Abstract
High risk human papilloma viruses cause several types of cancer. The HPV oncoproteins E6 and E7 are essential for oncogenic cell transformation. E6 mediates the degradation of the tumor suppressor p53, and E7 can form complexes with the retinoblastoma pRB tumor suppressor. Recently, it has been shown that HPV E7 can also interfere with the function of the DREAM transcriptional repressor complex. Disruption of DREAM-dependent transcriptional repression leads to untimely early expression of central cell cycle regulators. The p53-p21-DREAM pathway represents one important means of cell cycle checkpoint activation by p53. By activating this pathway, p53 can downregulate transcription of genes controlled by DREAM. Here, we present a genome-wide ranked list of genes deregulated by HPV E7 expression and relate it to datasets of cell cycle genes and DREAM targets. We find that DREAM targets are generally deregulated after E7 expression. Furthermore, our analysis shows that p53-dependent downregulation of DREAM targets is abrogated when HPV E7 is expressed. Thus, p53 checkpoint control is impaired by HPV E7 independently of E6. In summary, our analysis reveals that disruption of DREAM through the HPV E7 oncoprotein upregulates most, if not all, cell cycle genes and impairs p53’s control of cell cycle checkpoints.
Introduction
High risk human papilloma viruses (HPV) are oncogenic DNA viruses that can cause cancer of the cervix uteri, oropharynx, penis, vagina, vulva and anus1–4. The primary transforming capacities of HPV stem from the E6 and E7 proteins. These two oncoproteins cooperate in silencing the anti-proliferative control of the cell. The best-known target of E7 is the cell cycle regulator pRB5. Direct binding of E7 to the retinoblastoma tumor suppressor protein pRB impairs its function6–8. Furthermore, association of E7 with pRB leads to an increase in p53 levels9. The tumor suppressor p53 can either trigger checkpoints causing cell cycle arrest or lead to the induction of apoptosis. In the context of HPV infection, the E6 oncoprotein initiates degradation of p5310, 11.
Moreover, the E7 oncoprotein has additional functions aside from targeting pRB12, 13. These include means to impair p53 function even in the absence of E6. Thus, HPV E7 is sufficient to block cell cycle checkpoint control by p5314–18. The cyclin-dependent kinase (CDK) inhibitor p21 (CDKN1A) is a central mediator of p53 checkpoint control19, 20, and its function can be impaired by E718, 21, 22. This finding is particularly interesting given that p21 is required for downregulation of genes in response to p5323, 24.
Recently, another mechanism to impair p53 function that is independent of E6 was discovered. HPV E7 was found to disrupt the pRB-related transcriptional repressor complex DREAM (DP, RB-like, E2F4 and MuvB)25–27. The DREAM protein complex consists of E2F4, DP1 and p130/p107 in addition to RBBP4 and the LIN proteins LIN9, LIN37, LIN52 and LIN54 that form the MuvB core28–30. DREAM binds promoters through cell cycle-dependent elements (CDEs), cell cycle genes homology regions (CHRs), CHR-like elements (CLEs) and E2F sites31–35. In response to p53, DREAM is recruited to promoters of cell cycle genes, leading to their repression36, 37. While p53 itself is solely an activator of transcription, the p53-p21-DREAM pathway mediates indirect gene downregulation by p5338, 39. For example, Polo-like kinase 4 is an important target of this pathway. The mitotic kinase PLK4 is repressed through the p53-p21-DREAM-CDE/CHR pathway40, and its p53-dependent repression can be abrogated by HPV E741. Importantly, CDE/CHR elements are required for p53-dependent repression of PLK4, and the expression of HPV E7 impairs DREAM binding to the CDE/CHR elements in the PLK4 promoter41. As genome-wide expression profiling datasets of E7-expressing IMR90 lung fibroblasts42 and NIKS keratinocytes43 became recently available, we asked whether targets of the p53-p21-DREAM pathway are generally deregulated by HPV E7 on a genome-wide level.
Here, we integrate these new data with earlier genome-wide datasets that were derived from comparing HPV-16/18-infected cervical tumor samples with normal tissue44, 45, from CaSki cells expressing HPV E2C, a potent transcriptional repressor of E6 and E7 46, or from HeLa cells in which E6 and E7 were downregulated by RNAi47. Our analysis identifies genes that were observed as E7-regulated in most datasets, and we compared the results with lists of DREAM and pRB-E2F target genes23. We found that many DREAM targets are upregulated by E7 and that DREAM targets are the main class of genes deregulated by E7. Most importantly, p53-dependent downregulation of DREAM target genes is abrogated in HPV E7-expressing cells. In summary, our analysis provides a genome-wide high-confidence list of genes deregulated by HPV E7, most of which are DREAM targets. This study reveals the importance of E7-mediated DREAM disruption that interferes with p53-dependent gene downregulation. Thus, in HPV-infected cells, p53 function can be impaired by E7 independently of E6.
Materials and Methods
Computational analysis
A step-wise meta-analysis approach was employed to integrate multiple datasets23. This approach enables the integration of pre-analyzed datasets and does not require re-analysis of the raw data. Publicly available HPV E7 gene expression profiling datasets were curated42–47 and mapped to a collection of protein-coding genes23. Expression values of the analyzed genes were compiled and classified into downregulated (−1), upregulated (+1) and non-regulated genes (0).
Genes identified as significantly differentially regulated in HPV-16 E7 expressing NIKS cells were retrieved from Table 2 in Zhou et al.43. The pre-analyzed dataset of Rozenblatt-Rosen et al. from HPV-18 E7-expressing IMR90 cells was retrieved from the deposited Supplementary Table 19 in Rozenblatt-Rosen et al.42 and a gene was considered significantly differentially regulated if it passed the thresholds of adj. p-value ≤ 0.05 and absolute log2 (fold-change expression) ≥0.5. Genes identified as significantly differentially regulated in HPV-16/18 infected early stage cervical cancers compared to normal cervical epithelium were retrieved from the deposited Tables 2 and 3 in Santin et al.45. The pre-analyzed dataset from HeLa cells in which endogenous HPV-18 E6 and E7 expression was silenced by RNAi displays significantly differentially expressed genes and was retrieved from the Supplementary Table S1 in Kuner et al.47. Genes identified as significantly upregulated in HPV-16/18 infected primary cervical tumors compared to control cells were named “cervical cancer proliferation cluster” and were retrieved from Table 2 in Rosty et al.44. The pre-analyzed dataset from HPV E2C-expressing CaSki cells displays significantly differentially expressed genes and was retrieved from the Supplementary Table S1 in Pang et al.46. Of note, datasets by Rosty et al. and Pang et al. exclusively reported upregulated genes.
Table 2.
