Abstract
Prader–Willi syndrome (PWS) is caused by loss of paternally expressed genes in the 15q11-q13 region. To further characterize alterations in gene expression in this classical obesity syndrome we used whole genome microarrays to study a PWS mouse model resulting from a paternally derived imprinting center (IC) deletion (PWS IC deletion). These mice die generally within 2–3 days of life (reflective of failure to thrive in infants with PWS) and therefore, the analysis was performed on RNA extracted from the whole brain of PWS IC deletion mice and normal littermates at less than 24 hr after birth. Of more than 45,000 probes examined, 26,471 (59%) were detected for further analysis, and 69 had a significant change in expression of at least 1.5-fold and a false discovery rate (FDR) of 5%. Eight of the genes with differential expression were imprinted and from the PWS critical region (PWSCR). The three genes with the highest expression in the PWS IC mice were pro-opiomelanocortin (Pomc) and two transcripts of unknown function. Pomc knockout mice have been shown to develop obesity. Therefore, elevated Pomc RNA in PWS IC deletion neonatal mice may be an important genetic factor in the survival of these mice as it may affect eating behavior. Interestingly, Mc5r, a melanocortin receptor known to directly respond to Pomc expression changes, was upregulated as well. Mc5r is known to be involved with thermoregulation which is reportedly abnormal in PWS infants. These observations support a role for Pomc and the network of genes involved in regulating energy homeostasis in the early clinical findings of failure to thrive observed in PWS. Other notable patterns include three previously unstudied transcripts that are expressed only from the paternal allele under regulatory control of the IC and include AK013560, BB3144814, and BB182944 (whose genes are located in the mouse PWSCR on chromosome 7B). As expected, all the known paternally expressed genes from the PWSCR had detection signals below the threshold in the PWS IC deletion mice but were clearly detectable in control littermates. Several of the genes in this study were further examined by quantitative reverse transcription-PCR (RT-PCR) to confirm their expression status. Further analysis of gene expression in these mice may lead to novel pathways affected in PWS. These results, along with other recent reports, suggest that the cumulative effect of modest changes in expression of many genes, especially genes involved in energy metabolism, contribute to the failure to thrive of infants with PWS.
Keywords: Prader–Willi syndrome (PWS), PWS IC deletion mouse, microarray gene expression, pro-opiomelano-cortin (Pomc), quantitative RT-PCR
INTRODUCTION
Prader–Willi syndrome (PWS) is a complex neurodevelopmental disorder characterized by a two stage clinical sequence. Infants with PWS have severe hypotonia, feeding difficulties, hypogonadism, small hands and feet, respiratory problems, and genital hypoplasia [Cassidy, 1997; Butler and Thompson, 2000]. At about 2–4 years of age, children with PWS develop an insatiable appetite resulting in the onset of severe obesity if food intake is not strictly controlled. Subjects with PWS have a reduced metabolic rate and abnormal body composition with reduced lean mass and higher fat mass, particularly subcutaneous rather than visceral fat [Talebizadeh and Butler, 2005]. In addition, PWS subjects have moderate cognitive impairment often accompanied by behavioral difficulties [Cassidy et al., 1997; Butler and Thompson, 2000; Bittel and Butler, 2005].
The chromosome 15q11-q13 region contains imprinted sequences that are differentially expressed depending on the parent of origin. PWS results from the absence of expression of imprinted genes (paternally expressed) located on chromosome 15q11-q13. Imprinted expression is coordinately controlled in cis by an imprinting center (IC) which regulates the establishment of parental specific allelic differences in DNA methylation, chromatin structure, and expression [Brannan and Bartolomei, 1999; Nicholls and Knepper, 2001; Bittel and Butler, 2005].
Absence of paternally expressed genes is due to a paternally derived deletion of the 15q11-q13 region in about 70% of PWS subjects, maternal disomy 15 in approximately 25%, and an imprinting defect in 2–3% [Bittel and Butler, 2005]. The 15q11-q13 region contains about 4 million base pairs of DNA and as many as 50 genes/transcripts. To date, 10 genes have been located in this region that have been shown to be paternally expressed [Bittel and Butler, 2005; Stefan et al., 2005]. In addition, at least two genes in this region are maternally expressed in some tissues (UBE3A, ATP10C) and loss of expression of these genes causes Angelman syndrome, an entirely different clinical syndrome [Meguro et al., 2001a,b]. Analysis of gene expression in the 15q11-q13 region has identified several candidate genes which may play a role in PWS, including SNURF-SNRPN, NDN, MAGEL2, MKRN3, and one or more members of the snoRNA genes in the 15q11-q13 region; but the molecular basis of the PWS phenotype remains poorly understood [Nicholls, 1999; Lee et al., 2000, 2005; Hanel and Wevrick, 2001; Meguro et al., 2001a,b; Gallagher et al., 2002; Bittel et al., 2003, 2005; Ren et al., 2003].
Many of the genes in the 15q11-q13 region appear to have either RNA or protein processing functions, suggesting a wider role for these sequences in controlling gene expression. For example, the multi-cistronic SNURF-SNRPN-snoRNA locus codes for two nuclear localized proteins, SMN and SNURF, and multiple C/D small nucleolar RNAs (snoRNAs). SMN is a spliceosomal protein involved in RNA splicing [Gray et al., 1999a,b] and SNURF is rich in arginine residues suggesting that it may bind RNA but its exact function remains unknown [Gray et al., 1999a,b]. The function of the snoRNAs is unclear, but they likely play a role in mRNA splicing or methylation [Cavaille et al., 2000; de los Santos et al., 2000; Runte et al., 2001]. MKRN3 encodes a putative ribonucleoprotein, NDN, a MAGE protein, may act as a transcriptional repressor [Matsumoto et al., 2001; Ren et al., 2003; Lee et al., 2005] and the related MAGE gene, MAGEL2, encodes protein that is widely expressed in the central nervous system with unknown function [Boccaccio et al., 1999; Lee et al., 2000].
