Abstract
A well-studied RNA-binding protein Hu Antigen-R (HuR), controls post-transcriptional gene regulation and undergoes stress-activated caspase-3 dependent cleavage in cancer cells. The cleavage products of HuR are known to promote cell death however, the underlying molecular mechanisms facilitating caspase-3 activation and HuR cleavage remains unknown. Here, we show that HuR cleavage associated with active caspase-3 in oral cancer cells treated with ionizing radiation and chemotherapeutic drug, paclitaxel. We determined that oral cancer cells overexpressing cyclooxygenase-2 (COX-2) limited the cleavage of caspase-3 and HuR, which reduced the rate of cell death in paclitaxel resistant oral cancer cells. Specific inhibition of COX-2 by celecoxib, promoted apoptosis through activation of caspase-3 and cleavage of HuR in paclitaxel-resistant oral cancer cells, both in vitro and in vivo. In addition, oral cancer cells overexpressing cellular HuR increased the half-life of COX-2 mRNA, promoted COX-2 protein expression and exhibited enhanced tumor growth in vivo in comparison with cells expressing a cleavable form of HuR. Finally, our ribonucleoprotein immunoprecipitation and sequencing (RIP-seq) analyses of HuR in oral cancer cells treated with ionizing radiation (IR), determined that HuR cleavage product-1 (HuR-CP1) bound and promoted the expression of mRNAs encoding proteins involved in apoptosis. Our results indicated that, cellular non-cleavable HuR controls COX-2 mRNA expression and enzymatic activity. In addition, overexpressed COX-2 protein repressed the cleavage of caspase-3 and HuR to promote drug resistance and tumor growth. Altogether, our observations support the use of the COX-2 inhibitor celecoxib, in combination with paclitaxel, for the management of paclitaxel resistant oral cancer cells.
Keywords: Oral squamous cell carcinoma, Cyclooxygenase-2, caspase-3, HuR cleavage, mRNA turnover, cell death
INTRODUCTION
Understanding gene expression patterns in response to chemoradiotherapy is crucial to the successful treatment of cancer. Recent emerging technologies permitted the development of targeted therapies for a variety of cancers1. However, most of these therapies target DNA transcription initiation factors or signaling kinases. Interestingly, studies demonstrated that during chemotherapy, post-transcriptional control of messenger RNA (mRNA) stability/turnover is altered, this alteration offers a promising target for the development of new therapeutics2. For instance, RNA-binding proteins (RBPs) thought to be involved in controlling the expression of mRNAs that encode proteins responsible for cell death, consequently altering treatment efficacy in cancer3–5. RBP-mediated cell death is considered an additional checkpoint for cancer treatment. However, a mechanistic link between RBP-mediated halting of cancer cell growth and efficacy of treatment has not been thoroughly studied.
Human antigen R (HuR), belongs to the family of ELAV (embryonic lethal, abnormal vision) proteins, known to regulate cellular development and immune functions6, 7. In cancer, HuR regulates the mRNAs encoding proteins involved in cell proliferation, angiogenesis, apoptosis and cell migration8. Although HuR is an extensively studied RBP, HuR mediated control of apoptosis in cancer is poorly understood. Under lethal cellular stress like hypoxia or ionizing radiation, caspase-3 cleaves HuR and the resulting HuR cleavage products promote cell death9–11.
The cellular apoptotic response is controlled by the intrinsic or extrinsic pathway, depending on the stress response. The intrinsic pathway is triggered by disruption of the mitochondrial membrane potential by Bcl protein family members and dysregulation of this pathway induces cancer12. In the extrinsic pathway, extracellular death ligands like Fas L13 bind to cell-surface receptors stimulating caspase-8 cleavage and subsequently caspase-3 activation. Caspases are inhibited by several proteins involved in cell proliferation14. For example, cyclooxygenases (COX), also known as prostaglandin G/H synthases, enzymatically convert arachidonic acid to prostaglandins and thromboxane A2. There are two known isoforms of COX enzymes: COX-1 (alias PTGS1) and COX-2 (alias PTGS2)15. COX-2 is overexpressed in several cancers, including oral squamous cell carcinoma16–22. Interestingly, overexpressed COX-2 blocked caspase-3 activation, and subsequent inhibition of COX-2 activity by celecoxib induced apoptosis23. HuR is known to control the expression of COX-2 mRNA in multiple cancers through interaction with AU-rich elements in the 3' untranslated region24–27. Although these observations support that HuR controls COX-2 expression in cancer, the molecular mechanism of HuR-mediated COX-2 promotion of tumor growth under chemoradiotherapy remain elusive. In the present study, we define a previously unidentified role for COX-2 in regulating caspase-3 and HuR cleavage in oral cancer cells. Here we show that, inhibition of COX-2 activity by celecoxib, promotes caspase-3 activation and HuR cleavage, which destabilizes COX-2 mRNA. Our observations suggest that disruption of the COX-2/HuR reciprocal feedback loop, can sensitize paclitaxel-resistant oral cancer cells to treatment.
Results
Cleavage of HuR is cancer cell specific
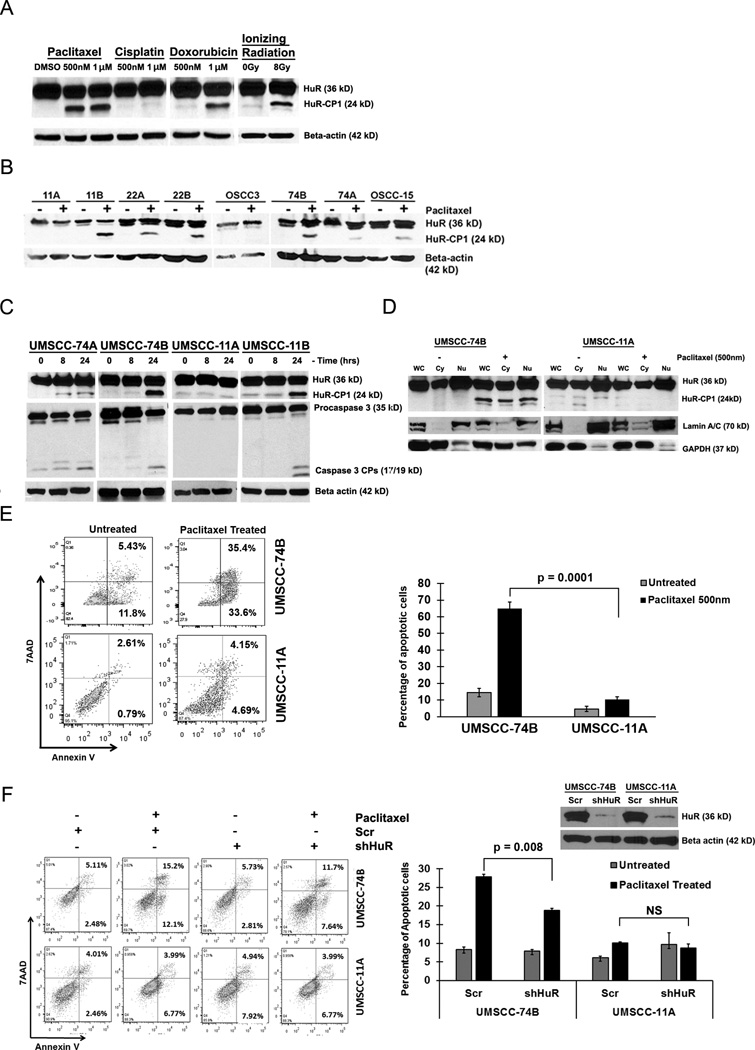
We previously showed that treatment of hypoxia and ionizing radiation (IR) promoted the cleavage of HuR through active caspase-39, 10. Here, in order to determine if chemo- and radio-therapy promoted the cleavage of HuR in drug resistant and sensitive oral cancer cells, we treated 74B oral cancer cells with a variety of cytotoxic agents and IR. Treatment of paclitaxel, doxorubicin and IR induced the cleavage of HuR; however, cisplatin did not promote HuR cleavage even at the highest concentration of 10µM (Figure 1A and S1A). Next, to extend our observation, we monitored the cleavage of HuR during paclitaxel treatment of primary (nomenclature of A) and recurrent (nomenclature of B) oral cancer cells. HuR was preferentially cleaved in the recurrent cell lines, compared to the primary cell lines; one exception was 22A cells which exhibited less cleavage of HuR compared to 22B cells (Figure 1B). Based on the pattern of cleavage of HuR, we selected two primary oral cancer cell lines UMSCC-11A and UMSCC-74A and two recurrent cell lines UMSCC-11B and UMSCC-74B for further analysis. To determine if the cleavage of HuR associated with active caspase-3, paclitaxel treated cells were probed for active caspase-3 and HuR. As shown in Figure 1C, recurrent cell lines 11B and 74B exhibited strong cleavage of HuR and caspase-3 compared to 11A cells. In contrast, primary 74A cells showed minimal HuR cleavage in association with activation of caspase-3 compared to 74B cells. Previous observations indicated that the cleavage of HuR is a cytoplasmic event controlled by active caspase-328, hence, we monitored the subcellular localization of HuR under paclitaxel. As shown in Figure 1D, presence of HuR was noted in both the cytoplasm and nucleus of 11A and 74B cells with or without treatment of paclitaxel. Although the cytoplasmic presence of HuR was observed in both 11A and 74B cells, the lack of active caspase-3 in 11A cells failed to cleave HuR compared to 74B cells under paclitaxel (Figure 1C–D). Collectively, these data suggest that paclitaxel mediated cleavage of HuR associated with active caspase-3 in cancer cells.
