Abstract
Aim: Most epidemiological and clinical studies calculated low-density lipoprotein-cholesterol (LDL-C) by Friedewald's formula which cannot be used in the postprandial samples. Although the homogeneous assays with poor analytical performance were withdrawn from the market, it remained unclear whether the currently available reagents for LDL-C and high-density lipoprotein-cholesterol (HDL-C) are as accurate for postprandial samples as for fasting samples.
Methods: Fresh blood samples were collected from 59 non-diseased and 109 diseased subjects. Postprandial samples constituted 72.9% and 39.4% of these samples. LDL-C and HDL-C concentrations were measured using the homogeneous assays of four manufacturers (Denka Seiken, Wako, Kyowa Medex, and Sekisui Medical). Simultaneously, LDL-C and HDL-C concentrations were determined using the reference measurement procedures (RMPs) of the Centers for Disease Control and Prevention (CDC). Total errors were calculated using a routine method (TEcom) and via error component analysis (TEeca).
Results: All homogeneous assays for LDL-C and HDL-C met the National Cholesterol Education Program (NCEP) requirements in terms of coefficient of variation, and TEcom in both non-diseased and diseased subjects. LDL-C and HDL-C values measured by the homogeneous assays were in good agreement with those measured by the RMPs in both fasting and postprandial samples. The TEcom and TEeca values of the postprandial samples were similar to those of fasting samples, although the TEeca values were up to 4.4-fold greater than the TEcom values.
Conclusions: In both non-diseased and diseased subjects, the homogeneous assays for LDL-C and HDL-C of four manufacturers are as accurate for postprandial samples as for fasting samples.
Keywords: Direct assay, Friedewald's formula, Standardization, Cholesterol Reference Method Laboratory Network (CRMLN), Dyslipidemia
See editorial vol. 24: 569–571
Introduction
The homogeneous assays for low-density lipoprotein-cholesterol (LDL-C) and high-density lipoprotein-cholesterol (HDL-C) are innovative because they require neither extensive pretreatment nor a long assay time1–4). Other strengths of such assays are excellent intra- and inter-assay reproducibility. These assays are also termed direct methods. To date, several original reagents have been developed in Japan by different manufacturers, based on various principles. Latecomer companies (distributors) purchase bulk reagents from the original manufacturers, and sell their products with different brand names5, 6). To ensure global standardization and harmonization of lipid laboratory tests, the Centers for Disease Control and Prevention (CDC) runs a manufacturer certification program with the aid of the Cholesterol Reference Method Laboratory Network (CRMLN)7). All commercial homogeneous assays produced by the original manufactures and their distributors have taken part in this program, and have passed the precision and accuracy requirements defined by the National Cholesterol Education Program (NCEP)8, 9).
Despite these efforts, earlier studies revealed that some homogenous assays exhibited poor analytical performance in patients with common diseases, and even in disease-free subjects5, 6, 10, 11). At the end of 2016, these reagents were withdrawn from the Japanese market. A common problem was that when these reagents were used, hypertriglyceridemia often triggered a positive assay bias, especially in LDL-C measurements5, 10). In contrast to the LDL-C estimations afforded by various equations, homogeneous assays are applicable to postprandial samples with triglyceride (TG)-rich lipoprotein levels greater than those of the fasting state. However, no study has yet directly compared the accuracy of homogeneous assays of fasting samples with those of postprandial samples using the reference measurement procedures (RMPs) of the CDC, employing fresh sera collected from non-diseased and diseased subjects.
Aim
Our aim was to clarify whether homogeneous assays for LDL-C and HDL-C are as accurate for post-prandial samples as for fasting samples.
Methods
Study Subjects
Study participants were volunteers and outpatients at Osaka University Hospital (Osaka, Japan) and the National Cerebral and Cardiovascular Center (NCVC, Osaka, Japan). Both normolipidemic and dyslipidemic subjects were eligible provided that their lipoprotein concentrations were in a certain range [20 mg/dL ≤ LDL-C; 20 mg/dL ≤ HDL-C < 100 mg/dL; TG < 1,000 mg/dL]. In line with the exclusion criteria of our previous studies5, 6), we did not enroll patients with serious infectious diseases, decompensated liver cirrhosis, or cholestatic liver diseases. A diagnosis of hyperlipidemia was made if the LDL-C level was > 160 mg/dL, and/or the TG level > 200 mg/dL, under conditions of ad libitum food intake. Normolipidemic subjects with no medical history were classified as non-diseased, whereas others were considered diseased. This study complied with the dictates of the latest version of the Declaration of Helsinki (thus, that amended at the 64th WMA General Assembly, Fortaleza, Brazil, October 201312)). At recruitment, written informed consent was obtained from all participants. The study protocol was approved by the Ethics Committees of the individual institutions.
Study Protocol
Fresh venous blood was drawn and serum was collected after centrifugation, dispensed into screw-capped tubes, and immediately stored at 4°C. The next morning, an aliquot of each sample was conveyed to the Kyoto Prefectural University where LDL-C and HDL-C were measured using homogeneous assays. The remaining samples were subjected (at NCVC) to LDL-C and HDL-C measurements using the RMPs described below. During sample transport, we used a special cooling container that held the samples at 2–4µC (without freezing)5). All measurements were performed on the day of sample arrival.
Assays for LDL-C and HDL-C
At Kyoto Prefectural University, we measured LDL-C and HDL-C concentrations using four homogeneous assays [LDL-Cha and HDL-Cha]. The reagents were manufactured by Denka Seiken, Wako, Kyowa Medex, and Sekisui Medical (Supplemental Table 1). All reagents, calibrators, and controls were provided by these companies. All reagents except the reagent of Kyowa Medex (used in the LDL-C assay) were the same as those described in our previous studies5, 6). The Kyowa Medex reagent (MetaboLead LDL-C) was a modified version of the previous formulation (Determiner L; LDL-C). The newer version exhibits an improved specificity for LDL particles; interference by TG-rich lipoproteins is reduced. TG concentrations were determined using an enzymatic method.
Supplemental Table 1. The reagents and calibrators used to measure LDL-C and HDL-C concentrations.
| Reagent |
||||
|---|---|---|---|---|
| Manufacturer | A | B | C | D |
| LDL-C | ||||
| Brand Name | LDL-EX (N) “SEIKEN” | L-Type LDL-C M | MetaboLead LDL-C | Cholestest LDL |
| Same reagent as in refs.#5, 6 | Yes | Yes | No (modified formula) | Yes |
| Principle | Selective elimination method | Selective elimination method | Selective solubilization method | Selective elimination method |
| Calibrator | Lipid Calibrator | Multi Calibrator Lipids | MetaboLead Standard Serum | Cholestest N Calibrator |
| Control | Lipid Control | Lipids Control Set | MetaboLead Control Serum | Cholestest Control |
| Sample vol. (µL) | 2.4 | 2.4 | 3.0 | 2.4 |
| Reagent vol. (µL) R1/R2 | 180/60 | 210/70 | 180/60 | 240/80 |
| Abs WL, 2nd/primary (nm) | 700/600 | 700/600 | 700/600 | 660/546 |
| Assay mode | 2 Point End | 2 Point End | 2 Point End | 2 Point End |
| Calibration | Linear | Linear | Linear | Linear |
| HDL-C | ||||
| Brand Name | HDL-EX “SEIKEN” | L-Type HDL-C M(2) | MetaboLead HDL-C | Cholestest N HDL |
| Principle | Selective elimination method | Selective elimination method | Selective inhibition method | Selective solubilization method |
| Calibrator | Lipid Calibrator | Multi Calibrator Lipids | MetaboLead Standard Serum | Cholestest N Calibrator |
| Control | Lipid Control | Lipids Control Set | MetaboLead Control Serum | Cholestest Control |
| Sample vol. (µL) | 2.4 | 2.4 | 3.6 | 2.4 |
| Reagent vol. (µL) R1/R2 | 180/60 | 210/70 | 180/60 | 240/80 |
| Abs WL, 2nd/primary (nm) | 700/600 | 700/600 | 700/600 | 700/600 |
| Assay mode | 2 Point End | 2 Point End | 2 Point End | 2 Point End |
| Calibration | Linear | Linear | Linear | Linear |
| TG | ||||
| Brand Name | L-Type Triglyceride H | |||
| Calibrator | Multi Calibrator Lipids | |||
| Control | QAP Trol | |||
| Sample Vol. (µL) | 2.0 | |||
| Reagent Vol. (µL) R1/R2 | 180/90 | |||
| Abs WL, 2nd/primary (nm) | 700/600 | |||
| Assay mode | 2 Point End | |||
| Calibration | Linear | |||
LDL-C, low-density lipoprotein-cholesterol; HDL-C, high-density lipoprotein-cholesterol; TG, triglyceride; R1, reagent 1; R2, reagent 2 Vol, volume; Abs WL, Absorption wavelength.
