Abstract
Circulating cell-free DNA (cfDNA) may be involved in immune response regulation. We studied the variations in abundance of telomeric sequences in plasma and serum in young healthy volunteers and the ability of cfDNA contained in these samples to co-activate the TNF-α m RNA expression in monocytes. We performed qPCR to determine relative telomere length (T/S ratios) in plasma, serum and whole blood of 36 volunteers. Using paired samples of plasma and serum and DNase treatment, we analysed the contribution of cfDNA to the co-activation of TNF-α mRNA expression in THP1 monocytic cell line. We found significant differences between paired plasma and serum samples in relative T/S ratios (median 1.38 ± 1.1 vs. 0.86 ± 0.25, respectively) and in total amounts of cfDNA and in estimated total amounts of telomeres which were significantly higher in serum than in plasma. TNF-α mRNA expression in THP1 cells increased significantly after DNase treatment of all samples used for stimulation. The highest TNF-α mRNA expressions were observed after stimulation with DNase treated serum samples. Our results suggest that the different content of telomeric sequences in plasma and serum may contribute to the tuning of immune response. Further studies of this interesting phenomenon are needed.
Introduction
Telomeres are known to prevent deleterious shortening of linear DNA of eukaryotic chromosomes during DNA replication. Telomere DNA contains G-rich repetitive sequences (TTAGGG in mammals). Telomeres progressively shorten after each cell division. They can be extended by the enzyme telomerase which is active mainly in embryonic stem cells1. The telomeres shorten not only with replication but also as the consequence of oxidative stress2. If the length of telomere sequences reaches a critical minimum, the telomeres are not more able to fulfil their tasks and the state of such cells may be described as replicative senescence. Therefore the length of telomeres might be used as an indicator of cellular aging and predictor of mortality. The shorter leukocyte telomere length (LTL) is reported to be associated with older age, male sex, Caucasian race, and possibly atherosclerosis but there is controversy with regard to association of LTL with other diseases of aging. Therefore the crucial question of whether shortening of the telomeres is a cause or a symptom of aging remains unanswered3. The length of telomere sequences is studied under wide clinical conditions with the goal to find markers distinguishing different health statuses of patients and allowing prognosis of their clinical outcomes4, 5. The comparison of different studies is difficult because different methodological approaches are used on various types of biological samples3. The attempt to compare and standardize the methodologies was already made6. The application of qPCR allows the estimation of the abundance of telomeric sequences in comparison with a single copy gene sequence.
Cell-free DNA (cfDNA) is recently studied with regard to its potential use as the source of useful biomarkers in various fields of medicine. The examination of foetal fraction of cfDNA in maternal circulation has been elaborated for non-invasive screening for foetal aneuploidies and chromosomal aberrations7. After establishment of methodologic approaches based on Next Generation Sequencing, the examination of cfDNA circulating in plasma became to be widely used for tumour detection and follow-up in oncologic patients8, 9. The elevated levels of cfDNA or the detection of specific gene sequences in its pool in circulation are reported in numerous clinical conditions associated with inflammation, tissue damage and cancer10–13. The detection of donor sequences in the cfDNA pool represents the important tool for monitoring of transplanted patients14. Levels of total cfDNA increase also after intensive physical activity15, 16.
Despite all above mentioned applications, the origin of cfDNA in circulation and the mode of its clearance are not fully understood. It is known that serum samples contain more cfDNA than the paired plasma samples17, examination of serum may be used to enhance the detection of cancer specific mutations in cfDNA18. The phenomenon of NETosis (Neutrophil Extracellular Trap) as quite recently described type of cell death19 could potentially provide an explanation of the different concentrations of cfDNA in paired plasma and serum samples as well as at least in some clinical situations associated with inflammation in which cfDNA levels in circulation are increased20. The main feature of NETosis is the formation of the so called extracellular traps which contain genomic DNA and wide spectrum of proteins including citrullinated histones and the enzymes released from neutrophil granules. The primary purpose of these nets formed preferentially by activated neutrophils is the entrapment of invading bacteria in circulation but NETosis may be stimulated by various signals also under sterile conditions and not only in neutrophils21. Platelet activation belongs to the well-known activators of NETosis21, 22.
Thrombi form on all known biomaterials partly due to platelet-mediated reactions and partly due to coagulation of blood plasma itself (the contact activates the blood zymogen FXII (Hageman factor) into an active enzyme form FXIIa23. Fuchs et al.24 found that NETs are liberated during storage of non-leukoreduced red blood cells (RBC).
Despite all these facts, the biological roles of cfDNA in circulation are very rarely studied25. In this study, we compared the relative amount of telomeric sequences in paired samples of plasma and serum in young healthy volunteers. We examined the effects of selected paired samples of plasma and serum on the stimulation of TNF-α (Tumor Necrosis Factor-α) mRNA expression in monocytic cell line THP1 before and after DNase treatment of these biological fluids.
