Abstract
Purpose
Fragile X syndrome, the most common inherited form of human mental retardation, arises as a consequence of a large expansion of a CGG trinucleotide repeat in 5′ untranslated region of the fragile X mental retardation 1 (FMR1) gene located on the X chromosome. Although the FMR1 gene was cloned 15 years ago, the mechanisms that cause fragile X syndrome remain to be elucidated. Multiple studies have identified proteins that potentially interact with FMRP, the product of FMR1, and differentially expressed genes in an Fmr1 knockout mouse. To assess the impact of fragile X syndrome on gene expression in humans and to attempt to identify disturbed genes and gene interactive pathways relevant to fragile X syndrome, we performed gene expression microarray analysis using RNA isolated from lymphoblastoid cells derived from males with fragile X syndrome with and similarly aged control males.
Methods
We used whole genome microarrays consisting of 57,000 probes to analyze global changes to the transcriptome in readily available lymphoblastoid cell lines derived from males with fragile X syndrome and healthy comparison males with normal intelligence. We verified the differential expression of several of these genes with known biological function relevant to fragile X syndrome using quantitative reverse transcription polymerase chain reaction using RNA from lymphoblastoid cells from fragile X syndrome and control males as well as RNA from human brain tissue (frontal cortex) of other affected fragile X syndrome males.
Results
We identified more than 90 genes that had significant differences in probe intensity of at least 1.5-fold with a false discovery rate of 5% in cells from males with fragile X syndrome relative to comparison males. The list of 90 differentially expressed genes contained an overrepresentation of genes involved in signaling (e.g., UNC13B [−3.3-fold change in expression in lymphoblasts by quantitative reverse transcription polymerase chain reaction), GABRD [+2.0-fold change] EEF1A2 [+4.3-fold change]), morphogenesis (e.g., MAP1B [−7.5-fold change], ACCN1 [−8.0-fold change]), and neurodevelopment and function (e.g., PPP1R9B [+3.5-fold change], HES1 [+2.8-fold change]).
Conclusions
These genes may represent members of candidate networks disturbed by the loss of FMR1 and consequently fragile X mental retardation protein function, thus lending support for altered fragile X mental retardation protein function resulting in an abnormal transcriptome. Further analyses of the genes, especially those that have been identified in multiple studies, are warranted to develop a more integrated description of the alterations in gene processing that lead to fragile X syndrome.
Keywords: microarray gene expression, quantitative reverse transcription polymerase chain reaction, lymphoblasts, frontal cortex, neurodevelopment, FMR1, gene fold change, pathway analysis
Fragile X syndrome (FXS) is the most common inherited form of human mental retardation, with an estimated prevalence of 1 in 4000 boys and 1 in 8000 girls.1 FXS is caused by a large expansion of the CGG trinucleotide repeat in the 5′ untranslated region of the fragile X mental retardation 1 (FMR1) gene located on the X chromosome. This triplet mutation results in the expansion of the CGG repeat sequence to >200 copies accompanied by hypermethylation of the repeated trinucleotide and a CpG island in the promoter.2 Individuals with FXS may display moderate to severe mental retardation with IQs between 20 and 60. Other clinical features include minor dysmorphic features, developmental delay during childhood, connective tissue dysplasia, and attention deficit hyperactivity disorder. Characteristic features during adolescence include an elongated face, prominent jaw, large ears, macroorchidism, and a range of behavioral anomalies.3
FMR1 was cloned in 19914 and is abundantly expressed during early embryonic development in multiple tissues including the brain and testes.5 The product of the FMR1 gene, fragile X mental retardation protein (FMRP), is an RNA binding protein that is a component of messenger ribonucleoproteins (mRNPs) that regulate translation and possibly RNA stability, which could have an impact on cellular mRNA levels. Inaccurate processing of mRNA of several genes may result in the clinical manifestation of FXS through defective regulatory and structural proteins, whereby disturbed protein networks may affect genome-wide transcription. If disturbed expression of genes is observed, representing members of candidate networks disrupted by the loss of FMR1 and consequently FMRP function in FXS subjects, then this evidence would support altered FMRP function resulting in an abnormal transcrip-tome. Some of the targets of the mRNP of which FMRP is a component have been elucidated, but a clear picture of the disruption of the transcriptome and proteome that produces FXS remains to be elucidated.
The development of an Fmr1 knockout mouse has facilitated the understanding of FXS neurobiology.6 The mice exhibit macroorchidism and subtle learning and memory deficits, which are reminiscent of the clinical phenotype. The Fmr1 knockout mice have elongated and thin dendritic spines analogous to profound abnormalities found in the brains of humans with FXS.7,8 Study of the development of the somatosensory cortical region in Fmr1 knockout mice indicate that normal dendritic regression is impaired, suggesting that FMRP may be required for the normal processes of maturation and elimination to occur in cerebral cortical development.9 The mice have been used to study FMRP interactions and search for target gene products. Studies of the knockout mice have identified candidate genes that may contribute to FXS.10,11
FMRP is involved in regulating translation including suppression of translation.12 Messenger RNAs encoding proteins involved in synaptic or developmental neurobiology that harbor FMRP-binding elements have been identified.13 It is hypothesized that FMRP may be involved in synaptic plasticity through regulation of mRNA transport and local protein synthesis at synapses.14,15 It also has been suggested that FMRP is involved in synaptic development and plasticity by regulating mRNA transportation and/or translation, thus regulating dendritic protein synthesis.12 Additionally, it appears that FMRP may be linked to micro-RNAs,15 adding another layer of complexity to the role that FMRP plays in regulating RNA transport and translation. A better understanding of the disruption of FMR1 gene expression and its impact on genome-wide expression is needed and becomes the central focus of our study.