Each reagent was evaluated on a Hitachi 917 platform, sold in Japan under the brand name “Hitachi 7170”.
A, Denka Seiken; B, Wako; C, Kyowa Medex; D, Sekisui Medical.
We employed the same automated analyzer used in previous studies (Hitachi-7170; overseas brand name Hitachi-917)5, 6). We prepared all reagents for the homogeneous assays in a defined order, and measured LDL-C, HDL-C, and TG concentrations once in each cycle. We repeated this process three times, yielding triplicate measurements. These procedures were the same as employed in earlier studies5, 6, 10, 11).
At NCVC, LDL-C and HDL-C concentrations were measured using the RMPs [LDL-Crmp and HDLCrmp] employed by the CRMLN5, 6). In brief, the top fraction of ultracentrifuged serum (d > 1.006, 18°C, 105,000 × g, 18.5 h), which contained VLDL and chylomicrons, was separated from the bottom fraction, which contained LDL and HDL. The volume of the bottom fraction was adjusted to the original volume (5 mL) with 0.15 mol/L NaCl (Fraction 1). Then, a precipitate was obtained by addition of 40 µL of a heparin solution (5,000 units/mL) and 50 µL MnCl2 (1.0 mol/L). The supernatant (Fraction 2, containing only HDL) was recovered after centrifugation of this mixture. After adjusting both volumes to the baseline levels, and measurement of cholesterol concentrations in both Fraction 1 [F1]chol and Fraction 2 [F2]chol using the Abell-Kendall method13), the LDL-Crmp concentration was defined as [F1]chol minus [F2]chol, and the HDL-Crmp concentration as [F2]chol.
Determination of Precision Using Pooled Sera
We prepared the pooled sera, dispensed the pool into screw-capped tubes, and stored the tubes at −80°C. After thawing, we measured LDL-C and HDL-C concentrations using all of the homogeneous assays (Supplemental Table 1) 30 times within a single day to determine among-run coefficients of variance (CVs), and on 21 different days to determine between-run CVs. Bias was assessed by subtracting the LDL-Crmp (or HDL-Crmp) from the LDL-Cha (or HDL-Cha). Bias percentages were calculated as (bias/LDL-Crmp or HDL-Crmp) × 100.
Statistical Analysis
We calculated two different total errors (Tecom and TEeca). TEcom was defined as |bias (%)| + 1.96 × between-run CV (%)14).
For error component analysis, we followed the modified methods of Nilsson15). The details have been described elsewhere5). Briefly, error component analysis is based on the hypothesis that analytical errors are derived from three components: the inter-assay CV (CVb), the intra-assay CV (CVe), and a CV derived from sample-specific effects (CVd). CVeca was calculated as the square root of [(CVb)2 + (CVe)2 + (CVd)2] where CVb equals the between-run CV of the log-transformed data from the pooled sera. We also calculated the mean bias [ECA], the among-run CV (CVe), and the subject-specific CV (CVd), using the log-transformed LDL-Crmp and LDL-Cha, or HDL-Crmp and HDL-Cha of fresh samples. From these log-transformed variations, TEeca was defined as |bias [ECA] (%)| + 1.96 × CVeca (%). It should be noted that the %bias [ECA] and CVeca differ from the %bias and between-run CV used to calculate the conventional TEcom. We evaluated the CVs and TEs as dictated by the NCEP (Supplemental Table 2)8, 9).
Supplemental Table 2. The NCEP requirements for LDL-C and HDL-C measurements.
| LDL-C | |
| Between-run CV (%) | ≤ 4.0% |
| |Mean bias| (%) | ≤ 4.0% |
| R2 | > 0.975 |
| TEcom | ≤ 12.0% |
| HDL-C | |
| Between-run CV (%) | ≤ 4.0% |
| |Mean bias| (%) | ≤ 5.0% |
| R2 | > 0.975 |
| TEcom | ≤ 13.0% |
LDL-C, low-density lipoprotein-cholesterol; CV, coefficient of variation; HDL-C, high-density lipoprotein-cholesterol
TEcom is defined as = |mean bias (%)|+1.96 × CV (= between-run CV).
Results
Background of Study Participants
We collected fresh blood samples from 183 subjects. A total of 15 samples were excluded on the basis of the exclusion criteria, and/or because some essential data were missing. Finally, 59 non-diseased and 109 diseased participants were selected for further analysis (Supplemental Fig. 1). Those in the diseased group had common diseases treated in the outpatient clinic, although one patient had type III hyperlipidemia and one a lipoprotein lipase deficiency (Table 1). More than half the samples were obtained in the postprandial state. The samples exhibited wide ranges of TG, LDL-C, and HDL-C concentrations (Supplemental Table 3). In the diseased group, 7.3% of patients had TG levels > 400 mg/dL.
Supplemental Fig. 1.
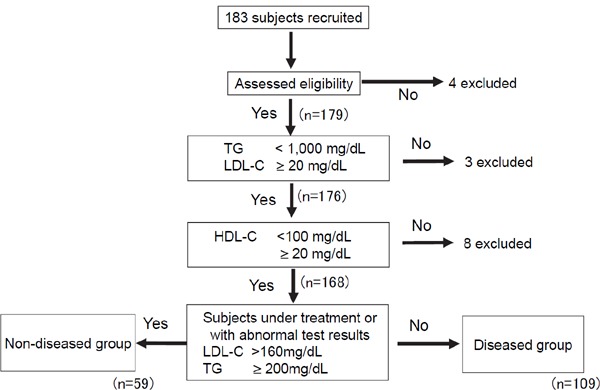
The flow chart summarizing enrollment of the study subjects.
Table 1. Study subjects.
| Non-diseased group | Diseased group | |
|---|---|---|
| (n = 59) | (n = 109) | |
| Age (years) | 41.7 ± 9.4 | 53.4 ± 15.9 |
| Gender (male, %) | 64.4 | 68.8 |
| Body height (cm) | 167.1 ± 7.5 | 166.4 ± 8.9 |
| Body weight (kg) | 65.3 ± 16.1 | 69.5 ± 17.1 |
| Body mass index (kg/m2) | 23.3 ± 5.3 | 24.8 ± 4.2 |
| TC (mg/dL) | 187.0 ± 24.3 [124.1–237.6] | 200.5 ± 38.9 [110.0–325.6] |
| LDL-Crmp (mg/dL) | 109.7 ± 20.9 [60.0–159.4] | 121.2 ± 35.9 [51.7–230.0] |
| HDL-Crmp (mg/dL) | 60.7 ± 12.7 [38.2–98.9] | 51.8 ± 13.3 [21.8–88.7] |
| TG (mg/dL) | 115.2 ± 45.3 [30.8–194.2] | 195.6 ± 144.6 [32.9–880.4] |
| Non-fasting subjects (%) | 72.9 | 39.4 |
| Time since last meal (h)1) | 2.8 ± 1.9 | 2.6 ± 1.8 |
| Hypolipidemic agents taken (statin) (%) | 0.0 | 56.9 (39.4) |
| Dyslipidemia (%)2) | 0.0 | 88.1 |
| Type I/V | 0.0 | 0.9/1.8 |
| Type IIa/IIb (FH) | 0.0 (0.0) | 64.2 (12.8) |
| Type III | 0.0 | 0.9 |
| Type IV | 0.0 | 20.2 |
| Cardiovascular disease (%) | 0.0 | 21.1 |
| Diabetes mellitus (%) | 0.0 | 8.3 |
| Fatty liver/alcoholic liver injury (%) | 0.0 | 1.8 |
| Renal disease (%) | 0.0 | 1.8 |
| Hyperthyroidism (%) | 0.0 | 0.9 |
Data are presented as means ± SD [minima to maxima], or as percentages.
TC, total cholesterol; LDL-C, low-density lipoprotein-cholesterol; HDL-C, high-density lipoprotein-cholesterol; RMP, reference measurement procedures; TG, triglyceride; FH, familial hypercholesterolemia
Values were calculated using the postprandial samples.
Phenotypes were determined by analysis of the pre-treatment lipid profiles in the fasting state, using the classification of Frederickson.
