Abstract
The NLRP3 inflammasome constitutes a major antiviral host defense mechanism during influenza virus infection. Inflammasome assembly in virus-infected cells facilitates autocatalytic processing of pro-caspase-1 and subsequent cleavage and secretion of proinflammatory cytokines IL-1β and IL-18. The NLRP3 inflammasome is critical for induction of both innate and adaptive immune responses during influenza virus infection. Inflammasome-dependent antiviral responses also regulate immunopathology and tissue repair in the infected lungs. The regulation of NLRP3 inflammasome assembly is an area of active research and recent studies have unraveled multiple cellular and viral factors involved in inflammasome assembly. Emerging studies have also identified the cross talk between inflammasome activation and programmed cell death pathways in influenza virus-infected cells. Here, we review the current literature regarding regulation and functions of NLRP3 inflammasome during influenza virus infection.
Keywords: NLRP3, Caspase-1, inflammasome, influenza virus, ZBP1, RIPK3
1. Innate immune sensing of influenza virus
Influenza virus, which commonly causes seasonal outbreaks of upper respiratory tract infection, is a single stranded, negative sense RNA virus of the family Orthomyxoviridae (WHO., 2003, Bouvier and Palese, 2008). The virus has a segmented genome composed of eight segments encoding eleven viral proteins: hemagglutinin (HA), neuraminidase (NA), matrix protein 1 (M1), matrix protein 2 (M2), nucleoprotein (NP), non-structural protein 1 (NS1), non-structural protein 2 (NS2) polymerase acidic protein (PA), polymerase basic protein 1 (PB1), polymerase basic protein 2 (PB2) and polymerase basic protein 1 – F2 (PB1-F2) (Bouvier and Palese, 2008, Chen et al., 2001). Based on antigenic differences, influenza virus is broadly classified into three antigenic types, Type A, B and C. Type A influenza viruses are distributed globally and can infect humans, animals and avian species whereas both Type B and C are mostly associated with milder infections in humans (WHO., 2003, Taubenberger and Morens, 2008). Type A influenza viruses are divided into various subtypes based on their surface glycoproteins, HA and NA. There are eighteen HA (H1 through H18) and 11 NA (N1 through N11) subtypes and these subtypes are further divided into strains based on HA and NA present on the viral envelope (eg: H1N1, H3N2) (WHO., 2003) (Bouvier and Palese, 2008). High mutation rates in the viral genome (antigenic drift) and reassortment of the segmented genome (antigenic shift) drive frequent genetic variation and emergence of antigenically new viruses, which in some instances cause pandemic influenza outbreaks (Taubenberger and Morens, 2008).
Airway epithelial cells are the primary targets of influenza virus and the infection is mostly confined within the lungs (Kuiken et al., 2012). Influenza virus in infected cells is sensed by specific intracellular receptors leading to activation of the host immune system, which mounts an antiviral response aimed at controlling virus replication and eliminating the infectious virus (Kawai and Akira, 2010, Kanneganti, 2010). Three major classes of pattern recognition receptors (PRRs) involved in influenza virus detection are toll-like receptors (TLRs), retinoic acid inducible gene-I (RIG-I)- like receptors (RLRs) and nucleotide and oligomerization domain, leucine-rich repeat-containing proteins (NLRs) (Figure 1) (Takeuchi and Akira, 2010, Kanneganti, 2010). Although virus sensing TLRs are primarily expressed by macrophages and dendritic cells (DCs) and recognize viral nucleic acids within the endosomal compartment, RLRs and NLRs are ubiquitously expressed and sense viral pathogen associated molecular patterns (PAMPs) within the cytoplasm of infected cells (Kanneganti, 2010). Recognition of influenza virus by PRRs typically occurs in respiratory epithelial cells, alveolar macrophages, conventional DCs and plasmacytoid DCs in a cell type-specific manner (Iwasaki and Pillai, 2014, Kato et al., 2005). The single-stranded viral RNA is recognized through endosomal TLR7 in pDCs in contrast to epithelial cells and cDCs where 5′-triphosphate of single-stranded RNA genome is sensed by the cytosolic sensor RIG-I (Diebold et al., 2004, Kato et al., 2005, Pichlmair et al., 2006). In addition to TLRs and RLRs, NLR family members are also important in mediating antiviral responses during influenza virus infection (Lupfer and Kanneganti, 2013). Among various NLR family members, nucleotide and oligomerization domain, leucine-rich repeat–containing protein family, pyrin domain containing 3 (NLRP3), which assembles a large multimeric protein complex called inflammasome, is the most widely studied NLR (Lupfer and Kanneganti, 2013). Recent studies have demonstrated a major role for NLRP3 inflammasome in promoting antiviral immune responses against a diverse array of viruses, including influenza virus (Lupfer et al., 2015). In this review, we provide a detailed overview on the regulation and functions of the NLRP3 inflammasome during influenza virus infection and specifically highlight recent advances in our understanding of NLRP3 inflammasome activation during influenza virus infection.
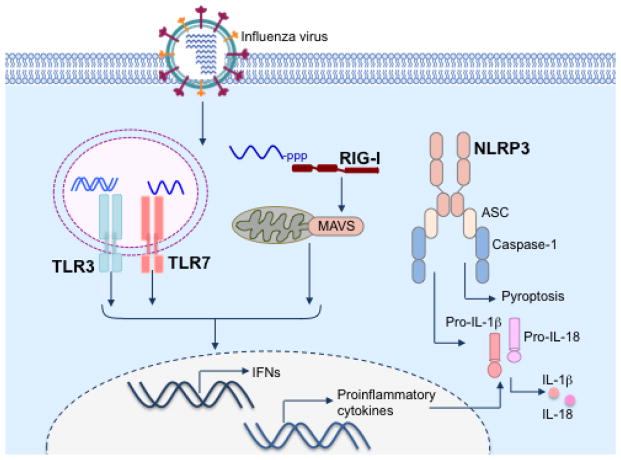
Figure 1. Sensing of influenza virus by innate immune receptors.
Influenza virus enters target cells via receptor-mediated endocytosis. Acidification of endosomes facilitates fusion of viral and endosomal membranes and release of virion contents into the cytoplasm. The genomic single stranded RNA of influenza virus is sensed by endosomal TLR7. Once the viral genome reaches the cytoplasm, the cytosolic sensor RIG-I recognizes 5′-triphosphate of the genomic RNA. Virus sensing by these receptors leads to induction of antiviral IFNs and other proinflammatory cytokines. Influenza virus infection also activates receptors of NLR family initiating assembly of NLRP3 inflammasome complex, which mediates processing and release of pro-IL-1β and pro-IL-18 and induces an inflammatory form of cell death called pyroptosis.
2. The NLRP3 inflammasome
Inflammasomes are multiprotein complexes whose assembly is dictated by cytoplasmic PRRs in response to microbial components or danger signals (Martinon et al., 2002, Man and Kanneganti, 2015b). Five distinct inflammasomes have been well characterized so far. In addition to the three NLR family members NLRP1, NLRP3 and NLRC4, the HIN-domain-containing family member AIM2 (absent in melanoma-2), and Pyrin can function as sensors to initiate assembly of the inflammasome complexes (Sharma and Kanneganti, 2016), Lamkanfi and Dixit, 2014). After recognition of PAMPs or damage-associated molecular patterns (DAMPs), the sensor oligomerizes and recruits adaptor protein apoptosis-associated speck-like protein containing a CARD domain (ASC). ASC bridges the inflammasome sensor to the inactive zymogen form of cysteine protease caspase-1 (Sharma and Kanneganti, 2016). Once assembled, inflammasomes mediate activation of caspase-1, which in turn mediate processing and release of proinflammatory cytokines IL-1β and IL-18 (Sharma and Kanneganti, 2016, Lamkanfi and Dixit, 2014). Active caspase-1 also induces an inflammatory form of cell death called pyroptosis, executed via its downstream substrate gasdermin D (Kayagaki et al., 2015, Shi et al., 2015, Aglietti et al., 2016, He et al., 2015, Liu et al., 2016, Man and Kanneganti, 2015a).