HPV E7-deregulated genes with an identification-overlap of at least four out of six datasets.
| Gene Symbol | Identified as upregulated by E7 in No. of datasets | Cell cycle gene | DREAM target | MMB-FOXM1 target | pRB-E2F target |
|---|---|---|---|---|---|
| MCM2 | 6 | G1/S | ✓ | × | ✓ |
| ZWINT | 6 | G1/S | ✓ | × | × |
| APOBEC3B | 5 | G2/M | ✓ | × | × |
| CDC6 | 5 | G1/S | ✓ | × | ✓ |
| KIF2C | 5 | G2/M | ✓ | ✓ | × |
| LMNB1 | 5 | G2/M | ✓ | ✓ | ✓ |
| MCM4 | 5 | G1/S | ✓ | × | ✓ |
| MYBL2 | 5 | G1/S | ✓ | × | ✓ |
| NUSAP1 | 5 | G2/M | ✓ | ✓ | ✓ |
| PRC1 | 5 | G2/M | ✓ | ✓ | × |
| RRM2 | 5 | G1/S | ✓ | ✓ | ✓ |
| SMC4 | 5 | G2/M | ✓ | ✓ | ✓ |
| STIL | 5 | G2/M | ✓ | ✓ | ✓ |
| TOP2A | 5 | G2/M | ✓ | ✓ | × |
| ASF1B | 4 | G1/S | ✓ | ✓ | ✓ |
| ASPM | 4 | G2/M | ✓ | ✓ | × |
| ATAD2 | 4 | G1/S | ✓ | ✓ | ✓ |
| BIRC5 | 4 | G2/M | ✓ | ✓ | × |
| BRCA1 | 4 | G1/S | ✓ | × | ✓ |
| CCNA2 | 4 | G2/M | ✓ | ✓ | × |
| CCNB1 | 4 | G2/M | ✓ | ✓ | × |
| CCNB2 | 4 | G2/M | ✓ | ✓ | × |
| CDC20 | 4 | G2/M | ✓ | ✓ | × |
| CDC25C | 4 | G2/M | ✓ | ✓ | × |
| CDC45 | 4 | G1/S | ✓ | × | ✓ |
| CDKN2A | 4 | × | × | × | × |
| CENPF | 4 | G2/M | ✓ | ✓ | × |
| DTL | 4 | G1/S | ✓ | ✓ | ✓ |
| E2F1 | 4 | G1/S | ✓ | × | ✓ |
| FANCI | 4 | G1/S | ✓ | × | ✓ |
| FKBP5 | 4 | UNKN | ✓ | × | × |
| FOXM1 | 4 | G2/M | ✓ | × | × |
| GINS2 | 4 | G1/S | ✓ | × | ✓ |
| KIF20A | 4 | G2/M | ✓ | ✓ | × |
| KIF23 | 4 | G2/M | ✓ | ✓ | × |
| MELK | 4 | G2/M | ✓ | × | × |
| NCAPG2 | 4 | G1/S | ✓ | × | ✓ |
| NEK2 | 4 | G2/M | ✓ | ✓ | × |
| POLQ | 4 | G2/M | ✓ | × | × |
| PRIM1 | 4 | G1/S | ✓ | × | ✓ |
| PTTG1 | 4 | G2/M | ✓ | ✓ | × |
| RAD51AP1 | 4 | G1/S | ✓ | × | ✓ |
| RFC3 | 4 | G1/S | ✓ | × | ✓ |
| SPAG5 | 4 | G2/M | ✓ | ✓ | × |
| TMPO | 4 | G2/M | ✓ | × | ✓ |
| TRIP13 | 4 | G2/M | ✓ | × | × |
| TTK | 4 | G2/M | ✓ | ✓ | × |
| WDHD1 | 4 | G1/S | ✓ | × | ✓ |
| WDR76 | 4 | G1/S | ✓ | × | ✓ |
49 genes were identified in at least 4 of the 6 datasets as being deregulated by HPV E7 (compiled from Table S1). Annotation of cell cycle genes, including the phase of peak expression and DREAM, MMB-FOXM1 or pRB-E2F targets were extracted from Fischer et al.23. UNKN, timing of peak expression of the cell cycle gene is unknown; X, cell cycle-dependent expression was not reported in the datasets.
Table 3.
Genes upregulated after HPV E7 expression with an identification-overlap of three in six datasets.
| Gene Symbol | Identified as upregulated by E7 in No. of datasets | Cell cycle gene | DREAM target | MMB-FOXM1 target | pRB-E2F target | Gene Symbol | Identified as upregulated by E7 in No. of datasets | Cell cycle gene | DREAM target | MMB-FOXM1 target | pRB-E2F target |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ANLN | 3 | G2/M | ✓ | ✓ | ✓ | KNTC1 | 3 | G1/S | ✓ | × | ✓ |
| ANP32E | 3 | G2/M | ✓ | ✓ | ✓ | MAD2L1 | 3 | G2/M | ✓ | ✓ | ✓ |
| ATAD5 | 3 | UNKN | ✓ | ✓ | ✓ | MASTL | 3 | G1/S | ✓ | × | × |
| AURKA | 3 | G2/M | ✓ | ✓ | ✓ | MCM10 | 3 | G1/S | ✓ | × | ✓ |
| BRCA2 | 3 | G1/S | ✓ | × | ✓ | MCM3 | 3 | G1/S | ✓ | × | ✓ |
| BRIP1 | 3 | G1/S | ✓ | × | ✓ | MCM5 | 3 | G1/S | ✓ | × | ✓ |
| BUB1 | 3 | G2/M | ✓ | ✓ | × | MCM6 | 3 | G1/S | ✓ | × | ✓ |
| BUB1B | 3 | G2/M | ✓ | ✓ | ✓ | MCM7 | 3 | UNKN | ✓ | × | ✓ |
| CCNE2 | 3 | G1/S | × | × | ✓ | MKI67 | 3 | G2/M | ✓ | ✓ | × |
| CCNF | 3 | G2/M | ✓ | ✓ | × | MMS22L | 3 | G1/S | ✓ | × | ✓ |
| CDC7 | 3 | G1/S | ✓ | × | ✓ | MSH2 | 3 | G1/S | ✓ | × | ✓ |
| CDCA3 | 3 | G2/M | ✓ | ✓ | × | MSH6 | 3 | G1/S | ✓ | × | ✓ |
| CENPA | 3 | G2/M | ✓ | ✓ | × | MTBP | 3 | G1/S | ✓ | × | ✓ |
| CENPE | 3 | G2/M | ✓ | ✓ | × | MTHFD1 | 3 | G1/S | ✓ | × | ✓ |
| CENPK | 3 | G1/S | ✓ | × | ✓ | MYBL1 | 3 | UNKN | ✓ | × | × |
| CENPN | 3 | UNKN | ✓ | ✓ | ✓ | NASP | 3 | G1/S | ✓ | × | ✓ |
| CENPQ | 3 | G1/S | ✓ | × | ✓ | NCAPG | 3 | G2/M | ✓ | ✓ | ✓ |
| CENPU | 3 | G1/S | ✓ | × | ✓ | NCAPH | 3 | G2/M | ✓ | ✓ | × |
| CHAF1A | 3 | G1/S | ✓ | × | ✓ | NDC1 | 3 | G2/M | ✓ | ✓ | × |
| CHAF1B | 3 | G1/S | × | × | ✓ | NEMP1 | 3 | G1/S | ✓ | × | ✓ |
| CKS1B | 3 | G2/M | ✓ | ✓ | × | OIP5 | 3 | G2/M | ✓ | ✓ | ✓ |
| DDIAS | 3 | G1/S | ✓ | × | × | ORC1 | 3 | G1/S | ✓ | × | ✓ |
| DHFR | 3 | G1/S | ✓ | x | × | PARP1 | 3 | G1/S | × | × | ✓ |
| DLGAP5 | 3 | G2/M | ✓ | ✓ | × | PBK | 3 | G2/M | ✓ | × | × |
| DNA2 | 3 | G1/S | ✓ | × | ✓ | PCNA | 3 | G1/S | ✓ | × | ✓ |
| DONSON | 3 | G1/S | × | × | ✓ | PKMYT1 | 3 | G1/S | ✓ | × | × |
| DSN1 | 3 | G1/S | ✓ | × | ✓ | PLK1 | 3 | G2/M | ✓ | ✓ | × |
| EMP2 | 3 | G1/S | × | × | ✓ | POLA1 | 3 | G1/S | ✓ | × | ✓ |
| EXO1 | 3 | G1/S | ✓ | × | ✓ | POLA2 | 3 | G1/S | ✓ | × | ✓ |
| EZH2 | 3 | G1/S | ✓ | ✓ | ✓ | POLD1 | 3 | G1/S | ✓ | × | ✓ |
| FAM111B | 3 | G1/S | ✓ | × | ✓ | POLD3 | 3 | G1/S | ✓ | × | ✓ |
| FEN1 | 3 | G1/S | ✓ | × | ✓ | POLE | 3 | G1/S | ✓ | ✓ | ✓ |
| FIGNL1 | 3 | G1/S | ✓ | × | × | RACGAP1 | 3 | G2/M | ✓ | ✓ | × |
| GINS1 | 3 | G1/S | ✓ | × | ✓ | RBL1 | 3 | G1/S | ✓ | × | ✓ |
| GMNN | 3 | G1/S | ✓ | × | ✓ | RFC4 | 3 | G1/S | ✓ | × | ✓ |
| GTSE1 | 3 | G2/M | ✓ | ✓ | × | RFC5 | 3 | G1/S | ✓ | × | ✓ |
| H2AFZ | 3 | UNKN | ✓ | ✓ | ✓ | RMI1 | 3 | G1/S | ✓ | × | ✓ |