Although the 15q11-q13 defect underlies the PWS phenotype, the genes within the region do not appear to be directly responsible for the complex phenotype. Rather, it seems likely that the PWS phenotype results from dysregulation of multiple interconnected neurological and metabolic pathways. We have previously used custom made microarrays to examine the expression patterns of the genes within and nearby the 15q11-q13 region [Bittel et al., 2003, 2005] in human lymphoblastoid cells. Our results indicated several genes/transcripts in the region had parent of origin allelic bias in expression.
The functional and regulatory mechanisms of this region have been conserved in mice. Postnatal survival of mice with a paternally derived deletion of the PWS critical region (PWSCR) caused by a fortuitous transgenic insert fail to survive beyond a few days, referred to here as TgPWS mice [Gabriel et al., 1999]. The TgPWS mice have been shown to have decreased levels of agouti-related protein (Agrp) and increased levels of pro-opiomelanocortin (Pomc) [Ge et al., 2002]. In addition, RNA from whole brain of this TgPWS mouse model analyzed by microarrays and quantitative reverse transcription-PCR (RT-PCR) revealed loss of expression of imprinted genes in the PWSCR as expected [Stefan et al., 2005]. Surprisingly, no other large changes in gene expression were identified. However, several genes located in close proximity to each other on mouse chromosome 18 had a modest increase in expression [Stefan et al., 2005]. The authors concluded that these genes were upregulated as a result of haploinsufficiency of a biallelically expressed (non-imprinted) gene(s) because upregulation occurred regardless of the parent of origin of the deletion.
Two other mouse models for PWS have been described including a model derived from uniparental maternal disomy [Cattanach et al., 1992] and an IC deletion model [Yang et al., 1998]. The maternal disomy mouse model dies within 2–7 days after birth due to failure to thrive. The mice with a targeted deletion of the IC that is paternally inherited (PWS IC deletion) also have failure to thrive and usually die within 2 days. The PWS IC deletion mice exhibit poor feeding, reduced activity, and small size. The PWS IC deletion mice were initially reported to lack expression of the imprinted genes, Snurf-Snrpn, Magel2, Ndn, Mkrn3 and the paternally expressed snoRNAs [Yang et al., 1998]. However, in a later report it was noted that the PWS IC deletion mice had a low level of expression of both alleles of the imprinted genes within the PWSCR [Chamberlain et al., 2004]. Interestingly, there is strain-dependent survival of these mice which appears to be the result of strain-specific maternally inherited modifiers of survival. Herein, we describe transcriptome analysis of RNA isolated from whole brains of PWS IC deletion mice [Yang et al., 1998] and compare the gene expression results with the reported findings seen in the TgPWS mouse model [Stefan et al., 2005].
MATERIALS AND METHODS
Production and Confirmation of Paternally Derived PWS IC Deletion Mice
PWS IC deletion mice were obtained from Dr. Camilynn Brannan at the University of Florida Medical Center [Yang et al., 1998]. The PWS IC deletion mice result from a targeted deletion of 35 kb which included exons 1 through 6 with part of exon 7 of the Snrpn gene and 16 kb of upstream sequence [Yang et al., 1998; Chamberlain et al., 2004]. A colony of C57BL/6J congenic PWS IC knockout mice was established and maintained by breeding female carriers of the PWS IC deletion with C57BL/6J male mice. Alternatively, paternally derived PWS IC deletion mice were produced by the mating of male PWS IC deletion carrier mice with C57BL/6J females. All mice for this study were maintained at the University of Missouri-Kansas City (UMKC) Lab Animal Center with animal usage guidelines and procedures approved by the UMKC Institutional Animal Care and Use Committee.
Genomic PCR was performed on DNA obtained from tail samples to identify mice carrying a maternal IC deletion which has no obvious phenotype. DNA extractions of tail clippings were performed as previously described [White et al., 2005]. Genotyping of the PWS IC deletion was accomplished using a forward primer (GTCACGTCCTGCACGACGCGAG) from the PGK-neo insert (used for the production of the deletion) and a reverse primer (CCGCATTTCAT-CATTCTCAGGCTC) from intron 7 of Snrpn. The product size visualized by 3% agarose gel electrophoresis was 423 base pairs. Wild-type alleles were identified by the amplification of a 218 base pair band using a forward primer from the Snrpn exon 7 (GGCATTGCTCGTGTGCCTCTTGC) and the same intron 5 reverse primer. Genomic PCR was completed using the Fail Safe PCR system (Epicentre Technologies, Madison, WI) using Fail Safe buffer D and the following thermal cycles: an initial denaturing at 94°C for 2 min and 35 cycles consisting of denaturing temperature of 94°C for 1 min, annealing temperature of 66°C for 1 min, and extension at 72°C for 1 min with a final extension of 72°C for 5 min.
Extraction of Whole Brain RNA for Microarray Analyses
Whole brains from normal and paternally derived PWS IC deletion mouse pups less than 24 hr old were collected, quick frozen in liquid nitrogen, and RNA extracted using the RNeasy Midi Kit (Qiagen, Valencia, CA) according to manufacturer’s protocol. The quality of RNA was assessed by gel electrophoresis to determine the relative amounts of 28s and 18s ribosomal markers and by quantitation via spectrophotometer. All RNAs were found to have a 260/280 OD ratio of greater than 1.8. The concentration of RNA samples was adjusted to 1.0 μg/μl.