Figure 1. Caspase-3 mediated cleavage of HuR is cancer cell specific.
A) Western blot analysis of HuR cleavage patterns in UMSCC-74B cells treated with either DMSO, paclitaxel (0.5µM-1µM), cisplatin (1µM), doxorubicin (1µM) or 8Gy of ionizing radiation. B) Western blot analysis of HuR in multiple oral cancer cells treated with DMSO and paclitaxel (0.5µM). C) Western blot analysis of HuR and caspase-3 oral cancer cell lines treated with 0.5µM paclitaxel at the indicated time points. β-actin: loading control. D) Western blot analysis of HuR expression in nuclear/cytoplasmic fractionation during paclitaxel treatment. Lamin A/C (nuclear marker) and GAPDH (cytoplasmic marker). Representative western blots from three independent experiments are shown. E) Apoptotic rate of 74B and 11A cells treated with DMSO or 0.5µM paclitaxel for 24hrs. The percentage of early (bottom right quadrant) and late (top right quadrant) apoptotic cells are depicted in the scatter plots and bar graphs. Data represented as mean ± SD; N=3. F) Apoptotic rate between HuR silenced 74B and 11A cells under DMSO or 0.5µM paclitaxel treatment for 16hrs measured by flow cytometry. Data represented as mean ± SD; N=3.
Next, to determine whether the observed changes in caspase-3 and HuR altered the rate of cell death under paclitaxel treatment, the percentage of apoptotic cells were quantified. As shown in Figure 1E, paclitaxel treated primary 11A and 74A cells (Figure S1B) exhibited reduced rate of apoptosis (from ~3% to 9%, 74A cells exhibits ~15 to 25%, Figure S1B) compared to 74B cells (~17 % to 70%; Figure 1E). These data indicated that paclitaxel induced apoptosis in recurrent 74B cells was significantly higher than primary 11A and 74A oral cancer cells. To test if the altered apoptotic rates were mediated through HuR, we silenced HuR and quantified the number of apoptotic cells under paclitaxel. As shown in Figure 1F, silencing of HuR by shRNA reduced the rate of cell death in 74B cells, indicating that HuR partly facilitated apoptosis in paclitaxel treated cells. We did not detect a significant change in the apoptotic rate of 11A cells (Figure 1F), in part due to inactivation of caspase-3 under paclitaxel. Altogether, our findings indicated that paclitaxel induced caspase-3 activation, promoted HuR cleavage and enhanced apoptosis in recurrent cancer cells compared to primary oral cancer cells.
HuR controls COX-2 expression in oral cancer cells
Our previous observation demonstrated that the non-cleavable isoform of HuR (HuR-D226A), increased cell proliferation compared to the full-length or cleavable isoforms of HuR10. The presence of cellular non-cleavable HuR and inactive caspase-3 in primary tumor cells compared to recurrent cells during paclitaxel treatment suggested that non-cleavable HuR and inactive caspase-3 may facilitate cell proliferation and drug resistance (Figure 1C). To further assess this, we determined the levels of HuR target mRNAs encoding proteins involved in cell proliferation such as COX-2, c-MYC, c-JUN and IL-8 in 11A and 74B cells. These mRNAs contain AU-rich elements, HuR recognition sequence motifs, in their 3' untranslated region (UTR). Most HuR target mRNAs were down regulated in 74B cells during paclitaxel treatment (Figure 2A). In contrast, 11A cells exhibited reduced levels of c-MYC, c-JUN and IL-8 mRNAs but significant upregulation of COX-2 mRNA under paclitaxel treatment (Figure 2B). Next, to determine whether the observed changes in COX-2 and other mRNAs were due to direct association with HuR, we performed ribonucleoprotein (RNP) IP on control and paclitaxel-treated cells using an antibody against HuR. The HuR-associating mRNAs were then measured by RT-qPCR. As shown in Figure 2A and 2B, we observed a strong association of COX-2 mRNA with HuR in 11A cells (~68 fold) compared to 74B cells (~18 fold) after treatment with paclitaxel (Figure 2C, 74B cells and 2D, 11A cells). Interestingly, the basal COX-2 protein levels in 11A cells is ~3.0 fold higher than 74B cells (Figure 2E). In addition, like mRNA expression, the COX-2 protein levels were increased 2.0 fold in paclitaxel treated 11A cells compared to control cells. In contrast, COX-2 protein level showed a moderate decrease from 1.0 fold 0.9 fold after paclitaxel treatment in 74B cells (Figure 2E). The remaining mRNAs c-myc, c-Jun and IL-8 mRNAs were also bound with HuR in IP samples.
Figure 2. HuR controls the expression of COX-2 mRNA in primary and recurrent oral cancer cells.
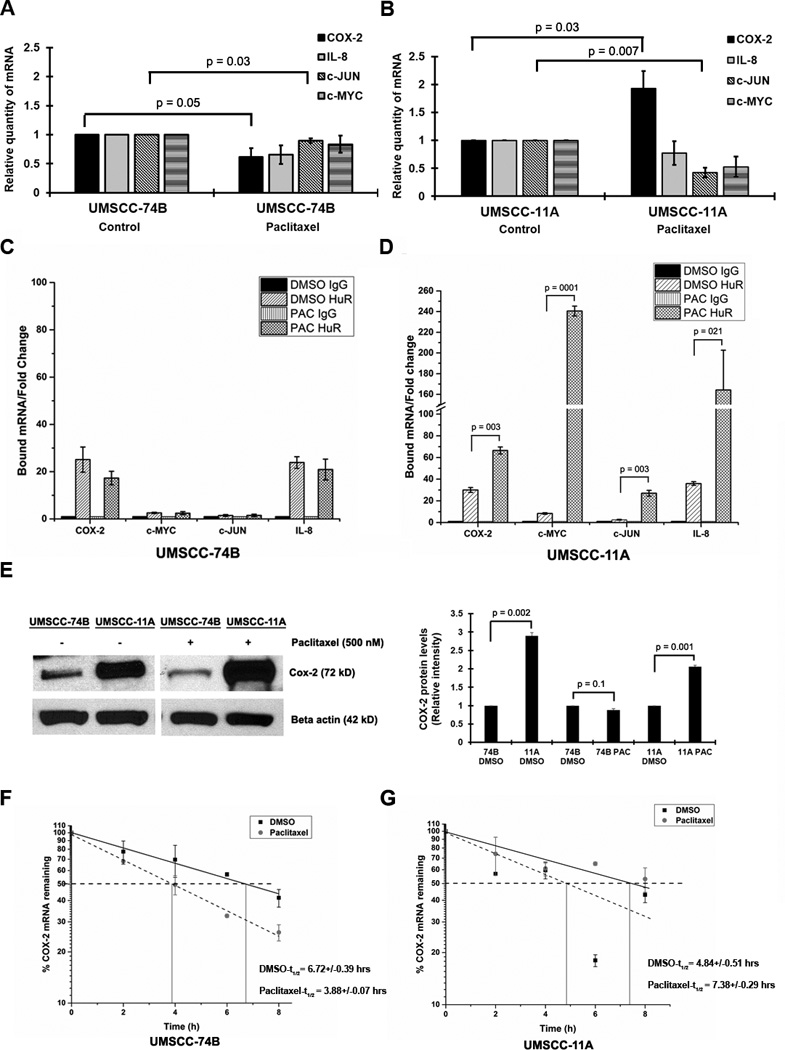
A–B) The relative quantity of indicated mRNAs measured by qRT-PCR after 0.5µM paclitaxel treatment (24hrs) of 74B and 11A cells. Data represented as mean ± SD; N=3. C–D) Protein lysates of 74B and 11A cells were subjected to RIP IP followed by qRT-PCR analysis to measure the relative quantities of indicated mRNAs in HuR IP compared control IgG IP. GAPDH served as a loading control. E) Cox-2 protein expression in 74B and 11A cells after DMSO or 0.5µM paclitaxel treatment for 24hrs β-actin: loading control. Quantification of COX-2 protein levels in DMSO or 0.5µM paclitaxel treated 74B and 11A cells. F–G) The decay rate of COX-2 mRNA in Paclitaxel treated 74B and 11A cells, respectively, was assessed by RT-qPCR after treatment with Actinomycin D. Values are the means ±SD (error bars) from three independent experiments.
