Abstract
Thyroid hormone (TH) is recognized for its role in cellular metabolism and growth and participates in homeostasis of the heart. T3 activates pro-survival pathways including Akt and mTOR. Treatment with T3 after myocardial infarction is cardioprotective and promotes elements of physiological hypertrophic response after cardiac injury. Although T3 is known to benefit the heart, very little about its regulation at the molecular level has been described to date. The ubiquitin proteasome system (UPS) regulates nuclear hormone receptors such as estrogen, progesterone, androgen, and glucocorticoid receptors by both degradatory and non-degradatory mechanisms. However, how the UPS regulates T3-mediated activity is not well understood. In this study, we aim to determine the role of the muscle-specific ubiquitin ligase muscle ring finger-1 (MuRF1) in regulating T3-induced cardiomyocyte growth. An increase in MuRF1 expression inhibits T3-induced physiological cardiac hypertrophy, whereas a decrease in MuRF1 expression enhances T3’s activity both in vitro and in cardiomyocytes in vivo. MuRF1 interacts directly with TRα to inhibit its activity by posttranslational ubiquitination in a non-canonical manner. We then demonstrated that a nuclear localization apparatus that regulates/inhibits nuclear receptors by sequestering them within a subcompartment of the nucleus was necessary for MuRF1 to inhibit T3 activity. This work implicates a novel mechanism that enhances the beneficial T3 activity specifically within the heart, thereby offering a potential target to enhance cardiac T3 activity in an organ-specific manner.
Keywords: cardiac hypertrophy, thyroid hormone, thyroid receptor α, ubiquitin, ubiquitin ligase
Introduction
Thyroid hormone (TH) is recognized for its role in cellular metabolism and growth and participates in homeostasis of the heart. Its biologically active form (T3) is involved in directing myocyte alignment and geometry (Pantos et al. 2007b), extracellular matrix remodeling (Ghose Roy et al. 2007), and cardiac function through the effects on contraction (Bengel et al. 2000). T3 mediates both direct influences on cardiac function by affecting calcium flux and calcium handling proteins (Belakavadi et al. 2010, Zarain-Herzberg 2006), in addition to voltage-gated potassium channels to affect contractility (Gassanov et al. 2009, Mansen et al. 2010), and through transcriptional regulation as TH is a ligand for the thyroid hormone receptor, predominantly the α isoform, TRα in the heart. A decrease in TH concentrations has been observed in acute myocardial infarction, and low circulating TH is commonly observed with severe heart failure (HF) (Gerdes & Iervasi 2010, Iervasi & Nicolini 2013). Treatment with TH following myocardial infarction is cardioprotective (Mourouzis et al. 2011, Pantos et al. 2007a) and promotes elements of physiological hypertrophic response after cardiac injury (Mourouzis et al. 2013a). T3 activates many of the physiological changes that would benefit the heart, activating pro-survival pathways Akt and mTOR and paralleling changes induced by exercise-mediated by IGF-1 (Ojamaa 2010). Despite our rich knowledge of the cardioprotective effects of T3, little is known about the intracellular regulation of its action.
Intracellular regulation of nuclear receptors by posttranslational modification by ubiquitin is well known and involves two distinct mechanisms: 1) targeted degradation of nuclear receptors by the proteasome and 2) ubiquitination-altering activity in non-degradatory ways (Ismail & Nawaz 2005). For example, estrogen, progesterone, androgen, and glucocorticoid nuclear receptors are degraded in a proteasome-dependent manner in some conditions (Hirotani et al. 2001, Hoeck et al. 1989, Lange et al. 2000, Li et al. 1999, Lin et al. 2002, Nawaz et al. 1999), with their heterodimeric binding partners RARγ and RXR also degraded by parallel mechanisms (Kopf et al. 2000). The muscle-specific ubiquitin ligase MuRF1 has recently been shown to specifically inhibit PPARα, but not PPARβ/δ or PPARγ1, activity in vivo (Rodriguez et al. 2015). In contrast to the previous studies on nuclear receptors, MuRF1 was found to inhibit PPARα in the cardiomyocyte through mono-ubiquitination that enhanced nuclear export without promoting PPARα degradation (Rodriguez et al. 2015). By mono-ubiquitinating one to three lysines adjacent to a newly identified nuclear export sequence in PPARα, MuRF1 inhibited PPARα-regulated fatty acid oxidation both in vitro and in vivo (Rodriguez et al. 2015). Complementary to these studies, the closely related muscle-specific ubiquitin ligase MuRF2 inhibited PPARγ1 (He et al. 2015), whereas MuRF3 inhibited PPARβ activity in vivo by mono-ubiquitination (Quintana et al. 2015). In this study, we identify the cardiac ubiquitin ligase MuRF1 as an inhibitor of T3-induced physiological cardiac hypertrophy in vivo. We determined that MuRF1 directly interacts with a specific thyroid receptor to mediate its posttranslational modification and inhibit its activity. These studies then determined a previously undescribed mechanism in which MuRF1 inhibits T3 activity, by altering TRα localization via posttranslational ubiquitination.
Materials and methods
Preparation of T3 and free T3 concentration estimates
3,3′,5-Tri-iodo-l-thyronine (T3, #T2877; Sigma-Aldrich) was weighed (0.003 g) on an analytical scale, dissolved in 20 ml sterile PBS, heated at 37°C for 10 min, and vortexed at the highest setting for 1 min. As a percentage of T3 remains insoluble using this method, we determined the resulting free T3 of this preparation compared with alkalizing T3 with 1 N NaOH (1 mL per mg) and putting in PBS, as described by the manufacturer. Analysis of free T3 heated/vortexed vs NaOH solubilized was made on the Vitros 5600 chemistry analyzer (Ortho Clinical Diagnostics, Rochester, NY, USA) in a matrix-compatible diluent to a final concentration of 50 ng/mL. With NaOH solubilization and dilution in PBS, 52.3 ± 8.4 ng/mL (N = 3) was recovered compared with 0.87 ± 0.012 ng/mL (N = 3). The recovery ratio (60.4 = 52.3/0.87) was used to estimate the concentrations of T3 in all the experiments described in vitro and in vivo.
Culturing of neonatal rat ventricular myocytes (NRVM), HL-1 cells, and in vitro experimental design
NRVMs were isolated using a commercial kit according to the manufacturer’s protocols, as described previously (#LK003300; Worthington Biochemical Corporation, Lakewood, NJ, USA) (Arya et al. 2004, Li et al. 2004). NRVMs cultured on fibronectin with medium 199 (M199) supplemented with 15% FBS were serum starved, transduced with plasmids, and treated with 16.6 nM T3. The cardiac-derived HL-1 cell line was maintained as described previously (Claycomb et al. 1998, White et al. 2004). COS-7 cells were cultured using the supplier’s protocols (CRL-1651, ATCC; Manassas, VA, USA). myc-tagged MuRF1/bicistronic GFP adenovirus (AdMuRF1), GFP adenovirus control, AdshRNA-MuRF1 (shMuRF1), or a control vector with scrambled AdshRNA (CTL) was used as described previously (Kedar et al. 2004, Wadosky et al. 2014). Cells were transduced with 25–60 MOI for 24–48 h and incubated with T3 in serum-free DMEM. Cellular fractionation was carried out using the Nuclear Extraction Kit (#AY2002, Panomics, Fremont, CA, USA) according to the manufacturer’s protocol.
CAP350 silencing and size measurement of HL-1 cells
HL-1 cells were treated with Accell mouse CAP350 (centrosome-associated protein 350) siRNA (GE Healthcare) using 1 μM Accell siRNA as per the manufacturer’s delivery protocol. Size measurement off HL-1 cells was performed using Image J.
TRα-mediated thyroid response element (TRE)-driven luciferase activity assay
Cos-7 cells were co-transfected with plasmids expressing β-galactosidase, growth hormone TRE luciferase reporter, as described previously (Liu et al. 2012), and the p3XFLAG-CMV-14 (Sigma-Aldrich) either empty or with the murine TRα sequence subcloned into the vector at the EcoRI and BamHI restriction sites.
Cell immunoblot analysis
Cells were lysed in Cell Lysis Buffer (Cell Signaling), supplemented with protease (#11697498001; Roche) and phosphatase inhibitors (#04906845001; Roche), in addition to 0.2 M glycerol-2-phosphate (Sigma-Aldrich) and resolved on NuPAGE gels (Novex; Life Technologies). Primary antibodies used in this study are β-actin (1:10,000, Cat. #A5441; Sigma-Aldrich), CAP350 (1:1000, Cat. #20022-1-AP; Acris Antibodies, San Diego, CA, USA), FLAG-TRα (1:1000, anti-FLAG #F7425; Sigma-Aldrich), GAPDH (1:10,000, Cat. #G8795; Sigma-Aldrich), MuRF1 (1:250–1:1000, #AF5366; R&D Systems), GAPDH (1:4000, G8795; Sigma-Aldrich), p-Rb (#sc-50; Santa Cruz Biotechnology), myc-tagged MuRF1 (1:4000, anti-myc, #C4439; or anti-myc-HRP-linked primary, 1:10,000, #A5598; Sigma-Aldrich), glutathione-S-transferase (GST)-tagged TRα (anti-GST; #G1160; Sigma-Aldrich, 1:10,000), TRα (PA1-211A; Thermo Scientific, 1:500), and HA-ubiquitin (anti-HA; #sc-57592, 1:1000). HRP-linked secondary antibodies used in this study are anti-mouse (#NA931V; GE Healthcare Life Sciences, 1:10,000), anti-rabbit (#A9169, Sigma-Aldrich, 1:20,000), and anti-goat (#A5420; Sigma-Aldrich, 1:10,000).
Constructs for confocal TRα lysine domain mutants and GST-TRα mapping i.p. studies
DNA constructs for the full sequence of TRα1 containing modification of all lysine codons to encode arginine (K→R) were synthesized for each domain separately (A/B, C, D, and E/F: Supplementary Fig. S2 (see section on supplementary data given at the end of this article)) and subcloned into the p3xFLAG-CMV-14 expression vector, sequence verified, and transfected into cells.
Confocal immunofluorescence analysis and co-localization analysis
Cells grown on Flexcell plates (Flexcell International, Hillsborough, NC) were transfected with plasmids indicated, transduced (AdGFP or AdMuRF1), and treated with 16.6 nM T3 (or vehicle) for 2 h. Cells were fixed in 4% formaldehyde, sectioned, and blocked in 0.4% Triton X-100/5% FBS. Primary antibodies were applied overnight followed by incubation with secondary antibodies. After washing, fixed cells were mounted onto glass slides with Fluoro-Gel II Mounting Media containing DAPI (17985-50; Electron Microscopy Services, Hatfield, PA, USA). A Leica DMIRB inverted fluorescence microscope (Leica Microsystems, Buffalo Grove, IL, USA) with a 40× objective lens and with a 10× Hamamatsu Orca ER camera (Bridgewater, NJ, USA) was used to analyze cell size.
Co-localization analysis was performed by determining the coefficient values representing the degree in which two different fluorescence channels (450, 488, 568, or 647) overlap in isolated regions of the cardiomyocyte using Volocity 3D Images Software Quantitation (PerkinElmer) (Lisby et al. 2003, Vandenbroucke St Amant et al. 2012). For TRα and nuclear co-localization, for every cell in each image, the nuclear region of the cell was selected by tracing around the DAPI fluorescent signal. Then, quantitative co-localization analysis within the selected region was analyzed between 450 (DAPI) and 568 (TRα). Measurements were made using the Costes’ Pearson’s coefficient correlation statistics function provided within the software that uses the threshold values for each channel to adjust the final coefficient data. For correlation coefficients, numeric range is from 0 to 1, where 0 indicates no correlation and 1 indicates perfect correlation. For at least 20 cells per condition in at least three images, correlation coefficients were averaged and final data were presented as the average ± SE.
