Abstract
The insulin-like growth factors IGF1 and IGF2 are closely related proteins that are essential for normal growth and development in humans and other species and play critical roles in many physiological and pathophysiological processes. IGF actions are mediated by transmembrane receptors and modulated by IGF-binding proteins. The importance of IGF actions in human physiology is strengthened by the rarity of inactivating mutations in their genes and by the devastating impact caused by such mutations on normal development and somatic growth. Large-scale genome sequencing has the potential to provide new insights into human variation and disease susceptibility. Toward this end, the availability of DNA sequence data from 60,706 people through the Exome Aggregation Consortium has prompted the analyses presented here. Results reveal a broad range of potential missense and other alterations in the coding regions of every IGF family gene, but the vast majority of predicted changes were uncommon. The total number of different alleles detected per gene in the population varied over an ∼15-fold range, from 57 for IGF1 to 872 for IGF2R, although when corrected for protein length the rate ranged from 0.22 to 0.59 changes/codon among the 11 genes evaluated. Previously characterized disease-causing mutations in IGF2, IGF1R, IGF2R, or IGFALS all were found in the general population but with allele frequencies of <1:30,000. A few new highly prevalent amino acid polymorphisms were also identified. Collectively, these data provide a wealth of opportunities to understand the intricacies of IGF signaling and action in both physiological and pathological contexts.
Keywords: functional genomics, genomics, growth factor, insulin, insulin-like growth factor (IGF), IGF binding proteins, IGF receptor, human evolution, human variation, population genetics
Introduction
The insulin-like growth factors, IGF12 and IGF2, are closely related, single-chain, secreted peptides that are essential for normal growth and development in mammals and other vertebrates, play critical roles in many physiological and pathophysiological processes in multiple species (1–4), and promote proliferation, differentiation, and/or survival of a variety of cell and tissue types (5, 6). Along with insulin (INS), the two IGFs define a conserved gene family that is found in many eukaryotes (7–9). Both IGFs exert their biological effects through binding to the IGF1 receptor (IGF1R), a transmembrane ligand-activated tyrosine-protein kinase related to the insulin receptor (INSR) in amino acid sequence and structure (6, 10, 11). IGF2 also can signal after binding to a variant INSR lacking 12 amino acids in its juxtamembrane region (12) and binds with high affinity to the IGF2R, which primarily acts to limit access of IGF2 to signaling receptors through its sequestration, internalization, and degradation (13–15). Along with the cation-dependent mannose 6-phosphate receptor (CDMPR), the IGF2R is additionally one of two receptors responsible for targeting lysosomal enzymes to the lysosome (13, 16).
In extracellular fluid and in the bloodstream, IGFs are normally bound to IGF-binding proteins (IGFBPs), which modulate IGF actions by regulating their half-life and access to cell surface signaling receptors (17–19). The six IGFBPs are secreted molecules of 201–289 amino acids in length that share ∼36% sequence identity (18, 19). Each IGFBP is composed of cysteine-rich conserved NH2- and COOH-terminal domains along with a less conserved central linker segment (18, 20, 21). Many studies have indicated that IGFBPs additionally control biological processes that are potentially independent of their IGF binding properties (18, 22–25).
The other key player in IGF biology is the IGF acid labile subunit (IGFALS), a 578-amino acid protein primarily produced by the liver that forms a ternary complex in the circulation with IGFBP3 or IGFBP5 when the latter binds either IGF1 or IGF2 (26, 27). IGFALS is unrelated in amino acid sequence or structure to other IGF family components and consists of a series of leucine-rich segments of ∼22 residues each flanked by NH2- and COOH-terminal cysteine-rich domains (28). The ternary complex greatly extends IGF half-life in the circulation (26, 27).
Human diseases caused by mutations or changes in levels of IGF family members are uncommon (29, 30). Only a handful of individuals with homozygous or heterozygous IGF1 gene defects have been reported (31–37), with homozygous individuals having very low IGF1 levels, severe intrauterine and post-natal growth deficiency, and other developmental and intellectual abnormalities (31–33), reflecting the multifunctional nature of IGF1 actions (35). A single heterozygous nonsense mutation has been described in the human IGF2 gene in four members of a family with severe pre- and post-natal growth retardation (38). Developmental disorders with altered expression of IGF2 include Silver-Russell syndrome, which is caused by abnormalities on chromosome 11p15 in the region of the parentally imprinted IGF2 locus (39, 40) (the IGF2 gene is normally expressed solely from the paternally derived chromosome (39, 40)). Individuals with Silver-Russell syndrome produce diminished amounts of IGF2, exhibit reduced fetal and post-natal somatic growth, and have a variety of dysmorphic features and bodily asymmetry (39, 40). Conversely, overexpression of IGF2 accompanies Beckwith-Wiedemann syndrome, which is characterized primarily by asymmetric overgrowth (39, 40). A handful of mostly heterozygous amino acid substitution mutations also have been identified in the IGF1R in children who were small for gestational age and who failed to exhibit post-natal “catch-up” growth (41–45). Frameshift and amino acid substitution mutations additionally have been detected in the IGFALS gene in a few children with moderate short stature but no other consistent physiological defects except a lack of IGFALS in the blood (46, 47). To date no disease-associated abnormalities have been described for any of the six human IGFBPs (Ref. 30; see OMIM). Amino acid substitutions have been identified in the IGF2R in a small number of presumably healthy adults (48), and others have been found in several individuals with liver cancer (49).
Population-based genome sequencing has the potential to provide new insights into human variation, disease susceptibility, and evolution (50–52). The recent release of DNA sequence data from >60,500 people via the efforts of the Exome Aggregation Consortium (ExAC) (53–57) has prompted dissection of this information to gain insights into the population genetics of the IGF system. Results reveal a broad range of potential missense and other alterations in the coding regions of every IGF family gene, with the vast majority of predicted changes being uncommon. Taken together, these data will provide new opportunities to understand the intricacies of IGF signaling and action in physiological and pathological contexts.
Results
Allelic variation in IGF family members in humans
ExAC contains DNA sequencing data from the exomes of 60,706 people derived from different population groups from around the world (53). One general conclusion from initial analysis of these 121,412 alleles is that there is substantial variation within the coding regions of genes (53). However, most predicted modifications were found to be uncommon, with more than half being detected in just a single allele and with >99% being observed in <1% of the entire study population (53). The vast majority of the variation found consisted of synonymous changes and amino acid substitutions (53).
