Abstract
Congenital Diaphragmatic Hernia (CDH) is a common and often lethal birth defect characterized by diaphragmatic structural defects and pulmonary hypoplasia. CDH is isolated in 60% of newborns, but may also be part of a complex phenotype with additional anomalies. We performed whole exome sequencing (WES) on 87 individuals with isolated or complex CDH and on their unaffected parents, to assess the contribution of de novo mutations in the etiology of diaphragmatic and pulmonary defects and to identify new candidate genes. A combined analysis with 39 additional trios with complex CDH, previously published, revealed a significant genome-wide burden of de novo variants compared to background mutation rate and 900 control trios. We identified an increased burden of likely gene disrupting (LGD, i.e. nonsense, frameshift, and canonical splice site) and predicted deleterious missense (D-mis) variants in complex and isolated CDH patients. Overall, an excess of predicted damaging de novo LGD and D-mis variants relative to the expected frequency contributed to 21% of complex cases and 12% of isolated CDH cases. The burden of de novo variants was higher in genes expressed in the developing mouse diaphragm and heart. Some overlap with genes responsible for congenital heart defects and neurodevelopmental disorders was observed in CDH patients within our cohorts. We propose that de novo variants contribute significantly to the development of CDH.
Keywords: congenital diaphragmatic hernia, de novo, insulin growth factor, cohesin, POGZ, NAA15
INTRODUCTION
Congenital Diaphragmatic Hernia (CDH) represents one of the most common birth defects (1:2500-3000) (Pober 2007; Stolar and Dillon 2012), with approximately 1600 children born every year in the United States with this condition. CDH is characterized by defective diaphragm development and pulmonary hypoplasia, and is associated with significant morbidity and mortality. Many patients require invasive interventions such as extracorporeal membrane oxygenation (ECMO) and assisted ventilation (Garcia et al. 2014). Long-term complications including pulmonary hypertension, asthma, gastroesophageal reflux, feeding disorders, and developmental delays are common in survivors. Hospitalizations are frequent and extended, resulting in economic and societal burden. In spite of considerable advances in treatment and improved outcomes, the overall mortality remains high.
Multiple lines of evidence support genetic contributions to the etiology of CDH, including the identification of diaphragm defects in monogenic model organisms, recurrent human copy number variants (“genomic hotspots”), and monogenic conditions associated with CDH in humans (Veenma et al. 2012; Wynn et al. 2014). More than 60 loci have been associated with CDH in rodents or humans, yet the cause of CDH for most patients remains elusive. Recently, WES studies have identified high priority candidate genes for CDH using systems biology integration of rare sequence variants with gene expression data and protein-protein interaction networks of previously known CDH genes (Longoni et al. 2014a).
De novo mutations are a major cause of birth defects and other conditions affecting reproductive fitness (Veltman and Brunner 2012). A fundamental role for de novo variants was demonstrated in congenital heart disease (Homsy et al. 2015), autism spectrum disorders (Neale et al. 2012; Michaelson et al. 2012), and schizophrenia (Xu et al. 2011; Gulsuner et al. 2013). Recently, de novo mutations were discovered in a large fraction of syndromic congenital heart disease cases, compared to a relative abundance of inherited high-risk variants with incomplete penetrance in isolated congenital heart disease (Sifrim et al. 2016). Trio studies designed to identify de novo and recessive mutations are emerging as a valuable tool for screening undiagnosed genetic conditions (Zhu et al. 2015).
Familial clustering of CDH has been reported in very rare kindreds with multiple affected individuals (Wolff 1980; Pober 2008). Despite the small number of familial cases, the majority of probands with CDH have no family history of CDH, leading to the hypothesis that de novo variants are an important and relatively frequent etiological mechanism, as CDH was nearly always lethal before the introduction of neonatal respiratory support and surgical repair in the modern era. A screen for de novo variants was therefore performed in a cohort of 39 sporadic CDH trios with multiple birth defects (Yu et al. 2015). Within this cohort, a minimum of 15% were estimated to have de novo predicted pathogenic variants; however, the sample size was limited, and larger studies are needed for replication and to better estimate the fraction of cases due to de novo mutations. In this report, we combine the results of these original 39 trios with 87 new trios, greatly expanding the power of this study, and we demonstrate significant genome-wide enrichment for likely deleterious de novo sequence variants in a cohort of CDH trios with no family history of CDH. The enrichment was greater for complex cases, but was also present in isolated CDH. Our study provides further insight into the genetic architecture of CDH and suggests novel genes and pathways for this condition.
RESULTS
De novo variants in complex and isolated CDH
WES was performed on a cohort of 87 parent proband trios, enrolled at the Massachusetts General Hospital (MGH) and Boston’s Children Hospital. Clinical assessment by a geneticist was conducted whenever possible and patients with known causative chromosomal anomalies or mutations were excluded from the study (clinical and genetic findings are summarized in Supplementary table S1). Familial relationships were confirmed by identity-by-descent estimation in PLINK (Purcell et al. 2007).
In this group, the gender distribution was M:F 1:0.6 (54:33). Two fetal samples did not have an assigned sex reported on their enrollment forms and were determined genetically. The distribution is consistent with previously observed male gender bias of 1:0.69 in CDH (McGivern et al. 2015). The type of diaphragmatic defect was predominantly left sided Bochdalek; however, patients with right or bilateral hernias were also present. Additionally, our series included four cases of eventration, three of diaphragm agenesis, one of Morgagni, and one of an anterior hernia in a Pentalogy of Cantrell case. Phenotypically, 33 (37.9%) of probands had a diagnosis of complex CDH, i.e. the co-occurrence of diaphragmatic defects and congenital anomalies except lung hypoplasia (Pober et al. 2010). The most frequent comorbid features were cardiac defects, including atrial and ventricular septal defects and hypoplastic left heart syndrome (HLHS). While some morphologic features and neurodevelopmental conditions can become apparent with time, the isolated cases in this study had no additional anomalies at the time of last contact with a study physician. Five cases with insufficient phenotypic characterization to make any conclusive determination are marked as ‘unclassified’ for the purpose of this study.
Probands and parents were sequenced in three batches: Yale, UW1, and UW2. Yale and UW1 probands were published as part of a separate study (Longoni et al. 2014). Exome quality and coverage metrics are available in Supplementary table S2. Among the CDH trios, 70% (61/87) of the probands carried at least one de novo variant. In 18.4% (16/87) of patients, de novo variants were predicted to be likely gene-disrupting (LGD), i.e. categorized as nonsense, frameshift, or splice site variants. One or more de novo missense variants were present in 50.5% (44/87).of the trios, and 34.5% (30/87) had at least one variant predicted to be damaging by MetaSVM (Dong et al. 2015). Two patients had in-frame deletions and an in-frame insertion. Severity measures and defect size, classified according to Ackerman et al. (2012), were collected in 31/87 probands; no correlation was observed between defect size and mutation status.
The complete annotated list of variants is provided in Supplementary table S3. The datasets generated analyzed during the current study are available in the dbGAP repository (accession No. phs000783.v2.p1). Among the genes with either de novo LGD or predicted damaging missense variants, no gene had more than one de novo event.
Excess of damaging de novo variants
In order to increase the power of the study, data from 39 previously published trios with complex CDH recruited by the DHREAMS study (www.cdhgenetics.com) were included in the subsequent analyses (Yu et al. 2015) (Figure 1). In the subset of patients with a complex phenotype, an average of 1.12 de novo variants per patient were identified in the Boston cohort and 1.02 in the DHREAMS cohort. The number of de novo events per patient was consistent with a Poisson distribution in the Boston cohort and in the combined groups (Supplementary figure S1).
