Abstract
The Na+/H+ antiporters (NHXs) are secondary ion transporters to exchange H+ and transfer the Na+ or K+ across membrane, they play crucial roles during plant development and stress responses. To gain insight into the functional divergence of NHX genes in poplar, eight PtNHX were identified from Populus trichocarpa genome. PtNHXs containing 10 transmembrane helices (TMH) and a hydrophilic C-terminal domain, the TMH compose a hollow cylinder to provide the channel for Na+ and H+ transport. The expression patterns and cis-acting elements showed that all the PtNHXs were response to single or multiple stresses including drought, heat, cold, salinity, MV, and ABA. Both the co-expression network and protein-protein interaction network of PtNHXs implying their functional divergence. Interestingly, although PtNHX7 and PtNHX8 were generated by whole genome duplication event, they showed significant differences in expression pattern, protein structure, co-expressed genes, and interacted proteins. Only PtNHX7 interact with CBL and CIPK, indicating PtNHX7 is the primary NHX involved in CBL-CIPK pathway during salt stress responses. Natural variation analysis based on 549 P. trichocarpa individuals indicated the frequency of SNPs in PtNHX7 was significantly higher than other PtNHXs. Our findings provide new insights into the functional divergence of NHX genes in poplar.
Introduction
Ion and pH homeostasis play important regulatory roles in cellular processes controlling plant growth and development. The optimal ion and pH gradients are dependent on H+-translocating enzymes (H+-pumps) and cation/H+ exchangers1, 2. Among the numerous transporters in monovalent cation/proton antiporter (CPA1) family, the Na+/H+ antiporters (NHXs) are secondary ion transporters to exchange H+ and transfer the Na+ or K+ across membrane3. To date, all the sequenced eukaryotes containing multiple NHX-like proteins, which were designated as Na+/H+ exchangers (NHEs), except for yeast only contains a single NHX4. Based on their subcellular localization, NHXs are classified into three major classes: plasma membrane (PM)-class, endosomal (Endo)-class, and vacuole (Vac)-class5. In mammalian, the specialized subcellular functions of NHEs are dependent on their organelle-specific distribution6.
In Arabidopsis, NHXs are consisting of six intracellular NHXs (1–6) and two plasma membrane bound NHXs (7 and 8). Among the six intracellular NHXs, four (AtNHX1–4) belong to Vac-class and two (AtNHX5, 6) belong to Endo-class NHX4. Based on the biochemical and kinetic analyses, NHEs containing 10–12 transmembrane domains and they may function as homodimers6. Although AtNHX1 differ from other known NHX-like antiporters in topological feature, it still contains 10–12 transmembrane domains according to the hydrophobicity analysis7.
NHXs involve in various biological processes such as salt stress response8, 9, pH homeostasis10, 11, K+ homeostasis12, 13, cell expansion9, 14, cellular vesicle trafficking15, 16. The Arabidopsis nhx1 nhx2 double mutant seedlings showed significantly reduced growth compared with either single mutant or wild-type plants. Microscopy observation indicated that cell expansion was reduced in all tissues of nhx1 nhx2 double mutant, especially in rapidly elongating organs, e.g. flower filaments and etiolated hypocotyls17. In addition, the nhx1 nhx2 double mutant sensitive to external K+ and showed curled root, poor expanded and yellow leaves17, 18. The Endo-class NHXs (NHX5 and NHX6) localized in the trans-Golgi network (TGN) and they played crucial regulatory roles in vesicle trafficking, especially to the vacuole9. Under salt stress, the high concentration of extracellular Na+ favouring Na+ influx into the cell. When the Na+ was accumulated to detrimental level, the PM-class NHX (NHX7/SOS1) actively extrude Na+ out of the cell, and the Vac-class NHX mediate the sequestration of Na+ into vacuole12, 19, 20.
Because of the economic importance in pulp and biofuel production21 and the completion of whole genome sequence22, Populus has been to hotspot for gene functional studies in woody species. In previous study, six putative Vac-class NHXs were identified from a high salinity tolerant poplar species, P. euphratica 23. To reveal the functional divergence of NHXs in P. trichocarpa, we performed a systematic analysis including phylogenetic relationships, gene structures, conserved protein domains, protein structures, cis-acting elements, expression compendium, co-expression network and protein-protein interaction network. Our results provide solid foundation for the study of the evolution and functions of NHX genes in poplar.
Materials and Methods
Characteristics and phylogenetic analysis of Populus NHXs
To identify Populus NHX genes, the published Arabidopsis and rice NHX protein sequences were searched against the P. trichocarpa (Pt) genome (http://phytozome.jgi.doe.gov/pz/portal.html#) through tBLASTn. All homologous protein sequences of the NHX candidates were accepted if they were satisfied with the expectation value (E) < 10–40. To further confirm the transmembrane helices (TMH) in NHX, the candidate sequences were scanned with TMHMM v.2.0 (http://www.cbs.dtu.dk/services/TMHMM/). Multiple sequences alignment of NHX protein sequences from P. trichocarpa and other species were performed using the Clustal X2.124. Phylogenetic tree was constructed using maximum likelihood (ML) method by PhyML V3.0 with 1,000 bootstrap replicates25.
Chromosome location and bioinformatics analyses
The chromosome locations of Populus NHX genes were analyzed based on the location sites on Populus chromosomes. Location information of Arabidopsis NHX genes were obtained from TAIR (https://www.arabidopsis.org/). The gene structure (exon and intron) was showed using Gene Structure Display Server (GSDS, http://gsds.cbi.pku.edu.cn). The duplication events were analyzed based on the synteny blocks from the Plant Genome Duplication Database (PGDD, http://chibba.agtec.uga.edu/duplication/). The conserved motifs were identified using MEME (http://meme-suite.org/tools/meme). For cis-acting elements, −1,500 nt upstream to + 500 nt downstream of the transcription start site (TSS) were analyzed by PlantCARE26. The PtNHX proteins were modelled using I-TASSER27.
Calculation of Ka/Ks
The PAL2NAL program (http://www.bork.embl.de/pal2nal/) was used to estimate synonymous (Ks) and nonsynonymous (Ka) substitution rates. The window size 60 bp and 90 bp were used for sliding window analysis. Divergence time (T) was calculated using the formula T = Ks/2λ (λ = 9.1 × 10–9 for Populus)28.
Plant material, abiotic stresses, and RT-PCR analysis
Two-month-old P. trichocarpa grown at 23–25 °C under long-day condition (16 h/8 h light/dark). Six tissues including shoot apical meristem (SAM), young leaves (YL), mature leaves (ML), primary stems (PS), secondary stems (SS), and roots (R) were collected for RT-PCR. For abiotic stresses or hormone treatment, the seedlings were treated with 10% polyethylene glycol (PEG, for drought stress), 37 °C (for heat stress), 4 °C (for cold stress), 150 mM NaCl (for salt stress), 100 μM methyl viologen (MV, for oxidative stress) or 100 μM abscisic acid (ABA) and the first fully expanded leaves were used for further study. The dosages of the abiotic stresses and hormone treatment were determined based on treatments in poplar29, 30. For different tissues and various stresses, three biological replicates were performed.
Total RNA was extracted using the RNeasy Plant Mini Kit (Qiagen), the RNase-free DNase I (Qiagen) was used to remove genomic DNA. First-strand cDNA was synthesized with ~1 μg RNA using the SuperScript III reverse transcription kit (Invitrogen). Primers were designed using Primer3 software (http://frodo.wi.mit.edu/primer3/input.htm) with melting temperature of 58–60 °C and production of 100–250 bp. The primers used in this study are listed in Supplementary Table S2. qRT-PCR was performed using SYBR Premix Taq Kit (TaKaRa, Dalian, China) and conducted on LightCycler 480 Detection System (Roche, Penzberg, Germany). The PtActin (Potri.019G010400) was used as internal control.
The expression data of Arabidopsis NHX genes was download from AtGenExpress database (http://jsp.weigelworld.org/expviz/expviz.jsp). Arabidopsis seedlings were treated under various stresses, such as heat (38 °C and recovered at 25 °C), cold (4 °C), salt (150 mM NaCl), osmotic stress (300 mM mannitol), genotoxic stress (1.5 µg/ml bleomycin +22 µg/ml mitomycin), oxidative stress (10 µM MV), wounding stress (punctured with pins), or 10 µM ABA. For drought stress, Arabidopsis seedlings were stressed by 15 min dry air stream (clean bench) until 10% loss of fresh weight, then incubation in closed vessels in the climate chamber.
Integrated network analyses
The co-expression data of PtNHXs and AtNHXs was obtained from Phytozome (https://phytozome.jgi.doe.gov/pz/portal.html). For the Populus genome-wide co-expression network construction, transcriptome data from 24 various P. trichocarpa tissues including five buds (predormant bud I, predormant bud II, early dormant bud, late dormant bud, and fully open bud), three leaves (immature leaf, young leaf, and first fully expanded leaf), five stems (node, inode, ammonia treatment, nitrate treatment, and urea treatment), five roots (root tip, standard root, ammonia treatment, nitrate treatment, and urea treatment), three male catkins (early from GW9592.ZK, early from GW9840.ZE, and mid from GW9911.ZK), and three female catkins (early from BESC423.ZL, late from BESC842, and receptive from BESC442.ZG) were used to construct the genome-wide co-expression network. The PtNHXs co-expression networks were constructed based on the Pearson correlations (>0.85). The protein-protein interaction (PPI) data of PtNHXs was obtained from the STRING database (http://string-db.org)31. Cytoscape32 was used to visualize the resulting networks.
Statistical analysis
The statistical analysis was performed using Duncan test and Fisher’s protected least significant difference (LSD) test of DPS (Zhejiang University, China) at 0.05 probability levels.
