Abstract
Programmed cell death or apoptosis of infected host cells is an important defense mechanism in response to viral infections. This process is regulated by proapoptotic and prosurvival members of the B-cell lymphoma 2 (Bcl-2) protein family. To counter premature death of a virus-infected cell, poxviruses use a range of different molecular strategies including the mimicry of prosurvival Bcl-2 proteins. One such viral prosurvival protein is the fowlpox virus protein FPV039, which is a potent apoptosis inhibitor, but the precise molecular mechanism by which FPV039 inhibits apoptosis is unknown. To understand how fowlpox virus inhibits apoptosis, we examined FPV039 using isothermal titration calorimetry, small-angle X-ray scattering, and X-ray crystallography. Here, we report that the fowlpox virus prosurvival protein FPV039 promiscuously binds to cellular proapoptotic Bcl-2 and engages all major proapoptotic Bcl-2 proteins. Unlike other identified viral Bcl-2 proteins to date, FPV039 engaged with cellular proapoptotic Bcl-2 with affinities comparable with those of Bcl-2's endogenous cellular counterparts. Structural studies revealed that FPV039 adopts the conserved Bcl-2 fold observed in cellular prosurvival Bcl-2 proteins and closely mimics the structure of the prosurvival Bcl-2 family protein Mcl-1. Our findings suggest that FPV039 is a pan-Bcl-2 protein inhibitor that can engage all host BH3-only proteins, as well as Bcl-2-associated X, apoptosis regulator (Bax) and Bcl-2 antagonist/killer (Bak) proteins to inhibit premature apoptosis of an infected host cell. This work therefore provides a mechanistic platform to better understand FPV039-mediated apoptosis inhibition.
Keywords: apoptosis, B-cell lymphoma 2 (Bcl-2) family, isothermal titration calorimetry (ITC), poxvirus, small-angle X-ray scattering (SAXS), X-ray crystallography
Introduction
Programmed cell death or apoptosis is a highly organized and tightly regulated mechanism of cell suicide (1, 2) and is evolutionary conserved in all multicellular organisms (3). Apoptosis allows the regulation of cell and tissue homeostasis as well as the removal of impaired, unwanted, or diseased cells (4). In addition to organogenesis, homeostasis, and roles in development, apoptosis also acts as a host defense mechanism to remove infected cells during viral infection, thus preventing viral survival and proliferation (5).
B-cell lymphoma 2 (Bcl-2)3 proteins are the main regulators of apoptosis executed by the intrinsic pathway (2). The Bcl-2 protein family is divided into two subfamilies, the proapoptotic Bcl-2 and prosurvival Bcl-2 proteins (6), which are characterized by the presence of one or more of the four conserved Bcl-2 homology (BH) motifs (7). Proapoptotic Bcl-2 proteins are further subdivided into members that contain multiple BH domains including Bax and Bak and the BH3-only proteins, which only harbor a BH3 motif. Upon activation, proapoptotic Bak and Bax oligomerize to permeabilize the outer mitochondrial membrane (8), leading to the release of apoptogenic mediators such as cytochrome c that ultimately activate caspases and lead to the destruction of the cell (4). Bak and Bax are activated by the expression of proapoptotic BH3-only proteins including Bim, Bid, Noxa, and Puma, which are able to directly bind and activate Bak and Bax, or by neutralizing prosurvival Bcl-2 proteins such as Bcl-2, Mcl-1, Bcl-xL, A1, Bcl-w, and Bcl-b (9). Structural studies have shown that the three subfamilies interact with each other in a BH3 motif-dependent manner where the BH3 motif of a proapoptotic protein will interact with an extended hydrophobic groove in a receiving prosurvival Bcl-2 protein (10).
Premature host cell apoptosis plays a significant role in combating viral infections, and many viruses have evolved strategies to counter host cell apoptotic defenses (5). For instance, many viruses express prosurvival factors to extend the lifespan of infected host cells (11). Numerous large DNA viruses utilize sequence and/or structural mimics of prosurvival Bcl-2 to inhibit apoptosis (7). Examples include γ-herpesviruses 68 M11 (12), adenovirus E1B19K (13), and African swine fever virus A179L (14). Among the poxviruses, a number of homologs of Bcl-2 have been identified including M11L from myxoma virus (15), F1L from vaccinia virus (16, 17) and variola virus (18), sheeppox virus SPPV14 (19), and deerpox virus DPV022 (20, 21). Interestingly, structural studies showed that M11L, F1L, and DPV022 adopt a Bcl-2 fold despite a lack of detectable sequence similarity to Bcl-2 (21–25). Functionally, M11L was shown to act by sequestering Bax and Bak (24), whereas F1L inhibits apoptosis by sequestering Bim (22).
Sequencing of the fowlpox virus genome and subsequent analysis identified the virus open reading frame 39 (FPV039) (26) as a putative prosurvival Bcl-2 protein due to the presence of recognizable BH1 and BH2 motifs as well as a C-terminal transmembrane domain (27). Subsequent studies revealed that FPV039 is a potent inhibitor of apoptosis and localizes to the mitochondrial membrane via a C-terminal transmembrane domain. Mechanistically, FPV039 suppresses apoptosis triggered by overexpression of proapoptotic proteins including Bak and Bax and all the BH3-only proteins and was shown to immunoprecipitate with the BH3-only proteins Bim and Bik as well as Bak and activated Bax (27, 28). However, the precise molecular and structural basis of how FPV039 inhibits apoptosis remains to be determined. To address these questions, we investigated the binding of FPV039 to peptides encoding the BH3 domain from all proapoptotic Bcl-2 family members including Bak, Bax, and Bok and all the BH3-only proteins. We then determined crystal structures of FPV039 bound to two of the identified interactors, Bmf and Bik BH3. Our studies showed that FPV039 is a highly promiscuous prosurvival viral Bcl-2 (vBcl-2) protein that binds to all BH3-only proteins as well as Bax and Bak and provide a mechanistic platform to understand FPV039-mediated apoptosis inhibition.
Results
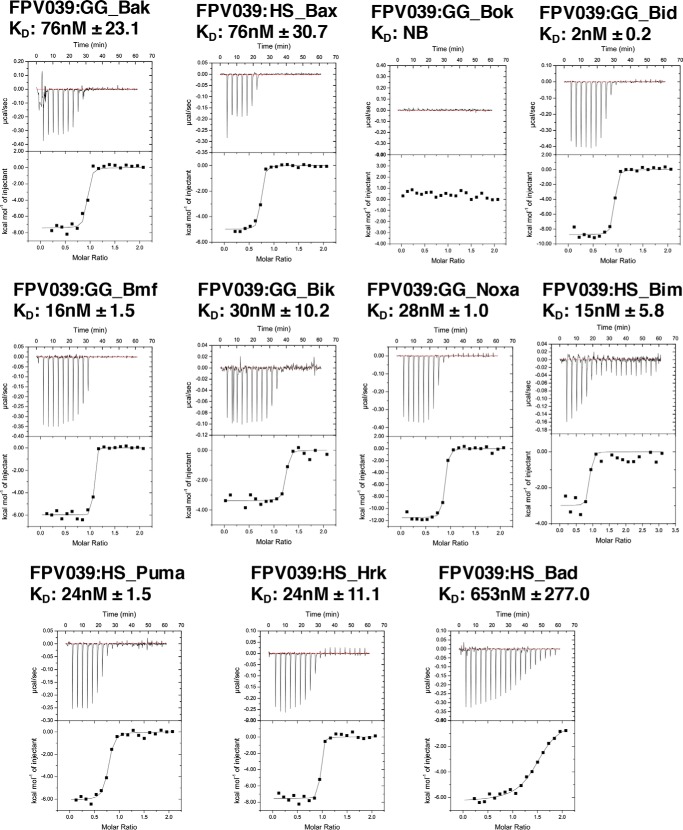
To understand how FPV039 interacts with proapoptotic members of the Bcl-2 family of proteins, truncated FPV039 comprising the first 143 residues was recombinantly expressed in Escherichia coli and purified using a two-step purification method using affinity chromatography followed by size-exclusion chromatography. FPV039 exhibited low solubility, necessitating sample buffer optimization to achieve concentrations of 30 μm in a final buffer comprising 20 mm trisodium citrate, pH 6.0, 200 mm sodium chloride. We then examined binding of FPV039 to peptides encompassing the BH3 domain of all proapoptotic Bcl-2 proteins using isothermal titration calorimetry (ITC). Unexpectedly, ITC revealed that FPV039 was able to bind to peptides of all BH3-only proteins as well as peptides from Bak and Bax but not Bok (Fig. 1). FPV039 engages several BH3-only proteins with high affinity including Bid (2 nm), Bim (15 nm), Hrk (24 nm), Bmf (16 nm), Puma (24 nm), Noxa (28 nm), and Bik (30 nm). In contrast, FPV039 only engaged Bad (653 nm) with moderate affinity. Furthermore, FPV039 bound Bak BH3 with 76 nm affinity and Bax BH3 with 76 nm affinity.
Figure 1.
FPV039 interacts with BH3 domains of all BH3-only proteins and Bak. Raw heats measured by ITC for FPV039 interactions with BH3 domain peptides of proapoptotic Bcl-2 proteins from H. sapiens (HS) and G. gallus (GG).