| HAT1 | 3 | UNKN | ✓ | × | ✓ | RNASEH2A | 3 | G1/S | ✓ | × | ✓ |
| HELLS | 3 | G1/S | ✓ | × | ✓ | SASS6 | 3 | UNKN | ✓ | × | × |
| HMMR | 3 | G2/M | ✓ | ✓ | × | SMC2 | 3 | UNKN | ✓ | × | × |
| ITGB3BP | 3 | UNKN | ✓ | × | ✓ | TICRR | 3 | G2/M | ✓ | ✓ | ✓ |
| KIAA0101 | 3 | G1/S | ✓ | × | ✓ | TIMELESS | 3 | UNKN | ✓ | × | ✓ |
| KIF11 | 3 | G2/M | ✓ | ✓ | × | TPX2 | 3 | G2/M | ✓ | ✓ | × |
| KIF15 | 3 | G2/M | ✓ | ✓ | ✓ | TYMS | 3 | × | × | × | × |
| KIF20B | 3 | G2/M | ✓ | ✓ | × | UBE2C | 3 | G2/M | ✓ | ✓ | × |
| KIF4A | 3 | G2/M | ✓ | ✓ | × | UHRF1 | 3 | G1/S | ✓ | × | ✓ |
92 genes were identified in 3 of the 6 datasets as being deregulated by HPV E7 (extracted from Table S1). Information whether the gene is a cell cycle gene, including the phase of peak expression, and whether it is a DREAM, MMB-FOXM1 or pRB-E2F target were extracted from Fischer et al.23. UNKN, timing of peak expression of the cell cycle gene is unknown; X, cell cycle-dependent expression was not reported in the datasets.
It is generally agreed that gene expression data from different experimental platforms are not directly comparable, and thus we used the stepwise meta-analysis approach instead that ranks genes by the number of datasets that find them significantly differentially regulated. Given that raw data were not re-analyzed, the approach does not include data points that were below the thresholds set in the individual studies. Differences in unprocessed data acquisition between several studies may reduce reproducibility, yet it minimizes the bias that would be introduced by using one particular analysis approach for all datasets. Following the stepwise meta-analysis approach23, genes were ranked by the number of datasets finding the gene to be significantly upregulated minus the number of datasets that find the gene to be downregulated (Supplementary Table S1).
Cell culture and drug treatment
HCT116 cells were grown in Dulbecco’s modified Eagle’s medium (DMEM; Lonza, Basel, Switzerland) supplemented with 10% fetal calf serum (FCS) (Biochrom, Berlin, Germany) and penicillin/streptomycin and maintained at 37 °C and 10% CO2. Stably transfected HCT116 cells were generated by transfection with pCMV-HPV16-E7 wt (kindly provided by Karl Münger48), and selection with G418/Geneticin (PAA Laboratories, Pasching, Austria) at a concentration of 0.5 mg/ml41. Wild-type mouse keratinocytes were isolated from C57BI6 mouse embryos as described previously49. Cells were grown on plates coated with collagen (Invitrogen, Darmstadt, Germany) and maintained at 10% CO2 and 32 °C in DMEM/Ham’s F12 (3.5:1.1) (PAN Biotech, Aidenbach, Germany). Cells were treated with doxorubicin (0.2 μg/ml; Medac, Wedel, Germany) or Nutlin-3a (10 μM; Cayman Chemicals, Ann Arbor, MI, USA) for 24 h. For cell sorting of transiently transfected wild-type mouse keratinocytes, pEGFP plasmid (Clontech, Mountain View, CA, USA) was co-transfected with pCMV-HPV16-E7 wt plasmid at a 1:3 ratio using GeneJuice (Merck, Darmstadt, Germany). Fluorescence-activated cell sorting was carried out on a FACS Aria SORP instrument (Becton Dickinson Biosciences, Franklin Lakes, NJ, USA).
RNA extraction, reverse transcription and semi-quantitative real-time PCR
Total cellular RNA was isolated using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s protocol. One-step reverse transcription and quantitative real-time PCR were performed with an ABI 7300 Real-Time PCR System (Applied Biosystems, Forster City, CA, USA) using QuantiTect SYBRGreen PCR Kit (Qiagen, Hilden, Germany) as described previously41. Primer sequences have been published previously34, 40, 41, 50, 51.
Sodium dodecyl sulphate-polyacrylamide gel electrophoresis and immunoblot
Sodium dodecyl sulphate-polyacrylamide gel electrophoresis and western blot were performed following standard protocols52. The following antibodies were used: E2F1 (sc-193, Santa Cruz Biotechnology, Santa Cruz, CA, 1:500 dilution), KIF23 (sc-136473, Santa Cruz Biotechnology, 1:200), CDC25C (sc-327, Santa Cruz Biotechnology, 1:1000), B-MYB (LX015.1, kindly provided by Roger Watson53, hybridoma media 1:5) and ß-actin (A5441, Sigma-Aldrich, Munich, Germany, 1:5000).
Results
The HPV E7 oncoprotein deregulates cell cycle genes targeted by the DREAM complex
Two recently published datasets identified genes deregulated upon expression of the HPV E7 oncoprotein on a genome-wide basis42, 43. Combined, those two datasets identified 753 genes deregulated by E7, including 453 upregulated and 300 downregulated genes (Table S1). A fraction of these genes was identified in both datasets, 66 upregulated and 2 downregulated genes (Table 1). The small number of genes downregulated upon E7 expression indicates that expression of E7 primarily causes gene upregulation. When comparing the overlap of the 66 upregulated genes with recently published lists of cell cycle genes and targets of the DREAM, MMB-FOXM1 and pRB-E2F complexes23, it becomes evident that most E7-upregulated genes are cell cycle genes and targets of the DREAM complex (Table 1).
Table 1.