A study of synaptic plasticity in the hippocampus of the Fmr1 knockout mouse suggested that the absence of FMRP leads to exaggerated metabotropic glutamate receptor (mGluR) activation of protein synthesis leading to altered synaptic development.16 A relatively large number of transcripts were shown to coprecipitate with FMRP in fractions isolated from mouse brain that represent candidate genes that may contribute to the FXS phenotype.10 The loss of FMRP on the expression of downstream genes is beginning to be elucidated primarily by using the knockout mouse. For example, a GABAA receptor subunit (Gabrd), a ρ guanine exchange factor and a regulator of G-protein signaling (Rgs4) are reportedly under-expressed in Fmr1 knockout mice relative to controls.17,18
Recently, lymphoblastoid cells from monozygotic twins with differing severity of autism and/or language impairment were used to identify novel candidate genes that may contribute to autism.19 These authors noted that mechanistic studies are more appropriately done in brain tissue, but access to sample tissue is limited, and, more importantly, brain tissue samples are inappropriate for diagnostic analysis. They identified candidate genes with known neurological functions and concluded that lymphoblastoid cells may exhibit biomarkers relevant to autism. To assess the impact of FXS on gene expression in humans and to attempt to identify disturbed genes and gene interactive pathways relevant to FXS, we performed genome wide microarrays using RNA isolated from available actively growing lymphoblastoid cells derived from males with FXS and similarly aged control males. These preliminary gene expression data representing the entire human transcriptome based on a small number of typical FXS males should stimulate additional and more extensive studies.
MATERIALS AND METHODS
Subjects
The study for FXS screening was approved by the local institutional review board (IRB) and consent procedures followed accordingly. The IRB identification number was 481. All males with FXS were diagnosed by Southern blot analysis from genomic DNA isolated from peripheral blood and found to have full mutations (i.e., CGG repeat number >200) without evidence of mosaicism. Lymphoblastoid cell lines were established from each subject with FXS as well as comparison subjects by transformation of lymphocytes with Epstein-Barr virus following standard protocols. Briefly, mononuclear cells were separated from red blood cells by layering on a bed of Ficoll and centrifuging for 45 minutes at 1000 rpm in a clinical centrifuge. The buffy coat was removed and washed twice with RPMI media. Approximately 3 × 106 cells were suspended in 0.5 mL of media supplemented with 20% serum, penicillin, and streptomycin and then placed in a single well of a 24-well culture plate with 0.5 mL of Epstein-Barr virus solution added along with 2 mg of cyclosporine. The cells were grown in an incubator maintained at 5% CO2 with 80% humidity until stably transformed. DNA isolated from the lymphoblastoid cell lines from each affected subject was also analyzed by Southern blot and polymerase chain reaction (PCR), and each cell line had a repeat number in the full mutation range consistent with the DNA findings from peripheral blood. Microarray analysis was performed on RNA isolated from actively growing lymphoblastoid cell lines established from each subject.
Four males with FXS (ages 16, 20, 28, and 38 years) and full mutations were similarly aged matched with four comparison males having normal intelligence (ages 15, 25, 28, and 30 years) and a pairwise analysis was performed based on age. In addition, frontal cortex brain samples were obtained from the National Institutes of Health (NIH)–funded Brain and Tissue Bank (BTB), Baltimore, MD. The brain samples were from 21-and 22-year-old males diagnosed with FXS (BTB 4751 and 1204, respectively) and from two 22-year-old control males (BTB 545 and 1442).
RNA isolation
Lymphoblastoid cells were chosen as a readily available tissue source to examine from the subject population. Lymphoblastoid cultures were grown in RPMI 1640 medium (JRH Biosciences, Lenexa, KS) supplemented with 2.05 mM of L-glutamine, 10% fetal calf serum and 1% Pen/Strep. Before RNA extraction, 2 mL of media containing 106 cells/mL were seeded into 10 mL of media in a T25 flask, and the cells were allowed to grow for 48 hours. Cells were pelleted by gentle centrifugation and media removed. The cells were resuspended in Trizol reagent (Invitrogen, Carlsbad, CA), and total RNA was extracted following the manufacturer’s instructions.
Microarrays
Codelink human whole genome microarrays (Amersham Biosciences, GE Health Care, Piscataway, NJ) consisting of 57,000 gene and transcript probes were used to compare gene expression from actively growing lymphoblastoid cells derived from our study subjects. Target preparation, hybridization, and initial data collection were done according to the manufacturer’s directions (detailed protocols are available at the GE Health Care Web site). Briefly, total RNA (1 μg) and control spike RNAs, together with an oligo dT primer, were incubated with first strand reaction kit components at 42°C for 2 hours to synthesize the first cDNA strand. The second strand reaction components were then added, followed by an additional 2-hour incubation at 16°C, which allowed for incorporation of a T7 promoter element in the resulting cDNA. The cDNA was then concentrated, purified, and combined with in vitro transcription reaction components including T7 polymerase and biotinylated UTP to produce cRNA. After incubating 14 hours at 37°C, the biotinylated cRNA was purified and chemically fragmented. The cRNA was combined with hybridization buffer components, heated to 90°C for 5 minutes and immediately chilled to 4°C. The hybridization reaction was injected into a hybridization chamber containing a single Codelink whole human microarray and incubated with shaking (300 rpm) for 18 hours at 37°C. The microarrays were then placed in a rack and simultaneously washed for 1 hour at 46°C. The microarrays were incubated with streptavidin-Cy5 dye conjugate in detection buffer at ambient temperature for 30 minutes. The microarrays were washed four times in wash buffer with a final rinse in 0.1 SSC/0.05% Tween and dried by centrifugation. The slides were scanned in a GenePix 4100A microarray scanner (Molecular Devices Corp., Sunnyvale CA).