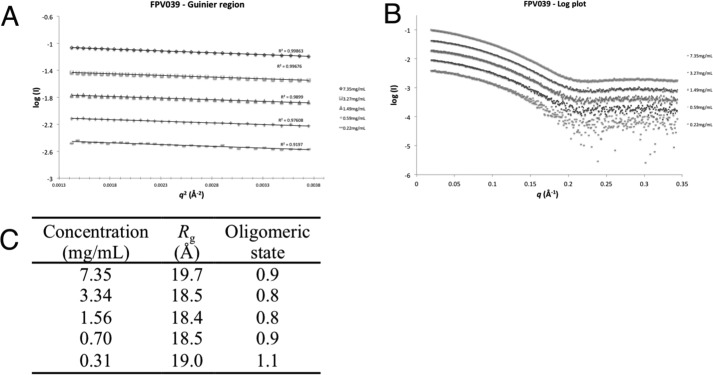
Because several viral prosurvival proteins from the Poxviridae have been shown to form dimers (18, 21, 23, 29), we next investigated the oligomeric state of FPV039 using small angle X-ray scattering (SAXS) (Table 1). In the presence of BH3 domain peptides, FPV039 could be concentrated to higher concentration, allowing us to perform SAXS analysis of FPV039·Bak BH3 domain peptide complex at concentrations ranging from 0.31 to 7.35 mg/ml. The scattering curve profile is conserved throughout the concentration range tested, suggesting an absence of interparticle interference (Fig. 2). The scattering conforms to a straight line in the low q region on the Guinier plot (Fig. 2A), and the calculated radius of gyration does not vary significantly with the measured concentration range, suggesting an absence of significant concentration effects from the highest concentration (Fig. 2). The molecular mass calculated from I(0) on the absolute scattering scale across the concentration range was ∼20.0 kDa, corresponding to an oligomerization state of ∼1 for the FPV039·Bak BH3 domain complex (Fig. 2C).
Table 1.
Small-angle X-ray scattering data collection and scattering-derived parameters
| FPV039·Bak BH3 domain | |
|---|---|
| Data collection parameters | |
| Instrument | SAXS/WAXS beamline, Australian Synchrotron |
| Beam geometry (μm) | 80 × 200 |
| Wavelength (keV) | 12 |
| q range (Å−1) | 0.011–0.619 |
| Exposure time (s) | 1 (per frame; 17 frames) |
| Concentration range (mg ml−1) | 0.31–7.35 |
| Temperature (K) | 293.15 |
| Structural parametersa | |
| I(0) (from Guinier) (cm−1) | 0.10 ± 0.000 |
| Rg (from Guinier) (Å) | 19.66 ± 0.06 |
| Molecular mass determinationa | |
| Partial specific volume (cm3 g−1)b | 0.737 |
| Contrast (Δρ × 1010 cm−2) | 2.900 |
| Mr (from I(0)) (Da) | 17,902 |
| Calculated monomeric Mr from sequence (Da) | 20,022 |
| Software used | |
| Primary data reduction | SAXS/WAXS beamline software |
| Data processing | PRIMUS |
a Reported for 7.35 mg/ml.
b Determined with MULCh (74).
Figure 2.
FPV039 is a monomer in solution. A, log plot of FPV039(1–143) raw SAXS data. Concentrations are in decreasing order starting at 7.35 mg/ml (top) followed by 3.34, 1.56, 0.70, and 0.31 mg/ml. B, Guinier plots of FPV039 SAXS data. Concentrations are as in A. C, summary of the SAXS data and analysis of the oligomeric state of FPV039 in solution.
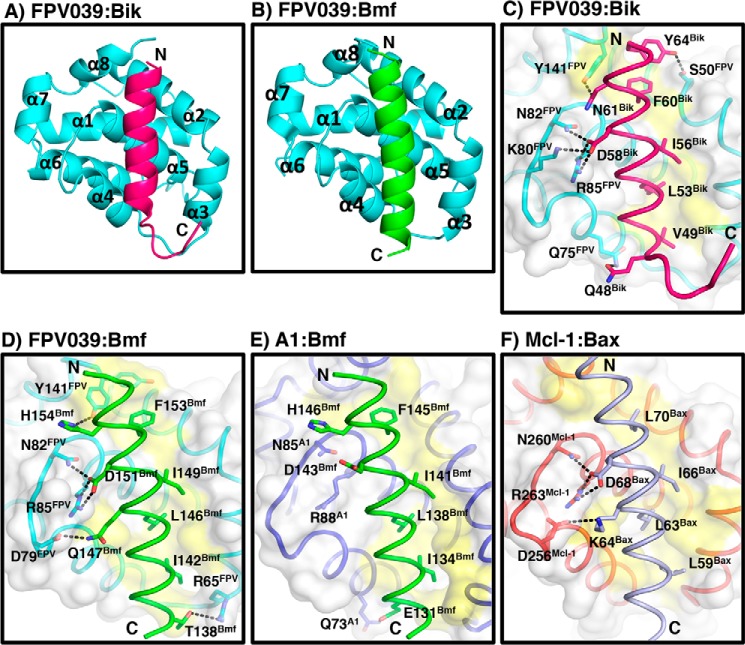
To understand the structural basis of FPV039 interaction with the BH3 domain of proapoptotic Bcl-2 proteins, we determined the structures of FPV039 in complex with Bmf (Fig. 3A and Table 2) and Bik BH3 domains (Fig. 3B). Although FPV039 has limited sequence identity to Bcl-2 proteins, FPV039 adopts a Bcl-2 fold comprising eight α-helices and harboring the conserved BH3 domain-binding groove observed in all the other prosurvival Bcl-2 members that is used to bind both Bmf and Bik BH3 domains (Fig. 4, A and B). Interestingly, FPV039 features an extended loop between α5 and α6 that is only visible in the FPV039·Bik BH3 complex structure but not in the FPV039·Bmf BH3 complex structure, suggesting considerable flexibility in the loop. The closest structural homolog as identified by a Dali search is Mcl-1 (root mean square deviation of 0.92 Å over 107 Cα carbon atoms with 25% sequence identity), whereas BHRF1 is the closest vBcl-2 homolog (30).
Figure 3.

2Fo − Fc electron density maps of FPV039·BH3 domain complexes. A, electron density map encompassing the hydrophobic binding groove of FPV039 in complex with Bmf BH3. FPV039 is shown as cyan sticks, and Bmf BH3 is shown as green sticks. The electron density map is shown as a blue mesh contoured at 2 σ. B, electron density map encompassing the hydrophobic binding groove of FPV039 in complex with Bik BH3. Molecules are shown as in A except for Bik BH3, which is shown as pink sticks.
Table 2.
Data collection and refinement statistics
Values in parentheses are for the highest resolution shell. r.m.s., root mean square. CC1/2 is the correlation coefficient between the average intensities of each subset.
| FPV039·Bmf BH3 | FPV039·Bik BH3 | |
|---|---|---|
| Data collection | ||
| Space group | C2 | P43 |
| No. of molecules in asymmetric unit | 2 + 2 | 1 + 1 |
| Cell dimensions | ||
| a, b, c (Å) | 123.60, 41.20, 67.82 | 47.85, 47.85, 74.30 |
| α, β, γ (°) | 90.00, 93.14, 90.00 | 90.00, 90.00, 90.00 |
| Wavelength (Å) | 0.9537 | 0.9537 |
| Resolution (Å) | 46.91–1.65 (1.68–1.65) | 47.85–1.35 (1.37–1.35) |
| No. unique reflections | 40,990 (1,971) | 36,750 (1,846) |
| Rsym or Rmerge | 0.050 (1.087) | 0.070 (1.592) |
| I/σI | 10.7 (1.1) | 25.79 (1.7) |
| CC1/2 | 0.99 (0.68) | 1.00 (0.60) |
| Wilson B-factor | 25.6 | 13.8 |
| Completeness (%) | 99.9 (98.6) | 100 (100) |
| Redundancy | 3.9 (3.9) | 14.5 (13.5) |
| Refinement | ||
| Resolution (Å) | 1.65 | 1.35 |
| No. reflections | 40,942 | 36,707 |
| Rwork/Rfree | 0.1729/0.2067 | 0.1444/0.1716 |
| No. atoms | ||
| Protein | 2,601 | 1,348 |
| Ligand/ion | 6 | 13 |
| Water | 298 | 202 |
| B-factors | ||
| Protein | 39.13 | 16.35 |
| Ligand/ion | 57.95 | 24.53 |
| Water | 46.53 | 29.94 |
| r.m.s. deviations | ||
| Bond lengths (Å) | 0.007 | 0.008 |
| Bond angles (°) | 0.77 | 0.87 |
| Ramachandran statistics (%) | ||
| Favored | 96.26 | 98.08 |
| Allowed | 3.74 | 1.92 |
| Disallowed | 0.00 | 0.00 |
Figure 4.