Examination of IGF family members in ExAC revealed a potentially wide range of coding variation in their exons, with most of the changes consisting of missense mutations (83–99% of modified alleles, depending on the gene; Tables 1–3). Second most common were alterations in the reading frame, including inserted stop codons (1% to ∼10%, Tables 1–3). The total number of different allelic variants per gene varied over an ∼15-fold range, from 57 for IGF1 to 872 for IGF2R, with genes encoding smaller proteins generally having fewer changes by a factor of 4–16 than those with larger proteins (compare IGF1, IGF2, and IGFBP1–6 with IGF1R, IGF2R, and IGFALS; Tables 1–3). However, when corrected for protein length, the overall range of variation was very similar for 9 of 11 genes (0.28–0.39 non-synonymous changes/codon), with the outliers being IGFBP4 (0.22) and IGFALS (0.58). Moreover, when examined for their prevalence, >97% of the missense alleles were detected in ≤0.1% of the study population, and 99.4% were found in ≤1.0% (Table 4). These results indicate that variation in human IGF family proteins is low in the population and are consistent with the overall conclusions from ExAC as noted above (53).
Table 1.
Human population variation in IGF1, IGF2, and insulin
| Protein | Number of codonsa | Missense and in-frame insertions-deletions | Frame shifts; stop codons | Splicing site changes | Loss of start codon | Loss of stop codon | Total number of different changes | Variants per codon | Total variant alleles in population |
|---|---|---|---|---|---|---|---|---|---|
| IGF1 | 195 | 52 | 2 | 1 | 0 | 2 | 57 | 0.39 | 0.6% |
| IGF2 | 236 | 78 | 4 | 2 | 1 | 0 | 85 | 0.36 | 2.5% |
| INS | 110 | 45 | 3 | 0 | 1 | 0 | 49 | 0.45 | <0.01%b |
a Based on transcripts used in ExAC database.
b Several predicted variants do not correspond with the amino acid sequence of insulin and are not included in the compiled data.
Table 2.
Human population variation in IGF and insulin receptors
| Protein | Number of codonsa | Missense and in-frame insertions-deletions | Frame shifts; stop codons | Splicing site changes | Loss of start codon | Loss of stop codon | Total number of different changes | Variants per codon | Total variant alleles in population |
|---|---|---|---|---|---|---|---|---|---|
| IGF1R | 1367 | 413 | 8 | 4 | 0 | 2 | 427 | 0.31 | 3.3% |
| INSR | 1382 | 316 | 10 | 5 | 0 | 0 | 331 | 0.24 | 2.1% |
| IGF2R | 2491 | 856 | 12 | 4 | 0 | 0 | 872 | 0.35 | 121.3% |
| CDMPR | 277 | 82 | 5 | 4 | 2 | 0 | 93 | 0.34 | 13.4% |
a Based on transcripts used in ExAC database.
Table 3.
Human population variation in IGFBPs
| Protein | Number of codonsa | Missense and in-frame insertions-deletions | Frame shifts; stop codons | Splicing site changes | Loss of start codon | Loss of stop codon | Total number of different changes | Variants per codon | Total variant alleles in population |
|---|---|---|---|---|---|---|---|---|---|
| IGFBP1 | 259 | 79 | 7 | 1 | 0 | 1 | 88 | 0.34 | 36.0% |
| IGFBP2 | 325 | 86 | 6 | 0 | 0 | 0 | 92 | 0.28 | 3.8% |
| IGFBP3 | 297 | 71 | 2 | 0 | 0 | 0 | 73 | 0.27 | 5.0% |
| IGFBP4 | 258 | 55 | 0 | 2 | 0 | 1 | 58 | 0.22 | 0.2% |
| IGFBP5 | 272 | 86 | 1 | 0 | 0 | 0 | 87 | 0.32 | 2.8% |
| IGFBP6 | 240 | 70 | 9 | 1 | 0 | 0 | 80 | 0.33 | 0.6% |
| IGFALS | 643 | 346 | 21 | 1 | 1 | 1 | 370 | 0.58 | 5.0%b |
a Based on transcript used in ExAC database.
b Three predicted major variants do not correspond with the amino acid sequence of IGFALS and are not included in the compiled data.
Table 4.
Prevalence in the population of IGF missense alleles
| Protein | ≤0.01% | ≤0.1% | ≤1 | ≤10% | >10% |
|---|---|---|---|---|---|
| IGF1R | 382 | 25 | 6 | 0 | 0 |
| IGF2R | 805 | 48 | 22 | 1 | 2 |
| IGFBP1 | 72 | 6 | 0 | 0 | 1 |
| IGFBP2 | 80 | 4 | 1 | 3 | 0 |
| IGFBP3 | 63 | 2 | 0 | 0 | 0 |
| IGFBP4 | 52 | 3 | 0 | 0 | 0 |
| IGFBP5 | 78 | 7 | 0 | 1 | 0 |
| IGFBP6 | 62 | 7 | 1 | 0 | 0 |
| IGFALS | 322 | 26 | 6 | 3 | 0 |
Splicing changes at exon-intron and intron-exon junctions, reading frame alterations, and the addition of stop codons each can contribute to loss of protein expression and thus a lack of function. The number of alleles showing these changes was very low among IGF family genes and ranged from 1 to 24 different instances, with most alterations being detected rarely in the population (0.002–0.2% allelic frequency), although IGFBP3, at 0.9%, and IGF2, at 2.0%, were exceptions (Table 5). Similarly, copy number variation, in which all or part of a gene is amplified in the genome, also was very low for IGF family members in the study population, as seen with most genes in ExAC (54), ranging from none for IGFALS to fewer than 7 instances for IGF1, IGF2, IGF1R and IGFBP1–6 to 26 for IGF2R.
Table 5.
Loss of expression alleles in IGF family members
| Protein | Number of alleles | Allelic frequency in population |
|---|---|---|
| IGF1 | 3 | 0.002% |
| IGF2 | 6 | 2.0% |
| IGF1R | 12 | 0.01% |
| IGF2R | 16 | 0.01% |
| IGFBP1 | 8 | 0.2% |
| IGFBP2 | 6 | 0.006% |
| IGFBP3 | 2 | 0.9% |
| IGFBP4 | 2 | 0.002% |
| IGFBP5 | 1 | 0.001% |
| IGFBP6 | 10 | 0.02% |
| IGFALS | 24 | 0.07% |
Population variation in IGF1 and IGF2
IGF1, IGF2, and INS is composed of a family of evolutionarily related secreted proteins (7–9) that bind to similar receptors (6, 10, 12) but are different in the overall topography of their protein precursors. Unlike INS, the progenitors of IGF1 and IGF2 contain COOH-terminal extensions or E domains, which are cleaved from IGF1 or IGF2 by proprotein convertases subsequent to protein secretion (58, 59). It is unknown whether or not there are distinct biological functions of the single E region in the IGF2 precursor or the two E domains of IGF1 (Refs. 60 and 61; see Fig. 1).
