Abstract
Asparagus kiusianus, an important wild relative of cultivated asparagus (A. officinalis), exhibits resistance to stem blight disease caused by Phomopsis asparagi. However, the mechanisms underlying this resistance are not understood and no transcriptomic or genetic resources are available for this species. De novo transcriptome sequencing of A. officinalis and A. kiusianus stems was performed 24 h after inoculation with P. asparagi. In total, 35,259 and 36,321 transcripts were annotated in A. officinalis and A. kiusianus, respectively. 1,027 up-regulated and 752 down-regulated transcripts were differentially expressed in the two Asparagus species. RNA sequencing data were validated using quantitative real-time reverse transcription PCR. Several defense-related genes including peroxidase 4, cationic peroxidase SPC4-like, pathogenesis-related protein-1-like, and jasmonic acid biosynthesis and signaling-related genes including phospholipase D alpha 1, 12-oxophytodienoate reductase and jasmonate-induced protein 23 KD were up-regulated in A. kiusianus relative to A. officinalis. In addition, infected A. kiusianuns exhibited a substantial increase in jasmonic acid and methyl jasmonate relative to A. officinalis. Peroxidase activity was significantly elevated in infected A. kiusianus compared with infected A. officinalis. Our transcriptomic database provides a resource for identifying novel genes and molecular markers-associated with Phomopsis disease resistance and will facilitate breeding and improvement of cultivated asparagus varieties.
Introduction
Asparagus officinalis L., a dioecious species of the family Asparagaceae, is an economically important horticultural crop worldwide because of its culinary and medicinal properties. The total worldwide asparagus production in 2013 was approximately 7.95 million tonnes, of which 89.4% was produced in Asia, 7.2% in the Americas, 3.2% in Europe, and 0.2% in Oceania1. Stem blight disease caused by P. asparagi is the most serious disease for asparagus production in many parts of the world, including Japan, China, Australia, New Zealand, Italy, Greece, and the United States2–4. Disease symptoms are first seen as small light brown lesions on the lower part of the stem. The primary lesions subsequently become extended, forming larger dark brown oval-shaped lesions that eventually lead to complete stem desiccation and stem death4, 5. Stem blight disease is mainly controlled using expensive chemical fungicides. However, concerns have been raised regarding the human and environmental impacts of fungicides, as well as their impacts on the capacity of P. asparagi to survive in various environments. Therefore, production of new asparagus cultivars with strong resistance to stem blight disease has become an urgent need in the context of sustainable crop production.
Asparagus is a large genus comprising 200–300 species distributed across the Old World Continents6. Diverse ecological niches led to the development of an extensive variety of different Asparagus species with different morphological and physiological traits. Wild Asparagus species represent a potential genetic resource for the development of disease-resistant Asparagus germplasm with desirable physiological attributes. Several wild Asparagus species exhibited a strong disease resistance phenotype in previous studies, but production of interspecific hybrids by crossing with cultivated A. officinalis was hampered by the genetic distance between species7. A. kiusianus is a wild diploid (2n = 2x = 20) species endemic to the coastal region of the Sea of Japan in the Kyushu area of Japan8. Analysis of the non-coding region of chloroplast DNA indicated that A. kiusianus was genetically closer to A. officinalis than many other wild Asparagus species9, and interspecific hybrids and backcross progenies were successfully obtained between A. kiusianus and A. officinalis 10. Evaluation of stem blight disease severity in A. kiusianus, A. officinalis, and their interspecific F1 hybrids revealed strong disease resistance characteristics in A. kiusianus and the F1 hybrids compared with A. officinalis 4. Although the molecular and physiological mechanisms underlying stem blight disease resistance remain unknown, these findings suggest that wild A. kiusianus could play a significant role in improving stem blight disease resistance in A. officinalis. Further development of Asparagus genetic resources and research to discover novel disease resistance alleles will help to improve germplasm utilization and facilitate breeding of new asparagus varieties.
Next-generation sequencing (NGS) for large-scale transcriptome analysis has become the technique of choice for generating large amounts of expression data in a relatively short time. Gene expression data have provided insights into the processes underlying gene expression and have facilitated gene discovery11–13. To identify the broad transcriptional network-associated with Phomopsis disease resistance in Asparagus, we conducted transcriptome analysis of susceptible A. officinalis and resistant wild A. kiusianus stems 24 h after inoculation with P. asparagi or mock inoculation with sterile distilled water (SDW). High-throughput Illumina HiSeq 2500 technology was used, and high-quality reads were de novo assembled into unique transcripts, which were then comprehensively evaluated and annotated. Several common and unique genes that were differentially expressed (DEG) between susceptible and resistant Asparagus species as a consequence of P. asparagi infection were detected. Selected candidate genes were validated using quantitative real-time reverse transcription PCR (qRT-PCR). qRT-PCR and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses of the DEG revealed many defense and stress-related genes including peroxidase 4, cationic peroxidase SPC4-like, chitinase-6, pathogenesis-related protein 1-like, and jasmonic acid (JA) biosynthesis and signaling-related genes are the major components of the A. kiusianus response to P. aspargi infection relative to A. officinalis. To dissect the significant role of JA pathway and other stress-related genes in A. kiusianus resistance, phytohormone accumulation including JA, methyl jasmonate (MeJA), salicylic acid (SA) and abscisic acid (ABA) in A. kiusianus and A. officinalis 24 h post-inoculation in comparison with SDW-treated control plants was carried out using liquid chromatography-tandem mass spectrometry (LC-MS/MS). In addition, enzyme assays for stress-related enzymes such as catalase (CAT, EC 1.11.1.6) and peroxidase (POX, EC 1.11.1.7) were performed. To our knowledge, RNA sequencing has not been used to examine Asparagus-P. asparagi interactions previously. Our transcriptome dataset is therefore a valuable and unique resource that will facilitate future functional genetics studies and molecular marker development for asparagus breeding.
Results
Stem blight disease occurrence in wild and cultivated Asparagus species
Differences in disease occurrence between susceptible cultivated A. officinalis ‘Mary Washington 500W’ and resistant wild A. kiusianus (AK0501 strain) were observed after inoculation with P. asparagi spores under greenhouse conditions. Primary signs of infection, seen as dark brown lesions were first visually detected on susceptible A. officinalis at 7 days post-inoculation. The fungus then spread through the stem, resulting in a fully diseased stem by 14 days post-inoculation (Fig. 1a). By contrast, typical disease symptoms were not seen on resistant A. kiusianus and the fungus was unable to spread (Fig. 1a), demonstrating that A. kiusianus exhibited resistance to P. asparagi. Disease severity was significantly (P < 0.001) different between the two Asparagus species, at 86.66% and 13.33% in A. officinalis and A. kiusianus, respectively (Fig. 1b).
Figure 1.
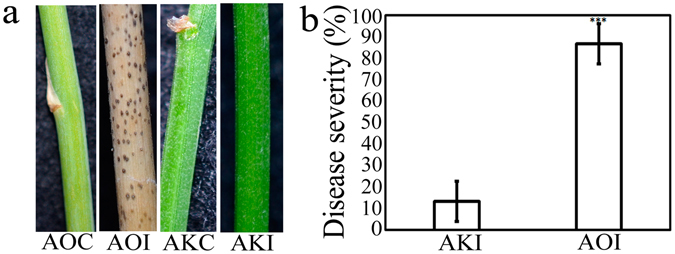
Stem blight disease symptoms in cultivated Asparagus officinalis and wild A. kiusianus 3 weeks after inoculation with Phomopsis asparagi. (a) Disease symptoms in A. officinalis treated with sterile distilled water (AOC), A. officinalis inoculated with P. asparagi (AOI), A. kiusianus treated with sterile distilled water (AKC), and A. kiusianus inoculated with P. asparagi (AKI). (b) Disease severity percentage in AKI and AOI plants. Values represent the mean ± standard error (SE) of three independent replicates (n = 3). Asterisks indicate significant difference between AOI and AKI plants as determined by Student’s t-test (***P < 0.001).
