1 Channel and the Purinergic P2X7 Pore to Mediate Pyroptosis and Endotoxic Shock. Immunity. 2015;43(5):923–32. Epub 2015/11/18. 10.1016/j.immuni.2015.10.009 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aachoui Y, Leaf IA, Hagar JA, Fontana MF, Campos CG, Zak DE, et al. Caspase-11 protects against bacteria that escape the vacuole. Science. 2013;339(6122):975–8. Epub 2013/01/26. 10.1126/science.1230751 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meunier E, Dick MS, Dreier RF, Schurmann N, Kenzelmann Broz D, Warming S, et al. Caspase-11 activation requires lysis of pathogen-containing vacuoles by IFN-induced GTPases. Nature. 2014;509(7500):366–70. Epub 2014/04/18. 10.1038/nature13157 . [DOI] [PubMed] [Google Scholar]
- 16.Guarda G, Braun M, Staehli F, Tardivel A, Mattmann C, Forster I, et al. Type I interferon inhibits interleukin-1 production and inflammasome activation. Immunity. 2011;34(2):213–23. Epub 2011/02/26. 10.1016/j.immuni.2011.02.006 . [DOI] [PubMed] [Google Scholar]
- 17.Rathinam VA, Vanaja SK, Waggoner L, Sokolovska A, Becker C, Stuart LM, et al. TRIF licenses caspase-11-dependent NLRP3 inflammasome activation by gram-negative bacteria. Cell. 2012;150(3):606–19. Epub 2012/07/24. 10.1016/j.cell.2012.07.007 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pilla DM, Hagar JA, Haldar AK, Mason AK, Degrandi D, Pfeffer K, et al. Guanylate binding proteins promote caspase-11-dependent pyroptosis in response to cytoplasmic LPS. Proc Natl Acad Sci U S A. 2014;111(16):6046–51. Epub 2014/04/10. 10.1073/pnas.1321700111 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abdul-Sater AA, Koo E, Hacker G, Ojcius DM. Inflammasome-dependent caspase-1 activation in cervical epithelial cells stimulates growth of the intracellular pathogen Chlamydia trachomatis. J Biol Chem. 2009;284(39):26789–96. Epub 2009/08/04. 10.1074/jbc.M109.026823 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abdul-Sater AA, Said-Sadier N, Padilla EV, Ojcius DM. Chlamydial infection of monocytes stimulates IL-1beta secretion through activation of the NLRP3 inflammasome. Microbes Infect. 2010;12(8–9):652–61. Epub 2010/05/04. 10.1016/j.micinf.2010.04.008 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shimada K, Crother TR, Karlin J, Dagvadorj J, Chiba N, Chen S, et al. Oxidized mitochondrial DNA activates the NLRP3 inflammasome during apoptosis. Immunity. 2012;36(3):401–14. Epub 2012/02/22. 10.1016/j.immuni.2012.01.009 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shimada K, Crother TR, Karlin J, Chen S, Chiba N, Ramanujan VK, et al. Caspase-1 dependent IL-1beta secretion is critical for host defense in a mouse model of Chlamydia pneumoniae lung infection. PLoS One. 2011;6(6):e21477 Epub 2011/07/07. 10.1371/journal.pone.0021477 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Finethy R, Jorgensen I, Haldar AK, de Zoete MR, Strowig T, Flavell RA, et al. Guanylate Binding Proteins Enable Rapid Activation of Canonical and Noncanonical Inflammasomes in Chlamydia-Infected Macrophages. Infect Immun. 2015;83(12):4740–9. Epub 2015/09/30. 10.1128/IAI.00856-15 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prantner D, Darville T, Nagarajan UM. Stimulator of IFN gene is critical for induction of IFN-beta during Chlamydia muridarum infection. J Immunol. 2010;184(5):2551–60. Epub 2010/01/29. 10.4049/jimmunol.0903704 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou R, Yazdi AS, Menu P, Tschopp J. A role for mitochondria in NLRP3 inflammasome activation. Nature. 2011;469(7329):221–5. Epub 2010/12/03. 10.1038/nature09663 . [DOI] [PubMed] [Google Scholar]
- 26.Bronner DN, Abuaita BH, Chen X, Fitzgerald KA, Nunez G, He Y, et al. Endoplasmic Reticulum Stress Activates the Inflammasome via NLRP3- and Caspase-2-Driven Mitochondrial Damage. Immunity. 2015;43(3):451–62. Epub 2015/09/06. 10.1016/j.immuni.2015.08.008 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Webster SJ, Ellis L, O'Brien LM, Tyrrell B, Fitzmaurice TJ, Elder MJ, et al. IRE1alpha mediates PKR activation in response to Chlamydia trachomatis infection. Microbes Infect. 2016. Epub 2016/03/30. S1286-4579(16)30020-X [pii] 10.1016/j.micinf.2016.03.010 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Snavely EA, Kokes M, Dunn JD, Saka HA, Nguyen BD, Bastidas RJ, et al. Reassessing the role of the secreted protease CPAF in Chlamydia trachomatis infection through genetic approaches. Pathog Dis. 2014;71(3):336–51. Epub 2014/05/20. 