Results
Genome-wide identification of NHX genes in P. trichocarpa
To identify the Populus NHX genes, a tBLASTn search was performed and total of eight NHX genes were identified from P. trichocarpa genome. These genes were named according to their sequence similarity to the NHX genes in Arabidopsis and rice. All the information of the eight PtNHX genes, including gene locus, gene length, and deduced amino acids, was shown in Table 1. The protein length PtNHXs varied from 528 (PtNHX5) to 1,147 (PtNHX8). The molecular weights (MW) varied from 58.7 (PtNHX5) to 127 (PtNHX8) kDa; while the predicted isoelectric points (pI) were from 5.47 (PtNHX6) to 8.76 (PtNHX4) (Table 1). To validate the sequence accuracy of PtNHX genes from P. trichocarpa genome database, three PtNHXs (−1, −6, and −8) were cloned from P. trichocarpa cDNA and they showed high identities (99.45%, 99.57%, and 99.54%, respectively) with the sequences from P. trichocarpa genome database (Supplementary Fig. S1).
Table 1.
NHX gene family in P. trichocarpa.
| Group | Gene Name | Gene Locus | CDS (bp) | ORF (aa) | pI | MW (kDa) | Arabidopsis orthologous locus |
|---|---|---|---|---|---|---|---|
| Vac | PtNHX1 | Potri.005G045100 | 1,635 | 544 | 7.30 | 60.4 | AT3G05030 |
| PtNHX2 | Potri.013G031700 | 1,638 | 545 | 7.67 | 60.2 | AT3G05030 | |
| PtNHX3 | Potri.013G026600 | 1,722 | 573 | 8.03 | 63.5 | AT5G55470 | |
| PtNHX4 | Potri.010G031500 | 1,611 | 536 | 8.76 | 59.6 | AT3G06370 | |
| PtNHX5 | Potri.014G134900 | 1,587 | 528 | 8.68 | 58.7 | AT3G06370 | |
| Endo | PtNHX6 | Potri.016G000200 | 1,614 | 537 | 5.47 | 58.9 | AT1G79610 |
| PM | PtNHX7 | Potri.008G140700 | 3,438 | 1,145 | 6.39 | 126.8 | AT2G01980 |
| PtNHX8 | Potri.010G100900 | 3,444 | 1,147 | 6.51 | 127.0 | AT2G01980 |
Phylogenetic and gene structure analyses of PtNHX genes
To explore the evolutionary relationship of NHXs in plant kingdom, we compared PtNHXs with NHXs from other 10 species. Except for Populus trichocarpa, five dicotyledonous angiosperms: Arabidopsis thaliana (At), Eucalyptus grandis (Eg), Medicago truncatula (Mt), Vitis vinifera (Vv), and Glycine max (Gm); four monocotyledonous angiosperms: Oryza sativa (Os), Sorghum bicolor (Sb), Brachypodium distachyon (Bd), and Zea mays (Zm); and one moss: Physcomitrella patens (Pp) were analyzed. The size of NHX families in these species varied from 7 to 12 (Fig. 1A). Then, we constructed a phylogenetic tree including 92 NHX members from the 11 species. As shown in Fig. 1B, the NHXs were grouped into three subfamilies (Vac-, Endo-, and PM-classes). Vac-class NHXs are the most abundant NHXs in all the detected species. Interestingly, no PM-class NHX was identified in Medicago.
Figure 1.
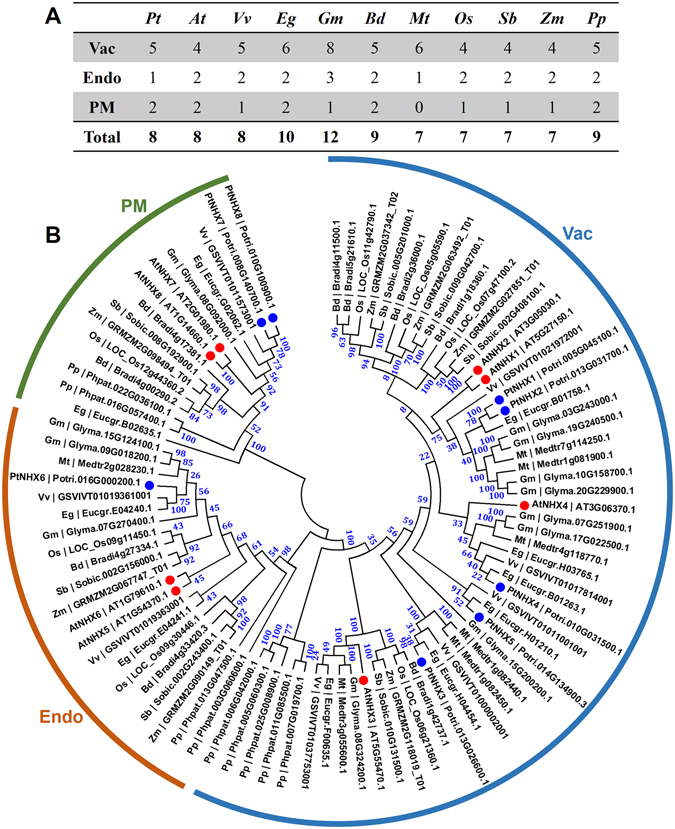
NHX members (A) and phylogenetic relationships (B) from 11 plant species. (A) NHX family members of P. trichocarpa (Pt), A. thaliana (At), V. vinifera (Vv), E. grandis (Eg), G. max (Gm), M. truncatula (Mt), B. distachyon (Bd), O. sativa (Os), S. bicolor (Sb), Z. mays (Zm), and P. patens (Pp). (B) Phylogenetic tree was constructed using the maximum likelihood (ML) method with 1,000 bootstrap replicates. The three major classes (Vac-, Endo-, and PM-) are marked with different colors. Details of NHXs from 11 plant species was listed in Supplementary Table S1.
To analyze the structural characteristics of the Populus NHX genes, we compared the exon-intron organizations of these genes (Fig. 2A). Among the PtNHX genes, Vac-class NHXs (PtNHX1–5) had 13 introns, Endo-class NHX (PtNHX6) had 21 introns, while PM-class NHXs (PtNHX7 and PtNHX8) had 22 introns. The intron number, exon length, and intron phase were relatively conserved among the members in the same subfamily. In addition, the sequence conservation among PtNHX proteins was also supported by amino acid sequence identity (Fig. 3A). Two PtNHX paralogous pairs exhibited high sequence identities in amino acid level (PtNHX1/PtNHX2 = 89.9% and PtNHX7/PtNHX8 = 88.6%), while the proteins in different PtNHX subfamilies showed low identity (7.4–25.8%).
Figure 2.
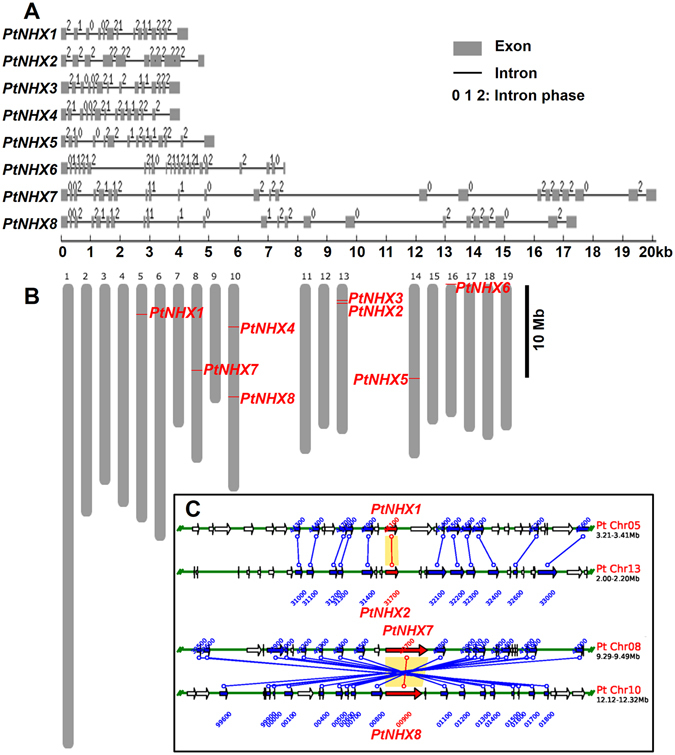
PtNHX gene structure, map position, and duplication analysis. (A) Gene structures of the PtNHXs. (B) Location of PtNHXs on P. trichocarpa chromosomes. (C) The duplicated PtNHXs (red labelled) from PGDD database.
Figure 3.