Transcriptome sequencing and de novo assembly
To identify genes involved in the response to stem blight disease, stems of susceptible cultivated A. officinalis ‘Mary Washington 500W’ and resistant wild A. kiusianus (AK0501 strain) were inoculated with P. asparagi. Samples were collected from SDW-treated control stems and inoculated stems 24 h after infection. Four RNA-Seq libraries were generated: A. officinalis SDW-treated control (AOC), A. officinalis-inoculated (AOI), A. kiusianus SDW-treated control (AKC), and A. kiusianus-inoculated (AKI). Libraries were sequenced using an Illumina HiSeq 2500 platform. In total, 44,65 and 41,55 Gbp nucleotide sequence were generated from A. officinalis and A. kiusianus, respectively. After removing adapter sequences and low-quality sequences, 94,440,525; 101,235,105; 90,926,718; and 94,362,439 high-quality cleaned reads with 100 bp were obtained from AOC, AOI, AKC, and AKI, respectively. Details of the Asparagus transcriptome data are shown in Supplementary Table S1. The high-quality reads were de novo assembled into unique transcripts using Trinity software. In total, 206,164 and 213,950 assembled transcripts (average length 973 bp) were obtained from A. officinalis and A. kiusianus, respectively. Many transcripts were short; nevertheless, 64,094 and 66,330 transcripts were assembled from A. officinalis and A. kiusianus, respectively, that were >1,000 bp in length. The quality of transcriptome assemblies was assessed, and the length distribution of the assembled transcripts in both Asparagus species is shown in Fig. 2a. The GC-content of the assembled Asparagus transcripts was similar in both species, with a distribution peak at ~35% GC (Fig. 2b).
Figure 2.
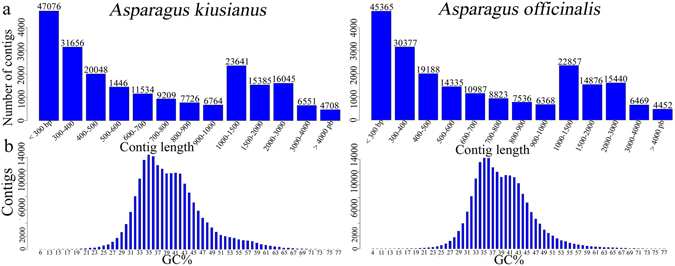
Transcript length and GC-content distribution graph in Asparagus officinalis and A. kiusianus. (a) Length distribution of assembled transcriptome fragments in A. officinalis and A. kiusianus. (b) GC-content percentage distribution in unique transcripts of A. officinalis and A. kiusianus.
Functional annotation of Asparagus transcriptomes
To functionally annotate the assembled Asparagus transcripts, translated sequences were searched against the NCBI non-redundant proteins (nr) database (http://www.ncbi.nlm.nih.gov) and the UniProt (Swiss-Prot) protein database using Blastp. Of the assembled sequences, 35,259 translated transcripts in A. officinalis and 36,321 in A. kiusianus matched known proteins in Swiss-Prot. A relatively large proportion of the Asparagus translated transcripts had no hits to any known proteins. Gene Ontology (GO) terms were assigned to the Asparagus transcripts. In total, 22,148 and 24,411 transcripts from A. officinalis and A. kiusianus, respectively, were assigned at least one GO term. Of these, 10,006 and 11,027 were assigned for biological process, 5,026 and 5,591 were assigned for cellular component, and 7,116 and 7,793 were assigned for molecular function, for A. officinalis and A. kiusianus, respectively. Within the biological process category (Fig. 3a), the most abundant groups were metabolic process (GO: 0008152, 36%), cellular process (GO: 0009987, 30%), localization and locomotion (GO: 0051179, 11%), cellular component organization (GO: 0071840, 7%), and response to stimulus (GO: 0050896, 6%). Within the cellular component category, cell part (GO: 0044464, 43%), organelle (GO: 0043226, 27%), macrocellular complex (GO: 0032991, 16%), and membrane (GO: 0016020, 12%) were the most highly represented groups (Fig. 3b). Catalytic activity (GO: 0003824, 46%), binding (GO: 0005488, 28%), transporter activity (GO: 0005215, 13%), receptor activity (GO: 0004872, 5%), and structural molecule activity (GO: 0005198, 4%) were the most highly represented groups in the molecular function category (Fig. 3c).
Figure 3.
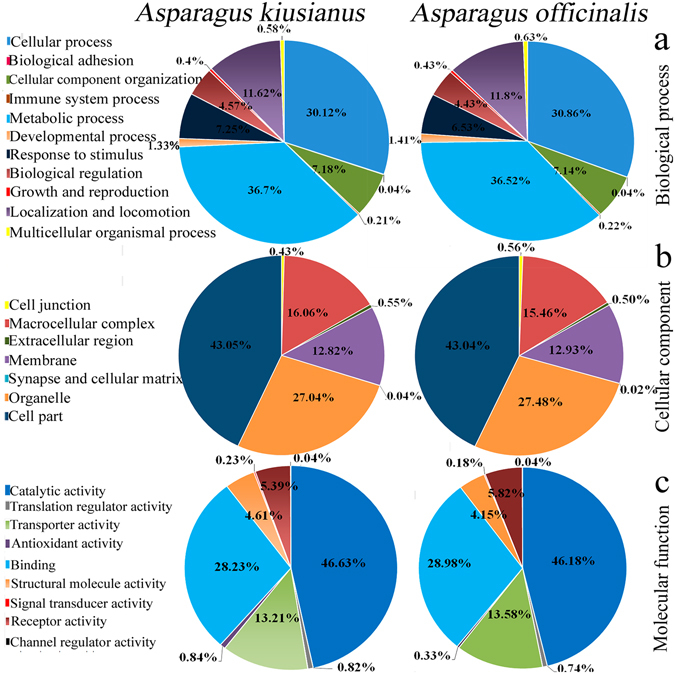
Pie chart of gene ontology (GO) analysis of Asparagus officinalis and A. kiusianus. Results are summarized for the three main GO categories: (a) biological process, (b) cellular component, and (c) molecular function.
Differentially Expressed Genes (DEGs)
Gene expression profiles were derived from the RNA-Seq data, and normalized DEGs were determined. To find genes that were induced by stem blight disease, pairwise comparisons were made between AOI and AOC and between AKI and AKC. A total of 1,779 differentially expressed transcripts were identified, of which 1,027 were up-regulated and 752 were down-regulated, in the two Asparagus species after inoculation with P. asparagi (Fig. 4a). Of the 1,027 up-regulated transcripts, 515 were up-regulated only in A. kiusianus, 352 were up-regulated only in A. officinalis, and 160 were up-regulated in both Asparagus species (Fig. 4b,c). These results demonstrated that the gene expression responses to P. asparagi infection differed between the two Asparagus species. GO term enrichment analysis indicated that genes involved in a range of biological processes such as transcriptional regulation, stress and defence response, protein kinase activity, phenylpropanoid and hormone biosynthesis and signaling, and cell wall assembly and organization were significantly enriched in the set of genes that was up-regulated in wild A. kiusianus but not in cultivated A. officinalis (Fig. 5).
Figure 4.