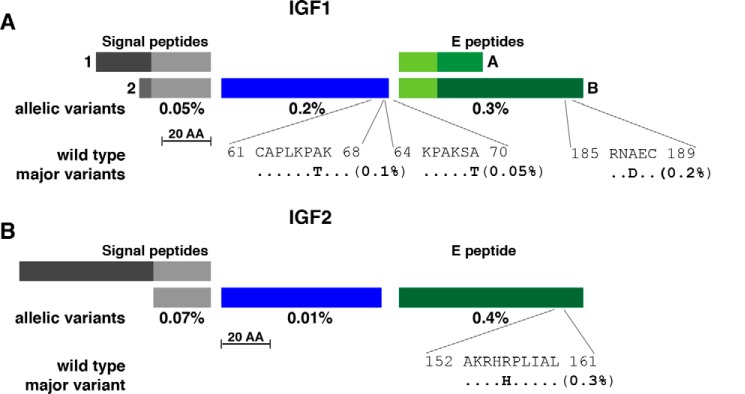
Figure 1.
Population variation in human IGF1 and IGF2. A, schematic of human IGF1 protein precursors, including two alternative signal peptides, a 70-residue mature IGF1 and the two COOH-terminal extension (E) peptides. Signal peptides 1 and 2 consist of 48 and 32 amino acids (AA), respectively, and E regions A and B of 35 and 77 residues, respectively, with the 16 NH2-terminal amino acids being identical. The overall population prevalence of variant alleles for each segment of pre-pro-IGF1 is listed below the map, and the location of the three most common variants is depicted in single-letter amino acid code. B, diagram of human IGF2 protein precursors, including alternative signal peptides, a 67-residue mature IGF2 and the COOH-terminal E peptide. The two signal peptides consist of 80 and 24 amino acids, respectively (the former has not been determined experimentally), and the E region is composed of 89 residues. The overall population prevalence of variant alleles for each segment of pre-pro-IGF2 is depicted below the map, and the location of the most common variant is shown. For A and B, the scale bar represents 20 amino acids.
Illustrated in Table 1 are comparisons of the extent of variation found within the coding segments of their three genes in the 60,706 individuals assessed in ExAC. The overall number of alterations that could change amino acids or modify production of the protein was similar among IGF1, IGF2, and INS, although the total representation of these variant alleles in the population was far lower for INS (<0.01%) than for IGF1 or IGF2 (0.6% and 2.5%, respectively). Amino acid substitutions in the IGF1 precursor were fairly evenly distributed among the signal peptides, 70-residue mature IGF1 and the COOH-terminal E peptides (Fig. 1A), although two alterations in mature IGF1, A67T and A70T, and one in the EB domain, A187D, accounted for most of the variation (Fig. 1A). Of note, there were no individuals identified with either a R36Q or V44M substitution or with a deletion within IGF1 exon 3 corresponding to mutations mapped in three children with severe short stature (31, 33, 35). Thus, if these alleles are present in the population, they have an ultra-low frequency.
Although population variation was 4-fold higher for the IGF2 gene (2.5%) than for IGF1 (0.6%, Table 1), most of this could be attributed to a single non-coding C to T change located 5′ to the adjacent INS gene (see single nucleotide polymorphism (SNP): rs14948363). Unlike what was observed for IGF1, most of the coding variation in IGF2 was concentrated in the COOH-terminal E peptide, with a single amino acid substitution, R157H, accounting for >75% of all changes (Fig. 1B). The functional consequences of this non-conservative change are unknown, because, as noted above, the specific roles of the IGF2 E segment have not been elucidated (60, 61).
Variability in IGF receptors in the population
The IGF1R and INSR are related ligand-activated tyrosine kinases that share a similar length and three-dimensional structure (6, 12). Both receptors bind their eponymous ligands with high affinity and also bind IGF2 with moderately high affinity, although a splicing variant for the INSR lacking the 12 codons of exon 11 (termed INSR-A (12)) binds IGF2 with far higher affinity than INSR-B (12). Population variation appears to occur with comparable frequency for the two receptors (3.3% for IGF1R and 2.1% for INSR; Table 2). Thirteen of 18 of the mutations in the IGF1R gene that have been associated with human growth deficiency syndromes are amino acid substitutions (41, 42). Alterations at 5 of these 13 sites are present in the ExAC database (Table 6), with one change, R511Q, detected in ∼0.15% of the population (Fig. 2A). Five other predicted amino acid substitution variants in the IGF1R gene have allelic frequencies of 0.1–0.7% (Fig. 2A). All six substitutions map to the extracellular part of the receptor protein (Fig. 2A) and collectively compose nearly 60% of the total population variation identified in the IGF1R in ExAC.
Table 6.
Disease-causing mutations in the IGF1R
| Mutation | Population variant | ExAC prevalencea |
|---|---|---|
| R40L | None | – |
| R89stop | None | – |
| E121K | None | – |
| R138Q | R138L | 1 allele |
| A140stop | None | – |
| K145N | None | – |
| E234K | None | – |
| Y387stop | None | – |
| R461L | R461L, R461H | 1, 4 alleles |
| R511W | R511W, R511Q | 1, 174 alleles |
| Y517F | None | – |
| S621L | None | – |
| V629E | V629L, V629M | 1, 1 allele |
| R739Q | None | – |
| E1046Kb | E1046K | 1 allele |
| M1123Rstop | None | – |
| G1155A | None | – |
| 3348_3366 dup19 | None | – |
a The dash represents no alleles identified with that variant.
b Listed incorrectly as E1050K in ExAC.
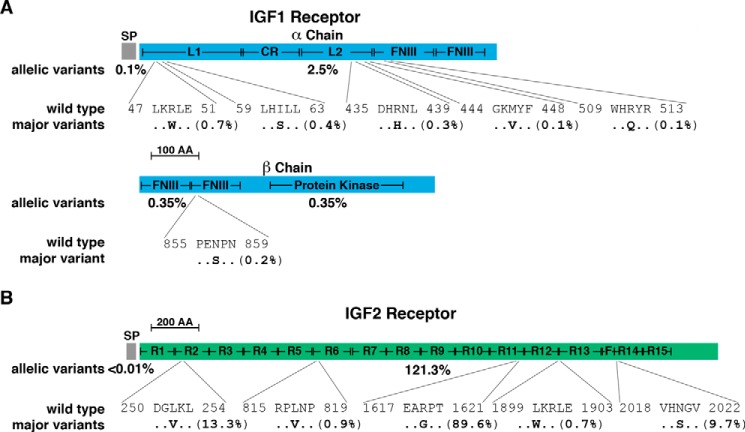
Figure 2.