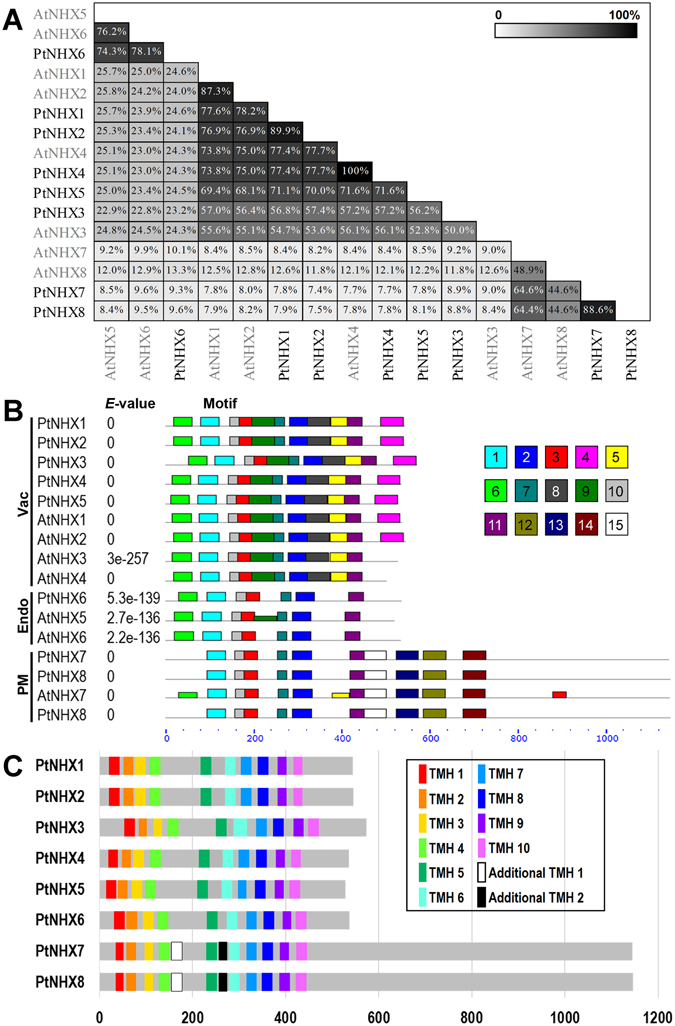
Sequence identity and conserved motifs in the NHX family members. (A) Pairwise sequence identity among Populus and Arabidopsis NHX proteins. (B) Conserved motifs of PtNHXs and AtNHXs identified by MEME. Details of different motifs indicated by different colors are shown in Fig. S4. (C) The transmembrane helices of PtNHX proteins were predicted using TMHMM2.
Chromosomal location and expansion analyses of PtNHX genes
We further analyzed the gene duplication events to understand the evolution of the PtNHX gene family. Eight PtNHX genes were mapped onto six of total 19 P. trichocarpa chromosomes unevenly (Fig. 2B). Two chromosomes (Chr10 and Chr13) contained two PtNHX genes and four chromosomes (Chr5, Chr8, Chr14, and Chr16) contained only one PtNHX gene on each (Fig. 2B). As a main mechanism of gene family expansion, gene duplication events provide opportunities for the new gene production and its functional divergence33. The paralogous genes were generated during the divergent evolution from a common ancestral gene through duplication events (segmental or tandem duplication)34. In poplar, only two PtNHX paralogous pairs (PtNHX1/PtNHX2 and PtNHX7/PtNHX8) were identified and both were generated by segmental duplication events (Fig. 2C), which was similar with the duplication events of NHXs in Arabidopsis (Supplementary Fig. S2). Therefore, the expansion of NHX genes in poplar was mainly attributed by segmental duplications.
In Populus, the recent large-scale genome duplication event was occurred in 13 million years ago (MYA)22. The non-synonymous (Ka) or synonymous (Ks) substitution rates and their ratio (Ka/Ks) of the two PtNHX paralogous gene pairs showed high similarities (Table 2). The Ka/Ks ratio could be used to evaluate whether Darwinian selection was involved in the duplication events. The ratio >1 or <1 implying that the genes underwent a positive Darwinian selection or a purifying selection34. The Ka/Ks of the two PtNHX pairs were less than 0.3, indicating that purifying selection played a dominative role in PtNHXs duplication. According to the divergence rate (λ = 9.1 × 10–9) for Populus 28, the dates of the two PtNHX paralogous pairs were estimated in 12.09 MYA (PtNHX1/PtNHX2) and 10.99 MYA (PtNHX7/PtNHX8), indicating that the two PtNHX paralogous pairs were likely generated during the stage of the recent Populus large-scale genome duplication event (~13 MYA).
Table 2.
Divergence between paralogous PtNHX gene pairs.
| Gene 1 | Gene 2 | Ka | Ks | Ka/Ks | Date (MYA) |
|---|---|---|---|---|---|
| PtNHX1 | PtNHX2 | 0.05 | 0.22 | 0.227 | 12.09 |
| PtNHX7 | PtNHX8 | 0.05 | 0.20 | 0.250 | 10.99 |
Notes: Synonymous (Ks) and nonsynonymous substitution (Ka) rates are presented for each pair.
In addition, the sliding window analysis was carried out to identify the Ka/Ks ratio across the full length of PtNHX paralogous pairs. As shown in Supplementary Fig. S3, the conserved domains of PtNHXs underwent strong purifying selection in both PtNHX1/PtNHX2 and PtNHX7/PtNHX8 paralogous pairs. While one exception region in C-terminus of PtNHX1/PtNHX2 pair showed high Ka/Ks ratio (Ka/Ks > 1), implying that the regions corresponding to conserved domains and non-conserved domains were evolved under different selective pressure (Supplementary Fig. S3). These results indicated that purifying selection played important roles in the evolution of NHX gene family in Populus.
Motifs characterization of PtNHX proteins
To further investigate the characteristic region of PtNHX proteins, the motif distributions in PtNHX and AtNHX proteins were analyzed using MEME and total of 15 individual motifs were identified (Fig. 3B and Supplementary Fig. S4). As predicted, the members with highly phylogenetic relationships had common motif composition. Six motifs (motif 1, 2, 3, 7, 10, and 11) were existed in all the members in the NHX family, and these motifs were enriched in the N-terminus of NHXs. Motif 6 was existed in Vac- and Endo-classes NHXs, three motifs (motif 5, 8, and 9) were only existed in Vac-class NHXs, and four motifs (motif 12, 13, 14, and 15) were only existed in PM-class NHXs. In addition, motif 4 was also Vac-class specific motif, but two AtNHXs (AtNHX3 and AtNHX4) in this subfamily were lost this motif (Fig. 3B).
Similar with other typical NHX proteins, PtNHXs containing 10 transmembrane helices (TMH1-TMH10) and a hydrophilic C-terminal domain (Fig. 3C and Supplementary Fig. S5). Despite the PM-class NHXs were significantly different from other two classes NHXs in length or sequence similarity (Fig. 3A), the 10 TMHs (TMH1-TMH10) could be detected in all of the PtNHXs. Moreover, two additional TMHs (Additional TMH1 and Additional TMH2) were identified in PM-class NHXs after TMH4 and TMH5, respectively (Fig. 3C and Supplementary Fig. S5).
Structural analysis of PtNHX proteins
In order to understand the functional mechanism of PtNHX proteins, we analyzed the 3D structures of PtNHXs. According to the best structural templates and crystal structures from Protein Data Bank (PDB), we constructed the best predicted model for the eight PtNHX proteins. To quantify the confidence of constructed model, C-score was used to evaluate the predicted protein model. Generally, the C-score range from −5 to 2, higher value means the model with higher confidence. In our study, the predicted PtNHX models were high accuracy with C-score ranged from −1.6 to 0.08 (Table 3), indicating that the protein structures were constructed with high accuracy. As shown in Fig. 4 and Supplementary Fig. S6, the TMHs compose a hollow cylinder and embedded in the membrane to provide the channel for Na+ and H+ transport.
Table 3.
Structural dependent modeling parameters for the PtNHX proteins.
| C-score | TM-score | RMSD (Å) | Best identified structural analogs in PDB | |||||
|---|---|---|---|---|---|---|---|---|
| PDB Hit | TM-scorea | RMSDa | IDENa | Cov | ||||
| PtNHX1 | −1.25 | 0.56 ± 0.15 | 10.4 ± 4.6 | 4cz9A | 0.711 | 1.16 | 0.213 | 0.724 |
| PtNHX2 | −1.15 | 0.57 ± 0.15 | 10.2 ± 4.6 | 4cz9A | 0.706 | 1.25 | 0.216 | 0.721 |
| PtNHX3 | −0.77 | 0.62 ± 0.14 | 9.4 ± 4.6 | 4cz8A | 0.668 | 1.34 | 0.217 | 0.682 |
| PtNHX4 | 0.08 | 0.72 ± 0.11 | 7.3 ± 4.2 | 4cz8A | 0.722 | 0.97 | 0.204 | 0.731 |
| PtNHX5 | −1.6 | 0.52 ± 0.15 | 11.2 ± 4.6 | 4cz8A | 0.723 | 1.48 | 0.199 | 0.742 |
| PtNHX6 | −0.37 | 0.67 ± 0.13 | 8.3 ± 4.5 | 4cz9A | 0.714 | 1.13 | 0.203 | 0.726 |
| PtNHX7 | −1.46 | 0.53 ± 0.15 | 12.9 ± 4.2 | 3gb8A | 0.771 | 1.04 | 0.097 | 0.777 |
| PtNHX8 | −0.9 | 0.60 ± 0.14 | 11.4 ± 4.5 | 3w3tA | 0.851 | 1.28 | 0.086 | 0.859 |
C-score [−5, 2] is the confidence of each model, higher value signifies a model with a higher confidence and vice-versa. TM-score and RMSD are estimated based on C-score and protein length following the correlation observed between these qualities. TM-scorea is TM-score of the structural alignment between the query structure and known structures in the PDB library. RMSDa is the RMSD between residues that are structurally aligned by TM-align. IDENa is the percentage sequence identity in the structurally aligned region. Cov represents the coverage of the alignment by TM-align and is equal to the number of structurally aligned residues divided by length of the query protein.
Figure 4.