Co-immunoprecipitation
Transduced Cos-7 cells (AdGFP or AdMuRF1) treated with T3 (or vehicle) were lysed in buffer (50 mM Tris, pH 7.5, 30 mM NaCl, 1 mM EDTA, and 0.5% (v/v) Triton X-100) supplemented with 2× protease inhibitor cocktail (Roche) and 1× phosphatase inhibitor cocktail (Roche), then pre-cleared with 5 μg rabbit IgG (I5006; Sigma-Aldrich) and 2% (v/v) Protein A/G PLUS Agarose beads (sc-2003; Santa Cruz Biotechnology) per 1 mg of protein lysate for 1 hour at 4°C with rotation. Myc-MuRF1 was immunoprecipitated using 5% (v/v) EZ View Red Anti-c-Myc Affinity Gel (E6654; Sigma-Aldrich) by incubating with anti-c-myc beads overnight at 4C with rotation.
GST pull down
Recombinant GST-TRα-mapping constructs were produced using BL21-CodonPlus Competent Cells transduced with pGEX-KG-TRα domain plasmids using standard protocols (Stratagene, La Jolla, CA, USA). Isolation of GST-tagged recombinant proteins was carried out using the BugBuster GST-Bind Purification Kit (70794-3; EMD Millipore Novagen, San Diego, CA, USA) according to the manufacturer’s protocol; the glutathione column with GST protein was then incubated with 1 mg protein lysate from Cos-7 cells transduced with AdMuRF1 overnight at 4C with rotation, allowing MuRF1 to bind TRα domains. Unbound proteins were then collected and elution steps were carried out according to the manufacturer’s protocol. Unbound and eluate fractions were immunoblotted for GST-tagged recombinant TRα (anti-GST (#G1160), 1:10,000; Sigma Aldrich) and myc-tagged MuRF1 (anti-myc (#C4439), 1:4000; Sigma Aldrich).
Cell-free ubiquitination assay
To evaluate direct ubiquitination of TRα by MuRF1, human recombinant E1 (50 nM, Cat. #E-305; Boston, Biochem, Cambridge, MA, USA), human recombinant UbcH5c/UBE2D3 (2.5 mM, Cat. #E2-627; Boston Biochem, Inc., Cambridge, MA), human recombinant ubiquitin (250 μM, Cat. #U-100 H; Boston Biochem), human recombinant his-6-MuRF1 protein (1 mg, Cat. #E3-100; Boston Biochem), and human TRα recombinant protein (500 ng; Abnova, Walnut, CA, USA) were added to the reaction buffer (50 mM HEPES, pH 7.5) containing 5 mM MgATP solution (Boston Biochem) and 0.6 mM DTT and were incubated overnight at 30°C. The reaction was stopped by SDS–PAGE sample buffer and heating, then resolved on a 4–12% Bis–Tris gel with MOPS running buffer (Invitrogen), and transferred to PVDF membranes for immunoblotting with rabbit polyclonal anti-TRα antibody (Thermo Scientific Pierce, Inc., Rockford, IL, USA) or rabbit polyclonal anti-MuRF1 antibody (R&D Systems).
Animals, treatment with T3, conscious echocardiography
Male and female adult mice (8–16 weeks) from a knockout (MuRF1−/−) and a cardiomyocyte-specific MuRF1 transgenic (αMHC promoter driven) mouse (MuRF1Tg+) with strain-matched wild-type mice (MuRF1+/+ or wild type MuRF1Tg+ respectively) were used as described previously (Willis et al. 2007, Willis et al. 2009a). Randomly assigned mice were treated with 16.6 μg/kg/day 3,3′,5-tri-iodo-l-thyronine (T3, #T2877; Sigma-Aldrich) dissolved in sterile PBS or PBS as a vehicle control via intraperitoneal injection (i.p.) for 2 weeks. Conscious echocardiographic analysis was performed at baseline, 1 week, and 2 weeks. After 2 weeks, mice were anesthetized to collect retro-orbital blood and then killed for tissue harvest. Hearts were quickly excised and frozen in liquid nitrogen and then stored at −80°C.
Histological analysis of mouse hearts
Fixed cardiac tissue was paraffin embedded, processed, sectioned, and stained with hematoxylin and eosin or Masson’s trichrome. Stained slides were scanned using an AperioScanscope and images were exported using AperioImagescope software (Aperio Technologies, Vista, CA, USA). Cardiomyocyte cross-sectional area was determined on MT-stained slides using ImageJ software (NIH, MD, USA) on a minimum of six randomly selected regions per image (100× magnification) from three different hearts per group.
Analysis of T3 and T4 in serum
T3 and T4 was quantitated in serum using the Luminex MAGPIX system (EMD Millipore) with the MILLIPLEX MAP magnetic bead-based Multi-Species Steroid/Thyroid Hormone assay (Cat. #STTHMAG-21 K) analyzed on the MAGPIX Multiplexing instrument (Life Technologies).
Tissue immunoblot analysis
Mouse heart tissue was lysed in 8 M urea buffer and stored at −80°C. Antibodies recognizing Rb (Cat. #MA5–11387) and p-Rb (Cat. #MA5-12584) were obtained from Life Technologies (Carlsbad, CA, USA) and were used at 1:200 dilution. Antibody recognizing TRα (Cat. #PA1-211A; Thermo Scientific) and antibody recognizing GAPDH (Cat. #G8795; Sigma-Aldrich) were used at 1:200 and 1:10,000 dilutions respectively.
RT-qPCR
Ventricular tissue (5 mg) was homogenized using the TissueLyser LT (Qiagen). Cells were homogenized by syringe and needle in TRIzol (Life Technologies, Invitrogen), and total RNA was isolated according to the manufacturer’s protocols. cDNA was generated from 1 μg RNA using a High-Capacity cDNA Archive Kit (Life Technologies, Invtirogen). One microliter of cDNA product was amplified in a LightCycler 480 Sequence Detection System (Roche) in 10 μl final volume using the LightCycler 480 Probes Master Mix (Roche). The PCR reaction mix included 0.5 μl of mouse-specific 20× Taqman probes (Life Technologies, Applied Biosystems): sarco/endoplasmic reticulum Ca2+-ATPase (SERCA) (Mm01201431_m1), MuRF1 (Mm01185221_m1), αMHC (Mm00440359_m1), mitochondrial adenine transporter (ANT) (Mm01207393_m1), ANF (Mm01255747_g1), BNP (Mm00435304_g1), α1 skeletal muscle actin (ACTA1) (Mm00808218_g1), monocarboxylate transporter 8 (MCT8) (Mm01232724_m1), or monocarboxylate transporter 10 (MCT10) (Mm00661045_m1). Tbp (Mm00446971_m1) or 18 S (Hs99999901_s1) served as reference genes. The −−Ct method was used to determine the relative expression values that were normalized to the wild-type control group.
Statistical analysis
A two-way ANOVA with the Holm–Sidak post hoc test for all pairwise comparisons was used to evaluate the statistical significance with SigmaPlot software (Systat Software, Inc., Chicago, IL, USA). In TRα KR domain mutant experiments, a one-way ANOVA test was applied with an all pairwise post-test. The criterion for significance was a P-value of ≤ 0.05 (Supplementary data, see section on supplementary data given at the end of this article).
Results
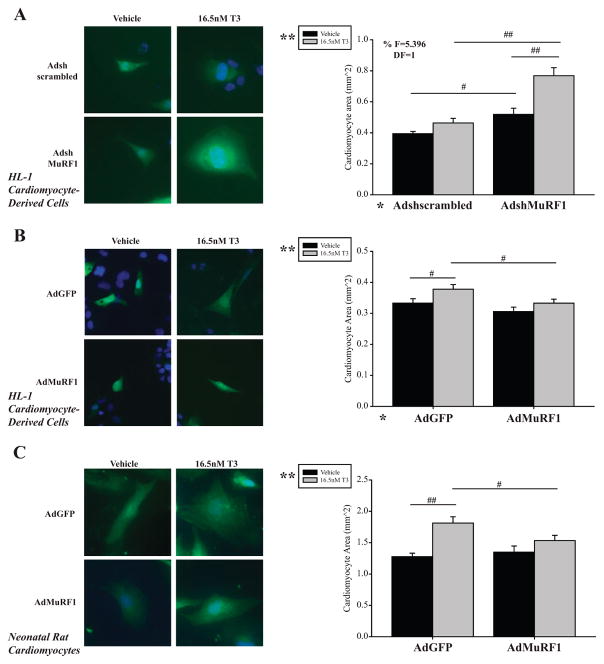
To identify the role of MuRF1 in regulating TH-induced physiological hypertrophy, HL-1 cardiomyocyte-derived cells and neonatal rat cardiomyocytes (NRVMs) were stimulated with tri-iodothyronine (T3). As expected, T3 induced cardiomyocyte hypertrophy, reflected by increased CSA (Fig. 1A, B and C, Supplementary Fig. S1A, B and C, uncropped). T3 stimulation of cardiomyocytes with MuRF1-knockdown (using AdshMuRF1, ~50% decrease in protein, Fig. 2C) resulted in a significant increase in CSA compared with T3 alone (Fig. 1A). Conversely, increasing MuRF1 expression (AdMuRF1) significantly inhibited T3-induced increase in CSA in both HL-1 cells and NRVM (B and C). Together, these studies indicated a role of endogenous MuRF1 in attenuating T3-induced cardiac hypertrophy.
Figure 1.
MuRF1 inhibits T3-induced cardiomyocyte hypertrophy in vitro. (A) Confocal fluorescent images of HL-1 cardiomyocytes with MuRF1 gene knock-down (shMuRF1) in the presence and absence of T3 (16.6 nM). Gene-silencing controls (scrambled Adsh) were run in parallel. (B) Confocal image of HL-1 cells increased MuRF1 expression (AdMuRF1) treated with T3. GFP-transfected controls were included (AdGFP). (C) Primary neonatal rat ventricular myocytes (NRVM) with increased MuRF1 expression (AdMuRF1) and T3 treatment. Fluorescent confocal images show cell volume highlighted by GFP (green) and nuclear staining with DAPI (blue). Graphs contain quantitative analysis of cell area (≥ 200 cardiomyocytes averaged per group from three independent experiments). A two-way ANOVA test was used to determine statistical significance using a pairwise post-test and significance between groups is denoted by #P < 0.05 or ##P < 0.001. The F statistic and degrees of freedom (DF) were reported when dependence between groups was found to be a significant source of variation. *Significance at the level of adenovirus group, **Significance on the level of treatment group, 400× final magnification.
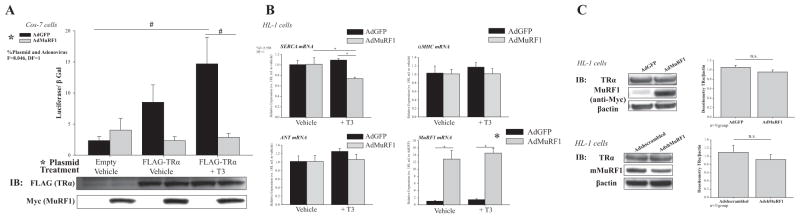
Figure 2.
MuRF1 inhibits TRα transcriptional activity following T3 treatment without affecting its steady-state levels. (A) TRE reporter assay results from Cos-7 cells co-transfected with luciferase plasmid containing TRE (or β-galactosidase control), and FLAG-TRα plasmid, transduced with AdMuRF1 (4 h), then treated with 16.6 nM T3 for 24 h (n = 3 independent experiments). Immunoblot verifying FLAG-TRα and myc-MuRF1 protein expression shown below graph. (B) Relative expression of TR transcriptionally regulated genes; HL-1 cells were transfected with AdMuRF1 (or AdGFP control) for 24 h and then treated with T3 for 30 min (C) Immunoblot of endogenous TR protein expression following MuRF1 knock-down (shMuRF1) or increased MuRF1 expression (AdMuRF1) in HL-1 cardiomyocytes (β-actin = loading control). Quantitative data shown in graphs below blots. Data are represented as mean ± S.E.M. (n = 3 independent experiments). A two-way ANOVA test was used to determine statistical significance. *Significance on the level of plasmid group. The F statistic and degrees of freedom (DF) were reported when dependence between groups was found to be a significant source of variation. Significance between groups is represented as #P < 0.05 as determined using a pairwise post-test; n.s. = not significant.