To our best knowledge, we present the first study demonstrating the different effects of plasma and serum of young healthy individuals on the regulation of immune response.
It seems that especially telomere sequences in the pool of cell-free DNA in circulation may play regulatory roles with regard to the fine tuning of immune system. Vertebrate DNA modified by apoptosis stimulates Toll-like receptor 9 (TLR9) which activates nuclear transcription factors including NFκB. These transcription factors promote increased expression and release of wide spectrum of immune-regulatory molecules26 but telomeric sequences are able to block TLR927, 28. Therefore we focused also on analysis of content of telomeric sequences in plasma and serum samples together with examination of their stimulatory effects on THP1 cells. Relative content of telomeric sequences in whole blood samples was examined due to the known fact that blood cells represent the main source of cfDNA in circulation29.
Results
Relative quantification of telomeric sequences
The construction of standard curves and the principles of calculations (total amount of cfDNA and T/S ratio) are described in Methods. The standard curves were set at the start of experiments and used for the entire study. Due to nonparametric data distribution, we report medians and standard deviations in the following text.
The total amount of cfDNA measured using qPCR on single copy gene was significantly (p < 0.0001, Wilcoxon test) higher in serum (148.13 ± 91.31 ng/ml, range 54.33–427.30 ng/ml) than in plasma (7.48 ± 5 ng/ml, range 3.16–26.05 ng/ml). Box plots are shown in Fig. 1. No correlations in total amounts of cfDNA between plasma and serum samples were detected in paired samples.
Figure 1.
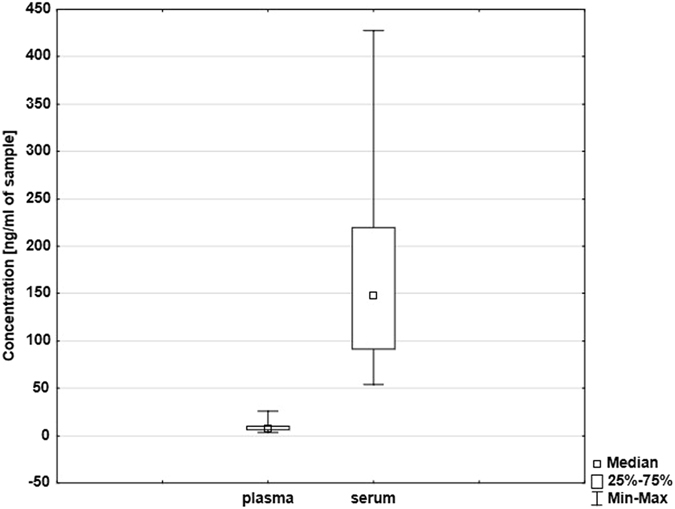
Box plot showing different concentrations of total cfDNA in plasma and serum samples [ng/ml] measured by qPCR for single copy gene 36B4 (n = 36).
Significant differences in the relative T/S ratios were found among whole blood, plasma and serum samples. These ratios were significantly (p < 0.0001, Dunn test) higher in plasma than in serum samples (1.38 ± 1.1 vs. 0.86 ± 0.25, respectively) and in plasma than in whole blood samples (1.38 ± 1.1 vs. 0.72 ± 0.25, respectively). In serum samples, we observed higher ratios than in whole blood samples too (0.86 ± 0.25 vs. 0.72 ± 0.25, respectively, p = 0.0225, Dunn test). All these results are documented in Figs 2 and 3.
Figure 2.
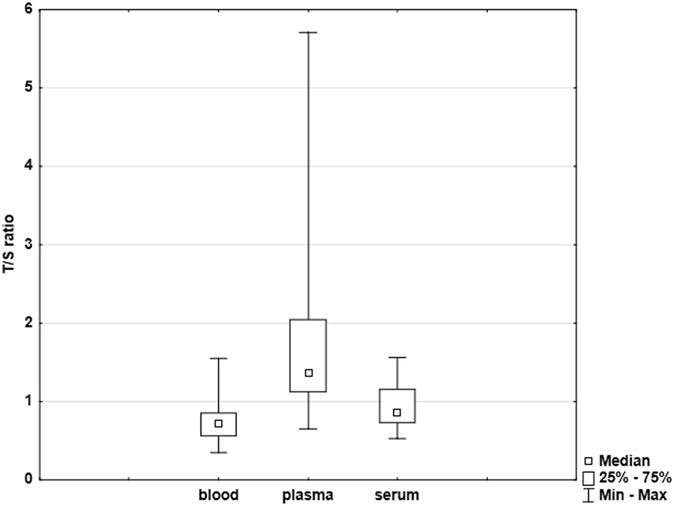
Box plot showing relative T/S ratios for whole blood, plasma and serum samples (n = 36).