10.1111/2049-632X.12179 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Segal BH, Grimm MJ, Khan AN, Han W, Blackwell TS. Regulation of innate immunity by NADPH oxidase. Free Radic Biol Med. 2012;53(1):72–80. Epub 2012/05/16. 10.1016/j.freeradbiomed.2012.04.022 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vazquez-Torres A, Jones-Carson J, Mastroeni P, Ischiropoulos H, Fang FC. Antimicrobial actions of the NADPH phagocyte oxidase and inducible nitric oxide synthase in experimental salmonellosis. I. Effects on microbial killing by activated peritoneal macrophages in vitro. J Exp Med. 2000;192(2):227–36. Epub 2000/07/19. ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abdul-Sater AA, Said-Sadier N, Lam VM, Singh B, Pettengill MA, Soares F, et al. Enhancement of reactive oxygen species production and chlamydial infection by the mitochondrial Nod-like family member NLRX1. J Biol Chem. 2010;285(53):41637–45. 10.1074/jbc.M110.137885 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Malireddi RK, Kanneganti TD. Role of type I interferons in inflammasome activation, cell death, and disease during microbial infection. Front Cell Infect Microbiol. 2013;3:77 Epub 2013/11/26. 10.3389/fcimb.2013.00077 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barker JR, Koestler BJ, Carpenter VK, Burdette DL, Waters CM, Vance RE, et al. STING-dependent recognition of cyclic di-AMP mediates type I interferon responses during Chlamydia trachomatis infection. MBio. 2013;4(3):e00018–13. Epub 2013/05/02. 10.1128/mBio.00018-13 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y, Yeruva L, Marinov A, Prantner D, Wyrick PB, Lupashin V, et al. The DNA sensor, cyclic GMP-AMP synthase, is essential for induction of IFN-beta during Chlamydia trachomatis infection. J Immunol. 2014;193(5):2394–404. Epub 2014/07/30. 10.4049/jimmunol.1302718 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Man SM, Karki R, Malireddi RK, Neale G, Vogel P, Yamamoto M, et al. The transcription factor IRF1 and guanylate-binding proteins target activation of the AIM2 inflammasome by Francisella infection. Nat Immunol. 2015;16(5):467–75. Epub 2015/03/17. 10.1038/ni.3118 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meunier E, Wallet P, Dreier RF, Costanzo S, Anton L, Ruhl S, et al. Guanylate-binding proteins promote activation of the AIM2 inflammasome during infection with Francisella novicida. Nat Immunol. 2015;16(5):476–84. Epub 2015/03/17. 10.1038/ni.3119 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Haldar AK, Saka HA, Piro AS, Dunn JD, Henry SC, Taylor GA, et al. IRG and GBP host resistance factors target aberrant, "non-self" vacuoles characterized by the missing of "self" IRGM proteins. PLoS Pathog. 2013;9(6):e1003414 Epub 2013/06/21. 10.1371/journal.ppat.1003414 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Haldar AK, Piro AS, Pilla DM, Yamamoto M, Coers J. The E2-like conjugation enzyme Atg3 promotes binding of IRG and Gbp proteins to Chlamydia- and Toxoplasma-containing vacuoles and host resistance. PLoS One. 2014;9(1):e86684 Epub 2014/01/28. 10.1371/journal.pone.0086684 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Haldar AK, Foltz C, Finethy R, Piro AS, Feeley EM, Pilla-Moffett DM, et al. Ubiquitin systems mark pathogen-containing vacuoles as targets for host defense by guanylate binding proteins. Proc Natl Acad Sci U S A. 2015;112(41):E5628–37. Epub 2015/09/30. 10.1073/pnas.1515966112 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Broz P, Ruby T, Belhocine K, Bouley DM, Kayagaki N, Dixit VM, et al. Caspase-11 increases susceptibility to Salmonella infection in the absence of caspase-1. Nature. 2012;490(7419):288–91. Epub 2012/08/17. 10.1038/nature11419 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.He WT, Wan H, Hu L, Chen P, Wang X, Huang Z, et al. Gasdermin D is an executor of pyroptosis and required for interleukin-1beta secretion. Cell Res. 2015;25(12):1285–98. Epub 2015/11/28. 10.1038/cr.2015.139 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jorgensen I, Rayamajhi M, Miao EA. Programmed cell death as a defence against infection. Nat Rev Immunol. 2017;17(3):151–64. 