Analysis of Whole Genome Mouse Microarrays
Affymetrix GeneChip® Mouse Genome 430 2.0 Array which assays more than 45,000 transcripts was used to examine the mouse brain transcriptome. Target preparation, hybridization, and initial data collection were done at the microarray facility, University of Kansas Medical Center according to standard protocols (Affymetrix, Inc., Santa Clara, CA). GeneSpring software (Silicon Genetics, Agilent Technologies, Palo Alto, CA) was used for microarray data characterization and analysis as previously described [Bittel and Butler, 2005].
Microarray analysis was performed using RNA from the brains of male mice with an IC deletion [Yang et al., 1998] and control littermates. The PWS mice on the C57BL/6J genetic background do not survive beyond 4 days so the brains were removed before the mice were 24 hr old. The brains of unaffected littermates were isolated at the same time.
Analysis and Statistics
The inclusion criteria for retention of probes for analysis required a “present” or “marginal” signal in at least three of the four microarrays done in each group (control or PWS IC deletion mice). In addition, within the PWS IC deletion group, all values for the gene had to be in the same direction (up or down) relative to the control group. Differences between mean gene expression values were evaluated using a Welsh t-test with Bonferroni correction without assuming equal variances and a false discovery rate (FDR) of 20% or less. We developed an a priori list of candidate genes associated with neurodevelopment using Ingenuity pathways analysis software (Ingenuity Systems, Inc., Redwood City, CA) which was used for small group analysis.
Quantitative RT-PCR
Quantitative RT-PCR was performed on a subset of genes/transcripts using a QuantiTect SYBR Green RT-PCR kit (Qiagen) according to the manufacturer’s directions. Total RNA was isolated from homogenized whole brain using Trizol reagent (Invitrogen, Inc., Carlsbad, CA) and evaluated by spectroscopy. An equal quantity of total RNA (500 ng) from each brain, together with gene-specific primers were added to a reaction mix containing all components necessary for reverse transcription and PCR (Superscript 3 Platinum SYBR Green 1 step quantitative RT-PCR kit with ROX, Invitrogen, Inc.). The reaction was carried out in an ABI 7000 system (Applied Biosystems, Foster City, CA) beginning with a 30 min step at 50°C to allow for reverse transcription, followed by 15 min at 95°C. PCR was followed for 45 cycles during which the intensity of the SYBR Green fluorescence was measured at the extension step of each PCR cycle. The point at which the intensity level crossed the PCR cycle threshold (CT) was used to compare individual reactions. At least five replicates were performed on each sample for each gene. A dissociation curve was then generated for all reactions and reactions were run on agarose gels to verify the presence of a single band. Amplification of GAPD served as a control gene for each sample. Normalization of expression was performed as previously described [Bittel et al., 2006] by dividing the mean GAPD gene expression (CT) value from each subject by the mean GAPD gene expression (CT) value of one of the comparison subjects to produce a correction value. Each mean CT value for the other genes was divided by the correction value to produce the normalized value. The normalized CT values were averaged to produce the mean CT value for each gene analyzed.
RESULTS
IC Knockout Mice Versus Control Mice
We analyzed total RNA expression isolated from four paternally derived PWS IC deletion male mice and four control male littermates. The PWS IC deletion pups do not survive more than 4 days on the mouse genetic strain used in this study. Therefore, the brains were removed prior to 24 hr to prevent changes in gene expression due to loss of viability as done in a previous analysis of a different PWS mouse model [Stefan et al., 2005]. Of more than 45,000 probes on the microarrays, 26,471 (about 59%) met the inclusion criteria for detectable expression. A total of 1,135 probes had changed expression of at least 1.5-fold, either increased or decreased, in the PWS IC deletion mice compared to the control mice. Of the 1,135 probes with at least a 1.5-fold or greater change, 424 had a statistically significant difference in expression with a FDR set at 20%. This group of genes was assigned to functional categories by Ingenuity pathways analysis and the top 10 categories with the number of genes in that group are shown in Table I. However, only 69 probes were significant with an FDR of 5% (Fig. 1 and Table II). Eight of the genes with differential expression were imprinted and from the PWSCR, including three transcripts not previously analyzed. Only 61 genes with a significant change in expression were located outside the PWSCR. Furthermore, with the exception of the dramatically reduced expression of the paternally expressed genes in the PWSCR, no genes had differential expression of more than threefold except Pomc. Differential gene expression identified by microarray analysis was validated by quantitative RT-PCR for several genes of interest with the greatest change in expression and because of their functional relevance to the PWS phenotype. These genes included Snrpn, Ndn, Pomc, Mc5r, and three imprinted transcripts from the PWSCR which have not been previously studied (Table III).
TABLE I.
Top 10 Significant Functional Categories (Assigned by Ingenuity Pathways Analysis Software) of Probes With a Significant Difference in Expression With a False Discovery Rate of 20% in Brain From PWS Imprinting Center Deletion Mice Compared to Control Littermates
| Category | Number of genes |
|---|---|
| Cancer | 16 |
| Cell death | 15 |
| Genetic disorder | 12 |
| Cellular development and organization | 10 |
| Molecular transport | 10 |
| Gene expression | 9 |
| Hemotological disease | 8 |
| Developmental disorder | 6 |
| Hepatic system development and function | 3 |
| Gastrointestinal disease | 2 |
There were 474 genes with P <0.05 by Welsh t-test.