We next aim to further define how MuRF1 inhibits T3-induced cardiomyocyte hypertrophy mechanistically. We focused our attention on MuRF1’s regulation of thyroid receptor (TR) TRα1 activity, as it is the only isoform in cardiomyocytes that stimulates growth (the less common TRα2, and TRβ1 are present but lack this activity) (, Sabatino et al. 2007). In these studies, we identified that increasing MuRF1 expression significantly inhibited TRα-induced TRE-luciferase activity (Fig. 2A). As another way to measure TRα activity, we next evaluated how increasing MuRF1 would affect TR target genes and identified that MuRF1 significantly inhibited sarco/endoplasmic reticulum Ca2+-ATPase (SERCA expression (Fig. 2B)). Other well-known TRα targets (αMHC and ANT) were not increased in these studies, which may be due to the limited 30-min T3 stimulation (Fig. 2B). As MuRF1’s inhibition of TRα could result from poly-ubiquitination and proteasome-dependent degradation (Kedar et al. 2004), we performed immunoblot analysis of TRα levels under conditions where MuRF1 inhibited its activity and found that increasing MuRF1 did not change TRα protein concentration (Fig. 2C). As both increasing and decreasing MuRF1 expression in T3-induced cardiomyocyte hypertrophy did not affect the steady-state levels of TRα (Fig. 2C), we were able to rule out MuRF1-mediated TRα degradation as a mechanism.
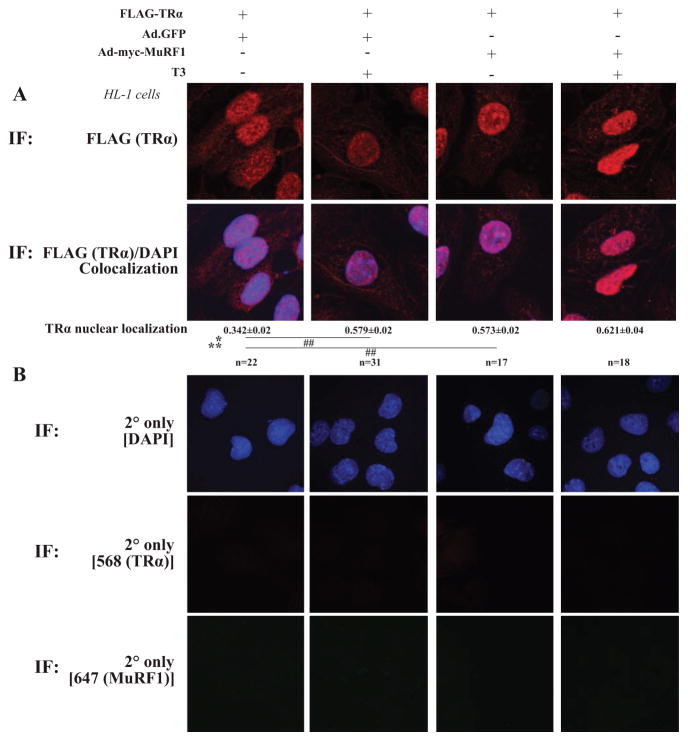
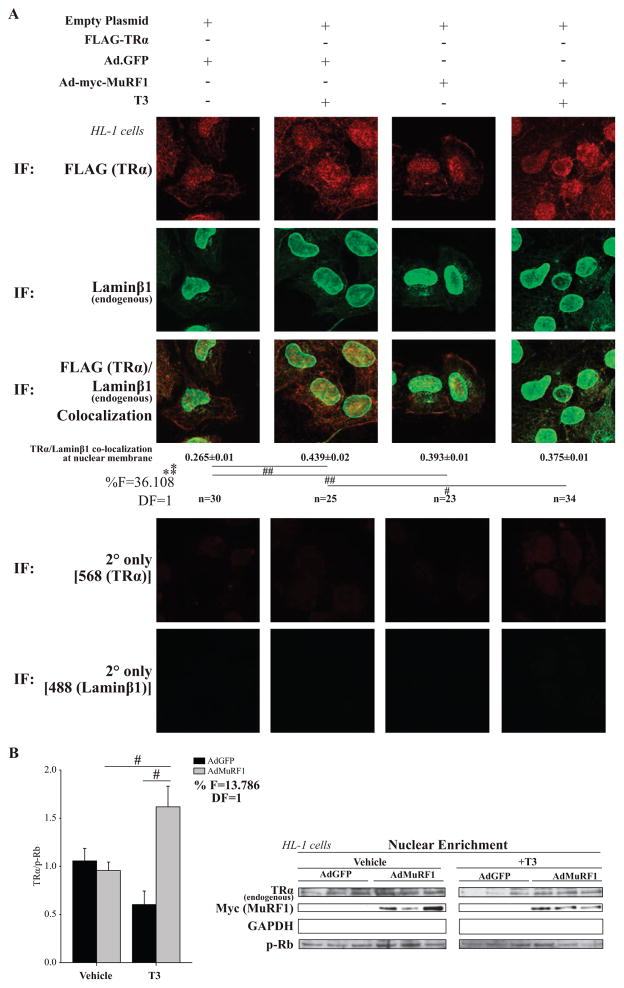
In recent studies, we demonstrated that MuRF1 inhibited PPARα activity similarly without affecting PPARα protein concentration (Rodriguez et al. 2015). Using confocal microscopy, we found that increasing MuRF1 removed nuclear PPARα in a majority of the cells that subsequent studies found was due to MuRF1-targeted increased in nuclear export (Rodriguez et al. 2015). Therefore, we next determined whether MuRF1 inhibits T3-induced TRα1 activity by altering TRα1’s nuclear localization. HL-1 cardiomyocytes were transfected with TRα1 and/or MuRF1 (+/−T3) and analyzed by confocal microscopy. Interestingly, T3 altered the nuclear localization of TRα1 (Fig. 3A, column 4) compared with controls (Fig. 3A, column 3). Specifically, it appeared that MuRF1 altered TRα1 nuclear localization by moving it to the nuclear periphery, a region closely aligned with the endoplasmic reticulum and microtubule anchoring complexes. To confirm this localization, we first identified the co-localization of the nuclear proteins lamin β1 and desmin with TRα1 using confocal immunofluorescent analysis (Fig. 4A). Increasing MuRF1 expression significantly increased the co-localization of TRα1 with nuclear lamin β1 (Fig. 4), while similarly increasing the co-localization of TRα1 with desmin (Supplementary Fig. S2). Lastly, we assessed MuRF1’s effects on TRα1 nuclear localization by isolating the cardiomyocyte nuclear fraction by differential centrifugation and measuring the amount of TRα1 by immunoblot (Fig. 4B). Together, these studies demonstrate that increasing MuRF1 sequestered TRα1 in the nucleus, to inhibit TRα1 nuclear activity.
Figure 3.
MuRF1 increases TRα localization to nuclear region in cells treated with T3. HL-1 cardiomyocytes were transfected with FLAG-TRα plasmid transduced with AdMuRF1 (or AdGFP) and treated with 16.6 nM T3 for 2 h. (A) Confocal immunofluorescent images used to define TRα (anti-FLAG, red) and nuclei (DAPI, blue) co-localization. (B) Secondary-only fluorescence controls. Data are represented as mean ± S.E.M. (n = 3 independent experiments). For TRα/DAPI co-localization, a two-way ANOVA test was used to determine statistical significance. *Significance at the level of adenovirus group, **Significance on the level of treatment group. Significance between groups is represented as ##P < 0.001 as determined using a pairwise post-test, 400× final magnification.
Figure 4.
MuRF1 inhibits T3-induced TRα localization to the nuclear envelope (lamin β1). HL-1 cardiomyocytes transfected with FLAG-TRα plasmid (empty FLAG = control), transduced with AdMuRF1 (AdGFP = control) for 4 h, and treated with 16.6 nM T3 for 2 h were assayed by confocal microscopy. (A) TRα (anti-FLAG, red) and endogenous lamin β1 (green) co-localization at the nuclear membrane. A two-way ANOVA test was used to determine statistical significance. *Significance on the level of adenovirus group, **Significance on the level of treatment group. The F statistic and degrees of freedom (DF) were reported when dependence between groups was found to be a significant source of variation. Significance between groups is represented as #P < 0.05, ##P < 0.001, as determined using a pairwise post-test. (B) HL-1 cardiomyocytes were transduced with AdMuRF1 (24 h) and treated with 16.6 nM T3 (2 h) followed by fractionation. Immunoblot of endogenous TR and myc-MuRF1 after T3 treatment (p-Rb used as a loading control) in nuclear fraction. Data represent three independent experiments. A two-way ANOVA test was used to determine statistical significance with a pairwise post-test. *Significance on the level of adenovirus group, % Significant interaction between adenovirus and treatment groups, #Difference from treatment control group (criterion for cutoff; P < 0.05). Data are presented as mean ± S.E.M., 400× final magnification.
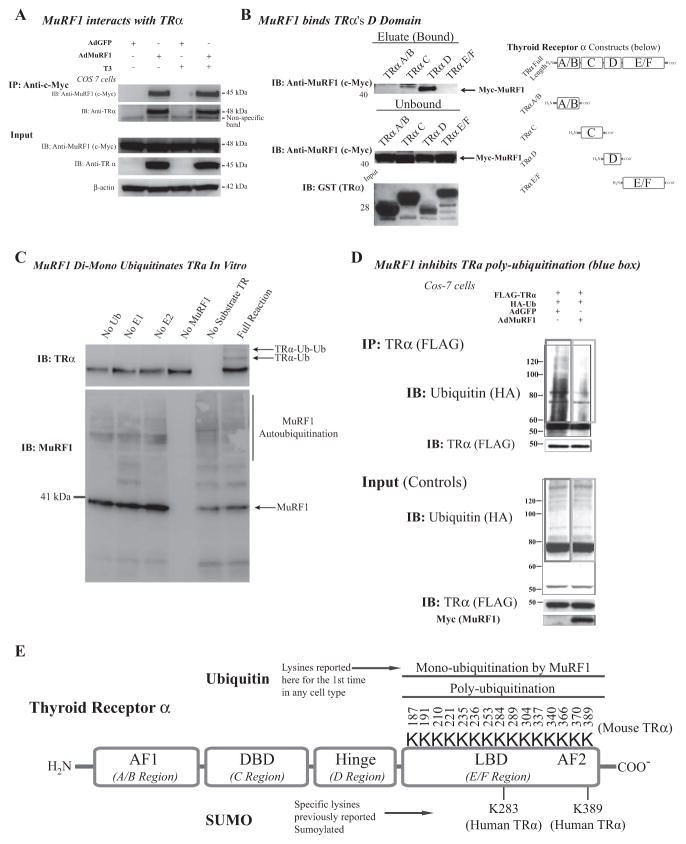
To assess whether TRα is a MuRF1 substrate, we next determined whether MuRF1 interacted directly with TRα by transfecting cells with MuRF1 and immunoprecipitating the myc-labeled MuRF1 (Fig. 5A). To determine the specific region of TRα that MuRF1 interacts, we next performed a pull-down assay using recombinant TRα domains to identify their ability to interact with full-length MuRF1 (Fig. 5B). We found that MuRF1 binds the TRα hinge (or D, as denoted on the diagram) domain of the receptor (Fig. 5B, right). With evidence that MuRF1 interacts with TRα, we next tested MuRF1’s ability to ubiquitinate TRα in vitro. We identified that MuRF1 catalyzes TRα mono-ubiquitination (Fig. 5C), a posttranslational modification that does not target substrate for degradation, consistent with our initial findings in cardiomyocytes.
Figure 5.