Analysis and statistics
GeneSpring software (Silicon Genetics, Agilent Technologies, Palo Alto, CA) was used for microarray data characterization and analysis as previously described.20 The microarray data were initially filtered by setting all values at <0.01 to 0.01 to prevent spurious values <0. All samples were normalized to the median value of the control samples such that each measurement for each gene in each microarray was divided by the median of that gene’s measurement in the corresponding control sample. Each measurement was then divided by the 50th percentile of all measurements in that sample to reduce chip-to-chip variation and to improve the normality of the distribution. The signal quality was used to filter the data requiring a present or marginal signal in at least three of the four members of either group (FXS or comparison male) to meet inclusion criteria for further analysis. In addition, genes were included for further analysis only if the mean change in signal intensity between subject groups was 1.5-fold or greater (i.e., log2 [ratio] greater than ±0.58). Furthermore, in the FXS group, all values for the gene had to be in the same direction (up or down) relative to the control group. This differential signal intensity cutoff was selected because reports in the literature suggest that 1.5-fold increases or decreases in gene signal intensity are generally reproducible when Lowess normalization is used19,21,22 and we are able to confirm expression changes of at least 1.5-fold by quantitative RT-PCR. Differences between normalized mean gene expression levels were evaluated using a Welch t test with Bonferroni correction without assuming equal variances and a false discovery rate (fdr) of ≤20%, as undertaken in other studies.22
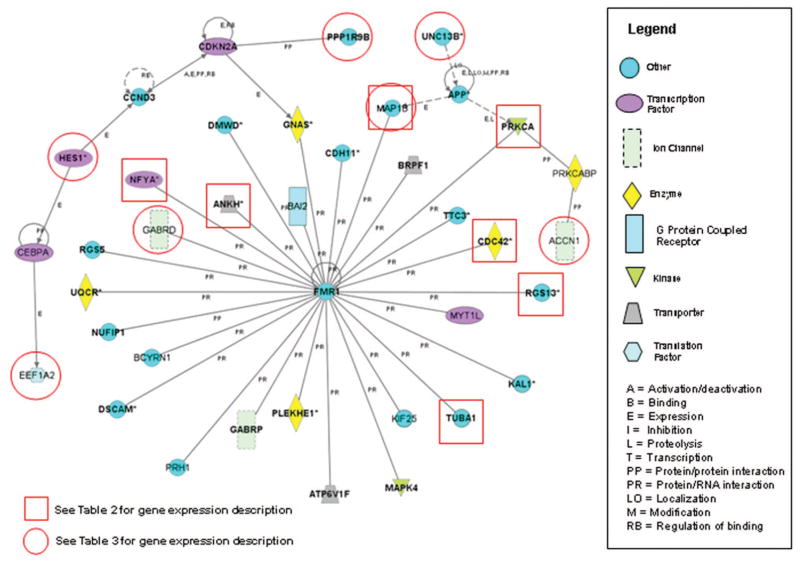
We used the Ingenuity Pathways Analysis software (Ingenuity Systems, Mountain View, CA, http://www.ingenuity.com/index.html) as previously described in similar studies19,20 to examine the gene network interactions of the FMR1 gene and derived a list of 121 genes known to directly interact with FMR1 or FMRP for small group analysis (Fig. 1). Ingenuity Pathways Analysis accesses a highly curated database allowing users to identify interacting components of gene networks, providing access to the primary literature supporting specific interactions.
Fig. 1.
Network of a subset of genes or gene products known to interact with FMR1. Symbol shape represents functional categories for each gene product. Lines between gene symbols indicate interactions. The type of interaction is indicated with a letter (e.g., B = binding) as noted in the legend inset. Genes with red squares directly interact with FMRP and had a significant change in expression of at least 1.5-fold with a false discovery rate of 5% (Table 2). Genes with red circles were analyzed by quantitative RT-PCR (Table 3). These data were generated using Ingenuity Pathways Analysis, Ingenuity Systems, Mountain View, CA, a Web-delivered application that enables the discovery, visualization, and exploration of potentially relevant gene interaction networks.
Quantitative RT-PCR
Quantitative RT-PCR was performed using primers specific to a subset of genes/transcripts identified as significantly differentially expressed by microarray analysis. Quantitative RT-PCR was performed using a QuantiTect SYBR Green RT-PCR kit (Qiagen, Valencia, CA) according to the manufacturer’s directions. Total RNA was isolated from cell lines and brain tissue (frontal cortex) using Trizol reagent (Invitrogen, Inc.) and quantified by spectroscopy. An equal quantity of total RNA (500 ng) from each subject, together with gene-specific primers were added to a reaction mix containing all components necessary for RT and PCR. The reaction was carried out in an ABI 7000 system (Applied Biosystems, Foster City, CA) beginning with a 30-minute step at 50°C to allow for RT, followed by 15 minutes at 95°C. The PCR followed for 45 cycles during which the intensity of the SYBR Green fluorescence was measured at the extension step of each PCR cycle. The point at which the intensity level crossed the PCR cycle threshold (CT) was used to compare individual reactions. At least five replicates were performed on each sample for each gene. A dissociation curve was generated for all reactions, and reactions were run on agarose gels to verify the presence of a single band. Amplification of GAPD served as a control for each sample. Normalization of gene expression from each subject (GeneSubj) was performed by dividing the mean GAPD gene expression (CT) value from each subject (GAPDSubj) by the mean GAPD gene expression (CT) value of one of the comparison males (GAPDRef) to produce a correction value. Each mean CT value for the other genes was multiplied by the correction value to produce the normalized value (GeneSubj[GAPDSubj/GAPDRef]). The normalized CT values were averaged to produce the mean CT value for each gene analyzed.
RESULTS
We used whole human genome microarrays containing 57,000 probes to analyze gene expression in actively growing lymphoblasts derived from males with FXS and similarly aged comparison males. Not surprisingly, microarray analysis showed a significant reduction of FMR1 expression in cells from FXS subjects relative to cells from comparison males (reduced to 25% of normal comparison subjects). Results from quantitative RT-PCR with primers specific to FMR1 showed a 39-fold reduction in signal intensity in the cells from FXS males relative to comparison males. However, FMR1 expression was readily detectable by quantitative RT-PCR in brain (frontal cortex) from two unaffected comparison males obtained from the Brain and Tissue Bank, but, as expected, we could not detect FMR1 expression by quantitative RT-PCR in the frontal cortex of two males with FXS.