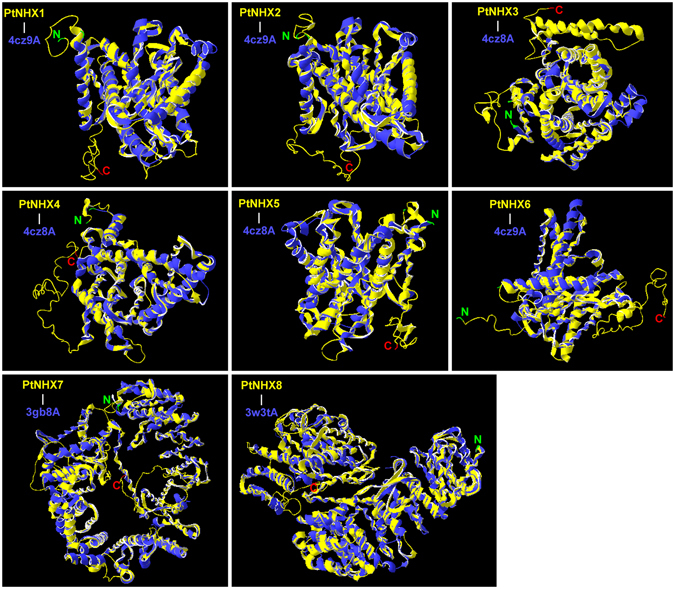
Structural analysis of PtNHX proteins. Yellow chains indicate the PtNHX proteins and blue chains indicate the best identified structural analogs in PDB for each PtNHX protein (Table 3). Details of secondary structure of PtNHX proteins are shown in Fig. S5.
Various cis-acting elements in the PtNHX promoters
To further understand the potential transcriptional regulatory mechanism of transcription factors to PtNHX genes, the sequences of −1,500 bp to +500 bp relative to the transcription start sites (TSS) of the eight PtNHXs were used to identify the cis-acting elements. As shown in Fig. 5, amount of stress-related (e.g. heat, low temperature, drought, wound, and defense) and hormone-related (e.g. auxin, ethylene, GA, SA, MeJA, and ABA) elements were identified in the PtNHX promoters. Among the stress-related cis-acting elements, total of 19 TC-rich repeats and 37 HSE were identified in the eight PtNHX promoters. Among the hormone-related cis-acting elements, the ABRE (cis-elements involved in ABA responsiveness) were found in seven of eight PtNHX promoters. Noticeably, the HSE were enriched in the promoter regions of PtNHX6, PtNHX7, and PtNHX8 (with eight, seven, and seven HSEs, respectively). The results suggested that PtNHXs might have potential roles in stress adaptation and various hormone signal responsiveness.
Figure 5.
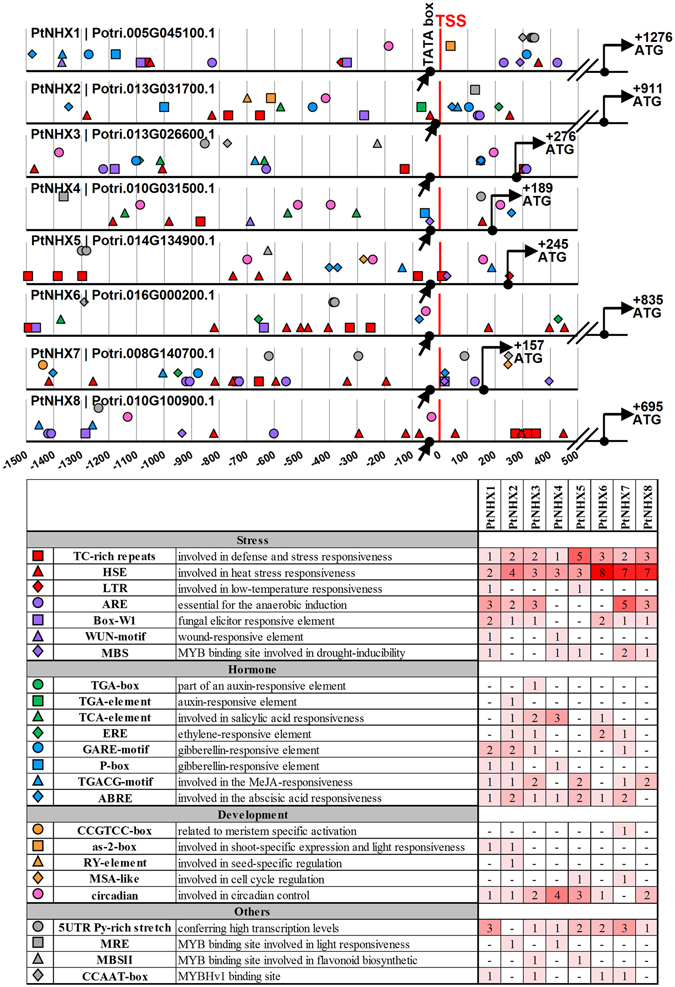
cis-acting elements in PtNHX promoters. Schematic representation of promoter regions of eight PtNHXs. Distances are relative to the transcription start site (TSS).
Expression patterns of PtNHXs across different tissues and response to stresses
To investigate the potential roles of PtNHXs in growth and/or development, their expression patterns across six poplar tissues were analyzed using qRT-PCR. Six tissues including shoot apical meristem (SAM), young leaves (YL), mature leaves (ML), primary stems (PS), secondary stems (SS), and roots (R) were selected for study. As shown in Fig. 6A, four genes (PtNHX1, PtNHX3, PtNHX6, and PtNHX8) were highly expressed in mature leaf, while two (PtNHX5 and PtNHX7) were highly expressed in root. PtNHX4 was highly expressed in both stem (including primary stem and secondary stem) and root; similarly, its homologous gene AtNHX4 was highly expressed in root and stem (Supplementary Fig. S7). We also compared the expression patterns between PtNHXs in each paralogous pair, the expression patterns of paralogous PtNHXs were significant different in both PtNHX1/PtNHX2 pair and PtNHX7/PtNHX8 pair, indicating that their expression were divergent during the evolution though the paralogous genes were duplicated from the same original gene.
Figure 6.
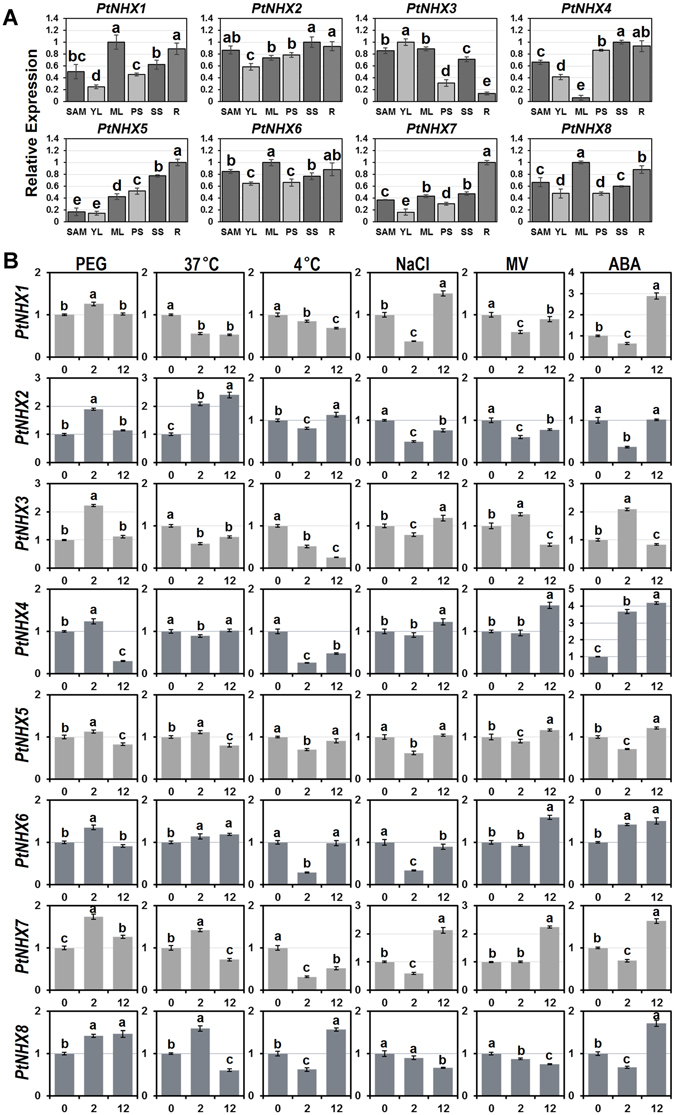
Expression patterns of PtNHX genes in different tissues (A) and under various abiotic stresses (B) using qRT-PCR. (A) The relative mRNA abundance of the PtNHXs were quantified in six tissues (SAM–shoot apical meristems, YL–young leaves, ML–mature leaves, PS–primary stem, SS–secondary stem, R–roots). (B) The expression patterns of the PtNHXs at 2 and 12 hours after treated with drought (10% PEG), heat (37 °C), cold (4 °C), salt (150 mM NaCl), oxidative stress (100 μM MV) or 100 μM ABA. Bars with the same letters are not significantly (P < 0.05).
Moreover, the expression patterns of eight PtNHXs under drought, heat, cold, salinity, oxidative stress, or ABA treatments were analyzed (Fig. 6B). Because all the eight PtNHXs could be detected in leaves, we selected the first fully expanded leaves to confirm their responses to various abiotic stresses or hormone signal. Under drought stress, all of PtNHX genes were up-regulated at 2 h after PEG treatment. Similarly, the expression of most AtNHXs were induced in Arabidopsis seedlings under drought and osmotic stresses (Supplementary Fig. S8). PtNHX2 was immediately up-regulated under heat stress, its expression pattern was consistent with its orthologous in Arabidopsis (AtNHX2, Supplementary Fig. S8). When the poplar seedlings were exposed to salinity stress, two PtNHXs (PtNHX1 and PtNHX7) were dramatically induced after 12 h treatment. The orthologous of PtNHX7 was well known as the salt tolerance locus SOS1 in Arabidopsis, which plays crucial role in root Na+ long distance transport to shoot20. As an important plant hormone, ABA is involved in not only plant development but also response to various stresses35. As shown in Fig. 6B, the expression of all of the eight PtNHXs were affected by ABA treatment, implying that the members in PtNHX family might be involved in ABA-dependent signal pathway. All the expression data indicated that most of the PtNHXs were responsive to certain abiotic stresses at the transcriptional level. Similar with that in poplar, the members of NHX family in Arabidopsis were also response to various abiotic stresses (Supplementary Fig. S8). NHX genes might be commonly involved in plant adapt to various environmental stress conditions in different species.
