Abstract
Intestinal tuft cells are a rare, poorly understood cell type recently shown to be a critical mediator of type 2 immune response to helminth infection. Here, we present advances in segmentation algorithms and analytical tools for multiplex immunofluorescence (MxIF), a platform that enables iterative staining of over 60 antibodies on a single tissue section. These refinements have enabled a comprehensive analysis of tuft cell number, distribution, and protein expression profiles as a function of anatomical location and physiological perturbations. Based solely on DCLK1 immunoreactivity, tuft cell numbers were similar throughout the mouse small intestine and colon. However, multiple subsets of tuft cells were uncovered when protein coexpression signatures were examined, including two new intestinal tuft cell markers, Hopx and EGFR phosphotyrosine 1068. Furthermore, we identified dynamic changes in tuft cell number, composition, and protein expression associated with fasting and refeeding and after introduction of microbiota to germ-free mice. These studies provide a foundational framework for future studies of intestinal tuft cell regulation and demonstrate the utility of our improved MxIF computational methods and workflow for understanding cellular heterogeneity in complex tissues in normal and disease states.
Keywords: Gastroenterology
Introduction
Multiplex immunofluorescence (MxIF) is a recently developed imaging platform whereby up to 60 directly labeled antibodies can be applied to a single tissue section (1–3). Following multiple rounds of staining, imaging, and dye inactivation, single-cell segmentation enables subcellular quantification of antibody staining as well as measurement of cell size, shape, and position. It was first used to study human colorectal cancer, in which it underscored the cellular heterogeneity that exists among individual cancer cells (1), including expression of unique markers at the invasive front (3). Other immunofluorescent quantification techniques have been developed from simple thresholding of images to more complex analyses, such as determining colocalization across subcellular domains for a few markers (4) and at the pixel level for many markers (5). Additional methods of multiplexed image acquisition have been proposed, including tyramide signal amplification and multispectral unmixing (6). However, none of these techniques have developed accompanying methods for single-cell analysis of tissues.
For analysis at the single-cell level, MxIF and other antibody-based techniques, such as multicolor flow cytometry (7) and mass cytometry (CyTOF) (8, 9), enable the analysis of large numbers of cells (104–106 cells), with a relatively modest number of protein analytes (15–100 analytical dimensions). Importantly, however, MxIF retains the tissue architecture that is lost when cells are dispersed for flow or mass cytometric analysis. While imaging mass cytometry (10) and multiplexed ion beam imaging (11) do retain some information regarding the spatial context from which the data are derived, limitations in the resolution of the technology have historically prohibited development of single-cell analysis algorithms to reconstruct the data at the cellular level. Recent advances in sequencing technology have enabled single-cell analysis of DNA and RNA (12). These sequencing techniques typically result in the study of a relatively small number of cells (102–103 cells) coupled with the measurement of large portions of the genome or transcriptome (104 analytical dimensions). The relatively limited number of cells analyzed by these methods does not allow for the study of rare cell types without extensive presorting. Recently, in situ barcoded sequential hybridization techniques for the detection of DNA and RNA have been developed (13) that should allow high-dimensional analysis at, in principle, the single-cell level in tissues. Here, we demonstrate the use of MxIF for the characterization of a rare cell type, the intestinal tuft cell.
Tuft, or caveolated, cells are a rare differentiated cell type found throughout the gastrointestinal tract. They comprise approximately 0.4% of all intestinal epithelial cells, exhibit a characteristic dense bundle of apical microfilaments, and serve a role in chemosensing (14, 15). They are thought to be derived from a secretory lineage (15), although this remains controversial (16). Recent elegant studies have demonstrated that helminth infection markedly increased tuft cell number and triggered a cytokine signaling cascade involving innate lymphoid group 2 cells (17–19). A number of markers have previously been identified for intestinal tuft cells (summarized by Gerbe et al., ref. 20), including structural (DCLK1, acetylated tubulin), taste-related (α-gustducin), and progenitor/stem (Sox9, Lgr5, ref. 21) markers. Applying newly developed MxIF analytical tools, we quantified tuft cell number and distribution throughout the mouse small intestine and colon. We identified two previously unreported intestinal tuft cell markers, Hopx and EGFR phosphotyrosine 1068 (p-EGFR), and characterized tuft cell heterogeneity based on combinatorial protein profiles in response to two physiological perturbations. The presence of p-EGFR in human duodenum and colon was confirmed by high-resolution microscopy.
Results
Improved MxIF analytical methods.
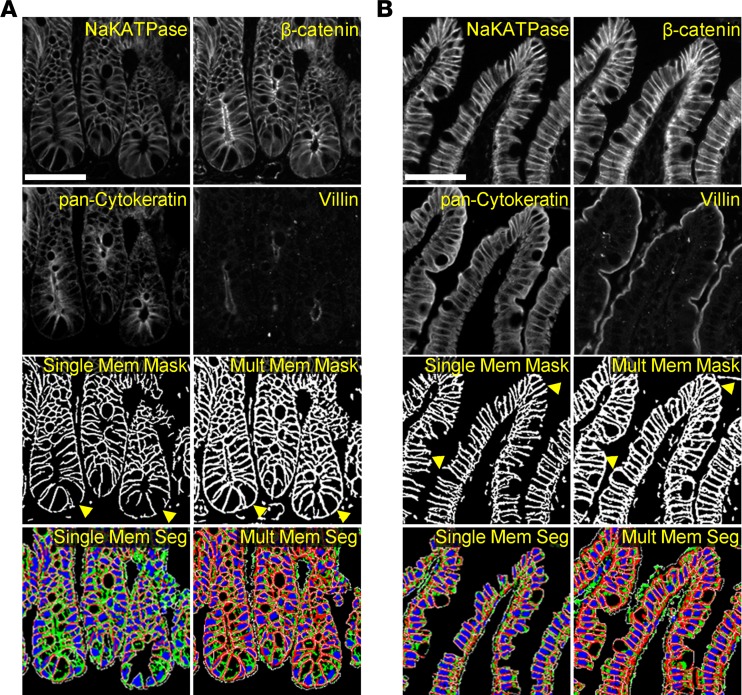
We observed that cell segmentation methods, previously developed for analysis of cancer-derived tissues (1, 2), performed suboptimally when applied to normal intestinal epithelium. In cancer, cells are typically symmetrically shaped, with uniform membrane protein expression due to loss of apico-basolateral polarity (22). In normal intestine, the epithelial population is made up of a mixture of cells at various stages of differentiation, resulting in varying cell shapes and sizes as well as nonuniform coverage of membrane markers due to separate apical and basolateral cell compartments. To overcome these issues, the antibody stains were reviewed with an emphasis on membrane or membrane-associated proteins to select a combination of markers that provided coverage of all intestinal epithelial cells. Multiple membrane markers were combined into a single mask to achieve this goal (Figure 1). Compared with using NaKATPase as a single membrane marker, the combined membrane mask derived from NaKATPase, β-catenin, pan-cytokeratin, and villin resulted in improved basolateral coverage at the crypt base (Figure 1A) and improved apical coverage in the villus compartment (Figure 1B). This combined mask ultimately resulted in greatly improved final cell segmentation and subsequently increased the quality of biomarker quantitation and cell morphological measurements. Additionally, advances in computational methods, such as cloud-based parallel computation using the open-source KNIME platform, allowed the rapid processing of the extremely large imaging data sets collected for this study.
Figure 1. Improvements in cell segmentation.