Among inflammasomes, the NLRP3 inflammasome is the most widely investigated complex owing to the diverse nature of its activators, which include both infectious and non-infectious agents (Lamkanfi and Dixit, 2014, Man and Kanneganti, 2015b). Activation of the NLRP3 inflammasome is a two-step process initiated after sensing of PAMPs and DAMPs by PRRs. In the first step, pathogen recognition by these PRRs activates various downstream signaling cascades including the NFκB pathway leading to transcriptional induction of pro-IL-1β and NLRP3. Inflammasome assembly happens in the second step after NLRP3 directly or indirectly senses the presence of its activators (Lamkanfi and Dixit, 2014, Man and Kanneganti, 2015b). The upstream mechanisms regulating NLRP3 inflammasome assembly is less characterized and recent studies have identified the serine-threonine kinase NIMA related kinase 7 (NEK7) as a selective regulator of NLRP3 inflammasome activation (He et al., 2016, Shi et al., 2016, Schmid-Burgk et al., 2016). NEK7 interacts with the leucine-rich repeat domain of NLRP3 and facilitate NLRP3 oligomerization in a kinase-independent manner. NEK7 interaction with NLRP3 is necessary for the assembly of the NLRP3-ASC complex (He et al., 2016, Shi et al., 2016).
After the identification and characterization of NLRP3 inflammasome complex (Kanneganti et al., 2006b, Mariathasan et al., 2006), our studies have demonstrated NLRP3-dependent activation of caspase-1 during influenza virus infection (Kanneganti et al., 2006a). Following this initial report of viral RNA-mediated NLRP3 inflammasome activation, we and others have demonstrated the relevance of NLRP3 inflammasome in regulating innate and adaptive immune responses during influenza virus infection in vivo (Ichinohe et al., 2009, Thomas et al., 2009, Allen et al., 2009). Multiple studies have also investigated the molecular mechanisms regulating inflammasome assembly and unraveled diverse cellular and viral factors involved in the regulation of NLRP3 inflammasome during influenza virus infection.
3. Activation of NLRP3 inflammasome during influenza virus infection
Caspase-1-dependent secretion of IL-1β and IL-18 during influenza virus infection in human macrophages was reported a few years after identifying caspase-1 as the IL-1β-converting enzyme (Pirhonen et al., 1999). Further studies demonstrated cleavage of caspase-1, caspase-3 and caspase-dependent proteolytic processing of IL-18 in human macrophages infected with influenza virus (Pirhonen et al., 2001). However, the mechanisms regulating caspase-1 activation were described only after the discovery of NLRP3 inflammasome (Martinon et al., 2002, Mariathasan et al., 2006, Kanneganti et al., 2006a). NLRP3-dependent processing of caspase-1 and secretion of IL-1β and IL-18 occurs in bone marrow derived macrophages (BMDMs) transfected with viral RNA or infected with influenza virus (Kanneganti et al., 2006a). Different strains of influenza virus, including Type A and B viruses, induce release of IL-1β from macrophages and human dendritic cells in response to infection (Ichinohe et al., 2009, Fernandez et al., 2016). NLRP3 inflammasome activation in mouse and human cells in response to synthetic RNA analogues has also been demonstrated (Kanneganti et al., 2006a). Moreover, intraperitoneal administration of purified influenza virus RNA or poly(I:C) induces NLRP3-dependent IL-1β secretion and inflammatory responses in mice (Kanneganti et al., 2006a). Although these studies suggested viral RNA as the ligand inducing NLRP3 assembly during influenza virus infection, the molecular mechanism by which viral RNA mediates inflammasome activation has remained unknown. Moreover, direct interaction of NLRP3 with viral RNA has not been demonstrated so far. Therefore, it is possible that viral RNA facilitates inflammasome assembly and release of inflammasome-dependent cytokines indirectly by interacting with known RNA sensors such as the TLRs and RIG-I as suggested by more recent studies (Ichinohe et al., 2009, Pothlichet et al., 2013, Kuriakose et al., 2016).
In addition to hematopoietic cells, NLRP3 inflammasome activation also occurs in non-immune cells like primary bronchial epithelial cells, lung fibroblasts and various epithelial cell lines infected with influenza virus (Allen et al., 2009, Ichinohe et al., 2009, Pothlichet et al., 2013). Influenza virus infection induces secretion of IL-1β from apical surfaces, but not from basolateral surface of primary human airway epithelial cells (Pothlichet et al., 2013, Bauer et al., 2012). Similarly, influenza virus infection also increases expression of inflammasome components in human nasal airway epithelial cell lines JME and A549 cells (Allen et al., 2009, Tiwari et al., 2014). In lung fibroblasts, caspase-1-dependent IL-1β secretion occurs independently of NLRP3 suggesting cell type-specific functions of NLRP3 in inflammasome activation (Ichinohe et al., 2009). Formation of ASC specks was also shown in tissue sections and lung fibroblasts from mice expressing fluorescent ASC protein further demonstrating inflammasome activity in non-hematopoietic cells (Tzeng et al., 2016). ASC speck formation was observed mainly in cells that stained positive for influenza virus nucleoprotein suggesting that inflammasome assembly occurs primarily in cells infected with influenza virus (Tzeng et al., 2016).
4. NLRP3 inflammasome in anti-viral immunity during influenza virus infection
After the demonstration of influenza virus-induced inflammasome activation in BMDMs, three independent studies have demonstrated the critical role of NLRP3 inflammasome in mediating antiviral immune responses during influenza virus infection in vivo (Thomas et al., 2009, Allen et al., 2009, Ichinohe et al., 2009). These studies reported NLRP3-, ASC- and caspase-1-dependent secretion of IL-1β and IL-18 in the lungs of mice infected with influenza virus. In addition, these studies also demonstrated inflammasome regulation of cellular recruitment into the lungs of influenza virus-infected mice. Studies from our lab and another group demonstrated a major role for NLRP3 in regulating mortality during influenza virus infection (Thomas et al., 2009, Allen et al., 2009). Deletion of NLRP3, ASC or caspase-1 decreases levels of IL-1β and IL-18 in bronchoalveolar lavage fluid (BALF) and serum. Deficiency of NLRP3, caspase-1 or ASC also decreases the levels of IL-6, TNF, KC and CXCL2 in the BALF three days after infection. Reduction in infiltration of neutrophils and monocytes into the lungs and increased mortality were also observed in these mice (Thomas et al., 2009, Allen et al., 2009). Interestingly, Ting and colleagues reported increased viral titers on day 7 after infection whereas we observed similar virus burden in WT and NLRP3-deficient mice on day 6 after infection (Allen et al., 2009, Thomas et al., 2009). We also found increased pulmonary necrosis and collagen deposition in Nlrp3−/− mice suggesting a role for NLRP3 in resolution of inflammation and wound repair (Thomas et al., 2009). Although both these studies demonstrate the importance of NLRP3 inflammasome in host protection, neither found a role for inflammasome in generation of adaptive immune responses in influenza virus-infected mice (Allen et al., 2009, Thomas et al., 2009).
In contrast to the studies described above, Iwasaki and colleagues reported an essential role for inflammasome in adaptive immune responses during influenza virus infection. According to this study, inflammasome activity regulates virus-specific IFNγ production by CD4+ and CD8+ T cells. Inflammasome activation is also needed for antibody class switching to generate IgA and IgG isotypes (Ichinohe et al., 2009). Whereas NLRP3 is necessary for secretion of IL-1β into the airways, IL-1β secretion and cellular recruitment into the lung parenchyma is not dependent on NLRP3, but requires ASC and caspase-1 (Ichinohe et al., 2009). The overall reduction in adaptive immune responses leads to impaired viral clearance and increased mortality in Caspase1−/− and Pycard−/− mice, but not in Nlrp3−/− mice, compared with WT mice infected with influenza virus (Ichinohe et al., 2009). Although some discrepancies regarding mechanisms and disease outcome exist, all these studies highlight the importance of inflammasome complex in antiviral host defense mechanisms.
Consistent with the observations from genetically altered mouse strains, a recent study using MCC950, a specific and potent inhibitor of NLRP3, demonstrated a temporal role for inflammasome activity in host protective responses during influenza virus infection (Tate et al., 2016). Inhibition of inflammasome activity by intranasal treatment with MCC950 from day 1 after infection decreases survival in mice infected with influenza virus (Tate et al., 2016). However, delayed treatment with MCC950 from day 3 or 7 after infection significantly improves the survival in infected mice (Tate et al., 2016). The increase in survival is associated with reduced levels of proinflammatory cytokines including IL-1β, IL-18, IL-6, TNF, CCL2 and CCL5 in BALF and decrease in recruitment of inflammatory cells into the airways (Tate et al., 2016). These observations suggest a detrimental role for NLRP3 inflammasome later during infection by enhancing lung inflammation, which is a major factor potentiating mortality during influenza virus infection (Ong et al., 2016).