Supplemental Table 3. Comparisons of TG, LDL-C, and HDL-C distributions among various studies.
| Principal investigator | Miller et al. #1 | Miida et al. #2 | Present study |
|---|---|---|---|
| [Reference] | [2010, ref.#10; 2011, ref.#11] | [2012, ref.#5; 2014, ref.#6] | |
| Number of samples (postprandial samples, %) | |||
| Non-diseased group | 37 (5, 13.5%) | 49 (15, 30.6%) | 59 (43, 72.9%) |
| Diseased group | 138 (43, 31.2%) | 124 (55, 44.4%) | 109 (43, 39.4%) |
| Total | 175 (48, 27.4%) | 173 (70, 40.5%) | 168 (86, 51.2%) |
| TG (mg/dL) (Non-diseased/diseased) | |||
| ≥ 1,000 | 3 (0/3) | (5, excluded) | (3, excluded) |
| 600 ∼ 999 | 1 (0/1) | 9 (0/9) | 3 (0/3) |
| 400 ∼ 599 | 3 (0/3) | 8 (0/8) | 5 (0/5) |
| 200 ∼ 399 | 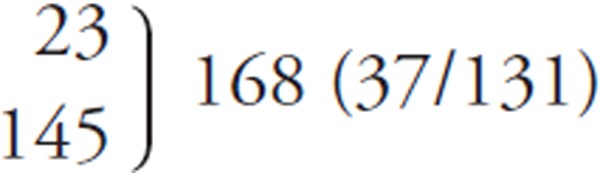 |
29 (0/29) | 34 (0/34) |
| < 199 | 127 (49/78) | 126 (59/67) | |
| LDL-Crmp (mg/dL) | |||
| ≥ 300 | 1 (0/1) | 1 (0/1) | 0 (0/0) |
| 200 ∼ 299 | 4 (0/4) | 4 (0/4) | 5 (0/5) |
| 100 ∼ 199 | 155 (36/119) | 119 (37/82) | 116 (43/73) |
| 50 ∼ 99 | 46 (12/34) | 47 (16/31) | |
| 20 ∼ 49 | 12 (1/11) | 3 (0/3) | 0 (0/0) |
| < 20 | 1 (0/1) | − | (1, excluded) |
| LDL-Crmp, not available | 2 (0/2) | − | − |
| HDL-Crmp (mg/dL) | |||
| ≥ 100 | 1 (0/1) | (1/5)#3 | (6, excluded) |
| 80 ∼ 99 | 7 (5/2) | 18 (10/8) | 7 (5/2) |
| 60 ∼ 79 |
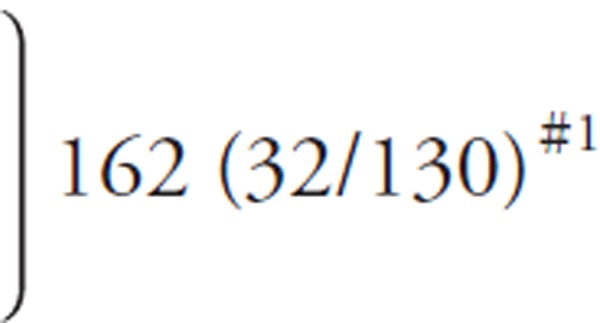 #1 #1
|
41 (22/19) | 52 (27/25) |
| 40 ∼ 59 | 87 (16/71) | 85 (23/62) | |
| 20 ∼ 39 | 21 (0/21) | 24 (4/20) | |
| < 20 | 4 (0/4) | − | (3, excluded) |
| HDL-Crmp, not available | 1 (0/1) | ||
LDL-Crmp, low-density lipoprotein-cholesterol concentration determined by the reference measurement procedure (RMP); HDL-Crmp, high-density lipoprotein-cholesterol concentration determined by the RMP.
The sample numbers given with the ranges of individual lipoproteins were determined using the data of the two publications, including supplemental data.
The sample numbers given with the ranges of individual lipoproteins were counted using original data.
Those with HDL-C > 100mg/dL were included in the study subjects in ref. #5, but excluded in ref. #6.
Precision of the Homogeneous Assays
Upon analyses of pooled sera, the homogeneous assays for LDL-Cha and HDL-Cha exhibited excellent reproducibilities. For all reagents, the among-run CVs for LDL-Cha and HDL-Cha were less than 1%. The between-run CVs for LDL-Cha and HDL-Cha were about 1.0% (Supplemental Table 4), lower than the acceptable imprecisions of the NCEP (LDL-C, ≤ 4%; HDL-C, ≤ 4%) (Supplemental Table 2).
Supplemental Table 4. Among-run and between-run CVs, determined using frozen pooled sera.
| Reagent |
||||
|---|---|---|---|---|
| Manufacturer | A | B | C | D |
| LDL-C | ||||
| Among-run CV (%)#1 | 0.51 | 0.71 | 0.38 | 0.54 |
| Between-run CV (%)#2 | 1.01 | 0.99 | 1.44 | 0.96 |
| HDL-C | ||||
| Among-run CV (%) | 0.86 | 0.69 | 0.44 | 0.77 |
| Between-run CV (%) | 1.27 | 0.77 | 1.27 | 1.29 |
| TC | ||||
| Among-run CV (%) | − | 0.44 | − | − |
| Between-run CV (%) | 1.02 | |||
| TG | ||||
| Among-run CV (%) | − | 0.75 | − | − |
| Between-run CV (%) | 1.62 | |||
LDL-C, low-density lipoprotein-cholesterol; CV, coefficient of variation; HDL-C, high-density lipoproteincholesterol; TC, total cholesterol; TG, triglyceride
We measured LDL-C and HDL-C concentrations of pooled sera, in triplicate, on 21 different days, to derive between-run CVs; or 30 times on the same day to derive among-run CVs.
A, Denka Seiken; B, Wako; C, Kyowa Medex; D, Sekisui Medical.
Accuracies of the Homogeneous Assays
1). Percentage Bias in Triplicate Measurements
Each bias was obtained by subtracting the LDL-Crmp from the mean of the triplicate LDL-Cha, or the HDL-Crmp from the mean of the triplicate HDL-Cha. For all reagents tested, both the non-diseased and diseased groups exhibited very low median %biases for LDL-Cha and HDL-Cha (Fig. 1). In the non-diseased group, the %biases for LDL-Cha and HDL-Cha lay close to the zero line for all reagents. Most individual %biases were ≤ 12% for LDL-C and ≤ 13% for HDLC. Even in diseased patients, only a small number of samples (slightly) exceeded these values.
Fig. 1.
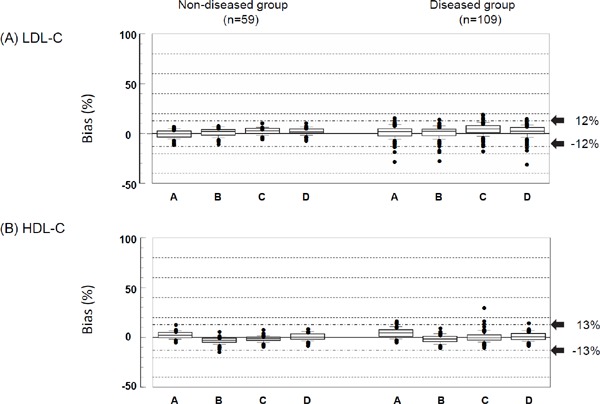
Box-and-whisker plots of the %bias of the low-density lipoprotein-cholesterol (LDL-C) and high-density lipoprotein-cholesterol (HDL-C) homogeneous assays (HAs) in the non-diseased and diseased groups
The LDL-C and HDL-C concentrations of fresh blood samples were simultaneously measured using HAs and the reference measurement procedures (RMPs) of the Centers for Disease Control and Prevention (CDC), as described in the Methods. %bias is the percentage deviation of the HA value from the RMP value.
A, Denka Seiken; B, Wako; C, Kyowa Medex; D, Sekisui Medical.
2). TE of Single Measurements
In this analysis, we used the first LDL-Cha or HDL-Cha obtained using triplicate measurements, and determined whether or not the calculated TEs of individual samples fulfilled the NCEP TE requirements. In the non-diseased group, all reagents for LDL-C and HDL-C surpassed the 95% acceptance criterion when fasting and postprandial samples were assayed together (Table 2). Little difference in analytical performance was evident between fasting and postprandial samples.