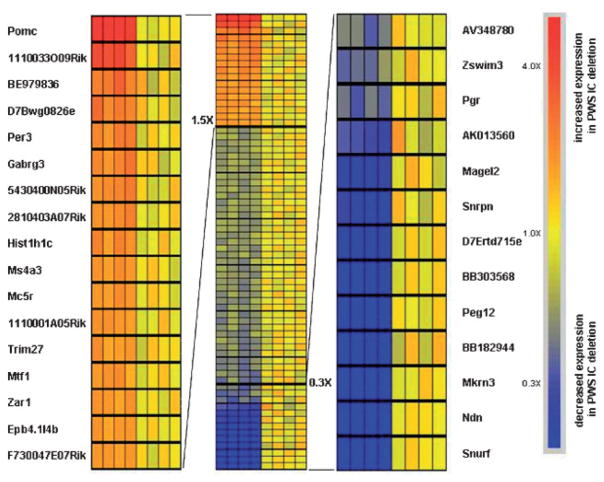
Fig. 1.
Probes with a significant difference in expression with a FDR of 5% in brain from PWS IC deletion mice compared to control littermates. There were 26,471 probes with a detectable signal and 1,135 had a difference in mean intensity of at least 1.5-fold between the two groups, of which 69 probes had a significant change of expression. The gene order is consistent with the order in Table II. The probes are arranged in order of descending gene expression ratio for PWS IC deletion mice relative to control littermates. The 17 probes with increased expression (1.5×) in PWS IC deletion mice are shown to the left. The 13 probes with lowest expression (0.3×) in the PWS IC deletion mice are shown to the right.
TABLE II.
Probes With a Significant Difference in Expression With a False Discovery Rate of 5% in Brain From PWS Imprinting Center (IC) Deletion Mice Compared to Control Littermates
| Gene | Ratioa | P-value | Genbank | Map | Description |
|---|---|---|---|---|---|
| Pomc | 14.4 | 0.001 | AV173741 | 12 A1.1 | Pro-opiomelanocortin-alpha |
| 1110033O09Rik | 3.74 | 0.001 | AW259452 | 7 D2 | RIKEN cDNA 1110033O09 gene |
| Transcript | 2.25 | 0.004 | BE979836 | 8 A2 | Transcribed sequences |
| D7Bwg0826e | 2.14 | 0.004 | R74675 | 7 B3 | RIKEN cDNA 2600013E07 gene |
| Per3 | 1.96 | 0.005 | AW553065 | 4 E2 | Period homolog 3 (Drosophila) |
| Gabrg3 | 1.92 | 0.005 | NM_008074 | 7 B4 | Gamma-aminobutyric acid (GABA-A) receptor, subunit gamma 3 |
| 5430400N05Rik | 1.89 | 0.002 | AK017247 | 7 D1 | RIKEN cDNA 5430400N05 gene |
| 2810403A07Rik | 1.78 | 0.002 | AK020778 | 3 F1 | RIKEN cDNA 2810403A07 gene |
| Hist1h1c | 1.77 | 0.007 | NM_015786 | 13 A3.1 | Histone 1, H1c |
| Ms4a3 | 1.61 | 0.002 | NM_133246 | 19 A | Membrane-spanning 4-domains, subfamily A, member 3 |
| Mc5r | 1.59 | 0.003 | NM_013596 | 18 E2 | Melanocortin 5 receptor |
| 1110001A05Rik | 1.57 | 0.001 | AK009464 | 3 H1 | ENH1 homolog [Mus musculus] |
| Trim27 | 1.55 | 0.004 | BB401251 | 13 A3.1 | Tripartite motif protein 27 |
| Mtf1 | 1.53 | 0.008 | AK012676 | 4 D2.2 | Metal response element binding transcription factor 1 |
| Zar1 | 1.53 | 0.004 | BG071693 | 5 C3.2 | Zygote arrest 1 |
| Epb4.1l4b | 1.52 | 0.003 | NM_019427 | 4 B3 | erythrocyte protein band 4.1-like 4b |
| F730047E07Rik | 1.51 | 0.000 | BG071041 | 4 A3 | RIKEN cDNA F730047E07 gene |
| Kalrn | 0.67 | 0.006 | BB662566 | 16 B3 | RIKEN cDNA E530005C20 gene |
| Transcript | 0.66 | 0.007 | BB272510 | 4 A5 | BB272510 RIKEN mRNA sequence. |
| AI854408 | 0.66 | 0.004 | AV274318 | 10 C2 | Expressed sequence AI854408 |
| 5830457O10Rik | 0.66 | 0.003 | BC023107 | 15 A1 | RIKEN cDNA 5830457O10 gene |
| Ankrd27 | 0.66 | 0.003 | BB401190 | 7 B1 | Similar to VPS9-ankyrin repeat-containing protein (LOC384745), mRNA |
| Transcript | 0.66 | 0.003 | BC027567 | 12 D1 | Clone IMAGE:3472070, mRNA |
| Msh2 | 0.65 | 0.001 | NM_008628 | 17 E4 | mutS homolog 2 (E. coli) |
| Dmn | 0.65 | 0.001 | AI594683 | 7 C | Desmuslin |
| Nlk | 0.65 | 0.005 | NM_008702 | 11 B5 | Nemo like kinase |
| Synj2bp | 0.65 | 0.002 | NM_025292 | 12 D1 | Synaptojanin 2 binding protein |
| Transcript | 0.65 | 0.009 | AV300514 | 9 A3 | Transcribed sequences |
| Cdc37l | 0.65 | 0.006 | BE952207 | 19 C1 | cell division cycle 37 homolog (S. cerevisiae)-like |
| Rab3d | 0.64 | 0.002 | BB349707 | 9 A3 | RAB3D, member RAS oncogene family |
| Fbxo18 | 0.64 | 0.005 | AF184275 | 2 A1 | F-box only protein 18 |
| Tardbp | 0.64 | 0.003 | BM935796 | 2 E2 | TAR DNA binding protein |
| Transcript | 0.63 | 0.003 | BB428304 | 9 A2 | Cysteine and histidine-rich domain (CHORD)-containing, zinc-binding protein 1 |
| Qk | 0.63 | 0.007 | AW060288 | 17 A1 | Quaking |
| C230094A16Rik | 0.63 | 0.005 | BC024726 | 11 A3.3 | RIKEN cDNA C230094A16 gene |
| 1700013G20Rik | 0.62 | 0.008 | AK005950 | 12 A1.1 | RIKEN cDNA 1700013G20 gene |