10.1038/nri.2016.147 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rusconi B, Greub G. Discovery of catalases in members of the Chlamydiales order. J Bacteriol. 2013;195(16):3543–51. 10.1128/JB.00563-13 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maltez VI, Tubbs AL, Cook KD, Aachoui Y, Falcone EL, Holland SM, et al. Inflammasomes Coordinate Pyroptosis and Natural Killer Cell Cytotoxicity to Clear Infection by a Ubiquitous Environmental Bacterium. Immunity. 2015;43(5):987–97. Epub 2015/11/18. 10.1016/j.immuni.2015.10.010 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yong EC, Klebanoff SJ, Kuo CC. Toxic effect of human polymorphonuclear leukocytes on Chlamydia trachomatis. Infect Immun. 1982;37(2):422–6. Epub 1982/08/01. ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.de Luca A, Smeekens SP, Casagrande A, Iannitti R, Conway KL, Gresnigt MS, et al. IL-1 receptor blockade restores autophagy and reduces inflammation in chronic granulomatous disease in mice and in humans. Proc Natl Acad Sci U S A. 2014;111(9):3526–31. Epub 2014/02/20. 10.1073/pnas.1322831111 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Elder MJ, Webster SJ, Williams DL, Gaston JS, Goodall JC. TSLP production by dendritic cells is modulated by IL-1beta and components of the endoplasmic reticulum stress response. Eur J Immunol. 2016;46(2):455–63. Epub 2015/11/18. 10.1002/eji.201545537 . [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) IL-1β secretion analysed by ELISA of supernatants wild-type BMDM infected with C. trachomatis (CT) presence of the mitochondrial anti-oxidant; MitoQ (1μM) for 24hrs (B) Pro IL-1β expression analysed by western blotting of lysates from unstimulated or C. trachomatis infected BMDM in the presence (+) or absence (-) of 1μM MitoQ for 24hrs. (C) Cell death analysed by LDH release from wild-type BMDM infected with C. trachomatis (CT) presence of the mitochondrial anti-oxidant; MitoQ (1μM) for 24hrs. (D) IL-1β secretion analysed by ELISA of supernatants from C. trachomatis (CT) infected wild-type (IRE-1+/+) or IRE1 knock-out (IRE-1-/-) BMDM for 24hrs. (E) IL-1β secretion analysed by ELISA of supernatants from wild-type BMDM infected with C. trachomatis (CT) in the presence of the IRE1 inhibitor 4μ8c (30μM) for 24hrs. Data represented as the mean of one experiment performed on cells from three individual mice, error bars indicate ±SEM. *p = <0.05, **p = <0.01 and ***p = <0.001. ns indicates no statistical significance between samples.
(TIF)
(A) IL-1β secretion analysed by ELISA of supernatants from wild-type BMDM infected with CPAF deficient (ΔCPAF) or CPAF sufficient control (CPAF WT) C. trachomatis. (B) IL-1β maturation and priming analysed by western blotting of cell lysates and supernatants from wild-type BMDM infected with CPAF deficient (ΔCPAF) or CPAF sufficient control (CPAF WT) C. trachomatis for 24hrs. (C) cell death analysed by LDH release from wild-type BMDM infected with CPAF deficient (ΔCPAF) or CPAF sufficient control (CPAF WT) C. trachomatis for 24hrs. Data represented as the mean of one experiment performed on cells from three individual mice, error bars indicate ±SEM. (D) Caspase-11 expression analysed by western blotting of lysates from BMDM infected with deficient (ΔCPAF) or CPAF sufficient control (CPAF WT) C. trachomatis for 24hrs.
(TIF)
(A) Intracellular staining of C. trachomatis LPS in BMDM analysed by FACS following infection with irradiated C. trachomatis (γ-CT) or non-attenuated C. trachomatis (CT). (B) Intracellular staining of C. trachomatis LPS in BMDM analysed by FACS following infection with CPAF deficient C. trachomatis (ΔCPAF) or wild-type C. trachomatis (WT CT).
(TIF)
C. trachomatis replication in wild type (Cybb+/+) or Cybb deficient (Cybb-/-) BMDM analysed by qRT-PCR of C. trachomatis 16s RNA expression following C. trachomatis infection for 6-hours. Data represented as the mean of one experiment performed on BMDM from three individual mice, error bars indicate ±SEM *p = <0.05.
(TIF)
Induction of IFNβ mRNA expression in wild type BMDM analysed by quantitative RT-PCR following infection with CPAF deficient (ΔCPAF) or CPAF sufficient control (CPAF WT) C. trachomatis for 8hrs. Data represented as the mean of one experiment performed on cells from three individual mice, error bars indicate ±SEM. *p = <0.05, **p = <0.01.
(TIF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.