Table 2. Percentage of samples fulfilling the NCEP total error requirements for single LDL-Cha and HDL-Cha determinations.
| Group | Non-diseased group | Diseased group | ||||||
|---|---|---|---|---|---|---|---|---|
| (fasting, n = 16/postprandial, n = 43) |
(fasting, n = 66/postprandial, n = 43) |
|||||||
| Manufacturer | A | B | C | D | A | B | C | D |
| LDL-Cha | ||||||||
| Fasting samples | 100.0 | 97.7 | 100.0 | 100.0 | 93.0 | 88.4 | 88.4 | 95.4 |
| Postprandial samples | 93.8 | 100.0 | 93.8 | 100.0 | 89.4 | 89.4 | 92.4 | 93.9 |
| All samples | 98.3 | 98.3 | 98.3 | 100.0 | 90.8 | 89.0 | 90.8 | 94.5 |
| HDL-Cha | ||||||||
| Fasting samples | 97.7 | 100.0 | 100.0 | 100.0 | 83.7 | 95.4 | 95.4 | 100.0 |
| Postprandial samples | 100.0 | 100.0 | 100.0 | 87.5 | 92.4 | 97.0 | 100.0 | 100.0 |
| All samples | 98.3 | 100.0 | 100.0 | 96.6 | 89.0 | 96.3 | 98.2 | 100.0 |
Percentages are based on the TEcom values of the first of triplicate LDL-Cha or HDL-Cha measurements on individual samples.
LDL-C, low-density lipoprotein-cholesterol; HDL-C, high-density lipoprotein-cholesterol; HA, homogeneous assay
A, Denka Seiken; B, Wako; C, Kyowa Medex; D, Sekisui Medical.
In the diseased group, the LDL-C reagents fulfilled the NCEP TE requirements in 90–95% of samples. The HDL-C reagents made by manufacturers B, C, and D met the acceptance criteria when used to assay all samples, independent of fasting status.
3). Comparison of Fasting and Postprandial Samples
For all reagents, the LDL-Cha concentrations agreed well with the LDL-Crmp concentrations in both fasting and postprandial samples, whether or not the subjects were diseased. In scatter plots, the postprandial data (Fig. 2, red circles) were well-superimposed on the fasting data (Fig. 2, yellow circles). Bland-Altman plots indicated an absence of systematic errors in the LDL-Cha concentrations. Similar findings were observed when fasting and postprandial HDL-Cha levels were assayed. In both the non-diseased and diseased groups, the postprandial data were compatible with the fasting data (Fig. 3).
Fig. 2.
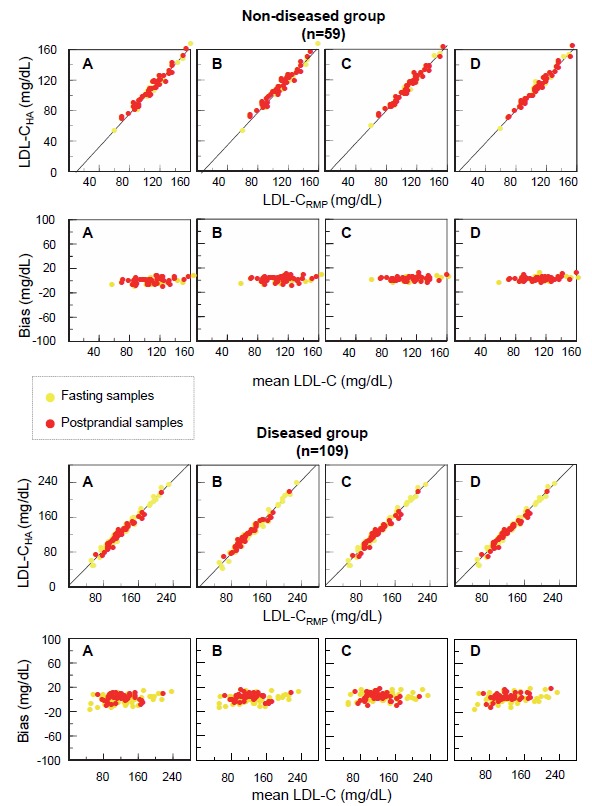
Scatter plots and Bland-Altman plots of the LDL-C values measured by four HAs, and the RMPs of the CDC, in the non-diseased and diseased groups
Fresh blood samples were obtained from non-diseased and diseased subjects of either fasting (yellow circles) or postprandial (red circles) status. LDL-C levels were determined using the HAs and the RMPs of the CDC. For each group, scatter plots are shown in the upper panels, and Bland-Altman plots in the lower panels. In Bland-Altman plots, the X-axis indicates the mean LDL-C values determined by the HAs and the RMPs of the CDC, and the Y-axis indicates the difference in LDL-C levels between the two methods.
A, Denka Seiken; B, Wako; C, Kyowa Medex; D, Sekisui Medical.
Fig. 3.
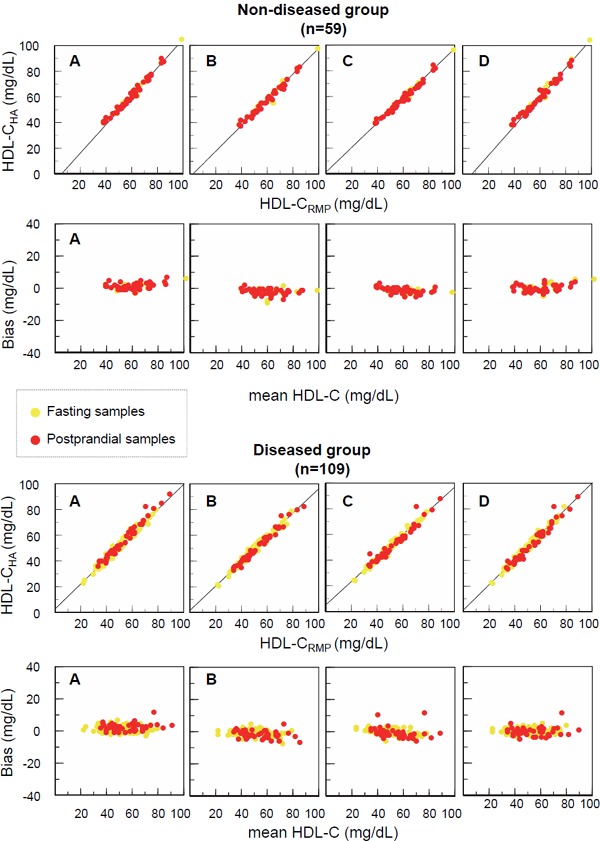
Scatter plots and Bland-Altman plots of HDL-C values measured by four Has, and the RMPs of the CDC in the non-diseased and diseased groups
Fasting (yellow circles) and postprandial (red circles) samples from non-diseased and diseased subjects were subjected to HDL-C measurement using four HAs and the RMPs of the CDC. Scatter plots (upper panels) and Bland-Altman plots (lower panels) were drawn, as described in the legend to Fig. 2.
A, Denka Seiken; B, Wako; C, Kyowa Medex; D, Sekisui Medical.
As in previous studies, we combined the data from fasting and postprandial subjects and evaluated the analytical performance of the LDL-C and HDL-C assay reagents using the TEeca approach. In addition, we calculated TEcom values to allow additional comparisons. For both LDL-Cha and HDL-Cha, the TEcom values were much lower than the NCEP TE requirements (LDL-C, ≤ 12%; HDL-C, ≤ 13%) for all reagents (Table 3). The maximum TEcom values were about 7% for both LDL-Cha and HDL-Cha. On the other hand, the TEeca was 1.1–3.6-fold greater than the TEcom in the non-diseased group, and 1.4–4.4-fold greater in the diseased group. With the exception of the reagents of mnufacturer C in the non-diseased group, the TEeca values of the LDL-C reagents were greater than those of the HDL-C reagents. Although the TEeca values of all HDL-C reagents met the NCEP TE requirements in both the non-diseased and diseased groups, the TEeca values for the LDL-C reagents fulfilled the NCEP TE requirements only in the non-diseased group. Although the TEeca values of the LDL-C reagents exceeded the cut-offs in the diseased group, the differences between the TEeca values and these cut-offs ranged from 0.76% to 1.84%.