| 2410015B03Rik | 0.62 | 0.004 | BC005637 | 18 B2 | RIKEN cDNA 2410015B03 gene |
| Ankhd1 | 0.62 | 0.008 | BM243710 | 18 B2 | RIKEN cDNA 9130019P20 gene |
| D330050P16Rik | 0.61 | 0.006 | BF140677 | 17 E2 | RIKEN cDNA A230048G03 gene |
| 1810063B05Rik | 0.61 | 0.002 | BG975168 | 8 E2 | RIKEN cDNA 1810063B05 gene |
| AW210570 | 0.61 | 0.007 | AK004129 | 2 F1 | Expressed sequence AW210570 |
| Transcript | 0.60 | 0.005 | AV308954 | 1 E2.1 | AV308954 RIKEN mRNA sequence. |
| D130027M04Rik | 0.60 | 0.003 | AW412503 | 3 F2.1 | Hypothetical protein D130027M04 |
| Fgd4 | 0.60 | 0.008 | AF402611 | 16 A1 | FYVE, RhoGEF and PH domain containing 4 |
| 6430604M11Rik | 0.60 | 0.008 | BB188841 | 6 B1 | RIKEN full-length clone:A330051C14 |
| Transcript | 0.58 | 0.005 | BB745175 | 19 C1 | RIKEN cDNA clone:A430010E21 |
| Trip4 | 0.58 | 0.004 | AV350958 | 9 C | Thyroid hormone receptor interactor 4 |
| Fancg | 0.57 | 0.007 | BG072083 | 4 A5 | Fanconi anemia, complementation group G |
| C030039L03Rik | 0.57 | 0.008 | BB359532 | 7 A3 | RIKEN cDNA clone C030039L03 |
| Cx3cr1 | 0.57 | 0.007 | BC012653 | 9 F4 | Chemokine (C-X3-C) receptor 1 |
| Pnma2 | 0.55 | 0.005 | BG072348 | 14 D1 | Paraneoplastic antigen MA2 |
| 2610528B01Rik | 0.54 | 0.002 | AK012160 | 4 D3 | RIKEN cDNA clone:2610528B01 |
| Chordc1 | 0.52 | 0.002 | NM_025844 | 9 A2 | Cysteine and histidine-rich domain (CHORD)-containing, zinc-binding protein 1 |
| 2810021B07Rik | 0.52 | 0.005 | AK021189 | 13 A2 | RIKEN cDNA 2810021B07 gene |
| C030032C09Rik | 0.52 | 0.005 | BB080832 | 10 C2 | RIKEN cDNA C030032C09 gene |
| D430028G21Rik | 0.48 | 0.006 | BC025825 | 2 F1 | RIKEN cDNA D430028G21 gene |
| Transcript | 0.47 | 0.006 | AV348780 | 2 H4 | Transcribed sequences |
| Zswim3 | 0.46 | 0.006 | AK014904 | 2 H3 | Zinc finger, SWIM domain containing 3 |
| Pgr | 0.41 | 0.003 | BB428874 | 9 A1 | Progesterone receptor |
| Transcript | 0.26 | 0.002 | AK013560 | 7 B5 | Musculus adult male hippocampus, RIKEN cDNA clone:2900019J01. |
| Magel2 | 0.11 | 0.000 | NM_013779 | 7 B5 | Melanoma antigen, family L, 2 |
| Snrpn | 0.08 | 0.004 | AK010671 | 7 B5 | Small nuclear ribonucleoprotein N |
| D7Ertd715e | 0.07 | 0.000 | BB314814 | 7 B5 | Adult male corpora quadrigemina, RIKEN cDNA clone:B230361E09 |
| Transcript | 0.04 | 0.002 | BB303568 | 12qB3 | Adult male corpora quadrigemina, RIKEN cDNA clone:B230105C16 |
| Peg12 | 0.02 | 0.000 | NM_013788 | 7 B5 | Paternally expressed 12 |
| Transcript | 0.02 | 0.002 | BB182944 | 7 B5 | Transcribed sequences |
| Mkrn3 | 0.01 | 0.000 | NM_011746 | 7 B5 | Makorin, ring finger protein, 3 |
| Ndn | 0.01 | 0.000 | AW743020 | 7 B5 | Necdin |
| Snurf | 0.01 | 0.000 | NM_033174 | 7 B5 | Small nuclear ribonucleoprotein N |
The shaded areas are sequences which map to the mouse PWS critical region on chromosome 7B.
There were 26,471 probes with a detectable signal and 1,135 had a difference in mean intensity of at least 1.5-fold between the two groups, of which 69 probes had a significant change of expression. The gene order is consistent with the order in Figure 1. The probes are arranged in order of descending gene expression ratio for PWS IC deletion mice relative to control littermates.
The ratio is based on the mean gene expression value of the PWS IC deletion mice relative to the control littermates.
TABLE III.
Quantitative RT-PCR Mean CT (+/−SD) Values and Fold Changea of Selected Probes From the Gene Expression Microarrays Comparing Gene Expression in Brain of Normal Littermates to PWS IC Deletion Mice
| Group | Pomc* | Mc5r** | AK013560 | BB314814 | BB182944 |
|---|---|---|---|---|---|
| Control littermates | 21.4 (0.5) | 24.7 (0.3) | 22.5 (0.9) | 38.2 (0.7) | 32.5 (0.7) |
| PWS IC deletion | 18.1 (0.6), 9.9-fold change | 23.6 (0.5), 2.1-fold change | nd | nd | nd |
Significantly different by t-test.
nd, not detected.