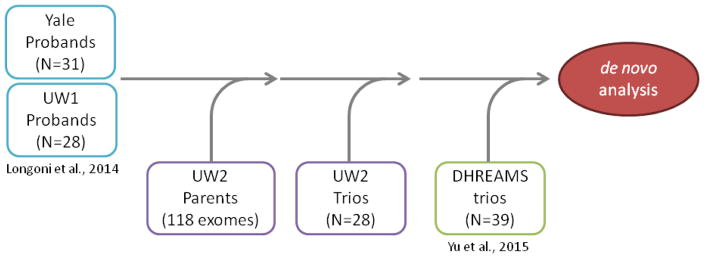
Fig. 1. Study flowchart.
Two groups of patients (UW1 and Yale) were sequenced as part of a previous study (Longoni et al. 2014). The corresponding parental samples and additional complete trios were sequenced in batch UW2. A cohort of 39 full trios (DHREAMS, Yu et al. 2015) was also used for the de novo mutation analysis and enrichment calculations.
In the combined cohorts, an excess of de novo variants was found in complex CDH cases compared to the expected background mutation rate (Samocha et al. 2014; Ware et al. 2015) (Table 1). A twofold enrichment (FE) in the frequency of de novo variants was observed for D-mis, determined by MetaSVM, and LGD variants (Poisson test p=0.008 and 0.02, respectively). A group of control trios (N=900) (De Rubeis et al. 2014; Iossifov et al. 2014), consisting of unaffected siblings and parents enrolled in the Simons Foundation Autism Research Initiative Simplex Collection (SSC) study (www.simonsfoundation.org), were jointly called and analyzed through the same pipeline as cases. In the control group, no significant differences with the background mutation frequency were detected. Similarly, no enrichment was identified for synonymous or for combined B-mis (benign missense) and D-mis de novo variants (All-mis). The enrichment of LGD and D-mis variants was also present when comparing CDH trios and control trios directly.
Table 1.
Fold enrichment of de novo variants in the CDH cohort
| Complex CDH (N=72) | Isolated CDH (N=49) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||||||
| Variant class | Obs. | Exp. | FE | p | Obs. | Exp. | FE | p | ||||
|
|
|
|
||||||||||
| N | Freq | N | Freq | N | Freq | N | Freq | |||||
| Silent | 12 | 0.17 | 22 | 0.31 | 0.54 | 0.99 | 15 | 0.31 | 15 | 0.31 | 0.99 | 0.55 |
| All-mis | 51 | 0.71 | 49 | 0.68 | 1.04 | 0.40 | 38 | 0.78 | 33 | 0.68 | 1.14 | 0.23 |
| D-Mis | 16 | 0.22 | 8 | 0.11 | 2.02 | 0.008 | 6 | 0.12 | 5 | 0.11 | 1.11 | 0.45 |
| LGD | 14 | 0.19 | 7 | 0.1 | 1.94 | 0.02 | 10 | 0.20 | 5 | 0.1 | 2.04 | 0.03 |
| LGD & D-Mis | 30 | 0.42 | 15 | 0.21 | 1.98 | 0.0005 | 16 | 0.33 | 10 | 0.21 | 1.55 | 0.06 |
| All CDH* (N=126) | Controls (N=900) | |||||||||||
|
|
|
|||||||||||
| Obs. | Exp. | FE | p | Obs. | Exp. | FE | p | |||||
|
|
|
|||||||||||
| N | Freq | N | Freq | N | Freq | N | Freq | |||||
| Silent | 31 | 0.25 | 39 | 0.31 | 0.79 | 0.92 | 226 | 0.12 | 279 | 0.31 | 0.81 | 1.00 |
| All-mis | 96 | 0.76 | 86 | 0.68 | 1.12 | 0.14 | 608 | 0.32 | 612 | 0.68 | 0.99 | 1.00 |
| D-Mis | 23 | 0.18 | 14 | 0.11 | 1.66 | 0.02 | 119 | 0.06 | 99 | 0.11 | 1.20 | 0.38 |
| LGD | 24 | 0.19 | 13 | 0.1 | 1.90 | 0.003 | 78 | 0.04 | 90 | 0.1 | 0.87 | 0.85 |
| LGD & D-Mis | 47 | 0.37 | 26 | 0.21 | 1.78 | 0.0002 | 197 | 0.10 | 189 | 0.21 | 1.04 | 0.69 |
includes 5 CDH patients with unclassified phenotype
The de novo frequency for the more common phenotype of isolated CDH phenotype was calculated for 49 Boston cases. In this group of patients, who represent the majority of patients with CDH, marginal enrichment was identified for LGD (FE=2.04, p=0.03), but not for D-mis variants (Table 1).
Based on the differential between the observed frequency of de novo variants in CDH cases and the expected mutation background frequency, we estimate that 20 (43%, or 0.37/0.21) (Table 1) of de novo LGD or D-mis reported in our study may contribute to the formation of diaphragmatic defects. Specifically, 21% of complex cases had an excess of LGD and D-mis variants, while 12% of isolated CDH patients had an excess of LGD variants.
Genes with de novo variants in CDH are expressed in the embryonic diaphragm and heart
We hypothesized that genes with predicted pathogenic mutations would be expressed during a critical stage in the developing diaphragm, comparable to murine E11.5 pleuro-peritoneal folds. Transcriptome data from this tissue are available in the Gene Expression Omnibus (GEO) database (Series GSE35243) (Russell et al. 2012). We defined high diaphragm expression as the top quartile of probe sets based on RMA (Robust Multi-Array Average)-normalized expression levels of microarray data (Yu et al. 2015). An increased burden of de novo variants in complex cases was detected in the group of highly expressed genes in the diaphragm. This burden was particularly striking for LGD variants (Figure 2 and Supplementary figure S2).
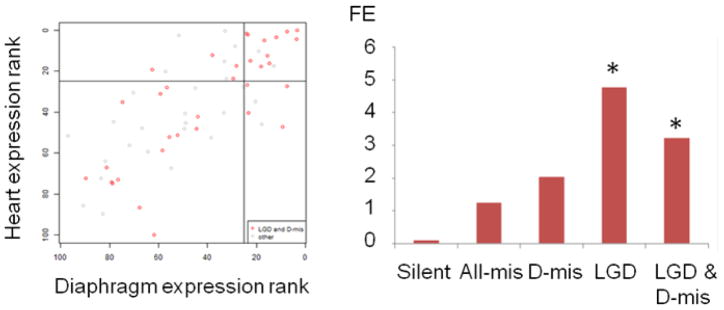
Fig 2. Diaphragm and heart expression of genes with de novo variants.
(a) LGD and D-mis variants (red) in complex CDH patients are plotted according to gene ranking by expression levels in the developing diaphragm (x axis) and in the developing heart (y axis) and compared to other variants in the same cohort (grey). (b) Complex CDH patients have an increased burden of LGD and D-mis variants in the first quartile of diaphragm expression relative to control trios. Bars depict the fold enrichment for each class of de novo sequence variant in the Boston and DHREAMS cohorts. Asterisks indicate p-values of 0.0003.
LGD and D-mis de novo variants expressed in the upper quartile of the developing diaphragm and confirmed by Sanger sequencing are listed in Table 2. The confirmation rate of de novo variants called bioinformatically was over 90% in the group of variants that underwent Sanger sequencing. An enrichment analysis (amp.pharm.mssm.edu/Enrichr, last accession on 10/21/2016) (Chen et al. 2013) indicated over-representation of several transcription factor targets (Lachmann et al. 2010), most notably targets of the CDH-associated CHD7 gene (Supplementary table S3).
Table 2.