Population variation in human IGF1 and IGF2 receptors. A, diagram of the human IGF1 receptor precursor, including the 30-residue signal peptide (SP) and the α and β chains of the mature 1337-amino acid (AA) IGF1 receptor protein. The α chain consists of 706 amino acids, and the β chain, 627, with 4 residues lost upon proteolytic cleavage of the precursor protein. Major domains of the mature receptor are indicated (L1 and L2, large domains 1 and 2; CR, cysteine-rich; FNIII, fibronectin type III elements 1–4; Protein Kinase, tyrosine kinase region). The overall population prevalence of variant alleles for each part of the receptor precursor is listed below the map (variation in the tyrosine kinase region is indicated separately), and the location of the six most common variants is depicted in single-letter amino acid code. B, schematic of the human IGF2 receptor precursor, including the 40-residue SP and the 2451-amino acid mature protein. Different domains of the mature receptor are indicated (R1–15, repeating units; F, fibronectin type-II element). The overall population prevalence of variant alleles for each part of the molecule is pictured below the map, and the location of five most common variants is shown. The scale bar represents 100 amino acids in A and 200 in B.
The IGF2R shares structural elements and functions with the CDMPR (13, 16). Both receptors contain units of ∼145 amino acids (15 in IGF2R and 1 in CDMPR), and both proteins are responsible for delivery of lysosomal enzymes from their intracellular sites of synthesis to the lysosome (13, 16), although only the IGF2R appears capable of internalizing these enzymes from the extracellular space (16). IGF2R also binds IGF2 with high affinity, a function that is mediated by repeat number 11 (13, 16).
Both the IGF2R and CDMPR genes appear to be highly polymorphic in the human genome, although the number of variants per codon is only marginally higher than for IGF1R (Table 2). However, in the IGF2R, one predicted amino acid substitution variant, R1619G, located in repeat 11, which contains the IGF2 binding unit, is far more prevalent in the ExAC study population than the amino acid found in the reference IGF2R gene in the Ensembl Genome Browser (∼90% of alleles; Fig. 2B), expanding on a prior observation using a much smaller study population (48). The functional differences between these two presumptive polymorphisms are unknown but now may be tested. Two additional substitutions, L252V, in repeat number 2, found in >13% of the population, and N2020S, in repeat 14, detected in nearly 10% (Fig. 2B), also were noted previously (48). Two other amino acid substitutions in IGF2R found in some individuals with liver cancer, G1449V and G1464E (49), are not present in the ExAC population. There is also variability within IGF2R repeats 3 and 9, the domains responsible for binding lysosomal enzymes, with changes detected in 0.3% and 0.7% of the population, respectively. A similar degree of polymorphism (0.3%) is found in the single repeat element in the CDMPR.
Population aspects of IGFBPs
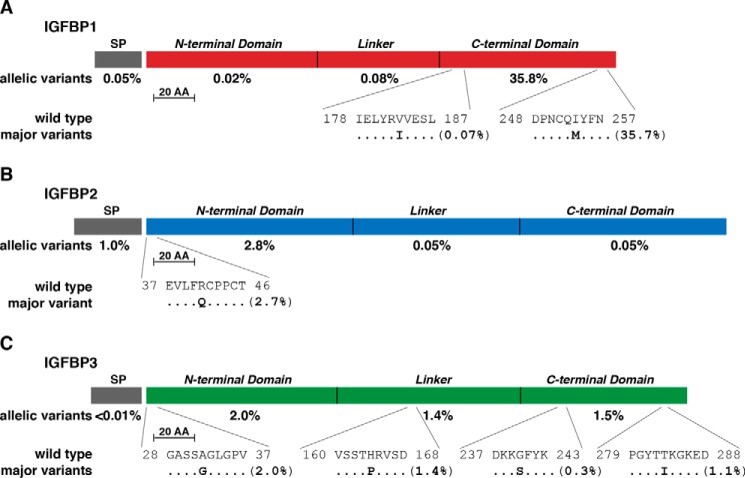
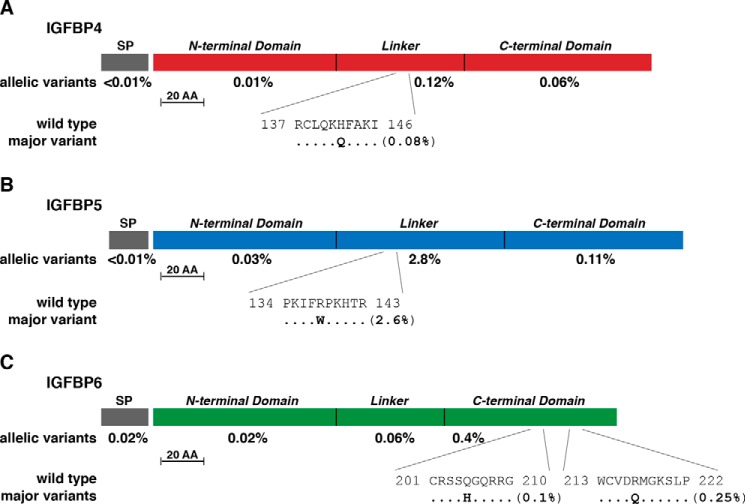
The IGFBP family in mammals consists of six proteins that arose during speciation by duplication and diversification of a progenitor gene (18, 20, 21). All six IGFBPs are secreted proteins that range in length from 201 to 289 amino acids and share a similar 3-domain structure (20, 21) (Figs. 3 and 4). The extent of variability in the exons of the genes encoding IGFBPs is generally similar to that of the two IGFs and two IGFRs, ranging from 0.27 to 0.34 changes per codon, with the exception of IGFBP4 at 0.22 (Table 3). However, total allelic variation ranged from very low in the population (0.2% for IGFBP4, 0.6% for IGFBP6) to moderate (2.8%, 3.8%, and 5.0% for IGFBP5, IGFBP2, and IGFBP3, respectively) to high (IGFBP1 at 36.0%, Table 3). Of note, most of the predicted changes consist of one or a few amino acid substitutions per IGFBP. For IGFBP1, the bulk of the variability is the result of a single change of I253M in the COOH-terminal domain (35.7%, Fig. 3A). For IGFBP2 it is R41Q in the NH2-terminal segment (2.7%, Fig. 3B), and for IGFBP3 it is A32G in the NH2-terminal domain, H164P in the linker, and T284I in the COOH-terminal region (2.0%, 1.4%, and 1.0%, respectively; Fig. 3C). Similarly, for IGFBP5, it is R138W in the linker segment (2.6%, Fig. 4B).
Figure 3.