The CT was set at the narrowest portion of the logarithmic phase of amplification in the quantitative RT-PCR reaction. Fold change = 2 |Control CT − PWS IC CT| (e.g., 2|21.4−18.1| = 23.3 = 9.9-fold change).
P=0.002.
P=0.019.
Genes on Chromosome 18B3
A small cluster of genes on chromosome 18B3 was reported to be upregulated by haploinsufficiency of the 7B region in the TgPWS mice [Stefan et al., 2005]. We examined all genes in this region on microarrays in our PWS IC deletion mice and could find no evidence of changed expression, even without correction for multiple sampling. However, this is perhaps an expected result since the lesion of the IC in our PWS IC deletion mice is significantly smaller than that in the TgPWS mice analyzed by Stefan et al. [2005]; therefore, providing support to their conclusion that the increased expression was the result of haploinsufficiency of a biallelically expressed gene(s). In addition, our mice were a different strain which may account for expression differences observed in our experiments.
DISCUSSION
Prader–Willi syndrome originates with the loss of expression of paternally expressed genes in the PWSCR, which directly or indirectly causes the dysregulation of genes that control neurodevelop-ment, metabolism, and behavior. While the underlying expression changes in the PWSCR have been delineated, little is known about the global effects on gene expression that result from disruption of this region. Whole genome expression analysis together with validation by other approaches such as quantitative RT-PCR may begin to identify the downstream transcriptional effects of the PWS chromosome 15 disruption.
A previous microarray analysis of a mouse model found no significant changes in gene expression between TgPWS mice with a large paternally inherited deletion encompassing the PWSCR and normal littermates [Stefan et al., 2005]. We identified 61 genes (excluding paternally expressed genes within the PWSCR) which are significantly differentially expressed in the PWS IC deletion mice and control littermates. Our microarrays contained more transcripts than those used by Stefan et al. [2005], >45,000 compared to their ~12,000, which together with strain differences may account for differences in the two studies. It is also possible that the gene expression in PWS IC deletion mice is more dramatically altered in response to this lesion compared to the TgPWS mice used by Stefan et al. [2005].
We identified three transcripts with little or no expression in the brains of the PWS IC deletion mice which are localized to chromosome 7B, the PWSCR. These three transcripts BB314814, AK013560, and BB182944 are located at nucleotide positions 55,723,161, 56,944,766, and 57,277,109 on chromosome 7B, respectively, according to the UCSC genome browser (http://genome.ucsc.edu/). Snrpn is located at 55,730,763 and Ndn is located at 58,098,171. Although the region is conserved between human and mouse, there does not appear to be a sequence of high similarity to any of these three sequences within the human EST database. Thus, these three transcripts may represent the product of retrotransposition into the mouse PWSCR as described for Peg12(Frat3) which is conserved in both the mouse and human genomes [Chai et al., 2001].
In addition, we identified two genes with increased expression in the PWS IC deletion mice which are of particular interest, Pomc and Mc5r, both of which were validated by QRT-PCR along with Snrpn, Ndn, and the three imprinted transcripts (Table III). Products of the Pomc gene are known to be anorexigenic. Upregulation of this gene was reported in the brains of PWS deletion mice [Ge et al., 2002] and our data are in agreement with this observation. Ge et al. [2002] speculated that upregu-lation of Pomc might contribute to failure of the deletion mice to survive. Our observation that Pomc is upregulated in the PWS IC deletion mice further support that Pomc overexpression might contribute to the inability of the PWS model mice to survive. Mc5r is known to directly respond to upregulation of Pomc [Mountjoy et al., 1999]. Mc5r is known to be involved with thermoregulation which is reportedly abnormal in PWS infants [Williams et al., 1994; Ince et al., 2005]. Mc5r is required for stress-regulated synthesis of porphyrins by the Harderian gland and ACTH/MSH-regulated protein secretion by the lacri-mal gland [Chen et al., 1997]. These observations support a role for Pomc and its derived peptides in the early clinical findings of failure to thrive (Stage I) observed in PWS. Additional microarray gene expression studies with PWS IC deletion mice from different strains are needed to confirm these findings, as well as to identify other genes that may be impacted by this abnormality. Other strains of mice with a paternally derived PWS IC deletion are known to be viable [Chamberlain et al., 2004]. These mice should prove particularly interesting for comparison with the mice in our study to allow examination of older mice for observation of expression of Pomc in Stage I and possibly during the hyperphagic Stage II and compare with Pomc expression in human subjects with PWS in Stage I and Stage II. These ongoing studies will identify additional genes and interacting networks that contribute to the PWS clinical presentation and may point the way toward improved therapeutic interventions.
Acknowledgments
Grant sponsor: NIH; Grant number: NICHD RO141672; Grant sponsor: Hall Foundation of Kansas City; Grant sponsor: Kansas City Area Life Science Institute.
This research was supported by the NIH (NICHD RO141672), the Hall Foundation of Kansas City (MGB), and the Kansas City Area Life Science Institute (DCB). The authors thank Dr. Stormy Chamberlain for having sent the mice from the Brannan lab and Dr. Stephen Simon for help with the statistical analysis.