De novo sequence variants in the CDH cohort
| ID | Cohort | Ph | Gene symbol | Dia | ExAC
|
Chr | Pos | Ref. | Alt. | cDNA | Protein | Category | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| pLI | mis_z | lof_z | ||||||||||||
| T2 | BOSTON | C | GRB10 | 3.4 | 0.367 | 1.854 | 3.918 | 7 | 50737473 | - | CT | NM_001001549: c.449_450insAG | p.S150fs | LGD |
| T33 | BOSTON | C | HSPD1 | 3.5 | 0.976 | 2.711 | 3.638 | 2 | 198362109 | G | A | NM_002156:c.C182T | p.T61I | D-mis |
| 01-0215 | DHREAMS | C | PTPN12 | 7.5 | 0.999 | −0.054 | 5.084 | 7 | 77221548 | C | T | NM_001131009:c.C77T | p.T26M | D-mis |
| T3 | BOSTON | C | ACTG1 | 7.6 | 0.219 | 5.131 | 2.413 | 17 | 79478517 | C | T | NM_001199954:c.G499A | p.E167K | D-mis |
| 01-0761 | DHREAMS | C | CDO1 | 9.4 | 0.020 | −0.088 | 1.586 | 5 | 115147001 | C | - | NM_001801:c.259delG | p.D87fs | LGD |
| T15 | BOSTON | C | ADD1 | 12.0 | 0.611 | 0.586 | 3.906 | 4 | 2906653 | C | T | NM_001119:c.C1324T | p.R442W | D-mis |
| T38 | BOSTON | I | NAA15 | 14.0 | 0.996 | 3.088 | 4.805 | 4 | 140258100 | AT | - | NM_057175:c.239_240del | p.H80fs | LGD |
| T57 | BOSTON | C | POGZ | 14.7 | 1.000 | 3.284 | 6.213 | 1 | 151378872 | GGAA | - | NM_145796:c.2350_2353del | p.F784fs | LGD |
| 01-0634 | DHREAMS | C | PRKACB | 15.6 | 0.612 | 2.547 | 3.179 | 1 | 84649798 | C | T | NM_001242862:c.C277T | p.R93X | LGD |
| 01-0562 | DHREAMS | C | GATA6 | 16.8 | - | - | - | 18 | 19761477 | C | T | NM_005257:c.C1366T | p.R456C | D-mis |
| 01-0147 | DHREAMS | C | STAG2 | 18.2 | 1.000 | 5.111 | 6.472 | X | 123197716 | C | T | NM_006603:c.C1840T | p.R614X | LGD |
| T49 | BOSTON | I | PLCG1 | 21.2 | 0.729 | 4.359 | 5.858 | 20 | 39797452 | G | A | NM_002660:c.G2425A | p.E809K | D-mis |
| T70 | BOSTON | C | FOXP4 | 22.4 | 0.672 | 1.628 | 3.652 | 6 | 41562624 | A | G | NM_001012426:c.A1553G | p.N518S | D-mis |
| T35 | BOS ON | C | ARL15 | 23.2 | 0.009 | 0.357 | 1.081 | 5 | 53450409 | C | T | sp | - | LGD |
| 01-0109 | DHREAMS | C | DLST | 23.5 | 0.950 | 0.277 | 3.734 | 14 | 75356621 | AG | - | NM_001244883:c.297_298del | p.E99fs | LGD |
| T36 | BOSTON | C | GINS3 | 23.8 | 0.236 | 0.613 | 1.888 | 16 | 58437236 | G | A | sp | - | LGD |
| 01-0136 | DHREAMS | C | PRR14 | 23.9 | 0.259 | 0.655 | 3.753 | 16 | 30667426 | C | T | NM_024031:c.C1552T | p.R518W | D-mis |
| 01-0083 | DHREAMS | C | TLN1 | 24.0 | 1.000 | 5.133 | 9.326 | 9 | 35725594 | C | T | NM_006289:c.G98A | p.R33H | D-mis |
Chr, chromosome. Ph, phenotype: C, complex; I, isolated CDH. Dia, expression rank in the embryonic mouse diaphragm. Allele: Ref., reference; Alt., alternative. Class: Sp, splice site variant. Category: LGD, likely gene disrupting; D-mis, predicted damaging missense variant by metaSVM, PolyPhen-2, SIFT, or CADD. DHREAMS variants are as reported in Yu et al. (2015). Human reference sequence: GRCh37/hg19.
As cardiac anomalies are the most common CDH-associated birth defects, accounting for 14.3% of patients without associated chromosome anomalies (McGivern et al. 2015), we investigated whether the de novo mutations identified in our cohorts were mapped to genes expressed in the developing heart derived from RNA-seq of E14.5 129SvEv mouse embryos (Zaidi et al. 2013). A significant enrichment in genes expressed in the top quartile of gene expression in the embryonic heart was confirmed.
Expression profile during lung development
We hypothesized that differential expression in specific phases of lung development is an indication of which genes are likely to affect branching morphogenesis and result in lung hypoplasia, an important component of the CDH phenotype (Donahoe et al. 2016). Primary defects of branching morphogenesis, resulting in lung hypoplasia, were shown in teratogenic (dual-hit hypothesis) (Keijzer et al. 2015) and genetic models of CDH (reviewed in Kinane et al. 2007).
Genome wide expression levels were previously assessed during lung development at 26 time points throughout murine lung development in three inbred strains (GEO Series GSE74243), providing stage-specific transcriptional profiles (Beauchemin et al. 2016). We used unsupervised hierarchical clustering of the 71 genes with de novo variants in our cohorts to identify their expression pattern during in the embryonic, pseudoglandular, and canalicular phases (Figure 3). Among the genes expressed during branching morphogenesis, three (NAA15, POGZ, and SIN3A) have LGD variants in our cohort and ExAC pLI scores of 1.0, suggesting intolerance to haploinsufficiency, i.e. genes in which heterozygous loss of function mutations are expected not to be tolerated (Lek et al. 2016).
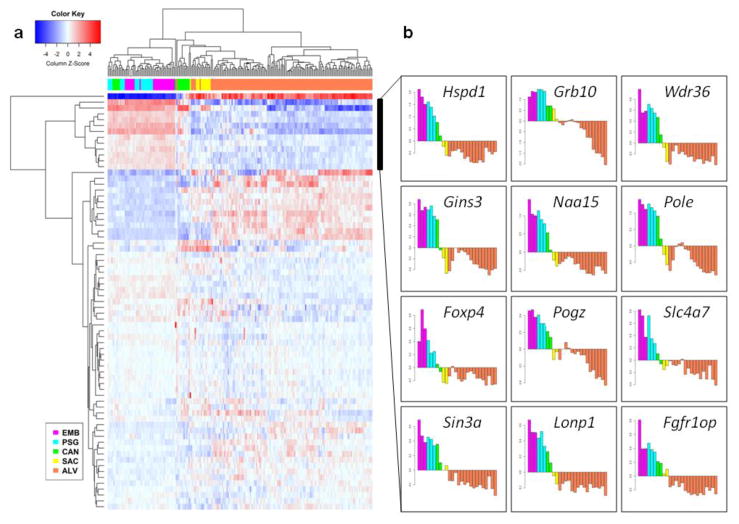
Fig. 3. Heatmap of lung expression data.