Segmentation of intestinal epithelial cells was improved by the use of multiple membrane markers. At the crypt (A) and tip of villus (B), the computational addition of membrane markers (NaKATPase, β-catenin, pan-cytokeratin, and villin) resulted in more complete membrane coverage than the use of NaKATPase as a single marker, as utilized in previous cancer studies. Membrane masks, used as one of the inputs to cellular segmentation, revealed a lack of coverage at the basal surface of cells in the bottom of the crypt and an inability to close the apical portion of the cell using NaKATPase as a single marker. These defects were largely ameliorated when the mask was generated using multiple markers (arrowheads). Similarly, the final cell segmentation (red: membrane; green: cytoplasm; blue: nucleus; white: cell border) derived from a single marker demonstrated substantial loss of cells due to the lack of robust membrane delineation that was vastly improved using the multiple marker input. Scale bar: 50 μm.
Using these optimized analytical methods, MxIF was performed with 19 markers, including the newly characterized ones, to identify tuft cell subpopulations over the entire small intestine (duodenum, jejunum, and ileum) and colon of 3 Lrig1-Apple; Lgr5-DTR-EGFP (23, 24) bigenic adult male mice at homeostasis (Supplemental Table 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.93487DS1). An RFP antibody and a GFP antibody allowed visualization of Lrig1- and Lgr5-positive cells, respectively. Lrig1-Apple staining was present at the crypt base and in interstitial cells of Cajal, as previously reported (25). However, it was not coexpressed with DCLK1 and thus was not included in this analysis (Supplemental Figure 1). The use of a comprehensive panel of markers to delineate the different cell populations allows for both isolation of cell populations for visual inspection as well as isolation within the quantitative data sets. Overall, 1,455,335 epithelial cells were analyzed from images of entire Swiss rolls of the duodenum, jejunum, ileum, and colon, of which 7,600 were classified as tuft cells by high DCLK1 staining intensity, defined by a clustering deconvolution method described below.
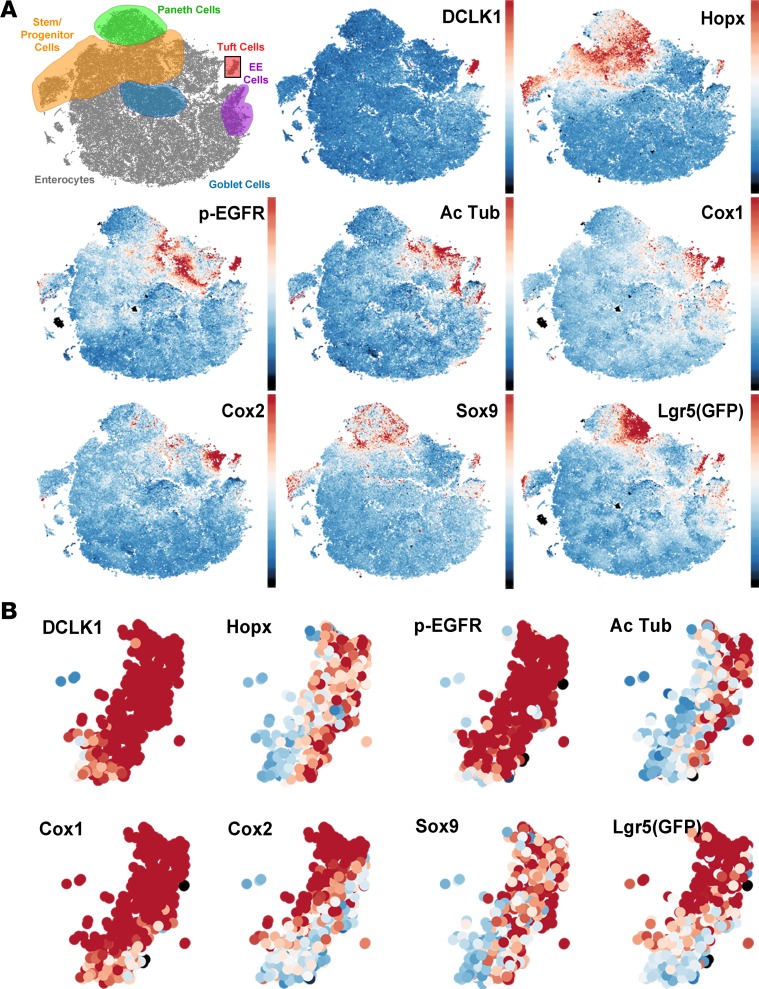
Previous MxIF studies analyzed aggregate cellular expression patterns mainly by clustering (1), with the emphasis placed on the most abundant populations, while masking rare cell types. Although a group of DCLK1-positive cells was identified using these methods, heterogeneity within this infrequent cell type was unresolvable due to the increasing underrepresentation of subpopulations in the analysis. To address this issue, we utilized a model-based clustering deconvolution method to identify intestinal tuft cells, defined here as cells with high DCLK1 expression, to assess their heterogeneity (Supplemental Figure 2). Without user input, a DCLK1 intensity threshold was determined for each sample to capture the intestinal tuft cell population (<1% of all epithelial cells). Subsequently, the deconvolution method was applied on only DCLK1-positive cells to determine intensity thresholds of positivity for the other markers within the DCLK1-positive population. Dimensionality reduction by t-distributed stochastic neighborhood embedding (t-SNE) of this multidimensional data for all epithelial cells clearly demonstrated segregation of the differentiated cell types (representative map shown in ileum, Figure 2A). Tuft cells were clearly distinguished from the other differentiated cell types using t-SNE analysis (Figure 2) and made up approximately 0.4% of epithelial cells (see below), consistent with prior reports (20).
Figure 2. t-SNE maps of epithelial cells in ileum reveal tuft cell heterogeneity.
(A) t-SNE maps of all epithelial cells in a mouse ileum (n = 129,379) reveal discrete localization of differentiated cell types. DCLK1 is constrained to a single “island,” while other tuft cell markers are expressed in other differentiated cell types. (B) Isolation of the tuft cell “island” demonstrates uniform DCLK1 expression and heterogeneous patterns of expression of other tuft cell markers.
Identification of tuft cell markers p-EGFR and Hopx.