Next, to determine if the alteration of COX-2 protein during paclitaxel treatment was due to changes in COX-2 mRNA stability, we treated both cell lines with paclitaxel and estimated the half-life (t1/2) of the mRNA under actinomycin-D (ActD). As shown in Figure 2F–G, after paclitaxel treatment COX-2 mRNA was unstable in 74B cells (t1/2 decreased from ~6:30 hrs to ~4 hrs) compared to 11A cells (t1/2 increased from ~5 hrs to ~ 7:20 hrs), indicating that paclitaxel promotes the stability of COX-2 mRNA in 11A cells. To directly assess whether COX-2 mRNA expression is controlled by HuR, we silenced HuR and evaluated COX-2 mRNA levels using RT-qPCR. The shRNA-mediated silencing of HuR reduced the relative expression of COX-2 mRNA and protein expression in both cell lines (Figure 3A–B). Next, the half-life of COX-2 mRNA was measured in HuR silenced 74B and 11A cells. In shcontrol 74B and 11A cells, COX-2 mRNA t1/2 was more than 6 hours, while in HuR silenced cells, the half-life was reduced to approximately 4 hours (Figure 3C–D), indicating that HuR directly controls COX-2 stability in both cell lines. Collectively, these data demonstrated that HuR is a dominant protein controlling COX-2 mRNA expression in cancer cells, but the observed difference in basal COX-2 protein expression between 74B and 11A cells may be due in part to additional regulatory pathways.
Figure 3. HuR regulates COX-2 expression.
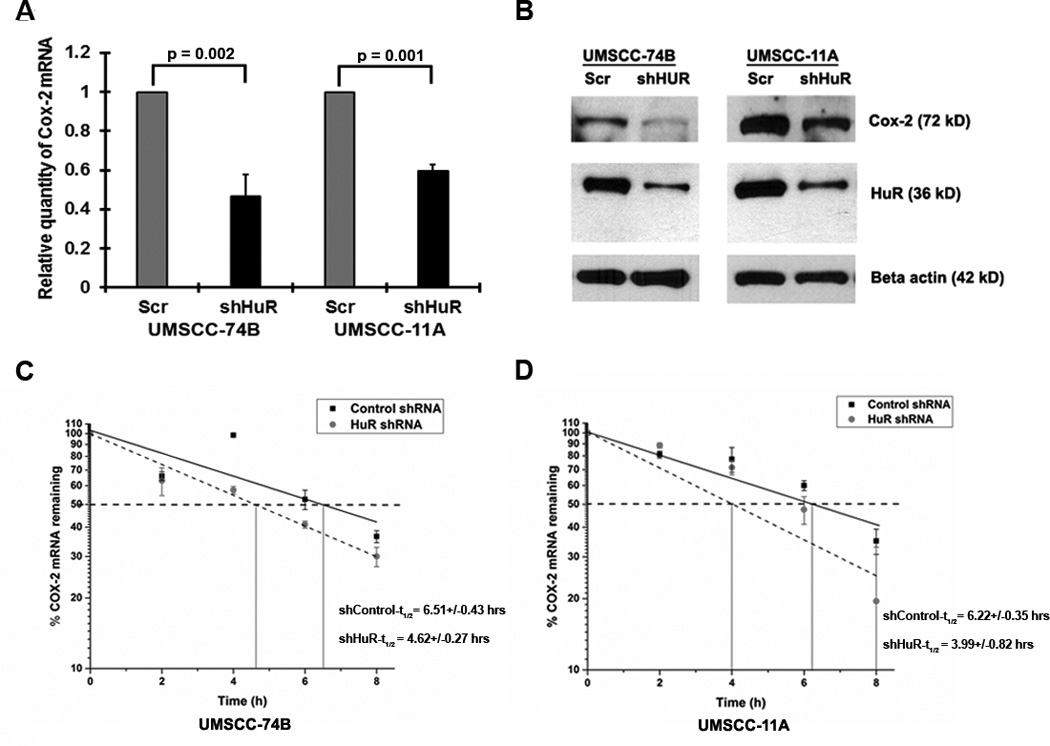
A) Relative quantity of COX-2 mRNA in 74B and 11A cells post-HuR knockdown. Data represented as mean ± SD; N=3. B) Cox-2 and HuR expression in 74B and 11A cells in HuR silenced cells. β-actin: loading control. C–D) The decay rate of COX-2 mRNA in HuR silenced 74B and 11A cells, respectively, was assessed by RT-qPCR after treatment with Actinomycin-D. Values are the means ±SD (error bars) from three independent experiments.
Inhibition of COX-2 by celecoxib promotes active caspase-3 and HuR cleavage
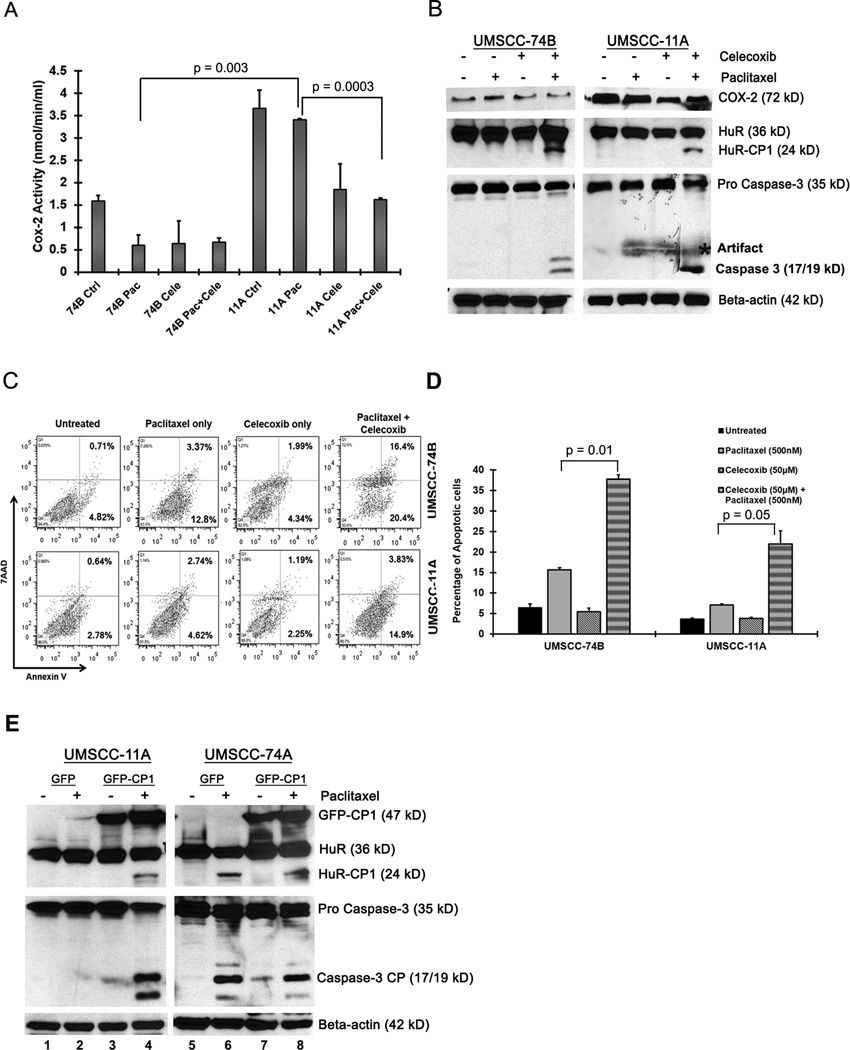
Increased COX-2 expression is correlated with tumor cell resistance to chemotherapeutic drugs29. Inhibition of COX-2 activity by celecoxib, resulted in increased apoptosis, activation of caspase-3, caspase-9 and PARP cleavage23. Hence, we sought to determine whether selective inhibition of COX-2 with celecoxib during paclitaxel treatment altered COX-2 activity, using a COX-2 enzymatic assay30. As shown in Figure 4A, basal COX-2 activity in 11A cells was significantly higher compared to 74B cells. Upon treatment with both celecoxib and paclitaxel, we observed a significant reduction in COX-2 activity in 11A cells in comparison to 11A cells treated with paclitaxel alone (Figure 4A). To determine if this combinatorial therapy in 11A cells induced caspase activation and HuR cleavage, the cells were treated for 2 hrs with paclitaxel and celecoxib, and protein expression was measured by Western blot. As expected, inhibition of COX-2 by celecoxib in association with paclitaxel treatment, promoted the cleavage of caspase-3 and HuR after 2 hours in 11A cells (Figure 4B). In addition, 74B cells treated with paclitaxel alone for 2 hours did not exhibit cleavage of HuR but both celecoxib and paclitaxel treatment promoted the cleavage of HuR (Figure 4B). In both cell lines, treatment of celecoxib alone failed to induce caspase-3 or HuR cleavage. To test whether the observed changes in the cleavage of caspase-3 and HuR altered the rate of apoptosis during celecoxib and paclitaxel treatment, we quantified the rate of apoptosis in both cell lines. As shown in Figure 4C, after 2 hours of celecoxib and paclitaxel treatment, the primary 11A cells exhibited increased apoptosis (~7% to ~18%) compared to paclitaxel alone (~3 % to ~7%; Figure 4C–D). In addition, the apoptotic rate in celecoxib and paclitaxel treated 74B cells increased (~15% to ~35%) compared to paclitaxel alone (Figure 4C–D). Collectively, these data suggest that combinatorial treatment with celecoxib and paclitaxel, is sufficient to induce HuR cleavage and activate apoptosis in COX-2 driven drug resistant oral cancer cells.