The signal intensity of 26,031 probes met our inclusion criteria (detectable signal, see “Materials and Methods”), which included 46% of the transcripts. We further analyzed the 26,031 probes and 3,841 had a change in signal intensity (i.e., differential expression) of at least 1.5-fold (i.e., 1910 increased by 1.5 times or greater and 1931 reduced to 0.7 times or less [which represents approximately a 1.5-time reduction]) in the subjects with FXS relative to comparison males. We analyzed the group of probes with differential expression of 1.5-fold using a small group analysis with a Bonferroni correction factor of 3841. Using an fdr of 20%, 168 genes were significantly different in expression between the FXS subjects and controls (data not shown). When the fdr was set at 5%, 43 genes had significant differences in expression (Table 1). The Ingenuity Pathways Analysis software was used to determine the network interactions of FMR1 with other genes (Fig. 1) and to assign this group of genes to functional categories.
Table 1.
Forty-three genes and transcripts found in lymphoblasts with a significant change in expression of at least 1.5-fold in FXS males relative to control males with an fdr of 5% not identified in previously reported studies on FXS
| Gene | Genbank | Fold | Chromosome | Description |
|---|---|---|---|---|
| PCNX | NM_014982 | 3.8 | 14q24.1 | Pecanex homolog |
| LYPLA3 | NM_012320 | 3.5 | 16q22.1 | Lysophospholipase 3 |
| NUTa | BX117628 | 3.2 | 15q13.2 | Nuclear protein in testis |
| AL833173 | 2.8 | 3 | Transcribed sequence | |
| MAP3K5 | NM_005923 | 2.5 | 6q22.33 | Mitogen-activated protein kinase kinase kinase 5 |
| ALEX1 | NM_016608 | 2.5 | Xq21.33-q22.2 | ALEX1 protein |
| AK023131 | 2.4 | 1 | cDNA FLJ13069 | |
| GJA4 | AL047476 | 2.4 | 1p35.1 | Gap junction protein, α4 (connexin 37) |
| PML | NM_033238 | 2.2 | 15q22 | Promyelocytic leukemia |
| BM509961 | 2.2 | 15q24.1 | Transcribed sequence | |
| BX089918 | 2.0 | 18 | Transcribed sequence | |
| AK094644 | 2.0 | 9 | cDNA FLJ37325 | |
| C6orf4 | NM_147200 | 1.9 | 6q21 | Chromosome 6 open reading frame 4 |
| AI760389 | 1.9 | 2 | Transcribed sequence | |
| CTSF | NM_003793 | 1.9 | 11q13 | Cathepsin F |
| MGC44287 | NM_182607 | 1.9 | Xq22.3 | Hypothetical protein MGC44287 |
| CCNA1 | NM_003914 | 1.9 | 13q12.3-q13 | Cyclin A1 |
| AI139765 | 1.9 | 11q13.1 | Transcribed sequence | |
| AMT | NM_000481 | 1.8 | 3p21.2-p21.1 | Aminomethyltransferase |
| T79869 | 1.8 | 1p13.1 | Transcribed sequence | |
| TRIM5 | NM_033034 | 1.8 | 11p15 | Tripartite motif-containing 5 |
| POU2F1 | NM_002697 | 1.8 | 1q22-q23 | POU domain, class 2, transcription factor 1 |
| MRPL54 | NM_172251 | 1.8 | 19p13.3 | Mitochondrial ribosomal protein L54 |
| BC042591 | 1.8 | 12 | Transcribed sequence | |
| BX109444 | 1.8 | 6 | Transcribed sequence | |
| IL4R | NM_000418 | 1.8 | 16p11.2-12.1 | Interleukin-4 receptor |
| AA151945 | 1.7 | 1p36.11 | Transcribed sequence | |
| MGC4170 | NM_024312 | 1.7 | 12q23.3 | MGC4170 protein |
| BM726893 | 1.7 | 9 | Transcribed sequence | |
| EMP3 | NM_001425 | 1.7 | 19q13.3 | Epithelial membrane protein 3 |
| AQP10 | NM_080429 | 1.6 | 1q22 | Aquaporin 10 |
| BC036409 | 1.6 | 7 | Transcribed sequence | |
| AW440157 | 1.5 | 7 | Transcribed sequence | |
| SLB | BM823579 | 0.7 | 2p23.3 | Selective LIM binding factor |
| UBE2J1 | NM_016021 | 0.7 | 6q16.1 | Ubiquitin-conjugating enzyme E2 |
| FLJ32356 | NM_144671 | 0.6 | 12q24.13 | Hypothetical protein FLJ32356 |
| UBQLN1a | NM_013438 | 0.6 | 9q22 | Ubiquilin 1 |
| AI961094 | 0.6 | 6p22.1 | Transcribed sequence | |
| CTNNA1 | AK022326 | 0.6 | 5q31 | Catenin (cadherin-associated protein), α1 |
| TRF4-2 | BP428534 | 0.6 | 16q12.1 | Topoisomerase-related function protein 4-2 |
| AA897664 | 0.5 | 5 | Transcribed sequence | |
| CA307826 | 0.2 | 8 | Transcribed sequence | |
| AI080026 | 0.1 | 9 | CDNA clone IMAGE:4800042 |
There were 3841 probes with a differential expression of 1.5 times, of which 168 were significant with an fdr of 20%, and 43 were significant with an fdr of 5%.
Validation by quantitative RT-PCR.