Co-expression network of PtNHX genes
To explore the potential molecular functions of PtNHXs, the co-expression network of PtNHXs was constructed based on the genome-wide gene expression patterns. As shown in Fig. 7, total of 6, 68, 51, and 7 genes were co-expressed with PtNHX1, PtNHX3, PtNHX4, and PtNHX5, respectively; and the genes co-expressed with the four PtNHXs are independent. No gene was co-expressed with PtNHX2. Different with those PtNHXs co-expressed with independent genes, three PtNHXs (PtNHX6, PtNHX7, and PtNHX8) with more co-expressed genes (199, 55, and 422, respectively) and they share several hub co-expressed genes. Among these hub genes, PIS (Potri.013G115300) was co-expressed with all the three PtNHXs, which is located in plasma membrane and required for growth (Table 4). Two salt stress related genes MKP1 (Potri.008G049900) and WD40 (Potri.009G139000) were co-expressed with both PtNHX6 and PtNHX7. In addition, seven genes were co-expressed with PtNHX6 and PtNHX8, six genes were co-expressed with PtNHX7 and PtNHX8 (Fig. 7A and Table 4).
Figure 7.
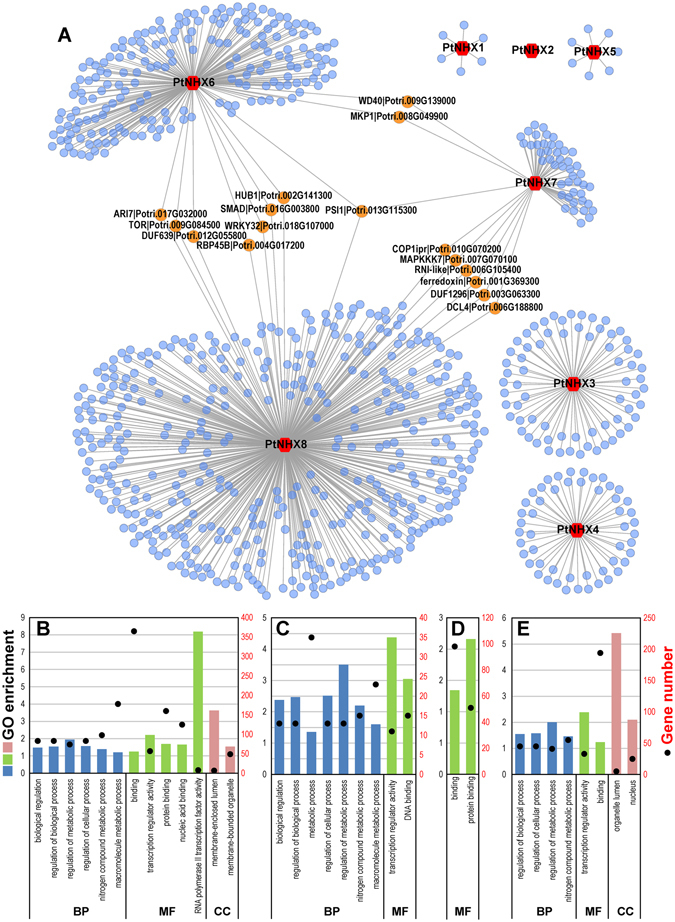
Co-expression network of PtNHX gene family. (A) PtNHXs are represented as red hexagon, hub co-expressed genes are represented as orange nodes. Partial results of GO enrichment and enriched gene number of all the genes co-expressed with the eight PtNHXs (B), PtNHX3 (C), PtNHX6 (D), and PtNHX8 (E) were shown. Details of GO enrichment are listed in Fig. S9. BP, biological process; MF, molecular function; CC, cellular component.
Table 4.
Annotation of hub genes in the PtNHX co-expression network.
| Gene Name | Gene ID | Arabidopsis orthologous | Functional Description | Co-expressed with PtNHXs |
|---|---|---|---|---|
| PSI1 | Potri.013G115300 | AT1G34320 | Located in plasma membrane, required for growth. | PtNHX6, PtNHX7, and PtNHX8 |
| MKP1 | Potri.008G049900 | AT3G55270 | Involved in protein dephosphorylation, inactivation of MAPK activity, response to salt stress, UV, and fungus. | PtNHX6 and PtNHX7 |
| WD40 | Potri.009G139000 | AT4G35140 | Regulated by NAC2 in salt stress response and lateral root development. | PtNHX6 and PtNHX7 |
| HUB1 | Potri.002G141300 | AT2G44950 | E3 uniquitin ligases involved in monoubiquitination of histone H2B. Involved in cell devision, cell growth and defense response. | PtNHX6 and PtNHX8 |
| SMAD | Potri.016G003800 | AT5G47790 | SMAD/FHA domain-containing protein | PtNHX6 and PtNHX8 |
| WRKY32 | Potri.018G107000 | AT4G30935 | Regulation of transcription. | PtNHX6 and PtNHX8 |
| RBP45B | Potri.004G017200 | AT1G11650 | RNA binding protein involved in mRNA processing, response to cytokinin and ozone. | PtNHX6 and PtNHX8 |
| TOR | Potri.009G084500 | AT1G50030 | Regulate cell growth in response to nutrient availability. | PtNHX6 and PtNHX8 |
| DUF639 | Potri.012G055800 | AT1G48840 | Plant protein of unknown function (DUF639) | PtNHX6 and PtNHX8 |
| ARI7 | Potri.017G032000 | AT2G31510 | Positive regulation of proteasomal ubiquitin-dependent protein catabolic process. | PtNHX6 and PtNHX8 |
| DCL4 | Potri.006G188800 | AT5G20320 | Rnase III-like enzyme catalyzes processing of trans-acting small interfering RNA precursors in a distinct small RNA biogenesis pathway. | PtNHX7 and PtNHX8 |
| COP1ipr | Potri.010G070200 | AT5G43310 | Located in plasma membrane, positive regulator of light-regulated genes. | PtNHX7 and PtNHX8 |
| ferredoxin | Potri.001G369300 | AT4G26620 | Sucrase/ferredoxin-like family protein | PtNHX7 and PtNHX8 |
| MAPKKK7 | Potri.007G070100 | AT3G13530 | Involved in cell cycle, cell division, plasma membrane organization, pollen development, regulation of cell division, regulation of embryonic development, regulation of extent of cell growth, regulation of unidimensional cell growth. | PtNHX7 and PtNHX8 |
| RNI-like | Potri.006G105400 | AT5G01720 | Involved in ubiquitin-dependent protein catabolic process. | PtNHX7 and PtNHX8 |
| DUF1296 | Potri.003G063300 | AT3G13990 | Kinase-related protein of unknown function (DUF1296) | PtNHX7 and PtNHX8 |
We then performed the GO enrichment analysis of the co-expressed genes to reveal their potential function. Among the genes co-expressed with the eight PtNHXs, 29 biological process (BP), 11 molecular function (MF), and 11 cellular component (CC) terms were significant enriched. For the BP, the enriched terms are related with “regulation of biological process”, “regulation of metabolic process”, “regulation of cellular process”, “nitrogen compound metabolic process”, and “macromolecule metabolic process”. The enriched MF terms are related with “binding”, “transcription regulation activity”, and “RNA polymerase II transcription factor activity”. And the enriched CC terms are related with “membrane-enclosed lumen” and “membrane-bounded organelle” (Fig. 7B and Supplementary Fig. S9). Separately GO enrichment analysis of co-expressed genes for each PtNHXs indicated only the genes co-expressed with three PtNHXs (PtNHX3, PtNHX6, and PtNHX8) enriched in specific GO terms. For PtNHX3, the co-expressed genes enriched in the BP terms “biological regulation” and “metabolic process” and the MF terms “transcription regulator activity”. The genes co-expressed with PtNHX6 were enriched in the MF terms “binding” and specifically in “protein binding”. For PtNHX8, the co-expressed genes were enriched in the BP term “regulation process”, the MF term “transcription regulator activity”, and the CC terms “organelle lumen” and “nucleus”.
To reveal the evolutionary divergence of NHX genes in model woody species Populus and model plant Arabidopsis, we then constructed the co-expression network of Arabidopsis NHX genes and compared the differences between PtNHX co-expression network and AtNHX co-expression network. Different with the co-expression pattern of PtNHXs, three AtNHXs (AtNHX4, AtNHX6, and AtNHX7) were connected by a series co-expressed genes in Arabidopsis NHX co-expression network (Supplementary Fig. S10). The co-expressed genes indicated that strong co-expression relationship between AtNHX6 and AtNHX7 while weak co-expression relationship between AtNHX4 and AtNHX6. To make the two co-expression networks (PtNHXs and AtNHXs co-expression networks) comparable, the GO enrichment were performed based on the genes co-expressed with each NHX gene in the two network. The GO enrichment analysis of AtNHX genes showed that the genes co-expressed with AtNHX4 were enriched in the BP terms “developmental process”, “cellular component organization” and “cellular signal pathway”; the genes co-expressed with AtNHX6 were enriched in the BP terms “biological regulation”, “developmental process”, and “reproduction”; while the genes co-expressed with AtNHX7 were enriched in the BP terms “biological regulation”, “developmental process”, “localization”, and “reproduction” (Supplementary Table S3). The co-expressed networks of PtNHXs and AtNHXs indicated that the NHX genes might be function in developmental processes and/or stress responses through cooperate with other functional genes.