The importance of NLRP3 inflammasome in antiviral responses has also been demonstrated in elderly mice infected with influenza virus (Stout-Delgado et al., 2012). DCs from aged mice show significantly reduced expression of Nlrp3, Asc and Caspase1 than DCs from young mice. Not surprisingly, IL-1β secretion in aged mice is reduced during in vitro and in vivo influenza virus infection (Stout-Delgado et al., 2012). Increased morbidity and mortality in aged mice during influenza virus infection can be reduced by nigericin treatment that augments inflammasome activation (Stout-Delgado et al., 2012). This study clearly demonstrates that decreased inflammasome activity contributes to waning immune responses and increased susceptibility to influenza virus infection in aged mice.
In addition to the observations from various murine models of infection, roles for NLRP3 inflammasome and inflammasome-dependent cytokines in immune and inflammatory responses have also been demonstrated during influenza virus infection in humans and primate models. A marked upregulation of both Nlrp3 and Il1b along with other genes associated with inflammation and cell death was observed in macaques infected with the reconstructed 1918 pandemic influenza virus (Cilloniz et al., 2009). The excessive activation of inflammasome is detrimental in this model because elevated inflammasome activation contributes to enhanced recruitment of inflammatory cells into the lungs resulting in a cytokine storm (Cilloniz et al., 2009). High levels of IL-1 were also induced in macaques infected with the highly pathogenic H5N1 virus (Baskin et al., 2009). Consistent with these findings, analysis of BAL samples from persons fatally infected with an avian origin H7N9 virus revealed a 1000 fold increase in some proinflammatory cytokines including IL-1β (Wang et al., 2014b). The greatly elevated cytokine/chemokine levels were also associated with severe lung damage in these patients (Wang et al., 2014b). Elevated levels of IL-1β were also noticed in plasma samples from children infected with H1N1 influenza virus (Chiaretti et al., 2013). Together, these studies demonstrate a role for inflammasome-dependent responses in the control of infection and disease outcome during acute influenza virus infection in both macaques and humans.
The observations from various murine and primate models as well as patients infected with influenza virus demonstrate both protective and damaging effects of inflammasome activation in the infected lungs. Whereas inflammasome-mediated responses are essential for the proper induction of both innate and adaptive immune responses, exaggerated inflammasome activation contribute to severe lung pathology during influenza virus infection. Therefore modulating NLRP3 inflammasome activity at different stages of influenza virus infection might serve as a potential therapeutic strategy for the clinical management of acute influenza virus infection.
5. Regulators of NLRP3 inflammasome activation during influenza virus infection
Although influenza virus is a potent activator of NLRP3 inflammasome, direct interaction of the virus with inflammasome components have not been demonstrated so far. Common mechanisms regulating NLRP3 inflammasome activation including perturbation of ionic balance, lysosomal rupture and generation of reactive oxygen species (ROS) have been shown to facilitate inflammasome assembly in influenza virus-infected cells (Pang and Iwasaki, 2011, Allen et al., 2009). Multiple host factors important in virus sensing and antiviral innate immune responses have also been shown to be critical for activation of NLRP3 inflammasome during influenza virus infection (Figure 2).
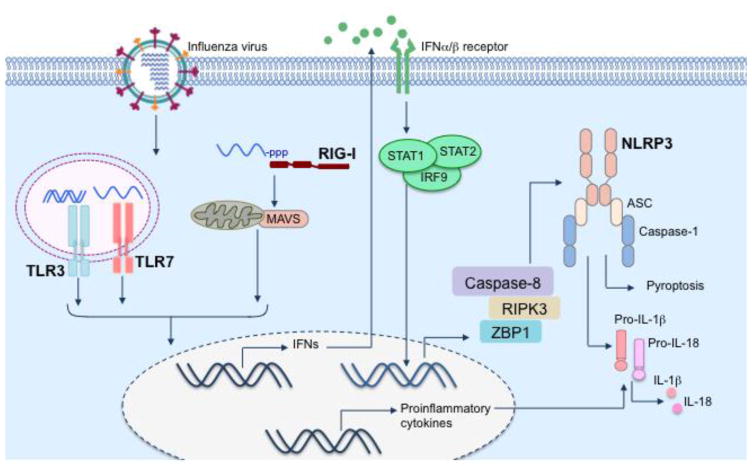
Figure 2. Regulation of NLRP3 inflammasome activation during influenza virus infection.
NLRP3 inflammasome activation is a two-step process that requires signal 1 (for upregulation of inflammasome components including NLRP3) and signal 2 (for inflammasome assembly). Virus sensing receptors TLR7 and RIG-I initiates transcriptional induction of NLRP3 and pro-IL-1β thus providing signal 1. Type I IFNs secreted in response to virus sensing induces expression of ZBP1, another sensor of influenza virus infection, via autocrine signaling. ZBP1 associates with RIPK3 in virus-infected cells and mediates inflammasome assembly via the RIPK3-caspase-8 axis. Although ZBP1 – RIPK3 – caspase-8 axis is essential for NLRP3 inflammasome activation in influenza virus-infected cells, the molecular mechanisms regulating inflammasome assembly is currently unknown.
Both TLR and RIG-I sensors of influenza virus are involved in regulation of inflammasome activation and release of IL-1β during infection (Ichinohe et al., 2009, Pothlichet et al., 2013). Virus sensing by these receptors activate various intracellular signaling cascades including NFκB activation pathways. Activated NFκB translocate to the nucleus and facilitate transcriptional induction of both Nlrp3 and Il1b (Elliott and Sutterwala, 2015). TLR7 sensing of viral RNA induces expression of Il1b and synthesis of pro-IL-1β in bone marrow derived dendritic cells (BMDCs) infected with influenza virus (Ichinohe et al., 2010). Secretion of IL-1β in influenza virus infected normal human bronchial epithelial cells (NHBE) cells is dependent on TLR3, demonstrating a role for TLR3 in inflammasome activation in primary lung epithelial cells (Pothlichet et al., 2013). In addition to TLRs, RIG-I-MAVS signaling is also involved in the priming step by upregulating Nlrp3 expression (Pothlichet et al., 2013).
RIG-I is regarded as one of the most upstream regulators of inflammasome assembly during influenza virus infection because it can either directly form inflammasome complex with ASC and caspase-1 or indirectly regulate inflammasome assembly via induction of type I IFNs (Pothlichet et al., 2013, Poeck et al., 2010). Pothlichet et al. demonstrated direct, RIG-I mediated inflammasome activation and IL-1 β secretion in primary lung epithelial cells infected with influenza virus (Pothlichet et al., 2013). By performing immunoprecipitation experiments, they showed formation of RIG-I/ASC complexes in influenza virus infected NHBE cells, but not in mock-infected cells. They further demonstrated increased interaction of RIG-I with caspase-1 in virus-infected epithelial cells compared with uninfected cells (Pothlichet et al., 2013). The formation of RIG-I/ASC and RIG-I-caspase-1 complex suggests assembly of RIG-I-dependent ASC/caspase-1 inflammasome independently of NLRP3. It is noteworthy that the expression of both Rig-i and Nlrp3 as well as inflammasome assembly is amplified by positive feed back signaling by type I IFNs induced in response to virus infection. Direct, RIG-I-dependent, IFNβ-independent inflammasome activation in influenza virus-infected cells was further confirmed in cells in which RIG-I expression was knocked down with siRNA treatment (Pothlichet et al., 2013). Although treatment with exogenous IFNβ increased IL-1β and caspase-1 p20 secretion from influenza virus-infected cells, the levels were reduced in RIG-I-deficient cells compared with cells having functional RIG-I (Pothlichet et al., 2013). The relative contribution of RIG-I interaction with ASC and caspase-1 versus RIG-I mediated IFN production in driving inflammasome activation in response to influenza virus infection warrants further investigation.