Population variation in human IGFBPs 1–3. A, diagram of the human IGFBP1 precursor, including the 25-residue SP and mature 234-amino acid (AA) IGFBP1. The overall population prevalence of variant alleles for each part of the molecule is listed below the map, and the location of the two most common variants is depicted in single-letter amino acid code. B, diagram of the human IGFBP2 precursor, including the 36-residue SP and mature 289-amino acid IGFBP2. The overall population prevalence of variant alleles for each part of the molecule is listed below the map, and the location of the most common amino acid variant is depicted in single-letter code. C, diagram of the human IGFBP3 precursor, including the 27-residue SP and mature 264-amino acid IGFBP3. The overall population prevalence of variant alleles for each part of the molecule is listed below the map, and the location of the four most common individual variants is depicted in single-letter amino acid code. The scale bar represents 20 amino acids for A–C. Protein segments for all three IGFBPs include NH2-terminal, linker, and COOH-terminal domains.
Figure 4.
Population variation in human IGFBPs 4–6. A, diagram of the human IGFBP4 precursor, including the 21-residue SP and mature 237-amino acid (AA) IGFBP4. The overall population prevalence of variant alleles for each part of the molecule is listed below the map, and the location of the most common variant is depicted in single-letter amino acid code. B, diagram of the human IGFBP5 precursor, including the 20-residue SP and mature 252-amino acid IGFBP5. The overall population prevalence of variant alleles for each part of the molecule is listed below the map, and the location of the most common amino acid variant is depicted in single-letter code. C, diagram of the human IGFBP6 precursor, including the 24-residue SP and mature 216-amino acid IGFBP6. The overall population prevalence of variant alleles for each part of the molecule is listed below the map, and the location of the two most common individual variants is depicted in single-letter amino acid code. The scale bar in A–C represents 20 amino acids. Protein segments for all three IGFBPs include NH2-terminal, linker, and COOH-terminal domains.
IGFALS is an outlier
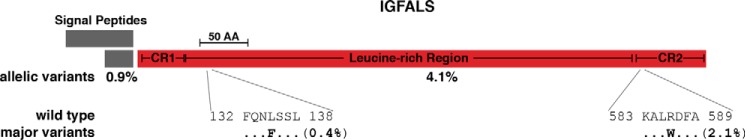
IGFALS, a 578-amino acid secreted liver protein, shares no structural or sequence similarity with IGFBPs, IGF receptors, or IGFs (27, 46). Its physiological role, in forming ternary complexes in the circulation with IGFBP3 or IGFBP5 and IGF1 or IGF2, is to prolong IGF half-life in the blood (27). From the perspective of the population genetics of IGF family members, IGFALS is an outlier, as its overall rate of variation was much higher than each of the other genes analyzed (0.58 non-synonymous changes/codon; Table 3). However, as seen with IGFBPs, 2 amino acid substitutions are responsible for half of the allelic variability in the population for IGFALS: L135F (0.4%) and R586W (2.1%; Fig. 5).
Figure 5.
Population variation in human IGFALS. Diagram of the human IGFALS precursor showing alternative 64 and 27 amino acid (AA) signal peptides (SP) (the former has not been determined experimentally) and the 578-amino acid mature protein. The cysteine-rich domains (CR1 and CR2) and the leucine-rich region are indicated in mature IGFALS. The overall population prevalence of variant alleles for each part of the molecule is listed below the map, and the location of the two most common variants is depicted in single-letter amino acid code. The scale bar represents 50 amino acids.
There are a small number of individuals identified with deficiency of IGFALS who have been ascertained because of moderate growth failure (46, 47), and of these, 10 are single amino acid substitutions, 2 are small in-frame duplications, 4 are frame shifts, and 1 is a stop codon (Table 7). Alterations at 5 of 10 substitution sites, at 1 of 2 internal duplications, at 3 frameshift locations (although 1 is composed of an amino acid substitution), and at the single stop codon are present in the ExAC database at the ultra low frequency of 1–4 alleles in the population, whereas the others are absent (Table 7).
Table 7.
Disease-causing mutations in IGFALS
| Mutation | Population variant | ExAC prevalencea |
|---|---|---|
| E73Gfs-stop | E73Gfs-stop | 1 allele |
| C98S | C98W | 1 allele |
| P111L | P111L, P111G112del | 3, 1 alleles |
| L165P | None | – |
| L172Q | None | – |
| L210F | L210F | 1 allele |
| A221Sfs-stop | A221E, A221V | 2, 2 alleles |
| S233-R235dup | S233-R235dup | 1 allele |
| L279P | None | – |
| L282F | None | – |
| N314S | N314S | 3 alleles |
| W469Gfs-stop | W469Gfs-stop | 2 alleles |
| Q358stop | Q358stop | 1 allele |
| L474–476dup | None | – |
| D478N | D478N | 4 alleles |
| L535fs-stop | None | – |
| C578R | None | – |
a The dash represents no alleles identified with that variant.
Discussion
Data from population-based genome sequencing of >60,500 people from ExAC (53) have been analyzed here to gain insights into the population genetics of the IGF system. Results identify a large number of possible missense alterations and other modifications in the coding regions of all 11 IGF family genes studied. The allelic frequency of the vast majority of these changes was very low, with most found in <0.1% of the population (Tables 4 and 5). However, 3 predicted amino acid substitutions were highly prevalent, as they were identified in 13.3% and 89.6% of IGF2R alleles (Fig. 2B) and in 34.8% of IGFBP-1 alleles (Fig. 3A). These modifications along with others found in the coding regions of IGFBP2 (Fig. 3B), IGFBP3 (Fig. 3C), IGFBP5 (Fig. 4B), and IGFALS (Fig. 5) in several percent of alleles, suggest that fairly common protein sequence variants are present in the population and that they thus have the potential to alter physiologically important aspects of IGF actions.
Limited population variability in IGF1 and IGF2
Coding variation in IGF1 and IGF2 genes is uncommon but is 60–250× more prevalent in the population that was detected in INS (Table 1), possibly reflecting both the essential role of INS in normal metabolic regulation and the important but less critical physiological functions of IGF1 and IGF2 in development and growth. Although population-based polymorphisms are 4× more frequent in IGF2 than in IGF1 genes (Table 1), 80% of the difference (∼2% of all alleles) is caused by a single non-coding G to C alteration in DNA that maps near the adjacent INS gene.
Three presumptive amino acid substitutions in the IGF1 precursor (Fig. 1A) are responsible for more than half of the variation detected in the ExAC population (Table 1). Two of these substitutions, found within mature the 70-residue IGF1, are changes of alanine to threonine at residues 68 and 70 in the D domain of the molecule (Fig. 1A) (7, 8). Although it is possible that one or both of these modifications might alter the stability or another aspect of IGF1 protein synthesis or half-life, the conservative nature and the locations of these modifications suggests that they are unlikely to interfere significantly with the ability of IGF1 to bind to the IGF1R or to IGFBPs. This segment of the protein, composing the last four residues of the eight-amino acid D domain, does not interact with other proteins or even with other parts of IGF1 and was not seen in three-dimensional structural determinations of IGF1 by protein crystallography (62). Substitutions and in-frame deletions occur throughout the IGF1 precursor protein but with allelic frequencies that are so low (from 1:3000 to 1:120,000) that they are unlikely to have a significant population impact on human physiology.