Protein-protein interaction (PPI) network of PtNHXs
To further reveal the function of PtNHXs during the interaction with other proteins, we constructed a PPI network. As shown in Fig. 8A, all the eight PtNHXs share most of the same interacted proteins. In addition, some of proteins were specifically interact with PtNHX7 and PtNHX8 separately. The proteins interacted with all the PtNHXs including CHX, CAX, KEA, HKT, and KUP proteins. Two CIPK and four CBL proteins specific interacted with PtNHX7, while six coatomer proteins specific interacted with PtNHX8. To detect the functional abundance of proteins interacted with PtNHXs, the GO enrichment analysis was performed based on the PPI network. The enriched BP terms of proteins interacted with PtNHXs including “cellular process”, “transport”, and “signal transduction” (Fig. 8B and Supplementary Fig. S11).
Figure 8.
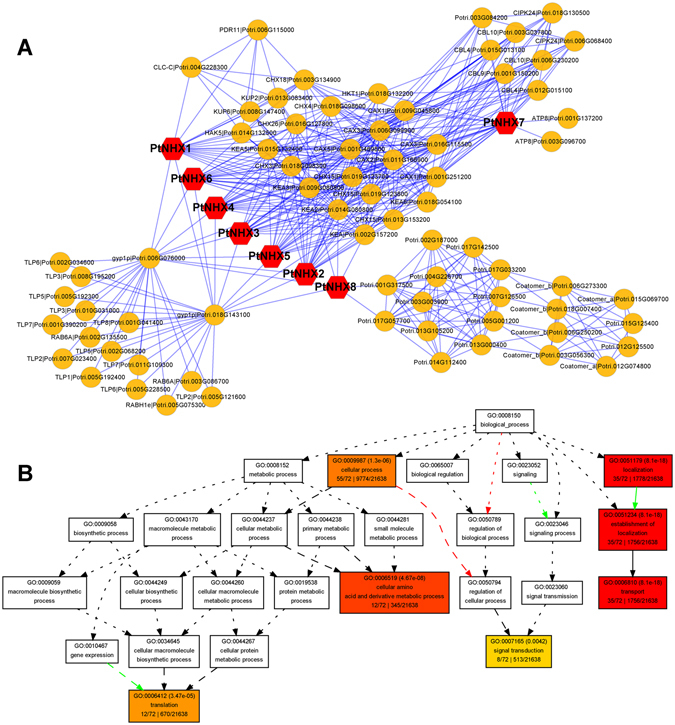
Protein-protein interaction (PPI) network of PtNHXs. (A) PPI network of PtNHX proteins. (B) Enriched BP terms of genes coding the proteins in the PtNHX PPI network. Complement GO enrichment data is shown Fig. S11.
Natural variation in PtNHX genes
Generally, the variation located in gene body might generate functional divergence or associate some traits. Based on the whole genome re-sequence data of 549 P. trichocarpa natural individuals in North America, we identified the single nucleotide polymorphisms (SNPs) in PtNHX genes. The SNPs located in gene region could be classified into different types according to their consequence, such as synonymous, non-synonymous, start gained, start lost, codon deletion, frame shift, splice site acceptor, splice site donor, and stop gained. Here we omit the synonymous SNPs because they will not affect the composition of their coding proteins. A total of 274 SNPs were detected in the eight PtNHX genes, among the detected SNPs 249 were non-synonymous coding SNPs (Table 5). The number of SNPs was significant different in the PtNHX genes, total of 77 SNPs were detected in PtNHX7 while only 8 SNPs were detected in PtNHX6. As membrane located proteins, the SNPs in the TMH regions might play important roles in their location. No SNP was located in the TMH regions of PtNHX6, while 15 and 17 SNPs were located in the TMH regions of PtNHX7 and PtNHX8. We then analyzed the SNP type frequency of each SNP based on the P. trichocarpa population. The frequency of SNPs of PtNHX7 was significantly higher than other PtNHX genes, especially in the C-terminal region of PtNHX7 (Fig. 9).
Table 5.
Summary of SNPs in PtNHX genes.
| PtNHXs | PtNHX1 | PtNHX2 | PtNHX3 | PtNHX4 | PtNHX5 | PtNHX6 | PtNHX7 | PtNHX8 | Total | |
|---|---|---|---|---|---|---|---|---|---|---|
| Gene length | 5680 | 6106 | 4351 | 4603 | 5865 | 8135 | 20568 | 17616 | ||
| Number of SNPs1 | 22 | 19 | 33 | 10 | 30 | 8 | 77 | 75 | 274 | |
| Different type of SNPs | non-synonymous coding | 16 | 17 | 32 | 10 | 24 | 6 | 71 | 73 | 249 |
| codon deletion | 1 | 1 | ||||||||
| frame shift | 2 | 2 | ||||||||
| start gained | 4 | 2 | 3 | 1 | 1 | 1 | 12 | |||
| start lost | 1 | 1 | 2 | |||||||
| stop gained | 1 | 1 | 2 | |||||||
| splice site acceptor | 1 | 1 | 2 | |||||||
| splice site donor | 1 | 1 | 1 | 1 | 4 | |||||
| SNPs located in TMH regions | 5 | 2 | 4 | 3 | 11 | 0 | 15 | 17 | 57 | |
| TMH SNPs/Total SNPs (%) | 22.7 | 10.5 | 12.1 | 30.0 | 36.7 | 0 | 19.5 | 22.7 | 22.8 | |
| Total SNPs/Gene length (%) | 3.87 | 3.11 | 7.58 | 2.17 | 5.12 | 0.98 | 3.74 | 4.26 |
1Only the SNPs affect the composition of the proteins were listed in here, i.e. synonymous SNPs were not analyzed in this study.
Figure 9.
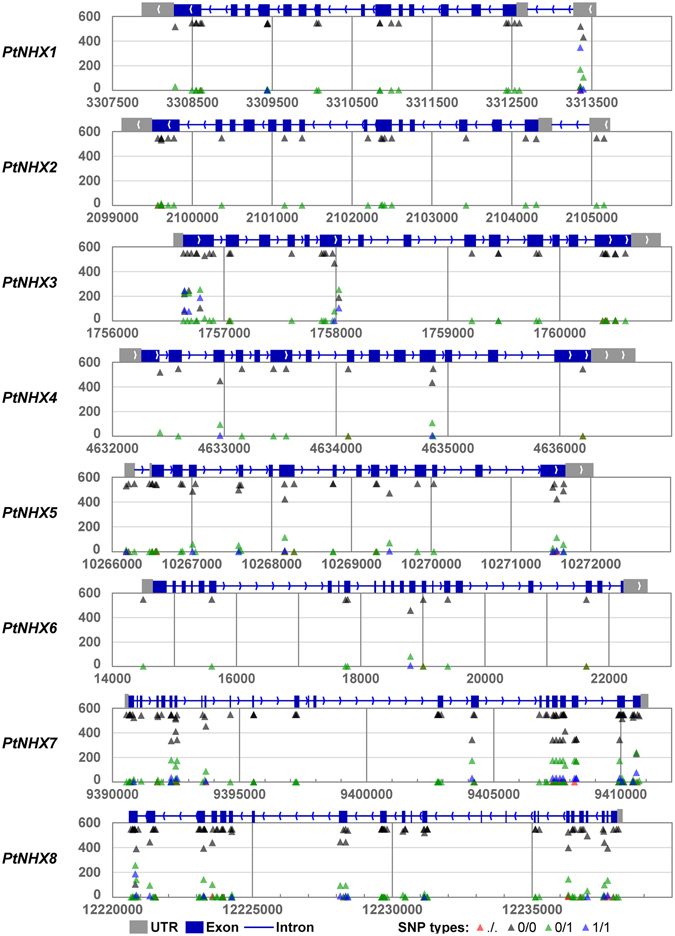
Identification of SNPs in PtNHX gene family from 549 P. trichocarpa individuals.
Discussion
Ion transports play crucial roles in many aspects of biological processes, such as ions uptake or sequestration, provide energy, and cell expansion36. In plants, Na+/H+ antiporters (NHXs) as important members in transporters, mediate the coupled exchange of Na+ or K+ for H+ in all cellular compartments36, 37. Based on their subcellular localizations, NHXs are grouped into three classes (Vac-, Endo-, and PM-class). The members in each class of NHX family were highly similar from algae to higher plants, indicating that NHXs retained conserved functions during the evolution38.
In this study, total of eight PtNHXs were identified from woody model species P. trichocarpa. In contrast with Arabidopsis, one more Vac-class NHX but one less Endo-class NHX were identified in poplar. According to comparative genomics studies, the genes in poplar are ~1.4–1.6 fold of Arabidopsis homologs22. But the NHX family did not expanded as other gene families in poplar. Overall, the size of NHX family was similar in all the other 10 species analyzed in this study, implying that the NHX family might be relatively conserved during the evolution. Gene duplication is the primary mechanism of new gene production, it plays important role in gene family expansion. In PtNHX family, two paralogous pairs were generated by segmental duplication events in 10.99 and 12.09 MYA. This stages were consist with the ~13 MYA the recent large-scale genome duplication event in Populus 22.
It has been reported that the membrane-spanning pore and cation-binding domain were highly conserved in plants5, 39, which comprises of the nonapeptide “FFIYLLPPI”40, 41. This nonapeptide was located in the N-terminus of motif 1 and it was existed in all the detected NHXs in Arabidopsis and poplar (Fig. 3B and Supplementary Fig. S4). In contrast with the conserved N-terminus, the C-terminus of NHX proteins were highly diverged even in the same class NHX. In Arabidopsis, the N-terminus of AtNHX1 is located in the cytosol while the hydrophilic C-terminus in the vacuolar lumen. The mutation deleted the C-terminal hydrophilic region resulted in an enhanced Na+/H+ transport activity, suggesting that the C-terminus play crucial roles in not only subcellular localization but also regulation of transport activity6, 7. The heterogeneous C-terminus of PtNHXs provide opportunity for their functional divergence. In addition, the members in PM-class had two additional TMHs after TMH4 and TMH5, where region was also considered critical for transport activity7.