MuRF1 interacts with and mono-ubiquitinates TRα. (A) Cos-7 cells transduced with AdMuRF1 and treated with 16.6 nM T3 for 2 h were used in analysis. Whole cell lysates were immunoprecipitated with myc-MuRF1 and probed for endogenous TRα to evaluate protein—protein interaction as shown in the blot (left). (B) GST-tagged recombinant peptides for each TR domain were used in an immunoprecipitation assay with myc-MuRF1 followed by immunoblot to establish the specific domain bound by MuRF1 (right). Blots shown on top right show the MuRF1-bound regions of TR (predominantly D); the top blot shows the bound myc-MuRF1 (eluate) for each domain construct. The middle blot shows unbound myc-MuRF1 and the bottom membrane is a Ponceau stain of each isolated peptide to confirm expression of the expected molecular weight protein (constructs were also sequenced to verify product, data not shown). At the far right is a diagram indicating the various domains, for reference. (C) An in vitro biochemical assay was used to assess ubiquitination with exogenous MuRF1 and was detected by immunoblot with HA-tagged Ub antibody. (D) Cos-7 cells co-transfected with HA-Ub and FLAG-TRα and transduced with AdMuRF1 (or AdGFP) at MOI 25 for 4 h; whole cell lysates were submitted to immunoprecipitation of FLAG-TRα followed by immunoblot for HA-Ub. The red box highlights ubiquitination in controls, whereas the blue box demonstrates the decrease in ubiquitination when MuRF1 expression is increased. Empty vector control (FLAG and HA) experiment blots are shown in left panel. (E) Diagram of TRα with known protein domains indicated in addition to the lysines within the E/F (ligand binding) domain with the corresponding (murine) amino acid sequence position (total of 15 residues). This region includes two lysines known to be targeted for regulation in the human isoform of TR for SUMOylation (K283 and K389).
With evidence that MuRF1 mono-ubiquitinated TR in a cell-free system, we next aim to identify these in cells. We observed an unexpected complication of identifying MuRF1’s mono-ubiquitination in cells where a strong background of TRpoly-ubiquitination occurs (Fig. 5D). As described in cancer cells, poly-ubiquitination is a common way in which thyroid hormone receptors have been described natively (Fig. 5E, as described in TRβ) (Dace et al. 2000, Kenessey & Ojamaa 2005). As immunoprecipitated TRα was polyubiquitinated in control cells (Figure 5D, red box), we hypothesized that the poly-ubiquitination competed for the sample lysines that MuRF1 mono-ubiquitinated. Support for this hypothesis came from studies with increased MuRF1 (AdMuRF1), where there was decreased poly-ubiquitination (Fig. 5D, blue box). While the identification of a mono-ubiquitinated TRα was not achieved, we did identify the common dichotomy competing mono- and poly-ubiquitin for substrates, known to be dependent on the types of E2 present and ratios of E3 to substrates (He et al. 2015, Kim et al. 2007, Lai et al. 2001).
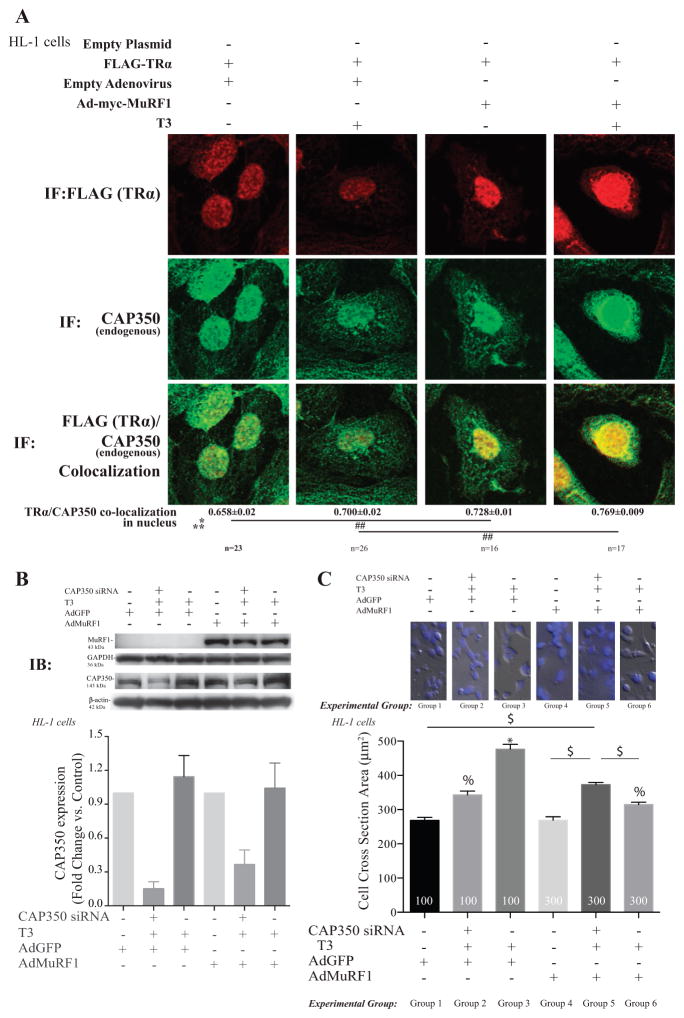
The centrosome-associated protein (CAP) CAP350 has recently been identified as protein involved in regulating nuclear receptor activity, including PPAR (Patel et al. 2005). Initially, we analyzed MuRF1’s effects on the co-localization of endogenous CAP350 and TRα in cardiomyocytes by confocal imaging (Fig. 6A). We found that increasing MuRF1 significantly increased nuclear CAP350 and the co-localization of CAP350 and TRα (Fig. 6A, right two columns). The increased co-localization of CAP350 and TRα driven by increasing MuRF1 was most apparent in the perinuclear region following T3 stimulation (Fig. 6A, in yellow). To solidify CAP350’s mechanistic relationship to MuRF1’s inhibition of TRα, we next knocked down CAP350 (to < 50%, Fig. 6B) and determined its effect on cardiomyocyte cross-sectional area (Fig. 6C). In control cells, CAP350 knock-down attenuated the T3-induced cardiomyocyte hypertrophy (experimental group 3 vs group 2, Fig. 6C). The MuRF1 attenuation of T3-stimulated cardiomyocyte growth (experimental group 6 vs group 3, Fig. 6C) was significantly overcome (i.e increased in cross-sectional area size) with CAP350 knock-down (experimental group 5, Fig. 6C). Together, these studies demonstrate a role of CAP350 in MuRF1’s regulation of TR in cardiomyocytes.
Figure 6.
MuRF1 inhibition of T3-induced cardiomyocyte hypertrophy is dependent on the centrosome-associated protein 350 (CAP350). (A) Confocal immunofluorescent images of TRα (anti-FLAG, red) and CAP350 (green) in cells transduced/transfected with vectors indicated and treated with T3 for 2 h (Fig. 3 for fluorescence controls). Data are reported as the mean ± S.E.M. (n = 3 independent experiments). A two-way ANOVA test was used to determine statistical significance. *Significance on the level of adenovirus group, **Significance on the level of treatment group. Significance between groups is represented as ##P < 0.001 as determined using a pairwise post-test. (B) Knock down of CAP350 in HL1 cardiomyocytes using siRNA achieved >50% reduction in protein by immunoblot analysis. (C) Cell cross-sectional area analysis of T3-stimulated HL-1 cells with increased MuRF1 (AdMuRF1) and CAP350 knock-down to identify the role of CAP350 in MuRF1-mediated inhibition of T3-induced hypertrophy. Data are reported as the mean ± S.E.M. (n = 2 independent experiments). A one-way ANOVA test was used to determine statistical significance. Significance between groups is represented as *P < 0.05 vs all other groups, % P < 0.05 vs groups 1, 4, and $P < 0.05 in indicated comparisons, 400× final magnification.
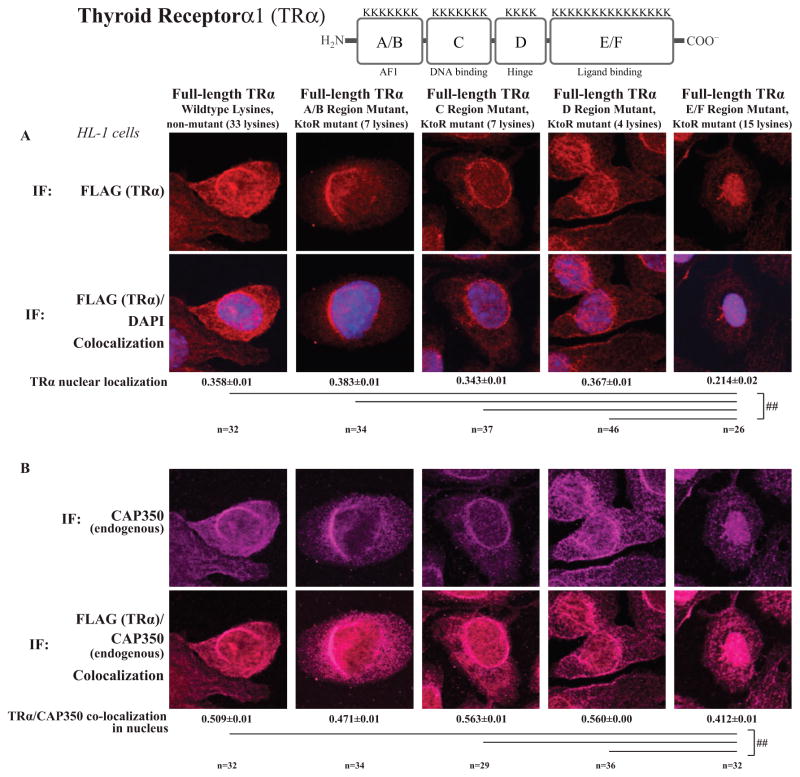
We next aim to identify the region within TRα protein that MuRF1 ubiquitinates to modify its activity. A well-known challenge to studying protein ubiquitination is its heterogeneous nature; when a major ubiquitination site (lysine) is mutated, it is common for nearby lysines to be alternatively targeted (Xu & Jaffrey 2013). In practice, this means that mutating single lysines does not alter ubiquitination, even if the major site is mutated, as alternative sites are targeted. As the goal of mutating a lysine would be to prevent specific ubiquitination, we took an alternative approach and mutated lysine residues in specific regions of the TR protein. Four full-length TRα constructs were created with lysine residues mutated to arginine (KR) (Supplementary Fig. S3): TRα A/B (AF1, N-terminus), TRα C (DNA-binding), TRα D (hinge), and TRα E/F (ligand-binding). These TRα constructs were then expressed in cardiomyocytes and assessed for their response to MuRF1 alterations in localization by confocal microscopy (). Increasing MuRF1 expression localized the TRA/B, C, and D lysine mutants to the perinuclear region as observed with the native receptor (Fig. 7A). In contrast, cells expressing the E/F TR domain mutant exhibit a distinctly nuclear localization of the receptor rather than a perinuclear one as observed with the native form implying that MuRF1 ubiquitinates residues in this domain to negatively regulate TR function (Fig. 7A, far right). Interestingly, when the E/F TR domain mutant was expressed, CAP350 localization was affected, moving from a perinuclear to a pan-nuclear distribution (Fig. 7B, far right). Together, these studies illustrate that MuRF1 inhibits TRα activity by mono-ubiquitinating it in the E/F region, resulting in an interaction with CAP350 and nuclear sequestration in cardiomyocytes.
Figure 7.
MuRF1-mediated TRα nuclear accumulation following T3 treatment is dependent on TRα ligand-binding domain (E/F region) lysines. HL-1 cardiomyocytes were transfected with plasmids containing FLAG-TRα with various lysine to arginine (KR) mutants (Supplementary Fig. S2), transduced with AdMuRF1 for 4 h, and treated with 16.6 nM T3 for 2 h. (A) Nuclear localization of TRα was assessed using confocal immunofluorescent imaging of TRα (anti-FLAG, red) and nuclei (DAPI, blue). (B) MuRF1 (anti-myc, green) and TRα at the nucleus was assessed TRα (anti-FLAG, red). (C) TRα (anti-FLAG, red) and endogenous CAP350 (green). Data represent the mean ± S.E.M. A one-way ANOVA test was used to determine statistical significance. ##P < 0.001 as determined by a pairwise post-test, 400× final magnification.