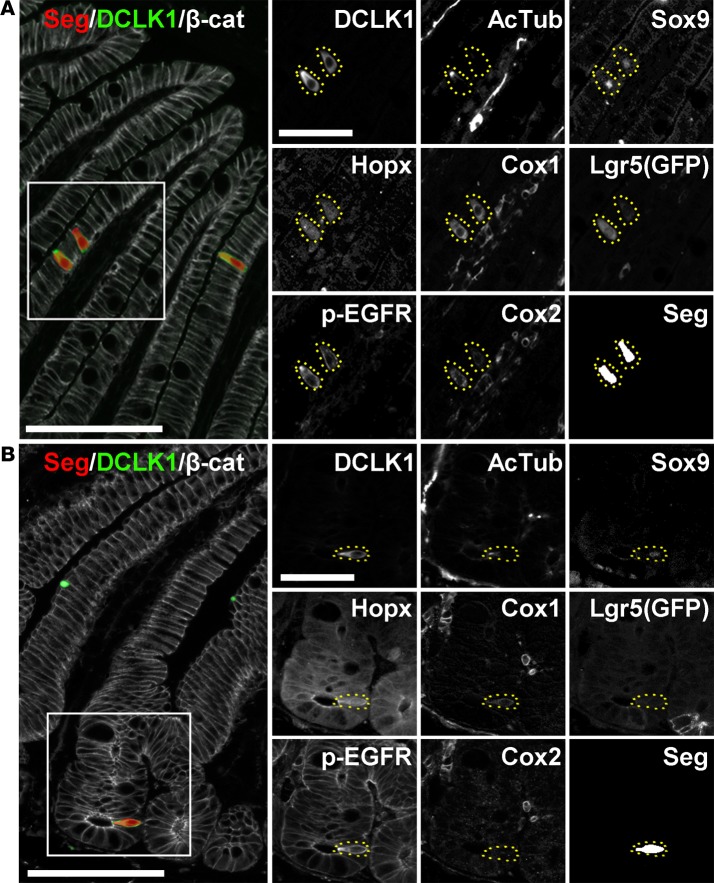
As part of a comprehensive survey of the normal mouse intestine using MxIF to analyze differentiated, progenitor/stem, and signaling cell states, along with a panel of segmentation markers, we discovered that both Hopx and p-EGFR were expressed in DCLK1-positive intestinal tuft cells. Visualization of single-cell expression data by t-SNE revealed a distinct tuft cell “island” characterized by high DCLK1 staining intensity (Supplemental Figure 3). Cells in this “island” did not express high levels of other specific differentiation markers (lysozyme in Paneth cells, Muc2 in goblet cells, and chromogranin A in enteroendocrine cells), and were negative for the proliferation marker PCNA. A subset of these cells expressed the previously recognized tuft cell marker Sox9 as well as p-EGFR and Hopx. While p-EGFR expression has been observed in tuft cells of the stomach (26) and pancreas (27), it has not been reported in intestinal tuft cells. Antibody staining for p-EGFR was observed in DCLK1-negative cells at the bottom of the crypt, but it was found at much higher levels in DCLK1-positive cells in the crypt and villus, especially at the apical “tuft” (Supplemental Figure 4). Hopx is an intestinal stem cell marker that labels mostly quiescent progenitor/stem cells (28). Staining for Hopx revealed expression throughout the crypt base progenitor/stem cell zone as well as tuft cells. Hopx antibody specificity was confirmed by the absence of staining in intestinal sections from Hopx-null mice (Supplemental Figure 5). Our staining was consistent with Hopx mRNA in situ patterns and staining with the same antibody (29, 30).
Characterization of intestinal tuft cells.
Additionally, substantial heterogeneity was observed in the tuft cell population for the 8 putative tuft cell markers analyzed (Figure 2B). Tuft cells were primarily localized in the villi throughout the small intestine (>80%, Supplemental Figure 6); they expressed known tuft cell markers, such as acetylated tubulin, Cox1, Cox2, Sox9, and Lgr5 (via Lgr5-EGFP reporter, ref. 24) as well as the two novel markers Hopx and p-EGFR (Figure 3A). Tuft cells in the crypt also expressed these markers; however, non-tuft epithelial cells in the progenitor/stem cell zone also expressed Sox9, Lgr5, Hopx, and p-EGFR (Figure 3B). DCLK1-positive cells never costained with the proliferative marker PCNA, even in the rare cells located in the proliferative crypt compartment (Supplemental Figure 7). Tuft cells represented a higher proportion of the total epithelial cell population in the ileum and jejunum than in the duodenum, but this did not reach statistical significance (Supplemental Figure 8). As expected, Hopx, Sox9, and Lgr5 were also highly expressed in stem and progenitor cells. At homeostasis, a higher proportion of DCLK1-positive tuft cells in the small intestine expressed high levels of Cox2 (Supplemental Figure 9) and Hopx (Supplemental Figure 10) than in the colon, but differences were not observed with the other tuft cell markers.
Figure 3. Expression of tuft cell markers in the small intestine.
Representative DCLK1-positive cells as shown in the villus (A) and crypt (B) of the ileum, along with segmentation of individual cells and β-catenin staining of the cell membrane (scale bar: 100 μm). Insets demonstrate heterogeneity in expression of tuft cell markers (scale bar: 50 μm).
Changes in tuft cell expression profiles after fasting and refeeding.
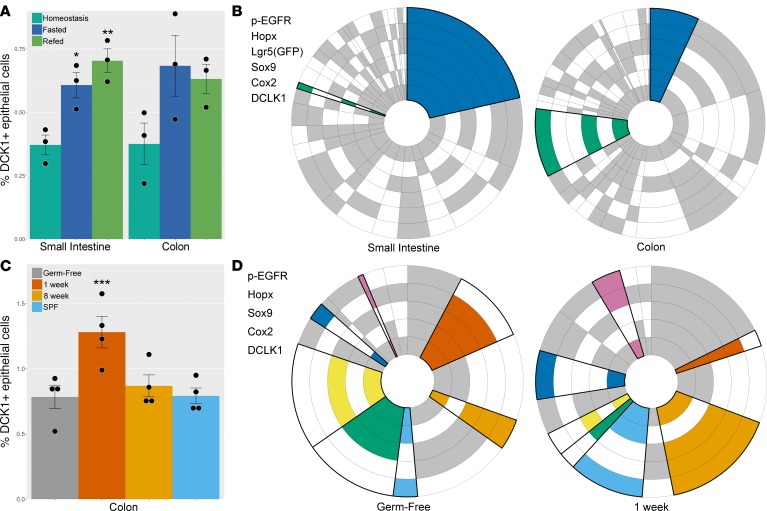
To assess the population dynamics within subsets of tuft cells, tissues were analyzed at homeostasis, after 48 hours of fasting, and after 24 hours of refeeding following the 48-hour fast. Intestinal atrophy was observed after fasting: villi in the small intestine shortened, crypts became thinner (Supplemental Figure 11), and the average cell size and total number of intestinal epithelial cells decreased (Supplemental Figure 12). Cell proliferation was markedly decreased in intestinal crypts after fasting (Supplemental Figure 13), consistent with prior studies (31, 32). After refeeding, a more normal tissue architecture was reestablished; as villi began to regrow, epithelial cell size and number increased and proliferation rebounded (Supplemental Figures 11–13). Using 3 mice for each condition, 32,488 tuft cells were identified from 4,368,519 epithelial cells over the entire intestine from images collected from the entire set of Swiss rolls of the duodenum, jejunum, ileum, and colon for each mouse. After fasting, the proportion of tuft cells was significantly increased in the small intestine and remained elevated after refeeding (Figure 4A). During this increase in overall tuft cell proportion, no change in tuft cell localization along the crypt-villus axis was observed (Supplemental Figure 6). With single-marker analysis, Cox2-positive tuft cells were increased after fasting (Supplemental Figure 9) in the small intestine and colon and returned to lower levels after refeeding. No changes were observed with the other markers analyzed.
Figure 4. Intestinal tuft cell number and expression patterns.
(A) The proportion of tuft cells in the small intestinal epithelium was increased following fasting and remained elevated after 24 hours of refeeding (*P = 0.0320, **P = 0.0064; n = 3). Similarly, the colon showed an increase in the fraction of tuft cells after fasting and refeeding; however, these results did not reach statistical significance. (B) Circular plots demonstrate the proportion of each cell expression profile for each tuft cell marker. Statistically significant changes in expression profiles between sites and conditions are denoted by bold outlines and colors. Two expression profiles were different between tuft cells in the small intestine and colon at homeostasis (blue: P = 0.0038, green: P < 0.0001; n = 3 for each condition). (C) The proportion of tuft cells in the colon was increased 1 week after introduction of microorganisms compared to germ-free mice (***P = 0.0024; n = 4) but was unchanged 8 weeks after introduction of microorganisms or in specific pathogen–free (SPF) mice. (D) Overall, 7 tuft cell expression profiles were significantly different between germ-free mice and mice inoculated with microbiota for 1 week. Statistically significant changes in expression profiles between sites and conditions are denoted by bold outlines and colors (clockwise from top of plot: P = 0.0197, P = 0.0415, P = 0.0137, P = 0.0291, P = 0.0006, P = 0.0074, P = 0.0005; n = 4 for each condition). Two-tailed Student’s t test was used for statistical comparisons.