IGF2 presents a picture broadly similar to IGF1 in terms of its population genetics. Most variants are uncommon, except for the non-coding change noted above, although in aggregate the IGF2 gene has the highest frequency of potential loss of function alleles of all IGF family members (Table 5). The most prevalent amino acid substitution in the IGF2 progenitor is located in the COOH-terminal part of the E-domain (Fig. 1B), a section of the molecule with limited known biological function (8, 61). A single truncation mutation in the IGF2 gene has been described in members of a family with severe growth deficiency and physical characteristics resembling Silver-Russell syndrome (38). Equally rare is an amino acid substitution at this same codon, which has been found on a single chromosome in the ExAC study population. The modification, A64P, is non-conservative, and it seems likely that it could significantly perturb IGF2 protein structure.
Rare and common polymorphisms in IGF receptors
Potential variations in amino acid sequence have been detected in nearly a third of the codons for both IGF receptors in the ExAC database, a frequency slightly higher than observed for INSR but similar to the CDMPR (Table 2). For the IGF1R, predicted alterations were found to be 3 times higher in the extracellular α chain than in the transmembrane and intracellular β chain, and 5 substitutions in the α chain accounted for ∼50% of all modified alleles in the population (Fig. 2A). One of these alterations, R437H, corresponds to a heterozygous change found in the IGF1R in a centenarian (63). In limited studies, lymphocytes from this individual were shown to exhibit reduced IGF1-mediated receptor signaling compared with control cells from another centenarian, and it was postulated that diminished IGF1R function caused by this allele might relate to extended lifespan (63). Another missense allele identified by the same investigators, A67T, was only found on 2 of 121,412 chromosomes in the ExAC database and is thus unlikely to have a significant physiological impact in the population.
The kinase domain extends over >60% of the β chain of the IGF1R (Fig. 2A). Substitutions or other modifications are predicted in ExAC for ∼40% of these ∼400 amino acids but are generally uncommon, having a collective population allelic frequency of ∼0.35% (Fig. 2A). Similarly, with one possible exception (R511Q, Table 6), mutations of the IGF1R at residues associated with growth deficiencies are rare, and most have not been detected in the ExAC study population (Table 6).
The IGF2R is a multifunctional protein involved in the clearance of IGF2 from the extracellular space and in the targeting of mannose 6-phosphate-containing lysosomal enzymes to the lysosome (16). Several predicted amino acid substitutions in the IGF2R appear frequently in the ExAC database, with one, R1619G, found in repeating unit 11, the IGF2-binding region (13, 16), detected in nearly 90% of alleles, indicating that it is ∼9× more prevalent in the population than the genomic reference residue (Fig. 2B). The comparative effect of either amino acid on the rate or extent of removal of IGF2 from the extracellular space and thus on IGF2 actions is now worth evaluating.
Predicted amino acid changes are also fairly common in the two domains of IGF2R that are responsible for binding lysosomal enzymes, 0.3% for repeat 3 and 0.7% for repeat 9, as are alterations in the single repeat in the CDMPR (0.3%). As none of this information had been available until now, an opportunity exists to determine whether changes in the sequence and/or structure of either of these proteins might alter lysosomal enzyme targeting and possibly lysosomal activity and function.
Wide range of variation in IGFBPs and IGFALS
The frequency of allelic modifications among the six IGFBP genes in the ExAC study population spans a 180-fold range, from 0.2% for IGFBP4 to 36% for IGFBP1 (Table 3). Only a few amino acid substitutions per IGFBP are responsible for most of this population variability (Figs. 3 and 4). At present, the biological significance of these modifications is unknown, as there are no specific diseases connected to IGFBP gene alterations in humans (Ref. 30; see OMIM). However, because several biochemical parameters of each IGFBP, including protein half-life and binding affinity for IGFs, can potentially influence IGF concentrations in the blood and extracellular fluid and thus modify IGF actions, one or more of these changes in individual IGFBPs could affect specific medical problems, including cancer susceptibility or others (22–25).
IGFALS is dissimilar to any other IGF family member. For example, its structure was shown recently to resemble Toll-like family receptors (28). Its primary function is as a carrier protein in blood for complexes of IGFBP3 and IGFBP5 and either IGF1 or IGF2 (26, 27). The total percentage of alternative alleles for the IGFALS gene in the population is similar to that of several IGFBPs, although the rate of variation is significantly higher (0.58 changes/codon; Table 3). One substitution, R586W, accounting for 40% of all variants, maps to the second cysteine-rich region of IGFALS (Fig. 5) and resides near the COOH terminus of the recently proposed horseshoe-shaped structural model for the protein (28). Mutations in IGFALS are a rare cause of post-natal growth failure (46). Several of these aberrant alleles are present at low frequency in the ExAC population, but half of the amino acid substitutions are absent (Table 7). Most of these changes have been predicted to severely alter IGFALS structure, thus providing a rational basis to understand disease-linked dysfunction (28).
Limitations and implications of population-based genome sequence data on understanding IGF actions in humans
As with any large-scale DNA sequencing endeavor, the ExAC database contains the raw material for novel biological insights as well as both errors and ambiguities. From the perspective of IGF family components, potential problems include the choice of some minor transcripts as the reference mRNA, although this problem can be resolved using databases such as Ensembl. Among the 11 IGF family genes, only IGF2 and IGFALS required explication, as in both cases the assigned mRNA encoded a signal peptide that was much longer than what was used by the major transcripts. More significantly, some of the proposed variants cannot be mapped to the respective gene or protein, as noted for IGFALS (Table 3), in which several identified highly polymorphic amino acid substitutions do not exist in the molecule. Other possible limitations of the data include the fact that even though many population groups are represented, >60% of study subjects are of European ancestry, and only ∼8% are either from Africa or Latin America (53). Thus, the true rate of protein variation among humans may not yet be realized. In addition, as a minor point, there is an unknown error rate associated with nucleotide changes that appears only once in 121,412 alleles evaluated.
Despite these limitations, analysis of these data points to new opportunities to reevaluate normal human IGF physiology and pathology. From a physiological perspective, IGF system components play major roles in the complex interactions that define normal somatic growth in children, including the relationships between genetic and environmental factors (64). The strength, duration, and durability of these interactions and their range of outcomes now may be re-examined in light of the many versions of IGF1, IGF2, IGFBPs and receptors, and IGFALS present in the population. As IGF signaling pathways have also been postulated to be involved in aging (4, 65) and in disease pathogenesis in adults (3, 11, 12, 22), it seems likely that some variants or combinations may enhance susceptibility and others may be neutral or protective.