Table 3. Comparisons of the analytical performances of homogeneous assays for LDL-C and HDL-C between the non-diseased and diseased groups.
| Group | Non-diseased group (n = 59) |
Diseased group (n = 109) |
||||||
|---|---|---|---|---|---|---|---|---|
| Manufacturer | A | B | C | D | A | B | C | D |
| LDL-Cha | ||||||||
| Between-run CV (%)#1 | 1.01 | 0.99 | 1.44 | 0.96 | − | − | − | − |
| Mean bias (%) | −0.34 | 1.26 | 2.31 | 2.38 | 1.16 | 1.06 | 4.41 | 2.22 |
| TEcom (%) | 2.31 | 3.20 | 5.13 | 4.26 | 3.13 | 3.00 | 7.23 | 4.11 |
| CVb (%)#1 | 0.22 | 0.22 | 0.31 | 0.21 | − | − | − | − |
| CVe (%)#2 | 0.59 | 0.58 | 0.53 | 0.51 | 0.70 | 0.59 | 0.61 | 0.55 |
| CVd (%)#2 | 4.00 | 3.25 | 3.00 | 3.97 | 6.92 | 5.81 | 6.66 | 6.40 |
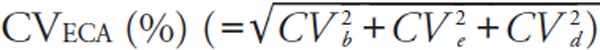
|
4.05 | 3.31 | 3.06 | 4.01 | 6.96 | 5.84 | 6.69 | 6.42 |
| Mean bias [ECA]#3 (%) | −0.11 | 0.46 | 0.49 | 0.26 | 0.20 | 0.90 | 0.40 | 0.18 |
| TEeca #4 (%) | 8.31 | 6.95 | 6.49 | 8.12 | 13.84 | 12.35 | 13.51 | 12.76 |
| HDL-Cha | ||||||||
| Between-run CV (%) | 1.27 | 0.77 | 1.27 | 1.29 | − | − | − | − |
| Mean bias (%) | 2.44 | −3.01 | −1.44 | 0.80 | 4.74 | −1.56 | 0.46 | 1.15 |
| TEcom (%) | 4.92 | 4.51 | 3.93 | 3.33 | 7.23 | 3.07 | 2.95 | 3.68 |
| CVb (%) | 0.27 | 0.17 | 0.27 | 0.28 | − | − | − | − |
| CVe (%) | 0.61 | 0.54 | 0.55 | 0.58 | 0.70 | 0.60 | 0.69 | 0.67 |
| CVd (%) | 3.09 | 2.38 | 3.21 | 2.97 | 4.61 | 4.37 | 4.37 | 3.99 |
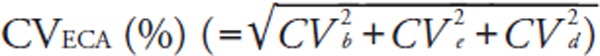
|
3.16 | 2.45 | 3.27 | 3.04 | 4.67 | 4.41 | 4.43 | 4.06 |
| Mean bias [ECA] (%) | 0.61 | −0.33 | 0.20 | −0.74 | 1.19 | 0.13 | 0.28 | −0.40 |
| TEeca (%) | 6.80 | 5.13 | 6.61 | 6.70 | 10.34 | 8.77 | 8.96 | 8.36 |
LDL-C, low-density lipoprotein-cholesterol; CV, coefficient of variance; HDL-C, high-density lipoprotein-cholesterol; HA, homogeneous assay
Between-run CVs were calculated using the LDL-Cha and HDL-Cha values of pooled sera measured on each of 21 days. These values differ from the CVb values used in error component analysis.
CVe and CVd were calculated by error component analysis, as described in ref. #5.
Mean bias [ECA] was calculated by error component analysis, and differs from the mean bias which was used for TEcom assessment.
TEeca was calculated using CVeca, whereas TEcom was calculated using between-run CV employing the following equation (TE= |mean bias (%)| + 1.96 × CV).
A, Denka Seiken; B, Wako; C, Kyowa Medex; D, Sekisui Medical.
Finally, we separately compared the analytical performances of the homogenous assays used to assess fasting and postprandial samples. Both the TEcom and TEeca values of fasting and postprandial data were very similar for all reagents in both groups (Table 4). For all reagents, except the LDL-C reagent of manufacturer C, the TEcom and TEeca values were less than the NCEP TE requirements.
Table 4. Comparison of the analytical performances of homogeneous assays for LDL-C and HDL-C between fasting and postprandial samples of non-diseased and diseased groups.
| Group | Non-diseased group | Diseased group | ||||||
|---|---|---|---|---|---|---|---|---|
| (fasting, n = 16/postprandial, n = 43) |
(fasting, n = 66/postprandial, n = 43) |
|||||||
| Manufacturer | A | B | C | D | A | B | C | D |
| LDL-Cha | ||||||||
| Mean bias (%) | −1.82/0.21 | −0.95/2.09 | 1.63/2.57 | 1.90/2.55 | 0.03/2.88 | −0.29/3.13 | 4.57/4.16 | 1.25/3.71 |
| TEcom (%) | 3.80/2.18 | 2.89/4.03 | 4.45/5.40 | 3.79/4.44 | 2.00/4.85 | 2.24/5.07 | 7.39/6.99 | 3.14/5.60 |
| CVe (%) | 0.60/0.56 | 0.58/0.57 | 0.54/0.50 | 0.52/0.47 | 0.67/0.72 | 0.48/0.64 | 0.60/0.62 | 0.46/0.61 |
| CVd (%) | 3.92/3.30 | 3.11/3.03 | 2.80/3.42 | 3.97/2.36 | 4.90/6.01 | 5.58/5.16 | 5.92/6.73 | 4.37/5.55 |
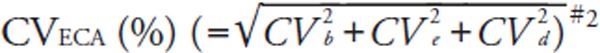
|
3.97/3.36 | 3.17/3.09 | 2.86/3.47 | 4.01/2.42 | 4.95/6.06 | 5.61/5.20 | 5.96/6.76 | 4.40/5.59 |
| Mean bias [ECA] (%) | 0.02/−0.44 | 0.51/0.32 | 0.55/0.36 | 0.46/−0.24 | 0.62/−0.07 | 0.88/0.91 | 0.72/0.20 | 0.67/−0.14 |
| TEeca (%) | 7.80/7.03 | 6.72/6.38 | 6.16/7.16 | 8.32/4.98 | 10.32/11.94 | 11.88/11.10 | 12.40/13.45 | 9.29/11.10 |
| HDL-Cha | ||||||||
| Mean bias (%) | 1.78/2.79 | −3.07/−2.99 | −1.59/−1.38 | 1.23/0.65 | 4.10/5.74 | −1.15/−2.21 | 0.94/−0.27 | 0.83/0.12 |
| TEcom (%) | 3.96/5.27 | 4.57/4.49 | 4.07/3.87 | 3.76/3.17 | 6.58/8.22 | 2.65/3.71 | 3.43/2.76 | 4.35/2.65 |
| CVe (%) | 0.61/0.62 | 0.49/0.65 | 0.50/0.65 | 0.58/0.60 | 0.64/0.74 | 0.57/0.61 | 0.75/0.64 | 0.57/0.72 |
| CVd (%) | 3.40/2.47 | 2.57/2.21 | 3.29/3.58 | 2.98/3.23 | 3.69/4.64 | 6.04/3.21 | 4.79/3.74 | 3.93/4.14 |
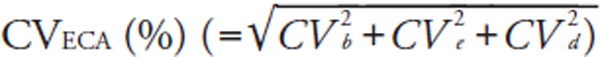
|
3.46/2.56 | 2.62/2.31 | 3.34/3.65 | 3.04/3.30 | 3.75/4.70 | 6.07/3.27 | 4.85/3.81 | 3.98/4.21 |
| Mean bias [ECA] (%) | 0.72/0.33 | −0.31/−0.39 | 0.17/0.26 | −0.73/−0.78 | 1.43/1.04 | −0.09/0.27 | 0.00/0.47 | −0.55/−0.31 |
| TEeca (%) | 7.50/5.35 | 5.45/4.92 | 6.72/7.41 | 6.69/7.25 | 8.78/10.25 | 11.99/6.68 | 9.51/7.94 | 8.35/8.56 |
LDL-C, low-density lipoprotein-cholesterol; CV, coefficient of variance; HDL-C, high-density lipoprotein-cholesterol; HA, homogeneous assay
Data in each cell are the calculated values for fasting (left) and postprandial (right) samples.
All parameters were calculated as described in the legend to Table 3.
A, Denka Seiken; B, Wako; C, Kyowa Medex; D, Sekisui Medical
Discussion
This study indicates that the homogeneous assays for LDL-C and HDL-C from all four manufacturers are as accurate for postprandial samples as for fasting samples. We found that the TEcom values for all reagents were less than 8% for the LDL-C reagents, and less than 9% for the HDL-C reagents, used to assay both fasting and postprandial samples in both the non-diseased and diseased groups (Table 4). Scatter plots and Bland-Altman plots showed that none of the homogeneous assays for LDL-C and HDL-C differed in terms of reactivity when used to assay fasting and postprandial samples (Figs. 2 and 3).
The LDL-C concentration is one of the most important risk markers of cardiovascular diseases, and is recognized as such in many countries16–22). Although LDL-C concentrations are often calculated using Friedewald's formula23), the accuracy depends on the TG concentration24). This is attributable to an increase in chylomicron levels25), and compositional changes of TG-rich lipoproteins, in patients with hypertriglyceridemia24, 26). To overcome these problems, several modified equations have been proposed by different groups24, 27–32). However, most methods, including that of Friedewald, require subjects to fast overnight to exclude a negative bias in LDL-C measurements27–33). In clinical practice, it is sometimes difficult to obtain fasting samples. For example, almost 90% of patients with acute coronary syndrome (ACS) have not fasted for a sufficiently long period to allow LDL-C levels to be calculated34). In outpatients with diabetes mellitus, postprandial samples are preferred, because postprandial hyperglycemia and hyperlipidemia are established risk factors for atherosclerotic disorders35–39). Homogeneous assays are useful to determine the lipid profiles of postprandial patients.