Extending single-cell analysis to focus on 6 tuft cell markers (DCLK1, Cox2, Sox9, Lgr5, Hopx, and p-EGFR) revealed distinct tuft cell expression profiles in the small intestine and colon. Acetylated tubulin was excluded from this analysis, as staining for this marker was only observed at the apical portion of the cell; it could be absent from other portions of the cell due to the plane of sectioning. However, all acetylated tubulin–positive epithelial cells observed in this study were also DCLK1 positive. Cox1 was excluded from analysis due to failure of staining in approximately half of the 36 samples. Patterns of expression were visualized by circular plots, as shown in Figure 4, B and D. Each concentric ring around the circumference of the plot represents a single tuft cell marker. Shaded sectors of the rings are positive, while white sectors are negative for staining, based on the clustering deconvolution method. Each sector or wedge of the circle, going from its center outward, represents subpopulations of tuft cells with different combinations of markers; the size of each wedge of the ring represents the percentage of cells containing those markers. Statistically significant changes in subpopulations between conditions are shown in color (Figure 4B and Supplemental Table 2). Two populations showed large changes in effect size when comparing small intestine to colon at homeostasis: the DCLK1/Cox2/Sox9/Lgr5/p-EGFR–positive population (blue wedge, Figure 4B) was significantly (P < 0.0038, n = 3 per condition) decreased in the colon and the DCLK1/p-EGFR/Sox9-positive population (green wedge, Figure 4B) was significantly increased (P < 0.0001, n = 3 per condition). Interestingly, in both the small intestine (Supplemental Figure 14) and colon (Supplemental Figure 15), only a few differences, with small effect sizes, in tuft cell expression profiles were observed when comparing homeostasis to the fasted or refed conditions.
Changes in colonic tuft cell expression profiles after introduction of microbiota.
Helminth infection increases tuft cell number (17–19), but the effect of commensal microbiota on tuft cells has not been examined. To address this question, we evaluated the effect of introduction of microorganisms on colonic tuft cells (Supplemental Table 3). Here, germ-free mice were inoculated with fecal microbiota and sacrificed after 1 or 8 weeks. The colons of these mice were compared with those from age-matched untreated germ-free mice or from mice raised in specific pathogen–free (SPF) housing conditions (Supplemental Table 4). Using 4 mice for each of the 4 conditions, 4,508 tuft cells were identified from 553,979 colonic epithelial cells from the 16 Swiss rolls of the mouse colons. A significant increase in colonic tuft cell number was observed in previously germ-free mice 1 week following inoculation with microbiota by gavage of fecal samples (notably, helminth free, Supplemental Table 4) compared with mice remaining in germ-free conditions (Figure 4C). Interestingly, the proportion of p-EGFR–positive tuft cells was increased in all conditions compared with germ-free mice (Supplemental Figure 16). Furthermore, substantial changes were observed in tuft cell marker expression profiles by 1 week following introduction of microbiota (Figure 4D), but, by 8 weeks, tuft cell numbers and expression profiles largely reverted to baseline conditions. The expression profiles for SPF mice were indistinguishable from germ-free mice (Supplemental Figure 17 and Supplemental Table 5). Additionally, no change in the number of Muc2-positive cells, representing the goblet cell population, was observed after the introduction of microbiota to germ-free mice (Supplemental Figure 18), as is observed after helminth infection (17–19).
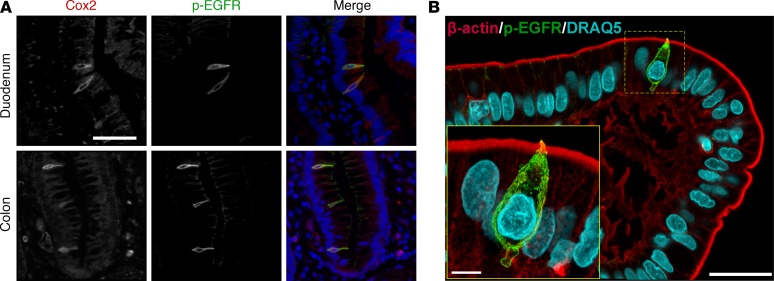
Identification of p-EGFR–positive tuft cells in human intestine.
Finally, we sought to use our mouse data to inform analysis of tuft cells in human tissues. Colocalization of Cox2 and p-EGFR was observed in human small intestine and colon (Figure 5A). Similar to mouse tissues, p-EGFR was concentrated at a narrow apical “tuft” in both anatomical locations. Furthermore, human duodenal tissue sections were stained for p-EGFR and β-actin and imaged at high resolution using confocal microscopy (Figure 5B). p-EGFR was expressed throughout the cell, with the exception of the nucleus, and was highly concentrated at the apical surface. This slender “tuft” of high p-EGFR expression extended into the lumen and colocalized with β-actin expression (Supplemental Figure 19). Tuft cells demonstrated a range of shapes (Supplemental Figure 20), as previously described (14), and frequently had membrane blebs (Supplemental Figure 19) and projections toward the basal lamina (Figure 3 and Supplemental Figure 20), which possibly interact with nerve fibers not visualized here (33).
Figure 5. p-EGFR expression in human intestinal tuft cells.
(A) Costaining with p-EGFR and Cox2 in tuft cells in the human duodenum and colon (scale bar: 50 μm). (B) Maximum intensity projection of confocal microscopy shows β-actin and p-EGFR in a tuft cell at the tip of the villus in the human duodenum (scale bar: 25 μm). Increased magnification (inset) shows projection of the “tuft” into the luminal space and a clearly defined projection at the tuft cell base (scale bar: 5 μm).
Discussion
Our data demonstrate the ability of MxIF to uncover heterogeneity and dynamic changes in protein expression patterns in a rare cell type, the intestinal tuft cell. Advances in cell segmentation and data analysis for MxIF enabled the results presented here. Previous MxIF studies focused entirely on cancer, where more uniform cell shape and loss of polarity allow for relatively straightforward cell segmentation (1, 2). This issue was solved by the addition of multiple membrane markers to provide coverage on all sides of the cell (Figure 1), similar to those described by Schüffler et al., using imaging mass cytometry (34). In addition, cloud processing facilitated rapid image processing and analysis of the large data sets collected for these studies (>1 terabyte raw image files). Furthermore, the use of the open-source KNIME platform permitted quick workflow prototyping and tuning. Importantly, MxIF allows the study of all cells present on a tissue slice, eliminating the potential bias involved when analysis is performed on a subset of the data (e.g., cells per field, cells per crypt/villus axis) and enabling the capture of enough cells to identify rare populations. Using MxIF allows the interrogation of spatial relationships and morphometrics among cells, including cell size (Supplemental Figure 12) and location (Supplemental Figure 6). We anticipate that further refinements in spatial analytical methods for MxIF will permit even more in-depth analysis of the complex spatial relationships among cells in tissue (35, 36).
Intestinal tuft cells provided an ideal case for rare cell detection, as they are characterized by expression of DCLK1, which is not expressed in other intestinal epithelial cell types, with the possible exception of an extremely rare subset of enteroendocrine cells (37). When all epithelial cells were viewed on a t-SNE map (Figure 2 and Supplemental Figure 2), tufts cells were readily isolated, with coincident expression of intestinal markers, p-EGFR and Hopx. Like many other reported tuft cell markers, such as Sox9 and Lgr5, these markers were not specific to tuft cells but were also expressed in the progenitor/stem compartment of the crypt (Supplemental Figures 3 and 4). However, cells that were DCLK1 positive, even at the crypt base, were not proliferative (Supplemental Figure 7), indicating that they are likely not actively cycling stem cells. However, our results cannot rule out that tuft cells could potentially act as a reserve stem cell population, as has been proposed (38), and, indeed, intestinal tuft cells express many putative stem cell markers (e.g., Lgr5, Sox9, Hopx).
