Abstract
There is growing evidence that hydrogen sulfide (H2S) is involved in many physiological processes in plants, but the role of H2S in dark-induced leaf senescence remains unknown. In this work, we found that H2S not only inhibited chlorophyll degradation but also caused the accumulation of photoreactive pheide a in detached leaves under extended darkness. Despite this, transcript levels of senescence-associated genes (SAGs) were less affected in H2S-treated detached leaves compared with those in H2S-untreated detached leaves. Furthermore, cell death/rapid bleaching occurred in both H2S-treated detached and attached leaves after transfer from extended darkness to light. Unlike the lack of effect of H2S on SAG transcripts in darkened detached leaves, exogenous H2S induced higher SAG transcript levels in attached leaves than untreated attached leaves. Genetic evidence further underlined the positive correlation between SAG expression in attached leaves and H2S. In addition, effects of H2S on SAG expression in attached leaves were compromised in the S-nitrosoglutathione reductase-deficient mutant, gsnor1. Taken together, our results suggest that H2S suppresses chlorophyll degradation of detached leaves by regulating a dark-dependent reaction, and that this gas positively modulates SAG expression in attached leaves under prolonged darkness in a GSNOR1-dependent manner.
Introduction
Hydrogen sulfide (H2S) is a pungent colorless gas with a distinctive rotten-egg odor, often regarded as an environmental pollutant and a toxin for almost all organisms. One of the well-known mechanisms for H2S toxicity involves inhibition of one of the key enzymes in the mitochondrial respiratory chain, cytochrome c oxidase1, 2. Despite the toxicity of H2S, it is well established that plants can themselves generate and release this gas, especially when exposed to external cysteine, sulfate, sulfite or SO2 3–5. This is thought to be a mechanism for dissipation of excess sulfur6, but certain adverse environmental stimuli such as pathogens and drought can also stimulate H2S emissions above basal, endogenously produced rates7, 8.
Plants can produce H2S through sulfite reductase, which catalyzes the reduction of sulfite to sulfide, or through two cysteine-dependent reactions involving members of the O-acetylserine(thiol)lyase (OAS-TL) gene family. L-cysteine desulfhydrase (DES, EC 4.4.1.1) converts L-cysteine to H2S, ammonia and pyruvate9 while β-cyanoalanine synthase produces H2S via detoxification of cyanide at the expense of cysteine10, 11. Another potential enzyme in plant H2S homeostasis is a D-cysteine desulfhydrase, which, similar to DES, produces H2S, ammonia and pyruvate12. However, the physiological function of D-cysteine desulfhydrase is completely unknown.
Many studies published since the late 1990s have shown that H2S can have signalling, defense and anti-apoptotic functions in mammalian systems13. The discovery of these novel functions of H2S in mammals stimulated work in plants, leading to an appreciation of the important and varied physiological functions of H2S6, 13–18. This gas has not only been implicated in seed germination, root development, and photosynthesis19–28, but can also enhance plant acclimation/tolerance to various stresses such as drought, heavy metals, salinity, cold, heat and osmotic stress8, 29–36. One notion is that the influence of H2S on stress responses is at least partly linked to enhanced antioxidant capacity6. Consistent with this notion, it has very recently been reported that several antioxidant components such as cytosolic ascorbate peroxidase1, 2-Cys peroxiredoxin A or B, and peroxisomal catalase3 in Arabidopsis plants, underwent S-sulfhydration in the presence of exogenous H2S, leading to enhanced enzyme activities37. H2S is also the end-product of assimilatory sulfate reduction, in which it is incorporated into OAS-TL to produce cysteine, the source of reduced sulphur including the redox buffer, glutathione38. Indeed, prolonged treatment with H2S leads to increased glutathione synthesis. This effect may have a direct protective effect during stress or, alternatively, may act to regulate defense genes that play important roles in adverse environmental conditions such as cadmium exposure or drought39–43. Thus, there are several mechanisms by which H2S could act to regulate stress responses by affecting antioxidant status, but their relative importance is still uncertain.
Apart from possible effects mediated by H2S modulation of cell thiol status, recent work has shown that this gas can have a signalling function through other pathways. For instance, genetic evidence revealed that DES1 deficiency leads to the accumulation and lipidation of ATG8 isoforms in Arabidopsis, which is associated with autophagy activation. Exogenous sulfide suppresses autophagy induction in Arabidopsis des1 mutants under nutrient-rich conditions and in wild type plants under nitrogen deprivation, whereas glutathione had no effect44. Interestingly, sulfide did not scavenge reactive oxygen species (ROS) triggered by nitrogen starvation, in contrast to glutathione. These results indicate that sulfide represses autophagy via mechanisms that are independent of redox conditions44, 45. However, Scuffi et al. (2014) found that the lack of cytosolic H2S in des1 significantly decreases endogenous nitric oxide (NO) levels and that NO acts downstream of H2S to close stomata via an ABA-dependent pathway26. These observations draw attention to the potential importance of H2S and its interactions with NO status in regulating various biological processes in plants. However, the signal mechanisms and direct downstream targets of H2S that regulate stomatal movement and autophagy remain to be identified.
The overall aim of the present work was to investigate the significance of H2S in modulation of processes involved in dark-induced senescence in plants. The specific objectives were (1) to assess the effect of H2S on dark-induced chlorophyll loss; (2) to establish whether H2S affects chlorophyll loss via alterations in autophagy and well-characterized senescence pathways; (3) to investigate the links between H2S and chlorophyll breakdown intermediates that are known to be implicated in cell death; and (4) to evaluate the role of cell redox components in mediating the effect of H2S. The results show that H2S favors a stay-green phenotype in detached leaves by affecting a dark-dependent reaction involved in chlorophyll degradation and that this gas regulates SAG expression in attached leaves through processes linked to NO homeostasis.
Results
Effect of H2S on dark-triggered leaf chlorophyll degradation in detached leaves
Prolonged darkness is often used to induce rapid and synchronous senescence in detached leaves. Hence, a dark-detached system has been widely used as a model to study senescence-associated regulatory mechanisms. Loss of chlorophyll has often been exploited as a well-characterized marker of dark-induced leaf senescence. To investigate the potential role of H2S in leaf chlorophyll metabolism, detached leaves were fumigated with H2S, released from 0.01 to 2 mM NaHS solution (see Materials and Methods), and chlorophyll content was assessed after extended darkness for 4d. Under normal growth conditions, leaf chlorophyll level was about 1.37 mg/g fresh weight. Extended darkness led to a loss of leaf color and a corresponding decrease in chlorophyll level in excised leaves of the wild type examined without treatment with H2S (Fig. 1a,b). In contrast, treatment with NaHS at external concentrations of 0.01, 0.1, 0.5, 1.0 and 2.0 mM significantly suppressed chlorophyll loss in a dose-dependent manner (Fig. 1b). Thus, H2S treatment caused a “stay-green” phenotype.
Figure 1.
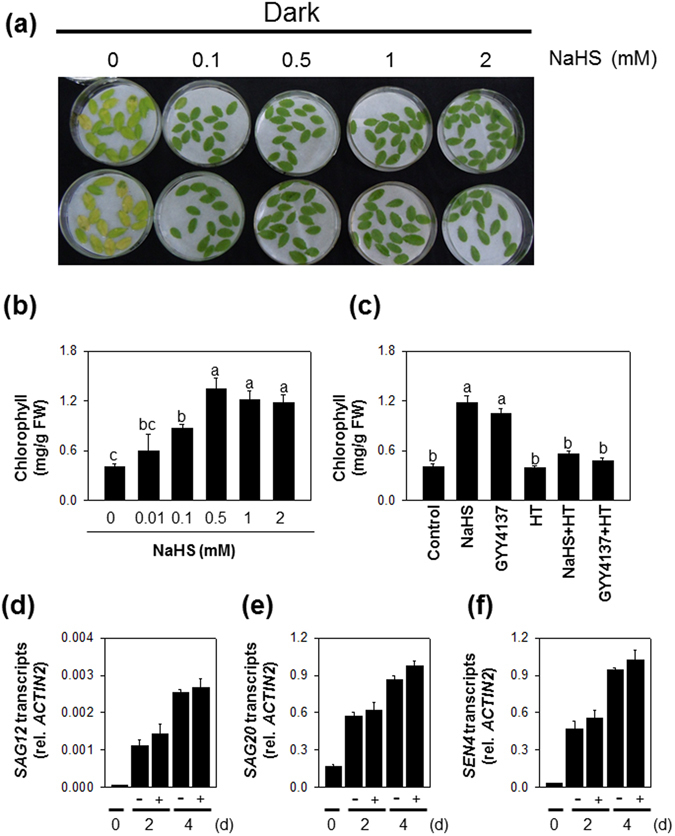
Effect of H2S exposure on chlorophyll breakdown and SAG expression in detached leaves during extended darkness for up to 4 d. (a,b), effects of H2S gas released from 0 to 2 mM H2S donor NaHS solution (see Materials and Methods) on leaf yellowing and chlorophyll content, respectively, at 4 d of darkness. Effect of another H2S donor GYY4137 and H2S scavenger HT (c) on chlorophyll degradation under extended darkness for 4 d. For GYY4137 and HT treatments, 3-week-old detached leaves were floated in petri dishes containing 3 mL solution of 0.1 mM GYY4137 alone, 0.1 mM HT alone or 0.1 mM GYY4137 plus 0.1 mM HT combined treatment. Transcript levels of SAG12 (d), SAG20 (e) and SEN4 (f) in detached leaves of wild type subjected to H2S or H2S-free treatment for up to 4 d of complete darkness. + and − indicate detached leaves of Col-0 fumigated with or without 0.5 mM NaHS respectively. Data are means ± SE of at least three independent samples from different plants. Letters indicates significant difference from the wild type at P < 0.05, using the Student’s t test.