In our previous study, we examined circadian changes in LDL-C and HDL-C concentrations determined by the homogeneous assays40). Although the mean TG concentration increased by 20.9% in the control group, and by 30.9% in the patients with coronary artery disease, LDL-C and HDL-C concentrations did not increase during the day. Instead, LDL-C and HDL-C decreased by 1.6 to 6.0%, and from 0.0% to 6.0% from the fasting state. These reductions were comparable to that in total cholesterol. Therefore, it is strongly suggested that homogeneous assays for LDL-C and HDL-C are not affected by postprandial increase in TG concentration. These results agree well with those of the present study.
To date, only two groups (including our group) have explored the accuracy of homogeneous assays for LDL-C and HDL-C using fresh sera from non-diseased and diseased subjects of various backgrounds5, 6, 10, 11). Both groups employed homogeneous assays to measure LDL-Cha and HDL-Cha levels, and also measured LDL-Crmp and HDL-Crmp using the RMPs of the CDC in a single central laboratory of a CRMLN institution. Both groups found that some homogeneous assays were of poor analytical performance5, 6, 10, 11). These studies prompted the manufacturers and distributors to withdraw defective reagents from the Japanese market. Therefore, all currently available homogeneous assays for LDL-C use the four original reagents (examined in the present study) or products derived therefrom. The LDL-C homogeneous assay of Kyowa Medex employs a modified version of the prior formulation5). Here, this new LDL-C assay was shown for the first time to exhibit satisfactory accuracy. In terms of HDL-C assays, six original reagents (including four examined in the present study) and their derivatives remain on the Japanese market. However, homogenous assays exhibit significant diversity in terms of LDL or HDL reactivities when sera from specific patients with extremely low or high lipoprotein concentrations are assayed, and when sera from those with highly abnormal lipoprotein compositions (such as patients with cholestatic liver diseases) are evaluated5, 6, 10, 11, 26, 41, 42). Therefore, homogeneous assays should only be used to screen for dyslipidemia in, and evaluate therapies for, subjects without disease and those with common disorders. The results of our two studies indicate that the reagents tested here in patients with TG concentrations < 1,000mg/dL are reliable.
In the present study, we calculated two distinct TE values: TEcom and TEeca. In general, TEcom is used for quality assurance surveillance14). Furthermore, TEcom was adopted by the CDC when establishing certification protocols for manufacturers of LDL-C and HDL-C assay reagents7–9). Total error is a concept reflecting precision and accuracy simultaneously. Precision refers to reproducibility upon multiple measurements, whereas accuracy refers to how close the measured values are to the true values14). In the present study, the TEcom values of all reagents were much lower than the NCEP requirements (12% for LDL-C and 13% for HDL-C), whether or not the study subjects were fasting or diseased. The TEcom data showed that all tested reagents exhibited satisfactory analytical performance.
It is rather puzzling that the TEeca values were much worse in the study of Miller than in our current work, even though both groups ran error component analysis using similar protocols5, 6, 10, 11). We speculate that the measurement errors recorded by Miller et al.10, 11) might have been accidentally exaggerated for the following reasons. The TEeca is based on the hypotheses that errors are exhibited by the log-transformed variations of three components; CVb, CVe, and CVd. CVd originates from patient-sample-specific effect, and is calculated as the mean square successive difference (MSSD) between the sorted measurement values, as follows:
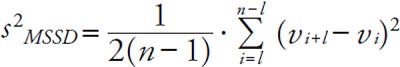 |
The major assumption of this equation is that Vi+1 − Vi is both very small and continuous5, 15). However, not every TEeca based on the MSSD is valid in diseased samples when LDL measurements exhibit discrete distributions. All previous publications, including ours5, 6, 10, 11), that employed the MSSD violated this continuity assumption (Supplemental Table 2). From the equation above, as the TE is a quadratic function of the MSSD, the TEeca is usually higher than the TEcom if the distributions are discrete. As shown in Table 3, this is in line with the fact that the TEcom and TEeca exhibited large discrepancies, although conventional assessments of LDL and HDL measurement accuracies were in the permissive ranges.
We should be aware that most epidemiological and clinical studies have used Friedewald's formula for LDL-C estimation although there are several methods to measure LDL-C concentrations. In the Lipid Research Clinics Coronary Primary Prevention Trial (LRC-CPPT), the first study to demonstrate the significant reduction in coronary artery disease by LDL-C lowering with cholestylamine, LDL-C concentrations were determined by ultracentrifugation and subsequent precipitation of apoB-containing lipoproteins43). The CDC's RMP of LDL-C measurement is based on this method with some modifications. In the early clinical trials with statins such as the West of Scotland Coronary Prevention Study (WOS-COPS)44) and the Scandinavian Simvastatin Survival Study (4S)45), LDL-C concentration was measured by the method of the LRC-CPPT43). Subsequently, many clinical trials and cohort studies used Friedewald's formula for LDL-C estimation. After the development of homogeneous assays in the late 1990s, some clinical trials including the Management of Elevated cholesterol in the primary prevention Group of Adult Japanese (MEGA) Study46), the Pravastatin or Atorvastatin Evaluation and Infection Therapy-Thrombolysis In Myocardial Infarction 22 (PROVE IT-TIMI 22) trial47), and the Study of Coronary Atheroma by Intravascular Ultrasound: Effect of Rosuvastatin versus Atorvastatin (SATURN)48), LDL-C concentrations were determined basically by the Friedewald's formula, but by the homogeneous assays when TG concentrations exceeded 300 or 400 mg/dL. More recently, some clinical and epidemiological studies used a homogeneous assay as the only method to measure LDL-C concentration49–53). Therefore, we need to accumulate more evidence that LDL-C concentration determined by homogeneous assays is a significant predictor of cardiovascular diseases.
We conclude that the homogeneous assays for LDL-C and HDL-C available from four manufacturers are as accurate for postprandial samples as for fasting samples. Although our series of studies on homogeneous assays has encouraged the withdrawal and improvement of reagents exhibiting poor analytical performance, future reagents require extensive preclinical evaluation to avoid unnecessary confusion.
Acknowledgments
This work was supported by a Health and Labour Sciences Research Grant (Japan; Comprehensive Research on Non-Communicable Diseases including Cardiovascular Diseases and Diabetes Mellitus; principal investigator Tamio Teramoto). The authors would like to thank the four Japanese manufacturers mentioned in the Methods section for providing the reagents used in the LDL-C and HDL-C homogeneous assays, calibrators, and controls.
Conflict of Interest
We identify all potential conflicts of interests associated with this study even when the total amount of research funding and/or honoraria received did not exceed that which the Japan Atherosclerosis Society requires members to disclose. Takashi Miida received research funding from Denka Seiken and Wako, and honoraria from Denka Seiken, Kyowa Medex, and Sekisui Medical. Satoshi Hirayama received research funding from Denka Seiken and Wako. Daisaku Masuda and Shizuya Yamashita received research funding from Kyowa Medex. However, the data presented in this article were not given to the manufacturers prior to submission, and the manufacturers were not involved in data analysis.