Our results suggest a broad heterogeneity of tuft cells not previously appreciated when considered as a single homogenous population. Lower dimensional analyses of discrete cell groups or specific markers have been very informative, for example, for enteroendocrine cells (39) or for gene expression of the claudin family of tight junctional proteins throughout the gastrointestinal tract (40). However, these studies did not provide the kind of in-depth analysis of these cells’ signaling and structural states provided by MxIF, in which coexpression of various markers in all cells is directly evaluated and not inferred through expression analyses of serial or different tissue sections. Notably, there were significant differences in tuft cell expression profiles between the small intestine and colon (Figure 4B). This may indicate differential tuft cell function, as has been recently speculated by Gerbe and Jay (41), or different developmental programs between the small intestine and colon. However, future work will be needed to determine if these differences in protein expression profiles are meaningful.
MxIF analysis also allows the observation of dynamic changes in protein expression profiles in rare cell types. Here, we have examined two physiological perturbations. Acute fasting has been shown to induce large structural changes in intestinal architecture as well as to transition intestinal stem cells to a dormant state primed for intestinal regeneration following the reintroduction of nutrients (32); however, the role of tuft cells has not been investigated in this setting. We observed a significant increase in the proportion of tuft cells in the small intestine along with large structural changes in the gut overall after fasting and refeeding. Despite this, little change was observed in tuft cell protein expression profiles. Tuft cell stability under these conditions is consistent with the long-lived nature of tuft cells relative to other differentiated intestinal cell types (16); tuft cell resistance to apoptosis following other insults, such as radiation (38), compared with other the other differentiated intestinal cell types; and the role of DCLK1-positive tuft cells in intestinal regeneration (42–44). Therefore, the enrichment of tuft cells in the intestinal epithelium after fasting is likely due to tuft cell persistence, compared with other intestinal cells, rather than proliferation of existing tuft cells. We did not observe PCNA-positive tuft cells in this study, and no increase in crypt-localized tuft cells was seen after fasting (Supplemental Figure 6). With single-marker analysis, only the Cox2-positive tuft cell population increased after fasting (Supplemental Figure 9) in the small intestine and colon and returned to lower levels after refeeding. Other studies have observed increased oxidative stress following fasting in other organ sites (45), which can lead to increased Cox2 expression (46). Furthermore, Chandrakesan et al. showed that tuft cells regulate Cox2 signaling through a DCLK1-dependent paracrine response to intestinal injury (44).
In a separate experiment, when helminth-free microbiota were inoculated into previously germ-free mice, we observed an increase in tuft cells, similar to that observed following infection by helminths, although less robust (17–19). In contrast to helminth infection, in which a concomitant increase in goblet cells is observed, no change in goblet cell number was detected in these studies (Supplemental Figure 18). Coupled with the increase in tuft cell number were large changes in tuft cell protein expression profiles at 1 week after inoculation that returned to near baseline by 8 weeks after inoculation (Figure 4, C and D, and Supplemental Figure 17). Of note was the sustained increase in p-EGFR–positive tuft cells after introduction of microbiota (Supplemental Figure 16), suggesting a potential role for EGFR signaling in luminal sensing for tuft cells. Previous studies have implicated commensal bacteria in activating EGFR (47). Recently, Basak et al. demonstrated that EGFR inhibition in mouse small intestinal organoids caused an increase in tuft cell numbers (48). We speculate that their results are most likely due to changes in differentiation trajectories as cells mature from a stem-like state, rather than changes in EGFR activation, as we present here. While the present work examined the short-term effects on colonic tuft cells of introducing nonpathogenic microbiota to germ-free mice, further studies on the effect of pathogenic bacteria on tuft cell number and function throughout the gastrointestinal tract are warranted.
We also demonstrated that, similar to that in the mouse, p-EGFR is strongly expressed in human intestinal tuft cells (Figure 5). The functional significance of p-EGFR in tuft cells, and the mechanism(s) underlying its regulation, should be fruitful areas of future investigation. As no reliable antibody for human Hopx could be identified, we were not able to assess Hopx staining in human intestinal tuft cells.
These results demonstrate the utility of MxIF to discover novel biomarkers and define rare and heterogeneous cellular populations. We showed that specific subsets of tuft cells express Hopx and p-EGFR, two previously unreported intestinal tuft cell markers. MxIF identified differing profiles of tuft cell expression in the small intestine and the colon and was able to identify changes in the colonic population following introduction of commensal microbiota to germ-free mice. These results demonstrate that there are discrete intestinal tuft cell populations with differing function(s) and regulation that can be probed under various physiological and pathophysiological conditions. Although this study focused on intestinal tuft cells, MxIF can be applied to any tissue, including immune infiltrates, providing a powerful tool for biomarker discovery in heterogeneous and dynamic tissues.
Methods
Mouse models, human samples, and tissue preparation.
Lrig1-Apple/+ mice (23) were crossed with Lgr5-EGFP-DTR mice (a gift from Frederic de Sauvage, Genentech, South San Francisco, California, USA, ref. 24) on a mixed C57BL/6-129/SvEv background. Intestinal tissues were collected from three 6- to 8-week-old male mice at homeostasis, after 48 hours of fasting, and after 24 hours of refeeding after a 48-hour fast. Germ-free WT 129/SvEv mice (mixed gender, 7 weeks old) were transferred to SPF housing conditions. Pathogens excluded from SPF housing are described in Supplemental Table 4; Trichuris muris was not detected by PCR (IDEXX Bioresearch). Germ-free mice were gavaged with WT SPF microbiota pooled from 4 WT 129/SvEv fecal samples and sacrificed after 1 or 8 weeks. WT 129/SvEv mice (born into SPF conditions) that acquired their microbiota via vertical transmission were also used (49). Upon sacrifice by CO2 inhalation, the intestinal tract was removed, washed with PBS, and fixed in 10% formalin overnight. Tissues were washed in PBS, Swiss rolled, and stored in 70% ethanol until processing and embedding in paraffin. Deidentified samples of human duodenum and colon were obtained from the Vanderbilt Cooperative Human Tissue Network (CHTN). These are defined as exempt, in this case meaning they are anonymous samples collected with IRB 160828 to JLF and 070166 to RJC. Tissues were sectioned (4-μm thickness) prior to deparaffinization, rehydration, and antigen retrieval using citrate buffer (pH 6.0) for 20 minutes in a pressure cooker at 105°C followed by a 10-minute bench cool down. Endogenous background signal was reduced by incubating slides in 3% H2O2 for 10 minutes, before blocking for 30 minutes in 3% BSA/10% donkey serum in PBS prior to antibody staining. Hopx-/- mouse tissue was a gift from Jonathan Epstein at the University of Pennsylvania, Philadelphia, Pennsylvania, USA.
Immunofluorescence imaging.