The extensive variability captured by ExAC should be traceable to our various recent and more distant ancestors, including extinct populations such as Neanderthals and Denisovans (66, 67). The genomes of modern humans contain traces of past contacts with these populations, and they have left us with DNA marks that clearly influence certain traits, such as hair color and skin pigmentation, and possibly certain disease predispositions (67). Although unlikely to occur, an ExAC-sized DNA database for Neanderthals, Denisovans, and other more distant human ancestors could lead to remarkable insights about human origins and how specific genetic influences shape human variation and biology. It is clear that new hypotheses inspired by these data can provide novel insights into the complex biology of IGF actions and those of other proteins in health and disease.
Experimental procedures
Data on variation in human IGF1, IGF2, IGF1, and IGF2 receptors, IGFBPs 1–6, IGFALS, INS, INSR, and the CDMPR were derived from information in the ExAc genome browser, consisting of compiled results from exome sequencing of 60,706 individuals (68). Human transcripts and genes were accessed from the Ensembl Genome Browser using genome assembly, GRCh38. Sources of human protein sequences and domains were the National Center for Biotechnology Information Consensus CDS Protein Set (www.ncbi.nlm.nih.gov) and the UniProt browser. Other databases consulted included Online Mendelian Inheritance in Man (OMIM) and the Growth Genetics Consortium.
Author contributions
P. R. conceived of the study, collected and interpreted the results, and wrote the manuscript.
Acknowledgment
I thank my colleagues for comments on the manuscript.
This work was supported by National Institutes of Health Research Grant R01 DK042748-26 (to P. R.). The author declares that he has no conflicts of interest with the contents of this article. The content is solely the responsibility of the author and does not necessarily represent the official views of the National Institutes of Health.
- IGF
- insulin (INS)-like growth factor
- IGFR
- insulin-like growth factor receptor
- INSR
- insulin receptor
- CDMPR
- cation-dependent mannose 6-phosphate receptor
- IGFBP
- IGF-binding protein
- IGFALS
- IGF acid labile subunit
- ExAC
- Exome Aggregation Consortium
- OMIM
- Online Mendelian Inheritance in Man
- SP
- signal peptide.
References
- 1. Le Roith D., Bondy C., Yakar S., Liu J. L., and Butler A. (2001) The somatomedin hypothesis: 2001. Endocr. Rev. 22, 53–74 [DOI] [PubMed] [Google Scholar]
- 2. LeRoith D. (2008) Clinical relevance of systemic and local IGF-I: lessons from animal models. Pediatr. Endocrinol. Rev. 5, 739–743 [PubMed] [Google Scholar]
- 3. Pollak M. (2012) The insulin and insulin-like growth factor receptor family in neoplasia: an update. Nat. Rev. Cancer 12, 159–169 [DOI] [PubMed] [Google Scholar]
- 4. Gems D., and Partridge L. (2013) Genetics of longevity in model organisms: debates and paradigm shifts. Annu. Rev. Physiol. 75, 621–644 [DOI] [PubMed] [Google Scholar]
- 5. Stewart C. E., and Rotwein P. (1996) Growth, differentiation, and survival: multiple physiological functions for insulin-like growth factors. Physiol. Rev. 76, 1005. [DOI] [PubMed] [Google Scholar]
- 6. Nakae J., Kido Y., and Accili D. (2001) Distinct and overlapping functions of insulin and IGF-I receptors. Endocr. Rev. 22, 818–835 [DOI] [PubMed] [Google Scholar]
- 7. Rotwein P. (1999) in The IGF System (Rosenfeld R. G., and Roberts C. T. Jr., eds) pp. 19–35, Humana Press, Totowa, NH [Google Scholar]
- 8. Wallis M. (2009) New insulin-like growth factor (IGF)-precursor sequences from mammalian genomes: the molecular evolution of IGFs and associated peptides in primates. Growth Horm. IGF Res. 19, 12–23 [DOI] [PubMed] [Google Scholar]
- 9. Schwartz T. S., and Bronikowski A. M. (2016) Evolution and function of the insulin and insulin-like signaling network in ectothermic reptiles: some answers and more questions. Integr. Comp. Biol. 56, 171–184 [DOI] [PubMed] [Google Scholar]
- 10. Kavran J. M., McCabe J. M., Byrne P. O., Connacher M. K., Wang Z., Ramek A., Sarabipour S., Shan Y., Shaw D. E., Hristova K., Cole P. A., and Leahy D. J. (2014) How IGF-1 activates its receptor. Elife 10.7554/eLife.03772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Baserga R. (2013) The decline and fall of the IGF-I receptor. J. Cell Physiol. 228, 675–679 [DOI] [PubMed] [Google Scholar]
- 12. Belfiore A., and Malaguarnera R. (2011) Insulin receptor and cancer. Endocr. Relat. Cancer 18, R125–R1247 [DOI] [PubMed] [Google Scholar]
- 13. Ghosh P., Dahms N. M., and Kornfeld S. (2003) Mannose 6-phosphate receptors: new twists in the tale. Nat. Rev. Mol. Cell Biol. 4, 202–212 [DOI] [PubMed] [Google Scholar]
- 14. Brown J., Jones E. Y., and Forbes B. E. (2009) Interactions of IGF-II with the IGF2R/cation-independent mannose-6-phosphate receptor mechanism and biological outcomes. Vitam. Horm. 80, 699–719 [DOI] [PubMed] [Google Scholar]
- 15. Scott C. D., and Kiess W. (2015) Soluble M6P/IGFIIR in the circulation. Best Pract. Res. Clin. Endocrinol. Metab. 29, 723–733 [DOI] [PubMed] [Google Scholar]
- 16. Kornfeld S. (1992) Structure and function of the mannose 6-phosphate/insulinlike growth factor II receptors. Annu. Rev. Biochem. 61, 307–330 [DOI] [PubMed] [Google Scholar]
- 17. Clemmons D. R. (1997) Insulin-like growth factor binding proteins and their role in controlling IGF actions. Cytokine Growth Factor Rev. 8, 45–62 [DOI] [PubMed] [Google Scholar]
- 18. Bach L. A., Headey S. J., and Norton R. S. (2005) IGF-binding proteins: the pieces are falling into place. Trends Endocrinol. Metab. 16, 228–234 [DOI] [PubMed] [Google Scholar]
- 19. Bach L. A. (2015) Insulin-Like growth factor binding proteins: an update. Pediatr. Endocrinol. Rev. 13, 521–530 [PubMed] [Google Scholar]