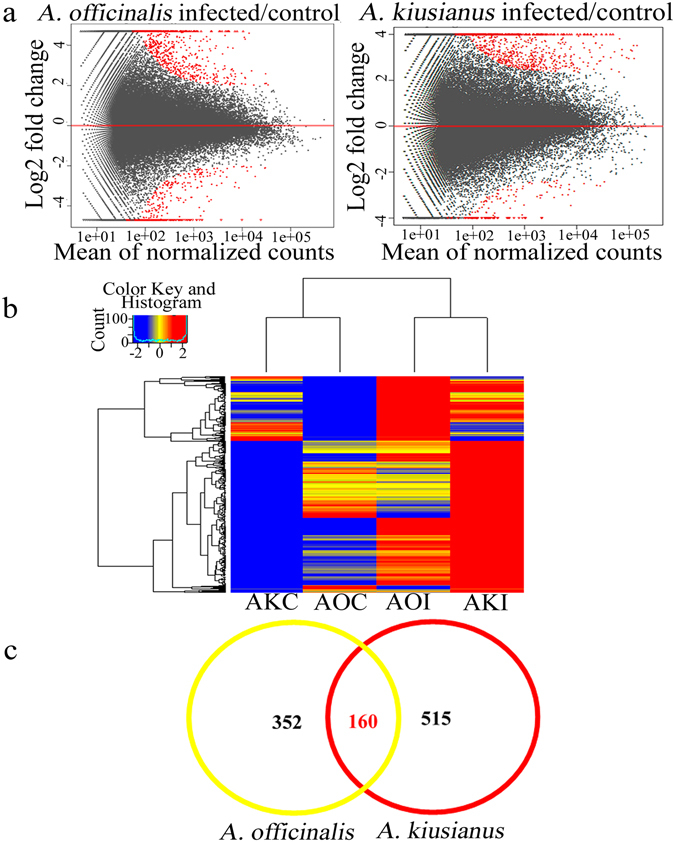
Differentially expressed genes (DEGs) in Asparagus officinalis and A. kiusianus stems 24 h after inoculation with Phomopsis asparagi. (a) MA scatter plot analysis showing log 2 fold change (y-axis) of pairwise comparisons between A. officinalis inoculated with P. asparagi (infected) versus A. officinalis treated with sterile distilled water (control), and A. kiusianus inoculated with P. asparagi (infected) versus A. kiusianus treated with sterile distilled water (control). DEGs are highlighted in red. (b) Heatmap clustering of up-regulated transcripts (fold change ≥ 2) in A. officinalis inoculated with P. asparagi (AOI) relative to A. officinalis treated with sterile distilled water (AOC), and A. kiusianus inoculated with P. asparagi (AKI) relative to A. kiusianus treated with sterile distilled water (AKC). (c) Venn diagram of transcripts up-regulated in A. officinalis and A. kiusianus 24 h after inoculation with P. asparagi.
Figure 5.
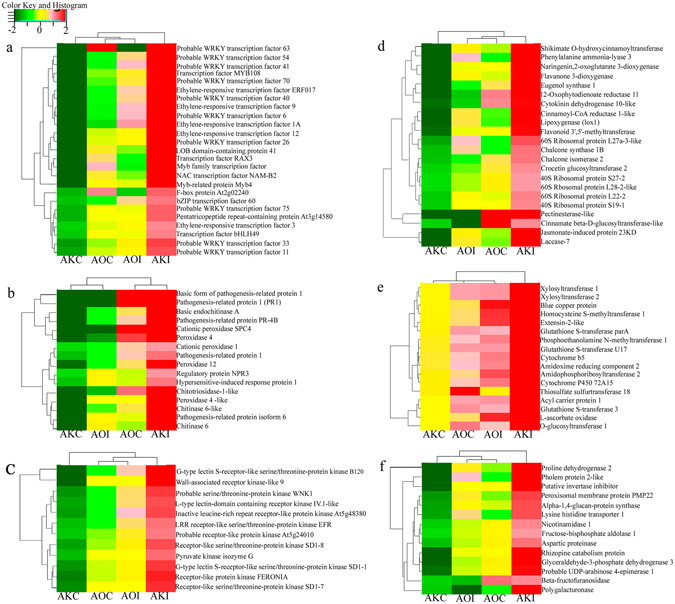
Heatmap clustering of most up-regulated (fold change ≥ 2) transcripts in Asparagus kiusianus inoculated with Phomopsis asparagi (AKI) and their relative expression levels in A. officinalis inoculated with P. asparagi (AOI). A. kiusianus and A. officinalis treated with sterile distilled water (AOC and AKC, respectively) served as controls. Genes are categorized according to those encoding (a) transcription factors, (b) Defence response factors, (c) kinase signaling proteins, (d) phenylpropanoid and ribosomal proteins, (e) transferase-related proteins, and (f) amino acid- and carbohydrate-related proteins.
In A. kiusianus, WRKY6, WRKY33, WRKY40, WRKY41, WRKY54, WRKY63, and WRKY70 exhibited 5.45-, 2.94-, 6.32-, 9.46-, 11.28-, 11.20-, and 8.11-fold higher expression in inoculated (AKI) plants than in control (AKC) plants, respectively. By contrast, in A. officinalis, these WRKY TFs were expressed at low levels in inoculated (AOI) plants compared with control (AOC) plants (Fig. 5a). These findings suggest that WRKY TFs are likely to play an important role in regulating transcription in A. kiusianus in response to P. asparagi infection. In A. kiusianus, expression of stress and defence response-related genes, including peroxidase 4, peroxidase 12, chitinase-6, chitotriosidase-1-like, and pathogenesis-related protein-1 (PR-1), was elevated in inoculated (AKI) plants, at levels 24.99-, 3.74-, 7.0-, 7.40-, and 114.2-fold higher, respectively, than seen in control (AKC) plants. By contrast, in A. officinalis, these genes were down-regulated in inoculated (AOI) plants relative to control (AOC) plants (Fig. 5b). In addition, the expression of JA biosynthesis-related genes including phospholipase D alpha 1 (PLDα1), lipoxygenase 1 (LOX1), 12-oxophytodienoate reductase 11 (OPR) and Jasmonate-induced 23 KD protein (JIP-23 KD) was evaluated in AKI plants at levels 2.22-, 5.25-, 6.02- and 36-fold higher expression than in AKC plants (Fig. 5d). However, these genes exhibited a significant reduction in AOI plants relative to AOC plants (Fig. 5d).
Metabolic pathways by KEGG analysis of differentially up-regulated genes in A. kiusianus
To characterize the metabolic pathways of the identified DEG in A. kiusianus, gene classification was performed based on Kyoto Encyclopedia of Genes and Genomes (KEGG) database14. Pathway categories were listed in the Supplementary Table S3. A total of 32 pathways were significantly identified. Genes involved in metabolic pathways, biosynthesis of secondary metabolites, plant-pathogen interaction, kinase signaling pathway and plant hormone signal transduction were the most represented up-regulated DEG in A. kiusianus in response to P. aspargi (Supplementary Table S3).
qRT-PCR
Fifteen up-regulated transcripts related to stress and defence, hormone biosynthesis and signaling, and ribosome were selected for validation of RNA-Seq data using qRT-PCR. Prior to qRT-PCR analysis, RT-PCR amplification was performed to confirm the suitability of the primer pairs. Single amplicons of the expected size were produced for each primer pair (Supplementary Fig. S1). The qRT-PCR results confirmed that the selected transcripts were all strongly up-regulated in AKI plants compared with AKC plants and down-regulated in AOI plants compared with AOC plants (Fig. 6a). In addition, the qRT-PCR results positively correlated (r = 0.78) with the RNA-Seq data (Fig. 6b), validating the sequencing results.
Figure. 6.
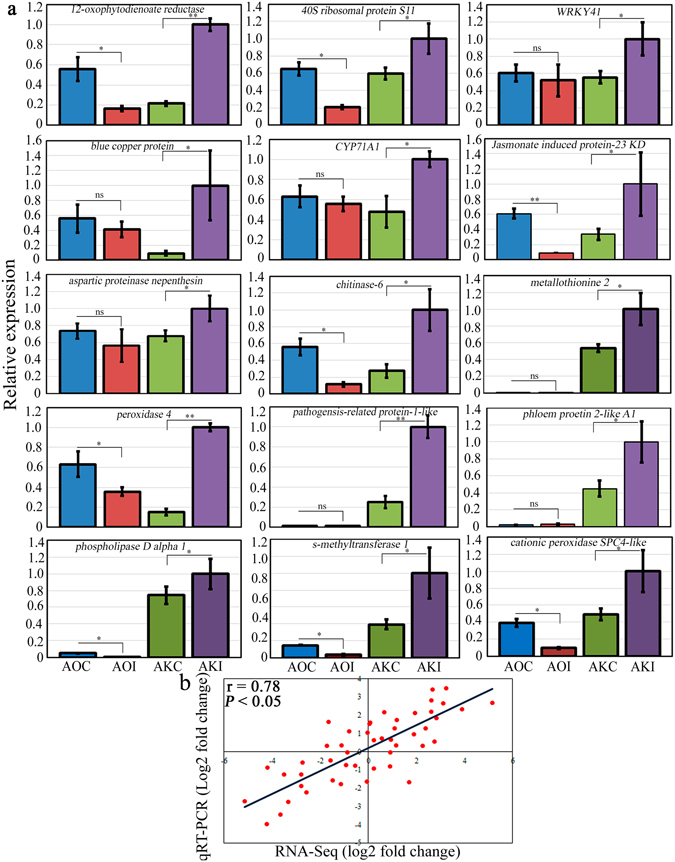
Validation of RNA-Seq data with quantitative real-time reverse transcription PCR (qRT-PCR). (a) Bar plot of qRT-PCR relative expression in Asparagus kiusianus and A. officinalis treated with distilled water (AKC and AOC, respectively) and inoculated with Phomopsis asparagi (AKI and AOI, respectively). (b) Scatter plot correlation between qRT-PCR and RNA-Seq log2 fold changes (Pearson’s correlation coefficient = 0.78). Values represent the mean on three independent biological replicates (n = 3) ± standard deviation (SD). Significance levels are given as: *(P < 0.05) and **(P < 0.01) and according to analysis of variance (ANOVA).