FPV039 interacts with BH3 domains from Bmf and Bik via a conserved hydrophobic groove. Shown are schematic representations of FPV039 (cyan) in complex with Bik BH3 domain (hot pink) (A) or Bmf BH3 domain (green) (B). The view is into the conserved hydrophobic ligand-binding groove formed by α-helices 2–5. C–F, surface representations of the FPV039·Bik BH3 domain complex (C), FPV039·Bmf BH3 domain complex (D), A1·Bmf BH3 domain complex (E), and Mcl-1·Bax BH3 domain complex (F) interfaces. Prosurvival Bcl-2 protein surfaces are shown in gray. Residues involved in interactions are shown as sticks and labeled. Interactions are denoted as black dotted lines. Protein Data Bank accession codes are as follows: A1, 2VOG; and Mcl-1, 3PK1.
FPV039 engages the BH3 domain of proapoptotic Bcl-2 proteins using the canonical ligand-binding groove found in other prosurvival Bcl-2 proteins. The ligand-binding groove of FPV039 is formed by α2–α5 with the α5 helix forming the floor of the groove. FPV039 bound to the BH3 domains of Bmf and Bik by utilizing the four conserved hydrophobic residues in the BH3 domain as well as a conserved ionic interaction between Asp-151 (for Bmf) and Asp-57 (for Bik) from the BH3 domain and Arg-85 from FPV039 (Figs. 4, C and D, and 5A).
Figure 5.
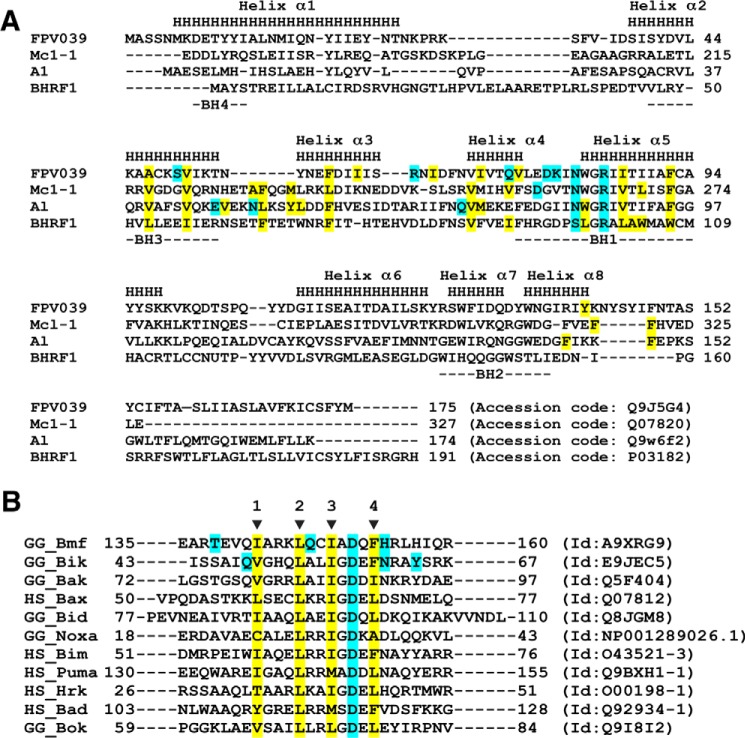
Sequence alignments of FPV039 with Bcl-2 family members. A, structure-based sequence alignment of FPV039 with related cellular Bcl-2 proteins Mcl-1 and A1 as well as the closest viral Bcl-2 protein BHRF1. The α-helical secondary structure elements indicated (denoted as H and labeled Helices 1–8) are based on FPV039. BH motifs 1–4 are marked underneath the aligned sequences. Hydrophobic residues involved in the interactions with BH3 domain peptides are highlighted in yellow, and residues that are involved in hydrogen bonds or ionic interactions are highlighted in cyan. Accession numbers are as follows: FPV039, Q9J5G4; Mcl-1, Q07820; A1, Q9W6F2; and BHRF1, P03182. B, sequence alignment of BH3 domains used in affinity measurements. The four conserved hydrophobic residues in the BH3 motif are indicated by arrows and numbered. The hydrophobic residues involved in hydrophobic interactions with FPV039 in the case of Bmf and Bik BH3 domains are highlighted in yellow. Residues that form polar contacts with FPV039 are highlighted in cyan for Bmf and Bik BH3 domains as is the conserved aspartic acid of the BH3 domains. Sequence accession numbers are shown next to each sequence.
Inspection of the FPV039·Bmf BH3 complex interface (Fig. 4C) revealed that Bmf residues Ile-142, Leu-146, Ile-149, and Phe-153 protrude into four hydrophobic pockets on the FPV039-binding groove. Furthermore, four hydrogen bonds and a salt bridge are found at the interface with the salt bridge formed by Arg-85FPV039 and Asp-151Bmf, and the hydrogen bonds are formed by Tyr-141FPV039 and His-154Bmf, Asn-82FPV039 and Asp-151Bmf, Asp-79FPV039 and Gln-147Bmf, and Arg-65FPV039 and Thr-138Bmf.
Similarly, the interface of FPV039 and the Bik BH3 domain (Fig. 4D) involves four hydrophobic residues of Bik (Phe-60, Ile-56, Leu-53, and Val-49), which protrude into four hydrophobic pockets on FPV039. Furthermore, the salt bridge observed in FPV039·Bmf is conserved and formed by Arg-85FPV039–Asp-57Bik in the FPV039·Bik BH3 domain complex. A total of five hydrogen bonds are formed between Bik BH3 domain and FPV039, two of which are also observed in FPV039·Bmf including Tyr-141FPV039–Asn-61Bik and Asn-82FPV039–Asp-57Bik. In contrast, three of the hydrogen bonds are unique to FPV039·Bik and not found in the Bmf complex, and these are formed by Ser-50FPV039–Tyr-64Bik, Lys-80FPV039–Asp-57Bik, and Gln-75FPV039–Gln-48Bik.
Discussion
Numerous viruses have been shown to express proteins to specifically inhibit apoptosis (5, 7), in particular the Bcl-2-mediated pathway, to ensure viral survival and proliferation. Some of these viral effector proteins harbor detectable sequence identity to members of the Bcl-2 family of proteins (7). Recognizable Bcl-2 mimics encoded by viruses include adenovirus E1B19K (13), Kaposi sarcoma herpesvirus KsBcl-2 (31), Epstein-Barr virus BHRF1 (32), γ-herpesvirus 68 M11 (12), African swine fever virus A179L (14, 33), turkey herpesvirus vNR13 (34), and orf virus ORFV125 (35) as well as the poxviral Bcl-2 proteins SPPV14 from sheeppox (19) and CNP058 from canarypox virus (36). However, many other virulence factors, in particular from poxviruses, share very limited sequence identity to prosurvival Bcl-2 but retain the Bcl-2 fold. Examples of such virulence factors include deerpox virus DPV022 (20, 21), vaccinia virus (22, 23) and variola virus F1L (18), and myxoma virus M11L (24).
FPV039 was identified as a putative vBcl-2 in the fowlpox virus genome as it shares limited sequence identity with mammalian Bcl-2 proteins (26). Cellular studies showed that FPV039 is a potent apoptosis inhibitor where FPV039 was able to inhibit apoptosis induced by Bak and Bax as well as all the BH3-only proteins (27, 28). However, the molecular basis of apoptosis inhibition remained unclear. To address this, we examined the ability of FPV039 to bind to BH3 domains of all proapoptotic Bcl-2 members (Figs. 1 and 5B). FPV039 exhibited binding to peptides of all BH3-only proteins with low nanomolar affinities except for Bad, which was only bound with modest affinity (653 nm). Furthermore, FPV039 bound the BH3 domain of the proapoptotic executor protein Bak with 76 nm affinity and Bax BH3 with 76 nm affinity but did not exhibit binding to Bok BH3. These findings suggest that FPV039 is able to neutralize all key Bcl-2-mediated aspects of the intrinsic apoptosis pathway by preventing initiation of apoptosis by directly sequestering all BH3-only proteins while also engaging proapoptotic Bak and Bax. Interestingly, Banadyga et al. (28) previously determined that FPV039 is only able to immunoprecipitate the BH3-only proteins Bim and Bik as well as proapoptotic Bak and activated Bax. Although this report also revealed some evidence for additional interactions with Noxa, Bid, and Bmf, ultimately the authors concluded that there was insufficient evidence for an interaction. We have now shown biochemically that peptides spanning the BH3 domain from all BH3-only proteins are able to engage FPV039. This discrepancy could be due to the use of peptides rather than full-length proteins, although we note that it has been previously shown that the isolated BH3 domain of BH3-only proteins recapitulates the bulk of the properties of their full-length counterparts (37). Considering the tight nanomolar affinity of FPV039 for some of the BH3-only proteins that were not observed to immunoprecipitate, this discrepancy is surprising. However, this issue is compounded by the observation that immunoprecipitation experiments for Bcl-2 protein interactions are influenced by the choice of detergent (38), and ultimately a different choice of detergent for the immunoprecipitation experiment may have yielded a different outcome.