References
- 1). Sugiuchi H, Uji Y, Okabe H, Irie T, Uekama K, Kayahara N, Miyauchi K: Direct measurement of high-density lipoprotein cholesterol in serum with polyethylene glycolmodified enzymes and sulfated alpha-cyclodextrin. Clin Chem, 1995; 41: 717-723 [PubMed] [Google Scholar]
- 2). Nakamura M, Taniguchi Y, Yamamoto M, Hino K, Manabe M: Homogenous assay of serum LDL-cholesterol on an automatic analyzer [Abstract]. Clin Chem, 1997; 43: S260 [Google Scholar]
- 3). Sugiuchi H, Irie T, Uji Y, Ueno T, Chaen T, Uekama K, Okabe H: Homogeneous assay for measuring lowdensity lipoprotein cholesterol in serum with triblock copolymer and alpha-cyclodextrin sulfate. Clin Chem, 1998; 44: 522-531 [PubMed] [Google Scholar]
- 4). Okada M, Matsui H, Ito Y, Fujiwara A: Direct measurement of HDL cholesterol: method eliminating apolipoprotein E-rich particles. J Clin Lab Anal, 2001; 15: 223-229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5). Miida T, Nishimura K, Okamura T, Hirayama S, Ohmura H, Yoshida H, Miyashita Y, Ai M, Tanaka A, Sumino H, Murakami M, Inoue I, Kayamori Y, Nakamura M, Nobori T, Miyazawa Y, Teramoto T, Yokoyama S: A multicenter study on the precision and accuracy of homogeneous assays for LDL-cholesterol: Comparison with a beta-quantification method using fresh serum obtained from non-diseased and diseased subjects. Atherosclerosis, 2012; 225: 208-215 [DOI] [PubMed] [Google Scholar]
- 6). Miida T, Nishimura K, Okamura T, Hirayama S, Ohmura H, Yoshida H, Miyashita Y, Ai M, Tanaka A, Sumino H, Murakami M, Inoue I, Kayamori Y, Nakamura M, Nobori T, Miyazawa Y, Teramoto T, Yokoyama S: Validation of homogeneous assays for HDL-cholesterol using fresh samples from healthy and diseased subjects. Atherosclerosis, 2014; 233: 253-259 [DOI] [PubMed] [Google Scholar]
- 7). Centers for Disease Control and Prevention: Manufacturer certification program. http://www.cdc.gov/labstandards/crmln_manufacturers.html (Accessed January 2017)
- 8). National Institutes of Health: Second report of the Expert Panel on Detection, Evaluation, and Treatment of High Blood Cholesterol in Adults. Bethesda, MD: National Institutes of Health, 1993. NIH publication no: 93-3095 [Google Scholar]
- 9). Bachorik PS, Ross JW: National Cholesterol Education Program recommendations for measurement of lowdensity lipoprotein cholesterol: executive summary. Clin Chem, 1995; 41: 1414-1420 [PubMed] [Google Scholar]
- 10). Miller WG, Myers GL, Sakurabayashi I, Bachmann LM, Caudill SP, Dziekonski A, Edwards S, Kimberly MM, Korzun WJ, Leary ET, Nakajima K, Nakamura M, Nilsson G, Shamburek RD, Vetrovec GW, Warnick GR, Remaley AT: Seven direct methods for measuring HDL and LDL cholesterol compared with ultracentrifugation reference measurement procedures. Clin Chem, 2010; 56: 977-986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11). van Deventer HE, Miller WG, Myers GL, Sakurabayashi I, Bachmann LM, Caudill SP, Dziekonski A, Edwards S, Kimberly MM, Korzun WJ, Leary ET, Nakajima K, Nakamura M, Shamburek RD, Vetrovec GW, Warnick GR, Remaley AT: Non-HDL cholesterol shows improved accuracy for cardiovascular risk score classification compared to direct or calculated LDL cholesterol in a dyslipidemic population. Clin Chem, 2011; 57: 490-501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12). World Medical Association: WMA declaration of Helsinki - Ethical principles for medical research involving human subjects. http://www.wma.net/en/30publications/10policies/b3/index.html (Accessed January 2017)
- 13). Abell LL, Levy BB, Brodie RB, Kendall FE: Simplified method for the estimation of total cholesterol in serum, and demonstration of its specificity. J Biol Chem, 1952; 195: 357-366 [PubMed] [Google Scholar]
- 14). Westgard JO: Useful measures and models for analytical quality management in medical laboratories. Clin Chem Lab Med, 2015; 54: 223-233 [DOI] [PubMed] [Google Scholar]
- 15). Nilsson G: Comparison of measurement methods based on a model for the error structure. J Chemometrics, 1991; 5: 523-536 [Google Scholar]
- 16). Reiner Z, Catapano AL, De Backer G, Graham I, Taskinen MR, Wiklund O, Agewall S, Alegria E, Chapman MJ, Durrington P, Erdine S, Halcox J, Hobbs R, Kjekshus J, Filardi PP, Riccardi G, Storey RF, Wood D: ESC/EAS guidelines for the management of dyslipidaemias: the task force for the management of dyslipidaemias of the European Society of Cardiology (ESC) and the European atherosclerosis Society (EAS). Eur Heart J, 2011; 32: 1769-1818 [DOI] [PubMed] [Google Scholar]
- 17).Expert Dyslipidemia Panel of the International Atherosclerosis Society Panel members: An International Atherosclerosis Society Position Paper: global recommendations for the management of dyslipidemia–full report. J Clin Lipidol, 2014; 8: 29-60 [DOI] [PubMed] [Google Scholar]
- 18). Teramoto T, Sasaki J, Ishibashi Sh, Birou S, Daida H, Dohi S, Egusa G, Hiro T, Hirobe K, Iida M, Kihara S, Kinoshita M, Maruyama C, Ohta T, Okamura T, Yamashita S, Yokode M, Yokote K: Executive summary of the Japan Atherosclerosis Society (JAS) guidelines for the diagnosis and prevention of atherosclerotic cardiovascular diseases in Japan-2012 version. J Atheroscler Thromb, 2013; 20: 517-523 [DOI] [PubMed] [Google Scholar]
- 19). Teramoto T, Kawamori R, Miyazaki S, Teramukai S, Sato Y, Okuda Y, Shirayama M. Lipid and blood pressure control for the prevention of cardiovascular disease in hypertensive patients: a subanalysis of the OMEGA study. J Atheroscler Thromb, 2015; 22: 62-75 [DOI] [PubMed] [Google Scholar]
- 20). Shimabukuro M1, Hasegawa Y, Higa M, Amano R, Yamada H, Mizushima S, Masuzaki H, Sata M. Subclinical carotid atherosclerosis burden in the Japanese: Comparison between Okinawa and Nagano residents. J Atheroscler Thromb, 2015; 22: 854-868 [DOI] [PubMed] [Google Scholar]
- 21). Wakabayashi K, Nozue T, Yamamoto S, Tohyama S, Fukui K, Umezawa S, Onishi Y, Kunishima T, Sato A, Miyake S, Morino Y, Yamauchi T, Muramatsu T, Hibi K, Terashima M, Suzuki H, Michishita I for TRUTH investigators Efficacy of statin therapy in inducing coronary plaque regression in patients with low baseline cholesterol levels. J Atheroscler Thromb, 2016; 23: 1055-1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22). Naito R, Miyauchi K, Daida H, Morimoto T, Hiro T, Kimura T, Nakagawa Y, Yamagishi M, Ozaki Y, Matsuzaki M for JAPAN-ACS Investigators Impact of total risk management on coronary plaque regression in diabetic patients with acute coronary syndrome - Sub Analysis of JAPAN-ACS Study -. J Atheroscler Thromb, 2016; 23: 922-931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23). Friedewald WT, Levy RI, Fredrickson DS: Estimation of the concentration of low-density lipoprotein cholesterol in plasma, without use of the preparative ultracentrifuge. Clin Chem, 1972; 18: 499-502 [PubMed] [Google Scholar]
- 24). Martin SS, Blaha MJ, Elshazly MB, Toth PP, Kwiterovich PO, Blumenthal RS, Jones SR: Comparison of a novel method vs the Friedewald equation for estimating low-density lipoprotein cholesterol levels from the standard lipid profile. JAMA, 2013; 310: 2061-2068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25). Nakajima K, Nakano T, Tokita Y, Nagamine T, Inazu A, Kobayashi J, Mabuchi H, Stanhope KL, Havel PJ, Okazaki M, Ai M, Tanaka A: Postprandial lipoprotein metabolism: VLDL vs chylomicrons. Clin Chim Acta, 2011; 412: 1306-1318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26). Kurosawa H, Yoshida H, Yanai H, Ogura Y, Hirowatari Y, Tada N: Comparative study between anion-exchange HPLC and homogeneous assay methods in regard to the accuracy of high- and low-density lipoprotein cholesterol measurement. Clin Biochem, 2007; 40: 1291-1296 [DOI] [PubMed] [Google Scholar]
- 27). DeLong DM, DeLong ER, Wood PD, Lippel K, Rifkind BM: A comparison of methods for the estimation of plasma low- and very low-density lipoprotein cholesterol. The Lipid Research Clinics Prevalence Study. JAMA, 1986; 256: 2372-2377 [PubMed] [Google Scholar]