Previous reports have identified candidate genes by coimmunoprecipitation of mRNA with FMRP in the mouse brain and by identifying genes with an abnormal polyribosomal profile in FXS cells.10 Additionally, microarray analysis comparing gene expression in the brain of Fmr1 knockout mice with that of normal mice identified genes differentially expressed by at least twofold.11 We examined the three sets of genes determined by coimmunoprecipitation, polyribosomal profile or mouse brain microarray by small group analysis using our microarray data to test for consensus. Brown et al.10 listed 432 mouse genes that immunoprecipitated with FMRP, of which our human microarrays contained 362 probes. We analyzed these 362 genes using a conservative small group analysis and 25 of them were differentially expressed at least 1.5-fold and statistically significant with an fdr of 5% in FXS lymphoblastoid cells relative to comparison cells. In addition, our microarrays contained 181 of the 282 genes identified by Brown et al.10 in the polyribosomal fraction. We could detect 139 of these genes on our microarray, and 15 were differentially expressed by at least 1.5-fold and statistically significant with an fdr of 5%. Our arrays also contained 24 of the 73 genes identified by D’Agata et al.11 as differentially expressed in the FXS mouse brains. We could detect 19 of these genes on our microarrays, and seven were significantly different in expression in the FXS cells with an fdr of 20%, and two were significant with an fdr of 5%. We then created a list of 121 genes that are known to directly interact with FMR1 using Ingenuity Pathways Analysis software. We could detect 74 of these genes, and seven of them had a significant difference in expression of at least 1.5-fold with an fdr of 5% (Table 2).
Table 2.
Significant change in expression (highest to lowest) of greater than 1.5 fold with a false discovery rate of 5% for seven genes which were identified using Ingenuity Pathways Analysis software as directly interacting with FMRP (there is evidence of 120 genes directly interacting with FMRP)
| Gene | Fold change | P | Genbank | Chromosome |
|---|---|---|---|---|
| RGS13 | 5.8 | 0.01 | NM_002927 | 1q31.1 |
| ANKH | 1.6 | 0.00 | NM_054027 | 5p15.1 |
| NFYA | 0.5 | 0.00 | NM_002505 | 6p21.3 |
| CDC42 | 0.4 | 0.01 | D53996 | 1p36.1 |
| PRKCA | 0.2 | 0.01 | AW888701 | 17q22-q23.2 |
| TUBA1 | 0.2 | 0.00 | AA551692 | 2q36.1 |
| MAP1Ba | 0.1 | 0.01 | NM_005909 | 5q13 |
Validated by quantitative RT-PCR.
We pooled the genes identified as having a significant change in expression in the FXS cells from the analysis of the detectable genes with 1.5 times differential expression and the genes from the three previously published lists with differential expression in our arrays (315 gene entries, 168 from our global analysis with an fdr of 20% and 147 from the published lists10,11 with an fdr of 20%) and analyzed them with the tools available at the Gene Ontology statistics site (GOstat site; http://gostat.wehi.edu.au,23). The GOstat site is a public-access Web site providing statistical tools to annotate and analyze the function of large numbers of genes of interest generated by analyses such as microarray experiments. The program provides functional annotation or Gene-Ontology groups, which are highly represented in the data. There were 27 gene ontologies with significant overrepresentation (P < 0.05) in our list of differentially expressed genes. These included mainly ontologies associated with regulation of the cell cycle and physiological processes, but one ontology categorized as protein binding contained 29 genes.
We selected nine differentially expressed genes with a change of at least 1.5-fold with an fdr of 5% from a total of approximately 90 genes to validate by quantitative RT-PCR. These included nuclear protein in the testis (NUT); ubiquilin 1 (UBQLN1); protein phosphatase 1, regulatory subunit 9B (PPP1R9B); eukaryotic translation elongation factor 1 α2 (EEF1A2); GABAA receptor δ (GABRD); amiloride-sensitive cation channel 1, neuronal (ACCN1); microtubule-associated protein 1B (MAP1B); unc-13 homolog B homolog (Caenorhabditis) (UNC13B); and hairy and enhancer of split 1 homolog (Drosophila) (HES1). They were differentially expressed in our microarray analysis using RNA isolated from lymphoblastoid cell lines established from our subjects as well as from the frontal cortex of males with and without FXS obtained from the NIH-funded BTB (University of Maryland, Baltimore, MD, and University of Miami, Miami, FL). These genes were chosen because of their relevance to the FXS phenotype such as neurodevelopment and function (e.g., PPP1R9B, HES1), signaling (e.g., UNC13B, GABRD, EEF1A2) and morphogenesis (e.g., MAP1B, ACCN1), as well as previous reports of an association with FXS and/or large expression differences between our males with FXS and comparison males (e.g., NUT, UNC13B, MAP1B, ACCN1, GABRD, EEF1A2, UBQLN1). Additionally, several of them are known to directly interact with FMR1 (e.g., GABRD24 and MAP1B25). Changes in expression of these genes in lymphoblastoid cells and frontal cortex in our males with FXS relative to the control males were in agreement with the microarrays in direction, although not necessarily in magnitude (Table 3). However, GABRD did not produce a signal sufficient for detection on our microarray, although it was reported recently to be underexpressed in the brain of a mouse model for FXS.17 Thus, we used quantitative RT-PCR to investigate its expression in our males with FXS. GABRD was increased in our subjects with FXS relative to controls in both lymphoblastoid cells and frontal cortex using quantitative RT-PCR in contrast to a previous report in mouse brain studies.17
Table 3.