At the structural level, FPV039 displays certain features that are unusual in the wider Bcl-2 family. Long loop regions are typically located in the loop connecting α1 and α2 such as in Bcl-2 and Bcl-xL, which both feature particularly long connecting loops. In contrast, FPV039 contains a long loop connecting α5 and α6, a feature only previously seen in Bcl-b and its mouse homolog Boo. However, despite the topological similarity to Bcl-b and Boo, FPV039 is highly promiscuous with respect to binding proapoptotic Bcl-2 proteins, whereas Bcl-b only binds Bim and Bax (39). For Boo, conflicting interaction studies have been published, suggesting that Boo is either unable to bind proapoptotic Bcl-2 family members (40) or engages all of them with weak micro- or millimolar affinities (41).
Among the vBcl-2 proteins, only BHRF1 displays a long connecting sequence, which is located between α1 and α2 and harbors a short additional helical sequence (30), with the remaining structures of vBcl-2 proteins displaying compact helical bundle folds with very short loops. In the cases of vaccinia and variola virus F1L (18, 22) and deerpox virus DPV022 (21), the loop connecting α1 and α2 is particularly short, resulting in a dimeric domain-swapped arrangement.
Although a number of structures of prosurvival Bcl-2 proteins bound to BH3-only proteins have been determined, the structural bases of prosurvival Bcl-2 protein interactions with Bik and Bmf are not well characterized. To date, no other structures of prosurvival Bcl-2 proteins with Bik have been solved, and the only complex structure involving Bmf that has been determined is the A1·Bmf BH3 complex (Fig. 4E). Interestingly, A1 binding to Bmf BH3 does not involve the hallmark ionic interaction between the conserved arginine from the NWGR sequence with the conserved aspartic acid from the BH3 domain that is observed in the FPV039·Bmf complex (42). Furthermore, the numerous additional hydrogen bond interactions seen in FPV039·Bmf complex are also absent in A1·Bmf complex (Fig. 4E) including interactions contributed by FPV039 Asp-79, Asn-82, and Arg-85, which are conserved between FPV039 and A1. Consistent with this, FPV039 engages Bmf with a higher affinity compared with A1 (17 and 160 nm, respectively) (42). In contrast, Mcl-1 utilizes the same interactions to bind Bax as FPV039 (Fig. 4F), consistent with the identification of Mcl-1 as the closest structural homolog of FPV039.
Our findings that FPV039 is able to bind to all BH3-only proteins as well as Bak and Bax has implications for the putative mechanism of apoptosis inhibition. The major cellular prosurvival Bcl-2 proteins Bcl-2, Bcl-xL, Bcl-w, Mcl-1, and A1 harbor distinct binding profiles for proapoptotic Bcl-2 proteins as shown by their ability to bind either the BH3-only protein Bad or Noxa. These overlapping but distinct binding profiles enable finely tuned and differential regulation of apoptosis in a cell (43). Interestingly, the binding profile for FPV039 for proapoptotic Bcl-2 proteins differs from its cellular counterparts in two significant aspects. FPV039 is able to engage both Bad and Noxa with measurable affinity, which is highly unusual among prosurvival Bcl-2 proteins (Table 3). In particular, the high affinity interaction with Noxa BH3 domain is striking. This feature is unique to FPV039. Although orf virus ORFV125 has been shown to engage Noxa by immunoprecipitation, the affinity for this interaction has never been determined (44). The only other Noxa-binding vBcl-2 protein is A179L, which displays only moderate affinity (33). Noxa has been shown to be a crucial contributor in defense mechanisms during viral infection where Noxa expression greatly increased in the cells exposed to double-stranded RNA, interferon (IFN), and virus (45, 46). The high-affinity interaction of FPV039 with Noxa that we found suggests that fowlpox virus is able to counter host defense mechanisms initiated via Noxa during viral infection. Furthermore, our data suggest that FPV039 is capable of preventing initiation of Bcl-2-mediated apoptosis by directly sequestering all BH3-only proteins while also engaging the proapoptotic executor proteins Bak and Bax to prevent perforation of the outer mitochondrial membrane. Consequently, FPV039 is in principle equipped to neutralize a vast range of apoptotic stimuli and constitutes a broad spectrum Bcl-2 inhibitor.
Table 3.
Binding affinities of prosurvival Bcl-2 proteins to BH3 domain peptides of proapoptotic Bcl-2 proteins
NB, no binding; n/a, not applicable.
| Poxviral Bcl-2 |
Asfarviridae Bcl-2 A179Lb | Herpesviral Bcl-2 |
Cellular Bcl-2 |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SPPV14a | M11La | MVAF1La | DPV022b | FPV039b | BHRF1b | KsBcl-2c | M11b | Bcl-2a | Bcl-wa | Bcl-xLa | Mcl-1a | A1b | ||
| Bad | >2,000 | >1,000 | NB | NB | 653 | 258 | >2,000 | >1,000 | n/a | 16 | 30 | 5.3 | >100,000 | 15,000 |
| Bid | 341 | 100 | NB | NB | 2 | 26 | 109 | 112 | 232 | 6,800 | 40 | 82 | 2,100 | 1 |
| Bik | >2,000 | >1,000 | NB | NB | 30 | 190 | >2,000 | >1,000 | n/a | 850 | 12 | 43 | 1,700 | 58 |
| Bim | 26 | 5 | 250 | 340 | 10 | 6 | 18 | 29 | 131 | 2.6 | 4.3 | 4.6 | 2.4 | 1 |
| Bmf | 67 | 100 | NB | NB | 16 | 254 | >2,000 | >1,000 | 300 | 3 | 9.8 | 9.7 | 1,100 | 180 |
| Hrk | 63 | >1,000 | NB | NB | 24 | 1,487 | >1,000 | >1,000 | 719 | 320 | 49 | 3.7 | 370 | 46 |
| Noxa | >2,000 | >1,000 | NB | NB | 28 | 1,575 | >2,000 | >1,000 | 132 | >100,000 | >100,000 | >100,000 | 24 | 20 |
| Puma | 65 | >1,000 | NB | NB | 24 | 31 | 70 | 69 | 370 | 3.3 | 5.1 | 6.3 | 5 | 1 |
| Bak | 46 | 50 | 4,300 | 6,930 | 76 | 29 | 150 | <50 | 76.3 | >1,000 | 500 | 50 | 10 | 3 |
| Bax | 32 | 75 | 1,850 | 4,040 | 76 | 26 | 1,400 | 980 | 690 | 100 | 58 | 130 | 12 | n/a |
a Affinity measured in nm by surface plasmon resonance (KD).
b Affinity measured in nm by isothermal calorimetry (KD).
c Affinity measured in nm by fluorescence polarization (IC50).
A number of studies on virus-encoded Bcl-2 proteins revealed that the range of proapoptotic Bcl-2 proteins that they interact with varies considerably across the different proteins and that a number of molecular mechanisms of action are utilized to inhibit premature host cell apoptosis (7, 21, 47). vBcl-2 members have been demonstrated to rely either on sequestration of Bax and Bak as shown for myxoma virus M11L (24), sequestration of Bim as shown for vaccinia virus F1L (22), or a mixed mode reliant on neutralization of both Bim and Bak as shown for Epstein-Barr virus BHRF1 (30, 48). In comparison with mammalian Bcl-2 proteins, vBcl-2 proteins typically interact with host proapoptotic proteins with considerably lower affinity (7) and tend to target only a smaller subset of proapoptotic regulators. Certain vBcl-2 members such as vaccinia and variola virus F1L (18, 23) and deerpox virus DPV022 (21) target only a very restricted subset of host proapoptotic proteins, whereas others are more promiscuous such as sheeppox virus SPPV14 (19) and murine γ-herpesvirus 68 M11 (49). Interestingly, FPV039 is only the second vBcl-2 proteins after A179L (33) that has been shown to interact with all major host proapoptotic Bcl-2 proteins. Furthermore, FPV039 engages host BH3-only proteins with high affinities, thus more closely resembling the behavior of cellular prosurvival Bcl-2 rather than other vBcl-2 members. We note that the affinity of FPV039 for Bak is lower compared with cellular Bcl-2 proteins; however, the relative importance of all the proapoptotic ligands identified for FPV039 remains to be clarified. Nonetheless, this remarkable promiscuity is in striking contrast to vaccinia virus F1L (22), which relies solely on the sequestration of Bim to prevent host cell apoptosis, underscoring the diversity of mechanisms utilized by viruses and their Bcl-2 homologs to subvert intrinsic apoptosis.
Interestingly, FPV039 is one of a small number of vBcl-2 proteins that is able to engage Bad BH3, albeit with modest affinity (Table 3). This raises the prospect of targeting FPV039 with first-generation BH3 mimetics such as ABT-737 and ABT-263, which were designed as Bad BH3 mimetics and engage the same mammalian prosurvival Bcl-2 proteins as Bad (50). Although the utility of targeting vBcl-2 proteins for therapy remains to be established for the Poxviridae, the efficacy of such a strategy was recently demonstrated for Epstein-Barr virus BHRF1 (51).