- 28). Anandaraja S, Narang R, Godeswar R, Laksmy R, Talwar KK: Low-density lipoprotein cholesterol estimation by a new formula in Indian population. Int J Cardiol, 2005; 102: 117-120 [DOI] [PubMed] [Google Scholar]
- 29). de Cordova CM, de Cordova MM: A new accurate, simple formula for LDL-cholesterol estimation based on directly measured blood lipids from a large cohort. Ann Clin Biochem, 2013; 50: 13-19 [DOI] [PubMed] [Google Scholar]
- 30). Rao A, Parker AH, el-Sheroni NA, Babelly MM: Calculation of low-density lipoprotein cholesterol with use of triglyceride/cholesterol ratios in lipoproteins compared with other calculation methods. Clin Chem, 1988; 34: 2532-2534 [PubMed] [Google Scholar]
- 31). Chen Y, Zhang X, Pan B, Jin X, Yao H, Chen B, Zou Y, Ge J, Chen H: A modified formula for calculating low-density lipoprotein cholesterol values. Lipids Health Dis, 2010; 9: 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32). Vujovic A, Kotur-Stevuljevic J, Spasic S, Bujisic N, Martinovic J, Vujovic M, Spasojevic-Kalimanovska V, Zeljkovic A, Pajic D: Evaluation of different formulas for LDL-C calculation. Lipids Health Dis, 2010; 9: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33). Chaen H1, Kinchiku S, Miyata M, Kajiya S, Uenomachi H, Yuasa T, Takasaki K, Ohishi M. Validity of a novel method for estimation of low-density lipoprotein cholesterol levels in diabetic patients. J Atheroscler Thromb, 2016; 23: 1355-1364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34). Fukushima Y, Hirayama S, Ueno T, Dohi T, Miyazaki T, Ohmura H, Mokuno H, Miyauchi K, Miida T, Daida H: Small dense LDL cholesterol is a robust therapeutic marker of statin treatment in patients with acute coronary syndrome and metabolic syndrome. Clin Chim Acta, 2011; 412: 1423-1427 [DOI] [PubMed] [Google Scholar]
- 35). DECODE Study Group, and the European Diabetes Epidemiology Group : Glucose tolerance and cardiovascular mortality: comparison of fasting and 2-hour diagnostic criteria. Arch Intern Med, 2001; 161: 397-405 [DOI] [PubMed] [Google Scholar]
- 36). Tominaga M, Eguchi H, Manaka H, Igarashi K, Kato T, Sekikawa A: Impaired glucose tolerance is a risk factor for cardiovascular disease, but not impaired fasting glucose. The Funagata Diabetes Study. Diabetes Care, 1999; 22: 920-924 [DOI] [PubMed] [Google Scholar]
- 37). Holme I, Tonstad S: Association of coronary heart disease mortality with risk factors according to length of follow-up and serum cholesterol level in men: the Oslo Study cohort. Eur J Prev Cardiol, 2013; 20: 168-175 [DOI] [PubMed] [Google Scholar]
- 38). Idei M, Hirayama S, Miyake N, Kon M, Horiuchi Y, Ueno T, Miyake K, Sato N, Yoshii H, Yamashiro K, Onuma T, Miida T: The mean postprandial triglyceride concentration is an independent risk factor of carotid atherosclerosis in patients with type 2 diabetes. Clin Chim Acta, 2014; 430: 134-139 [DOI] [PubMed] [Google Scholar]
- 39). Iso H, Naito Y, Sato S, Kitamura A, Okamura T, Sankai T, Shimamoto T, Iida M, Komachi Y: Serum triglycerides and risk of coronary heart disease among Japanese men and women. Am J Epidemiol, 2001; 153: 490-499 [DOI] [PubMed] [Google Scholar]
- 40). Miida T, Nakamura Y, Mezaki T, Hanyu O, Maruyama S, Horikawa Y, Izawa S, Yamada Y, Matsui H, Okada M: LDL-cholesterol and HDL-cholesterol decrease during the day. Ann Clin Biochem, 2002; 39: 241-249 [DOI] [PubMed] [Google Scholar]
- 41). Timón-Zapata J, Laserna-Mendieta EJ, Sáenz-Mateos LF, Ruiz-Trujillo L, Arpa-Fernández A, Palomino-Muñoz TJ, Loeches-Jiménez MP, Gómez-Serranillos M: A multicentre analysis of four low-density lipoprotein cholesterol direct assays in samples with extreme high-density lipoprotein cholesterol concentrations. Clin Chim Acta, 2014; 430: 71-76 [DOI] [PubMed] [Google Scholar]
- 42). Matsushima K, Sugiuchi H, Anraku K, Nishimura H, Manabe M, Ikeda K, Ando Y, Kondo Y, Ishitsuka Y, Irikura M, Irie T: Differences in reaction specificity toward lipoprotein X and abnormal LDL among 6 homogeneous assays for LDL-cholesterol. Clin Chim Acta, 2015; 439: 29-37 [DOI] [PubMed] [Google Scholar]
- 43). Hainline A, Jr, Karon J, Lippel K. Manual of laboratory operations: Lipid and lipoprotein analysis (2nd ed.) [HEW Pub. No. (NIH) 75-628 (rev.), U.S. Government Printing Office Publication No. 1982-361-132: 678.] Bethesda, MD: National Heart, Lung and Blood Institute, Lipid Research Clinics Program [Google Scholar]
- 44). Shepherd J, Cobbe SM, Ford I, Isles CG, Lorimer AR, MacFarlane PW, McKillop JH, Packard CJ: Prevention of coronary heart disease with pravastatin in men with hypercholesterolemia. N Engl J Med, 1995; 333: 1301-1307 [DOI] [PubMed] [Google Scholar]
- 45). Scandinavian Simvastatin Survival Study Group Randomised trial of cholesterol lowering in 4444 patients with coronary heart disease: the Scandinavian Simvastatin Survival Study (4S). Lancet, 1994; 344: 1383-1389 [PubMed] [Google Scholar]
- 46). Management of Elevated cholesterol in the primary prevention Group of Adult Japanese (MEGA) study group : Design and baseline characteristics of a study of primary prevention of coronary events with pravastatin among Japanese with mildly elevated cholesterol levels. Circ J, 2004; 68: 860-867 [DOI] [PubMed] [Google Scholar]
- 47). Miller M, Cannon CP, Murphy SA, Qin J, Ray KK, Braunwald E: Impact of triglyceride levels beyond lowdensity lipoprotein cholesterol after acute coronary syndrome in the PROVE IT-TIMI 22 trial. J Am Coll Cardiol, 2008; 51: 724-730 [DOI] [PubMed] [Google Scholar]
- 48). Nicholls SJ, Ballantyne CM, Barter PJ, Chapman MJ, Erbel RM, Libby P, Raichlen JS, Uno K, Borgman M, Wolski K, Nissen SE: Effect of two intensive statin regimens on progression of coronary disease. N Engl J Med, 2011; 365: 2078-2087 [DOI] [PubMed] [Google Scholar]
- 49). Masuda J, Tanigawa T, Yamada T, Nishimura Y, Sasou T, Nakata T, Sawai T, Fujimoto N, Dohi K, Miyahara M, Nishikawa M, Nakamura M, Ito M. Effect of combination therapy of ezetimibe and rosuvastatin on regression of coronary atherosclerosis in patients with coronary artery disease. Int Heart J, 2015; 56: 278-285 [DOI] [PubMed] [Google Scholar]
- 50). Tsujita K, Sugiyama S, Sumida H, Shimomura H, Yamashita T, Yamanaga K, Komura N, Sakamoto K, Ono T, Oka H, Nakao K, Nakamura S, Ishihara M, Matsui K, Sakaino N, Nakamura N, Yamamoto N, Koide S, Matsumura T, Fujimoto K, Tsunoda R, Morikami Y, Matsuyama K, Oshima S, Kaikita K, Hokimoto S, Ogawa H: Plaque REgression with Cholesterol absorption Inhibitor or Synthesis inhibitor Evaluated by IntraVascular UltraSound (PRECISE-IVUS Trial): Study protocol for a randomized controlled trial. J Cardiol, 2015; 66: 353-358 [DOI] [PubMed] [Google Scholar]
- 51). Otokozawa S, Ai M, Asztalos BF, White CC, Demissie-Banjaw S, Cupples LA, Nakajima K, Wilson PW, Schaefer EJ: Direct assessment of plasma low density lipoprotein and high density lipoprotein cholesterol levels and coronary heart disease: results from the Framingham Offspring Study. Atherosclerosis, 2010; 213: 251-255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52). Tanaka F, Makita S, Onoda T, Tanno K, Ohsawa M, Itai K, Sakata K, Omama S, Yoshida Y, Ogasawara K, Ogawa A, Ishibashi Y, Kuribayashi T, Okayama A, Nakamura M: Predictive value of lipoprotein indices for residual risk of acute myocardial infarction and sudden death in men with low-density lipoprotein cholesterol levels < 120 mg/dl. Am J Cardiol, 2013; 112: 1063-1068 [DOI] [PubMed] [Google Scholar]
- 53). Arai H, Kokubo Y, Watanabe M, Sawamura T, Ito Y, Minagawa A, Okamura T, Miyamato Y: Small dense low-density lipoproteins cholesterol can predict incident cardiovascular disease in an urban Japanese cohort: the Suita study. J Atheroscler Thromb, 2013; 20: 195-203 [DOI] [PubMed] [Google Scholar]