A role for IFN signaling in NLRP3 inflammasome activation is further supported by regulation of inflammasome activation during influenza virus infection by the 2′, 5′-oligoadenylate synthetase (OAS)/ribonuclease L (RNase L) system, a classical component of IFN-induced antiviral response (Chakrabarti et al., 2015). The 2′, 5′ OAS activates RNase L, which mediates its antiviral activity by cleaving cellular or viral ssRNA (Silverman, 2007). The cleavage products from RNase L induce IFNβ production via RIG-I/MDA5-MAVS signaling. The release of IL-1β and caspase-1 p20 is minimal in LPS-primed RNase L-deficient BMDCs infected with influenza virus (Chakrabarti et al., 2015). The enzymatic activity of RNase L is necessary for NLRP3 inflammasome activation because transfection of RNase L-cleaved cellular or viral RNA can induce similar levels of IL-1β in both WT and Rnasel−/− BMDCs (Chakrabarti et al., 2015). The cyclic phosphoryl groups at 2′, 3′ termini of RNase L-cleaved RNA facilitates formation of a complex containing RNA helicase DHX33, MAVS and NLRP3 in THP-1 macrophages (Chakrabarti et al., 2015). Consistent with association of DHX33 with NLRP3, activation of pro-caspase-1 and induction of IL-1β is impaired in DHX33-depleted THP-1 macrophages transfected with cleaved RNA. RNase L activity also regulates IL-1β production and promotes host survival during influenza virus infection further demonstrating the importance of RNase L in inflammasome-mediated antiviral responses (Chakrabarti et al., 2015). The relative contribution of DHX33-MAVS-NLRP3 complex formed in response to RNA cleavage products versus RNase L-mediated type I IFN production in facilitating inflammasome activation during influenza virus infection needs further investigation.
We have recently demonstrated the critical role of IFN-inducible protein Z-DNA binding protein 1 (ZBP1, also called as DNA-dependent activator of IFN regulatory factors (DAI)) in inflammasome activation during influenza virus infection (Kuriakose et al., 2016) (Figure 2). ZBP1 was initially identified as a tumor-associated protein and is highly expressed in lung, intestinal epithelium, liver and thymus in wild type mouse (Fu et al., 1999). Later studies demonstrated a role for ZBP1 in double stranded DNA-mediated innate immune responses and characterized the protein as a DNA sensor (Takaoka et al., 2007). However, the classification of ZBP1 as a DNA sensor is disputed because genetic deletion of ZBP1 did not impair dsDNA-mediated immune responses (Ishii et al., 2008). ZBP1 is induced in influenza virus infected cells in an IFNAR-, STAT1- and IRF9-dependent manner (Kuriakose et al., 2016) (Figure 2). ZBP1 functions as a key regulator of NLRP3 inflammasome activation during influenza virus infection. Genetic deletion of ZBP1 leads to lack of caspase-1 activation and release of IL-1β and IL-18 in BMDMs infected with influenza virus (Kuriakose et al., 2016). Another recent study by Balachandran and colleagues also demonstrated ZBP1-dependent secretion of IL-1β in influenza virus-infected cells (Thapa et al., 2016). ZBP1 regulation of inflammasome activation is specific to influenza virus infection because ZBP1 is dispensable for inflammasome activation and cytokine secretion in response to classic agonists of NLRP3 (including RNA virus vesicular stomatitis virus), NLRC4 and AIM2 inflammasomes (Kuriakose et al., 2016).
ZBP1 functions as a common upstream regulator of NLRP3 inflammasome and various programmed cell death pathways during influenza virus infection (Kuriakose et al., 2016, Thapa et al., 2016, Clancy and Martin, 2016). The receptor interacting protein homotypic interaction motif (RHIM) domains of ZBP1 is known to interact with other RHIM containing proteins, receptor interacting protein kinase 1 (RIPK1) and RIPK3 (Rebsamen et al., 2009, Kaiser et al., 2008). Consistent with this, ZBP1 – RIPK3 interaction occurs in influenza virus infected cells. The ZBP1 – RIPK3 complex mediates NLRP3 inflammasome activation during influenza virus infection (Kuriakose et al., 2016). Ablation of RIPK3 significantly reduces Caspase-1 cleavage and IL-1β/IL-18 release in response to influenza virus infection (Kuriakose et al., 2016, Wang et al., 2014a). Consistent with these in vitro data, secretion of both IL-1β and IL-18 into the airways is also regulated by RIPK3 during influenza virus infection in vivo (Wang et al., 2014a). In addition to regulation of NLRP3 inflammasome activation during RNA virus infection, RIPK3 also regulates necroptosis and apoptosis in response to diverse ligands via mixed lineage-kinase domain like (MLKL) and caspase-8 respectively (Newton et al., 2014, Mandal et al., 2014). RIPK3 regulation of NLRP3 inflammasome is not mediated via MLKL because caspase-1 activation occurs normally in Mlkl−/− BMDMs infected with influenza virus. Notably, the RIPK3 – caspase-8 axis controls NLRP3 inflammasome assembly, caspase-1 cleavage and release of IL-1β and IL-18 (Kuriakose et al., 2016) (Figure 2). Although a previous study reported a role for RIPK1 kinase activity in RNA virus induced NLRP3 inflammasome activation, we did not observe involvement of RIPK1 kinase in inflammasome activation during influenza virus infection (Wang et al., 2014a, Kuriakose et al., 2016). However, our data demonstrate ZBP1 – RIPK1 regulation of proinflammatory cytokine production in influenza virus-infected BMDMs (Kuriakose et al., 2016). The molecular mechanism by which ZBP1 – RIPK3 – caspase-8 axis facilitates NLRP3 inflammasome activation is not known yet. RIPK3-dependent activation of apoptotic and necroptotic pathways occurs only in cells harboring actively replicating influenza virus (Nogusa et al., 2016). It is not known whether the presence of replicating virus within the cell is also required for NLRP3 assembly and activation. Further studies are needed to unravel the cellular and biochemical processes leading to the assembly of NLRP3 inflammasome in influenza virus-infected cells.
6. Other host factors modulating influenza virus-induced NLRP3 inflammasome activation
Among the various cellular organelles, mitochondria have an important role in initiating antiviral immune responses including NLRP3 inflammasome activation (Gurung et al., 2015). Multiple studies have demonstrated the importance of mitochondrial membrane potential in NLRP3 inflammasome assembly in response to influenza and other RNA viruses (Ichinohe et al., 2013, Wang et al., 2014a). Loss of mitochondrial membrane potential by treatment with carbonyl cyanide m-chlorophenyl hydrazone (CCCP) suppressed IL-1β secretion and caspase-1 activation in BMDMs infected with influenza virus (Ichinohe et al., 2013). Moreover, overexpression of uncoupling protein-2, which reduces mitochondrial membrane potential, inhibits NLRP3 inflammasome activation (Ichinohe et al., 2013). Mitochondrial membrane potential facilitates inflammasome assembly by promoting interaction of mitochondrial outer membrane guanosine triphosphatase, mitofusin-2 with NLRP3 (Ichinohe et al., 2013). Another study reported a role for dynamin related protein 1 (DRP1), a GTPase that promotes mitochondrial fission, in NLRP3 inflammasome activation via induction of mitochondrial damage (Wang et al., 2014a). We have demonstrated a role for NOD2-RIPK2 signaling in inhibiting NLRP3 inflammasome activation during influenza virus infection by regulating mitophagy. Ripk2−/− cells exhibit defective mitophagy leading to enhanced production of mitochondrial superoxide and accumulation of damaged mitochondria, which ultimately induces NLRP3 inflammasome activation (Lupfer et al., 2013). Together, these studies highlight the physiological relevance of mitochondria in NLRP3 inflammasome activation during viral infections.
In addition to these cellular factors, commensal bacteria are also important in regulating inflammasome activation in the lungs of influenza virus-infected mice (Ichinohe et al., 2011). Antibiotic treatment of wild type mice reduces expression of Il1b, Il18 and Nlrp3 following influenza virus infection suggesting a role for commensal bacteria in providing signal 1 (priming) for inflammasome activation (Ichinohe et al., 2011). Antibiotic treatment also decreases IL-1β levels in the BAL to the levels comparable with that of ASC-deficient mice. The reduction in inflammasome dependent cytokines leads to impaired migration and function of lung DCs causing diminished adaptive immune responses (Ichinohe et al., 2011). Overall these observations indicate a role for commensal microbiota in promoting inflammasome-dependent antiviral responses during influenza virus infection.