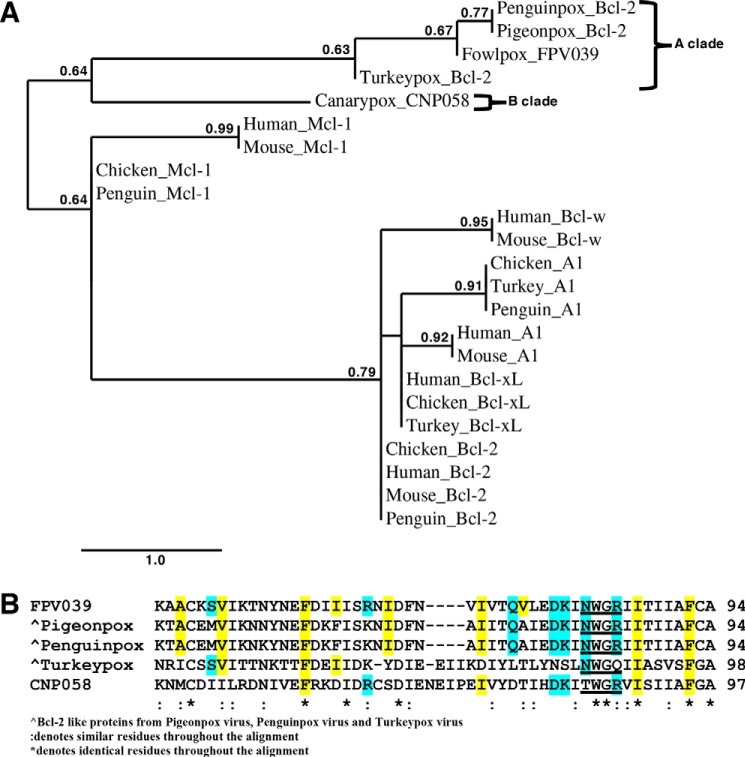
A phylogenetic analysis of FPV039 and its homologs among the avipoxviruses reveals that they can be divided into two clades (Fig. 6A) in agreement with previous analyses using the conserved core 4b protein (52) and more recently DNA polymerase (53). Furthermore, the overall protein sequence conservation between the canarypox and fowlpox Bcl-2 homologs is lower at 39% compared with overall conservation across homologous proteins (53%) (36), suggesting that the vBcl-2 proteins are subject to higher evolutionary pressure. This view would be consistent with the role of FPV039 as an immune evasion protein that is subject to higher selective pressure as part of the host-pathogen interface. Interestingly, mapping of conserved residues within the hydrophobic binding groove of FPV039 on avipoxviral homologs such as CNP058 reveals that several key residues in the groove are not conserved in the Clade B virus canarypox virus and even in the proximal Clade A virus turkeypox virus (Fig. 6B), suggesting that CNP058 is likely to harbor a different proapoptotic Bcl-2 ligand-binding profile compared with FPV039. Expansion of the phylogenetic analysis to include chicken prosurvival proteins reveals that the closest chicken prosurvival Bcl-2 protein is Mcl-1 (Fig. 6A). This finding is consistent with our identification of Mcl-1 as the closest structural homolog of FPV039 and the conservation of interactions between FPV039·BH3 ligand and Mcl-1·BH3 ligand complexes. Consequently, it is tempting to speculate that chicken Mcl-1 may have been the first prosurvival Bcl-2 protein to be acquired by an ancestral avipoxvirus. However, this hypothesis requires experimental verification.
Figure 6.
A, phylogenetic analysis of Bcl-2-like proteins in avipoxviruses. The tree comprises FPV039 encoded by fowlpox virus, CNP058 encoded by canarypox virus, and Bcl-2-like proteins from penguinpox virus, pigeonpox virus, and turkeypox virus. Avipoxvirus clades were classed as A and B according to the nomenclature of Jarmin et al. (52). Phylogeny analysis reveals that FPV039, pigeonpox Bcl-2-like, and penguinpox Bcl-2 locate to the same clade, whereas CNP058 and turkeypox Bcl-2 are found on a separate clade. The scale bar represents the number of substitutions per site, and the branch annotation is the branch support values where only branch support values of more than 0.5 are shown. B, multiple sequence alignment of FPV039 with the other avipoxvirus Bcl-2 proteins. Conserved NWGR motifs are underlined. Hydrophobic residues of FPV039 that interact with BH3 domains of Bik and Bmf are highlighted in yellow. Residues of FPV039 that form ionic interactions or hydrogen bonds with BH3 domains of Bik and Bmf are highlighted in cyan. Conserved residues in related Bcl-2-like proteins from avipoxviruses postulated to form hydrophobic interactions with cellular proapoptotic Bcl-2 proteins are highlighted in yellow, and residues postulated to form ionic interactions or hydrogen bonds are highlighted in cyan. Accession codes for sequences used are as follows: 1) FPV039, Q9J5G4; 2) pigeonpox Bcl-2-like, A0A068EEI8; 3) penguinpox Bcl-2-like, A0A068EHE4; 4) turkeypox Bcl-2-like, A0A0M3ZJZ2; and 5) CNP058, Q6VZT9.
In summary, our findings reveal that FPV039 adopts a Bcl-2 fold similar to the mammalian prosurvival protein Mcl-1, which allows engagement of all BH3-only proteins as well as Bak and Bax with high affinity. Our results will allow a detailed dissection of the interactions between FPV039 and host proapoptotic regulators to establish whether or not all of them are required as well as to pinpoint which interactions are ultimately key for suppression of premature host cell apoptosis.
Experimental procedures
FPV039 expression and purification
Synthetic codon-optimized cDNA encoding for FPV039 lacking the C-terminal transmembrane domain (residues 1–143) (Genscript) was cloned into the bacterial expression vector pGEX6P-1 (GE Healthcare). The plasmid was transformed into E. coli BL21 Codon Plus cells and grown in 2YT (yeast/tryptone) medium supplemented with 1 mg/ml ampicillin at 37 °C in an incubator shaking at 180 rpm until the culture reached an A600 of 0.6. Protein expression was induced using 0.5 mm isopropyl β-d-1-thiogalactopyranoside at 22 °C for 16 h. The cells were harvested by centrifugation at 3400 × g (JLA 9.1000 rotor, Beckman Coulter Avanti J-E) for 15 min, resuspended in 100 ml of lysis buffer (20 mm trisodium citrate, pH 6.0, 200 mm NaCl), and lysed using a sonicator at 50 kHz for four cycles (15 s) with 30-s rest intervals in the presence of lysozyme (Sigma-Aldrich) and DNase I (deoxyribonuclease I from bovine pancreas, Sigma-Aldrich). The resulting lysate was clarified by centrifugation at 31,000 × g (JA 25.50 rotor, Beckman Coulter Avanti J-E) for 30 min. The supernatant was filtered with a 0.45-μm filter (Millipore), loaded onto 5 ml of glutathione-Sepharose 4B beads (GE Healthcare) equilibrated with lysis buffer, and subsequently washed with 30 ml of lysis buffer. Human rhinovirus 3C protease was added to the column and incubated overnight at 4 °C to liberate the target protein from the GST affinity tag. The cleaved target protein was eluted and concentrated to 5 ml prior to being subjected to size-exclusion chromatography using a Superdex S75 16/600 column attached to an ÄKTAxpress system (GE Healthcare) equilibrated in 20 mm trisodium citrate, pH 6.0, 200 mm NaCl. The protein eluted as a single peak and displayed higher than 95% purity based on SDS-PAGE analysis.
Measurement of interactions with BH3 peptides
Affinities of FPV039 for different BH3 peptides were measured using a MicroCal iTC200 system (GE Healthcare) at 25 °C. The measurements were performed in 20 mm trisodium citrate, pH 6.0, 200 mm NaCl at a final protein concentration of 30 μm. BH3 domain peptides at concentration of 300 μm were titrated into the protein sample using 19 injections of 2 μl/injection. All assays were performed in triplicates. Protein concentrations were measured using a UV spectrophotometer (Thermo Scientific) at a wavelength of 280 nm. BH3 peptides concentrations were calculated from the dry weight of peptide. The BH3 peptides sequences used in this study are as follows: 1) Gallus gallus GG_Bak (UniProt accession number Q5F404), 72LGSTGSQVGRRLAIIGDDINKRYDAE97; 2) Homo sapiens HS_Bax (UniProt accession number Q07812), 50VPQDASTKKLSECLKRIGDELDSNMELQ77; 3) GG_Bid (UniProt accession number Q8JGM8), 77PEVNEAIVRTIAAQLAEIGDQLDKQIKAKVVNDL110; 4) GG_Bmf (UniProt accession number A9XRG9), 135EARTEVQIARKLQCIADQFHRLHIQR160; 5) GG_Bok (UniProt accession number Q9I8I2), 67VSAILLRLGDELEYIRPNVYRNIARQ92; 6) GG_Noxa (RefSeq accession number NP_001289026.1), ERDAVAECALELRRIGDKADLQQKVL; 7) GG_Bik (UniProt accession number E9JEC5), 43ISSAIQVGHQLALIGDEFNRAYSRK67; 8) HS_Bim (UniProt accession number O43521-3), 51DMRPEIWIAQELRRIGDEFNAYYARR76; 9) HS_Puma (UniProt accession number Q9BXH1-1), 130EEQWAREIGAQLRRM ADDLNAQYERR155; 10) HS_Hrk (UniProt accession number O00198-1), 26RSSAAQLTAARLKAIGDELHQRTMWR51; 11) HS_Bad (UniProt accession number Q92934-1), 103NLWAAQRYGRELRRMSDEFVDSFKKG128.