Figure 4. Celecoxib inhibits COX-2 enzymatic activity and promotes the cleavage of caspase-3 and HuR.
A) COX-2 enzyme activity was measured after treatment with DMSO, paclitaxel or paclitaxel/celecoxib (50µM) for 2 hrs. Data represented as mean ± SD; N=3. B) Western blot analysis of Cox-2, Caspase-3 and HuR in 74B and 11A cells treated with DMSO, paclitaxel or paclitaxel/celecoxib for 2 hrs. β-actin: loading control. Representative western blots from three independent experiments are shown. C–D) Measurement of apoptosis in 74B and 11A cells after DMSO, paclitaxel, celecoxib or paclitaxel/celecoxib treatment for 2 hrs. The percentage of early (bottom right quadrant) and late (top right quadrant) apoptotic cells are depicted in the scatter plots and bar graph. Data represented as mean ± SD; N=2. E) UMSCC-11A and 74A cells transiently over expressing GFP-CP1 were treated with DMSO or paclitaxel for 16 hrs. HuR and caspase-3 expression was analyzed by western blot. β-actin: loading control. Representative western blots of three independent experiments are shown.
It has been reported that overexpression of HuR cleavage products induced cell death under stress11. In order to investigate whether HuR cleavage product-1 and paclitaxel are sufficient to induce apoptosis by activating caspase-3 in primary oral cancer cells, we transiently overexpressed the cleaved HuR isoform GFP-CP1 in 11A and 74A cells. As expected, we observed cleavage of both HuR and caspase-3 in paclitaxel treated GFP-CP1 overexpressing 11A and 74A cells after 16hrs (lane 4 and 8 in Figure 4E); whereas, DMSO or paclitaxel treated GFP control cells and DMSO treated GFP-CP1 cells exhibited no cleavage of HuR and caspase-3 (lanes 1–3 and 5–7 in Figure 4E). These data demonstrate that overexpression of HuR-CP1 in combination with paclitaxel may overcome drug resistance in oral cancer cells.
COX-2 overexpression and non-cleavable HuR promotes oral cancer tumorigenesis
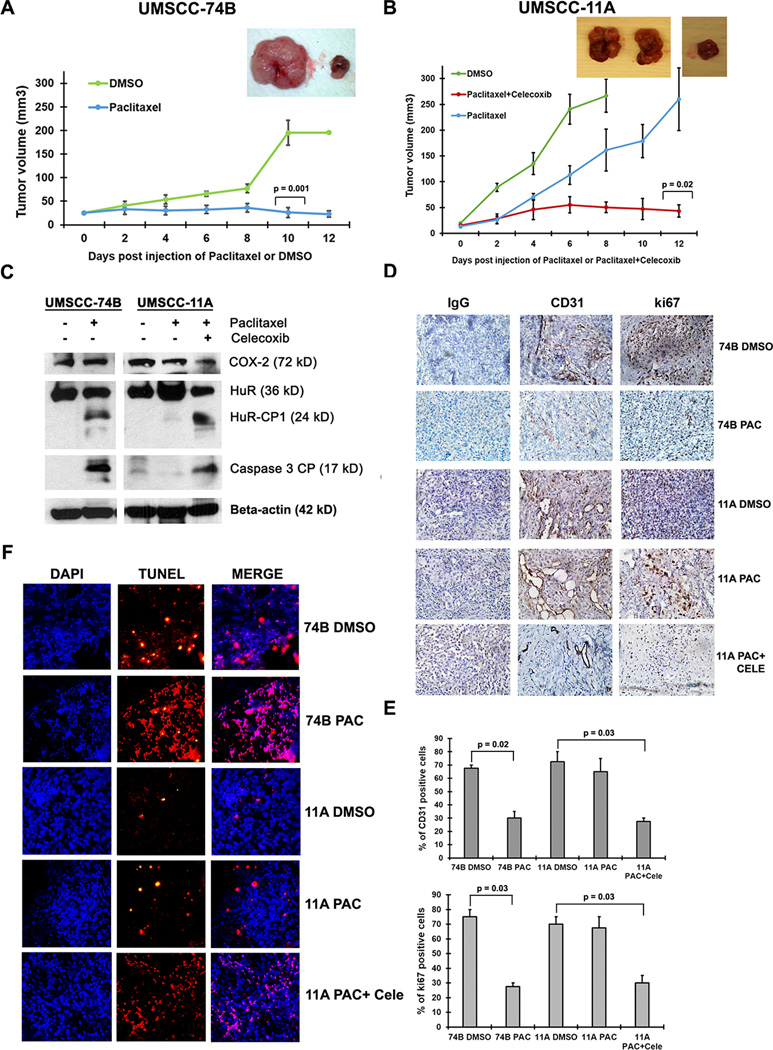
To gain insights into the cancer promoting and drug resistance function of COX-2 and HuR in vivo, we subcutaneously injected equal number of 11A or 74B cells into the right and left flank of 4 week old nude mice and monitored tumor growth. When the tumor volume reached 25 mm3, we injected 100µL of DMSO and 20mg/kg of paclitaxel in 100µL of DMSO intra-tumorally on the left and right flank of nude mice respectively, carrying either 11A or 74B cell derived xenograft tumors. The paclitaxel was injected once every two days for 2 weeks or until the tumors reached 200 to 250 mm3 in volume. Paclitaxel treated 74B derived xenograft tumors exhibited a reduction in growth after 10 days of treatment compared to DMSO treated tumors (Figure 5A). In contrast, there was no observed difference in tumor growth between DMSO and paclitaxel treated 11A derived xenograft tumors (Figure 5B). However, paclitaxel and celecoxib treated 11A xenograft tumors exhibited a reduction in growth after 12 days of treatment compared to paclitaxel alone treated tumors (Figure 5B). In agreement with our in vitro data, western blot analysis of paclitaxel treated 74B derived xenograft tumor samples showed HuR cleavage (Figure 5C), whereas, DMSO treated 74B or 11A tumors and paclitaxel treated 11A tumors exhibited no cleavage of HuR (Figure 5C). Next, we tested the cell proliferation marker Ki67 and angiogenesis marker CD31 in 74B and 11A derived xenograft tumors. Ki67 and CD31 staining was high in both 74B and 11A DMSO treated tumors and paclitaxel treated 11A tumors compared to paclitaxel treated 74B and paclitaxel/celecoxib treated 11A derived tumors (Figure 5D, quantitative representation illustrated in 5E). To detect apoptosis in the tumors, we stained the tissues with TUNEL and measured cell death. As shown in Figure 5F, both 74B paclitaxel and 11A paclitaxel/celecoxib treated tumors exhibited pronounced cell death compared to control tumors. Altogether, data from xenograft studies indicate that overexpression of COX-2 and non-cleavable HuR may control tumor growth and paclitaxel resistance of 11A cells in vivo.