(a) The 71 genes with de novo sequence variants in CDH patients cluster in three groups according to gene expression in the mouse pre- and post-natal lungs. (b) Gene expression profiles throughout lung development are shown for each of the 12 genes with high expression in the embryonic lung, including the NAA15, HSPD1, POGZ, and SIN3A mouse orthologs. Each bar corresponds to a different time point in mouse development from E9.5 to P56. Stages in lung development are indicated by purple (embryonic), cyan (pseudoglandular), green (canalicular), yellow (saccular), and orange (alveolar). Normalization methods are as in (Beauchemin et al. 2016)
Protein-protein interaction networks
GeNets (http://apps.broadinstitute.org/genets), a biological network analysis and visualization platform developed and housed at the Broad Institute, was used to analyze the de novo gene set discovered in our cohorts based on functional genomic data. A pathway analysis based on InWeb3 data (Lage et al. 2007) was performed on every gene with at least one de novo LGD or D-mis and expressed in the upper quartile of the developing diaphragm (Table 2), together with previously known CDH genes (Supplementary table S5). The results indicated that de novo sequence variants, although not recurring in any genes in our cohorts, form significant networks (p = 0.002), or communities, based on protein-protein interactions. Nine communities were identified, representing significant modules of pathway relationships among the genes of interest (Figure 4). Five communities, among them, contained one or more candidate genes with de novo variants.
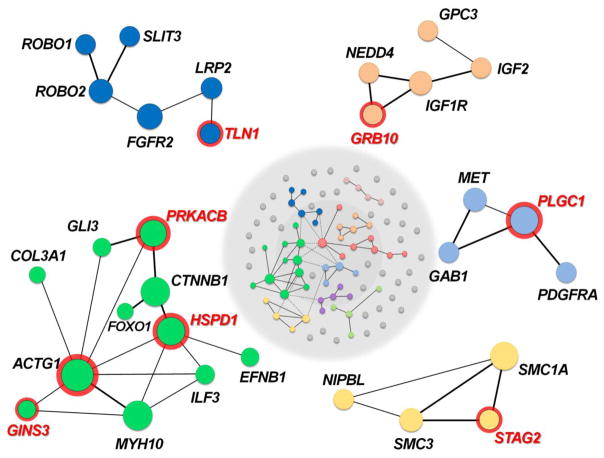
Fig. 4. Protein-protein network analysis.
Protein-protein interaction analysis based on the InWeb3 network (GeNets) revealed nine communities (center) of interacting gene products. Nodes represent CDH genes with genetic and functional evidence in Homo sapiens and/or Mus musculus, as well as candidate CDH genes with de novo mutations reported in this study. Communities with one or more de novo candidate genes (red) are magnified to provide greater detail. Larger circles indicate more central nodes. Thicker edges indicate interactions of the highest confidence, while dotted lines indicate interactions between nodes in different networks
A second pathway analysis, based only on genes with at least one de novo LGD or D-mis variant, revealed four communities. Regardless of the filtering strategy used, whether by high expression in the developing diaphragm or heart, three communities overlapped consistently around the key genes (nodes), ACTG1, HSPD1, and LONP1, suggesting their possible involvement in the pathogenesis of CDH (Supplementary figure S3).
Co-expression network analysis
The expression patterns of functionally related genes are expected to be similar temporally and across various perturbations, allowing for the identification of blocks of co-expressed genes. As an alternative way of integrating the gene expression data we therefore searched for co-expressed modules significantly enriched for CDH genes to prioritize candidate genes with de novo variants. We constructed a co-expression network and identified modules of genes with similar expression patterns based on the developing diaphragm expression data in 56 microarrays (Russell et al. 2012), using the R-package WGCNA (weighted gene correlation network analysis) (Langfelder and Horvath 2008). We then identified modules having significant overlap with the list of genes known to be involved in CDH pathology, manually curated from the literature and from available public data bases, followed by a Gene Ontology (GO) term enrichment analysis (Supplementary table S6 provides the GO terms of CDH enriched modules). The modules with significant overlap are involved in muscle differentiation, transcription regulation, cellular development, extracellular matrix, and cell division, among others.
Significantly, the expression modules identified overlapped with the protein-protein interaction pathways described above, substantiating their importance in CDH pathogenesis by an independent systems biology approach. The most suggestive similarities were the GenNets nodes and the WGCNA “pink” module, converging on IGF2/IGF1R signaling and muscle cell specification, and the “yellow” module, implicating cohesin complex function. The other modules, significantly enriched for CDH genes, are related to energy production and they are likely correlated with increased muscularization of diaphragm tissue and a miscellaneous collection of genes important in signal transduction and transcription factors (Supplementary table S6).
DISCUSSION
Genome wide burden of de novo variants
Historically, most individuals with CDH are the first members in their family to be diagnosed, partly because of the high morbidity and mortality resulting in impaired reproductive fitness. Conversely, the continuous emergence of sporadic cases suggests a role for causative mutations arising in the germline, as seen also in patients with congenital heart defects (Zaidi et al. 2013). De novo mutations leading to diaphragmatic defects have been reported anecdotally, as well as systematically, in smaller cohorts ((Yu et al. 2015) and Supplementary table S7). The present study was designed to confirm the contribution of de novo variation to human diaphragmatic defects and to provide a list of high priority CDH candidate genes.
Genetic contribution to the etiology of CDH has been previously demonstrated in cohorts of sporadic patients (Longoni et al. 2014a). A significant burden (p< 0.01, based on empirical distribution) was discovered for predicted pathogenic variants in well-characterized CDH genes. A previous trio study was performed on a small group of patients with complex, or syndromic, CDH (Yu et al. 2015), in which 15 de novo predicted deleterious sequence variants were identified. De novo variants in genes highly expressed in the developing diaphragm were significantly more frequent in CDH cases than in controls, a situation similar to what has been observed in congenital heart defects (Sanders et al. 2012; Zaidi et al. 2013). However, the same enrichment was not observed on a genome-wide level, likely because the study was underpowered to detect such an effect.
Here, we identified a significant enrichment of LGD or D-mis variants in complex CDH cases and enrichment of LGD variants in isolated cases, which represent the majority of CDH patients although they currently rarely receive a genetic diagnosis (Longoni et al. 2014b). Due to the extremely low frequency of de novo mutations, finding multiple de novo variants in the same gene associated with the same or overlapping phenotypes is considered strong evidence for causality (Samocha et al. 2014). Even combining all available exome data from CDH patients, no genes showed de novo predicted pathogenic variants in more than one individual with CDH; however, variants in some of these genes were identified in a cohort of CDH probands previously studied by WES (Longoni et al. 2014a). These findings underscore the genetic heterogeneity of CDH, and sequencing of additional trios will be necessary to enhance the discovery of causative genes.
Finally, in the combined cohorts examined in this study, we identified de novo variants in several genes previously reported in association with congenital heart disease and/or neurodevelopmental conditions, highlighting the complexity of phenotypes associated with diaphragmatic defects and the pleiotropic effects of CDH-associated genes. These phenotypes will be discussed below, with an emphasis on genes functioning as central nodes in the protein-protein interaction networks discovered in this study.
Comorbidity with neurodevelopmental conditions
The Pogo Transposable Element With Znf Domain (POGZ [OMIM:*614787]) has been identified as the causative gene for White-Sutton syndrome (OMIM:#616364) in a large-scale WES study of individuals with neurodevelopmental disorders (White et al. 2016). Proband T57 in our cohort is a male who presented with a right-sided Morgagni hernia, microcephaly, seizures, abnormal ears, micropenis, facial dysmorphism, and optic nerve hypoplasia. In this patient, we detected a p.F879Pfs (NM_015100:c.2635_2638del) de novo variant.
A patient with a p.T922Hfs mutation in POGZ was previously reported with microcephaly, short stature, global developmental delay, non-ocular visual impairment, failure to thrive, and multiple congenital abnormalities, including diaphragmatic hernia and a duplicated renal collecting system (White et al. 2016). All the frameshift mutations in POGZ, in addition to the mutation identified in patient T57, are predicted to disrupt three C-terminal-domains (CENP-B like DNA, DDE, and coiled-coil) (White et al. 2016).
The observed increased incidence of CDH in patients with this syndromic form of autism spectrum disorder and other neurobehavioral challenges may be due to the pleiotropic effects of POGZ on the development of many organs during embryogenesis because of its role in kinetochore assembly, mitotic sister chromatid cohesion, and mitotic chromosome segregation.