HPV E7 deregulates DREAM target genes.
| Gene Symbol | Cell cycle gene | DREAM target | Gene Symbol | Cell cycle gene | DREAM target |
|---|---|---|---|---|---|
| APOBEC3B | G2/M | ✓ | MCM2 | G1/S | ✓ |
| ASF1B | G1/S | ✓ | MCM3 | G1/S | ✓ |
| ATAD2 | G1/S | ✓ | MCM4 | G1/S | ✓ |
| ATAD5 | UNKN | ✓ | MCM5 | G1/S | ✓ |
| BRCA1 | G1/S | ✓ | MCM6 | G1/S | ✓ |
| BRCA2 | G1/S | ✓ | MCM7 | UNKN | ✓ |
| BRIP1 | G1/S | ✓ | MMS22L | G1/S | ✓ |
| CCNE2 | G1/S | × | MSH2 | G1/S | ✓ |
| CDC25A | G1/S | ✓ | MSH6 | G1/S | ✓ |
| CDC45 | G1/S | ✓ | MTBP | G1/S | ✓ |
| CDC6 | G1/S | ✓ | MYBL2 | G1/S | ✓ |
| CDC7 | G1/S | ✓ | NCAPG2 | G1/S | ✓ |
| CDK2 | G1/S | ✓ | NUSAP1 | G2/M | ✓ |
| CDKN2A | × | × | ORC1 | G1/S | ✓ |
| CENPK | G1/S | ✓ | PCNA | G1/S | ✓ |
| CENPQ | G1/S | ✓ | POLA1 | G1/S | ✓ |
| CENPU | G1/S | ✓ | POLD3 | G1/S | ✓ |
| CHAF1A | G1/S | ✓ | POLE | G1/S | ✓ |
| CHAF1B | G1/S | × | PRIM1 | G1/S | ✓ |
| DONSON | G1/S | × | RAD51AP1 | G1/S | ✓ |
| DSN1 | G1/S | ✓ | RBL1 | G1/S | ✓ |
| DTL | G1/S | ✓ | RFC3 | G1/S | ✓ |
| E2F1 | G1/S | ✓ | RFC5 | G1/S | ✓ |
| EMP2 | G1/S | × | RRM2 | G1/S | ✓ |
| FAM111B | G1/S | ✓ | SASS6 | UNKN | ✓ |
| FANCI | G1/S | ✓ | STIL | G2/M | ✓ |
| FANCL | G1/S | ✓ | TICRR | G2/M | ✓ |
| FIGNL1 | G1/S | ✓ | TMPO | G2/M | ✓ |
| GINS1 | G1/S | ✓ | UHRF1 | G1/S | ✓ |
| GINS2 | G1/S | ✓ | WDHD1 | G1/S | ✓ |
| GMNN | G1/S | ✓ | WDR76 | G1/S | ✓ |
| HELLS | G1/S | ✓ | ZWINT | G1/S | ✓ |
| KNTC1 | G1/S | ✓ | AMIGO2 | G1/S | × |
| MASTL | G1/S | ✓ | RHOB | × | × |
68 genes overlap in two datasets of genes deregulated upon HPV E7 expression42, 43. Two genes described in both datasets as downregulated are underlined. Annotation of genes as DREAM targets or cell cycle genes, including the cell cycle phase of peak expression, were extracted from Fischer et al.23. UNKN, timing of peak expression of the cell cycle gene is unknown; X, cell cycle-dependent expression was not reported in the datasets.
Next, we integrated additional datasets that identified genes that are deregulated by HPV E6 and E744–47, employing tools and data from a recent meta-analysis23. By combining six datasets and using stringent thresholds, this approach yields reliable target identification. In total, these six datasets identified 1,783 genes as deregulated by HPV E7 (Table S1). No gene was identified in all datasets as downregulated by E7, further supporting the notion that E7 expression primarily results in target gene induction. Fourteen genes were identified as upregulated by E7 in at least five of the six datasets. Remarkably, all of these genes are cell cycle genes and DREAM targets. Furthermore, when looking at the 49 genes identified in at least four of the six datasets, 34 are cell cycle genes and DREAM targets (Table 2). Only one gene from this group, CDKN2A (p16), is not a DREAM target. CDKN2A was previously reported to be upregulated by HPV E7 through a different mechanism, namely epigenetic derepression54.
To be considered a high confidence HPV E7-deregulated gene, we employed a threshold of at least three datasets that identify the gene as upregulated by E7. Remarkably, 139 of 141 genes (98.6%) that passed these criteria are predicted cell cycle genes, and 134 (95.0%) are DREAM targets (Tables 2 and 3). The cell cycle genes represent genes with peak expression during G1/S or G2/M phases. Although pRB is the best known target protein of E7, only 87 (61.7%) of the high confidence E7-deregulated genes are predicted pRB-E2F targets. It is important to note that pRB-E2F targets largely represent the G1/S subgroup of DREAM-targeted cell cycle genes23, 34. The finding that most HPV E7-deregulated genes are DREAM targets is consistent with the previous finding that disruption of the DREAM complex is critical to prevent cell cycle arrest in HPV-infected cells25. Together, our findings indicate that DREAM target genes are generally deregulated by HPV E7 expression.
High risk HPV E7 abrogates p53-p21-DREAM-mediated repression of cell cycle genes
Given that p53-p21-dependent downregulation of the DREAM target gene PLK4 was disturbed by HPV E741, we asked whether disruption of the p53-p21-DREAM pathway was a general phenomenon upon HPV E7 expression. The p53-p21-DREAM pathway is best characterized in the HCT116 colon carcinoma cell line37, 40, and thus, we employed HCT116 cells stably transfected with HPV-16 E7 expression plasmids41. We treated wild-type and HPV E7-expressing HCT116 cells with p53-stabilizing Nutlin-3a or the DNA intercalator doxorubicin and compared changes in mRNA levels to untreated control cells (Fig. 1). Consistent with earlier findings, the mRNA levels of the well-established DREAM target genes B-MYB (MYBL2)40, E2F1 23, CDC25C 51, Survivin (BIRC5)51, KIF23 50, ORC1 34 and RAD51 34 were downregulated in HCT116 wild-type cells treated with Nutlin-3a or doxorubicin compared to untreated cells (Fig. 1A). Most importantly, downregulation of these genes was abrogated upon HPV E7 expression (Fig. 1B). With B-MYB (MYBL2), E2F1, KIF23 and CDC25C serving as examples, western blot analyses showed that protein levels followed decreased mRNA levels. Nutlin-3a treatment led to reduced B-MYB, E2F1, KIF23 and CDC25C protein levels in HCT116 wild-type cells but not in E7-expression HCT116 cells (Fig. 1C). In contrast to the abrogated repression of cell cycle genes, p21 (CDKN1A) was still induced in response to p53 activation even when HPV E7 is present (Fig. 1B). Notably, HCT116 cells that express HPV E7 displayed an increased expression of DREAM target genes upon treatment with doxorubicin, but not in the presence of Nutlin-3a, when compared to untreated cells. Doxorubicin can induce G1/S and G2/M cell cycle arrest, while Nutlin-3a mainly induces G1/S arrest. A doxorubicin-induced increase in G2/M cell cycle population leads to increased mRNA levels of late cell cycle genes when the p53 pathway is not active or blocked, which has been observed previously41, 50, 51. To explore whether findings from HCT116 cancer cells are also observed in primary cells, we tested for mRNA expression changes following Nutlin-3a treatment in wild-type mouse keratinocytes compared to cells that were expressing HPV E7. Similar to the results from HCT116 cells, wild-type but not E7-expressing mouse keratinocytes displayed decreased mRNA levels of DREAM target genes upon Nutlin-3a treatment. Induction of Cdkn1a (p21), however, was not impaired by E7 expression (Fig. 2).