Figure 5. COX-2 and non-cleavable HuR promotes tumorigenesis in vivo.
A–B) Growth of xenografts derived from 74B or 11A cells (4 million) after intra-tumoral injection of DMSO, paclitaxel (20mg/kg) or paclitaxel (20mg/kg)/celecoxib (20mg/kg) for 8–12 days. Data represented as mean ± SD; N=5. Representative tumor images (insert) from UMSCC-74B or UMSCC-11A cell derived xenografts. C) Total protein (60µg) from 74B and 11A xenografts treated with DMSO, paclitaxel (20mg/kg) or paclitaxel (20mg/kg)/celecoxib (20mg/kg) for 8–12 days and blots were probed for COX-2, HuR, active caspase-3 and β-actin. D) Immunohistochemical analysis of CD31 and Ki67 expression in 74B or 11A derived tumor xenografts tissues. E) Quantification of CD31 and Ki67 expression in 74B or 11A derived tumor xenograft tissues. Data represented as mean ± SD; N=2. F) Tumor sections of 74B and 11A xenografts treated with DMSO, paclitaxel (20mg/kg) or paclitaxel (20mg/kg)/celecoxib (20mg/kg) for 8–12 days were immunostained for TUNEL positive cells (red) and counterstained with DAPI (blue).
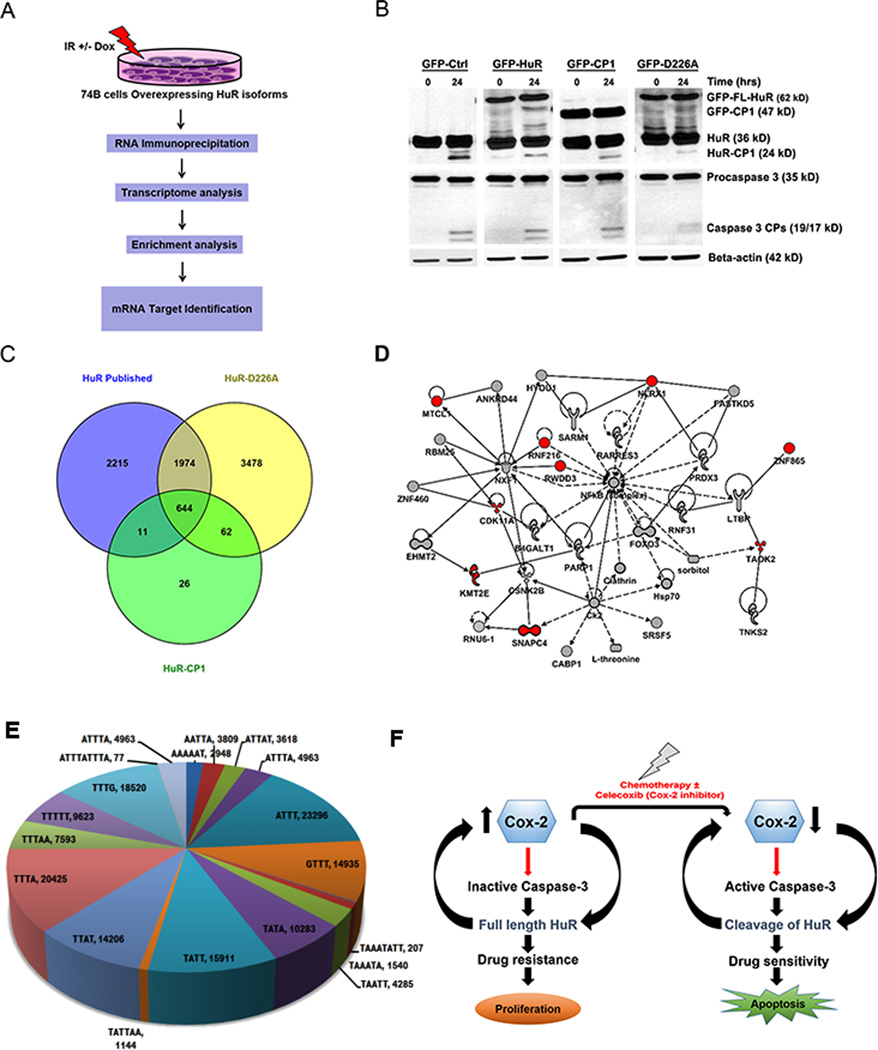
HuR cleavage product-1 (HuR-CP1) specifically associates with mRNAs encoding proteins involved in cell death
HuR-CP1 and CP2 are known to promote apoptosis by altering the expression of apoptosis-associated ARE containing mRNAs11. To determine whether full-length HuR, HuR-CP1 and non-cleavable form of HuR (HuR-D226A) forms a complex with target mRNAs under IR, we have used RNP IP assay followed by high-throughput sequencing (HTS). Figure 6A illustrates the methodology of RNP IP assays, HTS analysis and target enrichment analysis. First, 74B cells stably transfected with GFP-tagged HuR, HuR-CP1 and HuR-D226A isoforms were irradiated with ionizing radiation of 10Gy and cultured for 24 hours to promote cell death and cleavage of HuR (Figure 6B). IR induced the cleavage of GFP-HuR-FL but did not promote the cleavage of GFP-HuR-D226A compared to control GFP expressing cells (Figure 6B). Interestingly, overexpression of HuR-CP1 and HuR-FL minimizes the cleavage of cellular HuR possibly through competitive binding with caspase-3. Next, to determine if these isoforms of HuR bound with target mRNAs, we conducted RNP IP assay using anti-GFP antibody. The GFP-bound RNAs were extracted and subjected to HTS analysis. The mRNAs that were bound to GFP versus full-length HuR (Table S1), HuR-CP1 (Table S2) and HuR-D226A (Table S3) were compared with known HuR targets identified through PAR-CLIP31 (Figure 6C). As shown in Figure 6C, there are 644 target mRNAs bound to full-length HuR, HuR-CP1 and HuR-D226A isoforms, but more than 3000 targets associated with HuR-D226A isoform alone and only 26 targets were exclusively bound to HuR-CP1. These data suggest that HuR-D226A has an enhanced capability to bind RNA compared to HuR-CP1. Interestingly, HuR-CP1 associated with several target mRNAs encoding proteins noted to be involved in the cell death pathway (Table 1 and Figure 6D). Next, bioinformatic analysis revealed that mRNA sequences that bound with HuR contained several AREs in their 3'UTR (Figure 6E, S4). Thus, our findings indicate that HuR-CP1 specifically binds with mRNAs involved in cell death. Altogether, our model (Figure 6F) illustrates that the overexpression of COX-2, blocks the cleavage of caspase-3 and HuR, thus promoting drug resistance.
Figure 6. HuR-CP1 specifically associates with mRNAs encoding proteins involved in cell death.
A) Schematics showing the methodology used for identification of mRNA targets associated with different HuR isoforms in 74B cells post Ionizing radiation treatment. B) Western blot analysis of 74B cells stably expressing different isoforms of HuR post treatment with 10 Gy of Ionizing radiation. Total protein was extracted after 8hrs and the blots were probed for HuR, caspase 3 and β-actin. C) Venn diagram comparing mRNAs that are bound (more than 2 fold) to HuR-CP1, HuR-D226A and known HuR targets identified through PAR-CLIP47. D) Ingenuity Pathway analysis (IPA) network depicting HuR-CP1 associated mRNA targets involved in cell death pathway (as shown in Table 1). E) Pie-chart showing distribution of various AREs and HuR-binding sites in mRNA sequences. F) Model illustrating COX-2 control of HuR cleavage and drug resistance.
Table 1.