Nowadays, amounts of studies have indicated that NHXs play important roles in plant growth and development. For instance, the Arabidopsis nhx1 nhx2 double mutant showed severe reduction in vegetative growth due to the substantial decreased in cell size17. In this study, PtNHX genes were expressed in tissue-specific patterns and were induced by various stresses, indicating that the members in PtNHX family might be also involved in developmental processes or stress responses in different tissues. Duplicated PtNHXs showed different expression patterns in various tissues (Fig. 6A), indicating that the duplication events in PtNHX family provide opportunities to break the functional constraint from the original gene during the evolution. In Arabidopsis, GUS driven by the promoter of AtNHX7 was mainly expressed in epidermal cells of the root tips and parenchyma cells at the xylem/symplast boundary20. Similarly, its orthologous in poplar, PtNHX7, was also highly expressed in root. In addition, PtNHX4 was highly expressed in stem and root, which was consist with the expression of its orthologous in Arabidopsis (AtNHX4, Supplementary Fig. S8). The similar expression patterns of NHX ortholog in poplar and Arabidopsis indicating that the NHX genes might be retained the conserved function in different species.
Stress response analyses showed that each PtNHX was response to at least one abiotic stress of drought, heat, cold, salinity, oxidative stress, or ABA. Noticeably, PtNHX7 and PtNHX8 showed significant expression changes under all the abiotic stresses. As the binding sites of transcription factors, cis-acting elements play important role to determine genes’ expression patterns42. Compared with other PtNHXs, promoters of PtNHX7 and PtNHX8 harbored more stress-related cis-acting elements, including HSE (involved in heat stress responsiveness, seven in both PtNHX7 and PtNHX8), ARE (essential for the anaerobic induction, five in PtNHX7 and three in PtNHX8), TC-rich repeats (defense and stress response, two in PtNHX7 and three in PtNHX8), MBS (MYB binding site involved in drought-inducibility, two in PtNHX7 and one in PtNHX8), and Box-W1 (fungal elicitor responsive element, one in both PtNHX7 and PtNHX8), which might be the reason of PtNHX7 and PtNHX8 significant response to these stresses. Except for the stress-related cis-acting elements, amounts of hormone-related elements were also identified in the promoters of PtNHXs. In plants, various abiotic stresses could elicit the accumulation of ABA, then trigger a series of physiological and molecular responses to acclimate the environments. Seven PtNHX (PtNHX1–7) promoters containing one or two ABREs (ABA responsive cis-acting element), suggest PtNHX1–7 might be involved in ABA signal pathway. Although no ABRE was detected in the promoter of PtNHX8, it was also induced at 12 h under ABA treatment, suggesting that there are other regulatory mechanisms in ABA responsiveness of PtNHX8.
As the membrane proteins, NHXs might be cooperated with other proteins. The co-expression analysis provide systematic information on gene-to-gene association43. Based on the co-expression network, many stress-related and transport-related genes were co-expressed with PtNHX5. In Arabidopsis, its orthologous AtNHX4 (At3g06370) was localized in vacuole, and the nhx4 mutant showed enhanced tolerance to salt stress44. Vacuoles segment many cellular components (e.g. ions, sugars, proteins, and secondary metabolites) and play critical roles in plant stress responses45. The transport-related genes co-expressed with PtNHX5 suggest that PtNHX5 might be involved in the vacuolar trafficking processes in poplar. Compared to the other class PtNHXs, the PM-class PtNHX7 and PtNHX8 have a long C-terminal cytosolic tail (Fig. 3), which provide more possibility to interact with other proteins20. Based on the PtNHX PPI network, PtNHX7 and PtNHX8 were interacted with many other proteins different with the proteins interacted with PtNHX1–6 (Fig. 8). This was consistent with the structural characteristic of long C-terminal cytosolic tails of PtNHX7 and PtNHX8. Although PtNHX7 and PtNHX8 belong to paralogous pairs generated by WGD (Fig. 2 and Supplementary Fig. S3), the protein 3D structures of PtNHX7 and PtNHX8 were significant different (Fig. 4). The structural difference between PtNHX7 and PtNHX8 provide the physical foundation for they interact with different set of proteins (Fig. 8).
As one class of calcium sensors, the calcineurin B-like (CBL) proteins can interact with and regulate the CBL-interacting protein kinases (CIPK) to mediate the calcium signal transduction. During environmental adaptation reactions, the interaction between CBL and CIPKs provide flexible and specific signal response mechanism36, 46. It has been proved that NHX7 (SOS1) was regulated by CBL and CIPK mediated Ca2+ signaling pathway during salinity response. The kinase CIPK24 was recruitment to the membrane through physical interaction with CBL4 and then active the Ca2+-dependent NHX747. Noticeable, CBL4 is only expressed in root48. Similar with the expression pattern of CBL4, the PtNHX7 was highly expressed in root (Fig. 6A), the co-expression pattern of PtNHX7 and CBL4 in root provide opportunity of their functional interaction. The PPI network of PtNHXs indicated that PtNHX7 physically interact with CBL and CIPK proteins different with other PtNHX proteins. Recently, a meta-analysis indicates overexpression CPA1 (or SOS1), the ortholog of PtNHX7, had statistically significant impacts for 10 of the 19 plant characteristics examined, by 25% or more. Compared to transfer into or from other genera, the CPA1 transformed to or from Arabidopsis have led to smaller CPA1-induced increases. Heterogeneous expression of CPA1 led to greater increases in leaf chlorophyll and root length than homologous expression49. This is consistent with we identified PtNHX7 as the primary NHX involved in salt stress response. Our findings indicated that the members of PtNHX family play distinct roles in developmental processes and stress responses, and provided a comprehensive understanding of the function of PtNHXs in poplar.
Conclusions
In this study, eight NHXs from three subfamilies (Vac-, Endo-, and PM-classes) were identified in the P. trichocarpa genome. Comprehensive analysis including phylogeny, gene structures, duplications, conserved motifs, tissues-specific expression and stress responses, and integrated networks (co-expression network and protein-protein interaction network) were performed. In particular, we found that the whole genome duplication (WGD) event played a dominative role in expansion of NHX family in poplar, and purifying selection was the main force. Protein structural analysis indicated that the THMs compose a hollow cylinder and embedded in the membrane to provide the channel for Na+ and H+ transport. In addition, the tissue-specific expression patterns of PtNHXs provided functional information in poplar’s development processes. Moreover, responses of the PtNHXs to drought, heat, cold, salinity, oxidative stress, or ABA treatment indicated that the PtNHXs involved in single or multiple stress responses in poplar. Co-expression network of PtNHXs indicated that several hub genes were connected with the End- and PM-classes PtNHXs. Among the eight PtNHXs, only PtNHX7 interact with CBL and CIPK suggests that PtNHX7 might be the primary NHX involved in CBL-CIPK pathway during salt stress responses. Our results contribute valuable information for future functional investigations of PtNHX gene family.
Electronic supplementary material
Acknowledgements
This work was supported by the National Nonprofit Institute Research Grant of the Chinese Academy of Forestry (CAFYBB2014ZX001-4), the National Natural Science Foundation of China [31570669], and High-Level Talent Introduction Project of Xinjiang Uygur Autonomous Region (2014) to J.H.
Author Contributions
J.Z. contributed to the experimental design and management, data analysis, and manuscript preparation. J.H. contributed to proofreading and critical review of this manuscript. F.T. managed the experiments. E.C., Y.L. and P.S. contributed to gene expression analysis. All authors have read and approved the final manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-02894-8
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jianjun Hu, Email: hujj@caf.ac.cn.
Jin Zhang, Email: zhang007jin@163.com.
References
- 1.Martinoia E, Maeshima M, Neuhaus HE. Vacuolar transporters and their essential role in plant metabolism. J. Exp. Bot. 2007;58:83–102. doi: 10.1093/jxb/erl183. [DOI] [PubMed] [Google Scholar]
- 2.Amtmann, A. & Leigh, R. In Abiotic Stress Adaptation in Plants 245–262 (Springer, 2010).