Figure 3.
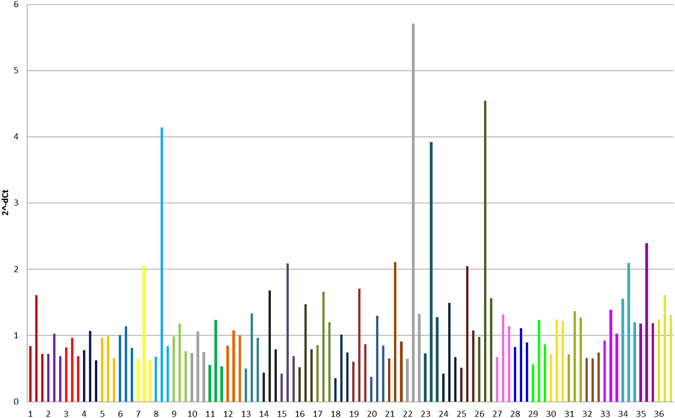
Differences in relative T/S ratios among whole blood, plasma and serum samples in 36 healthy young volunteers. Individuals are marked by different colours. The order of samples for each individual is following: whole blood, plasma and serum.
Figure 3 clearly demonstrates that the relative T/S ratios in all types of biological material in nearly all examined subjects follow the similar scenario with the highest values in plasma, intermediate values in serum and the lowest ones in whole blood.
Stimulations with paired plasma and serum samples with known T/S ratios and cfDNA concentrations
For stimulations, we selected randomly paired plasma and serum samples of individuals no. 12, 13, 16, 18, 21, 22, 27, 31, 33 and 36 (Fig. 3). Relative T/S ratios from previous experiments were reanalysed for this chosen samples and significant differences in the relative T/S ratios were confirmed again (plasma 1.38 ± 1.32 vs. serum 1.01 ± 0.2, p = 0.005, Wilcoxon test) (Table 1). Plasma and serum samples treated by DNase were examined by qPCR for single copy gene (36B4) and they do not revealed any amplification signals.
Table 1.
Selected characteristics of samples used for stimulatory experiments. Wilcoxon signed-rank test was used.
| Individual. No. | T/S ratio | ETAT [ng/ml] | ||
|---|---|---|---|---|
| Plasma | Serum | Plasma | Serum | |
| 12 | 1.08 | 1.00 | 10.15 | 163.24 |
| 13 | 1.33 | 0.96 | 34.29 | 73.99 |
| 16 | 1.48 | 0.80 | 10.98 | 85.64 |
| 18 | 1.01 | 0.75 | 7.69 | 69.78 |
| 21 | 2.11 | 0.9 | 16.51 | 248.43 |
| 22 | 5.71 | 1.33 | 47.79 | 150.37 |
| 27 | 1.32 | 1.14 | 8.01 | 357.47 |
| 31 | 1.37 | 1.27 | 15.17 | 307.79 |
| 33 | 1.39 | 1.03 | 10.37 | 71.04 |
| 36 | 1.61 | 1.31 | 25.38 | 188.26 |
| p values | 0.005 | 0.005 | ||
In case of stimulation by DNase treated and non-treated plasma, we found significant difference (p = 0.005) between the levels of TNF-α mRNA expression. DNase treatment of plasma samples led to statistically significant enhancement of their stimulatory activity in the term of increase of relative TNF-α mRNA expression (0.14 ± 0.02 vs. 0.07 ± 0.02, respectively) (Fig. 4). The same trend was observed between treated and non-treated serum samples (0.29 ± 0.05 vs. 0.21 ± 0.05 respectively, p = 0.013). Statistically significant (p = 0.005, Wilcoxon test) differences were observed in stimulation activity between non-treated plasma and serum samples. DNase non-treated serum samples stimulated the relative TNF-α mRNA expression in higher extent than non-treated plasma samples (0.21 ± 0.05 vs. 0.07 ± 0.02, respectively). DNase treated serum samples had also higher stimulatory effect than treated plasma samples T (0.29 ± 0.050 vs.14 ± 0.02, respectively). The results of stimulatory experiments are summarized in Fig. 4.
Figure 4.