To confirm the effect of H2S on chlorophyll degradation, another H2S donor (GYY4137) and a H2S scavenger (hypotaurine; HT) were employed. Consistently, it was found that H2S generated from 100 µM GYY4137 had the same effect on leaf chlorophyll content as NaHS treatment. In contrast, HT completely blocked the effects of both NaHS and GYY4137 treatment (Fig. 1c). Together, these results suggest that H2S plays a negative role in chlorophyll degradation.
Links between H2S and autophagy have recently been reported, and many autophagy-deficient mutants display an early senescence phenotype under extended darkness45–47. We therefore examined whether autophagy might be involved in the regulation of chlorophyll degradation by H2S. Consistent with previous studies, detached leaves from 3-week-old autophagy deficient mutants atg2 or atg18a kept in darkness for 2 days exhibited a much greater senescence-associated loss of green leaf color than those from the wild type (Supplementary Fig. S1a). However, leaf yellowing and chlorophyll degradation in the autophagy-deficient mutant were markedly suppressed in the presence of H2S at 2 or 4 d of extended darkness (Supplementary Fig. S1a–c). Furthermore, H2S contents and the activities of enzymes with H2S-releasing activity (LCD and DCD) were not decreased in the atg mutants compared with those in Col-0 (Supplementary Fig. S1d–f). These results suggest that the negative effect of H2S on chlorophyll loss under extended darkness is independent of the autophagic pathway.
H2S is the substrate for the biosynthesis of cysteine2, and cysteine and cysteine-containing compounds such as glutathione are key determinants of cell redox homeostasis48, 49. In a first step to study possible links between H2S and cellular thiols in the regulation of chlorophyll degradation, the effects of exogenous supply of either cysteine or glutathione on the loss of leaf color upon exposure to extended darkness were investigated. Unlike H2S, neither cysteine nor glutathione treatments affected the loss of leaf colour and chlorophyll degradation in Col-0 (Supplementary Fig. S2a). In a second approach, the importance of changes in endogenous cysteine and glutathione was examined in the cad2 mutant, which has higher cysteine contents than Col-042. In the absence of H2S, the cad2 mutant showed similar dark-induced loss of chlorophyll to Col-0, while H2S treatment produced a similar stay-green effect in both genotypes (Supplementary Fig. S2b). On the other hand, no visible difference in either glutathione levels or H2O2 contents was observed between H2S-treated wild type and the control treatment (Supplementary Fig. S2c,d). Thus, these observations provide little evidence that the inhibitory effect of H2S on chlorophyll degradation under extended darkness is linked to increased cysteine and glutathione.
Many genes that are up-regulated during the senescence processes have been defined as senescence-associated genes (SAGs). These notably include SAG12, SAG20 and SEN4 50. The transcript levels of these SAGs were determined in the wild type in the presence or absence of H2S treatment. Intriguingly, dark treatment led to significant increases in transcript levels of SAG12, SAG20 and SEN4 in wild type with or without H2S treatment (Fig. 1d–f). Thus, the expression patterns of SAGs are not correlated with an inhibition of chlorophyll metabolism in H2S-treated detached leaves under extended darkness. From this observation, we conclude that the stay-green effects of H2S are not occurring by a general modulation of dark-induced senescence pathways.
Effect of H2S on chlorophyll breakdown intermediates and cell death in detached leaves
To gain insight into the role of H2S in the regulation of chlorophyll degradation, green pigments were extracted and separated by HPLC. After 4 d of complete darkness, levels of chlorophyll a decreased faster in the control wild type than in H2S-treated plants (Fig. 2a). Moreover, H2S-treated plants accumulated increasing amounts of pheophytin a under 4 d of extended darkness (Fig. 2b) HPLC analyses also showed that the basal level of pheide a was very low (around 0.95 nmol.g−1 FW before dark treatment) and did not increase appreciably in detached leaves kept in the dark on water (Fig. 2c). In contrast, H2S treatment resulted in the accumulation of pheide a in a time-dependent manner (Fig. 2c) that correlated well with the “stay-green” phenotype (Fig. 1a).
Figure 2.
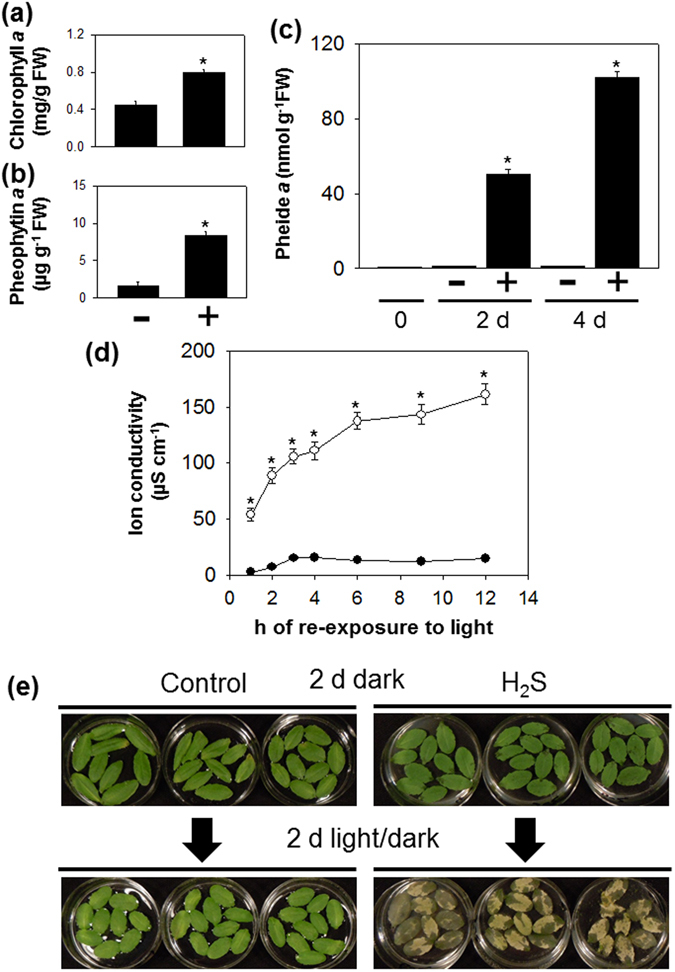
Effects of H2S exposure on accumulation of green intermediates of chlorophyll breakdown and cell death in detached leaves. Amounts of chlorophyll a (a) and pheophytin a (b) accumulation in detached leaves of wild type subjected to H2S or H2S-free treatment for up to 4 d of complete darkness. (c), Accumulation of pheide a in response to dark incubation of detached leaves of control and H2S-treated wild type for up to 4 d. + and − indicate detached leaves of Col-0 fumigated with or without 0.5 mM NaHS, respectively. (d), Determination of ion leakage as a measure for cell death in detached leaves treated with H2S (white bars) or without H2S (black bars). Before re-exposure to the light for up to 12 h, detached leaves were incubated in the presence or absence of 0.5 mM NaHS in the darkness for 2 d. (e), Photographs of detached wild type leaves treated with H2S released from 0.5 mM NaHS solution under extended darkness for 2 d and transfer to regular growth conditions for another 2 d. Data are means ± SE of at least three independent samples from different plants. Asterisks indicate significant difference between H2S-treated wild type and control wild type at the same time point at P < 0.05, using the Student’s t test.
Conversion of pheide a to red chlorophyll catabolite (RCC) by pheophorbide a oxygenase (PAO) is the critical step of loss of green pigment51, 52. Quantification of transcripts for enzymes involved in chlorophyll breakdown provided little evidence that H2S-mediated regulation of pheide a contents occurs at the transcriptional level (Supplementary Fig. S4). While PAO transcript abundance increased during darkness, this response was not significantly affected by H2S treatment. Transcript levels of other chlorophyll catabolic genes51, 53–56 were also similar in detached leaves treated or not with H2S during extended darkness (Supplementary Fig. S4). Exceptions were transcripts for CLH2 (CHLOROPHYLLASE 2) and NYC1 (NON-YELLOW COLORING 1), which were increased in H2S-treated leaves compared to controls at 2 d of darkness (Supplementary Fig. S4).
Light-dependent cell death could be induced by pheide a 52. While the water control showed very low ion leakage upon exposure to light, the stay-green phenotype associated with H2S treatment was accompanied by higher ion conductivity (Fig. 2d). Interestingly, ion leakage increased substantially on re-illumination whereas it stayed low in the water-treated controls (Fig. 2d). Moreover, on transfer to a standard light/dark regime, H2S-treatment induced a visible cell death phenotype that was not observed in water-treated controls or in H2S-treated detached leaves under standard growth conditions (16 h light/8 h dark; Fig. 2e; Supplementary Fig. S3). These results provide additional evidence that H2S-indcued suppression of chlorophyll degradation in detached leaves under extended darkness is unlikely to occur by regulation of known senescence-associated pathways.
Effect of H2S on phenotype and SAG expression in attached leaves subjected to extended darkness
H2S and dark treatments were performed on attached leaves. Neither the visual phenotype nor the chlorophyll contents of plants treated with different concentrations of NaHS was distinguishable from the untreated control after 2 d darkness (Fig. 3a; Supplementary Fig. S5a). However, within 1 d after transfer to the light/dark regime following exposure to 2 d of darkness, intact plants treated with NaHS at a concentration of 0.5 mM or above exhibited rapid loss of green pigments, including pheophytin a (Fig. 3b,c; Supplementary Fig. S5b). This effect is very similar to that observed in H2S-treated detached leaves after transfer from extended darkness to light (Fig. 2b). In contrast, effects of H2S on intact plants under complete darkness and regular light/dark cycles were less evident (Fig. 3d,e; Supplementary Fig. S5c,d). These data demonstrate that H2S triggers a phenotype of bleaching or cell death in both attached and detached leaves shifted from continuous darkness to light through one or more processes that require light.