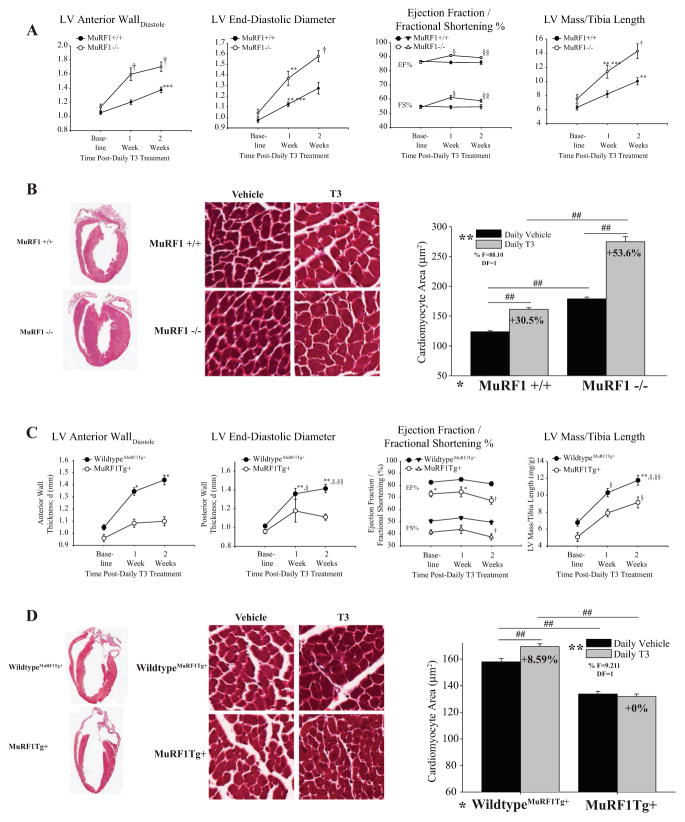
To determine the physiological relevance of these mechanistic findings in vivo, we next challenged MuRF1−/− and MHC-MuRF1 cardiomyocyte-specific transgenic mice (MuRF1Tg+) with daily T3 i.p. for 2 weeks. We followed the cardiac phenotype by conscious echocardiography and histological analysis. Echocardiographic and histological assessment identified that T3 induced an exaggerated cardiac hypertrophy in MuRF1−/− compared with sibling-matched wild-type controls, evidenced by increases in left ventricular anterior wall thickness and LV mass (Fig. 8A, Table 1 for full echocardiographic data) and cardiomyocyte area (Fig. 8B). Interestingly, MuRF1−/− had an enhanced systolic function, reflecting increases in contractility and reflective of the T3-induced enhancements driven by increases in SERCA and phospholamban (Periasamy et al. 2008). Overall, the MuRF1−/− mice had a 53.6% increase in cardiomyocyte area compared with the 30.5% increase found in the wild-type controls (Fig. 8B). Consistent with these findings, the MuRF1Tg+ mice challenged with T3 had an attenuated cardiac hypertrophy response in vivo (Fig. 8C). Echocardiographic and histological assessment identified that MuRF1Tg+ mice had significantly less LV anterior and posterior wall thickness compared with sibling-matched wild-type controls (Fig. 8A, Table 2 for full echocardiographic data) as well as cardiomyocyte area (Fig. 8D). Significant reductions in systolic function over time were observed (Fig. 8C), consistent with an increase in pathological hypertrophy, identified by increases in BNP, α1-skeletal muscle actin, and ANF (data not shown). The observed effects on TH-induced physiological hypertrophy in our mouse models were most likely attributable to MuRF1 activity as circulating concentrations of T3/T4 were not altered in MuRF1−/− or MuRF1Tg+ mice (Supplementary Fig. S5A and B). Furthermore, MCT8 and -10 expressions (transporters involved in the uptake and efflux of T3) were not significantly different across the genotypic groups (Supplementary Fig. S5C and D). All studies included parallel PBS-injected controls, with no alterations seen (as detailed in Supplementary Tables S1 and S2). Overall, these studies demonstrate that the MuRF1−/− mice exhibited an exaggerated cardiac hypertrophy, while MuRF1Tg+ hearts lacked the development of cardiac hypertrophy in response to T3, consistent with a role for MuRF1 in inhibiting TRα activity in vivo. With no evidence for alterations in steady-state TRα protein levels in vivo (Supplementary Fig. S4), like those found in vitro (Fig. 2C), cardiomyocyte MuRF1’s inhibition of TRα is relevant in the intact heart illustrating alternative mechanisms that could be targeted to protect against heart failure in a tissue-specific manner.
Figure 8.
MuRF1’s attenuation of TRα activity is identified in both MuRF1−/− and cardiac (αMHC)-MuRF1Tg+ mice in vivo after T3 stimulation. (A) Conscious echocardiographic data from wild-type (MuRF1+/+) and knockout (MuRF1−/−) receiving daily T3 i.p. (vehicle control = PBS) for 2 weeks (Table 1 for complete complementary echocardiographic data). (B) Gross and histological analysis of H&E and Masson’s trichrome hearts (100× magnification) from T3-treated MuRF1−/− mice. (C) Echocardiographic data from T3-treated wild-type MuRF1Tg+ and MuRF1Tg+ (Table 2 for complete complementary echocardiographic data). (D) Gross and histological analysis of H&E and Masson’s trichrome hearts (100× magnification) from T3-treated MuRF1Tg+ mice. A two-way ANOVA test was used to determine statistical significance using a pairwise post-test. For echocardiographic data: †P < 0.001 vs all other groups, ***P < 0.001 vs matched wild type, **P < 0.001 vs MuRF1+/+ and MuRF1−/− baseline; §P < 0.001 vs all other groups except MuRF1−/− +T3 at 2 weeks, §§P < 0.001 vs all other groups except MuRF1−/− +T3 at 1 week. Data on cardiomyocyte area were generated from three mice per group by averaging values from 400 cardiomyocytes per animal and statistical significance between indicated groups is designated by #P < 0.05, ##P < 0.001. The F statistic and degrees of freedom (DF) were reported when dependence between groups was found to be a significant source of variation, 20× final magnification.
Table 1.
High-resolution trans-thoracic echocardiography performed on conscious MuRF1−/− and age-matched wild-type mice at baseline, 1 week, and 2 weeks after daily i.p. T3 treatment. Data represent means ± S.E.M. Groups were first analyzed with a one-way analysis of variance, and if significant, analyzed with a Holm–Sidak test for pairwise comparisons.
| MuRF1+/+ Baseline n = 10 | MuRF1−/− Baseline n = 12 | MuRF1+/+ Thyroid hormone 1 week n = 10 | MuRF1−/− Thyroid hormone 1 week n = 12 | MuRF1+/+ Thyroid hormone 2 weeks n = 10 | MuRF1−/− Thyroid hormone 2 weeks n = 12 | |
|---|---|---|---|---|---|---|
| LVEDD (mm) | 3.03 ± 0.11 | 3.08 ± 0.12 | 3.13 ± 0.13 | 3.87 ± 0.15 | 3.11 ± 0.11 | 3.00 ± 0.14 |
| PWTD (mm) | 0.97 ± 0.02 | 1.04 ± 0.04 | 1.12 ± 0.02** | 1.37 ± 0.07**,*** | 1.28 ± 0.05 | 1.58 ± 0.05† |
| AWTS (mm) | 1.77 ± 0.04 | 1.82 ± 0.06 | 1.97 ± 0.02 | 2.23 ± 0.08 | 2.23 ± 0.07 | 2.44 ± 0.06 |
| PWTS (mm) | 1.58 ± 0.01 | 1.65 ± 0.07 | 1.81 ± 0.06 | 2.10 ± 0.08 | 1.93 ± 0.08 | 2.18 ± 0.07 |
| LV Vol; d (L) | 36.5 ± 3.3 | 38.2 ± 3.8 | 39.8 ± 3.7 | 32.6 ± 3.8 | 39.0 ± 3.4 | 36.3 ± 4.1 |
| LV Vol; s (L) | 4.8 ± 0.5 | 5.5 ± 0.8 | 5.8 ± 0.8 | 3.0 ± 0.5† | 5.7 ± 1.0 | 4.0 ± 0.7 |
| LV mass (mg) | 107.3 ± 6.1 | 124.8 ± 11.1 | 139.4 ± 7.6 | 187.8 ± 17.6**,*** | 170.3 ± 8.8** | 237.4 ± 19.8† |
| LV mass/BW (mg/g) | 4.1 ± 0.3 | 4.7 ± 0.2 | 5.2 ± 0.3 | 6.4 ± 0.2**,*** | 5.9 ± 0.3** | 8.0 ± 0.2† |
| BW (g) | 26.1 ± 0.8 | 26.7 ± 2.5 | 27.3 ± 0.7 | 29.0 ± 1.9 | 28.9 ± 1.1 | 29.5 ± 1.8 |
| HR (bpm) | 665 ± 9 | 670 ± 13 | 708 ± 21 | 727 ± 11** | 730 ± 14** | 729 ± 10** |
P < 0.001 vs all other groups.
P < 0.001 vs matched wild type.
P < 0.001 vs MuRF1+/+ and MuRF1−/− baseline.
P < 0.001 vs all other groups except MuRF1−/− with T3 at 2 weeks.
P < 0.001 vs all other groups except MuRF1−/− with T3 at 1 week. HR, heart rate; ExLVD, external left ventricular diameter; bpm, heart beats per minute; AWTD, anterior wall thickness in diastole; AWTS, anterior wall thickness in systole; PWTD, posterior wall thickness in diastole; PWTS, posterior wall thickness in systole; LVEDD, left ventricular end-diastolic dimension; LVESD, left ventricular end-systolic dimension; FS, fractional shortening, calculated as (LVEDD–LVESD)/LVEDD ×100; EF%, ejection fraction calculated as (end Simpson’s diastolic volume – end Simpson’s systolic volume)/end Simpson’s diastolic volume ×100, ND, not determined
Table 2.
High-resolution trans-thoracic echocardiography performed on conscious MuRF1Tg+ and age-matched wild-type mice at baseline, 1 week, and 2 weeks after daily i.p. T3 treatment. Data represent means ± S.E.M. Groups were first analyzed with a one-way ANOVA, and if significant, analyzed with a Holm–Sidak test for pairwise comparisons.
| MuRF1Tg+ Wild type Baseline n = 16 | MuRF1Tg+ Baseline n = 11 | MuRF1Tg+ wild type Thyroid hormone 1 week n = 16 | MuRF1Tg+ Thyroid hormone 1 week n = 10 | MuRF1Tg+ wild type Thyroid hormone 2 weeks n = 16 | MuRF1Tg+ Thyroid hormone 2 weeks n = 11 | |
|---|---|---|---|---|---|---|
| PWTD (mm) | 1.02 ± 0.02 | 0.96 ± 0.02 | 3.15 ± 0.10 | 3.27 ± 0.11 | 3.20 ± 0.11 | 3.54 ± 0.13 |
| AWTS (mm) | 1.70 ± 0.05 | 1.54 ± 0.06 | 1.48 ± 0.07§,§,** | 1.80 ± 0.09*** | 1.64 ± 0.10§,* | 2.23 ± 0.14 |
| LVESD (mm) | 1.53 ± 0.07 | 1.88 ± 0.14 | 1.36 ± 0.06 | 1.18 ± 0.12 | 1.41 ± 0.04 | 1.11 ± 0.03* |
| PWTS (mm) | 1.58 ± 0.04 | 1.33 ± 0.06 | 1.86 ± 0.05§ | 1.60 ± 0.03 | 1.92 ± 0.05§,§§,* | 1.50 ± 0.06 |
| LV Vol; d (L) | 37.4 ± 2.4 | 41.6 ± 4.6 | 40.3 ± 3.0 | 44.1 ± 3.7 | 42.1 ± 3.3 | 53.4 ± 4.6 |
| LV Vol; s (L) | 6.8 ± 0.8 | 12.0 ± 2.1 | 6.3 ± 0.9 | 10.4 ± 1.1 | 8.5 ± 1.4 | 17.9 ± 2.4†,§,§§ |
| LV mass (mg) | 116.6 ± 7.3 | 107.4 ± 9.3 | 178.5 ± 8.3 | 137.7 ± 7.4 | 201.8 ± 110.8§,§§,** | 155.0 ± 10.2 |
| LV mass/BW (mg/g) | 4.1 ± 0.3 | 5.1 ± 0.7 | 6.2 ± 0.2§ | 5.2 ± 0.3 | 6.7 ± 0.2§,§§,** | 5.5 ± 0.3 |
| BW (g) | 29.3 ± 1.3 | 27.7 ± 1.8 | 29.9 ± 1.1 | 26.9 ± 1.5 | 29.8 ± 0.9 | 28.1 ± 1.1 |
| HR (bpm) | 711 ± 13§§ | 679 ± 10§§ | 802 ± 9 | 802 ± 7 | 798 ± 13 | 768 ± 6§ |
P < 0.001 vs all other wild-type groups.