7. Viral effectors of NLRP3 inflammasome activation
Several studies have reported the contribution of viral factors in providing signals necessary for NLRP3 inflammasome activation during influenza virus infection. Inflammasome activation requires attachment, fusion and replication of the virus because either heat inactivated or UV-irradiated viruses fail to induce IL-1β secretion from BMDCs and lung fibroblasts (Ichinohe et al., 2009, Ichinohe et al., 2010). Although genomic RNA is sufficient to provide signal 1, other viral factors are also important in initiating inflammasome assembly. Perturbation in intracellular ionic concentration is regarded as a common mechanism of virus-induced NLRP3 inflammasome activation (Pang and Iwasaki, 2011, Lupfer et al., 2015). Consistent with this, the ion channel activity of influenza virus membrane protein M2 was shown to mediate NLRP3 inflammasome assembly (Ichinohe et al., 2010). M2 acts as a proton specific channel and facilitates uncoating and entry of virus into the cytoplasm from endosomal cavity (Pinto et al., 1992). Moreover, M2 also maintains the pH of trans-Golgi network to prevent premature conformational change of HA protein (Pang and Iwasaki, 2011). A mutant of influenza virus lacking H+ transport activity of M2 protein failed to induce IL-1β secretion from BMDMs and BMDCs (Ichinohe et al., 2010). The role of M2 in promoting inflammasome activation was further confirmed by ectopic expression of M2 from a lentivirus, which restored inflammasome activation (Ichinohe et al., 2010). In addition to murine cells, M2 and NLRP3 dependent IL-1β and IL-18 secretion also occurs in human dendritic cells infected with influenza virus (Fernandez et al., 2016). The M2-dependent IL-1β secretion from human cells can be attenuated by treatment with M2 inhibitors, amantadine and rimantadine. Infection with UV-irradiated virus incapable of de novo synthesis of M2 protein induces lower levels of IL-1β compared to the levels induced by live virus (Fernandez et al., 2016). The M2-mediated inflammasome activation is dependent on both intracellular K+ concentration and ROS production (Ichinohe et al., 2010). Whether M2 ion channel activity regulates K+ efflux and ROS production is not known. These studies suggest that M2 activity alone is sufficient for activating inflammasome in influenza virus-infected cells. However, our data demonstrating the complete absence of caspase-1 activation in Ripk3−/−/Caspase8−/− and Zbp1−/− cells support a central role for host factors rather than virus encoded proteins in initiating inflammasome assembly during influenza virus infection (Kuriakose et al., 2016).
The involvement of influenza virus NS1 protein in regulating caspase-1 activation and secretion of IL-1β and IL-18 was reported prior to the characterization of influenza virus-induced NLRP3 inflammasome (Stasakova et al., 2005). Influenza virus mutants lacking functional RNA binding and dimerization domains of NS1 induce much higher levels of biologically active IL-1β and IL-18 than wild type viruses (Stasakova et al., 2005). These mutant viruses also induce enhanced caspase-1 activation and induction of apoptosis in human macrophages. In contrast, NS1 mutants lacking C-terminal domains induce normal IL-1β or IL-18 secretion (Stasakova et al., 2005). These results suggest a role for N terminal domain of NS1 in regulating caspase-1 activation and IL-1β and IL-18 secretion during influenza virus infection. Of note, the N-terminal RNA-binding domain of NS1 is known to interact with RIG-I in virus-infected cells to suppress antiviral responses (Jureka et al., 2015). Another study reported direct interaction of NS1 with NLRP3, which blocks inflammasome assembly and decrease IL-1β and IL-18 secretion in THP-1 macrophages (Cheong et al., 2015). NS1-mediated inhibition of NLRP3 inflammasome activation is also demonstrated in lung epithelial cells (Pothlichet et al., 2013). A recent study demonstrated NS1 inhibition of NLRP3/ASC/caspase-1 complex formation and suppression of IL-1β secretion in J774A.1 macrophages (Moriyama et al., 2016). NS1 suppresses IL-1β secretion via its RNA-binding and TRIM25-binding domains (Moriyama et al., 2016). Although these studies suggest a direct role for NS1 in suppressing NLRP3 inflammasome activation during influenza virus infection, this inhibitory effect could also be due to the well-characterized, negative regulatory effects of NS1 on type I IFN responses (Kochs et al., 2007).
The influenza virus virulence protein PB1-F2 is another viral protein associated with NLRP3 inflammasome activation (McAuley et al., 2013). PB1-F2 is an 87aa protein encoded by the +1 reading frame of RNA polymerase subunit PB1 (Chen et al., 2001). Most of the low pathogenic seasonal strains of influenza virus contain a C-terminal truncated form of PB1-F2 whereas full-length protein is present in most highly pathogenic strains (Chen et al., 2001). Full length PB1-F2 is transported via translocase of the outer membrane (TOM)-40 channels and localizes to mitochondrial inner membrane and induces apoptosis in infected cells (Yoshizumi et al., 2014). PB1-F2-mediated NLRP3 inflammasome activation is associated with mitochondrial disruption and ROS production because localization of PB1-F2 to mitochondria leads to altered mitochondrial dynamics and attenuates mitochondrial membrane potential (Yoshizumi et al., 2014). Two studies investigating the role of PB1-F2 in inflammasome activation reported contrasting results (Yoshizumi et al., 2014, McAuley et al., 2013). In an in vivo study, secretion of IL-1β and cellular recruitment to lung is reduced in mice infected with genetically altered, PB1-F2-deficient virus compared with mice infected with wild type virus (McAuley et al., 2013). PB1-F2 peptide alone is sufficient to induce IL-1β secretion in mice suggesting that PB1-F2 regulation of NLRP3 inflammasome occurs independently of other viral factors like M2 ion channel protein (McAuley et al., 2013).
Consistent with its function as a virulence factor, PB1-F2 peptide from pathogenic strains induce higher levels of IL-1β compared to PB1-F2 peptide from seasonal influenza strains. PB1-F2-oligomers are phagocytosed and incorporated into lysosomal pathway and induce ASC speck formation and IL-1β secretion in a caspase-1 and NLRP3-dependent manner in macrophages (McAuley et al., 2013). The high molecular weight, aggregated form of PB1-F2, but not the low molecular weight species induce inflammasome activity. Consistent with its ability to induce NLRP3 inflammasome activation, infection with PB1-F2 expressing virus or treatment with the peptide leads to reduced IL-1β secretion, cellular recruitment and inflammatory responses in NLRP3-deficient mice compared with wild type mice (McAuley et al., 2013). PB1-F2 mediated IL-1β secretion also occurs in human PBMCs in a caspase-1-dependent manner (McAuley et al., 2013). A role for PB1-F2-mediated NLRP3 inflammasome activation in the pathogenesis of influenza virus infection is also demonstrated in another recent study. PB1-F2 from the recently emerged avian origin H7N9 influenza virus induces NLRP3-dependent IL-1β production, lung inflammation and cellular recruitment (Pinar et al., 2016). Importantly, treatment with NLRP3 inhibitor MCC950 significantly decreases the observed hyper inflammatory responses further suggesting NLRP3 inhibition as an effective treatment option during pathogenic influenza virus infection (Pinar et al., 2016).
In contrast to these in vivo studies, PB1-F2 inhibits NLRP3 inflammasome activation in an in vitro reconstitution system (Yoshizumi et al., 2014). Expression of full-length PB1-F2 decreases IL-1β secretion in HEK293T cells co-transfected with NLRP3, ASC, pro-caspase-1 and pro-IL-1β expression plasmids (Yoshizumi et al., 2014). This inhibition of inflammasome activation is also associated with mitochondrial localization of PB1-F2 because treatment with TOM-40 siRNA restores IL-1β secretion in the reconstituted system (Yoshizumi et al., 2014). This report suggests that PB1-F2 is a negative regulator of NLRP3 inflammasome activation. Despite reporting contrasting results, both these studies agree on the role of PB1-F2 in inflammasome activation and therefore warrant future studies investigating PB1-F2-mediated inflammasome activation. Of note, PB1-F2 is considered as the viral effector mediating cell death in influenza virus-infected cells (Chen et al., 2001). The interconnected nature of NLRP3 inflammasome activation and cell death pathways demonstrated in our study supports a role for this viral protein in promoting inflammasome-dependent immune responses (Kuriakose et al., 2016).
8. Conclusions
The importance of NLRP3 inflammasome in regulating host protective responses during influenza virus infection is well established. Recent studies have unraveled many cellular and viral factors involved in NLRP3 inflammasome activation; however, the molecular mechanism by which these diverse regulators mediate inflammasome assembly remains elusive. It is still unclear whether inflammasome activation occurs only within the infected cells or whether bystander cells can assemble and activate inflammasome in response to DAMPs released from other infected cells. Further studies are also warranted to fully understand how type I IFN signaling and various IFN-stimulated gens (ISGs) facilitate NLRP3 inflammasome activation. Type III IFNs are known to drive redundant amplification loops to induce ISGs in airway epithelial cells (Crotta et al., 2013) and are expressed normally in IFNAR-deficient mice infected with influenza virus (Kuriakose et al., 2015). However, the role of type III IFNs in regulating inflammasome-dependent responses has not been investigated so far. Another important aspect that warrants future studies is the cross talk between influenza virus-induced programmed cell death and inflammasome activation. Further insights into the regulatory mechanisms of NLRP3 inflammasome will enhance our understanding about the role of these innate immune complexes in antiviral immunity during influenza virus infection and may aid in the development of new therapeutic strategies that modulate inflammasome activation and functions.