Phytohormone accumulation in wild and cultivated Asparagus species 24 h post-inoculated with P. asparagi
Because JA biosynthesis and signaling-related genes were markedly elevated in AKI plants relative to AOC plants, we examined the JA, MeJA, SA and ABA levels in the AOC, AOI, AKC and AKI plants 24 h post-inoculation using LC-MS/MS (Fig. 7). JA and MeJA exhibited a significant (P < 0.001) increase at 65.92- and 14746.97-fold, respectively in AKI plants in comparison with AKC plants. However, there was no significant differences between AOI and AOC plants (Fig. 7a,b). SA levels exhibited non-significant increase in the two infected Asparagus species relative to control plants (Fig. 7c). In addition, ABA levels displayed a significant increase (P < 0.05) at 1.98-fold in AOI plants relative to AOC plants (Fig. 7d). However, non-significant differences were observed between AKI and AKC plants (Fig. 7d).
Figure 7.
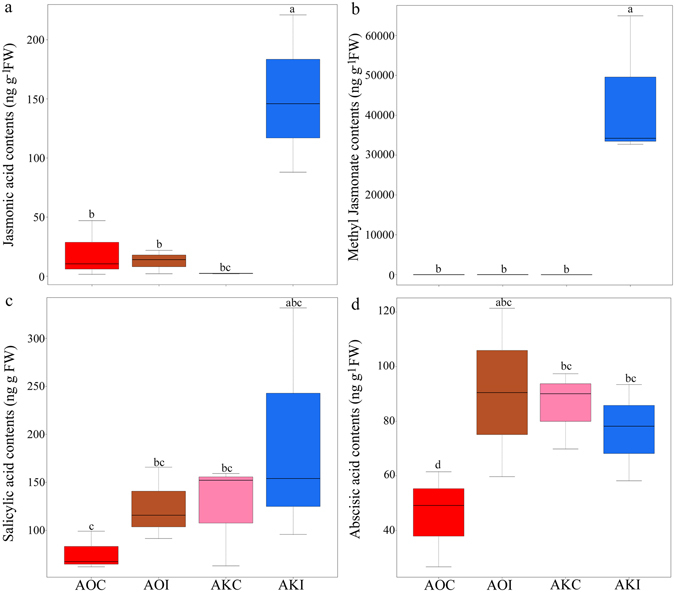
Box plot diagram showing the changes in (a) jasmonic acid (JA), (b) methyl jasmonate (MeJA), (c) salicylic acid (SA) and (d) abscisic acid (ABA) levels (ng g−1 FW) in Asparagus kiusianus inoculated with Phomopsis asparagi (AKI) and their relative levels in A. officinalis inoculated with P. asparagi (AOI). A. kiusianus and A. officinalis treated with sterile distilled water (AOC and AKC, respectively) served as controls. Values represent the maximum, third quartile, median, first quartile and minimum of three independent replicates (n = 3). Different letters indicate statistically significant difference at P < 0.05 according to Tukey’s honest significant difference (HSD) post hoc test.
Effects of P. asparagi on total protein content and stress-related enzyme activities in wild and cultivated Asparagus species
In A. kiusianus, total protein content was significantly higher (1.52-fold increase; P < 0.008) in plants inoculated with P. asparagi (AKI) than in control plants (AKC). Conversely, in A. officinalis, although protein content was slightly higher in inoculated plants, no significant difference in total protein content was seen between inoculated (AOI) and control (AOC) plants (Fig. 8a). POX and CAT enzymatic assays were carried out to investigate the nature of the defence response in the two Asparagus species. No significant difference in POX activity was detected between inoculated (AOI) and control (AOC) A. officinalis plants. However, in A. kiusianus, POX activity was significantly higher (1.32-fold increase; P < 0.002) in inoculated (AKI) plants than in control (AKC) plants (Fig. 8b). Inoculation stimulated significant increases in CAT activity in both A. officinalis and A. kiusianus. CAT activity was 1.64-fold higher (P < 0.001) in AOI plants than in AOC plants and 1.55-fold higher (P < 0.001) in AKI plants than in AKO plants (Fig. 8c).
Figure 8.
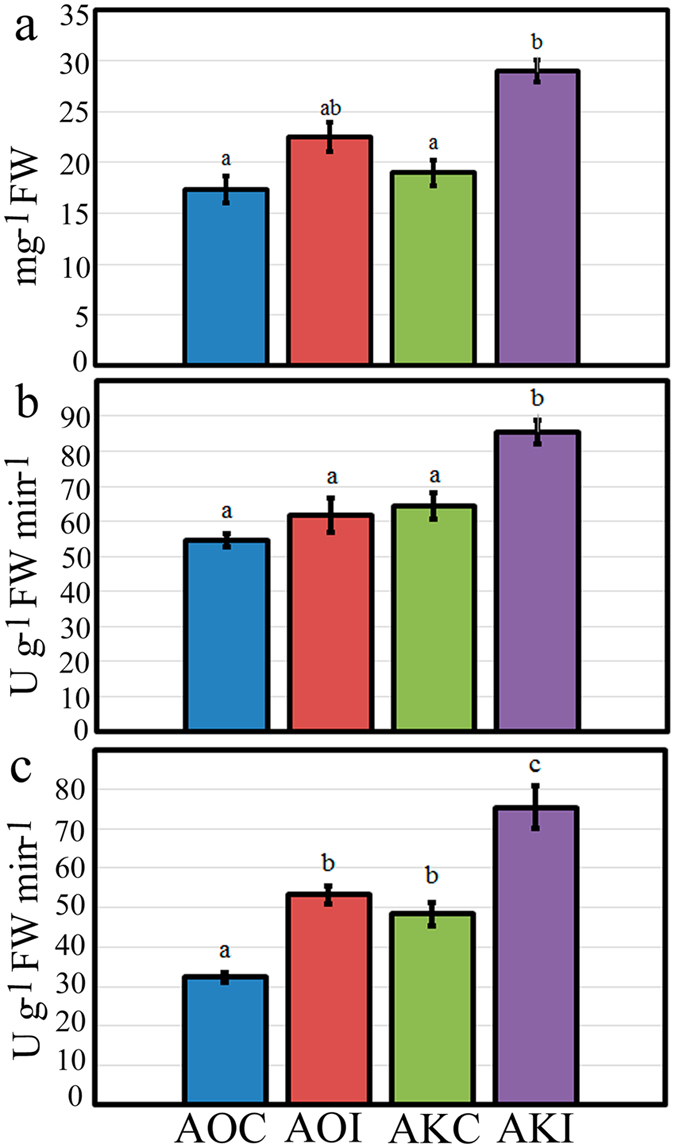
Accumulation of (a) total protein content, (b) peroxidase, and (c) catalase activities in Asparagus officinalis inoculated with Phomopsis asparagi (AOI) and A. kiusianus inoculated with P. asparagi (AKI). A. officinalis and A. kiusianus treated with sterile distilled water (AOC and AKC, respectively) served as controls. Values represent the mean ± standard error (SE) of three independent replicates (n = 3). Different letters indicate statistically significant difference at P < 0.05 according to Tukey’s honest significant difference (HSD) post hoc test.