References
- Bittel DC, Butler MG. Prader-Willi syndrome: Clinical genetics, cytogenetics and molecular biology. Expert Rev Mol Med. 2005;7:1–20. doi: 10.1017/S1462399405009531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bittel DC, Kibiryeva N, Talebizadeh Z, Butler MG. Microarray analysis of gene/transcript expression in Prader-Willi syndrome: Deletion versus UPD. J Med Genet. 2003;40:568–574. doi: 10.1136/jmg.40.8.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bittel DC, Kibiryeva N, Talebizadeh Z, Driscoll DJ, Butler MG. Microarray analysis of gene/transcript expression in Angelman syndrome: Deletion versus UPD. Genomics. 2005;85:85–91. doi: 10.1016/j.ygeno.2004.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bittel DC, Kibiryeva N, Dasouki M, Knoll JH, Butler MG. A 9-year-old male with a duplication of chromosome 3p25.3p26.2: clinical report and gene expression analysis. Am J Med Genet Part A. 2006;140A:573–579. doi: 10.1002/ajmg.a.31132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boccaccio I, Glatt-Deeley H, Watrin F, Roeckel N, Lalande M, Muscatelli F. The human MAGEL2 gene and its mouse homologue are paternally expressed and mapped to the Prader-Willi region. Hum Mol Genet. 1999;8:2497–2505. doi: 10.1093/hmg/8.13.2497. [DOI] [PubMed] [Google Scholar]
- Brannan CI, Bartolomei MS. Mechanisms of genomic imprinting. Curr Opin Genet Dev. 1999;9:164–170. doi: 10.1016/S0959-437X(99)80025-2. [DOI] [PubMed] [Google Scholar]
- Butler MG, Thompson T. Prader-Willi syndrome: Clinical and genetic findings. The Endocrinol. 2000;10:35–165. doi: 10.1097/00019616-200010041-00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassidy SB. Prader-Willi syndrome. J Med Genet. 1997;34:917–923. doi: 10.1136/jmg.34.11.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassidy SB, Forsythe M, Heeger S, Nicholls RD, Schork N, Benn P, Schwartz S. Comparison of phenotype between patients with Prader-Willi syndrome due to deletion 15q and uniparental disomy 15. Am J Med Genet. 1997;68:433–440. [PubMed] [Google Scholar]
- Cattanach BM, Barr JA, Evans EP, Burtenshaw M, Beechey CV, Leff SE, Brannan CI, Copeland NG, Jenkins NA, Jones J. A candidate mouse model for Prader-Willi syndrome which shows an absence of Snrpn expression. Nat Genet. 1992;2:270–274. doi: 10.1038/ng1292-270. [DOI] [PubMed] [Google Scholar]
- Cavaille J, Buiting K, Kiefmann M, Lalande M, Brannan CI, Horsthemke B, Bachellerie JP, Brosius J, Huttenhofer A. Identification of brain-specific and imprinted small nucleolar RNA genes exhibiting an unusual genomic organization. Proc Natl Acad Sci USA. 2000;97:14311–14316. doi: 10.1073/pnas.250426397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai JH, Locke DP, Ohta T, Greally JM, Nicholls RD. Retrotransposed genes such as Frat3 in the mouse Chromosome 7C Prader-Willi syndrome region acquire the imprinted status of their insertion site. Mamm Genome. 2001;12:813–821. doi: 10.1007/s00335-001-2083-1. [DOI] [PubMed] [Google Scholar]
- Chamberlain SJ, Johnstone KA, DuBose AJ, Simon TA, Bartolomei MS, Resnick JL, Brannan CI. Evidence for genetic modifiers of postnatal lethality in PWS-IC deletion mice. Hum Mol Genet. 2004;13:2971–2977. doi: 10.1093/hmg/ddh314. [DOI] [PubMed] [Google Scholar]
- Chen W, Kelly MA, Opitz-Araya X, Thomas RE, Low MJ, Cone RD. Exocrine gland dysfunction in MC5-R-deficient mice: Evidence for coordinated regulation of exocrine gland function by melanocortin peptides. Cell. 1997;91:789–798. doi: 10.1016/s0092-8674(00)80467-5. [DOI] [PubMed] [Google Scholar]
- de los Santos T, Schweizer J, Rees CA, Francke U. Small evolutionarily conserved RNA, resembling C/D box small nucleolar RNA, is transcribed from PWCR1, a novel imprinted gene in the Prader-Willi deletion region, which is highly expressed in brain. Am J Hum Genet. 2000;67:1067–1082. doi: 10.1086/303106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel JM, Merchant M, Ohta T, Ji Y, Caldwell RG, Ramsey MJ, Tucker JD, Longnecker R, Nicholls RD. A transgene insertion creating a heritable chromosome deletion mouse model of Prader-Willi and angelman syndromes. Proc Natl Acad Sci USA. 1999;96:9258–9263. doi: 10.1073/pnas.96.16.9258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher RC, Pils B, Albalwi M, Francke U. Evidence for the role of PWCR1/HBII-85 C/D box small nucleolar RNAs in Prader-Willi syndrome. Am J Hum Genet. 2002;71:669–678. doi: 10.1086/342408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Y, Ohta T, Driscoll DJ, Nicholls RD, Kalra SP. Anorexigenic melanocortin signaling in the hypothalamus is augmented in association with failure-to-thrive in a transgenic mouse model for Prader-Willi syndrome. Brain Res. 2002;957:42–45. doi: 10.1016/s0006-8993(02)03583-7. [DOI] [PubMed] [Google Scholar]