Figure 1.
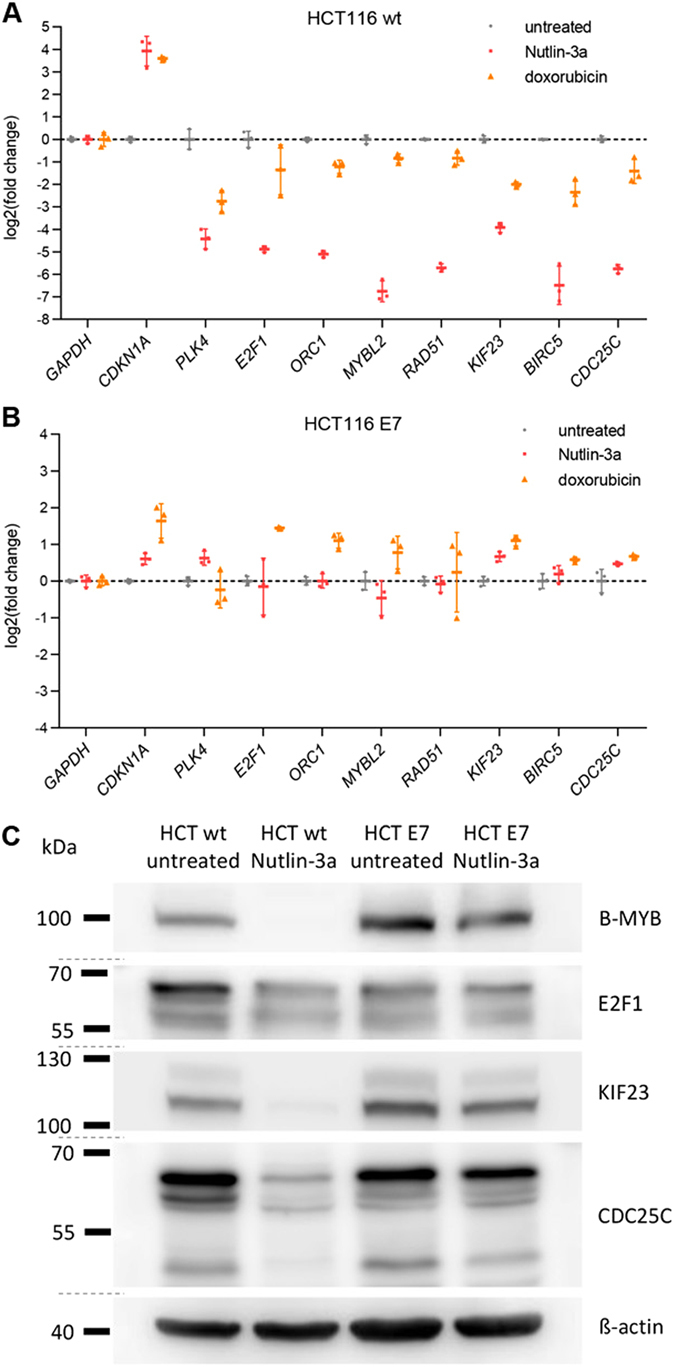
HPV E7 abrogates p53-mediated downregulation of DREAM target genes. (A) HCT116 wild-type and (B) HCT116 HPV E7-expressing cells were treated with Nutlin-3a or doxorubicin for 24 h. Untreated cells served as control. Semiquantitative RT-PCR was performed. The log 2-fold change of mRNA expression of treated compared to untreated cells is displayed. GAPDH served as a negative control for the p53 response, while p21 (CDKN1A) and PLK4 were tested as positive controls. (C) HCT116 wild-type and E7-expressing cells were treated with Nutlin-3a for 24 h or left untreated. Protein levels were analyzed through immunoblotting and ß-actin levels served as loading control. Cropped blot images are displayed; full images are included in Supplementary Figure S1.
Figure 2.
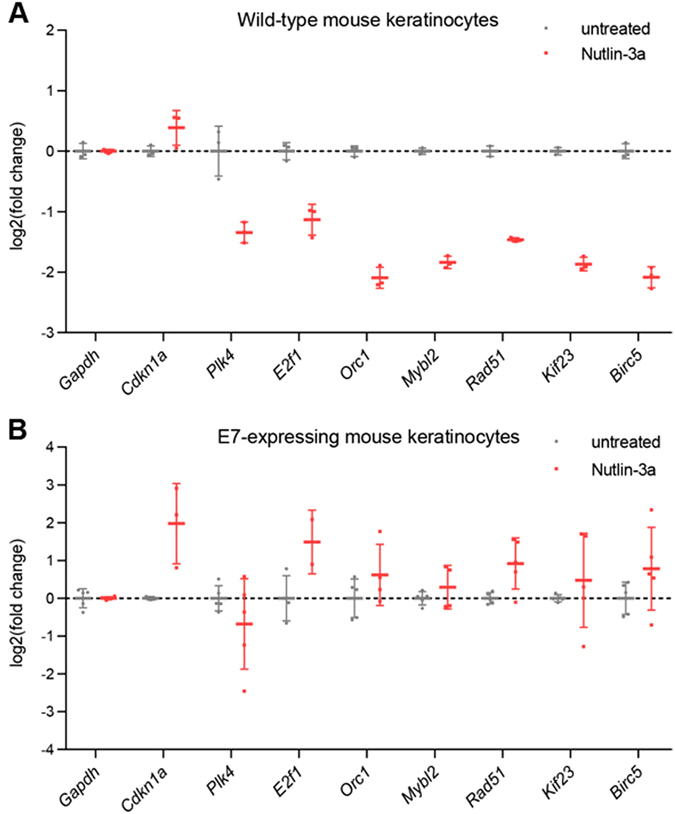
HPV E7 abrogates p53-mediated downregulation of DREAM target genes in wild-type keratinocytes. (A) Wild-type mouse keratinocytes were treated with Nutlin-3a for 24 h or left untreated. (B) Mouse keratinocytes were co-transfected with plasmids expressing HPV E7 and EGFP and treated with Nutlin-3a for 24 h or left untreated. Cells were sorted for green fluorescence followed by mRNA preparation. Relative mRNA expression was quantified by real-time RT-PCR and normalized to GAPDH RNA levels. The log2-fold change of mRNA expression is displayed for treated compared to untreated cells.
Discussion
A cell uses several mechanisms to control proliferation. Hypo-phosphorylated forms of the pRB tumor suppressor block E2F-mediated induction of cell cycle genes required for the G1/S transition55. In addition, activation of proliferation in cells with serious defects in replication leads to DNA damage and causes stabilization of p53, which triggers cell cycle arrest or apoptosis56. By employing E6 and E7 oncoproteins, human papilloma viruses have evolved two strategies to intercept the host’s control of proliferation and response to infection. HPV E6 causes destruction of p5310, 11, and E7 forms a complex with pRB, thereby interfering with pRB’s ability to form complexes with E2F transcription factors6–8. Recently, a third mechanism based on E7 preventing DREAM complex formation was discovered25, 26, 41. Here, we analyzed E7-mediated gene dysregulation using genome-wide data analysis and expression profiling of distinct cell cycle genes. Our analysis revealed that HPV E7 causes deregulation of a large number of cell cycle genes that are normally regulated by DREAM. Deregulation also affects p53 function through disruption of the p53-p21-DREAM pathway (Fig. 3). This mechanism is independent of HPV E6-mediated destruction of p53.
Figure 3.