Small angle X-ray scattering
SAXS data were collected at the Australian Synchrotron SAXS/WAXS beamline using protocols described previously (21, 54). Briefly, FPV039 in complex with Bak BH3 at concentrations of 7.35, 3.34, 1.56, 0.70, and 0.31 mg/ml in 20 mm trisodium citrate, pH 6.0, 200 mm NaCl was measured in a q range between 0.025 and 0.6 Å−1 at 12 keV with a 1.6-m camera length at 20 °C and collected on a Pilatus 1M detector (Dectris). The extinction coefficient of the complex was 1.872. Beam stop integration was used to achieve normalization. The data were absolute scaled using distilled water. The samples were measured in a 1.5-mm quartz capillary to minimize radiation damage and flowed past the beam while 30 × 1-s exposures were measured on samples and blanks. Software specific to the beamline was used to integrate, average, and calibrate the scattering images against water (55). The forward scattering (I(0)) and radius of gyration (Rg) were determined by the Guinier approximation (56, 57). Sample homogeneity and monodispersity were independently determined using SDS-PAGE analysis and size-exclusion chromatography. Details of the data collection are summarized in Table 1.
FPV039 complex crystallization and data collection
Complexes of FPV039 with Bik BH3 and Bmf BH3 were prepared as described previously (58). Briefly, FPV039 complexes were reconstituted by adding GG_Bik BH3 domain or GG_Bmf BH3 domain peptides at a 1:1.25 molar ratio to FPV039. The reconstituted complexes were concentrated to 10 mg/ml using a 3-kDa-molecular-mass-cutoff centrifugal concentrator (Millipore), flash cooled, and stored under liquid nitrogen. Crystallization trials were carried out using 96-well sitting-drop tray (Swissci) vapor diffusion at 20 °C either in-house or at the Commonwealth Scientific and Industrial Research Organisation (CSIRO) C3 Collaborative Crystallization Centre, Melbourne, Australia. 0.15 μl of FPV039·BH3 domain peptide complex was mixed with 0.15 μl of various crystallization condition solutions using a Phoenix nanodispenser robot (Art Robbins). Commercially available screening kits (Crystal Screen, PACT Suite, JCSG-plus Screen, PEG/Ion Screen, and ShotGun Screen) were used for the initial crystallization screening with hit optimization performed either in-house using 24-well hanging-drop plates (EasyXtal DG-Tool, Qiagen) of 1 μl protein mixture with 1 μl of reservoir solution or using a 96-well plate at the CSIRO C3 Collaborative Crystallization Centre.
Crystals of FPV039 in complex with the Bmf BH3 domain were obtained at 10 mg/ml in three conditions: 1) 0.1 m MMT (DL-malic acid, MES and Tris), pH 8.0, 25% PEG 1500; 2) 0.2 m sodium formate, pH 7.2, 20% PEG 3350; and 3) 0.2 m lithium chloride, 20% PEG 3350, pH 6.8. All three conditions produced hexagonal crystals of FPV039·Bmf BH3 domain complex belonging to space group C2 in the monoclinic crystal system. The final crystal contained two molecules of FPV039, each bound to one molecule of Bmf BH3 in the asymmetric unit, and had a solvent content of 41.9%. The crystals were cryoprotected using 20% (v/v) ethylene glycol and flash cooled at 100 K using liquid nitrogen. All diffraction data were collected on the MX2 beamline at the Australian Synchrotron using an ADSC Quantum 315r charge-coupled device detector (Area Detector Systems Corp., Poway, CA) with an oscillation range of 1.0°/frame using a wavelength of 0.9537 Å. Diffraction data were integrated using XDS (59) and scaled using AIMLESS (60, 61). The structure of FPV039·Bmf BH3 domain was solved using Phaser molecular replacement (62) with the structure of Mcl-1 (Protein Data Bank code 4HW4) as a search model (63). The final TFZ and LLG values were 9.6 and 86, respectively. The solution produced by Phaser was manually rebuilt over multiple cycles using Coot (64) and refined using PHENIX (65).
The crystals for FPV039 in complex with the Bik BH3 domain were obtained at 10 mg/ml in 0.2 m lithium chloride, 20% PEG 3350, pH 6.8. The crystal contained one molecule of FPV039 bound to one molecule of Bik BH3 in the asymmetric unit and had a solvent content of 42.2%. Diffraction data were collected and processed as described above, and the structure was solved by molecular replacement with Phaser (62) using the previously determined structure of FPV039 as a search model (TFZ = 62.1 and LLG = 3342). The structure was built and refined using Coot and PHENIX, respectively. Data collection and refinement statistics details are summarized in Table 2. Coordinate files have been deposited in the Protein Data Bank under the accession codes 5TZQ and 5TZP. All images were generated using the PyMOL Molecular Graphics System, version 1.8 (Schrödinger, LLC). All software was accessed using the SBGrid suite (66). All raw diffraction images were deposited on the SBGrid Data Bank (67) using accession numbers doi 10.15785/SBGRID/443 and 10.15785/SBGRID/442.
Phylogenetic analysis
Protein sequences of avipoxvirus Bcl-2-like proteins (fowlpox, canarypox, turkeypox, penguinpox, and pigeonpox) and cellular Bcl-2 proteins including Mcl-1, Bcl-2, A1, Bcl-xL, and Bcl-w from human, mouse, chicken, turkey, and penguin were aligned using T-Coffee (68). The multiple sequence alignment was modified using Gblocks where Gblocks selects conserved blocks of sequences that lack poorly aligned positions and divergent regions (69). Due to highly divergent sequences in the analysis, a less stringent setting that allows smaller final blocks chosen by Gblocks was used. The phylogenetic tree was generated using a maximum likelihood (ML) approach using the maximum likelihood (PhyML) program (70, 71). The ML analysis was bootstrapped with 100 replicates. The final representation of the tree was produced using the tree dynamics (TreeDyn) program (72). T-Coffee, Gblocks, PhyML, and TreeDyn were accessed via the phylogeny.fr (73). The accession numbers for the sequences used in the phylogenetic analysis are as follows: 1) fowlpox FPV039, Q9J5G4; 2) pigeonpox Bcl-2, A0A068EEI8; 3) penguinpox Bcl-2, A0A068EHE4; 4) turkeypox Bcl-2, A0A0M3ZJZ2; 5) canarypox CNP058, Q6VZT9; 6) human Mcl-1, Q07820; 7) mouse Mcl-1, P97287; 8) chicken Mcl-1, Q9W6F1; 9) penguin Mcl-1, A0A087RIT1; 10) human Bcl-w, Q92843; 11) mouse Bcl-w, P70345; 12) chicken A1, Q9W6F2; 13) turkey A1, A0A091LE25; 14) penguin A1, A0A093PIE7; 15) human A1, Q16548; 16) mouse A1, Q07440; 17) human Bcl-xL, Q07817; 18) chicken Bcl-xL, Q07816; 19) turkey Bcl-xL, G1N5N5; 20) chicken Bcl-2, Q00709; 21) mouse Bcl-2, P10417; 22) human Bcl-2, P10415; and 23) penguin Bcl-2, A0A093NW22.
Author contributions
M. I. A. contributed to acquisition of data, analysis and interpretation of data, and drafting and revising the article. S. C. contributed to acquisition of data, analysis and interpretation of data, and drafting and revising the article. M. A. S. contributed to analysis and interpretation of data and drafting and revising the article. M. K. contributed to study conception and design, acquisition of data, analysis and interpretation of data, and drafting and revising the article.
Acknowledgments
We thank staff at the MX and SAXS/WAXS beamlines at the Australian Synchrotron for help with X-ray data collection, the CSIRO C3 Collaborative Crystallization Centre for assistance with crystallization, and the Comprehensive Proteomics Platform at La Trobe University for core instrument support.
This work was supported in part by National Health and Medical Research Council Australia Project Grant APP1007918 (to M. K.), Australian Research Council Fellowship FT130101349 (to M. K.), and a La Trobe University scholarship (to M. I. A.). The authors declare that they have no conflicts of interest with the contents of this article.
The atomic coordinates and structure factors (codes 5TZQ and 5TZP) have been deposited in the Protein Data Bank (http://wwpdb.org/).
SAXS data were deposited at the SASBDB using accession code SASDC63.
- Bcl-2
- B-cell lymphoma 2
- BH
- Bcl-2 homology
- Bax
- Bcl-2-associated X, apoptosis regulator
- Bak
- Bcl-2 antagonist/killer
- FPV
- fowlpox virus
- vBcl-2
- viral Bcl-2
- SAXS
- small-angle X-ray scattering
- WAXS
- wide-angle X-ray scattering
- ITC
- isothermal titration calorimetry
- TFZ
- translation function Z-score
- LLG
- log-likelihood gain
- ML
- maximum likelihood
- HS
- H. sapiens
- GG
- G. gallus.