- Gray TA, Saitoh S, Nicholls RD. An imprinted, mammalian bicistronic transcript encodes two independent proteins. Proc Natl Acad Sci USA. 1999a;96:5616–5621. doi: 10.1073/pnas.96.10.5616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray TA, Smithwick MJ, Schaldach MA, Martone DL, Graves JA, McCarrey JR, Nicholls RD. Concerted regulation and molecular evolution of the duplicated SNRPB’/B and SNRPN loci. Nucleic Acids Res. 1999b;27:4577–4584. doi: 10.1093/nar/27.23.4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanel ML, Wevrick R. The role of genomic imprinting in human developmental disorders: Lessons from Prader-Willi syndrome. Clin Genet. 2001;59:156–164. doi: 10.1034/j.1399-0004.2001.590303.x. [DOI] [PubMed] [Google Scholar]
- Ince E, Ciftci E, Tekin M, Kendirli T, Tutar E, Dalgic N, Oncel S, Dogru U. Characteristics of hyperthermia and its complications in patients with Prader-Willi syndrome. Pediatr Int. 2005;47:550–553. doi: 10.1111/j.1442-200x.2005.02124.x. [DOI] [PubMed] [Google Scholar]
- Lee S, Kozlov S, Hernandez L, Chamberlain SJ, Brannan CI, Stewart CL, Wevrick R. Expression and imprinting of MAGEL2 suggest a role in Prader-willi syndrome and the homologous murine imprinting phenotype. Hum Mol Genet. 2000;9:1813–1819. doi: 10.1093/hmg/9.12.1813. [DOI] [PubMed] [Google Scholar]
- Lee S, Walker CL, Karten B, Kuny SL, Tennese AA, O’Neill MA, Wevrick R. Essential role for the Prader-Willi syndrome protein necdin in axonal outgrowth. Hum Mol Genet. 2005;14:627–637. doi: 10.1093/hmg/ddi059. [DOI] [PubMed] [Google Scholar]
- Matsumoto K, Taniura H, Uetsuki T, Yoshikawa K. Necdin acts as a transcriptional repressor that interacts with multiple guanosine clusters. Gene. 2001;272:173–179. doi: 10.1016/s0378-1119(01)00544-3. [DOI] [PubMed] [Google Scholar]
- Meguro M, Kashiwagi A, Mitsuya K, Nakao M, Kondo I, Saitoh S, Oshimura M. A novel maternally expressed gene, ATP10C, encodes a putative aminophospholipid translocase associated with Angelman syndrome. Nat Genet. 2001a;28:19–20. doi: 10.1038/ng0501-19. [DOI] [PubMed] [Google Scholar]
- Meguro M, Mitsuya K, Nomura N, Kohda M, Kashiwagi A, Nishigaki R, Yoshioka H, Nakao M, Oishi M, Oshimura M. Large-scale evaluation of imprinting status in the Prader-Willi syndrome region: An imprinted direct repeat cluster resembling small nucleolar RNA genes. Hum Mol Genet. 2001b;10:383–394. doi: 10.1093/hmg/10.4.383. [DOI] [PubMed] [Google Scholar]
- Mountjoy KG, Willard DH, Wilkison WO. Agouti antagonism of melanocortin-4 receptor: Greater effect with desacetyl-alpha-melanocyte-stimulating hormone (MSH) than with alpha-MSH. Endocrinology. 1999;140:2167–2172. doi: 10.1210/endo.140.5.6748. [DOI] [PubMed] [Google Scholar]
- Nicholls RD. Incriminating gene suspects, Prader-Willi style. Nat Genet. 1999;23:132–134. doi: 10.1038/13758. [DOI] [PubMed] [Google Scholar]
- Nicholls RD, Knepper JL. Genome organization, function, and imprinting in Prader-Willi and Angelman syndromes. Annu Rev Genomics Hum Genet. 2001;2:153–175. doi: 10.1146/annurev.genom.2.1.153. [DOI] [PubMed] [Google Scholar]
- Ren J, Lee S, Pagliardini S, Gerard M, Stewart CL, Greer JJ, Wevrick R. Absence of Ndn, encoding the Prader-Willi syndrome-deleted gene necdin, results in congenital deficiency of central respiratory drive in neonatal mice. J Neurosci. 2003;23:1569–1573. doi: 10.1523/JNEUROSCI.23-05-01569.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runte M, Huttenhofer A, Gross S, Kiefmann M, Horsthemke B, Buiting K. The IC-SNURF-SNRPN transcript serves as a host for multiple small nucleolar RNA species and as an antisense RNA for UBE3A. Hum Mol Genet. 2001;10:2687–2700. doi: 10.1093/hmg/10.23.2687. [DOI] [PubMed] [Google Scholar]
- Stefan M, Portis T, Longnecker R, Nicholls RD. A nonimprinted Prader-Willi syndrome (PWS)-region gene regulates a different chromosomal domain in trans but the imprinted PWS loci do not alter genome-wide mRNA levels. Genomics. 2005;85:630–640. doi: 10.1016/j.ygeno.2005.02.004. [DOI] [PubMed] [Google Scholar]
- Talebizadeh Z, Butler MG. Insulin resistance and obesity-related factors in Prader-Willi syndrome: Comparison with obese subjects. Clin Genet. 2005;67:230–239. doi: 10.1111/j.1399-0004.2004.00392.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White RA, McNulty SG, Nsumu NN, Boydston LA, Brewer BP, Shimizu K. Positional cloning of the Ttc7 gene required for normal iron homeostasis and mutated in hea and fsn anemia mice. Genomics. 2005;85:330–337. doi: 10.1016/j.ygeno.2004.11.008. [DOI] [PubMed] [Google Scholar]
- Williams MS, Rooney BL, Williams J, Josephson K, Pauli R. Investigation of thermoregulatory characteristics in patients with Prader-Willi syndrome. Am J Med Genet. 1994;49:302–307. doi: 10.1002/ajmg.1320490312. [DOI] [PubMed] [Google Scholar]
- Yang T, Adamson TE, Resnick JL, Leff S, Wevrick R, Francke U, Jenkins NA, Copeland NG, Brannan CI. A mouse model for Prader-Willi syndrome imprinting-centre mutations. Nat Genet. 1998;19:25–31. doi: 10.1038/ng0598-25. [DOI] [PubMed] [Google Scholar]