Figure 3.
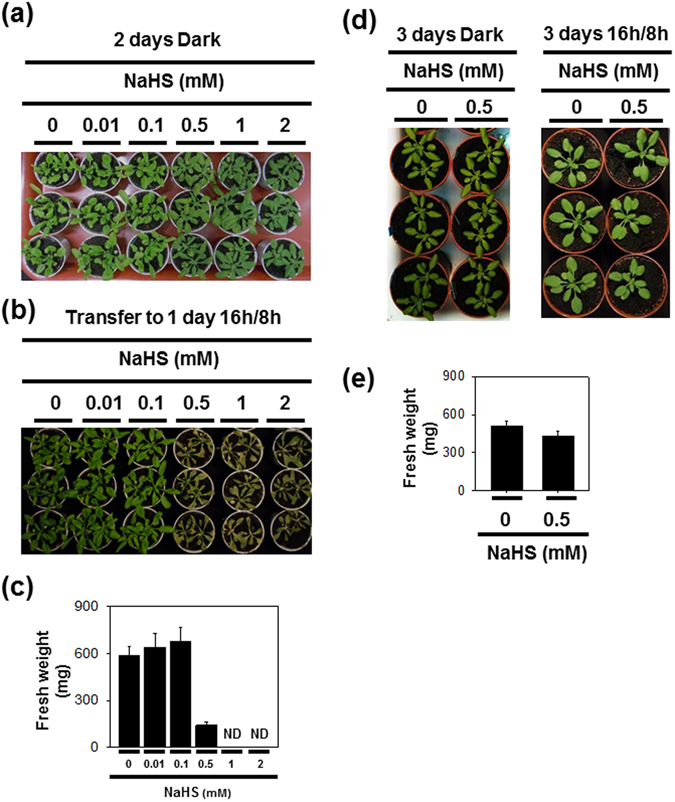
Effects of H2S exposure on phenotype of whole plants under prolonged darkness and regular growth conditions. Photographs of whole plants treated with different NaHS concentrations exposed to complete darkness for 2 d (a) and then transfer to regular growth conditions for another 1 d (b) (16 h light/8 h dark photoperiod). The fresh weight of plants treated with the indicated concentrations of NaHS was measured 6 d after transfer from continuous darkness to light-dark conditions (c). (d), Effects of 0.5 mM NaHS treatment under prolonged darkness or regular 16 h light/8 h dark conditions for 3 days. (e) Fresh weight were taken at 7 d after H2S treatment under 16 h light/8 h dark conditions. Data are means ± SE of at least 15 different plants. ND: not detected.
To investigate the influence of H2S on dark-induced senescence processes in attached leaves at the transcriptional level, expression analyses of SAG12 SAG20 and SEN4 were performed. As shown in Fig. 4a–c, H2S-treatment significantly increased expression of these genes in plants placed in darkness. In contrast, H2S treatment did not affect their transcript levels in plants in the repeated light/dark conditions at 2 d or 4 d (Supplementary Fig. S6). To directly test if endogenous H2S produces a similar effect on the expression of SAGs, transgenic plants expressing DES1 were generated. Two independent DES1 OE lines showed significant increases in both leaf total LCD activity and intracellular H2S contents (Supplementary Fig. S7a–c). As described above for exogenous H2S, DES1-dependent increases in leaf H2S significantly stimulated accumulation of SAG12, SAG20 and SEN4 transcripts above Col-0 values, when plants were kept in darkness for 2d or 4d (Fig. 4d–f).
Figure 4.
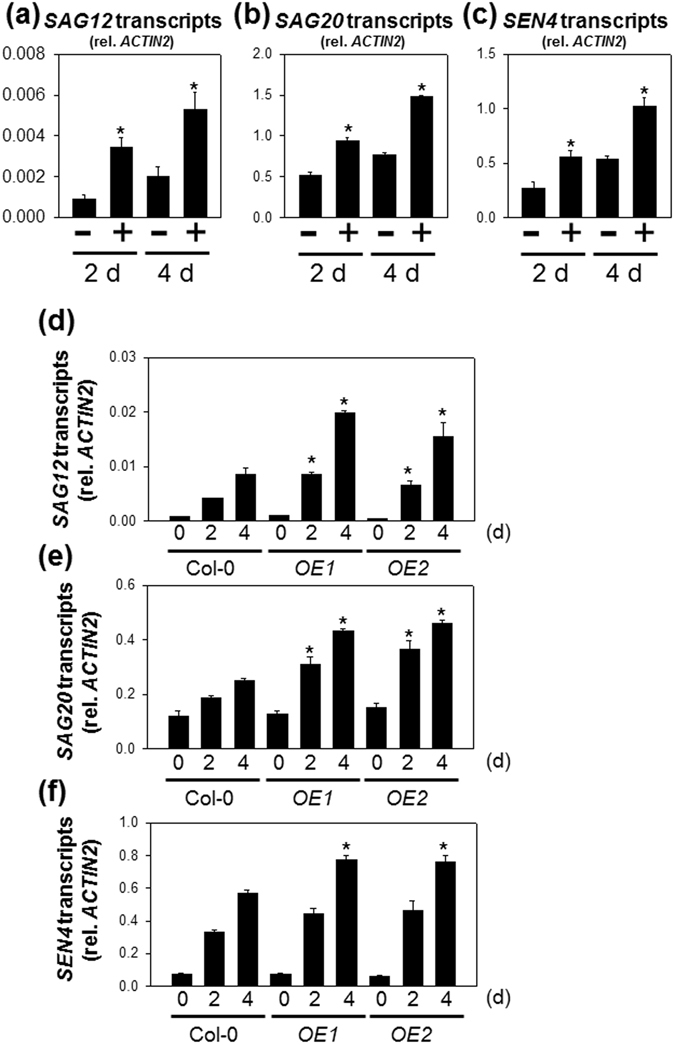
Effects of exogenously applied H2S and DES1 transgenic lines on SAG expression in attached leaves under extended darkness. Transcript levels of SAG12 (a), SAG20 (b) and SEN4 (c) in wild-type plants subjected to 0.5 mM NaHS treatment plus complete darkness. Samples were taken from the attached leaves at 2 or 4 d of darkness. + and − indicate intact plants fumigated with or without 0.5 mM NaHS, respectively. (d), SAG12. (e), SAG20. (f), SEN4. Samples were taken from the attached leaves at 2 and 4 d of darkness. OE1 and OE2 indicate two independent DES1 overexpression lines. Data are means ± SE of at least three independent samples from different plants. Asterisks indicate significant difference from the wild type at the same time point at P < 0.05, using the Student’s t test.
To analyze the role of DES1 further, we exploited the des1 mutant, which is impaired in LCD expression and activity (Supplementary Fig. S7d,e), and in H2S generation (Fig. 5)45. During extended darkness, increases in SAG transcript levels were significantly higher in the wild type than in des1 attached leaves (Fig. 5). In contrast to its effect on SAG expression, the des1 mutation did not affect transcript levels for the salicylic acid (SA)-dependent gene, PR1 (Fig. 5). Hence, evidence from both OE and loss-of-function lines (Figs 4 and 5) suggests that H2S is a regulator of SAG expression in attached leaves.
Figure 5.
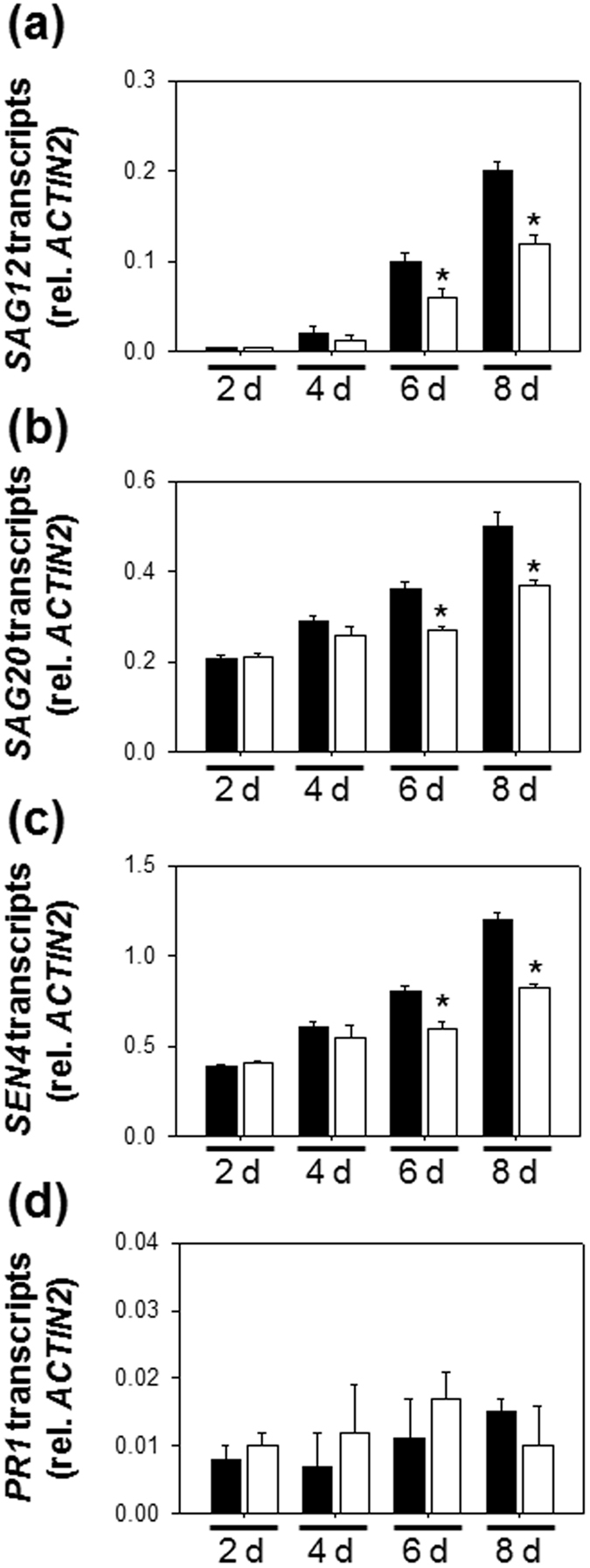
Expressions of of SAGs and PR1 gene in attached leaves of Col-0 and des1 mutant during extended darkness. (a), SAG12. (b), SAG20. (c), SEN4. (d), PR1. Ten-day-old seedlings of Col-0 and des1 were incubated under extended darkness for up to 8 d. White bars, des1 mutant. Black bars, Col-0. Asterisks indicate significant difference from the wild type at the same time point at P < 0.05, using the Student’s t test.