Highlights.
The NLRP3 inflammasome promotes host protective responses during influenza virus infection.
Host factors regulating inflammasome activation include TLR and RIG-I sensors and type I IFN signaling.
The IFN-inducible protein ZBP1 functions as a common upstream regulator of NLRP3 inflammasome and programmed cell death pathways during influenza virus infection.
Virus encoded proteins NS1, M2 and PB1-F2 facilitate inflammasome activation.
Acknowledgments
We thank P. Gurung, R.K.S. Malireddi and K. Sannula for critical reading of the manuscript. Research studies from our laboratory are supported by the US National Institutes of Health (AI101935, AI124346, AR056296 and CA163507 to T.D.K.), the American Lebanese Syrian Associated Charities (to T.D.K.).
Abbreviations
- HA
hemagglutinin
- NA
neuraminidase
- M1
matrix protein 1
- NP
nucleoprotein
- NS1
non-structural protein 1
- PA
polymerase acidic protein
- PB1
polymerase basic protein 1
- PRRs
pattern recognition receptors
- TLRs
Toll-like receptors
- RLRs
Retinoic acid inducible gene-I (RIG-I)- like receptors
- NLRs
nucleotide and oligomerization domain, leucine-rich repeat-containing proteins
- PAMPs
pathogen associated molecular patterns
- NLRP3
nucleotide and oligomerization domain, leucine-rich repeat–containing protein family, pyrin domain containing 3
- AIM2
absent in melanoma-2
- DAMPs
damage-associated molecular patterns
- ASC
apoptosis-associated speck-like protein containing a CARD domain
- NEK7
NIMA related kinase 7
- IFNs
interferons
- BMDMs
bone marrow derived macrophages
- BMDCs
bone marrow derived dendritic cells
- NHBE
normal human bronchial epithelial cells
- 2′,5′
OAS, 2′, 5′-oligoadenylate synthetase
- RNase L
ribonuclease L
- ZBP1
Z-DNA binding protein 1
- DAI
DNA-dependent activator of IFN regulatory factors
- RHIM
receptor interacting protein homotypic interaction motif
- RIPK1
receptor interacting protein kinase 1
- MLKL
mixed lineage-kinase domain like
- CCCP
carbonyl cyanide m-chlorophenyl hydrazine
- DRP1
dynamin related protein 1
- (TOM)-40
translocase of the outer membrane
- ISGs
IFN stimulated genes
Footnotes
Disclosures
The authors declare no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aglietti RA, Estevez A, Gupta A, Ramirez MG, Liu PS, Kayagaki N, Ciferri C, Dixit VM, Dueber EC. GsdmD p30 elicited by caspase-11 during pyroptosis forms pores in membranes. Proceedings of the National Academy of Sciences of the United States of America. 2016;113:7858–7863. doi: 10.1073/pnas.1607769113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen IC, Scull MA, Moore CB, Holl EK, McElvania-TeKippe E, Taxman DJ, Guthrie EH, Pickles RJ, Ting JP. The NLRP3 inflammasome mediates in vivo innate immunity to influenza A virus through recognition of viral RNA. Immunity. 2009;30:556–565. doi: 10.1016/j.immuni.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskin CR, Bielefeldt-Ohmann H, Tumpey TM, Sabourin PJ, Long JP, Garcia-Sastre A, Tolnay AE, Albrecht R, Pyles JA, Olson PH, et al. Early and sustained innate immune response defines pathology and death in nonhuman primates infected by highly pathogenic influenza virus. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:3455–3460. doi: 10.1073/pnas.0813234106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer RN, Brighton LE, Mueller L, Xiang Z, Rager JE, Fry RC, Peden DB, Jaspers I. Influenza enhances caspase-1 in bronchial epithelial cells from asthmatic volunteers and is associated with pathogenesis. The Journal of allergy and clinical immunology. 2012;130:958–967. e914. doi: 10.1016/j.jaci.2012.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvier NM, Palese P. The biology of influenza viruses. Vaccine. 2008;26(Suppl 4):D49–53. doi: 10.1016/j.vaccine.2008.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti A, Banerjee S, Franchi L, Loo YM, Gale M, Jr, Nunez G, Silverman RH. RNase L activates the NLRP3 inflammasome during viral infections. Cell host & microbe. 2015;17:466–477. doi: 10.1016/j.chom.2015.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Calvo PA, Malide D, Gibbs J, Schubert U, Bacik I, Basta S, O’Neill R, Schickli J, Palese P, et al. A novel influenza A virus mitochondrial protein that induces cell death. Nature medicine. 2001;7:1306–1312. doi: 10.1038/nm1201-1306. [DOI] [PubMed] [Google Scholar]
- Cheong WC, Kang HR, Yoon H, Kang SJ, Ting JP, Song MJ. Influenza A Virus NS1 Protein Inhibits the NLRP3 Inflammasome. PloS one. 2015;10:e0126456. doi: 10.1371/journal.pone.0126456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiaretti A, Pulitano S, Barone G, Ferrara P, Romano V, Capozzi D, Riccardi R. IL-1 beta and IL-6 upregulation in children with H1N1 influenza virus infection. Mediators of inflammation. 2013;2013:495848. doi: 10.1155/2013/495848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cilloniz C, Shinya K, Peng X, Korth MJ, Proll SC, Aicher LD, Carter VS, Chang JH, Kobasa D, Feldmann F, et al. Lethal influenza virus infection in macaques is associated with early dysregulation of inflammatory related genes. PLoS pathogens. 2009;5:e1000604. doi: 10.1371/journal.ppat.1000604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clancy DM, Martin SJ. Getting a gRIP on Flu by Casting the DAI. Cell host & microbe. 2016;20:552–554. doi: 10.1016/j.chom.2016.10.013. [DOI] [PubMed] [Google Scholar]
- Crotta S, Davidson S, Mahlakoiv T, Desmet CJ, Buckwalter MR, Albert ML, Staeheli P, Wack A. Type I and type III interferons drive redundant amplification loops to induce a transcriptional signature in influenza-infected airway epithelia. PLoS pathogens. 2013;9:e1003773. doi: 10.1371/journal.ppat.1003773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diebold SS, Kaisho T, Hemmi H, Akira S, Reis e Sousa C. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science. 2004;303:1529–1531. doi: 10.1126/science.1093616. [DOI] [PubMed] [Google Scholar]
- Elliott EI, Sutterwala FS. Initiation and perpetuation of NLRP3 inflammasome activation and assembly. Immunological reviews. 2015;265:35–52. doi: 10.1111/imr.12286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez MV, Miller E, Krammer F, Gopal R, Greenbaum BD, Bhardwaj N. Ion efflux and influenza infection trigger NLRP3 inflammasome signaling in human dendritic cells. Journal of leukocyte biology. 2016;99:723–734. doi: 10.1189/jlb.3A0614-313RRR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Comella N, Tognazzi K, Brown LF, Dvorak HF, Kocher O. Cloning of DLM-1, a novel gene that is up-regulated in activated macrophages, using RNA differential display. Gene. 1999;240:157–163. doi: 10.1016/s0378-1119(99)00419-9. [DOI] [PubMed] [Google Scholar]
- Gurung P, Lukens JR, Kanneganti TD. Mitochondria: diversity in the regulation of the NLRP3 inflammasome. Trends Mol Med. 2015;21:193–201. doi: 10.1016/j.molmed.2014.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He WT, Wan H, Hu L, Chen P, Wang X, Huang Z, Yang ZH, Zhong CQ, Han J. Gasdermin D is an executor of pyroptosis and required for interleukin-1beta secretion. Cell research. 2015;25:1285–1298. doi: 10.1038/cr.2015.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Zeng MY, Yang D, Motro B, Nunez G. NEK7 is an essential mediator of NLRP3 activation downstream of potassium efflux. Nature. 2016;530:354–357. doi: 10.1038/nature16959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichinohe T, Lee HK, Ogura Y, Flavell R, Iwasaki A. Inflammasome recognition of influenza virus is essential for adaptive immune responses. The Journal of experimental medicine. 2009;206:79–87. doi: 10.1084/jem.20081667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichinohe T, Pang IK, Iwasaki A. Influenza virus activates inflammasomes via its intracellular M2 ion channel. Nature immunology. 2010;11:404–410. doi: 10.1038/ni.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichinohe T, Pang IK, Kumamoto Y, Peaper DR, Ho JH, Murray TS, Iwasaki A. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:5354–5359. doi: 10.1073/pnas.1019378108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichinohe T, Yamazaki T, Koshiba T, Yanagi Y. Mitochondrial protein mitofusin 2 is required for NLRP3 inflammasome activation after RNA virus infection. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:17963–17968. doi: 10.1073/pnas.1312571110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii KJ, Kawagoe T, Koyama S, Matsui K, Kumar H, Kawai T, Uematsu S, Takeuchi O, Takeshita F, Coban C, Akira S. TANK-binding kinase-1 delineates innate and adaptive immune responses to DNA vaccines. Nature. 2008;451:725–729. doi: 10.1038/nature06537. [DOI] [PubMed] [Google Scholar]