References
- 1. Kerr J. F., Wyllie A. H., and Currie A. R. (1972) Apoptosis: a basic biological phenomenon with wide-ranging implications in tissue kinetics. Br. J. Cancer 26, 239–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Youle R. J., and Strasser A. (2008) The BCL-2 protein family: opposing activities that mediate cell death. Nat. Rev. Mol. Cell Biol. 9, 47–59 [DOI] [PubMed] [Google Scholar]
- 3. Vaux D. L., Haecker G., and Strasser A. (1994) An evolutionary perspective on apoptosis. Cell 76, 777–779 [DOI] [PubMed] [Google Scholar]
- 4. Green D. R., and Kroemer G. (2004) The pathophysiology of mitochondrial cell death. Science 305, 626–629 [DOI] [PubMed] [Google Scholar]
- 5. Galluzzi L., Brenner C., Morselli E., Touat Z., and Kroemer G. (2008) Viral control of mitochondrial apoptosis. PLoS Pathog. 4, e1000018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chipuk J. E., Moldoveanu T., Llambi F., Parsons M. J., and Green D. R. (2010) The BCL-2 family reunion. Mol. Cell 37, 299–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kvansakul M., and Hinds M. G. (2013) Structural biology of the Bcl-2 family and its mimicry by viral proteins. Cell Death Dis. 4, e909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Westphal D., Kluck R. M., and Dewson G. (2014) Building blocks of the apoptotic pore: how Bax and Bak are activated and oligomerize during apoptosis. Cell Death Differ. 21, 196–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shamas-Din A., Brahmbhatt H., Leber B., and Andrews D. W. (2011) BH3-only proteins: orchestrators of apoptosis. Biochim. Biophys. Acta 1813, 508–520 [DOI] [PubMed] [Google Scholar]
- 10. Kvansakul M., and Hinds M. G. (2014) The structural biology of BH3-only proteins. Methods Enzymol. 544, 49–74 [DOI] [PubMed] [Google Scholar]
- 11. Danthi P. (2016) Viruses and the diversity of cell death. Annu. Rev. Virol. 3, 533–553 [DOI] [PubMed] [Google Scholar]
- 12. Wang G. H., Garvey T. L., and Cohen J. I. (1999) The murine gammaherpesvirus-68 M11 protein inhibits Fas- and TNF-induced apoptosis. J. Gen. Virol 80, 2737–2740 [DOI] [PubMed] [Google Scholar]
- 13. Chiou S. K., Tseng C. C., Rao L., and White E. (1994) Functional complementation of the adenovirus E1B 19-kilodalton protein with Bcl-2 in the inhibition of apoptosis in infected cells. J. Virol. 68, 6553–6566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Brun A., Rivas C., Esteban M., Escribano J. M., and Alonso C. (1996) African swine fever virus gene A179L, a viral homologue of bcl-2, protects cells from programmed cell death. Virology 225, 227–230 [DOI] [PubMed] [Google Scholar]
- 15. Graham K. A., Opgenorth A., Upton C., and McFadden G. (1992) Myxoma virus M11L ORF encodes a protein for which cell surface localization is critical in manifestation of viral virulence. Virology 191, 112–124 [DOI] [PubMed] [Google Scholar]
- 16. Wasilenko S. T., Stewart T. L., Meyers A. F., and Barry M. (2003) Vaccinia virus encodes a previously uncharacterized mitochondrial-associated inhibitor of apoptosis. Proc. Natl. Acad. Sci. U.S.A. 100, 14345–14350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fischer S. F., Ludwig H., Holzapfel J., Kvansakul M., Chen L., Huang D. C., Sutter G., Knese M., and Häcker G. (2006) Modified vaccinia virus Ankara protein F1L is a novel BH3-domain-binding protein and acts together with the early viral protein E3L to block virus-associated apoptosis. Cell Death Differ. 13, 109–118 [DOI] [PubMed] [Google Scholar]
- 18. Marshall B., Puthalakath H., Caria S., Chugh S., Doerflinger M., Colman P. M., and Kvansakul M. (2015) Variola virus F1L is a Bcl-2-like protein that unlike its vaccinia virus counterpart inhibits apoptosis independent of Bim. Cell Death Dis. 6, e1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Okamoto T., Campbell S., Mehta N., Thibault J., Colman P. M., Barry M., Huang D. C., and Kvansakul M. (2012) Sheeppox virus SPPV14 encodes a Bcl-2-like cell death inhibitor that counters a distinct set of mammalian proapoptotic proteins. J. Virol. 86, 11501–11511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Banadyga L., Lam S. C., Okamoto T., Kvansakul M., Huang D. C., and Barry M. (2011) Deerpox virus encodes an inhibitor of apoptosis that regulates Bak and Bax. J. Virol. 85, 1922–1934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Burton D. R., Caria S., Marshall B., Barry M., and Kvansakul M. (2015) Structural basis of deerpox virus-mediated inhibition of apoptosis. Acta Crystallogr. D Biol. Crystallogr. 71, 1593–1603 [DOI] [PubMed] [Google Scholar]
- 22. Campbell S., Thibault J., Mehta N., Colman P. M., Barry M., and Kvansakul M. (2014) Structural insight into BH3 domain binding of vaccinia virus antiapoptotic F1L. J. Virol. 88, 8667–8677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kvansakul M., Yang H., Fairlie W. D., Czabotar P. E., Fischer S. F., Perugini M. A., Huang D. C., and Colman P. M. (2008) Vaccinia virus anti-apoptotic F1L is a novel Bcl-2-like domain-swapped dimer that binds a highly selective subset of BH3-containing death ligands. Cell Death Differ. 15, 1564–1571 [DOI] [PubMed] [Google Scholar]
- 24. Kvansakul M., van Delft M. F., Lee E. F., Gulbis J. M., Fairlie W. D., Huang D. C., and Colman P. M. (2007) A structural viral mimic of prosurvival Bcl-2: a pivotal role for sequestering proapoptotic Bax and Bak. Mol. Cell 25, 933–942 [DOI] [PubMed] [Google Scholar]
- 25. Douglas A. E., Corbett K. D., Berger J. M., McFadden G., and Handel T. M. (2007) Structure of M11L: a myxoma virus structural homolog of the apoptosis inhibitor, Bcl-2. Protein Sci. 16, 695–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Afonso C. L., Tulman E. R., Lu Z., Zsak L., Kutish G. F., and Rock D. L. (2000) The genome of fowlpox virus. J. Virol. 74, 3815–3831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Banadyga L., Gerig J., Stewart T., and Barry M. (2007) Fowlpox virus encodes a Bcl-2 homologue that protects cells from apoptotic death through interaction with the proapoptotic protein Bak. J. Virol. 81, 11032–11045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Banadyga L., Veugelers K., Campbell S., and Barry M. (2009) The fowlpox virus BCL-2 homologue, FPV039, interacts with activated Bax and a discrete subset of BH3-only proteins to inhibit apoptosis. J. Virol. 83, 7085–7098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cooray S., Bahar M. W., Abrescia N. G., McVey C. E., Bartlett N. W., Chen R. A., Stuart D. I., Grimes J. M., and Smith G. L. (2007) Functional and structural studies of the vaccinia virus virulence factor N1 reveal a Bcl-2-like anti-apoptotic protein. J. Gen. Virol. 88, 1656–1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kvansakul M., Wei A. H., Fletcher J. I., Willis S. N., Chen L., Roberts A. W., Huang D. C., and Colman P. M. (2010) Structural basis for apoptosis inhibition by Epstein-Barr virus BHRF1. PLoS Pathog. 6, e1001236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cheng E. H., Nicholas J., Bellows D. S., Hayward G. S., Guo H. G., Reitz M. S., and Hardwick J. M. (1997) A Bcl-2 homolog encoded by Kaposi sarcoma-associated virus, human herpesvirus 8, inhibits apoptosis but does not heterodimerize with Bax or Bak. Proc. Natl. Acad. Sci. U.S.A. 94, 690–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Henderson S., Huen D., Rowe M., Dawson C., Johnson G., and Rickinson A. (1993) Epstein-Barr virus-coded BHRF1 protein, a viral homologue of Bcl-2, protects human B cells from programmed cell death. Proc. Natl. Acad. Sci. U.S.A. 90, 8479–8483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Banjara S., Caria S., Dixon L. K., Hinds M. G., and Kvansakul M. (2017) Structural insight into African swine fever virus A179L-mediated inhibition of apoptosis. J. Virol. 91, e02228–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Aouacheria A., Banyai M., Rigal D., Schmidt C. J., and Gillet G. (2003) Characterization of vnr-13, the first alphaherpesvirus gene of the bcl-2 family. Virology 316, 256–266 [DOI] [PubMed] [Google Scholar]
- 35. Westphal D., Ledgerwood E. C., Hibma M. H., Fleming S. B., Whelan E. M., and Mercer A. A. (2007) A novel Bcl-2-like inhibitor of apoptosis is encoded by the parapoxvirus ORF virus. J. Virol. 81, 7178–7188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tulman E. R., Afonso C. L., Lu Z., Zsak L., Kutish G. F., and Rock D. L. (2004) The genome of canarypox virus. J. Virol. 78, 353–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wilson-Annan J., O'Reilly L. A., Crawford S. A., Hausmann G., Beaumont J. G., Parma L. P., Chen L., Lackmann M., Lithgow T., Hinds M. G., Day C. L., Adams J. M., and Huang D. C. (2003) Proapoptotic BH3-only proteins trigger membrane integration of prosurvival Bcl-w and neutralize its activity. J. Cell Biol. 162, 877–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hsu Y. T., and Youle R. J. (1997) Nonionic detergents induce dimerization among members of the Bcl-2 family. J. Biol. Chem. 272, 13829–13834 [DOI] [PubMed] [Google Scholar]