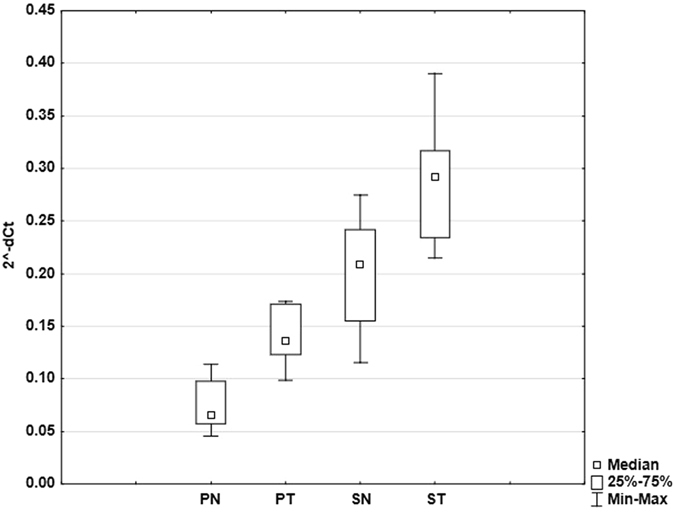
Graph showing the relative TNF-α mRNA expression after stimulation of THP1 cells with different types of sample (PN – non-treated plasma, PT – plasma treated with DNase, SN – non-treated serum, ST – serum treated with DNase). Bars show standard deviations (n = 10).
When we compared the performance of all ten quartets of samples in stimulatory experiments we observed again nearly the identical scenario in all examined individuals: The lowest values of relative TNF-α mRNA expression were found when the cells were stimulated using DNase non-treated plasma samples. After DNase treatment, the plasma samples enhanced their stimulatory capacity (fold change difference 1.86) which was still lower or almost identical as stimulatory capacity of DNase non-treated serum samples. DNase treatment of serum samples in most cases enhanced their stimulatory capacity (fold change difference 1.4) above the capacity of non-treated serum.
To estimate the content of telomeric sequences in each sample used for stimulatory experiment, we calculated ETAT values as described in Methods. We found significant differences (p = 0.005, Wilcoxon test) between ETAT values of plasma and serum samples (Table 1).
Discussion
In agreement with previously known facts17, we found statistically significant differences between the total amounts of cfDNA in plasma and serum with considerably higher levels in serum. Such results consistently found in most of individuals examined in this study provided the evidence that there were no pre-analytical events leading to more intensive degradation of cfDNA in serum samples.
Our comparison of relative T/S ratios between plasma and serum with significantly higher values in plasma demonstrates probably the different mode of physiological processing of cfDNA in these biological fluids. Serum contains higher content of telomeric sequences than plasma (higher ETAT values in this study) due to its higher amounts of total cfDNA.
Serum formation itself is a very complex process dependent on coagulation mechanisms which are associated with the activation of platelets and release of pro-inflammatory cytokines30. This fact is in good agreement with the higher stimulatory activity of native serum samples in comparison with plasma samples in our experiments on THP1 monocytic cell line. NETosis activated by complex stimuli during coagulation and serum formation21, 22 may be the main source of elevated levels of total cfDNA in serum.
Our comparison of T/S ratios in whole blood, plasma and serum provides the first evidence for the existence of specific mechanisms which are responsible for maintenance of telomeric sequences in circulation. The origin of cfDNA could determine the character and alternatively the mode of clearance of specific sequences in their pool in circulation - cfDNA released during NETosis may be decorated with specific proteins - e.g. citrullinated histones24 and therefore easily recognizable by molecular mechanisms.
It seems that especially telomeric sequences in circulation may play regulatory roles in the fine tuning of immune system. Some studies27, 28 provide the evidence that the telomeric sequences are able to block TLR9 and to inhibit immune response and that this activity is associated with the ability of telomeric repeat sequences to form 3-dimesional structures based on quainine-quadruplexes26.
For example, it is experimentally proven that artificially prepared telomeric sequences are able to inhibit proliferation of B-cells regulated via TLR28.
With regard to the interpretation of our stimulation experiment, it is necessary to keep in mind that cfDNA and telomeric sequences contained in its pool are not the only regulators of immune response in such complex biological fluids. Our experiments clearly demonstrated the difference in the stimulatory properties of paired DNase non-treated samples of plasma and serum of identical individuals. Stimulatory effects of non-treated serum samples were higher than the effect of non-treated plasma (Fig. 4). The experiments with the complete removal of DNA using DNase digestion demonstrated clearly the important role of cfDNA and its telomeric sequences in plasma and serum of healthy young individuals in the inhibition of immune response. The results are in full agreement with the known facts about the involvement of cell-free telomeric sequences in the regulation of immune functions26–28.