- 3.Mäser P, et al. Phylogenetic relationships within cation transporter families of Arabidopsis. Plant Physiol. 2001;126:1646–1667. doi: 10.1104/pp.126.4.1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brett CL, Donowitz M, Rao R. Evolutionary origins of eukaryotic sodium/proton exchangers. Am J Physiol-Cell Ph. 2005;288:C223–C239. doi: 10.1152/ajpcell.00360.2004. [DOI] [PubMed] [Google Scholar]
- 5.Yokoi S, et al. Differential expression and function of Arabidopsis thaliana NHX Na+/H+ antiporters in the salt stress response. Plant J. 2002;30:529–539. doi: 10.1046/j.1365-313X.2002.01309.x. [DOI] [PubMed] [Google Scholar]
- 6.Orlowski J, Grinstein S. Emerging roles of alkali cation/proton exchangers in organellar homeostasis. Curr. Opin. Cell Biol. 2007;19:483–492. doi: 10.1016/j.ceb.2007.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yamaguchi T, Apse MP, Shi H, Blumwald E. Topological analysis of a plant vacuolar Na+/H+ antiporter reveals a luminal C terminus that regulates antiporter cation selectivity. P Natl Acad Sci USA. 2003;100:12510–12515. doi: 10.1073/pnas.2034966100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hernández A, et al. Mutants of the Arabidopsis thaliana Cation/H+ Antiporter AtNHX1 Conferring Increased Salt Tolerance in Yeast: the endosome/prevacuolar compartment is a target for salt toxicity. J. Biol. Chem. 2009;284:14276–14285. doi: 10.1074/jbc.M806203200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bassil E, et al. The Arabidopsis intracellular Na+/H+ antiporters NHX5 and NHX6 are endosome associated and necessary for plant growth and development. Plant Cell. 2011;23:224–239. doi: 10.1105/tpc.110.079426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ohnishi M, et al. Characterization of a novel Na+/H+ antiporter gene InNHX2 and comparison of InNHX2 with InNHX1, which is responsible for blue flower coloration by increasing the vacuolar pH in the Japanese morning glory. Plant Cell Physiol. 2005;46:259–267. doi: 10.1093/pcp/pci028. [DOI] [PubMed] [Google Scholar]
- 11.Yoshida K, et al. Synchrony between flower opening and petal-color change from red to blue in morning glory, Ipomoea tricolor cv. Heavenly Blue. P Jpn Acad B-Phys. 2009;85:187–197. doi: 10.2183/pjab.85.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Apse MP, Blumwald E. Na+ transport in plants. FEBS Lett. 2007;581:2247–2254. doi: 10.1016/j.febslet.2007.04.014. [DOI] [PubMed] [Google Scholar]
- 13.Leidi EO, et al. The AtNHX1 exchanger mediates potassium compartmentation in vacuoles of transgenic tomato. Plant J. 2010;61:495–506. doi: 10.1111/j.1365-313X.2009.04073.x. [DOI] [PubMed] [Google Scholar]
- 14.Apse MP, Sottosanto JB, Blumwald E. Vacuolar cation/H+ exchange, ion homeostasis, and leaf development are altered in a T-DNA insertional mutant of AtNHX1, the Arabidopsis vacuolar Na+/H+ antiporter. Plant J. 2003;36:229–239. doi: 10.1046/j.1365-313X.2003.01871.x. [DOI] [PubMed] [Google Scholar]
- 15.Bowers K, Levi BP, Patel FI, Stevens TH. The sodium/proton exchanger Nhx1p is required for endosomal protein trafficking in the yeast Saccharomyces cerevisiae. Mol Biol Cell. 2000;11:4277–4294. doi: 10.1091/mbc.11.12.4277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brett CL, Tukaye DN, Mukherjee S, Rao R. The yeast endosomal Na+(K+)/H+ exchanger Nhx1 regulates cellular pH to control vesicle trafficking. Mol Biol Cell. 2005;16:1396–1405. doi: 10.1091/mbc.E04-11-0999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bassil E, et al. The Arabidopsis Na+/H+ antiporters NHX1 and NHX2 control vacuolar pH and K+ homeostasis to regulate growth, flower development, and reproduction. Plant Cell. 2011;23:3482–3497. doi: 10.1105/tpc.111.089581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barragán V, et al. Ion exchangers NHX1 and NHX2 mediate active potassium uptake into vacuoles to regulate cell turgor and stomatal function in Arabidopsis. Plant Cell. 2012;24:1127–1142. doi: 10.1105/tpc.111.095273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pardo JM, Cubero B, Leidi EO, Quintero FJ. Alkali cation exchangers: roles in cellular homeostasis and stress tolerance. J. Exp. Bot. 2006;57:1181–1199. doi: 10.1093/jxb/erj114. [DOI] [PubMed] [Google Scholar]
- 20.Shi H, Quintero FJ, Pardo JM, Zhu J-K. The putative plasma membrane Na+/H+ antiporter SOS1 controls long-distance Na+ transport in plants. Plant Cell. 2002;14:465–477. doi: 10.1105/tpc.010371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jansson S, Douglas CJ. Populus: a model system for plant biology. Annu. Rev. Plant Biol. 2007;58:435–458. doi: 10.1146/annurev.arplant.58.032806.103956. [DOI] [PubMed] [Google Scholar]
- 22.Tuskan GA, et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray) Science. 2006;313:1596–1604. doi: 10.1126/science.1128691. [DOI] [PubMed] [Google Scholar]
- 23.Ye CY, Zhang HC, Chen JH, Xia XL, Yin WL. Molecular characterization of putative vacuolar NHX-type Na+/H+ exchanger genes from the salt-resistant tree Populus euphratica. Physiol. Plant. 2009;137:166–174. doi: 10.1111/j.1399-3054.2009.01269.x. [DOI] [PubMed] [Google Scholar]
- 24.Larkin MA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 25.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 26.Lescot M, et al. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002;30:325–327. doi: 10.1093/nar/30.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang J, et al. The I-TASSER Suite: protein structure and function prediction. Nat. Methods. 2015;12:7–8. doi: 10.1038/nmeth.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- 29.Zhang, J. et al. Molecular evolution and expression divergence of the Populus euphratica Hsf genes provide insight into the stress acclimation of desert poplar. Scientific Reports6 (2016). [DOI] [PMC free article] [PubMed]
- 30.Zhang J, et al. Hsf and Hsp gene families in Populus: genome-wide identification, organization and correlated expression during development and in stress responses. BMC Genomics. 2015;16:181. doi: 10.1186/s12864-015-1398-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Szklarczyk D, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kong H, et al. Patterns of gene duplication in the plant SKP1 gene family in angiosperms: evidence for multiple mechanisms of rapid gene birth. Plant J. 2007;50:873–885. doi: 10.1111/j.1365-313X.2007.03097.x. [DOI] [PubMed] [Google Scholar]
- 34.Bowers JE, Chapman BA, Rong J, Paterson AH. Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature. 2003;422:433–438. doi: 10.1038/nature01521. [DOI] [PubMed] [Google Scholar]
- 35.Lee SC, Luan S. ABA signal transduction at the crossroad of biotic and abiotic stress responses. Plant, Cell Environ. 2012;35:53–60. doi: 10.1111/j.1365-3040.2011.02426.x. [DOI] [PubMed] [Google Scholar]
- 36.Bassil E, Blumwald E. The ins and outs of intracellular ion homeostasis: NHX-type cation/H+ transporters. Curr. Opin. Plant Biol. 2014;22:1–6. doi: 10.1016/j.pbi.2014.08.002. [DOI] [PubMed] [Google Scholar]
- 37.Qiu QS. Plant and yeast NHX antiporters: roles in membrane trafficking. J Integr Plant Biol. 2012;54:66–72. doi: 10.1111/j.1744-7909.2012.01097.x. [DOI] [PubMed] [Google Scholar]
- 38.Chanroj, S. et al. Conserved and diversified gene families of monovalent cation/H+ antiporters from algae to flowering plants. Frontiers in plant science3 (2012). [DOI] [PMC free article] [PubMed]
- 39.Aharon GS, Apse MP, Duan S, Hua X, Blumwald E. Characterization of a family of vacuolar Na+/H+ antiporters in Arabidopsis thaliana. Plant Soil. 2003;253:245–256. doi: 10.1023/A:1024577205697. [DOI] [Google Scholar]
- 40.Kinsella J, Aronson P. Amiloride inhibition of the Na+/H+ exchanger in renal microvillus membrane vesicles. Am J Physiol-Renal. 1981;241:F374–F379. doi: 10.1152/ajprenal.1981.241.4.F374. [DOI] [PubMed] [Google Scholar]
- 41.Blumwald E, Poole RJ. Na+/H+ antiport in isolated tonoplast vesicles from storage tissue of Beta vulgaris. Plant Physiol. 1985;78:163–167. doi: 10.1104/pp.78.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu Z, et al. Genome-wide identification, phylogeny, duplication, and expression analyses of two-component system genes in Chinese cabbage (Brassica rapa ssp. pekinensis) DNA Res. 2014;21:379–396. doi: 10.1093/dnares/dsu004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ogata Y, Suzuki H, Shibata D. A database for poplar gene co-expression analysis for systematic understanding of biological processes, including stress responses. J. Wood Sci. 2009;55:395–400. doi: 10.1007/s10086-009-1058-9. [DOI] [Google Scholar]
- 44.Li H-T, Liu H, Gao X-S, Zhang H. Knock-out of Arabidopsis AtNHX4 gene enhances tolerance to salt stress. Biochem. Biophys. Res. Commun. 2009;382:637–641. doi: 10.1016/j.bbrc.2009.03.091. [DOI] [PubMed] [Google Scholar]
- 45.Zhang C, Hicks GR, Raikhel NV. Plant vacuole morphology and vacuolar trafficking. Frontiers in plant science. 2013;5:476–476. doi: 10.3389/fpls.2014.00476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Luan S, Lan W, Lee SC. Potassium nutrition, sodium toxicity, and calcium signaling: connections through the CBL–CIPK network. Curr. Opin. Plant Biol. 2009;12:339–346. doi: 10.1016/j.pbi.2009.05.003. [DOI] [PubMed] [Google Scholar]
- 47.Weinl S, Kudla J. The CBL–CIPK Ca2+−decoding signaling network: function and perspectives. New Phytol. 2009;184:517–528. doi: 10.1111/j.1469-8137.2009.02938.x. [DOI] [PubMed] [Google Scholar]
- 48.Guo Y, Halfter U, Ishitani M, Zhu J-K. Molecular characterization of functional domains in the protein kinase SOS2 that is required for plant salt tolerance. The Plant Cell. 2001;13:1383–1400. doi: 10.1105/tpc.13.6.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ma, Y. C., Augé, R. M., Dong, C. & Cheng, Z. M. M. Increased salt tolerance with overexpression of cation/proton antiporter 1 genes: a meta‐analysis. Plant Biotechnol. J (2016). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