Confirmation of selected genes with differential RNA levels identified by microarray analysis using quantitative RT-PCR
| Gene | Lymphoblastoid cells (4 FXS subjects and 4 control subjects) | Frontal cortex (2 FXS subjects and 2 control subjects) | |||||||
|---|---|---|---|---|---|---|---|---|---|
|
|
|
||||||||
| Reason for selection | Interaction with FMR1 | Array fold changea | QRT-PCR fold changeb | P | Control mean | FXS mean | QRT-PCR fold changeb | P | |
| FMR1 | −4.0 | −39 | 0.00 | 25.2 | Not detected | ||||
| ACCN1 | Role in neurotransmission | Secondaryc | −50 | −8.0 | 0.00 | 27.2 | 30.8 | −12.1 | 0.01 |
| MAP1B | Role in neurodevelopment and function | Protein:RNA (see Ref. 25) | −10 | −7.5 | 0.00 | 14.3 | 16.6 | −4.9 | 0.07 |
| UNC13B | Promotes priming of synaptic vesicles | Secondaryc | −10 | −3.3 | 0.00 | 16.2 | 17.7 | −2.8 | 0.03 |
| UBQLN1 | Binds to multiple GABA receptor subunits | Secondaryc | −1.7 | −2.0 | 0.06 | 23.8 | 25.3 | −2.9 | 0.08 |
| GABRD | Underexpression in FXS mice | Protein:RNA (see Ref. 24) | Not detected | 2.0 | 0.02 | 29.6 | 27.1 | 5.7 | 0.07 |
| HES1 | Member of Notch pathway regulating CNS | Secondaryc | 3.4 | 2.8 | 0.00 | 21.2 | 20.3 | 1.9 | 0.03 |
| PPP1R9B | Enriched in dendritic spines receiving excitatory CNS signals | Secondaryc | 1.6 | 3.5 | 0.01 | 32.8 | 26.1 | 104.0 | 0.00 |
| EEF1A2 | Delivers aminoacyl tRNAs to the ribosome | Secondaryc | 1.8 | 4.3 | 0.00 | 24.8 | 23.9 | 1.9 | 0.09 |
| NUT | Highly expressed in testis (macroorchidism common in FXS) | Unknown | 3.2 | 4.5 | 0.15 | 31.1 | 28.8 | 4.8 | 0.06 |
Array fold change is generally reported as a log value but has been converted to an arithmetic value for comparison purposes with the QRT-PCR data.
Quantitative RT-PCR (QRT-PCR) fold change calculated using the formula: 2|Comp CT − FXS CT| (Comp CT = mean CT of the comparison males and FXS = mean CT of the FXS males). See “Materials and Methods” for further description.
Secondary interaction indicates genes or gene products that do not interact directly with FMR1 but are either upstream or downstream in a connected pathway (as illustrated in Figure 1).
DISCUSSION
We analyzed microarrays containing more than 57,000 probes and approximately 46% of the probes had a detectable signal using RNA isolated from lymphoblastoid cells. Lymphoblastoid cells represent a viable model system for studying gene expression patterns as reported in other studies.19,26–28 The expression of FMR1 was significantly reduced in our study but detectable in agreement with recent reports of FMR1 expression in lymphoblastoid cells derived from individuals with FXS.29 Similarly, FMR1 message was detected using quantitative RT-PCR in peripheral blood leukocytes of FXS males with methylated full mutations.30 Approximately one third of FXS males had FMR1 mRNA levels in peripheral blood of ≤1% compared with normal males; one third had levels between 1% and 10%, and the remaining males with FXS had FMR1 mRNA levels between 10% and 100%. Despite the presence of FMR1 mRNA detectable in most of the FXS males with full mutations, no FMRP production was detected by either immunocytochemistry or Western blotting.30 The average FMR1 mRNA level was significantly reduced in our FXS subjects relative to controls as assessed by both microarray analysis and quantitative RT-PCR, which is in general agreement with FMR1 mRNA levels reported by Tassone et al.30
Genes and transcripts meeting the inclusion criteria and with a change in expression of at least 1.5-fold were analyzed in our study by Welch t tests with Bonferroni correction for multiple observations following established protocols as used in other studies.19 We selected genes from our microarray analysis and from the literature including mice gene knockout and immunoprecipitation studies to undertake quantitative RT-PCR analysis and validated 10 genes (Table 3). There was concordance between each of these genes in direction of altered expression; either up or down in the FXS subjects relative to the comparison subjects, although not necessarily in magnitude in lymphoblastoid cells and frontal cortex.
Several recent reports have identified sets of genes that interact with FMRP or are differentially expressed in FXS cells in mouse and/or in human cells.10,11,17,18 We collected these lists of genes and cross-referenced them with our microarrays. We analyzed the resulting lists of genes using small group analysis, allowing a smaller Bonferroni correction coefficient, because of their previous association with FXS. We were able to detect 228 sequences previously reported to coimmunoprecipitate with FMRP in mouse brain,10 25 of which had a significant difference in expression of at least 1.5-fold. Additionally, Brown et al.10 identified genes with a differential profile in polyribosomes from FXS lymphoblasts compared with control cells. We identified 15 genes on our arrays with a significant difference of at least 1.5-fold from this list of genes. Our arrays contained 24 genes reported by D’Agata et al.11 to have differential expression in Fmr1 knockout mice compared with controls. Seven of these genes (SND1, EEF1A2, TRPV, RNASEP1, ADRB2, INCENP, DAPK2) had significant differences in expression on our microarrays with an fdr of 20% and two (INCENP, DAPK2) were significant with an fdr of 5%. The remaining genes either did not have sufficiently strong signals to be detected or did not have a significant difference in signal intensity.
Several genes have recently been reported to have reduced expression in Fmr1 knockout mice including GABAA receptor δ subunit (Gabrd), a ρ guanine exchange factor and a regulator of G-protein signaling (Rgs4).17,18 Unfortunately, none of these genes had a reliable detection signal on our microarrays. We then examined the expression of GABRD with quantitative RT-PCR in lymphoblastoid cells and frontal cortex from our males. In contrast to the report of reduced expression of Gabrd in brain of Fmr1 knockout mice, an increase in expression was observed for GABRD in lymphoblastoid cells and the frontal cortex of our males with FXS (Table 3). However, the RT-PCR produced multiple fragments that were sequenced. These fragments are apparently splice variants of GABRD, and therefore it is difficult to make any conclusions regarding changed expression of GABRD without further clarification using different assays. Additionally, we examined genes known to directly interact with FMRP (from the Ingenuity Pathways Analysis database) and found a significant difference in expression of seven of these genes between the FXS cells and control cells (Table 2).