P < 0.001 vs matched wild type.
P < 0.001 vs MuRF1Tg+ 2 weeks after T3 treatment.
P < 0.001 vs MuRF1Tg+ baseline.
P < 0.001 vs baseline groups
P < 0.001 vs 1 week T3 groups. HR, heart rate; ExLVD, external left ventricular diameter; bpm, heart beats per minute; AWTD, anterior wall thickness in diastole; AWTS, anterior wall thickness in systole; PWTD, posterior wall thickness in diastole; PWTS, posterior wall thickness in systole; LVEDD, left ventricular end-diastolic dimension; LVESD, left ventricular end-systolic dimension; FS, fractional shortening, calculated as (LVEDD–LVESD)/LVEDD ×100; EF%, ejection fraction calculated as (end Simpson’s diastolic volume – end Simpson’s systolic volume)/end Simpson’s diastolic volume ×100, ND, not determined
Discussion
Understanding how the thyroid hormone receptor α is regulated is critical to our understanding of heart disease. Inhibiting TRα in myocardial infarction experimentally impairs post-ischemic performance due to activation of p38 and an increased phospholamban (Mourouzis et al. 2013b). Conversely, T3 therapy protects the heart against ischemia—reperfusion injury in a TRα-dependent manner (Pantos et al. 2011). T3 also seems to counteract the maladaptive responses implicated in adrenergic induced cardiomyocyte remodeling. Stimulation of cardiomyocytes with phenylephrine results in increased fetal gene expression (βMHC); co-stimulating cardiomyocytes with phenylephrine and T3 blocks these maladaptive changes, while increasing TRα expression in the nucleus and cytosol (Pantos et al. 2008). In models of myocardial infarction (acute coronary artery ligation) leading to compensated heart failure, a distinct pattern of TRα expression was found during the course of post-ischemic remodeling (Pantos et al. 2010). TRα was upregulated initially, then markedly decreased during the transition to decompensated heart failure, resulting in tissue hypothyroidism (Pantos et al. 2010). While circulating T3 and T4 levels remained unchanged, a link between mTOR and TRα was identified (Pantos et al. 2010). Together, these studies illustrate the critical role the TRα plays in heart failure and how regulating TRα expression can critically affect disease progression and severity.
The regulation of nuclear hormones by the ubiquitin proteasome system occurs in a myriad of ways (Ismail & Nawaz 2005). Some nuclear hormone receptors are degraded in both the presence and absence of their cognate ligands, such as the estrogen, progesterone, and glucocorticoid receptors (Hoeck et al. 1989, Lange et al. 2000 ; Nawaz et al. 1999). Alternatively, the androgen receptor and the PPARα are targeted to the proteasome in the absence of ligand (Hirotani et al. 2001, Li et al. 1999, Lin et al. 2002). The closely related receptors that heterodimerize with PPARs, such as RARγ and RXR, are also degraded by the ubiquitin proteasome system with fewer details available (Kopf et al. 2000). We have recently extended these findings to demonstrate that the muscle-specific ubiquitin ligase MuRF1 specifically inhibits PPARα, but not PPARβ/δ or PPARγ1 activity in vivo (Rodriguez et al. 2015). In contrast to previous studies on nuclear receptors, we identified that MuRF1 had an inhibitory effect on PPARα in cardiomyocytes by enhancing nuclear export, not degradation. By multi-mono-ubiquitination of lysines adjacent to the newly identified nuclear export sequence in PPARα, MuRF1 inhibited PPARα-regulated fatty acid oxidation both in vitro and in vivo (Rodriguez et al. 2015). Complementary to these studies, the closely related muscle-specific ubiquitin ligase MuRF2 was found to inhibit PPARγ1 (He et al. 2015), while MuRF3 inhibits predominantly PPARβ activity in vivo through mono-ubiquitination of substrates (Quintana et al. 2015).
While cardiac MuRF1 has multiple roles in the heart, a consistent theme of its activity has been its anti-hypertrophic activity (Wadosky et al. 2014, Willis et al. 2007, Willis et al. 2009a, Willis et al. 2014) and regulation of metabolism. In pressure overload-induced cardiac hypertrophy, MuRF1 inhibits pathological hypertrophy by inhibiting the serum response transcription factor (Willis et al. 2007, Willis et al. 2009a, Willis et al. 2014). In physiological hypertrophy induced by exercise (IGF-1), MuRF1 transcriptionally inhibits cardiomyocyte growth by inhibiting cJun-mediated increases in IGF-1 signaling proteins (Wadosky et al. 2014), by preferentially recognizing phospho-cJun, poly-ubiquitinating, and targeting it for proteasome-dependent degradation in vivo (Li et al. 2011). Metabolically, MuRF1 has been found to ubiquitinate oxidized creatine kinase, mediating its turnover in vivo. Increasing MuRF1 decreases creatine kinase activity significantly in the intact heart, resulting in a reduction of this critical transporter of ATP from the mitochondria to the sarcomere, resulting in heart failure (Willis et al. 2009b, Zhao et al. 2007). MuRF1 also inhibits PPARα (Rodriguez et al. 2015), interacts with a host of energy-related proteins (Witt et al. 2005), and alters the cardiac metabolome in vivo (Banerjee et al. 2015). Building upon this larger theme, this study demonstrates MuRF1’s anti-hypertrophic role in T3-mediated cardiomyocyte growth, with broader implications in T3-regulated metabolic rates critical to cardiac function in health and disease.
The link between MuRF1 and the centrosome-associated protein 350 (CAP350) stems from the observation that MuRF1 alters nuclear TRα distribution in a similar manner as CAP350 does with PPARα (Patel et al. 2005). Although the role of CAP350 in regulating nuclear hormone receptors (such as TRα) is established (Lelievre et al. 2008, Patel et al. 2005), the mechanistic details have yet to be elucidated. In previous studies, the recently defined centrosome- and cytoskeleton-associated protein CAP350 was found to inhibit PPARα activity by redirecting PPARα’s nuclear distribution to the perinuclear area with punctate distribution (Patel et al. 2005). In this study, we similarly observed that MuRF1 altered the nuclear distribution of TR (Fig. 3A), which led to subsequent studies to delineate the underlying mechanistic relationship between MuRF1’s anti-hypertrophic activity with T3 stimulation and MuRF1’s reliance upon CAP350 for preventing hypertrophic growth (Fig. 6C). The present studies illustrate that MuRF1’s attenuation of T3-induced cardiomyocyte growth is dependent on TRα mono-ubiquitination of the ligand-binding domain (Fig. 7) and interaction with CAP350 (with significantly smaller cardiomyocytes with MuRF1 and CAP350 present, Fig. 6C, group 6). Our working hypothesis on the role of MuRF1 in this process is that MuRF1 mono-ubiquitinates TRα to enhance its affinity for CAP350, which sequesters TRα-Ub in nuclear compartments making it inaccessible to DNA promoters to diminish transcriptional activity.
Protein mono-ubiquitination that results in an enhanced affinity to other proteins is a common molecular process noted in the biology of cancer. An established example is the ubiquitin ligase MDM2, which mediates p53 mono-ubiquitination. In this system, p53 mono-ubiquitination alters its conformation to preferentially bind CRM1 (a subunit of the nuclear export apparatus), which then enhances p53 nuclear export resulting in activity inhibition (Brooks et al. 2007, Lee & Gu 2010). This is an emerging, common theme, further illustrated with the estrogen receptor (ER), when the BRCA1 ubiquitin ligase mono-ubiquitinates it to inhibit its activity (Ma et al. 2010) and the suppression of DNA polymerase Eta, when mono-ubiquitinated by the Pirh2 ubiquitin ligase (Jung et al. 2011). The present studies demonstrate a novel role for CAP350 activity in the heart and an emerging role in regulating TRα activity that has not been previously reported.
Posttranslational modification of TRα by ubiquitin has not previously been described; however, its regulation by the closely related small ubiquitin-related modifier (SUMO) has been in cancer. Paralleling the ubiquitination process, SUMOylation involves the addition of a single SUMO modification, added by the cooperation of the SUMO E1, E2, and E3 (SUMO ligase) enzymes. Interestingly, SUMO modification alters cellular processes, including nuclear transport, transcriptional regulation, apoptosis, and protein stability without degrading its substrate. SUMOylated TRα has been reported in both liver (HepG2) and pituitary (GH3) cell lines (Liu et al.2012), mediated by the SUMO ligase PIAS1. In the absence of ligand, PIAS1 SUMOylates TRα, whereas the SUMO ligase PIASx SUMOylates TRβ only in the presence of ligand (Liu et al. 2012). Both SUMO1 and SUMO3 (two different types of SUMO modification) have been reported in T3-dependent gene regulation, closely matching TR activity and cofactor recruitment to the TRE in ChIP assays (Liu et al. 2012).
This study describes a non-canonical ubiquitination (mono-ubiquitination) of TRα, which parallels the process of TRα SUMOylation. Both SUMOylation and mono-ubiquitination occur at specific lysines and do not result in degradation of TRα. SUMOylation and mono-ubiquitination also inhibit T3-induced TRα activity. Interestingly, TRα is SUMOylated at lysines 284 and 389 (Liu et al. 2012), both of which are within the ligand-binding domain (E/F region) of TRα, consistent with our findings that MuRF1 interacts with and catalyzes the addition of ubiquitin in the E/F domain of TRα are critical to regulation by MuRF1 (Fig. 7, lysines 187 through 389, Supplementary Fig. S3 for additional information). The physiological importance of SUMOylation in health and disease is not known; however, the common T3-mediated growth functions of TRα make it likely that SUMOylation in hepatocytes and pituitary parallel the critical regulation of mono-ubiquitination in cardiomyocytes reported here.
The exaggerated hypertrophy in the MuRF1−/− hearts is consistent with an enhanced cardiomyocyte TRα activity (Fig. 1A, Fig. 8A). Similarly, the lack of cardiac hypertrophy in the MuRF1Tg+ hearts was consistent with in vitro findings (Fig. 1B and C, Fig. 8D). However, the baseline MuRF1Tg+ heart function worsens after 2 weeks of T3 treatment (Fig. 8C). One explanation for this may be the energetic defects seen in the MuRF1Tg+ mice, such as decreased creatine kinase activity and altered fatty acid oxidation (Rodriguez et al. 2015, Willis et al. 2009b). Conversely, the enhanced MuRF1−/− function seen with T3 stimulation (Fig. 8A) may reflect an athletic phenotype previously identified in the MuRF1−/− hearts that may be related (Hwee et al. 2014, Wadosky et al. 2014), but not fully due to enhanced T3 activity resulting from decreased ubiquitination.