The telomere sequences in plasma samples were examined till today only in two studies31, 32. Wu and Tanaka found significantly shortened telomeres in cfDNA in plasma of breast cancer patients with no prior treatment32. Idei et al. reported similar results in patients with ovarian cancer31. Two studies were focused also on analysis of telomere sequences in serum. Fu et al. conducted a study to determine the relative telomere length (RTL) in serum samples from hepatitis B virus (HBV) -related hepatocellular carcinoma patients and cancer-free HBV controls33. Cancer patients had a significantly longer RTL in cfDNA in serum. Wan et al. found significantly longer RTL in cfDNA in serum in cirrhotic cases than in non-cirrhotic controls and proposed the determination of RTL in serum as suitable non-invasive marker for detection of patients with the risk of cirrhosis34. The terms “longer” or”shorter” telomeres in the texts of these publications are inappropriate because the methodologies used by these researchers examine rather the abundance of telomeric sequences in samples and are not able to provide the results documenting the lengths of the longest and/or shortest telomeres in examined samples. The results of such studies analysing the abundance of telomeric sequences in plasma or serum in cancer patients should be interpreted in association with the selected markers describing the functions of immune system of examined individuals to allow better evaluation of observed changes in the context of immune response associated with tumour development.
Our results document the existence of molecular mechanisms which are responsible for maintenance of appropriate levels of free telomeric sequences in human circulation and the role of cfDNA in plasma and serum in the regulation of immune response. According to our results, it seems that telomeric sequences in native plasma and serum samples may contribute to the inhibition of TNF-α mRNA expression in monocytes. Further studies are needed to elucidate the character of these physiological regulatory events and the consequences of their disturbances for human health.
Methods
Subjects
Healthy volunteers (n = 36, 17 men and 19 women) – participants of the study – were 20 to 25 years old students. Blood withdraws were performed during the everyday student’s activities without any excessive physical load. The study was approved by the Ethical committee of the 1st Faculty of Medicine of Charles University and General Faculty Hospital in Prague. Informed consent was obtained from all participants included in the study. All methods were performed in accordance with the relevant guidelines and regulations.
Blood, serum and plasma sampling
This study compares content of telomere sequences in three types of biological samples – whole blood, plasma and serum. In case of plasma and whole blood, the blood was collected into tubes containing EDTA (BD Vacutainer), serum samples were collected into tubes with clot activator and polymer gel (BD Vacutainer). The whole blood (300 μl) for DNA isolation was taken from EDTA tubes before the processing of samples for plasma isolation. For plasma and serum isolation, the whole blood was centrifuged at 2600 rpm for 10 min at 10 °C. After this procedure, the collected material was centrifuged once again in 2 ml low-bind tubes at 14500 rpm for 10 min to remove residual cells. All samples were stored at −20 °C until DNA and cfDNA isolation.
DNA isolation and qPCR
CfDNA from 1 ml plasma or serum was isolated by QIAamp® Circulating Nucleic Acid Kit (QIAGEN, Germany) according to the manufacturer’s instructions and eluted in 50 μl of elution buffer and 5 μl of this solution was used for each amplification reaction. Whole blood DNA was isolated by QIAamp® DNA Blood Mini Kit (QIAGEN, Germany) and eluted in 200 μl of elution buffer and then diluted 15-fold and also 5 μl of this diluted solution was used for each amplification reaction.
DNA isolations were immediately followed by qPCR which was performed at QuantStudio™ 12 K Flex Real-Time PCR System (Applied Biosystems, USA). Final volume of a PCR reaction was 25 μl, one half of this volume consisted of Power SYBR® Green PCR Master Mix (Life Technologies, USA). Primers for amplification of telomere sequences (“telomeres”) and single copy gene 36B4 (coding acidic ribosomal phosphoprotein PO) were chosen from literature35, 36. Final concentrations of primers were 300 nM for single copy gene (36B4) and 900 nM for telomeres (tel). The primer sequences were as follow: 36B4u: 5′-CAG CAA GTG GGA AGG TGT AAT CC-3′; 36b4d: 5′-CCC ATT CTA TCA TCA ACG GGT ACA A-3′35, 36; tel 1 1b: 5′-CGG TTT GTT TGG GTT TGG GTT TGG GTT TGG GTT TGG GTT-3′; tel 1 2b: 5′-GGC TTG CCT TAC CCT TAC CCT TAC CCT TAC CCT TAC CCT-3′36. All reactions were run in triplicates. The hold stage which activated the enzyme included 2 steps: 50 °C for 2 min and 95 °C for 2 min. This stage was followed by 40 cycles of PCR: 95 °C 30 s and 54 °C 1 min. At the end of each run, there was a dissociation step (default software setting). Ramp rates between all steps were 1.6 °C/s. The standard curves for both amplification reactions were constructed before the processing of samples. Standard DNA was diluted and used at concentration 5–0.312 ng per reaction. Amplification efficiency was 90.85% for 36B4 (slope −3.562, R2 0.998 and error 0.041), and 97.94% for “telomeres” (slope −3.372, R2 0.998 and error 0,043).