Sequential staining and dye inactivation was performed as described previously (1). Briefly, MxIF imaging was performed on an Olympus X81 inverted microscope with a motorized stage and with filter sets specific for DAPI, GFP, CY3, CY5, and CY7. Images were acquired at ×200 magnification, and exposure times were determined for each antibody stain (typically, DAPI = 20 ms, GFP = 200 ms, CY3 = 300 ms, CY5 = 750 ms, and CY7 = 750 ms). Antibody reagents are listed in Supplemental Table 6, and antibody conjugations were performed as previously described (1). All antibody staining was performed overnight at 4°C, and the antibody staining and dye inactivation sequence is described in Supplemental Tables 1 and 3. Dye inactivation was performed using an alkaline peroxide solution as previously described (1), and background images were collected between staining rounds to ensure complete inactivation of the fluorochromes. After slide clearing and antigen retrieval, the samples were DAPI stained and scanned at ×4 followed by image stitching and the generation of a pseudo-H&E image, from which regions were selected for subsequent analysis and imaging. Typically, 80–100 image positions were collected for each Swiss roll.
Confocal imaging of human duodenal tissue sections was performed using a Nikon TiE inverted microscope platform outfitted with an A1R-plus point-scanning confocal ×100 1.49 NA Apo TIRF objective as well as 405 nm, 488 nm, 561 nm, and 640 nm solid-state lasers. Additionally, an encoded, motorized stage (Nikon Instruments Inc.) and piezo stage insert (Mad City Labs) enabled both multichannel stitching as well as high precision axial image stacking, respectively. Subsequent to image stack acquisition, data sets were deconvolved using Richardson-Lucy 3D deconvolution. Both image acquisition and processing of confocal images were accomplished through the use of NIS-Elements software (Nikon Instruments Inc.).
Image processing, single-cell quantification, and analysis.
Immunofluorescent images were processed and cells were segmented and quantified as described previously (1). Briefly, for each round of staining, images were first aligned using rigid transformations (i.e., translation and rotation), which were estimated by comparing the DAPI images from the successive rounds to those from the first round. The registered images were then corrected for uneven illumination, and autofluorescence was removed. Subsequently, single-cell segmentation was performed as follows. First, DAPI-stained nuclei were segmented using a wavelet-based nuclei detection algorithm followed by shape-based watershed (50). Second, an epithelial segmentation algorithm was used to identify the epithelial cells in the image using an epithelial-specific marker (pan-cytokeratin). Third, the cell cytoplasm and membrane were segmented using an algorithm that detects tubular structures in the image, based on computing Frangi’s vesselness (51) followed by image thresholding. In this work, however, no single marker was able to provide complete membrane staining for all of the cells. Therefore, we linearly combined 4 different markers (NaKATPase, β-catenin, pan-cytokeratin, and villin). Membrane markers were first normalized to the same dynamic range before they were combined, and linear combination coefficients/weights were estimated empirically to maximize the staining coverage of the cell membrane. The membrane segmentation algorithm produced a probability map and a binary segmentation mask. A multilevel watershed with a shape-ranking algorithm was applied on the membrane probability map to extract initial cell contours (52). Finally, initial cell segmentation results were combined with the 3 individual compartment segmentations (i.e., nucleus, cytoplasm, and plasma membrane) as well as the epithelial mask to generate single-cell segmentation masks (52, 53). Image analysis algorithms were deployed on a cloud-based platform (54) using Amazon Web Services (AWS), and Hadoop was used to execute the algorithms on the computer clusters. MxIF data were stored in the cloud (in encrypted form, for data protection) and were processed using computing resources provisioned on-demand within the cloud. This framework integrates a number of managed AWS services, including the Simple Storage Service (S3), the Relational Database Service, and the Elastic MapReduce service with General Electric Healthcare services for secure, high-speed upload and download of data sets to and from S3. Furthermore, open-source KNIME software (http://www.knime.org/) was used as an orchestration tool to quickly build customized image analysis workflows or to make quick adjustments. To do that, KNIME nodes for the different image analysis algorithms/steps were developed. Once the single-cell segmentation was completed, the cells were filtered to remove partial cells. From the single-cell segmentation masks, the mean, standard deviation, median, and maximum staining intensity for each protein were quantified with respect to the whole cell, cell membrane, cytoplasm, and nucleus as well as cell location, area, and shape. Localization of tuft cells in the crypt or villus was determined by manual counting of tuft cells in each compartment in 10 fields of view in 3 mice in the duodenum, jejunum, and ileum.
Following quantification, slides were normalized for batch effects and exposure time for each channel/marker analyzed. A common reference exposure was chosen for all slides for each channel to minimize slide-to-slide variation due to this factor. As slides were processed in 2 batches per round, the median normalization composed of (a) log2 transformation of marker intensity and (b) equalization of median intensity of all cells from one batch to median intensity of all cells of the other batch was performed. Due to variability across slides, a slide-specific threshold was determined for each marker by a model-based clustering deconvolution method, assuming the observed intensity distribution of a marker to be a Gaussian mixture of multiple components (55). The likelihood was constructed based on the normal density, and the parameters were estimated using an expectation-maximization algorithm. Bayesian information criterion was used to choose the optimal number of components. The number of components was specified to be two, resulting in reduced computation time, and captured the overall distribution patterns. Slide-specific thresholds were determined from the top 3% of DCLK1 intensity, containing both DCLK1-negative and -positive cells in a bimodal distribution (Supplemental Figure 2). Once the DCLK1-positive cells were determined for each slide, the deconvolution method was again applied to determine thresholds of positivity for the other tuft cell markers (Cox2, Sox9, Lgr5 [GFP], Hopx, and p-EGFR). Acetylated tubulin and Cox1 were excluded due to variability in sectioning (i.e., apical “tuft” may be excluded, see Figure 3) and inconsistent staining across slides, respectively. Quantification algorithms were deployed in R (https://cran.r-project.org/). Multidimensional image visualization was accomplished by t-SNE in R (56).
Statistics.
Pairwise condition effects (e.g., homeostasis to fasting, small intestine to colon, germ-free to 1 week) were compared by 2-tailed t test. A P value of less than 0.05 was considered significant. All data are presented as mean ± SEM. All statistical tests were performed in R.
Study approval.
All mouse studies were approved by the Vanderbilt University Institutional Animal Care and Use Committee (fasting/refeeding) or the University of Florida Institutional Animal Care and Use Committee (germ free), and all efforts were made to minimize animal suffering. All research involving human samples was approved by the Vanderbilt University Medical Center IRB. Deidentified samples of human duodenum and colon were obtained from the Vanderbilt CHTN. These are defined as exempt, in this case meaning they are anonymous samples collected with IRB 160828 to JLF and 070166 to RJC.
Author contributions
ETM, JTR, CJ, MJT, JLF, KSL, MJG, and RJC conceived of the study and designed experiments. ETM, YS, YAK, BAM, ASP, and CLO conducted experiments and analyzed data. ETM, JLF, KSL, MJG, and RJC wrote and edited the manuscript.
Supplementary Material
Acknowledgments
Special thanks to Brion Sarachan and Vijay Kumar (General Electric Global Research Center) for cloud implementation for single-cell analysis, Sean Dinn and Elizabeth McDonough (General Electric Global Research Center) for technical assistance with MxIF, and Jonathan Epstein (University of Pennsylvania) for providing Hopx-null tissue. Research reported in this publication was supported by the National Cancer Institute (NCI) of the NIH under award R01CA194600 to MJG and RJC. RJC was funded by the NCI under awards P50CA095103 and R01CA046413. ETM was supported by a training grant funded by the NIH under award R25CA092043. KSL was funded by the NIH under award R01DK103831, an Innovator Award from the American Association for Cancer Research–Landon Foundation (15-20-27-LAUK), a Crohn’s & Colitis Foundation of America Career Development Award (308221), and pilot grants from the NIH (P30DK058404) and the NCI (P50CA095103). CJ acknowledges support from the NIH under awards R01DK073338 and R01AT08623 and the University of Florida Department of Medicine Gatorade Fund. MJT acknowledges support from the NIH under awards R01DK075555, R01DK095811, and R01DK111949. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
YS’s present address is: Data and Statistical Sciences, AbbVie Inc., North Chicago, Illinois, USA.