Effect of H2S on oxidative stress and the ascorbate-glutathione pathway in attached leaves
Leaf senescence-linked events are often associated with pronounced accumulation of ROS47. Thus, levels of hydrogen peroxide (H2O2) were monitored in H2S-treated attached leaves under dark incubation. Although no difference in H2O2 contents was observed between H2S-treated and and -untreated whole plants in normal growth conditions (Supplementary Fig. S8), dark-induced H2O2 generation was further enhanced by H2S treatment (Fig. 6a). Because the glutathione pool is in close correspondence to H2O2 availability40, 57, we analyzed this key intracellular thiol-disulfide compound. Treatment with NaHS for 2 d resulted in a dramatic decrease in the content of GSH (Fig. 6b). Like glutathione, ascorbate is an abundant and stable redox buffer required for H2O2 metabolism, and is considered to be partly regenerated by glutathione48. Despite this, the effects of H2S on glutathione pools were not associated with marked perturbation of leaf ascorbate pools (Fig. 6b,c).
Figure 6.
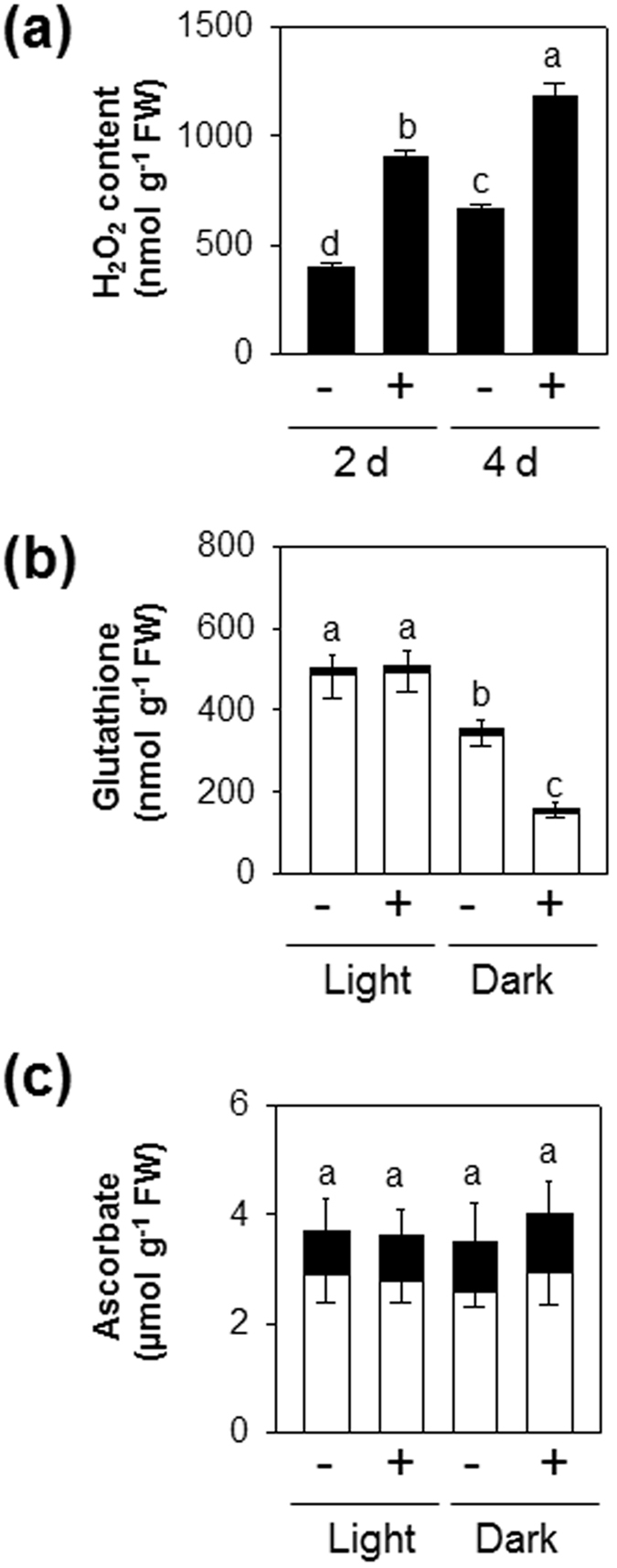
Effects of H2S exposure on leaf H2O2, glutathione and ascorbate in attached leaves of Col-0 under extended darkness and normal growth conditions. (a), H2O2 content. Samples were taken from the attached leaves at 2 and 4 d of darkness. (b), reduced glutathione (white bars) and oxidized glutathione (black bars). (c), ascorbate (white bars) and dehydroascorbate (black bars). Samples were taken from the attached leaves at 2 d of darkness and regular growth conditions within 16 h light/8 h dark photoperiod. + and − indicate intact plants fumigated with or without 0.5 mM NaHS, respectively. Light indicates regular growth conditions within 16 h light/8 h dark photoperiod. Data are means ± SE of at least three independent samples from different plants. Letters indicates significant difference from the wild type at P < 0.05, using the Student’s t test.
To further assess the effect of the H2S on the ROS-antioxidant interaction, we measured the extractable activities of ascorbate peroxidase (APX), catalase (CAT), glutathione reductase (GR), and dehydroascorbate reductase (DHAR), the enzyme linking glutathione and ascorbate pools. Activities of APX and DHAR were significantly decreased in darkened plants treated with H2S compared with those treated with H2S or darkness alone, although CAT and GR activities were not greatly affected by any of the treatments (Fig. 7).
Figure 7.
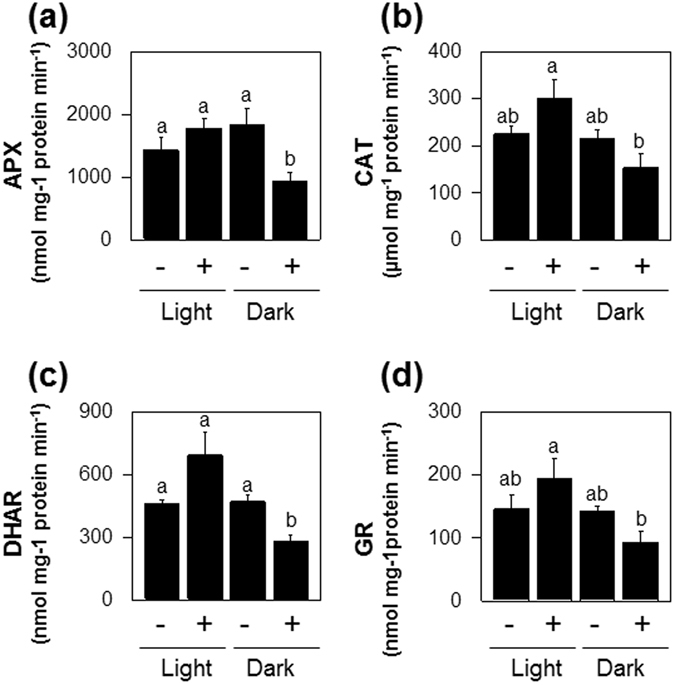
Effects of H2S exposure on major antioxidative enzyme in attached leaves of Col-0 under extended darkness and normal growth conditions. (a) APX. (b), CAT. (c), DHAR. (d), GR. Samples were taken from the attached leaves at 2 d of darkness and light/dark growth conditions. + and − indicate intact plants fumigated with or without 0.5 mM NaHS, respectively, during 2 d of dark incubation and light/dark growth conditions. Data are means ± SE of at least three independent samples from different plants. Letters indicates significant difference from the wild type at P < 0.05, using the Student’s t test.