After manual curation of our trio dataset, two additional patients were discovered to carry rare inherited and predicted pathogenic POGZ variants. Patient T62 had an isolated left-sided Bochdalek hernia, and has a heterozygous c.4086A>C transversion (rs374364810), inherited from an apparently unaffected mother, resulting in the p.Glu1362Asp missense change, predicted to be damaging by PolyPhen-2 (URL: http://genetics.bwh.harvard.edu/pph2/) and MutationTaster (URL: http://www.mutationtaster.org/). The second patient, T46, had a left-sided Bochdalek hernia, atrial septal defect, and congenital hip dysplasia. She is heterozygous for the paternally inherited p.Arg1285Gln missense variant caused by the c.3854G>A substitution, predicted damaging by MutationTaster. Both variants are extremely rare in ExAC with allele frequencies of 6.59×10−5 (allele count: 8/121400) and 7.417×10−5 (9/121350), respectively. Reported pathogenic mutations in POGZ are consistently de novo (White et al. 2016) and larger studies will be necessary to determine whether inherited variants increase the risk for CDH. In these cases, parents were not examined for subclinical phenotypes which have been recently reported in CDH families (Yu et al. 2013).
Among the candidates with de novo mutations, the Cytoplasmic γ-actin gene (ACTG1) protein is at the center of a sub-network predicted based on de novo variants identified in the CDH cohorts, which could indicate a role for actin-related function in the development of diaphragmatic and lung defects.
We identified the p.E167K de novo missense variant in ACTG1 in a CDH patient with multiple minor anomalies and autism. Missense mutations in ACTG1 (NM_001614.2), which is expressed in the top 10th percentile of transcripts in the developing diaphragm, or in its paralog ACTB have been previously associated with the rare Baraitser-Winter syndrome (BRWS [OMIM #614583]) (Rivière et al. 2012). Characteristic findings in BRWS include congenital ptosis, high-arched eyebrows, ocular colobomata, and lissencephaly (Verloes et al. 2015). Other missense mutations in the same gene cause dominant progressive deafness (DFNA20/26) (Zhu et al. 2003) and are considered to be milder than those reported in BRWS. Functional studies suggest that BRWS mutations are gain-of-function, as they were shown to increase F-actin content and to impact F-actin dynamics when stimulated with latrunculin A, which induces cytoskeletal depolymerization (Rivière et al. 2012). Deletions of 7p22 (ACTB) or 17q25.3 (ACTG1), however, result in a different phenotype than BWRS. Notably, a patient with 7p22.1-p22.3 deletion (chr7:1,174,615-5,819,144, hg19) displayed right-sided CDH and various cardiac defects (Verloes et al. 2015).
Comorbidity with congenital heart defects
The N-Alpha-Acetyltransferase 15, NatA Auxiliary Subunit (NAA15, [OMIM:*608000]) encodes an acetyltransferase subunit and has a role in maintaining cell proliferation (Sugiura et al. 2003). Its downregulation by retinoic acid results in neuronal differentiation (Sugiura et al. 2003). Two LGD variants were discovered in an exome study of 1213 trios recruited in the Pediatric Cardiac Genetics Consortium or the Pediatric Heart Network (P=4.67 × 10−5, Poisson test against expectation) (Homsy et al. 2015). In the congenital heart defect study, a nonsense variant was discovered in a patient with pulmonary stenosis, single left coronary and tetralogy of Fallot. The second patient, with a frameshift mutation, had presented with a complex cardiac phenotype which included heterotaxy, hypoplastic right ventricle, and pulmonary stenosis. In our cohort, we identified a patient with isolated CDH and a frameshift variant (NM_057175:c.239_240del, p.H80fs).
Genes with de novo variants in CDH networks
Network analysis allowed us to identify functional subnetworks containing genes with de novo variants in the Boston and DHREAMS cohorts.
The insulin growth factor (IGF) signaling controls cell proliferation, differentiation and survival, and it is important for organogenesis. Insulin-like growth factor-1 receptor (Igf1r) (Epaud et al. 2012) and its ligands Igf1 (Liu et al. 1993) and Igf2 (Borensztein et al. 2013) have been associated with lung and diaphragm defects, the latter probably due to abnormal terminal muscle differentiation. Further, IGF signaling promotes alveolar and vascular maturation during late phases of lung development (Nagata et al. 2007), which are critical for the postnatal course of CDH patients. The regulation of Igf1r-mediated signaling is complex and includes Nedd4 (Cao et al. 2008) and Grb10 (Morrione et al. 1999), which affect stability of the receptor. In the Boston cohort, we identified a patient with complex CDH and a frameshift mutation in GRB10 that could alter IGF1R turnover.
Cohesin complexes are required for proper sister chromatid cohesion during meiosis and mitosis, and have additional roles in DNA repair and transcription (Peters et al. 2008). Smc1 and Smc3 directly at their hinge domains and at the opposite end through the mediation of Scc1, thus forming a ring-like structure around chromatin. Mutations in components of the cohesin complex cause autosomal dominant (NIPBL, SMC3) and X-linked recessive (SMC1A) Cornelia de Lange syndrome, which is frequently associated with CDH (Schrier et al. 2011). A patient with a de novo variant in the Stromal Antigen 2 (STAG2) gene, encoding a structural component of the complex, was identified in the DHREAMS cohort. The mechanism whereby defects in cohesin biology affects diaphragm development is unknown.
Conclusions
Because of the high mortality of CDH, the emergence of sporadic cases is consistent with a role for de novo mutations. Combining cohorts from two separate studies, it was possible to gain supporting evidence that de novo variants contribute significantly to the development of CDH.
METHODS
Patients
The study subjects were enrolled in the “Gene Mutation and Rescue in Human Diaphragmatic Hernia” (1 P01 HD068250) study according to the Partners Human Research Committee and Boston Children’s Hospital clinical investigation standards (Protocol 2000P000372 and 05-07-105R, respectively). Standard patient evaluation included a thorough genetic examination by our study geneticists or the referring geneticist/physician, analysis of the family pedigree, a questionnaire (87/87), and clinical and/or research chromosome microarrays (65/87) (Supplementary table 1). Medical records and operative notes were reviewed for phenotypic classification. Sporadic cases without a known chromosomal or genetic cause for their diaphragmatic defect were chosen, but variants of uncertain function were not considered exclusion criteria. Parents are not clinically affected with CDH. Exome results on some of the probands (N=33) had been reported as part of a previous cohort study (Longoni et al. 2014a) (dbGAP accession no. phs000783.v1.p1). Procedures relative to samples enrolled in the DHREAMS study are outlined in the relevant publications (Yu et al. 2014; Yu et al. 2015).
Sample processing
The DNA samples were extracted using a standard phenol-chloroform protocol (Epstein-Barr Virus transformed Lymphoblastoid Cell Lines, LCLs) or dedicated Qiagen kits (whole blood samples or primary cultured fibroblasts). After extraction, the DNA was measured by Qubit® 2.0 Fluorometric Quantitation (Life Technologies), and gel electrophoresis was performed to assess DNA quality.
Whole Exome Sequencing (WES)
Library construction and exome capture were automated (Perkin-Elmer Janus II) in 96-well plate format. 1 ug of genomic DNA was subjected to a series of shotgun library construction steps, including fragmentation through acoustic sonication (Covaris), end-polishing and A-tailing, ligation of sequencing adaptors, and PCR amplification with 8 bp barcodes for multiplexing. Libraries underwent exome capture using the Roche/Nimblegen SeqCap EZ v2.0 or v3.0. The library concentration is determined by triplicate qPCR and molecular weight distributions verified on the Agilent Bioanalyzer (consistently 125 ± 15 bp). Lastly, 100 bp paired-end sequencing was performed on the IlluminaHiSeq2000 platform (Illumina, Inc, San Diego, California, USA). The sequencing reads generated were combined with previously sequenced probands (Longoni et al. 2014a) and 38 trios (Yu et al. 2015). Sequencing metrics are provided in Supplementary table S3. Linked genotype and phenotype data was deposited in dbGAP (accession no. phs000783.v2.p1).