- 39. Rautureau G. J., Yabal M., Yang H., Huang D. C., Kvansakul M., and Hinds M. G. (2012) The restricted binding repertoire of Bcl-B leaves Bim as the universal BH3-only prosurvival Bcl-2 protein antagonist. Cell Death Dis. 3, e443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rautureau G. J., Day C. L., and Hinds M. G. (2010) The structure of Boo/Diva reveals a divergent Bcl-2 protein. Proteins 78, 2181–2186 [DOI] [PubMed] [Google Scholar]
- 41. Santiveri C. M., Sborgi L., and de Alba E. (2012) Nuclear magnetic resonance study of protein-protein interactions involving apoptosis regulator Diva (Boo) and the BH3 domain of proapoptotic Bcl-2 members. J. Mol. Recognit. 25, 665–673 [DOI] [PubMed] [Google Scholar]
- 42. Smits C., Czabotar P. E., Hinds M. G., and Day C. L. (2008) Structural plasticity underpins promiscuous binding of the prosurvival protein A1. Structure 16, 818–829 [DOI] [PubMed] [Google Scholar]
- 43. Shamas-Din A., Kale J., Leber B., and Andrews D. W. (2013) Mechanisms of action of Bcl-2 family proteins. Cold Spring Harb. Perspect. Biol. 5, a008714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Westphal D., Ledgerwood E. C., Tyndall J. D., Hibma M. H., Ueda N., Fleming S. B., and Mercer A. A. (2009) The orf virus inhibitor of apoptosis functions in a Bcl-2-like manner, binding and neutralizing a set of BH3-only proteins and active Bax. Apoptosis 14, 1317–1330 [DOI] [PubMed] [Google Scholar]
- 45. Sun Y., and Leaman D. W. (2005) Involvement of Noxa in cellular apoptotic responses to interferon, double-stranded RNA, and virus infection. J. Biol. Chem. 280, 15561–15568 [DOI] [PubMed] [Google Scholar]
- 46. Eitz Ferrer P., Potthoff S., Kirschnek S., Gasteiger G., Kastenmüller W., Ludwig H., Paschen S. A., Villunger A., Sutter G., Drexler I., and Häcker G. (2011) Induction of Noxa-mediated apoptosis by modified vaccinia virus Ankara depends on viral recognition by cytosolic helicases, leading to IRF-3/IFN-β-dependent induction of pro-apoptotic Noxa. PLoS Pathog. 7, e1002083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kvansakul M., and Hinds M. G. (2015) The Bcl-2 family: structures, interactions and targets for drug discovery. Apoptosis 20, 136–150 [DOI] [PubMed] [Google Scholar]
- 48. Desbien A. L., Kappler J. W., and Marrack P. (2009) The Epstein-Barr virus Bcl-2 homolog, BHRF1, blocks apoptosis by binding to a limited amount of Bim. Proc. Natl. Acad. Sci. U.S.A. 106, 5663–5668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ku B., Woo J. S., Liang C., Lee K. H., Hong H. S., E X., Kim K. S., Jung J. U., and Oh B. H. (2008) Structural and biochemical bases for the inhibition of autophagy and apoptosis by viral BCL-2 of murine γ-herpesvirus 68. PLoS Pathog. 4, e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Oltersdorf T., Elmore S. W., Shoemaker A. R., Armstrong R. C., Augeri D. J., Belli B. A., Bruncko M., Deckwerth T. L., Dinges J., Hajduk P. J., Joseph M. K., Kitada S., Korsmeyer S. J., Kunzer A. R., Letai A., et al. (2005) An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature 435, 677–681 [DOI] [PubMed] [Google Scholar]
- 51. Procko E., Berguig G. Y., Shen B. W., Song Y., Frayo S., Convertine A. J., Margineantu D., Booth G., Correia B. E., Cheng Y., Schief W. R., Hockenbery D. M., Press O. W., Stoddard B. L., Stayton P. S., et al. (2014) A computationally designed inhibitor of an Epstein-Barr viral Bcl-2 protein induces apoptosis in infected cells. Cell 157, 1644–1656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Jarmin S., Manvell R., Gough R. E., Laidlaw S. M., and Skinner M. A. (2006) Avipoxvirus phylogenetics: identification of a PCR length polymorphism that discriminates between the two major clades. J. Gen. Virol. 87, 2191–2201 [DOI] [PubMed] [Google Scholar]
- 53. Gyuranecz M., Foster J. T., Dán Á., Ip H. S., Egstad K. F., Parker P. G., Higashiguchi J. M., Skinner M. A., Höfle U., Kreizinger Z., Dorrestein G. M., Solt S., Sós E., Kim Y. J., Uhart M., et al. (2013) Worldwide phylogenetic relationship of avian poxviruses. J. Virol. 87, 4938–4951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Caria S., Marshall B., Burton R. L., Campbell S., Pantaki-Eimany D., Hawkins C. J., Barry M., and Kvansakul M. (2016) The N terminus of the vaccinia virus protein F1L is an intrinsically unstructured region that is not involved in apoptosis regulation. J. Biol. Chem. 291, 14600–14608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kirby N. M., and Cowieson N. P. (2014) Time-resolved studies of dynamic biomolecules using small angle X-ray scattering. Curr. Opin. Struct. Biol. 28, 41–46 [DOI] [PubMed] [Google Scholar]
- 56. Petoukhov M. V., Franke D., Shkumatov A. V., Tria G., Kikhney A. G., Gajda M., Gorba C., Mertens H. D., Konarev P. V., and Svergun D. I. (2012) New developments in the program package for small-angle scattering data analysis. J. Appl. Crystallogr. 45, 342–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Konarev P. V., Volkov V. V., Sokolova A. V., Koch M. H., and Svergun D. I. (2003) PRIMUS: a Windows PC-based system for small-angle scattering data analysis. J. Appl. Crystallogr. 36, 1277–1282 [Google Scholar]
- 58. Kvansakul M., and Czabotar P. E. (2016) Preparing samples for crystallization of Bcl-2 family complexes. Methods Mol. Biol. 1419, 213–229 [DOI] [PubMed] [Google Scholar]
- 59. Kabsch W. (2010) XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Evans P. (2006) Scaling and assessment of data quality. Acta Crystallogr. D Biol. Crystallogr. 62, 72–82 [DOI] [PubMed] [Google Scholar]
- 61. Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., et al. (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. McCoy A. J. (2007) Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr. D Biol. Crystallogr. 63, 32–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Friberg A., Vigil D., Zhao B., Daniels R. N., Burke J. P., Garcia-Barrantes P. M., Camper D., Chauder B. A., Lee T., Olejniczak E. T., and Fesik S. W. (2013) Discovery of potent myeloid cell leukemia 1 (Mcl-1) inhibitors using fragment-based methods and structure-based design. J. Med. Chem. 56, 15–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Emsley P., Lohkamp B., Scott W. G., and Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Afonine P. V., Grosse-Kunstleve R. W., Echols N., Headd J. J., Moriarty N. W., Mustyakimov M., Terwilliger T. C., Urzhumtsev A., Zwart P. H., and Adams P. D. (2012) Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 68, 352–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Morin A., Eisenbraun B., Key J., Sanschagrin P. C., Timony M. A., Ottaviano M., and Sliz P. (2013) Collaboration gets the most out of software. Elife 2, e01456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Meyer P. A., Socias S., Key J., Ransey E., Tjon E. C., Buschiazzo A., Lei M., Botka C., Withrow J., Neau D., Rajashankar K., Anderson K. S., Baxter R. H., Blacklow S. C., Boggon T. J., et al. (2016) Data publication with the structural biology data grid supports live analysis. Nat. Commun. 7, 10882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Notredame C., Higgins D. G., and Heringa J. (2000) T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 302, 205–217 [DOI] [PubMed] [Google Scholar]
- 69. Castresana J. (2000) Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17, 540–552 [DOI] [PubMed] [Google Scholar]
- 70. Anisimova M., and Gascuel O. (2006) Approximate likelihood-ratio test for branches: a fast, accurate, and powerful alternative. Syst. Biol. 55, 539–552 [DOI] [PubMed] [Google Scholar]
- 71. Guindon S., and Gascuel O. (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52, 696–704 [DOI] [PubMed] [Google Scholar]
- 72. Chevenet F., Brun C., Bañuls A. L., Jacq B., and Christen R. (2006) TreeDyn: towards dynamic graphics and annotations for analyses of trees. BMC Bioinformatics 7, 439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Dereeper A., Guignon V., Blanc G., Audic S., Buffet S., Chevenet F., Dufayard J. F., Guindon S., Lefort V., Lescot M., Claverie J. M., and Gascuel O. (2008) Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36, W465–W469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Whitten A. E., Cai S., and Trewhella J. (2008) MULCh: modules for the analysis of small-angle neutron contrast variation data from biomolecular assemblies. J. Appl. Crystallogr. 41, 222–226 [Google Scholar]