Discussion
Asparagus stem blight, which is caused by P. asparagi, is a serious disease that affects asparagus production worldwide, and there is an urgent need to produce asparagus cultivars with strong resistance to this disease. After artificial inoculation with P. asparagi, A. kiusianus, a wild relative of cultivated A. officinalis, exhibited significantly (P < 0.001) reduced disease severity compared with A. officinalis (Fig. 1), consistent with a previous study examining disease in these Asparagus species4. These results suggest that wild A. kiusianus is a potential genetic reservoir for Phomopsis disease resistance improvement in cultivated A. officinalis. Indeed, interspecific hybridization between A. kiusianus and A. officinalis resulted in F1 hybrids with strong Phomopsis disease resistance4. However, the mechanisms underlying Phomopsis disease resistance in A. kiusianus remain unknown. Candidate A. kiusianus genes involved in Phomopsis disease resistance were identified by comparing transcription in susceptible A. officinalis and resistant A. kiusianus 24 h after infection with P. asparagi. Samples from inoculated and control A. officinalis and A. kiusianus plants were sequenced using the Illumina HiSeq 2500 platform, and de novo transcriptome assemblies were generated and compared. In total, 1,779 differentially expressed transcripts, 1,027 of which were up-regulated and 752 of which were down-regulated, were detected in the two Asparagus species in response to P. asparagi infection (Fig. 4). GO term enrichment and KEGG metabolic pathway analyses revealed that transcripts that were highly expressed in A. kiusianus and exhibited low expression in A. officinalis were enriched for metabolic pathways, biosynthesis of secondary metabolites, plant-pathogen interaction, protein kinase signaling and plant hormone biosynthesis and signaling transduction (Fig. 5 and Supplementary Table S3).
Expression of stress and defence response-related genes was strongly up-regulated in resistant wild A. kiusianus in comparison with susceptible A. officinalis (Figs 5 and 6) in response to P. aspargi infection. Up-regulation of PR genes, including PR-1, is a prerequisite for activation of systemic acquired resistance (SAR), which plays a central role in plant basal defence against pathogen infection15, 16. Plant chitinases and POXs, which are also classified as PR proteins, play direct roles in plant resistance by hydrolyzing fungal cell walls and oxidating phenolic residues in the infected tissues, respectively17–19. A similar pattern of up-regulation of stress and defence response-related genes was reported in leaves of Withania somnifera after stimulation of salicylic acid-induced defence mechanisms19. Up-regulation of POX, chitinase, and PR transcripts was also reported in Fusarium-resistant wheat (Triticum aestivum) compared with a susceptible genotype20. Similarly, rice (Oryza sativa) variety ‘Selenio’, which was resistant to Fusarium fujikuroi, exhibited up-regulation of several PR genes compared with the susceptible variety ‘Dorella’21. Our data and results from previous studies clearly suggest that POX, chitinase, and PR-1 play important roles in plant basal defenses and SAR induction during early plant responses to infection in resistant genotypes22. SAR is responsible for triggering host defence mechanisms to increase de novo expression of defense-related genes, leading to enhanced expression and de novo synthesis and accumulation of chitinases, POXs, and other pathogenesis-related proteins in uninfected tissues, thereby defending them against any further pathogen attack23. It is therefore likely that similar defence pathways are involved in responses to P. asparagi infection in wild A. kiusianus.
Our results also indicate a possible role for genes encoding ribosomal proteins (RPLs) in Asparagus defence mechanisms. Several genes encoding RPLs were strongly up-regulated in inoculated A. kiusianus (AKI) plants and suppressed in inoculated A. officinalis (AOI) plants (Fig. 5d). RPLs have established roles in facilitating protein synthesis and preserving the stability of the ribosomal complex24. The involvement of RPLs in plant defence was recently reported in non-host resistance against bacterial pathogenic attack in Nicotiana benthamiana 25. The study suggested that RPL12 and RPL19 played substantial roles in the development of non-host disease resistance through the induction of the hypersensitive response (HR)25. Similarly, strong up-regulation of 80% of rice RPL-related genes was observed in response to exposure to Xanthomonas oryzae 24. The putative promoter regions of these RPL-related genes carry cis-elements that respond specifically to stress, suggesting that genes in the RPL family might be useful targets in strategies to develop stress tolerance in rice and other crops24. Large-scale analysis of differential gene expression in coffee (Coffea arabica) genotypes that were resistant or susceptible to leaf miner revealed up-regulation of RPL-encoding genes in the resistant genotype compared with the susceptible genotype26. Our results indicated that genes encoding RPLs may be involved in disease resistance in A. kiusianus in response to P. asparagi exposure, and that this response may be species-specific. Further molecular and biochemical analyses of RPLs identified in this study will be needed to understand the functional role of RPLs in the Asparagus defence mechanism.
In the present study the expression level of several JA biosynthesis-related genes including PLDα1, LOX1, OPR and JIP-23 KD was elevated in the resistant wild A. kiusianus in comparison with susceptible A. officinalis in response to P. aspargi infection (Figs 5 and 6). PLD and LOX genes are necessary for the initial steps of JA biosynthesis in Arabidopsis 27. PLD release linolenic and α-linolenic acids from chloroplast membranes and these substrates are subsequently catalyzed by LOX, leading to the formation of hydroperoxy octadecadienoic acids27. During the early stage of plant-pathogen interaction the activation of PLD and LOX is essential for the production of important defense signaling molecules, such as oxylipins and JA which has been shown to modulate the activity of variety of proteins involved in defense signaling28. RNA-Seq analysis of F. fujikuroi- resistant rice variety ‘Selenio’ showed up-regulation in PLD and LOX genes in comparison with susceptible rice ‘Dorella’, indicating a crucial role of JA pathway in the resistance of rice to F. fujikuroi infection21. JA signaling has broadly been associated to the defense against necrotrophic pathogens, inducing the accumulation of secondary metabolites and PR proteins29. However, recent studies have revealed that certain biotrophic fungal species can also trigger JA-mediated responses29. In this context, Phomopsis pathogens are nectrotrophic at least for the latent phase of infection and are therefore named hemi-biotrophs30. In A. kisuainus a substantial increase in OPR expression was observed (Figs 5 and 6). OPR catalyzes the final step in JA biosynthesis by reducing 12-oxophytodienoic acid to 3-oxo-2(2′-pentenyl)-cyclopentane-1-octanoic acid28. OPR was previously proposed as a disease resistance marker in tomato (Solanum lycopersicum)31. A cleaved amplified polymorphic sequences (CAPS) marker derived from tomato OPR showed putative co-segregation with a potato (S. tuberosum) quantitative trait locus (QTL) for late blight disease resistance. Likewise, gene expression profile of JA biosynthesis-related genes (LOX, AOC and OPR) in Plasmopara viticola disease resistance and susceptible grapevine (Vitis vinifera) cultivars, revealed a strong up-regulation of these genes in the resistant cultivar in comparison with susceptible cultivar32. Additionally, expression of OPR and LOX was substantially higher in a Fusarium head blight-resistant wheat variety ‘Wangshuibai’ than in a susceptible variety ‘NAUH117’ 24–48 h after infection33. These results suggest that OPR and LOX have potentially important roles in the early defence response in wheat against Fusarium head blight33. Another JA biosynthesis-related gene, JIP-23 KD, was expressed at substantially higher levels in inoculated A. kiusianus (AKI) plants than in AKC plants. In A. officinalis, JIP-23 KD exhibited reduced expression in inoculated plants (AOI) compared with control plants (AOC) (Fig. 5d). An early study examining the effect of JA on the interactions between barley (Hordeum vulgare) and powdery mildew suggested a possible role for JIP-23 KD in cell wall modification or in pathogen defence34. However, the specific roles of JIP-23 KD in pathogen defence remain to be determined34. Recently, JIP-23 KD was shown to be up-regulated in cadmium-tolerant barley genotype compared with a cadmium-susceptible genotype35. Collectively, these results suggest that JIP-23 KD may have a role in plant stress responses. The up-regulation of PLDα1, LOX1, OPR and JIP-23 KD genes in A. kiusianus inoculated with P. asparagi suggests that JA signal transduction may play a crucial role in Phomopsis disease resistance in A. kiusianus. Phytohormone analysis revealed a significant increase in JA and MeJA contents in wild-resistant A. kiusianus relative to cultivated-susceptible A. officinalis in response to P. asparagi infection (Fig. 7), providing an additional support for our hypothesis regarding the important role of JA-dependent signaling pathway in the P. asparagus disease resistance.