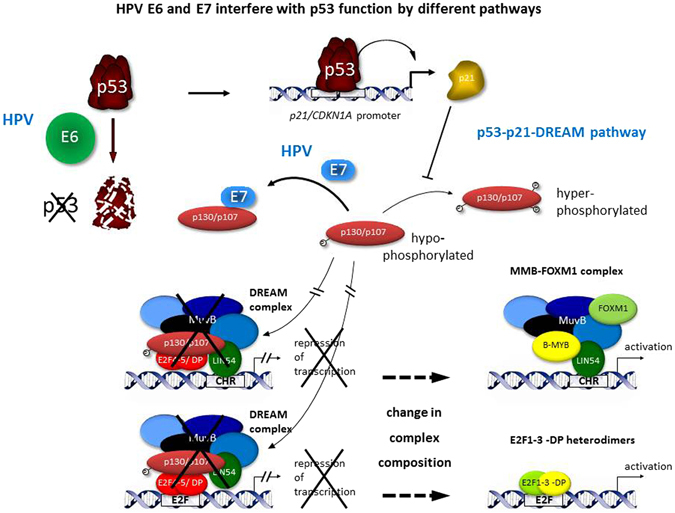
Both HPV E6 and E7 interfere with p53 function. HPV E6-mediated degradation of p53 is well established. By an independent mechanism, the HPV E7 oncoprotein interferes with the p53-p21-DREAM pathway. Interference is caused by the abrogation of indirect p53-dependent transcriptional repression of many genes required for cell cycle progression. E7 sequesters hypo-phosphorylated p130 and p107 proteins, thereby preventing them from forming the DREAM transcriptional repressor complex. In general, DREAM can bind to four combinations of promoter elements: CHR sites, E2F sites, CDE/CHR or E2F/CLE tandem elements34. For clarity only genes with CHR or E2F sites are depicted as examples. When p130/p107 pocket proteins are sequestered and not available for DREAM repressor formation, protein complexes on E2F or CHR sites change their composition from repressor to activator complexes. CHR elements then bind MMB-FOXM1 and E2F sites bind activating E2F1-3-DP complexes, respectively. In conclusion, sequestration of p130 and p107 by HPV E7 abrogates p53-dependent repression of cell cycle genes and thus impairs cell cycle checkpoint control by p53.
It is widely accepted that pRB controls the G1/S checkpoint and that it is required for G1/S transition5. However, HPV-induced proliferation additionally requires deregulation of the G2/M checkpoint. The DREAM complex contributes to the G2/M checkpoint through downregulation of cell cycle genes in response to p53 activation40. The HPV E7 oncoprotein deregulates target genes of the DREAM complex that comprise G1/S and G2/M cell cycle genes (Tables 1, 2 and 3 and Figs 1 and 2). Furthermore, the G1/S subgroup of DREAM target genes is also bound by pRB-E2F complexes23. DREAM binds to these genes either through E2F promoter elements or through a combination of an E2F site and a CHR-like element (CLE)34. E2F1 and ORC1 are examples for G1/S genes that are controlled through an E2F promoter element, while B-MYB (MYBL2) and RAD51 are examples for G1/S genes controlled through E2F/CLE tandem elements34. The p53-mediated downregulation of genes from both groups is abrogated by HPV E7 (Figs 1, 2 and 3). In contrast to G1/S genes, the G2/M subgroup of DREAM target genes is additionally regulated by MMB-FOXM1 complexes23. DREAM binds to these genes either through CHR promoter elements or through CDE/CHR tandem sites34. KIF23 and Survivin (BIRC5) are examples for G2/M genes that are controlled through a CHR element37, 50, 51, while CDC25C and PLK4 are examples for G2/M genes controlled through CDE/CHR sites41, 51, and the p53-mediated downregulation of these genes is abrogated by E7 (Figs 1 and 2). Thus, the data suggest that HPV E7 interferes not only with pRB function but also with DREAM to impair cell cycle checkpoints (Fig. 3).
Except for RAD51, all experimentally tested DREAM target genes were correctly predicted by the meta-analysis to be deregulated by HPV E7 (Tables 1, 2 and 3). These observations indicate that threshold settings were so stringent that the computational analysis rather missed candidates than to include false-positive genes. This suggests that the genes in Tables 2 and 3 are indeed high confidence targets deregulated by HPV E7, but that some additional target genes may have been missed. Taken together, our findings provide evidence that DREAM target genes are generally deregulated by HPV E7 expression.
It is important to note that pRB differs in its function from the pRB-like pocket proteins p107 and p130. While all pocket proteins pRB, p107 and p130 bind to LxCxE motifs, only p107 and p130 can be recruited to the MuvB core through an LxSxExL motif in LIN52 to form the DREAM complex57. HPV E7 possesses an LxCxE motif through which it binds pocket proteins8, and binding of E7 to p107 and p130 inhibits their interaction with the LxSxExL motif in LIN5257. Several other viral oncoproteins target the pocket proteins through LxCxE motifs, including adenovirus early-region 1A (E1A) and large T antigens of several polyomaviruses, such as SV40, JCV and BKV58. Consistent with this notion, also SV40 large T was reported to impair DREAM function59, 60.
It is established that HPV destroys p53 function through marking it for degradation by the E6 oncoprotein10, 11. This mechanism may be sufficient to block p53 activity completely. However, p21 is a central effector of the p53 response, and p21 can be activated independently of p53, for example through the MAPK and TGFβ pathways61. Also in the absence of HPV E6, we observe that the p53-p21-DREAM pathway is intercepted further downstream by E7 interfering with DREAM function and host cell cycle arrest (Tables 1, 2 and 3 and Figs 1 and 2). The data indicate that HPV employs several means to disrupt cell cycle checkpoints (Fig. 3).
In summary, the data reveal that deregulation of DREAM function by the HPV E7 oncoprotein may contribute substantially to the development of the many cancer types caused by HPV.
Electronic supplementary material
Acknowledgements
The authors are indebted to Carola Koschke, Andrea Rothe and Aileen Wingenfeld for expert technical assistance, Dr. Andreas Lösche at the IZKF Leipzig core unit for performing cell sorting, Dr. Karl Münger for the kind gift of HPV-16 E7 plasmids, Dr. Roger Watson for the kind gift of B-MYB antibody and Dr. Christine E. Engeland for comments on the manuscript. This work was supported through a junior research grant by the Medical School, University of Leipzig and an Add-On Fellowship for Interdisciplinary Sciences in Systems Biology by the Joachim Herz Stiftung (to M.F.); a scholarship by the German National Merit Foundation (Studienstiftung des deutschen Volkes) and a fellowship by the Christiane Nüsslein-Volhard Foundation (to S.U.). We acknowledge support from the German Research Foundation (DFG) and the University of Leipzig within the program of Open Access Publishing.
Author Contributions
M.F. and K.E. conceived the study. M.F., S.U. and C.S. performed the experiments. M.F. performed the computational analysis. T.M.M. contributed reagents and advice. M.F. and K.E. interpreted the data and wrote the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-02831-9
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Martin Fischer, Email: Martin.Fischer@medizin.uni-leipzig.de.
Kurt Engeland, Email: Engeland@medizin.uni-leipzig.de.