De novo variant calling
Raw sequencing data for each individual were aligned to the human reference genome (build hg19) using Burrows-Wheeler Aligner (BWA 0.7.5a) (Li and Durbin 2010). The alignment files of probands and parental samples were converted from a sequence alignment map (SAM) format to a sorted, indexed, binary alignment map (BAM) file (SAMtools version 0.1.19) (Li et al. 2009). To improve alignments and genotype calling, BAM files were re-aligned with the GATK IndelRealigner, base quality scores recalibrated, and duplicate reads removed by the GATK base quality recalibration tool. Variants were called using GATK HaplotypeCaller with recommended best practices for germline SNP & Indel discovery in whole genome and exome sequence (URL: https://software.broadinstitute.org/gatk/best-practices/bp_3step.php?case=GermShortWGS).
Rare variants were classified for downstream analysis based on their predicted effect on protein function. Sequence variants (SNVs and indels) were filtered to identify Mendelian errors, i.e. false positives, and potentially de novo rare variants. We used an established approach to distinguish candidate de novo mutations from false positives by calculating the Bayesian score(s) of the data for all three subjects of a trio and the allele frequency in the reference data bases (Yu et al. 2015; Homsy et al. 2015). Missense variants were labeled as D-mis if predicted to be damaging by MetaSVM or by the combination of CADD > 15, SIFT and polyphen-2 predicted damaging. Gene-level constraint metrics were as in Lek et al. (2016) using cutoffs of Z-score=3 for intolerance to missense variants and pLI=0.9 for intolerance to LGD variants.
Burden test
The expected number of de novo mutations in each class was calculated for CDH patients based on the background mutation frequency (Homsy et al. 2015) and compared to the observed number in the combined Boston and DHREAMS cohorts. The excess is determined by enrichment rate (r), which is defined as the average number of de novo LGD or D-mis variants per case / average number of variants per control. The percentage of cases with excess of LGD or D-mis variants is then (r-1)/r. Exact Poisson test, implemented in R, was used to test of the null hypothesis. The most severe predicted functional effect for each variant was used in the burden test calculations. D-mis was defined as predicted to be damaging by metaSVM in the burden analysis (Homsy et al. 2015).
Sanger sequencing
Sequence variants reported in the manuscript (Table 2) were confirmed in independent proband and parental samples. Fragments were obtained by PCR amplification using the Qiagen Taq PCR Master Mix, the cycle sequencing reaction was carried out using the Applied Biosystems BigDye v3.1 Cycle Sequencing Kit, with separation on an ABI3730XL DNA Analyzer. Primers (18–22-mer oligos) were designed within 300 bases on either side of the variant using the NCBI Primer-BLAST (http://www.ncbi.nlm.nih.gov/tools/primer-blast/) (Ye et al. 2012).
Pathway analysis and network integration
Microarray gene expression data was transformed into a diaphragm developmental network using the Weighted Gene Correlation Network Analysis as implemented in the R-package WGCNA (Langfelder and Horvath 2008). In this network edges represent correlation of gene expression trends across the developmental time points. GeNets (http://apps.broadinstitute.org/genets) was used for biological network analysis and visualization, implementing machine learning to identify previously unknown pathway relationships and discover functional modules based on protein-protein interaction data from InWeb3 (Lage et al. 2007).
Supplementary Material
Additional network analysis of proteins with de novo variants: entire set (A), top quartile of expression rank in the developing diaphragm (B) and heart (C). GeNets (http://apps.broadinstitute.org/genets) was used for biological network analysis and visualization, implementing machine learning to identify previously unknown pathway relationships and discover functional modules based on the GeNets Metanetwork v1.0 combining protein-protein interaction information from InWeb3 and Gene Expression Networks from GEO.
The distribution of de novo variants is shown according to gene expression ranks in the developing diaphragm (x-axis) and in the developing heart (y-axis). Lines indicate the top quartile of expression rank. Genes are color coded according to functional consequence of the de novo variants (top left), cohort and patient phenotype (top right), type of LGD variant (bottom left), and functional prediction of missense variants compared to silent variants (bottom right).
If previously published in the Longoni et al. 2014 publication, a conversion matrix to the alternative study ID is provided. Samples necessitating with whole genome amplification prior to library construction are indicated. Patient data include self-reported ethnicity, gender, isolated or complex (syndromic) phenotype, side of the diaphragmatic defect, type of the diaphragmatic defect (Bochdalek, Morgagni, eventration, agenesis, or not otherwise specified).
Read length, reads per sample, median and mean coverage at each targeted base, and percent of targeted bases with at least 15X reads are indicated for each of the three sequencing batches (Yale, UW1, UW2) along with number of samples per each batch. Mean of samples and standard deviation are reported.
The complete and annotated list of variants is provided. Proband ID, study cohort and symbols of genes with de novo variants are indicated, as are position of the variants and official nomenclature according to Human Genome Variation Society (HGVS) recommendations.
The list of genes with de novo variants was associated to functional biological terms in a systematic way for data exploration and interpretation using the integrative web-based application Enrichr. Significant enrichments in three libraries are shown (ChEA for transcription factor regulation, 2016 update; Biocarta for metabolic and signaling pathways, 2016 update; Reactome for curated biological pathways, 2016 update).
Nodes are indicated in alphabetical order. Evidence for their implication in CDH is provided: de novo (genes with de novo variants in human cohorts), human (genetic variants in human cohorts except de novo), mouse (mouse models with diaphragmatic defects), bioinformatics (identified by expanding protein-protein networks to first and second order interactors). Community numbers indicate which interaction subnetwork, if any, the proteins belong to.
Modules are listed by arbitrary names (pink, yellow, brown, and blue). Total number of genes, number of CDH genes, and number of genes with de novo variants are shown for each module. Hypergeometric enrichment p-values indicate whether each module contains more CDH genes than expected by random distribution.
Table S7 Literature review of genes with de novo variants in CDH patients.
Number of patients with 0~7 de novo variants in the MGH/CHB (purple) and DHREAMS (cyan) cohorts are indicated. The superimposed lines indicate the expected distribution.
Acknowledgements
We thank the surgeons at MassGeneral Hospital for Children and Boston Children’s Hospital for their continued support: T. Buchmiller, C. C. Chen, D. Doody, S. J. Fishman, A. Goldstein, L. Holmes, T. Jaksic, R. Jennings, C. Kelleher, D. Lawlor, C.W. Lillehei, P. Masiakos, D. P. Mooney, K. Papadakis, R. Pieretti, M. Puder, D. P. Ryan, R. C. Shamberger, C. Smithers, J. Vacanti, and C. Weldon. We are also grateful to all of the families at the participating Simons Simplex Collection (SSC) sites, as well as the principal investigators (A. Beaudet, R. Bernier, J. Constantino, E. Cook, E. Fombonne, D. Geschwind, R. Goin-Kochel, E. Hanson, D. Grice, A. Klin, D. Ledbetter, C. Lord, C. Martin, D. Martin, R. Maxim, J. Miles, O. Ousley, K. Pelphrey, B. Peterson, J. Piggot, C. Saulnier, M. State, W. Stone, J. Sutcliffe, C. Walsh, Z. Warren, E. Wijsman).
Footnotes
Conflicts of interest.