The fluctuation in POX and CAT activities between resistance and susceptible genotypes has been previously reported36. Therefore, stress-response enzyme activities were assessed in the present study. A significant (P < 0.002) increase in POX activity was seen in A. kiusianus plants after P. asparagi infection (AKI) compared with control plants (AKC). No significant difference was seen in POX activity between inoculated (AOI) and control (AOC) A. officinalis plants (Fig. 7). This suggested that POX played a substantial role in A. kiusianus defence via suppression of P. asparagi spread. The accumulation pattern of POX in inoculated A. kiusianus plants correlated with the up-regulation of several POX-related genes in the RNA-Seq data (Fig. 5c). Similar findings were reported in cabbage (Brassica oleracea var. capitata), where strong POX activities and up-regulation of POX-related genes was seen in cabbage resistant to black rot disease37. CAT activity was significantly (P < 0.001) induced by P. asparagi inoculation in both A. kiusianus (AKI) and A. officinalis (AOI) (Fig. 7c). Similar findings were previously reported in A. officinalis inoculated with P. asparagi 2: CAT activities increased at all time points after infection whereas POX activities increased only initially, and declined thereafter2. These data indicate that CAT is involved in basal defence mechanisms in both resistant and susceptible Asparagus species.
This is the first study to examine the molecular responses of Asparagus species to P. asparagi infection. RNA sequencing was used to identify DEGs in resistant wild A. kiusianus and susceptible cultivated A. officinalis 24 h after infection with P. asparagi. Functional annotation and KEGG pathway analysis showed that the group of up-regulated genes in A. kiusianus was enriched for metabolic pathways, biosynthesis of secondary metabolites, plant-pathogen interaction, transcriptional regulation, protein kinase signaling, phenylpropanoid and hormone biosynthesis and signaling. Results from RNA-Seq data and qRT-PCR were correlated, confirming the reliability of the transcriptome data. Activity of the stress-related enzyme POX was elevated in A. kiusianus compared with A. officinalis. Overall, comparative transcription profiling provided valuable insights into the mechanisms underlying Phomopsis disease resistance in Asparagus. These findings will be valuable in the future development of disease-resistant asparagus varieties. The RNA-Seq datasets generated in this study will be mined for sequence variations associated with gene structure and function, which will facilitate genetic trait mapping and marker-assisted selection in asparagus breeding programs.
Methods
Plant materials
Cultivated A. officinalis ‘Mary Washington 500W’ (AO0060 strain) and wild A. kiusianus (AK0501 strain) for RNA-Seq analysis were grown under standard greenhouse conditions (25 ± 2 °C and 14 h light/10 h dark) at Kagawa Prefectural Agricultural Experiment Station (Kagawa, Japan). Male A. kiusianus and female A. officinalis were kept to grow for 4-year-old in a commercial soil consisted of loam, well-drained soil, leaf mold and garden soil with fertilizers (2:1:1:2 and 2:1:1:4 for A. kiusianus and A. officinalis, respectively v:v) containing 50 mg N kg−1, 500 mg P kg−1, and 100 mg K kg−1 [pH 6–6.5, water-holding capacity ~70%, Nippi Engei Baido No.1 (Nihon Hiryo Co. Ltd)] as a root-supporting medium (Supplementary Fig. S2).
Inoculation of A. officinalis and A. kiusianus with P. asparagi for RNA-Seq analysis
Plants were artificially inoculated with spores of P. asparagi at a final concentration of 107 CFU ml−1 according to the vinyl cotton (VC) method38 (Supplementary Fig. 2). Minimal methodological modifications were made: silicone tubes were used instead of vinyl tubes and water-retentive polyester fiber sheeting was used instead of cotton. Stems from three independent biological replicates (n = 3) were harvested 24 h after infection, immediately frozen in liquid nitrogen, and stored at −80 °C until used. Plants treated with SDW under the same conditions served as a control.
Determination of stem blight disease severity in cultivated and wild Asparagus species
In a separate experiment, A. officinalis and A. kiusianus plants cultivated and inoculated as described above were kept for 3 weeks after inoculation and disease severity on the infected stems was recorded. Disease severity was scored on a 1–5 scale, where 1 = healthy and 5 = heavily infected. Disease severity percentage was calculated based on the following formula:
| 1 |
RNA extraction, cDNA library construction, and Illumina sequencing
Total RNA was extracted from three independent biological replicates (n = 3) using a QIAGEN RNeasy Plant Mini Kit (QIAGEN Sciences, Maryland, USA) following the manufacturer’s instructions. The quality and quantity of RNA were assessed using 2% agarose gel electrophoresis and an Agilent 2100 Bioanalyzer (Agilent Technologies Inc., USA) with a minimum RNA integrated number of eight. Sequencing libraries were generated using a TruSeq Stranded RNA Sample Preparation Kit (Illumina). The cDNA libraries were sequenced using an Illumina HiSeq 2500 instrument (Illumina Inc., USA), and both ends of the inserts were sequenced.
RNA-Seq data processing and de novo transcriptome assembly
The quality of the raw sequences was inspected using the open-source software FastQC (www.bioinformatics.babraham.ac.uk/projects/fastqc). After quality checking, adapter sequences were removed by Cutadapt v.1.339, and low-quality (phred score < 30) and short (length < 50 bp) reads were trimmed by Sickle v.1.200 (www.github.com/najoshi/sickle). The resulting high-quality trimmed and size-selected reads were then de novo assembled using Trinity package v.2.0.640, 41. The resulting assembled unique transcripts were annotated by alignment against the NCBI nucleotide (nt) database (http://www.ncbi.nlm.nih.gov), using Blastn algorithms with a significance threshold of E-value Ie-3. The likely open reading frame (ORF) for each transcript was extracted using the TransDecoder program (www.transdecoder.sourceforge.net). Predicted ORFs were searched against the UniProt (Swiss-Prot) database using Blastp with a significance threshold of E-value Ie-642. To annotate transcripts with GO terms, the top hit from the NCBI nt database for each transcript was submitted to the Blast2GO platform43, and GO terms for each transcript were retrieved based on the relationship between gene names and GO terms.
Differential gene expression analysis
The expression level of each transcript was measured with a FPKM (fragments per kilobase of exon per million fragments mapped) method. FPKM values were calculated using RSEM v.1.2.1944. Genes that were differentially expressed (DEGs) between A. officinalis and A. kiusianus 24 h after inoculation with P. asparagi were determined using BESeq R Bioconductor45, 46. Genes were classified as DEG if they exhibited ≥2-fold changes between the two samples with statistical significance adjusted-P < 0.0547.
KEGG pathway analysis
The metabolic pathways of the up-regulated DEG in A. kiusianus in response to P. aspargi infection was constructed based on KEGG database. Initially, all the DEG uniprot ID was converted into KEGG ID by using retrieve/ID mapping (http://www.uniprot.org/uploadlists). Further, the KO number for each metabolic pathway was obtained by using KEGG mapper web-based tool (http://www.genome.Jp/keg/tool/map_pathway2.html).