References
- 1.zur Hausen H. Papillomaviruses and cancer: From basic studies to clinical application. Nat. Rev. Cancer. 2002;2:342–350. doi: 10.1038/nrc798. [DOI] [PubMed] [Google Scholar]
- 2.Moody CA, Laimins LA. Human papillomavirus oncoproteins: pathways to transformation. Nat. Rev. Cancer. 2010;10:550–560. doi: 10.1038/nrc2886. [DOI] [PubMed] [Google Scholar]
- 3.Bosch FX, et al. Comprehensive control of human papillomavirus infections and related diseases. Vaccine. 2013;31(Suppl 8):I1–31. doi: 10.1016/j.vaccine.2013.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gillison ML, Chaturvedi AK, Anderson WF, Fakhry C. Epidemiology of Human Papillomavirus-Positive Head and Neck Squamous Cell Carcinoma. J. Clin. Oncol. 2015;33:3235–42. doi: 10.1200/JCO.2015.61.6995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dyson NJ. RB1: a prototype tumor suppressor and an enigma. Genes Dev. 2016;30:1492–502. doi: 10.1101/gad.282145.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dyson N, Howley PM, Munger K, Harlow E. The human papilloma virus-16 E7 oncoprotein is able to bind to the retinoblastoma gene product. Science (80-). 1989;243:934–937. doi: 10.1126/science.2537532. [DOI] [PubMed] [Google Scholar]
- 7.Münger K, et al. Complex formation of human papillomavirus E7 proteins with the retinoblastoma tumor suppressor gene product. EMBO J. 1989;8:4099–105. doi: 10.1002/j.1460-2075.1989.tb08594.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee JO, Russo AA, Pavletich NP. Structure of the retinoblastoma tumour-suppressor pocket domain bound to a peptide from HPV E7. Nature. 1998;391:859–865. doi: 10.1038/36038. [DOI] [PubMed] [Google Scholar]
- 9.Demers GW, Halbert CL, Galloway DA. Elevated wild-type p53 protein levels in human epithelial cell lines immortalized by the human papillomavirus type 16 E7 gene. Virology. 1994;198:169–74. doi: 10.1006/viro.1994.1019. [DOI] [PubMed] [Google Scholar]
- 10.Scheffner M, Werness BA, Huibregtse JM, Levine AJ, Howley PM. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell. 1990;63:1129–36. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]
- 11.Martinez-Zapien D, et al. Structure of the E6/E6AP/p53 complex required for HPV-mediated degradation of p53. Nature. 2016;529:541–5. doi: 10.1038/nature16481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McLaughlin-Drubin ME, Münger K. The human papillomavirus E7 oncoprotein. Virology. 2009;384:335–344. doi: 10.1016/j.virol.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Songock WK, Kim S, Bodily JM. The human papillomavirus E7 oncoprotein as a regulator of transcription. Virus Res. 2017;231:56–75. doi: 10.1016/j.virusres.2016.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vousden KH, Vojtesek B, Fisher C, Lane D. HPV-16 E7 or adenovirus E1A can overcome the growth arrest of cells immortalized with a temperature-sensitive p53. Oncogene. 1993;8:1697–702. [PubMed] [Google Scholar]
- 15.Demers GW, Foster SA, Halbert CL, Galloway DA. Growth arrest by induction of p53 in DNA damaged keratinocytes is bypassed by human papillomavirus 16 E7. Proc. Natl. Acad. Sci. USA. 1994;91:4382–6. doi: 10.1073/pnas.91.10.4382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Slebos RJ, et al. p53-dependent G1 arrest involves pRB-related proteins and is disrupted by the human papillomavirus 16 E7 oncoprotein. Proc. Natl. Acad. Sci. USA. 1994;91:5320–4. doi: 10.1073/pnas.91.12.5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hickman ES, Bates S, Vousden KH. Perturbation of the p53 response by human papillomavirus type 16 E7. J. Virol. 1997;71:3710–3718. doi: 10.1128/jvi.71.5.3710-3718.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones DL, Münger K. Analysis of the p53-mediated G1 growth arrest pathway in cells expressing the human papillomavirus type 16 E7 oncoprotein. J. Virol. 1997;71:2905–12. doi: 10.1128/jvi.71.4.2905-2912.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Waldman T, Kinzler KW, Vogelstein B. p21 is necessary for the p53-mediated G1 arrest in human cancer cells. Cancer Res. 1995;55:5187–90. [PubMed] [Google Scholar]
- 20.Bunz F, et al. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science (80-). 1998;282:1497–1501. doi: 10.1126/science.282.5393.1497. [DOI] [PubMed] [Google Scholar]
- 21.Funk JO, et al. Inhibition of CDK activity and pcna-dependent DNA replication p21 is blocked by interaction with the HPV-16 E7 oncoprotein. Genes Dev. 1997;11:2090–2100. doi: 10.1101/gad.11.16.2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shin MK, Balsitis S, Brake T, Lambert PF. Human papillomavirus E7 oncoprotein overrides the tumor suppressor activity of p21Cip1 in cervical carcinogenesis. Cancer Res. 2009;69:5656–5663. doi: 10.1158/0008-5472.CAN-08-3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fischer M, Grossmann P, Padi M, DeCaprio JA. Integration of TP53, DREAM, MMB-FOXM1 and RB-E2F target gene analyses identifies cell cycle gene regulatory networks. Nucleic Acids Res. 2016;44:6070–6086. doi: 10.1093/nar/gkw523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fischer M. p21 governs p53’s repressive side. Cell Cycle. 2016;15:2852–2853. doi: 10.1080/15384101.2016.1205393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rashid NN, Yusof R, Watson RJ. Disruption of repressive p130-DREAM complexes by human papillomavirus 16 E6/E7 oncoproteins is required for cell-cycle progression in cervical cancer cells. J. Gen. Virol. 2011;92:2620–2627. doi: 10.1099/vir.0.035352-0. [DOI] [PubMed] [Google Scholar]
- 26.Rashid NN, Yusof R, Watson RJ. Disruption of pocket protein dream complexes by E7 proteins of different types of human papillomaviruses. Acta Virol. 2013;57:447–51. doi: 10.4149/av_2013_04_447. [DOI] [PubMed] [Google Scholar]
- 27.DeCaprio JA. Human papillomavirus type 16 E7 perturbs DREAM to promote cellular proliferation and mitotic gene expression. Oncogene. 2013;33:1–3. doi: 10.1038/onc.2013.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Litovchick L, et al. Evolutionarily Conserved Multisubunit RBL2/p130 and E2F4 Protein Complex Represses Human Cell Cycle-Dependent Genes in Quiescence. Mol. Cell. 2007;26:539–551. doi: 10.1016/j.molcel.2007.04.015. [DOI] [PubMed] [Google Scholar]
- 29.Schmit F, et al. LINC, a human complex that is related to pRB-containing complexes in invertebrates regulates the expression of G2/M genes. Cell Cycle. 2007;6:1903–1913. doi: 10.4161/cc.6.15.4512. [DOI] [PubMed] [Google Scholar]
- 30.Sadasivam S, DeCaprio JA. The DREAM complex: master coordinator of cell cycle-dependent gene expression. Nat. Rev. Cancer. 2013;13:585–595. doi: 10.1038/nrc3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Müller GA, Engeland K. The central role of CDE/CHR promoter elements in the regulation of cell cycle-dependent gene transcription. FEBS J. 2010;277:877–893. doi: 10.1111/j.1742-4658.2009.07508.x. [DOI] [PubMed] [Google Scholar]