GSNOR1 is required for H2S-mediated SAG expression under extended darkness
NO has been proposed to act as a regulator of leaf senescence58, 59. A major route for the transmission of NO signaling is S-nitrosylation, a reversible post-translational modification involving the covalent addition of NO to a protein cysteine thiol to form an S-nitrosothiol60 (NO). Total cellular levels of protein S-nitrosylation are controlled predominantly by S-nitrosoglutathione reductase 1 (GSNOR1) which removes GSNO61. Recently, several publications reported that H2S interacts with NO to regulate diverse plant processes in response to adverse environmental clues31, 35, 36. Interestingly, many of the protein sites in Arabidopsis reported to undergo endogenous S-nitrosylation have also been found to undergo S-sulfydration. This latter reaction involves interaction of H2S with the thiol groups of specific proteins to form a persulfide group (R-SSH)37, 62. Hence, the potential role of GSNOR1 in H2S-regulated SAG expression was investigated in attached leaves. After 4 d of dark treatment, H2S-mediated induction of SAG12, SAG20 and SEN4 in H2S-treated Col-0 were compromised in H2S-treated gsnor1 mutant (Fig. 8a–c). Taken together, these results demonstrate that GSNOR1 is involved in the H2S-induced expression of SAGs in attached leaves under extended darkness.
Figure 8.
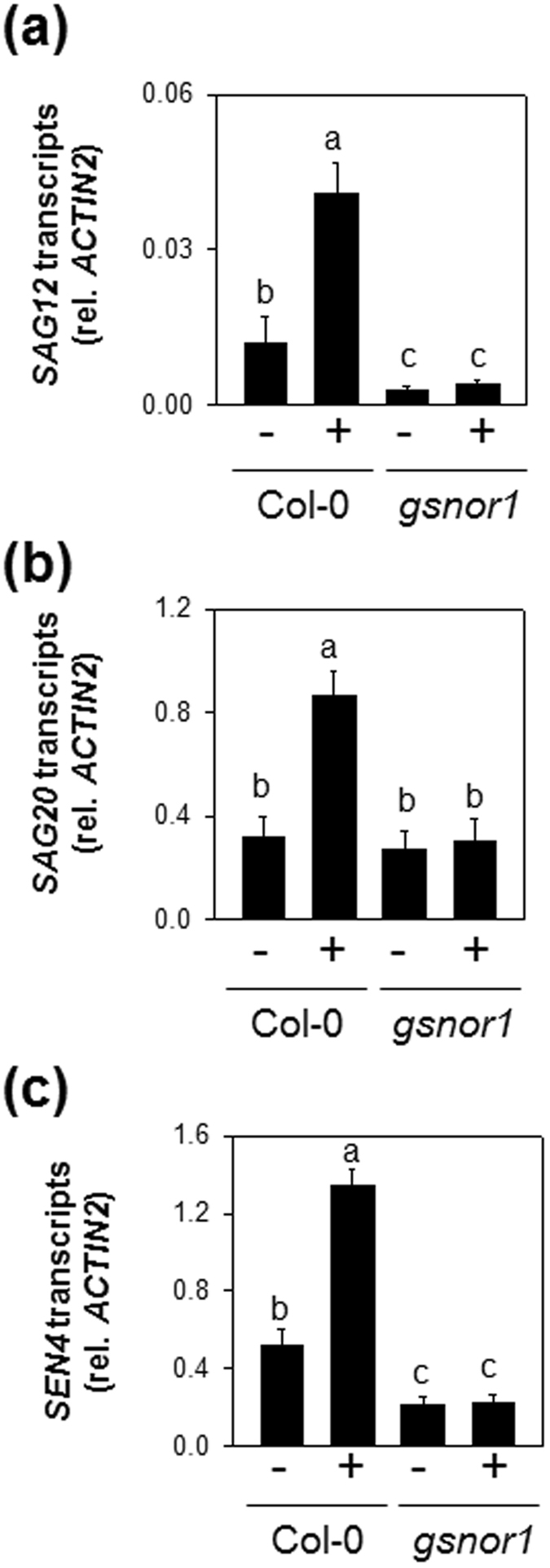
SAG expression in attached leaves of Col-0 and gsnor1 treated with or without H2S under extended darkness. (a), SAG12 expression. (b), SAG20 expression. (c), SEN4 expression. Samples were taken from the attached leaves at 4 d of darkness treatment. + and − indicate intact plants fumigated with or without 0.5 mM NaHS, respectively, during 4 d of dark incubation. Data are means ± SE of at least three independent samples from different plants. Letters indicates significant difference from the wild type at P < 0.05, using the Student’s t test.
Discussion
It is accepted that H2S can affect plant defense and development either by acting as a toxic molecule or as a precursor of reduced sulphur required to produce cysteine and glutathione38. An increasing number of reports point to regulatory functions for H2S in plants6, but the role of H2S in the regulation of dark-induced leaf senescence is largely unknown. In this study, a variety of approaches were exploited to understand the action of H2S in leaf senescence-dependent and senescence-independent processes under extended darkness. To this aim, we exploited two H2S donors, NaHS and GYY4137, that have been widely applied for experimental purposes in both plants and animals13. The concentration of H2S detected in plants is reported to range from 1 to 100 µM8. The level of gaseous H2S generated from 100 µM NaHS solution is close to 100 µM22, which is within the range of concentrations that modulate physiological processes in plants (10 to 200 µM). The concentration of fumigated H2S released from 0.5 mM NaHS (200 mL) in 3 L sealed containers is around 33 µM. Therefore, most experiments in this work were conducted using this physiologically relevant concentration range.
H2S represses chlorophyll breakdown via a mechanism that is independent of anti-senescence processes
Darkness is often used to induce rapid and synchronous senescence in detached leaves, and chlorophyll catabolism is an integral process of leaf senescence50. In the present study, our findings demonstrate that H2S has a negative effect on chlorophyll degradation under extended darkness, but that this effect is uncoupled from the expression of SAGs (Fig. 1). Furthermore, like in the stay-green mutant pao1, our results show that the presence of H2S results in the accumulation of pheide a during dark incubation (Fig. 2c), further supporting the existence of a feedback mechanism that limits metabolism of chlorophyll in H2S-treated detached leaves or in mutants that are unable to degrade chlorophyll beyond pheide a 53. Hence, H2S probably inhibits chlorophyll breakdown at the level of pheide a under extended darkness. Additionally, the accumulation of pheide a is reported to be responsible for the cell death phenotype on leaves in a light-dependent way52. Consistent with this notion, cell death or rapid bleaching after transfer to light is apparently observed in both detached and attached leaves treated with H2S (Figs 2 and 3). The results presented here imply (1) that pheide a metabolism is important in linking H2S to a downstream “stay-green” phenotype under extended darkness, (2) that pheide a is required for the cell death reaction observed in H2S-treated leaves shifted from extended darkness to light, (3) and that H2S suppresses chlorophyll degradation of detached leaves through regulating unidentified dark-dependent reactions rather than modulating anti-senescence processes. In addition, formation of the colorless primary fluorescent chl catabolite (pFCC) from RCC a is responsible for the loss of green pigment in chlorophyll breakdown, while RCC accumulation causes leaf cell death54. Therefore it would be interesting to understand if RCC mediates H2S-associated responses.
H2S potentiates dark-induced expression of SAGs in attached leaves
Recently, Álvarez et al. (2012) reported that mutation of DES1 led to a 30% reduction in endogenous sulfide and early age-associated senescence as evidenced at the cellular and transcriptional levels45. DES1 deficiency promoted accumulation of de novo senescence-associated vacuoles and the expression of SAG12 and NAP 9, 45. This is markedly different from what we observed. Our results show that exogenously applied H2S promotes higher transcript levels of several SAGs compared with H2S-untreated attached leaves (Fig. 4a–c). Moreover, des1 mutants and two independent DES1 transgenic lines show, respectively, decreased or enhanced expression of SAGs during extended darkness (Figs 4b–d and 5). This apparent discrepancy appears to be the cause of the difference between age-triggered senescence and dark-induced senescence. It is possible that the SA pathway is specifically involved in age-dependent leaf senescence50. SA is not only a key plant hormone mediating the plant response to pathogens but also functions in leaf senescence. Higher SA levels have been reported in senescing Arabidopsis leaves, and this observation is accompanied by the induction of genes such as SAG12 63. Consistent with this possibility, the levels of SA and SA-responsive defense markers such as PR1 are significantly increased in the des1 mutant, correlating with the up-regulation of several SAG genes including SAG12 and SAG21 during age-related senescence9, 45, 64. However, we observed no difference in PR1 expression in attached leaves in the presence or absence of H2S during extended darkness (Fig. 5d).
Elevated levels of H2O2, either through enhanced H2O2 generation or down-regulation of antioxidant levels, could promote senescence47, 65. In agreement, increases in SAG expression in H2S-treated attached leaves under extended darkness were accompanied by increased H2O2 and decreased GSH (Figs 4 and 6). Failure of H2S-treated detached leaves to further enhance the expression levels of SAGs was perhaps due to decreased accumulation of oxidants (Supplementary Fig. S2c,d). Thus, it would be interesting to investigate further the role of redox regulation in H2S-mediated senescence processes.
GRSNOR1 is required for H2S-mediated expression of SAGs in attached leaves under extended darkness through modulating SNO level
Although S-sulfhydration has been proposed as a likely mechanism of H2S signaling in mammalian systems, evidence for this process has only been very recently reported in plants37. One hundred and six S-sulfhydrated proteins were identified in Arabidopspis, many of which also underwent S-nitrosylation62. Moreover, recent work found that H2S treatment can suppress the accumulation of SNO by enhancing GSNOR enzyme activity66. These reports are consistent with several lines of evidence that point to an interaction between H2S and NO in plant growth and defenses17. S-nitrosylation typically inhibits protein function. In contrast, S-sulfhydration can activate enzymatic activities. For instance, S-nitrosylation negatively regulates the activities of a cytosolic ascorbate peroxidase, APX167, and a cytosolic glyceraldehyde-3-phosphate dehydrogenase, GAPC168, in plants. Both of these enzymes can also be S-sulfhydrated by H2S, which increases their activities37. If H2S regulates dark-induced senescence through S-nitrosylation mechanisms, enhanced SNO levels may attenuate the effects of H2S. This is a possible explanation of why H2S-mediated induction of SAG expression was compromised by the gsnor1 mutation (Fig. 8). Thus, appropriate modulation of SNO levels by GSNOR1 is crucial to H2S-regulated SAG expression triggered in darkened attached leaves. Although studies on the downstream targets of H2S signal functioning in plant responses to stress are still quite limited, the effects reported here clearly point to interplay between H2S and NO in post-translationally determining the status of protein thiols37, 62.