In each PCR run, the inter-run calibration reaction containing 20 ng of standard DNA was included. For construction of standard curves, for inter-run calibration and for calculation of relative T/S ratio, the standard DNA (Taq Man Control Genomic DNA, Applied Biosystems, USA) was used.
The T/S ratio was calculated as 2−[Ct (telomeres)/Ct (36B4)] = 2−∆Ct. To determine the relative T/S ratio of the sample, its T/S ratio was normalized to ∆Ct of the standard. The final formula for relative T/S ratio was 2−(∆CtSample−∆CtStandard).
The logarithm of the total amount of cfDNA (x) was calculated using the experimental data for 36B4 and following equation: y = Ax + B where A is slope, B is Y-inter, and y is Ct.
Cell cultivation and stimulation
For stimulations, paired samples of plasma and serum from 10 young healthy volunteers were selected from the sample set analysed in previous experiments leading to the determination of T/S ratios and total cfDNA in these samples. At least 5 × 105 cells of THP-1 monocytic cell line were cultivated in 24 well plate (Orange Scientific, Belgium) in 1 ml of RPMI 1640 medium (Sigma Aldrich, USA) supplemented with 10% plasma or serum of healthy volunteer, 200 mM L-glutamine and 1% penicillin/streptomycin (Sigma Aldrich, USA).
Each sample of plasma and serum was divided into two technical replicates. One of each replicate was treated by DNase Turbo DNA-freeTM (Ambion by Life Technologies, USA) and also added in 10% concentration to the cultivation medium and used in parallel with its non-treated replicate in the stimulatory experiment. In plasma samples, the DNases are inhibited by EDTA immediately after blood withdraws. EDTA chelates divalent ions which are essential for the activity of these enzymes37. The DNase Turbo kit contains a specific buffer for proper supplementations of reaction with ions to allow the DNase activity. All four samples (DNase – treated and non-treated plasma, DNase – treated and non-treated serum) from one healthy volunteer were stimulated at 37 °C in 5% CO2 for 3 hours. After this interval, samples were centrifuged at 14 000 g for 6 min. Supernatant was removed and cells were resuspended in 500 μl Lysis Solution (GenEluteTM Mammalian Total RNA Miniprep Kit, Sigma Aldrich, USA).
RNA isolation, reverse transcription and qPCR
RNA was isolated by GenEluteTM Mammalian Total RNA Miniprep Kit (Sigma Aldrich, USA). Extracted RNA was reverse-transcribed into cDNA by HyperScriptTM One-step RT-PCR Master mix (GeneAll, Korea). QPCR was performed in LightCycler®480 System (Roche Applied, USA) with Assays-on-Demand TaqMan® Gene Expression Assay Mix (Life Technologies-Applied Biosystems,USA) for PGK1 Hs99999906_m1 (Phosphoglycerate Kinase-1) as a reference gene and TNF-α Hs01113624_g1. The expressions of both genes were examined in all samples, the levels of TNF-α mRNA expression were given as = 2−∆Ct related to the reference gene PGK1.
Statistical analysis
We used nonparametric statistical tests due to small sample sizes. We employed Wilcoxon test (STATISTICA, StatSoft, USA) for paired samples and Friedman tests for multiple samples (comparison of whole blood, plasma and serum). We used significance level α = 0.05 for all comparisons.
cfDNA analysis
We performed Friedman rank-sum at R environment to reveal any differences among whole blood, plasma and serum. Then we used Dunn post hoc test with Bonferroni correction to find out T/S ratio differences among blood-plasma, blood-serum and plasma-serum samples.
Analysis of stimulation experiments
Wilcoxon signed-rank tests were used for all comparisons. To study the amount of telomeric sequences in samples used for stimulation, ETAT (Estimated Total Amount of Telomeres) was calculated as T/S ratio of the sample multiplied by its total cfDNA concentration determined by qPCR on single copy gene using calibration curve for this gene.
Acknowledgements
This study was funded by the grant no. PRVOUK P25/LF1/2 of the Ministry of Education, Youth and Sport of the Czech Republic, by the grant RVO-VFN 64165 of the Ministry of Health of the Czech Republic and by the grants no. SVV 260 263 and SVV 260 312 of the Ministry of Education, Youth and Sport of the Czech Republic. We thank Ales Horinek from Department of Biology and Medical Genetics, First Faculty of Medicine, Charles University and General Faculty Hospital in Prague for helpful technical assistance, Dr. Pavlina Dankova from Department of Anthropology and Human Genetics, Faculty of Science of Charles University for kind support and Dr. Jan Hanzelka from Institute for Environmental Studies Faculty of Science of Charles University for statistics consultations.