Ingenuity Pathways Analysis (IPA) summary. The interactions of differently regulated genes obtained from the datasets representing HuR, HuR-D226A and HuR-CP1 bound mRNA profiles obtained from the RNAseq imported into the Ingenuity Pathway Analysis.
| Biological Process | Diseases or Functions Annotations |
p-value | Target molecules |
|---|---|---|---|
| Cell Death and Survival |
apoptosis of basal epithelial prostate cells |
9.26E-04 | ING4 |
| Cell Death and Survival, Cellular Assembly and Organization |
formation of apoptotic bodies | 8.31E-03 | TAOK2 |
| Cell Death and Survival |
cell viability of myeloma cell lines | 2.97E-03 | CDK11A, TNK2 |
| Cell Death and Survival |
cell death of breast cell lines | 2.58E-03 | ING4, TNK2 |
| Cell Death and Survival |
anoikis of fibroblast cell lines | 1.29E-02 | CDK11A |
| Cell Death and Survival |
cell death of epithelial cell lines | 3.79E-02 | ING4, RNF216 |
Discussion
The regulation of post-transcriptional gene expression is always tightly associated with mRNA processing, transport, stability and translation. These processes are critical for precise control of gene expression and regulate several cancer biological pathways including cell death and proliferation. RBPs are the major determining factors that facilitate post-transcriptional gene expression. During cancer progression RBPs are post translationally modified and are involved in drug resistance. Here, we show that RBP HuR undergoes a post-translational modification which is integral to chemotherapeutic drug susceptibility in oral cancer cells. HuR control of COX-2 mRNA expression is well-known. Multiple studies demonstrated that HuR directly associates with AU-rich elements present in the 3' UTR of COX-2 and controls COX-2 stability and translation32. Overexpressed COX-2 protein is reported in multiple cancers, including colon,33 stomach,16 lung,17 esophagus,18 pancreas,18 bladder,34 prostate35 and oral cavity19–22. In oral cancer, expression of COX-2 is associated with tumor size and poor patient response to chemoradiation26. Based on our findings in this report, we establish that HuR controls COX-2 mRNA stability in both 11A and 74B cells. In addition, activated caspase-3 promoted the cleavage of HuR during paclitaxel treatment in 74B cells; however, 11A cells, which exhibits increased levels of COX-2 protein, failed to cleave both caspase-3 and HuR. Interestingly, in support of these observations, HuR was reported to enhance COX-2 mRNA stability in mammary epithelial cells treated with taxanes36. Altogether, our observations indicate that overexpression of COX-2 may play a positive role in cancer progression by promoting resistance to paclitaxel treatment, through blocking the activation and cleavage of caspase-3 and HuR, respectively.
Caspases are defined as either tumor suppressors or oncogenes by executing cell death in normal cells or undergo severe suppression in some cancers. For example, absence of caspase-3 was reported in multiple cancers including breast cancer tissues37, hepatocellular, and prostate carcinoma38. In addition, inactive caspase-3 was reported in both oral carcinoma and differentiated normal oral epithelial tissues39, but the molecular basis for this inactivation of caspase-3 was not known. Our results clearly indicate that overexpressed COX-2 inhibits the cleavage of caspase-3 and HuR in oral cancer cells. Taken together, the level of COX-2 and HuR can be used as an index for oral cancer patient prognosis and survival. Moreover, COX-2 and HuR may also be therapeutic targets for oral malignancy, particularly in patients who underwent chemotherapy that resulted in drug resistance.
HuR alters many biological functions in cancer cells which includes mediating apoptotic or anti-apoptotic responses.9, 40, 41. Post-translational modification of HuR may provide a mechanism to control the stability of mRNAs encoding proteins involved in the apoptosis pathway. For instance, HuR cleavage products are known to target several genes, such as Caspase-9, and control their mRNA stability and expression11. We assume that the apoptotic function of cleaved HuR in the form ofHuR-CP1, its associated target mRNAs and their coding proteins, may provide an opportunity to develop novel therapeutics. Based on our RNP IP analysis, we were able to identify HuR targets such as ING442, RNF21643 and CDK11A44, which are associated with apoptosis suggesting a novel mechanism whereby HuR-CP1 initiates cell death by controlling a subset of mRNAs encoding apoptotic proteins. Furthermore, we have previously shown that HuR-CP1, inhibits c-myc mRNA translation under hypoxic lethal stress10.- In addition, reports postulate that HuR-CP1 binds with multiple protein partners and controls the cell death pathway28, 40. As HuR-CP1 impacts the cellular HuR cleavage (Figure 6B), there is a strong possibility that HuR-CP1 could play a role in apoptotic pathway either as an initiator or competitor full length HuR to alter the cell death machinery. Hence, it would be interesting to identify HuR-CPs and their role in post-transcriptional gene regulation. Based on our RNP IP and transcriptome analysis, we were able to identify several novel HuR-CP1 regulons compared to full length HuR that could potentially impact apoptosis pathway.
Materials and Methods
Cell lines and constructs
The oral cancer cell lines were genotyped and authenticated as described45. The HuR isoforms tagged with GFP such as full-length HuR, HuR-D226A, HuR-CP1 (gift from Dr. Gallouzi laboratory, McGill University) have been previously described (27). The transfections of plasmids were conducted by using Lipofectamine-2000 (Life Technologies). For HuR transient silencing experiments, cells were transduced with either a scrambled shRNA or two different shRNAs targeting CDS and 3'UTR of HuR (Sigma clones: TRCN0000017274 and TRCN0000276186).
Immunoblotting and Apoptosis assay
The cell lysates were separated by SDS–PAGE and followed by immunoblotting using antibodies against HuR (Santa Cruz, sc-5261), caspase-3 (Cell Signaling, 8G10) and COX-2 (One World Lab, 43977). Apoptosis assay was carried out based on manufacturer’s protocol (Annexin V-PE apoptosis detection kit; Biovision, K128-100).
Cox-2 enzyme activity assay
Cox-2 enzyme activity was assayed according to the manufacturer’s protocol (Cayman Chemical Company, Michigan, USA). Briefly, either untreated, paclitaxel treated or paclitaxel and celecoxib treated cells were washed with HBSS and harvested with a rubber policeman. Cell pellets were sonicated in the presence of cold buffer (0.1M Tris-HCl, pH 7.8 containing 1mM EDTA) and centrifuged at 10,000 xg for 15 minutes at 4°C. The supernatant was used for the assay.
RNA Extraction, qRT-PCR and RNP analysis
Total RNA was prepared from mice tissues and HNSCC cell lines using TRIzol (Qiagen). The qRT-PCR was conducted using ABI Step One Plus system with the SYBR Green qRT-PCR kit. The sequences of the primers were shown in supplemental Table S1. HuR RNP IP was performed as previously described46.
RNA sequencing (RNA-seq) and analyses
100 ng of total RNA from RIP samples was used to prepare RNA-seq libraries using the TruSeq RNA Sample Prep Kit (Illumina, San Diego, CA), following the protocol described by the manufacturer. High throughput sequencing (HTS) was performed on an Illumina HiSeq2500 with each sample sequenced to a minimum depth of ~50 million reads using a paired end approach. The primary data analysis demanded that >80% of the sequence data had a Phred score of 30 in accordance with Illumina quality control (QC) procedures. Secondary data analysis involved an RNAseq workflow to process the data. This enabled data validation and quality control, read alignment to the human genome (hg19) using TopHat247, which indicated >93% read mapping. Gene-level count data was extracted with HTSeq and DEseq248 was utilized to uncover differential expression with robust statistical power. The workflow was executed on a OnRamp Bioinformatics Genomics Research Platform. Software versioning included FastQValidator v0.1.1a, Fastqc v0.11.3, Bowtie2 v2.1.0, TopHat2 v2.0.9, HTSeq v0.6.0 and DEseq v1.8.0. Transcript count data from the DESeq2 analysis of the samples was ranked based on the q-value from lowest to highest. FDR is the expected fraction of false positive tests among significant tests and was calculated using the Benjamini-Hochberg multiple testing adjustment procedure. Statistical analysis of pathways and gene ontology (GO) terms was carried as described by us previously49, 50. Venn diagrams were created using BioVenn52.
Accession numbers
Transcriptome fastq files have been deposited into GEO (Gene Expression Omnibus) and available using the GEO number GSE79477.