Methods
Plant material and growth conditions
Arabidopsis thaliana wild-type Columbia-0 (Col-0), atg2 (SALK_076727), atg5 (SAIL_129B07), atg18a (GABI_651D08), cad2 69, des1 (SALK_103855) and gsnor1 (GABI_315D11) lines were used in this work. Seeds were incubated for 2 d at 4 °C and then sown in soil. Plants were grown in soil in a controlled-environment growth chamber in a 16 h photoperiod and an irradiance of 120 μmol m−2 s−1 at leaf level, 22 °C day/20 °C night, 65% humidity and given nutrient solution twice per week. Samples were rapidly frozen in liquid nitrogen and stored at −80 °C until analysis. Unless otherwise stated, data are means SE of at least three independent samples from different plants.
Hydrogen sulfide fumigation and dark treatment
Solutions of sodium hydrosulfide (NaHS•3H2O) were used as one of the hydrogen sulfide (H2S) donors. To examine the dose effect of H2S on leaf yellowing, aqueous solutions (200 mL) of 0 (control), 0.01, 0.1, 0.5, 1 or 2 mM NaHS were prepared, from which H2S gas was released in a sealed glass desiccator (volume 3 L). For SAG expression analysis, excised leaves from 3- to 4-week-old plants incubated on wet filter paper or attached leaves from either 10-day-old or 3-week-old seedlings were kept in the presence or absence of H2S released from 0.5 mM NaHS solution in darkness for several days. For cell death assay, detached and attached leaves of 3- to 4-week-old plants were kept in darkness in combination with 0.5 mM NaHS for 2 or more days and then transfer to 16 h/8 h photoperiod conditions. The NaHS solutions were renewed each two days and treated leaves were collected at designated time intervals for analyses.
To confirm the effects of gaseous H2S on the senescence of Arabidopsis leaves, 0.1 mM Morpholin-4-ium 4-methoxyphenyl (morpholino) phosphinodithionate (GYY4137) was used as a second H2S donor, while 0.1 mM hypotaurine (HT) was used as an H2S scavenger.
Thiol treatment
To study the effect of cysteine and glutathione on dark-triggered leaf senescence, detached leaves from 3- to 4-week-old plants were placed in petri dishes containing 3 mL solution of 0.1 mM cysteine or 0.1 mM glutathione under extended darkness.
Generation of DES1 transgenic plants
DES1 cDNA was amplified from Arabidopsis with primer pairs of DES1-F2/DES1-R2 by RT-PCR. After verifying the sequence fidelity by sequencing, these products were cloned into the XbaI and XhoI sites of pBI121 under the control of 35 S promoter. The 35 S::DES1 construct was introduced into the Agrobacterium tumefaciens GV3101 strain, which was then used to transform Col-0 using the flower infiltration method. Two independent lines overexpressing DES1 were identified and characterized for further analyses. All transgenic lines used in this study were T3 homozygous plants.
RT-qPCR analyses
Total RNA was extracted from the designated tissues using Trizol (Invitrogen). 2 μg of total RNA was used for the synthesis of the first-strand cDNA using the All-in-One cDNA Synthesis Super Mix and oligo dT as primers (Biotool). Quantitative PCR was performed in a StepOnePlusTM Real-Time PCR System (Applied Biosystems) using 2x SYBR Green qPCR Master Mix (High ROX) (Biotool). Transcript levels of target genes were normalized to that of the housekeeping gene ACTIN2 (AT3G18780) using the equation of 2−ΔCT, where CT is the threshold cycle for each gene in the sample. The primers used are listed in Supplementary Table S1.
Measurement of DES activity and H2S content
L-Cysteine desulfhydrase (DES; EC 4.4.1.1) activity was determined according to the method of Riemenschneider et al.12. This method is based on the measurement of catalytic release of sulfide from cysteine. The soluble proteins were extracted by adding 1 mL of 20 mM Tris-HCl (pH 8.0), and centrifuged at 15,000 g for 15 min at 4 °C. The reaction mixture (1 mL) consisted of 2.5 mM dithiothreitol (DTT), 0.8 mM L-Cysteine, 100 mM Tris-HCl (pH 9.0), and enzyme extract. The reaction was initiated by the addition of L-Cysteine. The reaction mixture was incubated at 37 °C for 15 min, and then the reaction was terminated by the addition of 0.1 mL of 30 mM FeCl3 dissolved in 1.2 N HCl and 0.1 mL 20 mM N, N-dimethyl-p-phenylenediaminedihy drochloride dissolved in 7.2 N HCl. The formation of methylene blue was determined at 670 nm. DES enzymatic activity was calculated using a standard curve prepared with NaHS. D-Cysteine desulfhydrase activity was determined in the same way, but D-Cysteine was used instead of L-Cysteine.
The determination of H2S was carried out according to the method of Singh et al.70. 0.5 g plant leaves was ground into fine powder with a mortar and pestle under liquid nitrogen and then homogenized in 1 ml of the following extraction buffer: 20 mM Tris-HCl buffer (pH 8.0), 10 mM EDTA, 20 mM Zn(OAc)2. The homogenate was centrifuged at 15,000 g for 15 min at 4 °C. The reaction mixture (2 mL) consisted of 0.1 mL supernatant, 1.88 mL extraction buffer and 0.02 mL of 20 mM 5,5′-dithiobis(2-nitrobenzoic acid). The reaction mixture was incubated at room temperature for 2 min and absorbance was recorded at 412 nm. The level of H2S was calculated according to a standard curve of NaHS.
Analyses of chlorophyll and green catabolites
For spectrophotometric determination of chlorophyll level, chlorophyll was extracted from leaf tissue by homogenization in liquid nitrogen and subsequent threefold extraction into 80% (v/v) acetone containing 1 mM KOH. After centrifugation (10 min, 12,000 g), supernatants were combined and used for analysis. The absorbance of the supernatant was read at 663 and 645 nm, and the amount of total chlorophyll (μg/mL) was calculated as 8.02 × A663 + 20.2 × A645 71.
For HPLC analyses of green chlorophyll a catabolites (pheophytin a and pheide a), liquid nitrogen-homogenized tissue was extracted in 10% (v/v) 0.2 M Tris-HCl (pH 8.0) in acetone, and incubated at −20 °C for 2 h in the dark. After removal of insoluble material by centrifugation (10 min, 12,000 g), supernatants were analyzed by reverse-phase HPLC as previously described52.
Ion Leakage
For ion conductivity analysis, detached leaves of 3- to 4 weeks plant were incubated in the presence or absence of H2S released from 0.5 mM NaHS solution in the dark for 2 d. Eight leaves for each treatment were then soaked in 10 ml of distilled water in a test tube. After re-exposure to light (120 μmol m−2 s−1) for up to 12 h, ion leakage as a measure of cellular damage was determined by measuring the conductivity of the solution with a FiveGo F3 meter (Mettler Toledo).
Antioxidant enzyme assays metabolite, and H2O2 analyses
Extractable enzyme activities were measured as described in Noctor et al.72. Oxidized and reduced forms of glutathione and ascorbate were measured by plate-reader assay as described in Queval and Noctor57. H2O2 content was determined by the method of titanium oxidation with hydroperoxide-titanium complex formed73.
Statistical analysis
The statistical analysis of data was based on Student’s t tests. Calculations were performed on a minimum of three independent datasets, assuming two samples equal variance and a two-tailed distribution. Unless stated otherwise, significant difference was assessed using multiple pair wise t test comparisons at P < 0.05.
Electronic supplementary material
Acknowledgements
The authors thank the Salk Institute Genomic Analysis Laboratory for providing the sequence-indexed Arabidopsis T-DNA insertion mutants and the Arabidopsis Biological Resource Center, USA, for the supply of seed stocks. They are grateful to Dr. Fangming Xiao (University of Idaho) for helpful discussions. This work was supported by the National Natural Science Foundation of China (31300225 to Y Han).
Author Contributions
B.W. and Y.H. designed and interpreted the experiments; B.W., W.Z., J.C., T.Z., T.Z. and Y.H. conducted the experiments; Y.H. and Y.L. wrote the article. Y.H. and G.N. revised the article.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-02872-0
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Yongsheng Liu, Email: liuyongsheng1122@hfut.edu.cn.
Yi Han, Email: yi.han@hfut.edu.cn.