Author Contributions
A.Z. and I.B. performed the experiments and statistical analysis, A.S. was responsible for clinical part of study (obtaining and management of samples). M.K. and M.J. initiated the study. M.K. interpreted the data and participated on manuscript preparation. All authors read the manuscript critically.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Lenart P, Krejci L. DNA, the central molecule of aging. Mutat Res. 2016;786:1–7. doi: 10.1016/j.mrfmmm.2016.01.007. [DOI] [PubMed] [Google Scholar]
- 2.Kepinska M, Szyller J, Milnerowicz H. The influence of oxidative stress induced by iron on telomere length. Environ Toxicol Pharmacol. 2015;40:931–935. doi: 10.1016/j.etap.2015.10.002. [DOI] [PubMed] [Google Scholar]
- 3.Glei DA, Goldman N, Weinstein M, Risques RA. Shorter Ends, Faster End? Leukocyte Telomere Length and Mortality Among Older Taiwanese. J Gerontol A Biol Sci Med Sci. 2015;70:1490–1498. doi: 10.1093/gerona/glu191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mather KA, Jorm AF, Parslow RA, Christensen H. Is telomere length a biomarker of aging? A review. J Gerontol A Biol Sci Med Sci. 2011;66:202–213. doi: 10.1093/gerona/glq180. [DOI] [PubMed] [Google Scholar]
- 5.Sanders JL, Newman AB. Telomere length in epidemiology: a biomarker of aging, age-related disease, both, or neither? Epidemiol Rev. 2013;35:112–131. doi: 10.1093/epirev/mxs008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martin-Ruiz CM, et al. Reproducibility of telomere length assessment: an international collaborative study. Int J Epidemiol. 2015;44:1673–1683. doi: 10.1093/ije/dyu191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lefkowitz, R. B. et al. Clinical validation of a noninvasive prenatal test for genomewide detection of fetal copy number variants. Am J Obstet Gynecol, doi:10.1016/j.ajog.2016.02.030 (2016). [DOI] [PubMed]
- 8.Lanman RB, et al. Analytical and Clinical Validation of a Digital Sequencing Panel for Quantitative, Highly Accurate Evaluation of Cell-Free Circulating Tumor DNA. PLoS One. 2015;10:e0140712. doi: 10.1371/journal.pone.0140712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Francis G, Stein S. Circulating Cell-Free Tumour DNA in the Management of Cancer. Int J Mol Sci. 2015;16:14122–14142. doi: 10.3390/ijms160614122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clementi A, et al. The Role of Cell-Free Plasma DNA in Critically Ill Patients with Sepsis. Blood Purif. 2016;41:34–40. doi: 10.1159/000440975. [DOI] [PubMed] [Google Scholar]
- 11.Wang L, et al. Plasma nuclear and mitochondrial DNA levels in acute myocardial infarction patients. Coron Artery Dis. 2015;26:296–300. doi: 10.1097/MCA.0000000000000231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Purhonen AK, et al. Human plasma cell-free DNA as a predictor of infectious complications of neutropenic fever in hematological patients. Infect Dis (Lond) 2015;47:255–259. doi: 10.3109/00365548.2014.985711. [DOI] [PubMed] [Google Scholar]
- 13.Lehmann-Werman R, et al. Identification of tissue-specific cell death using methylation patterns of circulating DNA. Proc Natl Acad Sci USA. 2016;113:E1826–1834. doi: 10.1073/pnas.1519286113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oellerich M, et al. Graft-Derived Cell-Free DNA as a Marker of Transplant Graft Injury. Ther Drug Monit. 2016;38(Suppl 1):S75–79. doi: 10.1097/FTD.0000000000000239. [DOI] [PubMed] [Google Scholar]
- 15.Atamaniuk J, et al. Effects of ultra-marathon on circulating DNA and mRNA expression of pro- and anti-apoptotic genes in mononuclear cells. Eur J Appl Physiol. 2008;104:711–717. doi: 10.1007/s00421-008-0827-2. [DOI] [PubMed] [Google Scholar]
- 16.Atamaniuk J, et al. Cell-free plasma DNA and purine nucleotide degradation markers following weightlifting exercise. Eur J Appl Physiol. 2010;110:695–701. doi: 10.1007/s00421-010-1532-5. [DOI] [PubMed] [Google Scholar]
- 17.Nie K, Jia Y, Zhang X. Cell-free circulating tumor DNA in plasma/serum of non-small cell lung cancer. Tumour Biol. 2015;36:7–19. doi: 10.1007/s13277-014-2758-3. [DOI] [PubMed] [Google Scholar]