A subset of genes identified by microarray analysis as having differential mRNA levels in subjects with FXS was studied using quantitative RT-PCR of RNA isolated from lymphoblastoid cells and frontal cortex that were in general concordance with the microarray results (Table 3). These 10 genes (plus FMR1) were chosen for quantitative RT-PCR because of potential biological relevance to FXS and had significant changes in signal intensity on our microarrays. The lack of FMRP leads to elevated and abnormal dendritic spine formation in both humans and mice.31 Furthermore, FMRP is synthesized at the synapse in response to glutamate and synaptic activity, suggesting a role in synapse maturation and function. PPP1R9B is a regulatory subunit of protein phosphatase-1 and is highly enriched in dendritic spines, which receive most of the excitatory input in the central nervous system. EEF1A2 encodes the α subunit of the elongation factor-1 complex that delivers aminoacyl tRNAs to the ribosome. Mice homozygous for defects in Eef1a2 develop neurological and immunological defects and die by 28 days. HES1 has been found to be essential to neurogenesis, myogenesis, hematopoiesis, and sex determination in mammals. UNC13B is a presynaptic protein that promotes the priming of synaptic vesicles by acting through syntaxin. MAP1B is involved in microtubule assembly, an essential step in neurogenesis, and the RNA is known to interact with FMRP.25 ACCN1 is an amiloride-sensitive sodium channel that may play a role in neurotransmission. NUT is highly expressed in the testis and subjects with FXS have macroorchidism. UBQLN1 ubiquilins are structurally similar to ubiquitin and thought to link the ubiquitination machinery to the proteasome to affect in vivo protein degradation. Thus, these genes all have diverse functions in the central nervous system or with potential relevance to the FXS phenotype (OMIM http://www.ncbi.nlm.nih.gov/omim/) and could potentially contribute to the etiology of FXS.
In summary, we found >90 genes and transcripts that differed in expression by at least 1.5-fold with an fdr of 5% between lymphoblastoid cells derived from males with FXS and lymphoblastoid cells from sex and similarly aged control individuals. Additionally, genes involved in signaling (e.g., kinases), morphogenesis, and neurodevelopment were significantly overrepresented in this list of genes. These genes represent candidate networks that were apparently disturbed by the incorrect functioning of the FMR1 gene in FXS. Most of the gene expression analyses done to date have used brain tissue from the Fmr1 knockout mice, whereas we used RNA isolated from actively growing lymphoblastoid cells and frontal cortex from human males with FXS. Therefore, it is not surprising that we do not have complete concordance across species, tissue type, and/or analysis platform, considering the additional layers of complexity associated with RNA processing and protein translation between species, particularly humans and mice. Further analyses of these genes, especially those that have been identified in multiple studies, are warranted to develop a more integrated description of the alterations in gene processing that lead to FXS and possibly to produce new treatment modalities.
Acknowledgments
This research was supported by the Hall Foundation of Kansas City. The authors thank the individuals who participated in this research project. They also acknowledge the NICHD Brain and Tissue Bank for Developmental Disorders (University of Maryland and University of Miami) for providing brain tissue on subjects used in our study.
Footnotes
Disclosure: The authors declare no conflict of interest.
References
- 1.Terracciano A, Chiurazzi P, Neri G. Fragile X syndrome. Am J Med Genet C Semin Med Genet. 2005;137:32–37. doi: 10.1002/ajmg.c.30062. [DOI] [PubMed] [Google Scholar]
- 2.Lim JH, Booker AB, Fallon JR. Regulating fragile X gene transcription in the brain and beyond. J Cell Physiol. 2005;205:170–175. doi: 10.1002/jcp.20401. [DOI] [PubMed] [Google Scholar]
- 3.Jin P, Warren ST. Understanding the molecular basis of fragile X syndrome. Hum Mol Genet. 2000;9:901–908. doi: 10.1093/hmg/9.6.901. [DOI] [PubMed] [Google Scholar]
- 4.Verkerk AJ, Pieretti M, Sutcliffe JS, Fu YH, et al. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell. 1991;65:905–914. doi: 10.1016/0092-8674(91)90397-h. [DOI] [PubMed] [Google Scholar]
- 5.Hinds HL, Ashley CT, Sutcliffe JS, Nelson DL, et al. Tissue specific expression of FMR-1 provides evidence for a functional role in fragile X syndrome. Nat Genet. 1993;3:36–43. doi: 10.1038/ng0193-36. [DOI] [PubMed] [Google Scholar]
- 6.Bakker CE. Fmr1 knockout mice: a model to study fragile X mental retardation. The Dutch-Belgian Fragile X Consortium. Cell. 1994;78:23–33. [PubMed] [Google Scholar]
- 7.Irwin SA, Galvez R, Greenough WT. Dendritic spine structural anomalies in fragile-X mental retardation syndrome. Cereb Cortex. 2000;10:1038–1044. doi: 10.1093/cercor/10.10.1038. [DOI] [PubMed] [Google Scholar]
- 8.Irwin SA, Patel B, Idupulapati M, Harris JB, et al. Abnormal dendritic spine characteristics in the temporal and visual cortices of patients with fragile-X syndrome: a quantitative examination. Am J Med Genet. 2001;98:161–167. doi: 10.1002/1096-8628(20010115)98:2<161::aid-ajmg1025>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 9.Greenough WT, Klintsova AY, Irwin SA, Galvez R, et al. Synaptic regulation of protein synthesis and the fragile X protein. Proc Natl Acad Sci U S A. 2001;98:7101–7106. doi: 10.1073/pnas.141145998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brown V, Jin P, Ceman S, Darnell JC, et al. Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in fragile X syndrome. Cell. 2001;107:477–487. doi: 10.1016/s0092-8674(01)00568-2. [DOI] [PubMed] [Google Scholar]