- 32.Müller GA, et al. The CHR promoter element controls cell cycle-dependent gene transcription and binds the DREAM and MMB complexes. Nucleic Acids Res. 2012;40:1561–1578. doi: 10.1093/nar/gkr793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Müller GA, et al. The CHR site: Definition and genome-wide identification of a cell cycle transcriptional element. Nucleic Acids Res. 2014;42:10331–10350A. doi: 10.1093/nar/gku696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Müller, G. A., Stangner, K., Schmitt, T., Wintsche, A. & Engeland, K. Timing of transcription during the cell cycle: Protein complexes binding to E2F, E2F/CLE, CDE/CHR, or CHR promoter elements define early and late cell cycle gene expression. Oncotarget, doi:10.18632/oncotarget.10888 (2016). [DOI] [PMC free article] [PubMed]
- 35.Marceau AH, et al. Structural basis for LIN54 recognition of CHR elements in cell cycle-regulated promoters. Nat. Commun. 2016;7:12301. doi: 10.1038/ncomms12301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mannefeld M, Klassen E, Gaubatz S. B-MYB is required for recovery from the DNA damage-induced G2 checkpoint in p53 mutant cells. Cancer Res. 2009;69:4073–4080. doi: 10.1158/0008-5472.CAN-08-4156. [DOI] [PubMed] [Google Scholar]
- 37.Quaas M, Müller GA, Engeland K. p53 can repress transcription of cell cycle genes through a p21 WAF1/CIP1-dependent switch from MMB to DREAM protein complex binding at CHR promoter elements. Cell Cycle. 2012;11:4661–4672. doi: 10.4161/cc.22917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fischer M, Steiner L, Engeland K. The transcription factor p53: Not a repressor, solely an activator. Cell Cycle. 2014;13:3037–3058. doi: 10.4161/15384101.2014.949083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fischer, M. Census and evaluation of p53 target genes. Oncogene, doi:10.1038/onc.2016.502 (2017). [DOI] [PMC free article] [PubMed]
- 40.Fischer M, Quaas M, Steiner L, Engeland K. The p53-p21-DREAM-CDE/CHR pathway regulates G2/M cell cycle genes. Nucleic Acids Res. 2016;44:164–174. doi: 10.1093/nar/gkv927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fischer M, Quaas M, Wintsche A, Müller GA, Engeland K. Polo-like kinase 4 transcription is activated via CRE and NRF1 elements, repressed by DREAM through CDE/CHR sites and deregulated by HPV E7 protein. Nucleic Acids Res. 2014;42:163–180. doi: 10.1093/nar/gkt849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rozenblatt-Rosen O, et al. Interpreting cancer genomes using systematic host network perturbations by tumour virus proteins. Nature. 2012;487:491–5. doi: 10.1038/nature11288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou Y, et al. Role of WDHD1 in Human Papillomavirus-Mediated Oncogenesis Identified by Transcriptional Profiling of E7-Expressing Cells. J. Virol. 2016;90:6071–6084. doi: 10.1128/JVI.00513-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rosty C, et al. Identification of a proliferation gene cluster associated with HPV E6/E7 expression level and viral DNA load in invasive cervical carcinoma. Oncogene. 2005;24:7094–7104. doi: 10.1038/sj.onc.1208854. [DOI] [PubMed] [Google Scholar]
- 45.Santin AD, et al. Gene expression profiles of primary HPV16- and HPV18-infected early stage cervical cancers and normal cervical epithelium: Identification of novel candidate molecular markers for cervical cancer diagnosis and therapy. Virology. 2005;331:269–291. doi: 10.1016/j.virol.2004.09.045. [DOI] [PubMed] [Google Scholar]
- 46.Pang CL, et al. A functional interaction of E7 with B-Myb-MuvB complex promotes acute cooperative transcriptional activation of both S- and M-phase genes. Oncogene. 2014;33:4039–4049. doi: 10.1038/onc.2013.426. [DOI] [PubMed] [Google Scholar]
- 47.Kuner R, et al. Identification of cellular targets for the human papillomavirus E6 and E7 oncogenes by RNA interference and transcriptome analyses. J. Mol. Med. 2007;85:1253–1262. doi: 10.1007/s00109-007-0230-1. [DOI] [PubMed] [Google Scholar]
- 48.Gonzalez SL, Stremlau M, He X, Basile JR, Münger K. Degradation of the retinoblastoma tumor suppressor by the human papillomavirus type 16 E7 oncoprotein is important for functional inactivation and is separable from proteasomal degradation of E7. J. Virol. 2001;75:7583–91. doi: 10.1128/JVI.75.16.7583-7591.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Loschke F, Homberg M, Magin TM. Keratin Isotypes Control Desmosome Stability and Dynamics through PKCα. J. Invest. Dermatol. 2016;136:202–213. doi: 10.1038/JID.2015.403. [DOI] [PubMed] [Google Scholar]
- 50.Fischer M, et al. p53 and Cell Cycle Dependent Transcription of kinesin family member 23 (KIF23) Is Controlled Via a CHR Promoter Element Bound by DREAM and MMB Complexes. PLoS One. 2013;8:e63187. doi: 10.1371/journal.pone.0063187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fischer M, Quaas M, Nickel A, Engeland K. Indirect p53-dependent transcriptional repression of Survivin, CDC25C, and PLK1 genes requires the cyclin-dependent kinase inhibitor p21/CDKN1A and CDE/CHR promoter sites binding the DREAM complex. Oncotarget. 2015;6:41402–17. doi: 10.18632/oncotarget.3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kirschner RD, Sänger K, Müller GA, Engeland K. Transcriptional activation of the tumor suppressor and differentiation gene S100A2 by a novel p63-binding site. Nucleic Acids Res. 2008;36:2969–2980. doi: 10.1093/nar/gkn132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tavner F, Frampton J, Watson RJ. Targeting an E2F site in the mouse genome prevents promoter silencing in quiescent and post-mitotic cells. Oncogene. 2007;26:2727–2735. doi: 10.1038/sj.onc.1210087. [DOI] [PubMed] [Google Scholar]
- 54.McLaughlin-Drubin ME, Park D, Munger K. Tumor suppressor p16INK4A is necessary for survival of cervical carcinoma cell lines. Proc. Natl. Acad. Sci. USA. 2013;110:16175–80. doi: 10.1073/pnas.1310432110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Narasimha AM, et al. Cyclin D activates the Rb tumor suppressor by mono-phosphorylation. Elife. 2014;3:1068–1080. doi: 10.7554/eLife.02872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bester AC, et al. Nucleotide deficiency promotes genomic instability in early stages of cancer development. Cell. 2011;145:435–446. doi: 10.1016/j.cell.2011.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Guiley KZ, et al. Structural mechanisms of DREAM complex assembly and regulation. Genes Dev. 2015;29:961–974. doi: 10.1101/gad.257568.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Felsani, A., Mileo, A. M. & Paggi, M. G. Retinoblastoma family proteins as key targets of the small DNA virus oncoproteins. Oncogene25, 5277–5285 (2006). [DOI] [PubMed]
- 59.Hauser S, et al. Loss of LIN9, a member of the DREAM complex, cooperates with SV40 large T antigen to induce genomic instability and anchorage-independent growth. Oncogene. 2012;31:1859–1868. doi: 10.1038/onc.2011.364. [DOI] [PubMed] [Google Scholar]
- 60.Fine DA, et al. Identification of FAM111A as an SV40 Host Range Restriction and Adenovirus Helper Factor. PLoS Pathog. 2012;8:e1002949. doi: 10.1371/journal.ppat.1002949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Abbas T, Dutta A. P21 in Cancer: Intricate Networks and Multiple Activities. Nat. Rev. Cancer. 2009;9:400–414. doi: 10.1038/nrc2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