Experimental validation of candidate gene expression by qRT-PCR
To confirm DEG results, qRT-PCR was performed on six selected up-regulated disease resistance candidate genes from A. kiusianus libraries. Details of gene annotations and primer sets are shown in Supplementary Table S2. Gene-specific primers and internal standard primers were designed using the aligned sequence of A. officinalis and A. kiusianus cDNA obtained from the Illumina RNA-Seq data. First, real time reverse transcription-PCR (RT-PCR) was carried out using a Takara PCR Thermal Cycler and their PCR products were checked using 2% agarose gel electrophoresis. Second, qRT-PCR was performed using a SYBR Green Supermix Kit (Bio-Rad Laboratories, Inc.) with a Mini Option Real-Time PCR system (Bio-Rad). Relative expression values were calculated using the 2−ΔΔCt method, with elongation factor as an internal standard. RNA pools used in the RT-PCR and qRT-PCR analyses were extracted from samples used for RNA-Seq. All reactions were performed with three biological replicates.
Quantification of plant hormones
For analysis of JA, MeJA, SA and ABA, 100 mg from AKC, AKI, AOC and AOI stem was homogenized separately in liquid nitrogen and placed in 5 ml of 80% methanol containing 0.1% acetic acid. [2H2]JA, [2H2]MeJA (Tokyo Chemical Industry), [2H4]SA (Cambridge Isotope Laboratories) and [2H6]ABA (OlchemIm) were added to the extracts to serve as internal standards. After overnight extraction at 4 °C, solids were separated by centrifugation and re-extracted for 30 min in 5 ml of the same extraction solution. The extracts were combined, and passed through BOND ELUT C18 column (500 mg, Agilent Technologies), equilibrated with 1% acetic acid. The eluate was evaporated and dissolved in water:methanol:acetic acid (89.9:10:0.1, v-v:v) and analyzed by LC-MS/MS system. The LC-MS/MS system consisted of a Prominence 20A Series HPLC (Shimadzu) equipped with a 3200 QTrap LC/MS/MS System (AB Sciex), using an electrospray interface. For quantification of JA, SA and ABA, the purified samples were injected onto a Cadenza CD-C18 column (3 µm, 150 × 3.0 mm; Imtakt) at 45 °C and eluted at a flow rate of 0.2 ml min−1. Hormones were separated with a gradient of mobile phase A (water:methanol:acetic acid, 89.9:10:0.1) and B (methanol). The initial conditions were 100% A, maintaining for 2 min, changing linearly to 40% A and 60% B in 5 min, changing to 100% B in 10 min, and finally maintained at 100% B for 6 min. The column was equilibrated with the starting composition of the mobile phase for 12 min. For quantification of MeJA, the purified samples were injected onto a Shim-pack XR-ODS column (2.2 μm, 75 × 2.0 mm; Shimadzu). The mobile phases and gradient conditions were described above. Quantification was obtained by multiple reaction monitoring (MRM) of the selected precursor ions and a specific product ions as described in Supplemental Table S4.
Preparation of crude enzyme extract
Stem tissue (200 mg) was ground to a fine powder in liquid nitrogen using a pre-cooled mortar and pestle, and 2 ml of extraction buffer [0.2 M phosphate buffer (pH 7.2), 0.1 mM EDTA, 1 mM DTT, and 2 U protease inhibitor cocktail] was added. The macerated suspension was centrifuged at 10,000 rpm for 5 min at 4 °C. The supernatant was collected and used as the source of enzyme. Total protein content of the crude enzyme extract was determined using a Bradford assay48.
Determination of stress-related enzyme activities
POX activity was estimated using a UV/Vis Beckman DU 730 spectrophotometer (Beckman Coulter Inc. California, USA) as described previously49. The reaction mixture, which consisted of 0.8 ml of 0.2 M phosphate buffer (pH 7.2), 1 ml of 15 mM guaiacol, 1 ml of 3 mM hydrogen peroxide, and 0.2 ml of crude enzyme extract, was incubated at room temperature for 3 min. Absorbance of the colored product was monitored at 470 nm. POX activity expressed as ∆470 g−1 fresh weight (FW) min−1 was calculated using the following formula:
| 2 |
CAT activity was determined spectrophotometrically at 240 nm as described previously50. CAT activity expressed as ∆240 g−1 FW min−1 was calculated using the following formula, modified with hydrogen peroxide coefficient ε240:
| 3 |
Electronic supplementary material
Acknowledgements
This work was supported in part by a Grant-in-Aid for Science and technology research promotion program for agriculture, forestry, fisheries and food industry from the Ministry of Agriculture, Forestry and Fisheries (MAFF) (Japan). We would like to thank Pranali S. for some bioinformatic analysis.
Author Contributions
M.A. and A.K. conceived and designed the study. N.S., T.I., M.M., K.M., Y.O., M.M., A.U., and A.K. carried out plant cultivation, Phomopsis inoculation, and sampling. M.A. performed RNA-Seq data analysis and data interpretation. M.A., N.S. and S.M. carried out experiments. M.A. and A.K. wrote the manuscript. All authors read and approved the final manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-02566-7
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Mostafa Abdelrahman, Email: meettoo2000@ige.tohoku.ac.jp.
Akira Kanno, Email: kanno@ige.tohoku.ac.jp.
References
- 1.Food and Agriculture Organization of the United Nations Statistics. FAOSTAT. Availabe at: http://faostat3.fao.org (2015).
- 2.Lu G, Jian W, Zhang J, Zhou Y, Cao J. Suppressive effect of silicon nutrient on Phomopsis stem blight development in asparagus. HortSci. 2008;43:811–817. [Google Scholar]
- 3.Udayanga D, et al. The genus Phomopsis: biology, applications, species concepts and names of common pathogens. Fungal Div. 2011;50:189–225. doi: 10.1007/s13225-011-0126-9. [DOI] [Google Scholar]
- 4.Iwato M, et al. Stem blight resistance of Asparagus kiusianus and its hybrid with A. officinalis. Adv. Hort. Sci. 2014;28:202–207. [Google Scholar]
- 5.Elena K. First report of Phomopsis asparagi causing stem blight of asparagus in Greece. Plant Pathol. 2006;55:300. doi: 10.1111/j.1365-3059.2005.01265.x. [DOI] [Google Scholar]
- 6.Kanno, A. & Yokoyama, J. Asparagus. In Wild Crop Relatives: Genomic and Breeding Resources, Vegetables (ed. Kole, C.) 23–42 (Springer, 2011).
- 7.Marcellán ON, Camadro EL. Self- and cross-incompatibility in Asparagus officinalis and Asparagus densiflorus cv. Sprengeri. Can. J. Bot. 1996;74:1621–1625. doi: 10.1139/b96-196. [DOI] [Google Scholar]
- 8.Ohwi, J. Asparagus L. In Flora of Japan (eds Meyer, F. G. & Walker, E. H.) 300 (Smithsonian Institution, 1965).