Experimental animals and xenograft studies
Four week old athymic nude mice (Medical University of South Carolina Animal Husbandry) were used for the transplantation studies. The animal research was handled in accordance with the guidelines of the Institutional Animal Care and Use Committee. Both UMSCC-74B and UMSCC-11A cells were harvested a brief exposure to 0.02% EDTA and 0.05% trypsin. After blocking trypsinization the cells were washed with HBSS, then resuspended in complete medium and counted with a hemocytomer. Only suspensions containing viable cells (>90%) were used for transplantation experiments. Approximately 4 million UMSCC-74B or UMSCC-11A cells were suspended in 100µl of complete medium containing 30% Matrigel (Corning) /and injected into the flank of the nude mice using a tuberculin syringe and 30-gauge needle, as described previously. A total of 16 mice (8 mice for 74B oral cancer cells group and 8 mice for 11A oral cancer cells group) received injection. Once in every two days, the weight of the mice and size of the tumors were measured. The formula V=1/2(L×W^2) was used to calculate and measure the tumor volume. The maximum tumor volume of 25 mm3 as a cut off for the injection of paclitaxel and celecoxib. Mice with same sized tumors were randomized into two groups (n=5) group 1 (control) received 100µl DMSO and group 2 (paclitaxel treatment) received 20mg/kg paclitaxel dissolved in DMSO and injected intra-tumorally. Using a 5% level of significance, 80% power, and a standard deviation in tumor volume within a group, the two-sided t-test states those 5 animals per group is sufficient to detect a difference in tumor volume between any groups.
ARE motif analysis
A basic R script based on the biomart R library was used to retrieve 3´UTR sequences. The longest 3´UTR sequences retrieved for each gene was scanned with a custom PERL script. The script performed a basic string match in scalar mode for ARE motifs in each 3´UTR. The script tracked the total number of 3´UTR matches and also counted the number of matches for each of the ARE motifs. These latter numbers were then charted with the standard Microsoft EXCEL 3D pie chart function.
Statistics
The graphical data are represented as mean ± S.D. We used student’s t tests with equal variances to assess differences between means. p values <0.05 were considered significant.
Supplementary Material
Acknowledgments
We acknowledge the funding source from National Institutes of Health R01DE022776 and R01DE022776S1 to VP. We acknowledge the Center for Oral Health Research (L-COHR) which is supported by the NIH grant P30 GM103331.
The abbreviations used are
- H&E
hematoxylin and eosin
- HuR-CP1
Hu Antigen R cleavage product-1
- HuR-D226A
non-cleavable isoform of HuR
- RBP
RNA-binding protein
- ARE
AU-rich elements
- 3′ UTR
untranslated region
- HNSCC
head and neck squamous cell carcinoma
- IR
ionizing radiation.
Footnotes
Conflict of Interest
We have no conflict of interest to declare.
References
- 1.Bombard Y, Bach PB, Offit K. Translating genomics in cancer care. J Natl Compr Canc Netw. 2013;11:1343–1353. doi: 10.6004/jnccn.2013.0158. [DOI] [PubMed] [Google Scholar]
- 2.Brody JR, Gonye GE. HuR's role in gemcitabine efficacy: an exception or opportunity? Wiley interdisciplinary reviews RNA. 2011;2:435–444. doi: 10.1002/wrna.62. [DOI] [PubMed] [Google Scholar]
- 3.Williams TK, Costantino CL, Bildzukewicz NA, Richards NG, Rittenhouse DW, Einstein L, et al. pp32 (ANP32A) expression inhibits pancreatic cancer cell growth and induces gemcitabine resistance by disrupting HuR binding to mRNAs. PloS one. 2010;5:e15455. doi: 10.1371/journal.pone.0015455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Costantino CL, Witkiewicz AK, Kuwano Y, Cozzitorto JA, Kennedy EP, Dasgupta A, et al. The role of HuR in gemcitabine efficacy in pancreatic cancer: HuR Up-regulates the expression of the gemcitabine metabolizing enzyme deoxycytidine kinase. Cancer Res. 2009;69:4567–4572. doi: 10.1158/0008-5472.CAN-09-0371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Filippova N, Yang X, Wang Y, Gillespie GY, Langford C, King PH, et al. The RNA-binding protein HuR promotes glioma growth and treatment resistance. Molecular cancer research : MCR. 2011;9:648–659. doi: 10.1158/1541-7786.MCR-10-0325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Papadaki O, Milatos S, Grammenoudi S, Mukherjee N, Keene JD, Kontoyiannis DL. Control of thymic T cell maturation, deletion and egress by the RNA-binding protein HuR. Journal of immunology. 2009;182:6779–6788. doi: 10.4049/jimmunol.0900377. [DOI] [PubMed] [Google Scholar]
- 7.Katsanou V, Milatos S, Yiakouvaki A, Sgantzis N, Kotsoni A, Alexiou M, et al. The RNA-binding protein Elavl1/HuR is essential for placental branching morphogenesis and embryonic development. Molecular and cellular biology. 2009;29:2762–2776. doi: 10.1128/MCB.01393-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abdelmohsen K, Gorospe M. Posttranscriptional regulation of cancer traits by HuR. Wiley interdisciplinary reviews RNA. 2010;1:214–229. doi: 10.1002/wrna.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Talwar S, House R, Sundaramurthy S, Balasubramanian S, Yu H, Palanisamy V. Inhibition of caspases protects mice from radiation-induced oral mucositis and abolishes the cleavage of RNA-binding protein HuR. J Biol Chem. 2014;289:3487–3500. doi: 10.1074/jbc.M113.504951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Talwar S, Jin J, Carroll B, Liu A, Gillespie MB, Palanisamy V. Caspase-mediated cleavage of RNA-binding protein HuR regulates c-Myc protein expression after hypoxic stress. J Biol Chem. 2011;286:32333–32343. doi: 10.1074/jbc.M111.255927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.von Roretz C, Lian XJ, Macri AM, Punjani N, Clair E, Drouin O, et al. Apoptotic-induced cleavage shifts HuR from being a promoter of survival to an activator of caspase-mediated apoptosis. Cell Death Differ. 2013;20:154–168. doi: 10.1038/cdd.2012.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fulda S, Debatin KM. Extrinsic versus intrinsic apoptosis pathways in anticancer chemotherapy. Oncogene. 2006;25:4798–4811. doi: 10.1038/sj.onc.1209608. [DOI] [PubMed] [Google Scholar]
- 13.Nagata S. Fas ligand-induced apoptosis. Annu Rev Genet. 1999;33:29–55. doi: 10.1146/annurev.genet.33.1.29. [DOI] [PubMed] [Google Scholar]
- 14.McIlwain DR, Berger T, Mak TW. Caspase functions in cell death and disease. Cold Spring Harb Perspect Biol. 2013;5:a008656. doi: 10.1101/cshperspect.a008656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang D, Dubois RN. Eicosanoids and cancer. Nat Rev Cancer. 2010;10:181–193. doi: 10.1038/nrc2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ristimaki A, Honkanen N, Jankala H, Sipponen P, Harkonen M. Expression of cyclooxygenase-2 in human gastric carcinoma. Cancer Res. 1997;57:1276–1280. [PubMed] [Google Scholar]
- 17.Hida T, Yatabe Y, Achiwa H, Muramatsu H, Kozaki K, Nakamura S, et al. Increased expression of cyclooxygenase 2 occurs frequently in human lung cancers, specifically in adenocarcinomas. Cancer Res. 1998;58:3761–3764. [PubMed] [Google Scholar]
- 18.Zimmermann KC, Sarbia M, Weber AA, Borchard F, Gabbert HE, Schror K. Cyclooxygenase-2 expression in human esophageal carcinoma. Cancer Res. 1999;59:198–204. [PubMed] [Google Scholar]
- 19.Itoh S, Matsui K, Furuta I, Takano Y. Immunohistochemical study on overexpression of cyclooxygenase-2 in squamous cell carcinoma of the oral cavity: its importance as a prognostic predictor. Oral Oncol. 2003;39:829–835. doi: 10.1016/s1368-8375(03)00105-2. [DOI] [PubMed] [Google Scholar]