- Iwasaki A, Pillai PS. Innate immunity to influenza virus infection. Nature reviews Immunology. 2014;14:315–328. doi: 10.1038/nri3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jureka AS, Kleinpeter AB, Cornilescu G, Cornilescu CC, Petit CM. Structural Basis for a Novel Interaction between the NS1 Protein Derived from the 1918 Influenza Virus and RIG-I. Structure. 2015;23:2001–2010. doi: 10.1016/j.str.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser WJ, Upton JW, Mocarski ES. Receptor-interacting protein homotypic interaction motif-dependent control of NF-kappa B activation via the DNA-dependent activator of IFN regulatory factors. Journal of immunology. 2008;181:6427–6434. doi: 10.4049/jimmunol.181.9.6427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanneganti TD. Central roles of NLRs and inflammasomes in viral infection. Nature reviews Immunology. 2010;10:688–698. doi: 10.1038/nri2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanneganti TD, Body-Malapel M, Amer A, Park JH, Whitfield J, Franchi L, Taraporewala ZF, Miller D, Patton JT, Inohara N, Nunez G. Critical role for Cryopyrin/Nalp3 in activation of caspase-1 in response to viral infection and double-stranded RNA. The Journal of biological chemistry. 2006a;281:36560–36568. doi: 10.1074/jbc.M607594200. [DOI] [PubMed] [Google Scholar]
- Kanneganti TD, Ozoren N, Body-Malapel M, Amer A, Park JH, Franchi L, Whitfield J, Barchet W, Colonna M, Vandenabeele P, et al. Bacterial RNA and small antiviral compounds activate caspase-1 through cryopyrin/Nalp3. Nature. 2006b;440:233–236. doi: 10.1038/nature04517. [DOI] [PubMed] [Google Scholar]
- Kato H, Sato S, Yoneyama M, Yamamoto M, Uematsu S, Matsui K, Tsujimura T, Takeda K, Fujita T, Takeuchi O, Akira S. Cell type-specific involvement of RIG-I in antiviral response. Immunity. 2005;23:19–28. doi: 10.1016/j.immuni.2005.04.010. [DOI] [PubMed] [Google Scholar]
- Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nature immunology. 2010;11:373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- Kayagaki N, Stowe IB, Lee BL, O’Rourke K, Anderson K, Warming S, Cuellar T, Haley B, Roose-Girma M, Phung QT, et al. Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling. Nature. 2015;526:666–671. doi: 10.1038/nature15541. [DOI] [PubMed] [Google Scholar]
- Kochs G, Garcia-Sastre A, Martinez-Sobrido L. Multiple anti-interferon actions of the influenza A virus NS1 protein. Journal of virology. 2007;81:7011–7021. doi: 10.1128/JVI.02581-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuiken T, Riteau B, Fouchier RA, Rimmelzwaan GF. Pathogenesis of influenza virus infections: the good, the bad and the ugly. Current opinion in virology. 2012;2:276–286. doi: 10.1016/j.coviro.2012.02.013. [DOI] [PubMed] [Google Scholar]
- Kuriakose T, Man SM, Subbarao Malireddi RK, Karki R, Kesavardhana S, Place DE, Neale G, Vogel P, Kanneganti T-D. ZBP1/DAI is an innate sensor of influenza virus triggering the NLRP3 inflammasome and programmed cell death pathways. Science Immunology. 2016;1:aag2045–aag2045. doi: 10.1126/sciimmunol.aag2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuriakose T, Tripp RA, Watford WT. Tumor Progression Locus 2 Promotes Induction of IFNlambda, Interferon Stimulated Genes and Antigen-Specific CD8+ T Cell Responses and Protects against Influenza Virus. PLoS pathogens. 2015;11:e1005038. doi: 10.1371/journal.ppat.1005038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamkanfi M, Dixit VM. Mechanisms and functions of inflammasomes. Cell. 2014;157:1013–1022. doi: 10.1016/j.cell.2014.04.007. [DOI] [PubMed] [Google Scholar]
- Liu X, Zhang Z, Ruan J, Pan Y, Magupalli VG, Wu H, Lieberman J. Inflammasome-activated gasdermin D causes pyroptosis by forming membrane pores. Nature. 2016;535:153–158. doi: 10.1038/nature18629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupfer C, Kanneganti TD. The expanding role of NLRs in antiviral immunity. Immunological reviews. 2013;255:13–24. doi: 10.1111/imr.12089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupfer C, Malik A, Kanneganti TD. Inflammasome control of viral infection. Current opinion in virology. 2015;12:38–46. doi: 10.1016/j.coviro.2015.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupfer C, Thomas PG, Anand PK, Vogel P, Milasta S, Martinez J, Huang G, Green M, Kundu M, Chi H, et al. Receptor interacting protein kinase 2-mediated mitophagy regulates inflammasome activation during virus infection. Nature immunology. 2013;14:480–488. doi: 10.1038/ni.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man SM, Kanneganti TD. Gasdermin D: the long-awaited executioner of pyroptosis. Cell research. 2015a;25:1183–1184. doi: 10.1038/cr.2015.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man SM, Kanneganti TD. Regulation of inflammasome activation. Immunological reviews. 2015b;265:6–21. doi: 10.1111/imr.12296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal P, Berger SB, Pillay S, Moriwaki K, Huang C, Guo H, Lich JD, Finger J, Kasparcova V, Votta B, et al. RIP3 induces apoptosis independent of pronecrotic kinase activity. Molecular cell. 2014;56:481–495. doi: 10.1016/j.molcel.2014.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariathasan S, Weiss DS, Newton K, McBride J, O’Rourke K, Roose-Girma M, Lee WP, Weinrauch Y, Monack DM, Dixit VM. Cryopyrin activates the inflammasome in response to toxins and ATP. Nature. 2006;440:228–232. doi: 10.1038/nature04515. [DOI] [PubMed] [Google Scholar]
- Martinon F, Burns K, Tschopp J. The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Molecular cell. 2002;10:417–426. doi: 10.1016/s1097-2765(02)00599-3. [DOI] [PubMed] [Google Scholar]
- McAuley JL, Tate MD, MacKenzie-Kludas CJ, Pinar A, Zeng W, Stutz A, Latz E, Brown LE, Mansell A. Activation of the NLRP3 inflammasome by IAV virulence protein PB1-F2 contributes to severe pathophysiology and disease. PLoS pathogens. 2013;9:e1003392. doi: 10.1371/journal.ppat.1003392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriyama M, Chen IY, Kawaguchi A, Koshiba T, Nagata K, Takeyama H, Hasegawa H, Ichinohe T. The RNA- and TRIM25-Binding Domains of Influenza Virus NS1 Protein Are Essential for Suppression of NLRP3 Inflammasome-Mediated Interleukin-1beta Secretion. Journal of virology. 2016;90:4105–4114. doi: 10.1128/JVI.00120-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton K, Dugger DL, Wickliffe KE, Kapoor N, de Almagro MC, Vucic D, Komuves L, Ferrando RE, French DM, Webster J, et al. Activity of protein kinase RIPK3 determines whether cells die by necroptosis or apoptosis. Science. 2014;343:1357–1360. doi: 10.1126/science.1249361. [DOI] [PubMed] [Google Scholar]
- Nogusa S, Thapa RJ, Dillon CP, Liedmann S, Oguin TH, 3rd, Ingram JP, Rodriguez DA, Kosoff R, Sharma S, Sturm O, et al. RIPK3 Activates Parallel Pathways of MLKL-Driven Necroptosis and FADD-Mediated Apoptosis to Protect against Influenza A Virus. Cell host & microbe. 2016;20:13–24. doi: 10.1016/j.chom.2016.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong JD, Mansell A, Tate MD. Hero turned villain: NLRP3 inflammasome-induced inflammation during influenza A virus infection. Journal of leukocyte biology. 2016 doi: 10.1189/jlb.4MR0616-288R. [DOI] [PubMed] [Google Scholar]