- 9.Kubota S, Konno I, Kanno K. Molecular phylogeny of the genus Asparagus (Asparagaceae) explains interspecific crossability between the garden asparagus (A. officinalis) and other Asparagus species. Theor. Appl. Genet. 2012;124:345–354. doi: 10.1007/s00122-011-1709-2. [DOI] [PubMed] [Google Scholar]
- 10.Ito T, et al. Production and characterization of interspecific hybrids between Asparagus kiusianus Makino and A. officinalis L. Euphytica. 2011;182:285–294. doi: 10.1007/s10681-011-0508-9. [DOI] [Google Scholar]
- 11.Mutz KO, Heilkenbrinker A, Lönne M, Walter JG, Stahl F. Transcriptome analysis using next-generation sequencing. Curr. Opin. Biotechnol. 2013;24:22–30. doi: 10.1016/j.copbio.2012.09.004. [DOI] [PubMed] [Google Scholar]
- 12.Abdelrahman M, et al. Integrating transcriptome and target metabolome variability in doubled haploids of Allium cepa for abiotic stress protection. Mol. Breeding. 2015;35:195. doi: 10.1007/s11032-015-0378-2. [DOI] [Google Scholar]
- 13.Unamba CIN, Nag A, Sharma RK. Next generation sequencing technologies: the doorway to the unexplored genomics of non-model plants. Front. Plant Sci. 2015;6:1074. doi: 10.3389/fpls.2015.01074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Du H, Wang Y, Yang J, Yang W. Comparative transcriptome analysis of resistant and susceptible tomato lines in response to infection by Xanthomonas perforans race T3. Front. Plant Sci. 2015;6:1173. doi: 10.3389/fpls.2015.01173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Durrant WE, Dong X. Systemic acquired resistance. Annu. Rev. Phytopathol. 2004;42:185–209. doi: 10.1146/annurev.phyto.42.040803.140421. [DOI] [PubMed] [Google Scholar]
- 16.An C, Mou Z. Salicylic acid and its function in plant immunity. J. Integr. Plant Biol. 2011;53:412–428. doi: 10.1111/j.1744-7909.2011.01043.x. [DOI] [PubMed] [Google Scholar]
- 17.van Loon LC, van Strien EA. The families of pathogenesis-related proteins, their activities, and comparative analysis of PR-1 type proteins. Physiol. Mol. Plant Pathol. 1999;55:85–97. doi: 10.1006/pmpp.1999.0213. [DOI] [Google Scholar]
- 18.Golubenko Z, et al. Induction of peroxidase as a disease resistance response in resistant (Hibiscus trionum) and susceptible (Althea armeniaca) species in the family Malvaceae. Phytoparasitica. 2007;35:401–413. doi: 10.1007/BF02980704. [DOI] [Google Scholar]
- 19.Ghosh Dasgupta M, George BS, Bhatia A, Sidhu OP. Characterization of Withania somnifera leaf transcriptome and expression analysis of pathogenesis–related genes during salicylic acid signaling. PLoS One. 2014;9:e94803. doi: 10.1371/journal.pone.0094803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dhokane D, Karre S, Kushalappa AC, McCartney C. Integrated metabolo-transcriptomics reveals Fusarium head blight candidate resistance genes in wheat QTL-Fhb2. PLoS One. 2016;11:e0155851. doi: 10.1371/journal.pone.0155851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matić S, et al. Comparative transcriptome profiling of resistant and susceptible rice genotypes in response to the seedborne pathogen Fusarium fujikuroi. BMC Genomics. 2016;17:608. doi: 10.1186/s12864-016-2925-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taheri P, Tarighi S. Genetic and virulence analysis of Rhizoctonia spp. associated with sugar beet root and crown rot in the northeast region of Iran. Plant Dis. 2012;96:398–408. doi: 10.1094/PDIS-08-11-0661. [DOI] [PubMed] [Google Scholar]
- 23.Govind SR, Jogaiah S, Abdelrahman M, Shetty HS, Tran LS. Exogenous trehalose treatment enhances the activities of defense-related enzymes and triggers resistance against downy mildew disease of pearl millet. Front. Plant Sci. 2016;7:1593. doi: 10.3389/fpls.2016.01593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moin M, et al. Rice ribosomal protein large subunit genes and their spatio-temporal and stress regulation. Front. Plant Sci. 2016;7:1284. doi: 10.3389/fpls.2016.01284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nagaraj S, Senthil-Kumar M, Ramu VS, Wang K, Mysore KS. Plant ribosomal proteins, RPL12 and RPL19, play a role in nonhost disease resistance against bacterial pathogens. Front. Plant Sci. 2015;6:1192. doi: 10.3389/fpls.2015.01192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cardoso DC, et al. Large-scale analysis of differential gene expression in coffee genotypes resistant and susceptible to leaf miner–toward the identification of candidate genes for marker assisted-selection. BMC Genomics. 2014;15:66. doi: 10.1186/1471-2164-15-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lyons R, Manners JM, Kazan K. Jasmonate biosynthesis and signaling in monocots: a comparative overview. Plant Cell Rep. 2013;32:815–827. doi: 10.1007/s00299-013-1400-y. [DOI] [PubMed] [Google Scholar]
- 28.Biesgen C, Weiler EW. Structure and regulation of OPR1 and OPR2, two closely related genes encoding 12-oxophytodienoic acid-10, 11-reductases from Arabidopsis thaliana. Planta. 1999;208:155–165. doi: 10.1007/s004250050545. [DOI] [PubMed] [Google Scholar]
- 29.Guerreiro A, Figueiredo J, Sousa Silva M, Figueiredo A. Linking jasmonic acid to grapevine resistance against the biotrophic oomycete Plasmopara viticola. Front. Plant Sci. 2016;7:565. doi: 10.3389/fpls.2016.00565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rosskopf EN, Charudattan R, DeValerio JT, Stall WM. Field evaluation of Phomopsis amaranthicola, a biological control agent of Amaranthus spp. Plant Dis. 2000;84:1225–1230. doi: 10.1094/PDIS.2000.84.11.1225. [DOI] [PubMed] [Google Scholar]
- 31.Madsen DK, Sorensen KK, Madsen MH, Kirk HGA. CAPS marker derived from tomato12-oxophytodienoate reductase shows putative co-segregation with potato late blight resistance. Amer. J. Potato Res. 2006;83:349–355. doi: 10.1007/BF02871596. [DOI] [Google Scholar]
- 32.Figueiredo A, Monteiro F, Sebastiana M. First clues on a jasmonic acid role in grapevine resistance against the biotrophic fungus Plasmopara viticola. Eur. J. Plant Pathol. 2015;142:645–652. doi: 10.1007/s10658-015-0634-7. [DOI] [Google Scholar]
- 33.Xiao J, et al. Transcriptome-based discovery of pathways and genes related to resistance against Fusarium head blight in wheat landrace Wangshuibai. BMC Genomics. 2013;14:197. doi: 10.1186/1471-2164-14-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schweizer P, Cees R, Mosinger E. Effect of jasmonic acid on the interaction (Hordeum vulgare L) with the powdery Erysiphe graminis f sp. hordei of barley mildew. Plant Physiol. 1993;102:503–511. doi: 10.1104/pp.102.2.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cao F, et al. Genome-wide transcriptome and functional analysis of two contrasting genotypes reveals key genes for cadmium tolerance in barley. BMC Genomics. 2014;15:611. doi: 10.1186/1471-2164-15-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Taggar GK, Gill RS, Gupta AK, Sandhu JS. Fluctuations in peroxidase and catalase activities of resistant and susceptible black gram (Vigna mungo (L.) Hepper) genotypes elicited by Bemisia tabaci (Gennadius) feeding. Plant Signal Behav. 2012;7:1321–1329. doi: 10.4161/psb.21435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roohie RK, Umesha S. Identification of genes associated with black rot resistance in cabbage through suppression subtractive hybridization. 3 Biotech. 2015;5:1089–1100. doi: 10.1007/s13205-015-0311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sonoda T, Uragami A, Kaji K. Evaluation of Asparagus officinalis cultivars for resistance to stem blight by using a novel inoculation method. HortSci. 1997;32:1085–1086. [Google Scholar]
- 39.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011;1:10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 40.Grabherr MG, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Haas BJ, et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protocol. 2013;8:1494–1512. doi: 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Apweiler R, et al. Activities at the universal protein resource (UniProt) Nucleic Acids Res. 2014;42:D191–D198. doi: 10.1093/nar/gkt1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Conesa A, et al. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- 44.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;4:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li B, Ruotti V, Stewart RM, Thomson JA, Dewey CN. RNA-Seq gene expression estimation with read mapping uncertainty. Bioinformatics. 2010;26:493–500. doi: 10.1093/bioinformatics/btp692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leng N, et al. EBSeq: An empirical Bayes hierarchical model for inference in RNA-seq experiments. Bioinformatics. 2013;29:1035–1043. doi: 10.1093/bioinformatics/btt087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li H, et al. RNA-Seq derived identification of differential transcription in the chrysanthemum leaf following inoculation with Alternaria tenuissima. BMC Genomics. 2014;15:9–10. doi: 10.1186/1471-2164-15-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bradford MM. Rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 49.Kim Y, Yoo JY. Peroxidase production from carrot hairy root cell culture. Enzyme Mircob. Technol. 1996;18:531–535. doi: 10.1016/0141-0229(95)00168-9. [DOI] [Google Scholar]
- 50.Aebi H. Catalase in vitro. Methods Enzymol. 1984;105:121–126. doi: 10.1016/S0076-6879(84)05016-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