References
- 1.Dorman DC, et al. Cytochrome oxidase inhibition induced by acute hydrogen sulfide inhalation: correlation with tissue sulfide concentrations in the rat brain, liver, lung, and nasal epithelium. Toxicol Sci. 2002;65:18–25. doi: 10.1093/toxsci/65.1.18. [DOI] [PubMed] [Google Scholar]
- 2.Birke H, De Kok LJ, Wirtz M, Hell R. The role of compartment-specific cysteine synthesis for sulfur homeostasis during H2S exposure in Arabidopsis. Plant Cell Physiol. 2015;56:358–367. doi: 10.1093/pcp/pcu166. [DOI] [PubMed] [Google Scholar]
- 3.Wilson LG, Bressan RA, Filner P. Light-dependent emission of hydrogen sulfide from plants. Plant Physiol. 1978;61:184–189. doi: 10.1104/pp.61.2.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sekiya J, Schmidt A, Wilson LG, Filner P. Emission of Hydrogen Sulfide by Leaf Tissue in Response to L-Cysteine. Plant Physiol. 1982;70:430–436. doi: 10.1104/pp.70.2.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sekiya J, Wilson LG, Filner P. Resistance to injury by sulfur dioxide: correlation with its reduction to, and emission of, hydrogen sulfide in Cucurbitaceae. Plant Physiol. 1982;70:437–441. doi: 10.1104/pp.70.2.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Calderwood A, Kopriva S. Hydrogen sulfide in plants: from dissipation of excess sulfur to signaling molecule. Nitric Oxide. 2014;41:72–78. doi: 10.1016/j.niox.2014.02.005. [DOI] [PubMed] [Google Scholar]
- 7.Bloem E, et al. Sulphur supply and infection with Pyrenopezizabrassicae influence L-cysteine desulphydrase activity in Brassica napus L. J Exp Bot. 2004;55:2305–2312. doi: 10.1093/jxb/erh236. [DOI] [PubMed] [Google Scholar]
- 8.Jin Z, et al. Hydrogen sulfide improves drought resistance in Arabidopsis thaliana. Biochem Biophys Res Commun. 2011;414:481–486. doi: 10.1016/j.bbrc.2011.09.090. [DOI] [PubMed] [Google Scholar]
- 9.Álvarez C, Calo L, Romero LC, García I, Gotor C. An O-acetylserine(thiol)lyase homolog with L-cysteine desulfhydrase activity regulates cysteine homeostasis in Arabidopsis. Plant Physiol. 2010;152:656–669. doi: 10.1104/pp.109.147975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Akopyan TN, Braunstein AE, Goryachenkova EV. Beta-cyanoalanine synthase: purification and characterization. Proc Natl Acad Sci USA. 1975;72:1617–1621. doi: 10.1073/pnas.72.4.1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hatzfeld., et al. beta-Cyanoalanine synthase is a mitochondrial cysteine synthase-like protein in spinach and Arabidopsis. Plant Physiol. 2000;123:1163–1171. doi: 10.1104/pp.123.3.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Riemenschneider A, Wegele R, Schmidt A, Papenbrock J. Isolation and characterization of a D-cysteine desulfhydrase protein from Arabidopsis thaliana. FEBS J. 2005;272:1291–1304. doi: 10.1111/j.1742-4658.2005.04567.x. [DOI] [PubMed] [Google Scholar]
- 13.Yamasaki H, Cohen MF. Biological consilience of hydrogen sulfide and nitric oxide in plants: Gases of primordial earth linking plant, microbial and animal physiologies. Nitric Oxide. 2016;55–56:91–100. doi: 10.1016/j.niox.2016.04.002. [DOI] [PubMed] [Google Scholar]
- 14.García-Mata C, Lamattina L. Gasotransmitters are emerging as new guard cell signaling molecules and regulators of leaf gas exchange. Plant Sci. 2013;201–202:66–73. doi: 10.1016/j.plantsci.2012.11.007. [DOI] [PubMed] [Google Scholar]
- 15.Gotor C, García I, Crespo JL, Romero LC. Sulfide as a signaling molecule in autophagy. Autophagy. 2013;9:609–611. doi: 10.4161/auto.23460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lisjak M, Teklic T, Wilson ID, Whiteman M, Hancock JT. Hydrogen sulfide: environmental factor or signalling molecule? Plant Cell Environ. 2013;36:1607–1616. doi: 10.1111/pce.12073. [DOI] [PubMed] [Google Scholar]
- 17.Hancock JT, Whiteman M. Hydrogen sulfide and cell signaling: team player or referee? Plant Physiol Biochem. 2014;78:37–42. doi: 10.1016/j.plaphy.2014.02.012. [DOI] [PubMed] [Google Scholar]
- 18.Jin, Z. & Pei, Y. Physiological implications of hydrogen sulfide in plants: pleasant exploration behind its unpleasant odour. Oxid Med Cell Longev 397502 (2015). [DOI] [PMC free article] [PubMed]
- 19.Zhang H, et al. Hydrogen sulfide promotes wheat seed germination and alleviates oxidative damage against copper stress. J Integr Plant Biol. 2008;50:1518–1529. doi: 10.1111/j.1744-7909.2008.00769.x. [DOI] [PubMed] [Google Scholar]
- 20.Zhang H, et al. Hydrogen sulfide promotes root organogenesis in Ipomoea batatas, Salix matsudana and Glycine max. J Integr Plant Biol. 2009;51:1086–1094. doi: 10.1111/j.1744-7909.2009.00885.x. [DOI] [PubMed] [Google Scholar]
- 21.Lisjak M, et al. A novel hydrogen sulfide donor causes stomatal opening and reduces nitric oxide accumulation. Plant Physiol Biochem. 2010;48:931–935. doi: 10.1016/j.plaphy.2010.09.016. [DOI] [PubMed] [Google Scholar]
- 22.Chen J, et al. Hydrogen sulphide enhances photosynthesis through promoting chloroplast biogenesis, photosynthetic enzyme expression, and thiol redox modification in Spinaciaoleracea seedlings. J Exp Bot. 2011;62:4481–4493. doi: 10.1093/jxb/err145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dooley FD, Nair SP, Ward PD. Increased growth and germination success in plants following hydrogen sulfide administration. PLoS One. 2013;8:e62048. doi: 10.1371/journal.pone.0062048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hou Z, Wang L, Liu J, Hou L, Liu X. Hydrogen sulfide regulates ethylene-induced stomatal closure in Arabidopsis thaliana. J Integr Plant Biol. 2013;5:277–289. doi: 10.1111/jipb.12004. [DOI] [PubMed] [Google Scholar]
- 25.Fang T, Cao Z, Li J, Shen W, Huang L. Auxin-induced hydrogen sulfide generation is involved in lateral root formation in tomato. Plant Physiol Biochem. 2014;76:44–51. doi: 10.1016/j.plaphy.2013.12.024. [DOI] [PubMed] [Google Scholar]
- 26.Scuffi D, et al. Hydrogen sulfide generated by L-cysteine desulfhydrase acts upstream of nitric oxide to modulate abscisic acid-dependent stomatal closure. Plant Physiol. 2014;166:2065–2076. doi: 10.1104/pp.114.245373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jia H, Hu Y, Fan T, Li J. Hydrogen sulfide modulates actin-dependent auxin transport via regulating ABPs results in changing of root development in Arabidopsis. Sci Rep. 2015;5:8251. doi: 10.1038/srep08251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baudouin E, et al. The Significance of hydrogen sulfide for Arabidopsis seed germination. Front Plant Sci. 2016;7:930. doi: 10.3389/fpls.2016.00930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang BL, Shi L, Li YX, Zhang WH. Boron toxicity is alleviated by hydrogen sulfide in cucumber (Cucumissativus L.) seedlings. Planta. 2010;231:1301–1309. doi: 10.1007/s00425-010-1134-9. [DOI] [PubMed] [Google Scholar]
- 30.Christou A, Manganaris GA, Papadopoulos I, Fotopoulos V. Hydrogen sulfide induces systemic tolerance to salinity and non-ionic osmotic stress in strawberry plants through modification of reactive species biosynthesis and transcriptional regulation of multiple defence pathways. J Exp Bot. 2013;64:1951–1953. doi: 10.1093/jxb/ert055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li ZG, Yang SZ, Long WB, Yang GX, Shen ZZ. Hydrogen sulphide may be a novel downstream signal molecule in nitric oxide-induced heat tolerance of maize (Zea mays L.) seedlings. Plant Cell Environ. 2013;36:1564–1572. doi: 10.1111/pce.12092. [DOI] [PubMed] [Google Scholar]
- 32.Shen. J, et al. Hydrogen sulfide improves drought tolerance in Arabidopsis thaliana by microRNA expressions. PLoS One. 2013;8:e77047. doi: 10.1371/journal.pone.0077047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shi H, Ye T, Chan Z. Exogenous application of hydrogen sulfide donor sodium hydrosulfide enhanced multiple abiotic stress tolerance in bermudagrass (Cynodondactylon(L). Pers.) Plant Physiol Biochem. 2013;71:226–234. doi: 10.1016/j.plaphy.2013.07.021. [DOI] [PubMed] [Google Scholar]
- 34.Sun J, et al. Hydrogen sulfide alleviates cadmium toxicity through regulations of cadmium transport across the plasma and vacuolar membranes in Populuseuphratica cells. Plant Physiol Biochem. 2013;65:67–74. doi: 10.1016/j.plaphy.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 35.Chen J, et al. Hydrogen sulfide enhances salt tolerance through nitric oxide-mediated maintenance of ion homeostasis in barley seedling roots. Sci Rep. 2015;5:12516. doi: 10.1038/srep12516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ziogas V, et al. Roles of sodium hydrosulfide and sodium nitroprusside as priming molecules during drought acclimation in citrus plants. Plant Mol Biol. 2015;89:433–450. doi: 10.1007/s11103-015-0379-x. [DOI] [PubMed] [Google Scholar]