- 11.D’Agata V, Warren ST, Zhao W, Torre ER, et al. Gene expression profiles in a transgenic animal model of fragile X syndrome. Neurobiol Dis. 2002;10:211–218. doi: 10.1006/nbdi.2002.0506. [DOI] [PubMed] [Google Scholar]
- 12.Jin P, Warren ST. New insights into fragile X syndrome: from molecules to neurobehaviors. Trends Biochem Sci. 2003;28:152–158. doi: 10.1016/S0968-0004(03)00033-1. [DOI] [PubMed] [Google Scholar]
- 13.Darnell JC, Jensen KB, Jin P, Brown V, et al. Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell. 2001;107:489–499. doi: 10.1016/s0092-8674(01)00566-9. [DOI] [PubMed] [Google Scholar]
- 14.Jin P, Alisch RS, Warren ST. RNA and microRNAs in fragile X mental retardation. Nat Cell Biol. 2004;6:1048–1053. doi: 10.1038/ncb1104-1048. [DOI] [PubMed] [Google Scholar]
- 15.Jin P, Zarnescu DC, Ceman S, Nakamoto M, et al. Biochemical and genetic interaction between the fragile X mental retardation protein and the microRNA pathway. Nat Neurosci. 2004;7:113–117. doi: 10.1038/nn1174. [DOI] [PubMed] [Google Scholar]
- 16.Bear MF. Therapeutic implications of the mGluR theory of fragile X mental retardation. Genes Brain Behav. 2005;4:393–398. doi: 10.1111/j.1601-183X.2005.00135.x. [DOI] [PubMed] [Google Scholar]
- 17.Gantois I, Vandesompele J, Speleman F, Reyniers E, et al. Expression profiling suggests underexpression of the GABA(A) receptor subunit delta in the fragile X knockout mouse model. Neurobiol Dis. 2005 doi: 10.1016/j.nbd.2005.07.017. [DOI] [PubMed] [Google Scholar]
- 18.Tervonen T, Akerman K, Oostra BA, Castren M. Rgs4 mRNA expression is decreased in the brain of Fmr1 knockout mouse. Brain Res Mol Brain Res. 2005;133:162–165. doi: 10.1016/j.molbrainres.2004.09.023. [DOI] [PubMed] [Google Scholar]
- 19.Hu VW, Frank BC, Heine S, Lee NH, et al. Gene expression profiling of lymphoblastoid cell lines from monozygotic twins discordant in severity of autism reveals differential regulation of neurologically relevant genes. BMC Genomics. 2006;7:118. doi: 10.1186/1471-2164-7-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bittel DC, O’Brien JE, Lofland GK, Kibiryeva N, et al. Gene expression in pediatric heart disease with emphasis on conotruncal defects. Prog Pediatr Cardiol. 2005;20:127–141. doi: 10.1016/j.ppedcard.2005.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang IV, Chen E, Hasseman JP, Liang W, et al. Within the fold: assessing differential expression measures and reproducibility in microarray assays. Genome Biol. 2002;3:research0062. doi: 10.1186/gb-2002-3-11-research0062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stefan M, Portis T, Longnecker R, Nicholls RD. A nonimprinted Prader-Willi syndrome (PWS)-region gene regulates a different chromosomal domain in trans but the imprinted PWS loci do not alter genome-wide mRNA levels. Genomics. 2005;85:630–640. doi: 10.1016/j.ygeno.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 23.BeiBbarth T, Speed TP. GOstat; find statistically overrepresented Gene Ontologies within a group of genes. Bioinformatics. 2004;20:1464–1465. doi: 10.1093/bioinformatics/bth088. [DOI] [PubMed] [Google Scholar]
- 24.Miyashiro KY, Beckel-Mitchener A, Purk TP, Becker KG, et al. RNA cargoes associating with FMRP reveal deficits in cellular functioning in Fmr1 null mice. Neuron. 2003;37:417–431. doi: 10.1016/s0896-6273(03)00034-5. [DOI] [PubMed] [Google Scholar]
- 25.Zalfa F, Giorgi M, Primerano B, Moro A, et al. The fragile X syndrome protein FMRP associates with BC1 RNA and regulates the translation of specific mRNAs at synapses. Cell. 2003;112:317–327. doi: 10.1016/s0092-8674(03)00079-5. [DOI] [PubMed] [Google Scholar]
- 26.Tassone F, Hagerman RJ, Chamberlain WD, Hagerman PJ. Transcription of the FMR1 gene in individuals with fragile X syndrome. Am J Med Genet. 2000;97:195–203. doi: 10.1002/1096-8628(200023)97:3<195::AID-AJMG1037>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 27.Bittel DC, Kibiryeva N, Dasouki M, Knoll JH, et al. A 9-year-old male with a duplication of chromosome 3p25.3p26. 2: clinical report and gene expression analysis. Am J Med Genet A. 2006;140:573–579. doi: 10.1002/ajmg.a.31132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baron CA, Liu SY, Hicks C, Gregg JP. Utilization of lymphoblastoid cell lines as a system for the molecular modeling of autism. J Autism Dev Disord. 2006 doi: 10.1007/s10803-006-0134-x. [DOI] [PubMed] [Google Scholar]
- 29.Bonilla V, Sobrino F, Lucas M, Pintado E. Epstein-Barr virus transformation of human lymphoblastoid cells from patients with fragile X syndrome induces variable changes on CGG repeats size and promoter methylation. Mol Diagn. 2003;7:163–167. doi: 10.1007/BF03260033. [DOI] [PubMed] [Google Scholar]
- 30.Tassone F, Hagerman RJ, Taylor AK, Hagerman PJ. A majority of fragile X males with methylated, full mutation alleles have significant levels of FMR1 messenger RNA. J Med Genet. 2001;38:453–456. doi: 10.1136/jmg.38.7.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hagerman RJ, Hagerman PJ. Fragile X syndrome; diagnosis, treatment and research. Baltimore: Johns Hopkins University Press; 2002. [Google Scholar]