- 20.Sakurai K, Urade M, Noguchi K, Hashitani S, Takaoka K, Segawa E, et al. Prognostic significance of cyclooxygenase-2 and DNA topoisomerase IIalpha expression in oral carcinoma. Head Neck. 2007;29:1002–1009. doi: 10.1002/hed.20627. [DOI] [PubMed] [Google Scholar]
- 21.Renkonen J, Wolff H, Paavonen T. Expression of cyclo-oxygenase-2 in human tongue carcinoma and its precursor lesions. Virchows Arch. 2002;440:594–597. doi: 10.1007/s00428-002-0616-y. [DOI] [PubMed] [Google Scholar]
- 22.Sudbo J, Ristimaki A, Sondresen JE, Kildal W, Boysen M, Koppang HS, et al. Cyclooxygenase-2 (COX-2) expression in high-risk premalignant oral lesions. Oral Oncol. 2003;39:497–505. doi: 10.1016/s1368-8375(03)00012-5. [DOI] [PubMed] [Google Scholar]
- 23.Dandekar DS, Lopez M, Carey RI, Lokeshwar BL. Cyclooxygenase-2 inhibitor celecoxib augments chemotherapeutic drug-induced apoptosis by enhancing activation of caspase-3 and-9 in prostate cancer cells. Int J Cancer. 2005;115:484–492. doi: 10.1002/ijc.20878. [DOI] [PubMed] [Google Scholar]
- 24.Barbisan F, Mazzucchelli R, Santinelli A, Lopez-Beltran A, Cheng L, Scarpelli M, et al. Overexpression of ELAV-like protein HuR is associated with increased COX-2 expression in atrophy, high-grade prostatic intraepithelial neoplasia, and incidental prostate cancer in cystoprostatectomies. Eur Urol. 2009;56:105–112. doi: 10.1016/j.eururo.2008.04.043. [DOI] [PubMed] [Google Scholar]
- 25.Do SI, Araujo ES, Kalil RK, Bacchini P, Bertoni F, Unni KK, et al. Expression of embryonic lethal abnormal vision (ELAV)-like protein HuR and cyclooxygenase-2 (COX-2) in Ewing sarcoma. Tumori. 2008;94:347–350. doi: 10.1177/030089160809400310. [DOI] [PubMed] [Google Scholar]
- 26.Mohammad S, Ram H, Gupta PN, Husain N, Bhatt ML. Overexpression of COX-2 in oral squamous cell carcinoma patients undergoing chemoradiotherapy. Natl J Maxillofac Surg. 2011;2:17–21. doi: 10.4103/0975-5950.85848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Niesporek S, Kristiansen G, Thoma A, Weichert W, Noske A, Buckendahl AC, et al. Expression of the ELAV-like protein HuR in human prostate carcinoma is an indicator of disease relapse and linked to COX-2 expression. Int J Oncol. 2008;32:341–347. [PubMed] [Google Scholar]
- 28.Mazroui R, Di Marco S, Clair E, von Roretz C, Tenenbaum SA, Keene JD, et al. Caspase-mediated cleavage of HuR in the cytoplasm contributes to pp32/PHAP-I regulation of apoptosis. J Cell Biol. 2008;180:113–127. doi: 10.1083/jcb.200709030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ferrandina G, Ranelletti FO, Martinelli E, Paglia A, Zannoni GF, Scambia G. Cyclo-oxygenase-2 (Cox-2) expression and resistance to platinum versus platinum/paclitaxel containing chemotherapy in advanced ovarian cancer. BMC Cancer. 2006;6:182. doi: 10.1186/1471-2407-6-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kulmacz RJ, Lands WE. Requirements for hydroperoxide by the cyclooxygenase and peroxidase activities of prostaglandin H synthase. Prostaglandins. 1983;25:531–540. doi: 10.1016/0090-6980(83)90025-4. [DOI] [PubMed] [Google Scholar]
- 31.Mukherjee N, Corcoran DL, Nusbaum JD, Reid DW, Georgiev S, Hafner M, et al. Integrative regulatory mapping indicates that the RNA-binding protein HuR couples pre-mRNA processing and mRNA stability. Mol Cell. 2011;43:327–339. doi: 10.1016/j.molcel.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Young LE, Dixon DA. Posttranscriptional Regulation of Cyclooxygenase 2 Expression in Colorectal Cancer. Curr Colorectal Cancer Rep. 2010;6:60–67. doi: 10.1007/s11888-010-0044-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kargman SL, O'Neill GP, Vickers PJ, Evans JF, Mancini JA, Jothy S. Expression of prostaglandin G/H synthase-1 and-2 protein in human colon cancer. Cancer Res. 1995;55:2556–2559. [PubMed] [Google Scholar]
- 34.Shirahama T. Cyclooxygenase-2 expression is up-regulated in transitional cell carcinoma and its preneoplastic lesions in the human urinary bladder. Clin Cancer Res. 2000;6:2424–2430. [PubMed] [Google Scholar]
- 35.Yoshimura R, Sano H, Masuda C, Kawamura M, Tsubouchi Y, Chargui J, et al. Expression of cyclooxygenase-2 in prostate carcinoma. Cancer. 2000;89:589–596. [PubMed] [Google Scholar]
- 36.Subbaramaiah K, Marmo TP, Dixon DA, Dannenberg AJ. Regulation of cyclooxgenase-2 mRNA stability by taxanes: evidence for involvement of p38, MAPKAPK-2 and HuR. J Biol Chem. 2003;278:37637–37647. doi: 10.1074/jbc.M301481200. [DOI] [PubMed] [Google Scholar]
- 37.Devarajan E, Sahin AA, Chen JS, Krishnamurthy RR, Aggarwal N, Brun AM, et al. Down-regulation of caspase 3 in breast cancer: a possible mechanism for chemoresistance. Oncogene. 2002;21:8843–8851. doi: 10.1038/sj.onc.1206044. [DOI] [PubMed] [Google Scholar]
- 38.Olsson M, Zhivotovsky B. Caspases and cancer. Cell Death Differ. 2011;18:1441–1449. doi: 10.1038/cdd.2011.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hague A, Eveson JW, MacFarlane M, Huntley S, Janghra N, Thavaraj S. Caspase-3 expression is reduced, in the absence of cleavage, in terminally differentiated normal oral epithelium but is increased in oral squamous cell carcinomas and correlates with tumour stage. J Pathol. 2004;204:175–182. doi: 10.1002/path.1630. [DOI] [PubMed] [Google Scholar]
- 40.Beauchamp P, Nassif C, Hillock S, van der Giessen K, von Roretz C, Jasmin BJ, et al. The cleavage of HuR interferes with its transportin-2-mediated nuclear import and promotes muscle fiber formation. Cell Death Differ. 2010 doi: 10.1038/cdd.2010.34. [DOI] [PubMed] [Google Scholar]
- 41.von Roretz C, Gallouzi IE. Protein kinase RNA/FADD/caspase-8 pathway mediates the proapoptotic activity of the RNA-binding protein human antigen R (HuR) J Biol Chem. 2010;285:16806–16813. doi: 10.1074/jbc.M109.087320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li M, Zhu Y, Zhang H, Li L, He P, Xia H, et al. Delivery of inhibitor of growth 4 (ING4) gene significantly inhibits proliferation and invasion and promotes apoptosis of human osteosarcoma cells. Sci Rep. 2014;4:7380. doi: 10.1038/srep07380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen D, Li X, Zhai Z, Shu HB. A novel zinc finger protein interacts with receptor-interacting protein (RIP) and inhibits tumor necrosis factor (TNF)- and IL1-induced NF-kappa B activation. J Biol Chem. 2002;277:15985–15991. doi: 10.1074/jbc.M108675200. [DOI] [PubMed] [Google Scholar]
- 44.Lahti JM, Xiang J, Heath LS, Campana D, Kidd VJ. PITSLRE protein kinase activity is associated with apoptosis. Mol Cell Biol. 1995;15:1–11. doi: 10.1128/mcb.15.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brenner JC, Graham MP, Kumar B, Saunders LM, Kupfer R, Lyons RH, et al. Genotyping of 73 UM-SCC head and neck squamous cell carcinoma cell lines. Head Neck. 2010;32:417–426. doi: 10.1002/hed.21198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lopez de Silanes I, Zhan M, Lal A, Yang X, Gorospe M. Identification of a target RNA motif for RNA-binding protein HuR. Proc Natl Acad Sci U S A. 2004;101:2987–2992. doi: 10.1073/pnas.0306453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lebedeva S, Jens M, Theil K, Schwanhausser B, Selbach M, Landthaler M, et al. Transcriptome-wide Analysis of Regulatory Interactions of the RNA-Binding Protein HuR. Mol Cell. 2011;43:340–352. doi: 10.1016/j.molcel.2011.06.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.