- 37.Aroca Á, Serna A, Gotor C, Romero LC. S-sulfhydration: a cysteine posttranslational modification in plant systems. Plant Physiol. 2015;168:334–342. doi: 10.1104/pp.15.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takahashi H, Kopriva S, Giordano M, Saito K, Hell R. Sulfur assimilation in photosynthetic organisms: molecular functions and regulations of transporters and assimilatory enzymes. Annu Rev Plant Biol. 2011;62:157–184. doi: 10.1146/annurev-arplant-042110-103921. [DOI] [PubMed] [Google Scholar]
- 39.Noctor G, Foyer CH. Ascorbate and Glutathione: Keeping active oxygen under control. Annu Rev Plant Biol. 1998;49:249–279. doi: 10.1146/annurev.arplant.49.1.249. [DOI] [PubMed] [Google Scholar]
- 40.Mhamdi A, et al. Arabidopsis Glutathione Reductase1 plays a crucial role in leaf responses to intracellular hydrogen peroxide and in ensuring appropriate gene expression through both salicylic acid and jasmonic acid signaling pathways. Plant Physiol. 2010;153:1144–1160. doi: 10.1104/pp.110.153767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Han Y, et al. Functional analysis of Arabidopsis mutants points to novel roles for glutathione in coupling H2O2 to activation of salicylic acid accumulation and signaling. Antioxid Redox Sign. 2013;18:2106–2121. doi: 10.1089/ars.2012.5052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Han Y, Mhamdi A, Chaouch S, Noctor G. Regulation of basal and oxidative stress-triggered jasmonic acid-related gene expression by glutathione. Plant Cell Environ. 2013;36:1135–1146. doi: 10.1111/pce.12048. [DOI] [PubMed] [Google Scholar]
- 43.Cheng MC, et al. Increased glutathione contributes to stress tolerance and global translational changes in Arabidopsis. Plant J. 2015;83:926–939. doi: 10.1111/tpj.12940. [DOI] [PubMed] [Google Scholar]
- 44.Laureano-Marín AM, Moreno I, Romero LC, Gotor C. Negative regulation of autophagy by sulfide is independent of Reactive Oxygen Species. Plant Physiol. 2016;171:1378–1391. doi: 10.1104/pp.16.00110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Álvarez C, et al. Cysteine-generated sulfide in the cytosol negatively regulates autophagy and modulates the transcriptional profile in Arabidopsis. Plant Cell. 2012;24:4621–4634. doi: 10.1105/tpc.112.105403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thompson AR, Doelling JH, Suttangkakul A, Vierstra RD. Autophagic nutrient recycling in Arabidopsis directed by the ATG8 and ATG12 conjugation pathways. Plant Physiol. 2005;138:2097–2110. doi: 10.1104/pp.105.060673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yoshimoto K, et al. Autophagy negatively regulates cell death by controlling NPR1-dependent salicylic acid signaling during senescence and the innate immune response in Arabidopsis. Plant Cell. 2009;21:2914–2927. doi: 10.1105/tpc.109.068635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Noctor G, et al. Glutathione in plants: an integrated overview. Plant Cell Environ. 2012;35:454–484. doi: 10.1111/j.1365-3040.2011.02400.x. [DOI] [PubMed] [Google Scholar]
- 49.Romero LC, et al. Cysteine and cysteine-related signaling pathways in Arabidopsis thaliana. Mol Plant. 2014;7:264–276. doi: 10.1093/mp/sst168. [DOI] [PubMed] [Google Scholar]
- 50.Lim PO, Kim HJ, Nam HG. Leaf senescence. Annu Rev Plant Biol. 2007;58:115–136. doi: 10.1146/annurev.arplant.57.032905.105316. [DOI] [PubMed] [Google Scholar]
- 51.Rodoni S, et al. Chlorophyll breakdown in senescent chloroplasts (Cleavage of Pheophorbide a in Two Enzymic Steps. Plant Physiol. 1997;115:669–676. doi: 10.1104/pp.115.2.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pružinská A, et al. Chlorophyll breakdown in senescent Arabidopsis leaves. Characterization of chlorophyll catabolites and of chlorophyll catabolic enzymes involved in the degreening reaction. Plant Physiol. 2005;139:52–63. doi: 10.1104/pp.105.065870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tsuchiya T, et al. Cloning of chlorophyllase, the key enzyme in chlorophyll degradation: finding of a lipase motif and the induction by methyl jasmonate. Proc Natl Acad Sci USA. 1999;96:15362–15367. doi: 10.1073/pnas.96.26.15362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pružinská A, et al. In vivo participation of red chlorophyll catabolite reductase in chlorophyll breakdown. Plant Cell. 2007;19:369–387. doi: 10.1105/tpc.106.044404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ren G, et al. Identification of a novel chloroplast protein AtNYE1 regulating chlorophyll degradation during leaf senescence in Arabidopsis. Plant Physiol. 2007;144:1429–1441. doi: 10.1104/pp.107.100172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Horie Y, et al. Participation of chlorophyll b reductase in the initial step of the degradation of light-harvesting chlorophyll a/b-protein complexes in Arabidopsis. J Biol Chem. 2009;284:17449–17456. doi: 10.1074/jbc.M109.008912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Queval G, Noctor G. A plate reader method for the measurement of NAD, NADP, glutathione, and ascorbate in tissue extracts: Application to redox profiling during Arabidopsis rosette development. Anal. Biochem. 2007;363:58–69. doi: 10.1016/j.ab.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 58.Guo FQ, Crawford NM. Arabidopsis nitric oxide synthase1 is targeted to mitochondria and protects against oxidative damage and dark-induced senescence. Plant Cell. 2005;17:3436–3450. doi: 10.1105/tpc.105.037770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mishina TE, Lamb C, Zeier J. Expression of a nitric oxide degrading enzyme induces a senescence programme in Arabidopsis. Plant Cell Environ. 2007;30:39–52. doi: 10.1111/j.1365-3040.2006.01604.x. [DOI] [PubMed] [Google Scholar]
- 60.Jaffrey SR, Erdjument-Bromage H, Ferris CD, Tempst P, Snyder SH. Protein S-nitrosylation: a physiological signal for neuronal nitric oxide. Nat Cell Biol. 2001;3:193–197. doi: 10.1038/35055104. [DOI] [PubMed] [Google Scholar]
- 61.Feechan A, et al. A central role for S-nitrosothiols in plant disease resistance. Proc Natl Acad Sci USA. 2005;102:8054–8059. doi: 10.1073/pnas.0501456102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hu J, et al. Site-specific nitrosoproteomic identification of endogenously S-nitrosylated proteins in Arabidopsis. Plant Physiol. 2015;167:1731–1746. doi: 10.1104/pp.15.00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Morris K, et al. Salicylic acid has a role in regulating gene expression during leaf senescence. The Plant J. 2000;23:677–685. doi: 10.1046/j.1365-313x.2000.00836.x. [DOI] [PubMed] [Google Scholar]
- 64.Álvarez C, Bermúdez MÁ, Romero LC, Gotor C, García I. Cysteine homeostasis plays an essential role in plant immunity. New Phytol. 2012;193:165–177. doi: 10.1111/j.1469-8137.2011.03889.x. [DOI] [PubMed] [Google Scholar]
- 65.Smykowski A, Zimmermann P, Zentgraf U. G-Box binding factor1 reduces CATALASE2 expression and regulates the onset of leaf senescence in Arabidopsis. Plant Physiol. 2010;153:1321–1331. doi: 10.1104/pp.110.157180. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 66.Ma L, et al. Comparative proteomic analysis reveals the role of hydrogen sulfide in the adaptation of the alpine plant Lamiophlomis rotata to altitude gradient in the Northern Tibetan Plateau. Planta. 2015;241:887–906. doi: 10.1007/s00425-014-2209-9. [DOI] [PubMed] [Google Scholar]
- 67.de Pinto MC, et al. S-nitrosylation of ascorbate peroxidase is part of programmed cell death signaling in tobacco Bright Yellow-2 cells. Plant Physiol. 2013;163:1766–1775. doi: 10.1104/pp.113.222703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zaffagnini M, Michelet L, Massot V, Trost P, Lemaire SD. Biochemical characterization of glutaredoxins from Chlamydomonas reinhardtii reveals the unique properties of a chloroplastic CGFS-type glutaredoxin. J Biol Chem. 2008;283:8868–8876. doi: 10.1074/jbc.M709567200. [DOI] [PubMed] [Google Scholar]
- 69.Cobbett CS, May MJ, Howden R, Rolls B. The glutathione-deficient, cadmium-sensitive mutant, cad2-1, of Arabidopsis thaliana is deficient in gamma-glutamylcysteine synthetase. Plant J. 1998;16:73–78. doi: 10.1046/j.1365-313x.1998.00262.x. [DOI] [PubMed] [Google Scholar]
- 70.Singh VP, Singh S, Kumar J, Prasad SM. Hydrogen sulfide alleviates toxic effects of arsenate in pea seedlings through up-regulation of the ascorbate-glutathione cycle: Possible involvement of nitric oxide. J Plant Physiol. 2015;181:20–29. doi: 10.1016/j.jplph.2015.03.015. [DOI] [PubMed] [Google Scholar]
- 71.Lichtenthaler FWK, Freudenberg B, Helferich HOL. Fischer: a centennial tribute. Carbohydr Res. 1987;164:1–22. doi: 10.1016/0008-6215(87)80114-3. [DOI] [PubMed] [Google Scholar]
- 72.Noctor G, Mhamdi A, Foyer CH. Oxidative stress and antioxidative systems: recipes for successful data collection and interpretation. Plant Cell Environ. 2016;39:1140–1160. doi: 10.1111/pce.12726. [DOI] [PubMed] [Google Scholar]
- 73.Brennan T, Frenkel C. Involvement of hydrogen peroxide in the regulation of senescence in pear. Plant Physiol. 1977;59:411–416. doi: 10.1104/pp.59.3.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