This study may offer several clinically relevant insights into approaches to treating heart failure. TH induces molecular changes that benefit the failing heart, including the stimulation of pro-survival pathways such as Akt and mTOR (Mourouzis et al. 2012) that is accompanied by improved cardiac function, paralleling changes induced by exercise, mediated by IGF-1 (Ojamaa 2010), to drive increases in contractile proteins critical to cardiac function and performance (Periasamy et al. 2008). Exercise has also been shown to increase myocardial TR expression and T3 levels in a small cohort of HF patients with ventricular assist devices (Adamopoulos et al. 2013), underscoring the role for TH in promoting positive adaptive remodeling of the injured heart. Exogenous approaches to supplementing TH also hold promise as low-dose T3 replacement has been shown to restore cardiac T3 levels and ameliorate dysfunction in preclinical models of heart failure (Weltman et al. 2014, Zhang et al. 2014). Additionally, long-term physiological T3 supplementation as an adjunct therapy in hypertensive heart disease has been beneficial in preclinical studies (Weltman et al. 2014) and may enhance ventricular remodeling in humans (Rajagopalan & Gerdes 2015). The use of T3 in heart failure has been a topic of discussion for many decades, with a growing number of animal and human studies suggesting that TH treatment may contribute to improved cardiovascular outcomes (Gerdes 2015), although off-target effects may limit application, particularly in patients who appear systemically euthyroid. The present studies offer insight into how one might specifically enhance the actions of TH by inhibiting MuRF1’s antagonism, thereby targeting muscle-specific activation without necessarily supplementing T3 systemically.
Supplementary Material
Acknowledgments
Funding
This study was supported by grants from the American Heart Association Mid-Atlantic Affiliate (12PRE11480009 to K W), Jefferson-Pilot Corporation Fellowship in Academic Medicine (to M W), the NIH National Heart, Lung, and Blood Institute (R01HL104129 to M W), and the Leducq Foundation Grant (to M W).
The authors would like to thank William Claycomb for gifting the HL-1 cells used in this study and for his guidance in detailing their care and use. We also thank Michael Chua and Neal Kramarcy, PhD, of the Michael Hooker Microscopy Facility at University of North Carolina at Chapel Hill for their advice and expertise in obtaining fluorescent cell images and Janice Weaver (University of North Carolina Animal Histopathology Laboratory) for assistance in preparing histological specimens.
Footnotes
This is linked to the online version of the paper at http://dx.doi.org/10.1530/JME-15-0283.
Declaration of interest
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
Author contribution
K W, J B, M P, M G, and M W conceived the concept and detailed the research design and were involved in all aspects of interpretation, discussion, writing, and editing. K W and M W performed the animal experiments. K W, W T, and M Z conceived, planned, and performed in vitro studies, then statistically analyzed and plotted, and interpreted in vitro studies. K W, J B, W T, and M W statistically analyzed, interpreted, and plotted data, wrote, and edited the manuscript.
References
- Adamopoulos S, Gouziouta A, Mantzouratou P, Laoutaris ID, Dritsas A, Cokkinos DV, Mourouzis I, Sfyrakis P, Iervasi G, Pantos C. Thyroid hormone signalling is altered in response to physical training in patients with end-stage heart failure and mechanical assist devices: Potential physiological consequences? Interactive CardioVascular and Thoracic Surgery. 2013;17:664–668. doi: 10.1093/icvts/ivt294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arya R, Kedar V, Hwang JR, McDonough H, Li HH, Taylor J, Patterson C. Muscle ring finger protein-1 inhibits PKC{epsilon} activation and prevents cardiomyocyte hypertrophy. Journal of Cell Biology. 2004;167:1147–1159. doi: 10.1083/jcb.200402033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee R, He J, Spaniel C, Quintana MT, Wang Z, Bain JR, Newgard CB, Muehlbauer MJ, Willis MS. Non-targeted metabolomics analysis of cardiac muscle ring finger-1 (MuRF1), MuRF2, and MuRF3 in vivo reveals novel and redundant metabolic changes. Metabolomics. 2015;11:312–322. doi: 10.1007/s11306-014-0695-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belakavadi M, Saunders J, Weisleder N, Raghava PS, Fondell JD. Repression of cardiac phospholamban gene expression is mediated by thyroid hormone receptor-{alpha}1 and involves targeted covalent histone modifications. Endocrinology. 2010;151:2946–2956. doi: 10.1210/en.2009-1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengel FM, Nekolla SG, Ibrahim T, Weniger C, Ziegler SI, Schwaiger M. Effect of thyroid hormones on cardiac function, geometry, and oxidative metabolism assessed noninvasively by positron emission tomography and magnetic resonance imaging. Journal of Clinical Endocrinology & Metabolism. 2000;85:1822–1827. doi: 10.1210/jcem.85.5.6520. [DOI] [PubMed] [Google Scholar]
- Brooks CL, Li M, Gu W. Mechanistic studies of MDM2-mediated ubiquitination in p53 regulation. Journal of Biological Chemistry. 2007;282:22804–22815. doi: 10.1038/onc.2009.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claycomb WC, Lanson NA, Jr, Stallworth BS, Egeland DB, Delcarpio JB, Bahinski A, Izzo NJ., Jr HL-1 cells: A cardiac muscle cell line that contracts and retains phenotypic characteristics of the adult cardiomyocyte. PNAS. 1998;95:2979–2984. doi: 10.1073/pnas.95.6.2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dace A, Zhao L, Park KS, Furuno T, Takamura N, Nakanishi M, West BL, Hanover JA, Cheng S. Hormone binding induces rapid proteasome-mediated degradation of thyroid hormone receptors. PNAS. 2000;97:8985–8990. doi: 10.1073/pnas.160257997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillmann W. Cardiac hypertrophy and thyroid hormone signaling. Heart Failure Reviews. 2010;15:125–132. doi: 10.1007/s10741-008-9125-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gassanov N, Er F, Michels G, Zagidullin N, Brandt MC, Hoppe UC. Divergent regulation of cardiac KCND3 potassium channel expression by the thyroid hormone receptors alpha1 and beta1. Journal of Physiology. 2009;587:1319–1329. doi: 10.1113/jphysiol.2008.168385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes AM. Restoration of thyroid hormone balance: A game changer in the treatment of heart failure? American Journal of Physiology: Heart and Circulatory Physiology. 2015;308:H1–10. doi: 10.1152/ajpheart.00704.2014. [DOI] [PubMed] [Google Scholar]
- Gerdes AM, Iervasi G. Thyroid replacement therapy and heart failure. Circulation. 2010;122:385–393. doi: 10.1161/CIRCULATIONAHA.109.917922. [DOI] [PubMed] [Google Scholar]
- Ghose Roy S, Mishra S, Ghosh G, Bandyopadhyay A. Thyroid hormone induces myocardial matrix degradation by activating matrix metalloproteinase-1. Matrix Biology. 2007;26:269–279. doi: 10.1016/j.matbio.2006.12.005. [DOI] [PubMed] [Google Scholar]
- He J, Quintana MT, Sullivan J, TLP, TJG, Schisler, Hill JC, Yates JA, Mapanga CC, Essop RFMF, et al. MuRF2 regulates PPARgamma1 activity to protect against diabetic cardiomyopathy and enhance weight gain induced by a high fat diet. Cardiovascular Diabetology. 2015;14:97. doi: 10.1186/s12933-015-0252-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirotani M, Tsukamoto T, Bourdeaux J, Sadano H, Osumi T. Stabilization of peroxisome proliferator-activated receptor alpha by the ligand. Biochemical and Biophysical Research Communications. 2001;288:106–110. doi: 10.1006/bbrc.2001.5739. [DOI] [PubMed] [Google Scholar]
- Hoeck W, Rusconi S, Groner B. Down-regulation and phosphorylation of glucocorticoid receptors in cultured cells. Investigations with a monospecific antiserum against a bacterially expressed receptor fragment. Journal of Biological Chemistry. 1989;264:14396–14402. [PubMed] [Google Scholar]
- Hwee DT, Baehr LM, Philp A, Baar K, Bodine SC. Maintenance of muscle mass and load-induced growth in muscle RINGfinger 1 null mice with age. Aging Cell. 2014;13:92–101. doi: 10.1111/acel.2014.13.issue-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iervasi G, Nicolini G. Thyroid hormone and cardiovascular system: From basic concepts to clinical application. Internal and Emergency Medicine. 2013;8(Supplement 11):S71–74. doi: 10.1007/s11739-013-0911-4. [DOI] [PubMed] [Google Scholar]
- Ismail A, Nawaz Z. Nuclear hormone receptor degradation and gene transcription: An update. International Union of Biochemistry and Molecular Biology Life. 2005;57:483–490. doi: 10.4103/1735-1995.172796. [DOI] [PubMed] [Google Scholar]
- Jung YS, Hakem A, Hakem R, Chen X. Pirh2 E3 ubiquitin ligase monoubiquitinates DNA polymerase eta to suppress translesion DNA synthesis. Molecular and Cellular Biology. 2011;31:3997–4006. doi: 10.1371/journal.pgen.1002360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedar V, McDonough H, Arya R, Li HH, Rockman HA, Patterson C. Muscle-specific RING finger 1 is a bona fide ubiquitin ligase that degrades cardiac troponin I. PNAS. 2004;101:18135–18140. doi: 10.1073/pnas.0404341102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenessey A, Ojamaa K. Ligand-mediated decrease of thyroid hormone receptor-alpha1 in cardiomyocytes by proteosome-dependent degradation and altered mRNA stability. American Journal of Physiology: Heart and Circulatory Physiology. 2005;288:H813–821. doi: 10.1152/ajpheart.00595.2012. [DOI] [PubMed] [Google Scholar]
- Kim HT, Kim KP, Lledias F, Kisselev AF, Scaglione KM, Skowyra D, Gygi SP, Goldberg AL. Certain pairs of ubiquitin-conjugating enzymes (E2s) and ubiquitin-protein ligases (E3s) synthesize nondegradable forked ubiquitin chains containing all possible isopeptide linkages. Journal of Biological Chemistry. 2007;282:17375–17386. doi: 10.1074/jbc.M609659200. [DOI] [PubMed] [Google Scholar]
- Kopf E, Plassat JL, Vivat V, de The H, Chambon P, Rochette-Egly C. Dimerization with retinoid X receptors and phosphorylation modulate the retinoic acid-induced degradation of retinoic acid receptors alpha and gamma through the ubiquitin-proteasome pathway. Journal of Biological Chemistry. 2000;275:33280–33288. doi: 10.1074/jbc.M002840200. [DOI] [PubMed] [Google Scholar]
- Lai Z, Ferry KV, Diamond MA, Wee KE, Kim YB, Ma J, Yang T, Benfield PA, Copeland RA, Auger KR. Human mdm2 mediates multiple mono-ubiquitination of p53 by a mechanism requiring enzyme isomerization. Journal of Biological Chemistry. 2001;276:31357–31367. doi: 10.1074/jbc.M011517200. [DOI] [PubMed] [Google Scholar]