- 18.Jackson JB, et al. Multiplex Preamplification of Serum DNA to Facilitate Reliable Detection of Extremely Rare Cancer Mutations in Circulating DNA by Digital PCR. J Mol Diagn. 2016;18:235–243. doi: 10.1016/j.jmoldx.2015.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brinkmann V, et al. Neutrophil extracellular traps kill bacteria. Science. 2004;303:1532–1535. doi: 10.1126/science.1092385. [DOI] [PubMed] [Google Scholar]
- 20.Korabecna, M. & Tesar, V. NETosis provides the link between activation of neutrophils on hemodialysis membrane and comorbidities in dialyzed patients. Inflamm Res, doi:10.1007/s00011-016-1010-6 (2016). [DOI] [PMC free article] [PubMed]
- 21.Fadini GP, et al. A perspective on NETosis in diabetes and cardiometabolic disorders. Nutr Metab Cardiovasc Dis. 2016;26:1–8. doi: 10.1016/j.numecd.2015.11.008. [DOI] [PubMed] [Google Scholar]
- 22.Clark SR, et al. Platelet TLR4 activates neutrophil extracellular traps to ensnare bacteria in septic blood. Nat Med. 2007;13:463–469. doi: 10.1038/nm1565. [DOI] [PubMed] [Google Scholar]
- 23.Vogler EA, Siedlecki CA. Contact activation of blood-plasma coagulation. Biomaterials. 2009;30:1857–1869. doi: 10.1016/j.biomaterials.2008.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fuchs TA, et al. Neutrophils release extracellular DNA traps during storage of red blood cell units. Transfusion. 2013;53:3210–3216. doi: 10.1111/trf.12203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Atamaniuk J, Kopecky C, Skoupy S, Saemann MD, Weichhart T. Apoptotic cell-free DNA promotes inflammation in haemodialysis patients. Nephrol Dial Transplant. 2012;27:902–905. doi: 10.1093/ndt/gfr695. [DOI] [PubMed] [Google Scholar]
- 26.Phillippe M. Cell-Free Fetal DNA, Telomeres, and the Spontaneous Onset of Parturition. Reprod Sci. 2015;22:1186–1201. doi: 10.1177/1933719115592714. [DOI] [PubMed] [Google Scholar]
- 27.Gursel I, et al. Repetitive elements in mammalian telomeres suppress bacterial DNA-induced immune activation. J Immunol. 2003;171:1393–1400. doi: 10.4049/jimmunol.171.3.1393. [DOI] [PubMed] [Google Scholar]
- 28.Sackesen C, et al. Suppression of B-cell activation and IgE, IgA, IgG1 and IgG4 production by mammalian telomeric oligonucleotides. Allergy. 2013;68:593–603. doi: 10.1111/all.12133. [DOI] [PubMed] [Google Scholar]
- 29.Lui YY, et al. Predominant hematopoietic origin of cell-free DNA in plasma and serum after sex-mismatched bone marrow transplantation. Clin Chem. 2002;48:421–427. [PubMed] [Google Scholar]
- 30.Yu Z, et al. Differences between human plasma and serum metabolite profiles. PLoS One. 2011;6:e21230. doi: 10.1371/journal.pone.0021230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Idei T, Sakamoto H, Yamamoto T. Terminal restriction fragments of telomere are detectable in plasma and their length correlates with clinical status of ovarian cancer patients. J Int Med Res. 2002;30:244–250. doi: 10.1177/147323000203000304. [DOI] [PubMed] [Google Scholar]
- 32.Wu X, Tanaka H. Aberrant reduction of telomere repetitive sequences in plasma cell-free DNA for early breast cancer detection. Oncotarget. 2015;6:29795–29807. doi: 10.18632/oncotarget.5083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fu X, et al. Relative telomere length: a novel non-invasive biomarker for the risk of non-cirrhotic hepatocellular carcinoma in patients with chronic hepatitis B infection. Eur J Cancer. 2012;48:1014–1022. doi: 10.1016/j.ejca.2012.02.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wan S, et al. Telomere length in circulating serum DNA as a novel non-invasive biomarker for cirrhosis: a nested case-control analysis. Liver Int. 2012;32:1233–1241. doi: 10.1111/j.1478-3231.2012.02801.x. [DOI] [PubMed] [Google Scholar]
- 35.Cawthon RM. Telomere measurement by quantitative PCR. Nucleic Acids Res. 2002;30:e47–47. doi: 10.1093/nar/30.10.e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gil ME, Coetzer TL. Real-time quantitative PCR of telomere length. Mol Biotechnol. 2004;27:169–172. doi: 10.1385/MB:27:2:169. [DOI] [PubMed] [Google Scholar]
- 37.Macanovic M, Lachmann PJ. Measurement of deoxyribonuclease I (DNase) in the serum and urine of systemic lupus erythematosus (SLE)-prone NZB/NZW mice by a new radial enzyme diffusion assay. Clin Exp Immunol. 1997;108:220–226. doi: 10.1046/j.1365-2249.1997.3571249.x. [DOI] [PMC free article] [PubMed] [Google Scholar]