Conflict of interest: Y. Sui, Y. Al-Kofahi, A. Santamaria-Pang, and M.J. Gerdes are employed by and receive salary from General Electric Corporation. Research support for the development of the single-cell algorithms and methodology was provided by the General Electric Corporation. The authors are inventors on numerous patents (US20080032321A, US7629125B2, US20090245611A1, US20090245598A1, US20110053171A1, and US20090141959A1) both issued and in review for the single-cell analysis approaches.
Reference information:JCI Insight. 2017;2(11):e93487. https://doi.org/10.1172/jci.insight.93487.
Contributor Information
Yunxia Sui, Email: yunxia.sui@gmail.com.
Yousef Al-Kofahi, Email: alkofahi@ge.com.
Alberto Santamaria-Pang, Email: santamar@ge.com.
Christian Jobin, Email: christian.jobin@medicine.ufl.edu.
Robert J. Coffey, Email: robert.coffey@vanderbilt.edu.
References
- 1.Gerdes MJ, et al. Highly multiplexed single-cell analysis of formalin-fixed, paraffin-embedded cancer tissue. Proc Natl Acad Sci USA. 2013;110(29):11982–11987. doi: 10.1073/pnas.1300136110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sood A, et al. Multiplexed immunofluorescence delineates proteomic cancer cell states associated with metabolism. JCI Insight. 2016;1(6):e87030. doi: 10.1172/jci.insight.87030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li C, et al. Excess PLAC8 promotes an unconventional ERK2-dependent EMT in colon cancer. J Clin Invest. 2014;124(5):2172–2187. doi: 10.1172/JCI71103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McCabe A, Dolled-Filhart M, Camp RL, Rimm DL. Automated quantitative analysis (AQUA) of in situ protein expression, antibody concentration, and prognosis. J Natl Cancer Inst. 2005;97(24):1808–1815. doi: 10.1093/jnci/dji427. [DOI] [PubMed] [Google Scholar]
- 5.Schubert W, et al. Analyzing proteome topology and function by automated multidimensional fluorescence microscopy. Nat Biotechnol. 2006;24(10):1270–1278. doi: 10.1038/nbt1250. [DOI] [PubMed] [Google Scholar]
- 6.Stack EC, Wang C, Roman KA, Hoyt CC. Multiplexed immunohistochemistry, imaging, and quantitation: a review, with an assessment of tyramide signal amplification, multispectral imaging and multiplex analysis. Methods. 2014;70(1):46–58. doi: 10.1016/j.ymeth.2014.08.016. [DOI] [PubMed] [Google Scholar]
- 7.O’Donnell EA, Ernst DN, Hingorani R. Multiparameter flow cytometry: advances in high resolution analysis. Immune Netw. 2013;13(2):43–54. doi: 10.4110/in.2013.13.2.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Simmons AJ, et al. Impaired coordination between signaling pathways is revealed in human colorectal cancer using single-cell mass cytometry of archival tissue blocks. Sci Signal. 2016;9(449):rs11. doi: 10.1126/scisignal.aah4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Simmons AJ, et al. Cytometry-based single-cell analysis of intact epithelial signaling reveals MAPK activation divergent from TNF-α-induced apoptosis in vivo. Mol Syst Biol. 2015;11(10):835. doi: 10.15252/msb.20156282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giesen C, et al. Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry. Nat Methods. 2014;11(4):417–422. doi: 10.1038/nmeth.2869. [DOI] [PubMed] [Google Scholar]
- 11.Angelo M, et al. Multiplexed ion beam imaging of human breast tumors. Nat Med. 2014;20(4):436–442. doi: 10.1038/nm.3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eberwine J, Sul JY, Bartfai T, Kim J. The promise of single-cell sequencing. Nat Methods. 2014;11(1):25–27. doi: 10.1038/nmeth.2769. [DOI] [PubMed] [Google Scholar]
- 13.Chen KH, Boettiger AN, Moffitt JR, Wang S, Zhuang X. RNA imaging. Spatially resolved, highly multiplexed RNA profiling in single cells. Science. 2015;348(6233):aaa6090. doi: 10.1126/science.aaa6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sato A. Tuft cells. Anat Sci Int. 2007;82(4):187–199. doi: 10.1111/j.1447-073X.2007.00188.x. [DOI] [PubMed] [Google Scholar]
- 15.Gerbe F, et al. Distinct ATOH1 and Neurog3 requirements define tuft cells as a new secretory cell type in the intestinal epithelium. J Cell Biol. 2011;192(5):767–780. doi: 10.1083/jcb.201010127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Westphalen CB, et al. Long-lived intestinal tuft cells serve as colon cancer-initiating cells. J Clin Invest. 2014;124(3):1283–1295. doi: 10.1172/JCI73434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Howitt MR, et al. Tuft cells, taste-chemosensory cells, orchestrate parasite type 2 immunity in the gut. Science. 2016;351(6279):1329–1333. doi: 10.1126/science.aaf1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.von Moltke J, Ji M, Liang HE, Locksley RM. Tuft-cell-derived IL-25 regulates an intestinal ILC2-epithelial response circuit. Nature. 2016;529(7585):221–225. doi: 10.1038/nature16161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gerbe F, et al. Intestinal epithelial tuft cells initiate type 2 mucosal immunity to helminth parasites. Nature. 2016;529(7585):226–230. doi: 10.1038/nature16527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gerbe F, Legraverend C, Jay P. The intestinal epithelium tuft cells: specification and function. Cell Mol Life Sci. 2012;69(17):2907–2917. doi: 10.1007/s00018-012-0984-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Itzkovitz S, et al. Single-molecule transcript counting of stem-cell markers in the mouse intestine. Nat Cell Biol. 2011;14(1):106–114. doi: 10.1038/ncb2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Royer C, Lu X. Epithelial cell polarity: a major gatekeeper against cancer? Cell Death Differ. 2011;18(9):1470–1477. doi: 10.1038/cdd.2011.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Poulin EJ, Powell AE, Wang Y, Li Y, Franklin JL, Coffey RJ. Using a new Lrig1 reporter mouse to assess differences between two Lrig1 antibodies in the intestine. Stem Cell Res. 2014;13(3 Pt A):422–430. doi: 10.1016/j.scr.2014.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tian H, et al. A reserve stem cell population in small intestine renders Lgr5-positive cells dispensable. Nature. 2011;478(7368):255–259. doi: 10.1038/nature10408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kondo J, et al. LRIG1 regulates ontogeny of smooth muscle-derived subsets of interstitial cells of cajal in mice. Gastroenterology. 2015;149(2):407–19.e8. doi: 10.1053/j.gastro.2015.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Choi E, et al. Dynamic expansion of gastric mucosal doublecortin-like kinase 1-expressing cells in response to parietal cell loss is regulated by gastrin. Am J Pathol. 2015;185(8):2219–2231. doi: 10.1016/j.ajpath.2015.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Delgiorno KE, et al. Identification and manipulation of biliary metaplasia in pancreatic tumors. Gastroenterology. 2014;146(1):233–44.e5. doi: 10.1053/j.gastro.2013.08.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takeda N, Jain R, LeBoeuf MR, Wang Q, Lu MM, Epstein JA. Interconversion between intestinal stem cell populations in distinct niches. Science. 2011;334(6061):1420–1424. doi: 10.1126/science.1213214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li N, et al. Single-cell analysis of proxy reporter allele-marked epithelial cells establishes intestinal stem cell hierarchy. Stem Cell Reports. 2014;3(5):876–891. doi: 10.1016/j.stemcr.2014.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Muñoz J, et al. The Lgr5 intestinal stem cell signature: robust expression of proposed quiescent ‘+4’ cell markers. EMBO J. 2012;31(14):3079–3091. doi: 10.1038/emboj.2012.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hagemann RF, Stragand JJ. Fasting and refeeding: cell kinetic response of jejunum, ileum and colon. Cell Tissue Kinet. 1977;10(1):3–14. doi: 10.1111/j.1365-2184.1977.tb00278.x. [DOI] [PubMed] [Google Scholar]