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Financial Disclosure.
Sequencing services were provided through the RS&G Service by the Northwest Genomics Center at the University of Washington, Department of Genome Sciences, under U.S. Federal Government contract number HHSN268201100037C from the National Heart, Lung, and Blood Institute (r223 to M. Longoni). Funding was provided by the National Institute of Child Health and Human Development (NICHD/NIH, www.nichd.nih.gov) P01HD068250. Partial funding was provided by the NICHD/NIH grant HD057036, by the Columbia University’s Clinical and Translational Science Award (CTSA), and by the National Center for Advancing Translational Sciences/National Institutes of Health (NCATS-NCRR/NIH, ncats.nih.gov) grant UL1 RR024156. Philanthropic funding was obtained by CHERUBS, the National Greek Orthodox Ladies Philoptochos Society, Inc., and generous donations from The Wheeler foundation, Vanech Family Foundation, Larsen Family, Wilke Family, and many other families. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- Ackerman KG, Vargas SO, Wilson JA, et al. Congenital diaphragmatic defects: proposal for a new classification based on observations in 234 patients. Pediatr Dev Pathol. 2012;15(4):265–74. doi: 10.2350/11-05-1041-OA.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beauchemin KJ, Wells JM, Kho AT, et al. Temporal dynamics of the developing lung transcriptome in three common inbred strains of laboratory mice reveals multiple stages of postnatal alveolar development. PeerJ. 2016;4:e2318. doi: 10.7717/peerj.2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borensztein M, Monnier P, Court F, et al. Myod and H19-Igf2 locus interactions are required for diaphragm formation in the mouse. Development. 2013;140(6):1231–9. doi: 10.1242/dev.084665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen EY, Tan CM, Kou Y, et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao XR, Lill NL, Boase N, et al. Nedd4 controls animal growth by regulating IGF-1 signaling. Sci Signal. 2008;1(38):ra5. doi: 10.1126/scisignal.1160940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rubeis S, He X, Goldberg AP, et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515:209–15. doi: 10.1038/nature13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donahoe PK, Longoni M, High FA. Polygenic Causes of Congenital Diaphragmatic Hernia Produce Common Lung Pathologies: A Multimodal War on Congenital Diaphragmatic Hernia. Am J Pathol. 2016 doi: 10.1016/j.ajpath.2016.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C, Wei P, Jian X, et al. Comparison and integration of deleteriousness prediction methods for nonsynonymous SNVs in whole exome sequencing studies. Hum Mol Genet. 2015;24:2125–2137. doi: 10.1093/hmg/ddu733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epaud R, Aubey F, Xu J, et al. Knockout of insulin-like growth factor-1 receptor impairs distal lung morphogenesis. PLoS One. 2012;7(11):e48071. doi: 10.1371/journal.pone.0048071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia AV, Thirumoorthi AS, Stolar CJH. Extracorporeal Membrane Oxygenation. In: Saunders L, editor. Ashcraft’s Pediatric Surgery. 6. London: Saunders; 2014. pp. 80–93. [Google Scholar]
- Gulsuner S, Walsh T, Watts AC, et al. Spatial and temporal mapping of de novo mutations in schizophrenia to a fetal prefrontal cortical network. Cell. 2013;154:518–29. doi: 10.1016/j.cell.2013.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homsy J, Zaidi S, Shen Y, et al. De novo mutations in congenital heart disease with neurodevelopmental and other congenital anomalies. Science. 2015;350:1262–6. doi: 10.1126/science.aac9396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iossifov I, O‘roak BJ, Sanders SJ, et al. The contribution of de novo coding mutations to autism spectrum disorder. November. 2014;13:216–221. doi: 10.15154/1149697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keijzer R, Liu J, Deimling J, et al. Dual-hit hypothesis explains pulmonary hypoplasia in the nitrofen model of congenital diaphragmatic hernia. Am J Pathol. 2000;156(4):1299–306. doi: 10.1016/S0002-9440(10)65000-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinane TB. Lung development and implications for hypoplasia found in congenital diaphragmatic hernia. Am J Med Genet C Semin Med Genet. 2007;145C(2):117–24. doi: 10.1002/ajmg.c.30124. [DOI] [PubMed] [Google Scholar]
- Lachmann A, Xu H, Krishnan J, et al. ChEA: Transcription factor regulation inferred from integrating genome-wide ChIP-X experiments. Bioinformatics. 2010;26:2438–2444. doi: 10.1093/bioinformatics/btq466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lage K, Karlberg EO, Størling ZM, et al. A human phenome-interactome network of protein complexes implicated in genetic disorders. Nat Biotechnol. 2007;25:309–16. doi: 10.1038/nbt1295. [DOI] [PubMed] [Google Scholar]
- Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lek M, Karczewski KJ, Minikel EV, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536(7616):285–91. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–95. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–9. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JP, Baker J, Perkins AS, et al. Mice carrying null mutations of the genes encoding insulin-like growth factor I (Igf-1) and type 1 IGF receptor (Igf1r) Cell. 1993;75(1):59–72. [PubMed] [Google Scholar]
- Longoni M, High FA, Russell MK, et al. Molecular pathogenesis of congenital diaphragmatic hernia revealed by exome sequencing, developmental data, and bioinformatics. Proc Natl Acad Sci U S A. 2014a;111:12450–5. doi: 10.1073/pnas.1412509111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longoni M, Russell MK, High FA, et al. Prevalence and penetrance of ZFPM2 mutations and deletions causing congenital diaphragmatic hernia. Clin Genet. 2014b doi: 10.1111/cge.12395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGivern MR, Best KE, Rankin J, et al. Epidemiology of congenital diaphragmatic hernia in Europe: a register-based study. Arch Dis Child Fetal Neonatal Ed. 2015;100:F137–44. doi: 10.1136/archdischild-2014-306174. [DOI] [PubMed] [Google Scholar]
- Michaelson JJ, Shi Y, Gujral M, et al. Whole-genome sequencing in autism identifies hot spots for de novo germline mutation. Cell. 2012;151:1431–42. doi: 10.1016/j.cell.2012.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrione A. Grb10 adapter protein as regulator of insulin-like growth factor receptor signaling. J Cell Physiol. 2003;197(3):307–11. doi: 10.1002/jcp.10363. [DOI] [PubMed] [Google Scholar]
- Nagata K, Masumoto K, Uesugi T, et al. Effect of insulin-like-growth factor and its receptors regarding lung development in fetal mice. Pediatr Surg Int. 2007;23(10):953–9. doi: 10.1007/s00383-007-1977-8. [DOI] [PubMed] [Google Scholar]
- Neale BM, Kou Y, Liu L, et al. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature. 2012;485:242–5. doi: 10.1038/nature11011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters JM, Tedeschi A, Schmitz J. The cohesin complex and its roles in chromosome biology. Genes Dev. 2008;22(22):3089–114. doi: 10.1101/gad.1724308. [DOI] [PubMed] [Google Scholar]