- Lange CA, Shen T, Horwitz KB. Phosphorylation of human progesterone receptors at serine-294 by mitogen-activated protein kinase signals their degradation by the 26 S proteasome. PNAS. 2000;97:1032–1037. doi: 10.1128/MCB.21.18.6122-6131.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JT, Gu W. The multiple levels of regulation by p53 ubiquitination. Cell Death And Differentiation. 2010;17:86–92. doi: 10.1038/cdd.2009.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lelievre H, Chevrier V, Tassin AM, Birnbaum D. Myeloproliferative disorder FOP-FGFR1 fusion kinase recruits phosphoinositide-3 kinase and phospholipase Cgamma at the centrosome. Molecular Cancer. 2008;7:30. doi: 10.1186/1476-4598-7-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li HH, Du J, Fan YN, Zhang ML, Liu DP, Li L, Lockyer P, Kang EY, Patterson C, Willis MS. The ubiquitin ligase MuRF1 protects against cardiac ischemia/reperfusion injury by its proteasome-dependent degradation of phospho-c-Jun. American Journal of Pathology. 2011;178:1043–1058. doi: 10.1016/j.ajpath.2010.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li HH, Kedar V, Zhang C, McDonough H, Arya R, Wang DZ, Patterson C. Atrogin-1/muscle atrophy F-box inhibits calcineurin-dependent cardiac hypertrophy by participating in an SCF ubiquitin ligase complex. Journal of Clinical Investigation. 2004;114:1058–1071. doi: 10.1172/JCI200422220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XY, Boudjelal M, Xiao JH, Peng ZH, Asuru A, Kang S, Fisher GJ, Voorhees JJ. 1,25-Dihydroxyvitamin D3 increases nuclear vitamin D3 receptors by blocking ubiquitin/proteasome-mediated degradation in human skin. Molecular Endocrinology. 1999;13:1686–1694. doi: 10.1210/mend.13.10.0362. [DOI] [PubMed] [Google Scholar]
- Lin HK, Altuwaijri S, Lin WJ, Kan PY, Collins LL, Chang C. Proteasome activity is required for androgen receptor transcriptional activity via regulation of androgen receptor nuclear translocation and interaction with coregulators in prostate cancer cells. The Journal of Biological Chemistry. 2002;277:36570–36576. doi: 10.1074/jbc.M204751200. [DOI] [PubMed] [Google Scholar]
- Lisby M, Mortensen UH, Rothstein R. Colocalization of multiple DNA double-strand breaks at a single Rad52 repair centre. Nature Cell Biology. 2003;5:572–577. doi: 10.1016/j.dnarep.2009.10.001. [DOI] [PubMed] [Google Scholar]
- Liu YY, Kogai T, Schultz JJ, Mody K, Brent GA. Thyroid hormone receptor isoform-specific modification by small ubiquitin-like modifier (SUMO) modulates thyroid hormone-dependent gene regulation. Journal of Biological Chemistry. 2012;287:36499–36508. doi: 10.1074/jbc.M112.344317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Fan S, Hu C, Meng Q, Fuqua SA, Pestell RG, Tomita YA, Rosen EM. BRCA1 regulates acetylation and ubiquitination of estrogen receptor-alpha. Molecular Endocrinology. 2010;24:76–90. doi: 10.1210/me.2009-0218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansen A, Tiselius C, Sand P, Fauconnier J, Westerblad H, Rydqvist B, Vennstrom B. Thyroid hormone receptor alpha can control action potential duration in mouse ventricular myocytes through the KCNE1 ion channel subunit. Acta physiologica (Oxford, England) 2010;198:133–142. doi: 10.1111/aps.2010.198.issue-2. [DOI] [PubMed] [Google Scholar]
- Mourouzis I, Forini F, Pantos C, Iervasi G. Thyroid hormone and cardiac disease: From basic concepts to clinical application. Journal of Thyroid Research. 2011;2011:958626. doi: 10.4061/2011/958626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourouzis I, Giagourta I, Galanopoulos G, Mantzouratou P, Kostakou E, Kokkinos AD, Tentolouris N, Pantos C. Thyroid hormone improves the mechanical performance of the post-infarcted diabetic myocardium: A response associated with up-regulation of Akt/mTOR and AMPK activation. Metabolism. 2013a;62:1387–1393. doi: 10.1016/j.metabol.2013.05.008. [DOI] [PubMed] [Google Scholar]
- Mourouzis I, Kostakou E, Galanopoulos G, Mantzouratou P, Pantos C. Inhibition of thyroid hormone receptor alpha1 impairs post-ischemic cardiac performance after myocardial infarction in mice. Molecular and Cellular Biochemistry. 2013b;379:97–105. doi: 10.1177/1074248414524481. [DOI] [PubMed] [Google Scholar]
- Mourouzis I, Mantzouratou P, Galanopoulos G, Kostakou E, Roukounakis N, Kokkinos AD, Cokkinos DV, Pantos C. Dose-dependent effects of thyroid hormone on post-ischemic cardiac performance: Potential involvement of Akt and ERK signalings. Molecular and Cellular Biochemistry. 2012;363:235–243. doi: 10.1007/s11010-011-1175-9. [DOI] [PubMed] [Google Scholar]
- Nawaz Z, Lonard DM, Dennis AP, Smith CL, O’Malley BW. Proteasome-dependent degradation of the human estrogen receptor. PNAS. 1999;96:1858–1862. doi: 10.1073/pnas.96.5.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojamaa K. Signaling mechanisms in thyroid hormone-induced cardiac hypertrophy. Vascular Pharmacology. 2010;52:113–119. doi: 10.1016/j.vph.2009.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantos C, Mourouzis I, Galanopoulos G, Gavra M, Perimenis P, Spanou D, Cokkinos DV. Thyroid hormone receptor alpha1 downregulation in postischemic heart failure progression: The potential role of tissue hypothyroidism. Hormone And Metabolic Research. 2010;42:718–724. doi: 10.1055/s-0030-1255035. [DOI] [PubMed] [Google Scholar]
- Pantos C, Mourouzis I, Markakis K, Dimopoulos A, Xinaris C, Kokkinos AD, Panagiotou M, Cokkinos DV. Thyroid hormone attenuates cardiac remodeling and improves hemodynamics early after acute myocardial infarction in rats. European Journal Cardio-Thoracic Surgery. 2007a;32:333–339. doi: 10.1016/j.ejcts.2007.05.004. [DOI] [PubMed] [Google Scholar]
- Pantos C, Mourouzis I, Saranteas T, Brozou V, Galanopoulos G, Kostopanagiotou G, Cokkinos DV. Acute T3 treatment protects the heart against ischemia-reperfusion injury via TRalpha1 receptor. Molecular and Cellular Biochemistry. 2011;353:235–241. doi: 10.1007/s11010-011-0791-8. [DOI] [PubMed] [Google Scholar]
- Pantos C, Xinaris C, Mourouzis I, Malliopoulou V, Kardami E, Cokkinos DV. Thyroid hormone changes cardiomyocyte shape and geometry via ERK signaling pathway: Potential therapeutic implications in reversing cardiac remodeling? Molecular and Cellular Biochemistry. 2007b;297:65–72. doi: 10.1007/s11010-006-9323-3. [DOI] [PubMed] [Google Scholar]
- Pantos C, Xinaris C, Mourouzis I, Perimenis P, Politi E, Spanou D, Cokkinos DV. Thyroid hormone receptor alpha 1: A switch to cardiac cell metamorphosis? Journal of Physiology and Pharmacology. 2008;59:253–269. [PubMed] [Google Scholar]
- Patel H, Truant R, Rachubinski RA, Capone JP. Activity and subcellular compartmentalization of peroxisome proliferator-activated receptor alpha are altered by the centrosome-associated protein CAP350. Journal of Cell Science. 2005;118:175–186. doi: 10.1242/jcs.01600. [DOI] [PubMed] [Google Scholar]
- Periasamy M, Bhupathy P, Babu GJ. Regulation of sarcoplasmic reticulum Ca2+ ATPase pump expression and its relevance to cardiac muscle physiology and pathology. Cardiovascular Research. 2008;77:265–273. doi: 10.1016/j.yjmcc.2009.07.014. [DOI] [PubMed] [Google Scholar]
- Quintana MT, He J, Sullivan J, Grevengoed T, Schisler J, Han Y, Hill JA, Yates CC, Stansfield WE, Mapanga RF, et al. Muscle ring finger-3 protects against diabetic cardiomyopathy induced by a high fat diet. BMC Endocrine Disorders. 2015;15:36. doi: 10.1186/s12902-015-0028-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopalan V, Gerdes AM. Role of thyroid hormones in ventricular remodeling. Current Heart Failure Reports. 2015;12:141–149. doi: 10.1007/s11897-014-0246-0. [DOI] [PubMed] [Google Scholar]
- Rodriguez JE, Liao JY, He J, Schisler JC, Newgard CB, Drujan D, Glass DJ, Frederick CB, Yoder BC, Lalush DS, et al. The ubiquitin ligase MuRF1 regulates PPARalpha activity in the heart by enhancing nuclear export via monoubiquitination. Molecular and Cellular Endocrinology. 2015;413:36–48. doi: 10.1016/j.mce.2015.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatino L, Gliozheni E, Molinaro S, Bonotti A, Azzolina S, Popoff G, Carpi A, Iervasi G. Thyroid hormone receptor and IGF1/IGFR systems: Possible relations in the human heart. Biomedicine & Pharmacotherapy. 2007;61:457–462. doi: 10.1016/j.biopha.2007.04.002. [DOI] [PubMed] [Google Scholar]
- Vandenbroucke St Amant E, Tauseef M, Vogel SM, Gao XP, Mehta D, Komarova YA, Malik AB. PKCalpha activation of p120-catenin serine 879 phospho-switch disassembles VE-cadherin junctions and disrupts vascular integrity. Circulation Research. 2012;111:739–749. doi: 10.1161/CIRCRESAHA.112.269654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadosky KM, Rodriguez JE, Hite RL, Min JN, Walton B, Willis MS. Muscle RING Finger-1 Attenuates IGF-1-dependent cardiomyocyte hypertrophy by inhibiting JNK signaling. American Journal of Physiology – Endocrinology and Metabolism. 2014;306:E732–E739. doi: 10.1152/ajpendo.00326.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weltman NY, Ojamaa K, Schlenker EH, Chen YF, Zucchi R, Saba A, Colligiani D, Rajagopalan V, Pol CJ, Gerdes AM. Low-dose T(3) replacement restores depressed cardiac T(3) levels, preserves coronary microvasculature and attenuates cardiac dysfunction in experimental diabetes mellitus. Molecular Medicine. 2014;20:302–312. doi: 10.2119/molmed.2013.00040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White SM, Constantin PE, Claycomb WC. Cardiac physiology at the cellular level: Use of cultured HL-1 cardiomyocytes for studies of cardiac muscle cell structure and function. American Journal of Physiology: Heart and Circulatory Physiology. 2004;286:H823–829. doi: 10.1152/ajpheart.00986.2003. [DOI] [PubMed] [Google Scholar]
- Willis MS, Ike C, Li L, Wang DZ, Glass DJ, Patterson C. Muscle ring finger 1, but not muscle ring finger 2, regulates cardiac hypertrophy in vivo. Circulation Research. 2007;100:456–459. doi: 10.1161/01.RES.0000259559.48597.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis MS, Rojas M, Li L, Selzman CH, Tang RH, Stansfield WE, Rodriguez JE, Glass DJ, Patterson C. Muscle ring finger 1 mediates cardiac atrophy in vivo. American Journal of Physiology: Heart and Circulatory Physiology. 2009a;296:H997–H1006. doi: 10.1152/ajpheart.00660.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis MS, Schisler JC, Li L, Rodriguez JE, Hilliard EG, Charles PC, Patterson C. Cardiac muscle ring finger-1 increases susceptibility to heart failure in vivo. Circulation Research. 2009b;105:80–88. doi: 10.1161/CIRCRESAHA.109.194928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis MS, Wadosky KM, Rodriguez JE, Schisler JC, Lockyer P, Hilliard EG, Glass DJ, Patterson C. Muscle ring finger 1 and muscle ring finger 2 are necessary but functionally redundant during developmental cardiac growth and regulate E2F1-mediated gene expression in vivo. Cell Biochemistry and Function. 2014;32:39–50. doi: 10.1002/cbf.v32.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witt SH, Granzier H, Witt CC, Labeit S. MURF-1 and MURF-2 target a specific subset of myofibrillar proteins redundantly: Towards understanding MURF-dependent muscle ubiquitination. Journal of Molecular Biology. 2005;350:713–722. doi: 10.1016/j.jmb.2005.05.021. [DOI] [PubMed] [Google Scholar]
- Xu G, Jaffrey SR. Proteomic identification of protein ubiquitination events. Biotechnology & Genetic Engineering Reviews. 2013;29:73–109. doi: 10.5582/bst.2016.01033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarain-Herzberg A. Regulation of the sarcoplasmic reticulum Ca2+-ATPase expression in the hypertrophic and failing heart. Canadian Journal of Physiology and Pharmacology. 2006;84:509–521. doi: 10.1139/y06-023. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Dedkov EI, Lee B, 3rd, Li Y, Pun K, Gerdes AM. Thyroid hormone replacement therapy attenuates atrial remodeling and reduces atrial fibrillation inducibility in a rat myocardial infarction-heart failure model. Journal of Cardiac Failure. 2014;20:1012–1019. doi: 10.1016/j.cardfail.2014.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao TJ, Yan YB, Liu Y, Zhou HM. The generation of the oxidized form of creatine kinase is a negative regulation on muscle creatine kinase. Journal of Biological Chemistry. 2007;282:12022–12029. doi: 10.1016/j.bbapap.2008.08.018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.