- 32.Richmond CA, et al. Dormant intestinal stem cells are regulated by PTEN and nutritional status. Cell Rep. 2015;13(11):2403–2411. doi: 10.1016/j.celrep.2015.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morroni M, Cangiotti AM, Cinti S. Brush cells in the human duodenojejunal junction: an ultrastructural study. J Anat. 2007;211(1):125–131. doi: 10.1111/j.1469-7580.2007.00738.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schüffler PJ, Schapiro D, Giesen C, Wang HA, Bodenmiller B, Buhmann JM. Automatic single cell segmentation on highly multiplexed tissue images. Cytometry A. 2015;87(10):936–942. doi: 10.1002/cyto.a.22702. [DOI] [PubMed] [Google Scholar]
- 35. Margolis D, Santamaria-Pang A, Rittscher J. Tissue segmentation and classification using graph-based unsupervised clustering. In: 2012 9th IEEE International Symposium on Biomedical Imaging (ISBI). 2012:162–165. [Google Scholar]
- 36.Ding H, Wang C, Huang K, Machiraju R. GRAPHIE: graph bas ed histology image explorer. BMC Bioinformatics. 2015;16 Suppl 11:S10. doi: 10.1186/1471-2105-16-S11-S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bjerknes M, et al. Origin of the brush cell lineage in the mouse intestinal epithelium. Dev Biol. 2012;362(2):194–218. doi: 10.1016/j.ydbio.2011.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.May R, Riehl TE, Hunt C, Sureban SM, Anant S, Houchen CW. Identification of a novel putative gastrointestinal stem cell and adenoma stem cell marker, doublecortin and CaM kinase-like-1, following radiation injury and in adenomatous polyposis coli/multiple intestinal neoplasia mice. Stem Cells. 2008;26(3):630–637. doi: 10.1634/stemcells.2007-0621. [DOI] [PubMed] [Google Scholar]
- 39.Gunawardene AR, Corfe BM, Staton CA. Classification and functions of enteroendocrine cells of the lower gastrointestinal tract. Int J Exp Pathol. 2011;92(4):219–231. doi: 10.1111/j.1365-2613.2011.00767.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Holmes JL, Van Itallie CM, Rasmussen JE, Anderson JM. Claudin profiling in the mouse during postnatal intestinal development and along the gastrointestinal tract reveals complex expression patterns. Gene Expr Patterns. 2006;6(6):581–588. doi: 10.1016/j.modgep.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 41.Gerbe F, Jay P. Intestinal tuft cells: epithelial sentinels linking luminal cues to the immune system. Mucosal Immunol. 2016;9(6):1353–1359. doi: 10.1038/mi.2016.68. [DOI] [PubMed] [Google Scholar]
- 42.Qu D, et al. Ablation of Doublecortin-Like Kinase 1 in the colonic epithelium exacerbates dextran sulfate sodium-induced colitis. PLoS ONE. 2015;10(8):e0134212. doi: 10.1371/journal.pone.0134212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.May R, et al. Brief report: Dclk1 deletion in tuft cells results in impaired epithelial repair after radiation injury. Stem Cells. 2014;32(3):822–827. doi: 10.1002/stem.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chandrakesan P, et al. Intestinal tuft cells regulate the ATM mediated DNA Damage response via Dclk1 dependent mechanism for crypt restitution following radiation injury. Sci Rep. 2016;6:37667. doi: 10.1038/srep37667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Domenicali M, et al. Food deprivation exacerbates mitochondrial oxidative stress in rat liver exposed to ischemia-reperfusion injury. J Nutr. 2001;131(1):105–110. doi: 10.1093/jn/131.1.105. [DOI] [PubMed] [Google Scholar]
- 46.Kiritoshi S, et al. Reactive oxygen species from mitochondria induce cyclooxygenase-2 gene expression in human mesangial cells: potential role in diabetic nephropathy. Diabetes. 2003;52(10):2570–2577. doi: 10.2337/diabetes.52.10.2570. [DOI] [PubMed] [Google Scholar]
- 47.Yan F, et al. Colon-specific delivery of a probiotic-derived soluble protein ameliorates intestinal inflammation in mice through an EGFR-dependent mechanism. J Clin Invest. 2011;121(6):2242–2253. doi: 10.1172/JCI44031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Basak O, Beumer J, Wiebrands K, Seno H, van Oudenaarden A, Clevers H. Induced quiescence of lgr5+ stem cells in intestinal organoids enables differentiation of hormone-producing enteroendocrine cells. Cell Stem Cell. 2017;20(2):177–190.e4. doi: 10.1016/j.stem.2016.11.001. [DOI] [PubMed] [Google Scholar]
- 49.McCafferty J, et al. Stochastic changes over time and not founder effects drive cage effects in microbial community assembly in a mouse model. ISME J. 2013;7(11):2116–2125. doi: 10.1038/ismej.2013.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Padfield D, Rittscher J, Roysam B. Spatio-temporal cell segmentation and tracking for automated screening. In: 2008 5th IEEE International Symposium on Biomedical Imaging: From Nano to Macro. 2008:376–379. [Google Scholar]
- 51. Frangi AF, Niessen WJ, Vincken KL, Viergever MA. Multiscale vessel enhancement filtering. In: Medical Image Computing and Computer-Assisted Intervention — MICCAI’98. Springer, Berlin, Heidelberg; 1998:130–137. [Google Scholar]
- 52. Santamaria-Pang A, Huangy Y, Rittscher J. Cell segmentation and classification via unsupervised shape ranking. In: 2013 IEEE 10th International Symposium on Biomedical Imaging. 2013:406–409. [Google Scholar]
- 53. Santamaria-Pang A, Huang Y, Pang Z, Qing L, Rittscher J. Epithelial Cell Segmentation via Shape Ranking. In: Li S, João Manuel R eds. Shape Analysis in Medical Image Analysis. Springer International Publishing; 2014:315–338. [Google Scholar]
- 54. Kumar VS, Williams JW, Aggour KS, Sarachan B, Al-Kofahi Y, Santamaria-Pang A. Collaborative Analysis of High-Content Image Data. Paper presented at: NIST BioImage Informatics Conference 2015; October 14, 2015; Gaithersburg, MD. https://isg.nist.gov/BII_2015/webPages/pages/2015_BII_program/program.htm Accessed May 3, 2017.
- 55.Fraley C, Raftery AE. Model-based clustering, discriminant analysis, and density estimation. J Am Stat Assoc. 2002;97(458):611–631. doi: 10.1198/016214502760047131. [DOI] [Google Scholar]
- 56.Jamieson AR, Giger ML, Drukker K, Li H, Yuan Y, Bhooshan N. Exploring nonlinear feature space dimension reduction and data representation in breast Cadx with Laplacian eigenmaps and t-SNE. Med Phys. 2010;37(1):339–351. doi: 10.1118/1.3267037. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.