- Pober BR. Genetic aspects of human congenital diaphragmatic hernia. Clin Genet. 2008;74:1–15. doi: 10.1111/j.1399-0004.2008.01031.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pober BR. Overview of epidemiology, genetics, birth defects, and chromosome abnormalities associated with CDH. Am J Med Genet C Semin Med Genet. 2007;145C:158–71. doi: 10.1002/ajmg.c.30126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pober BR, Russell MK, Ackerman KG. Congenital Diaphragmatic Hernia Overview [GeneReviewsTM. 1993] In: Pagon RA, Adam MP, Bird TD, Dolan CR, Fong CT, Smith RJH, Stephens K, editors. SourceGeneReviewsTM. Seattle Univ; Washington, Seattle: 2010. pp. 1993–2013. [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivière J-B, van Bon BWM, Hoischen A, et al. De novo mutations in the actin genes ACTB and ACTG1 cause Baraitser-Winter syndrome. Nat Genet. 2012;44:440–4. S1–2. doi: 10.1038/ng.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdóttir H, Winckler W, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell MK, Longoni M, Wells J, et al. Congenital diaphragmatic hernia candidate genes derived from embryonic transcriptomes. Proc Natl Acad Sci U S A. 2012;109:2978–83. doi: 10.1073/pnas.1121621109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samocha KE, Robinson EB, Sanders SJ, et al. A framework for the interpretation of de novo mutation in human disease. Nat Genet. 2014;46:944–50. doi: 10.1038/ng.3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders SJ, Murtha MT, Gupta AR, et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature. 2012;485:237–41. doi: 10.1038/nature10945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrier SA, Sherer I, Deardorff MA, et al. Causes of death and autopsy findings in a large study cohort of individuals with Cornelia de Lange syndrome and review of the literature. Am J Med Genet A. 2011;155A(12):3007–24. doi: 10.1002/ajmg.a.34329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sifrim A, Hitz M-P, Wilsdon A, et al. Distinct genetic architectures for syndromic and nonsyndromic congenital heart defects identified by exome sequencing. Nat Genet. 2016:1–9. doi: 10.1038/ng.3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolar CJH, Dillon PW. Pediatric Surgery. 7. 2012. Congenital Diaphragmatic Hernia and Eventration; pp. 809–824. [Google Scholar]
- Sugiura N, Adams SM, Corriveau RA. An evolutionarily conserved N-terminal acetyltransferase complex associated with neuronal development. J Biol Chem. 2003;278:40113–40120. doi: 10.1074/jbc.M301218200. [DOI] [PubMed] [Google Scholar]
- Veenma DCM, de Klein A, Tibboel D. Developmental and genetic aspects of congenital diaphragmatic hernia. Pediatr Pulmonol. 2012;47:534–45. doi: 10.1002/ppul.22553. [DOI] [PubMed] [Google Scholar]
- Veltman JA, Brunner HG. De novo mutations in human genetic disease. Nat Rev Genet. 2012;13:565–75. doi: 10.1038/nrg3241. [DOI] [PubMed] [Google Scholar]
- Verloes A, Di Donato N, Masliah-Planchon J, et al. Baraitser-Winter cerebrofrontofacial syndrome: delineation of the spectrum in 42 cases. Eur J Hum Genet. 2015;23:292–301. doi: 10.1038/ejhg.2014.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ware JS, Samocha KE, Homsy J, Daly MJ. Interpreting de novo Variation in Human Disease Using denovolyzeR. Curr Protoc Hum Genet. 2015;87:7.25.1–7.25.15. doi: 10.1002/0471142905.hg0725s87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J, Beck CR, Harel T, et al. POGZ truncating alleles cause syndromic intellectual disability. Genome Med. 2016;8:3. doi: 10.1186/s13073-015-0253-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff G. Familial congenital diaphragmatic defect: review and conclusions. Hum Genet. 1980;54:1–5. doi: 10.1007/BF00279041. [DOI] [PubMed] [Google Scholar]
- Wynn J, Yu L, Chung WK. Genetic causes of congenital diaphragmatic hernia. Semin Fetal Neonatal Med. 2014;19:324–330. doi: 10.1016/j.siny.2014.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B, Roos JL, Dexheimer P, et al. Exome sequencing supports a de novo mutational paradigm for schizophrenia. Nat Genet. 2011;43:864–8. doi: 10.1038/ng.902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Coulouris G, Zaretskaya I, et al. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics. 2012;13:134. doi: 10.1186/1471-2105-13-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, Bennett JT, Wynn J, et al. Whole exome sequencing identifies de novo mutations in GATA6 associated with congenital diaphragmatic hernia. J Med Genet. 2014 doi: 10.1136/jmedgenet-2013-101989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, Sawle AD, Wynn J, et al. Increased burden of de novo predicted deleterious variants in complex congenital diaphragmatic hernia. Hum Mol Genet. 2015;24:4764–73. doi: 10.1093/hmg/ddv196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, Wynn J, Cheung YH, et al. Variants in GATA4 are a rare cause of familial and sporadic congenital diaphragmatic hernia. Hum Genet. 2013;132:285–92. doi: 10.1007/s00439-012-1249-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi S, Choi M, Wakimoto H, et al. De novo mutations in histone-modifying genes in congenital heart disease. Nature. 2013;498:220–3. doi: 10.1038/nature12141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu M, Yang T, Wei S, et al. Mutations in the gamma-actin gene (ACTG1) are associated with dominant progressive deafness (DFNA20/26) Am J Hum Genet. 2003;73:1082–91. doi: 10.1086/379286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Petrovski S, Xie P, et al. Whole-exome sequencing in undiagnosed genetic diseases: interpreting 119 trios. Genet Med. 2015;17:774–781. doi: 10.1038/gim.2014.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional network analysis of proteins with de novo variants: entire set (A), top quartile of expression rank in the developing diaphragm (B) and heart (C). GeNets (http://apps.broadinstitute.org/genets) was used for biological network analysis and visualization, implementing machine learning to identify previously unknown pathway relationships and discover functional modules based on the GeNets Metanetwork v1.0 combining protein-protein interaction information from InWeb3 and Gene Expression Networks from GEO.
The distribution of de novo variants is shown according to gene expression ranks in the developing diaphragm (x-axis) and in the developing heart (y-axis). Lines indicate the top quartile of expression rank. Genes are color coded according to functional consequence of the de novo variants (top left), cohort and patient phenotype (top right), type of LGD variant (bottom left), and functional prediction of missense variants compared to silent variants (bottom right).
If previously published in the Longoni et al. 2014 publication, a conversion matrix to the alternative study ID is provided. Samples necessitating with whole genome amplification prior to library construction are indicated. Patient data include self-reported ethnicity, gender, isolated or complex (syndromic) phenotype, side of the diaphragmatic defect, type of the diaphragmatic defect (Bochdalek, Morgagni, eventration, agenesis, or not otherwise specified).
Read length, reads per sample, median and mean coverage at each targeted base, and percent of targeted bases with at least 15X reads are indicated for each of the three sequencing batches (Yale, UW1, UW2) along with number of samples per each batch. Mean of samples and standard deviation are reported.
The complete and annotated list of variants is provided. Proband ID, study cohort and symbols of genes with de novo variants are indicated, as are position of the variants and official nomenclature according to Human Genome Variation Society (HGVS) recommendations.
The list of genes with de novo variants was associated to functional biological terms in a systematic way for data exploration and interpretation using the integrative web-based application Enrichr. Significant enrichments in three libraries are shown (ChEA for transcription factor regulation, 2016 update; Biocarta for metabolic and signaling pathways, 2016 update; Reactome for curated biological pathways, 2016 update).
Nodes are indicated in alphabetical order. Evidence for their implication in CDH is provided: de novo (genes with de novo variants in human cohorts), human (genetic variants in human cohorts except de novo), mouse (mouse models with diaphragmatic defects), bioinformatics (identified by expanding protein-protein networks to first and second order interactors). Community numbers indicate which interaction subnetwork, if any, the proteins belong to.
Modules are listed by arbitrary names (pink, yellow, brown, and blue). Total number of genes, number of CDH genes, and number of genes with de novo variants are shown for each module. Hypergeometric enrichment p-values indicate whether each module contains more CDH genes than expected by random distribution.
Table S7 Literature review of genes with de novo variants in CDH patients.
Number of patients with 0~7 de novo variants in the MGH/CHB (purple) and DHREAMS (cyan) cohorts are indicated. The superimposed lines indicate the expected distribution.