- Pang IK, Iwasaki A. Inflammasomes as mediators of immunity against influenza virus. Trends in immunology. 2011;32:34–41. doi: 10.1016/j.it.2010.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichlmair A, Schulz O, Tan CP, Naslund TI, Liljestrom P, Weber F, Reis e Sousa C. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science. 2006;314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- Pinar A, Dowling JK, Bitto NJ, Robertson AB, Latz E, Stewart CR, Drummond GR, Cooper MA, McAuley JL, Tate MD, Mansell A. PB1-F2 derived from avian influenza a virus H7N9 induces inflammation via activation of the NLRP3 inflammasome. The Journal of biological chemistry. 2016 doi: 10.1074/jbc.M116.756379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto LH, Holsinger LJ, Lamb RA. Influenza virus M2 protein has ion channel activity. Cell. 1992;69:517–528. doi: 10.1016/0092-8674(92)90452-i. [DOI] [PubMed] [Google Scholar]
- Pirhonen J, Sareneva T, Julkunen I, Matikainen S. Virus infection induces proteolytic processing of IL-18 in human macrophages via caspase-1 and caspase-3 activation. European journal of immunology. 2001;31:726–733. doi: 10.1002/1521-4141(200103)31:3<726::aid-immu726>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- Pirhonen J, Sareneva T, Kurimoto M, Julkunen I, Matikainen S. Virus infection activates IL-1 beta and IL-18 production in human macrophages by a caspase-1-dependent pathway. Journal of immunology. 1999;162:7322–7329. [PubMed] [Google Scholar]
- Poeck H, Bscheider M, Gross O, Finger K, Roth S, Rebsamen M, Hannesschlager N, Schlee M, Rothenfusser S, Barchet W, et al. Recognition of RNA virus by RIG-I results in activation of CARD9 and inflammasome signaling for interleukin 1 beta production. Nature immunology. 2010;11:63–69. doi: 10.1038/ni.1824. [DOI] [PubMed] [Google Scholar]
- Pothlichet J, Meunier I, Davis BK, Ting JP, Skamene E, von Messling V, Vidal SM. Type I IFN triggers RIG-I/TLR3/NLRP3-dependent inflammasome activation in influenza A virus infected cells. PLoS pathogens. 2013;9:e1003256. doi: 10.1371/journal.ppat.1003256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebsamen M, Heinz LX, Meylan E, Michallet MC, Schroder K, Hofmann K, Vazquez J, Benedict CA, Tschopp J. DAI/ZBP1 recruits RIP1 and RIP3 through RIP homotypic interaction motifs to activate NF-kappaB. EMBO reports. 2009;10:916–922. doi: 10.1038/embor.2009.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid-Burgk JL, Chauhan D, Schmidt T, Ebert TS, Reinhardt J, Endl E, Hornung V. A Genome-wide CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) Screen Identifies NEK7 as an Essential Component of NLRP3 Inflammasome Activation. The Journal of biological chemistry. 2016;291:103–109. doi: 10.1074/jbc.C115.700492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma D, Kanneganti TD. The cell biology of inflammasomes: Mechanisms of inflammasome activation and regulation. The Journal of cell biology. 2016;213:617–629. doi: 10.1083/jcb.201602089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Wang Y, Li X, Zhan X, Tang M, Fina M, Su L, Pratt D, Bu CH, Hildebrand S, et al. NLRP3 activation and mitosis are mutually exclusive events coordinated by NEK7, a new inflammasome component. Nature immunology. 2016;17:250–258. doi: 10.1038/ni.3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Zhao Y, Wang K, Shi X, Wang Y, Huang H, Zhuang Y, Cai T, Wang F, Shao F. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature. 2015;526:660–665. doi: 10.1038/nature15514. [DOI] [PubMed] [Google Scholar]
- Silverman RH. Viral encounters with 2′,5′-oligoadenylate synthetase and RNase L during the interferon antiviral response. Journal of virology. 2007;81:12720–12729. doi: 10.1128/JVI.01471-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stasakova J, Ferko B, Kittel C, Sereinig S, Romanova J, Katinger H, Egorov A. Influenza A mutant viruses with altered NS1 protein function provoke caspase-1 activation in primary human macrophages, resulting in fast apoptosis and release of high levels of interleukins 1beta and 18. The Journal of general virology. 2005;86:185–195. doi: 10.1099/vir.0.80422-0. [DOI] [PubMed] [Google Scholar]
- Stout-Delgado HW, Vaughan SE, Shirali AC, Jaramillo RJ, Harrod KS. Impaired NLRP3 inflammasome function in elderly mice during influenza infection is rescued by treatment with nigericin. Journal of immunology. 2012;188:2815–2824. doi: 10.4049/jimmunol.1103051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaoka A, Wang Z, Choi MK, Yanai H, Negishi H, Ban T, Lu Y, Miyagishi M, Kodama T, Honda K, et al. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature. 2007;448:501–505. doi: 10.1038/nature06013. [DOI] [PubMed] [Google Scholar]
- Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- Tate MD, Ong JD, Dowling JK, McAuley JL, Robertson AB, Latz E, Drummond GR, Cooper MA, Hertzog PJ, Mansell A. Reassessing the role of the NLRP3 inflammasome during pathogenic influenza A virus infection via temporal inhibition. Scientific reports. 2016;6:27912. doi: 10.1038/srep27912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taubenberger JK, Morens DM. The pathology of influenza virus infections. Annual review of pathology. 2008;3:499–522. doi: 10.1146/annurev.pathmechdis.3.121806.154316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thapa RJ, Ingram JP, Ragan KB, Nogusa S, Boyd DF, Benitez AA, Sridharan H, Kosoff R, Shubina M, Landsteiner VJ, et al. DAI Senses Influenza A Virus Genomic RNA and Activates RIPK3-Dependent Cell Death. Cell host & microbe. 2016;20:674–681. doi: 10.1016/j.chom.2016.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas PG, Dash P, Aldridge JR, Jr, Ellebedy AH, Reynolds C, Funk AJ, Martin WJ, Lamkanfi M, Webby RJ, Boyd KL, et al. The intracellular sensor NLRP3 mediates key innate and healing responses to influenza A virus via the regulation of caspase-1. Immunity. 2009;30:566–575. doi: 10.1016/j.immuni.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari N, Kapoor P, Dhole TN. Antibody and inflammatory response-mediated severity of pandemic 2009 (pH1N1) influenza virus. Journal of medical virology. 2014;86:1034–1040. doi: 10.1002/jmv.23877. [DOI] [PubMed] [Google Scholar]
- Tzeng TC, Schattgen S, Monks B, Wang D, Cerny A, Latz E, Fitzgerald K, Golenbock DT. A Fluorescent Reporter Mouse for Inflammasome Assembly Demonstrates an Important Role for Cell-Bound and Free ASC Specks during In Vivo Infection. Cell reports. 2016;16:571–582. doi: 10.1016/j.celrep.2016.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Jiang W, Yan Y, Gong T, Han J, Tian Z, Zhou R. RNA viruses promote activation of the NLRP3 inflammasome through a RIP1-RIP3-DRP1 signaling pathway. Nature immunology. 2014a;15:1126–1133. doi: 10.1038/ni.3015. [DOI] [PubMed] [Google Scholar]
- Wang Z, Zhang A, Wan Y, Liu X, Qiu C, Xi X, Ren Y, Wang J, Dong Y, Bao M, et al. Early hypercytokinemia is associated with interferon-induced transmembrane protein-3 dysfunction and predictive of fatal H7N9 infection. Proceedings of the National Academy of Sciences of the United States of America. 2014b;111:769–774. doi: 10.1073/pnas.1321748111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO. Fact sheet Number 211. Influenza. 2003 ( http://www.who.int/mediacentre/factsheets/fs211/)
- Yoshizumi T, Ichinohe T, Sasaki O, Otera H, Kawabata S, Mihara K, Koshiba T. Influenza A virus protein PB1-F2 translocates into mitochondria via Tom40 channels and impairs innate immunity. Nature communications. 2014;5:4713. doi: 10.1038/ncomms5713. [DOI] [PubMed] [Google Scholar]