- 20. Duan C., and Xu Q. (2005) Roles of insulin-like growth factor (IGF) binding proteins in regulating IGF actions. Gen. Comp. Endocrinol. 142, 44–52 [DOI] [PubMed] [Google Scholar]
- 21. Forbes B. E., McCarthy P., and Norton R. S. (2012) Insulin-like growth factor binding proteins: a structural perspective. Front. Endocrinol. (Lausanne) 3, 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Baxter R. C. (2014) IGF binding proteins in cancer: mechanistic and clinical insights. Nat. Rev. Cancer 14, 329–341 [DOI] [PubMed] [Google Scholar]
- 23. Hoeflich A., and Russo V. C. (2015) Physiology and pathophysiology of IGFBP-1 and IGFBP-2: consensus and dissent on metabolic control and malignant potential. Best Pract. Res. Clin. Endocrinol. Metab. 29, 685–700 [DOI] [PubMed] [Google Scholar]
- 24. Ranke M. B. (2015) Insulin-like growth factor binding-protein-3 (IGFBP-3). Best Pract. Res. Clin. Endocrinol. Metab. 29, 701–711 [DOI] [PubMed] [Google Scholar]
- 25. Bach L. A. (2015) Insulin-like growth factor binding proteins 4–6. Best Pract. Res. Clin. Endocrinol. Metab. 29, 713–722 [DOI] [PubMed] [Google Scholar]
- 26. Baxter R. C. (2001) Changes in the IGF-IGFBP axis in critical illness. Best Pract. Res. Clin. Endocrinol. Metab. 15, 421–434 [DOI] [PubMed] [Google Scholar]
- 27. Baxter R. C. (2001) Inhibition of the insulin-like growth factor (IGF)-IGF-binding protein interaction. Horm. Res. 55, 68–72 [DOI] [PubMed] [Google Scholar]
- 28. David A., Kelley L. A., and Sternberg M. J. (2012) A new structural model of the acid-labile subunit: pathogenetic mechanisms of short stature-causing mutations. J. Mol. Endocrinol. 49, 213–220 [DOI] [PubMed] [Google Scholar]
- 29. Savage M. O., Burren C. P., and Rosenfeld R. G. (2010) The continuum of growth hormone-IGF-I axis defects causing short stature: diagnostic and therapeutic challenges. Clin. Endocrinol. (Oxf) 72, 721–728 [DOI] [PubMed] [Google Scholar]
- 30. Dauber A., Rosenfeld R. G., and Hirschhorn J. N. (2014) Genetic evaluation of short stature. J. Clin. Endocrinol. Metab. 99, 3080–3092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Woods K. A., Camacho-Hübner C., Savage M. O., and Clark A. J. (1996) Intrauterine growth retardation and postnatal growth failure associated with deletion of the insulin-like growth factor I gene. N. Engl. J. Med. 335, 1363–1367 [DOI] [PubMed] [Google Scholar]
- 32. Bonapace G., Concolino D., Formicola S., and Strisciuglio P. (2003) A novel mutation in a patient with insulin-like growth factor 1 (IGF1) deficiency. J. Med. Genet. 40, 913–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Walenkamp M. J., Karperien M., Pereira A. M., Hilhorst-Hofstee Y., van Doorn J., Chen J. W., Mohan S., Denley A., Forbes B., van Duyvenvoorde H. A., van Thiel S. W., Sluimers C. A., Bax J. J., de Laat J. A., Breuning M. B., Romijn J. A., and Wit J. M. (2005) Homozygous and heterozygous expression of a novel insulin-like growth factor-I mutation. J. Clin. Endocrinol. Metab. 90, 2855–2864 [DOI] [PubMed] [Google Scholar]
- 34. van Duyvenvoorde H. A., van Setten P. A., Walenkamp M. J., van Doorn J., Koenig J., Gauguin L., Oostdijk W., Ruivenkamp C. A., Losekoot M., Wade J. D., De Meyts P., Karperien M., Noordam C., and Wit J. M. (2010) Short stature associated with a novel heterozygous mutation in the insulin-like growth factor 1 gene. J. Clin. Endocrinol. Metab. 95, E363–E367 [DOI] [PubMed] [Google Scholar]
- 35. Netchine I., Azzi S., Le Bouc Y., and Savage M. O. (2011) IGF1 molecular anomalies demonstrate its critical role in fetal, postnatal growth and brain development. Best Pract. Res. Clin. Endocrinol. Metab. 25, 181–190 [DOI] [PubMed] [Google Scholar]
- 36. Fuqua J. S., Derr M., Rosenfeld R. G., and Hwa V. (2012) Identification of a novel heterozygous IGF1 splicing mutation in a large kindred with familial short stature. Horm. Res. Paediatr. 78, 59–66 [DOI] [PubMed] [Google Scholar]
- 37. Batey L., Moon J. E., Yu Y., Wu B., Hirschhorn J. N., Shen Y., and Dauber A. (2014) A novel deletion of IGF1 in a patient with idiopathic short stature provides insight Into IGF1 haploinsufficiency. J. Clin. Endocrinol. Metab. 99, E153–E159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Begemann M., Zirn B., Santen G., Wirthgen E., Soellner L., Büttel H. M., Schweizer R., van Workum W., Binder G., and Eggermann T. (2015) Paternally inherited IGF2 mutation and growth restriction. N. Engl. J. Med. 373, 349–356 [DOI] [PubMed] [Google Scholar]
- 39. Eggermann T., Begemann M., Spengler S., Schröder C., Kordass U., and Binder G. (2010) Genetic and epigenetic findings in Silver-Russell syndrome. Pediatr. Endocrinol. Rev. 8, 86–93 [PubMed] [Google Scholar]
- 40. Azzi S., Abi Habib W., and Netchine I. (2014) Beckwith-Wiedemann and Russell-Silver Syndromes: from new molecular insights to the comprehension of imprinting regulation. Curr. Opin. Endocrinol. Diabetes Obes. 21, 30–38 [DOI] [PubMed] [Google Scholar]
- 41. Klammt J., Kiess W., and Pfäffle R. (2011) IGF1R mutations as cause of SGA. Best Pract. Res. Clin. Endocrinol. Metab. 25, 191–206 [DOI] [PubMed] [Google Scholar]
- 42. Walenkamp M. J., Losekoot M., and Wit J. M. (2013) Molecular IGF-1 and IGF-1 receptor defects: from genetics to clinical management. Endocr. Dev. 24, 128–137 [DOI] [PubMed] [Google Scholar]
- 43. Gannagé-Yared M. H., Klammt J., Chouery E., Corbani S., Mégarbané H., Abou Ghoch J., Choucair N., Pfäffle R., and Mégarbané A. (2013) Homozygous mutation of the IGF1 receptor gene in a patient with severe pre- and postnatal growth failure and congenital malformations. Eur. J. Endocrinol. 168, K1–K7 [DOI] [PubMed] [Google Scholar]
- 44. Burkhardt S., Gesing J., Kapellen T. M., Kovacs P., Kratzsch J., Schlicke M., Stobbe H., Tönjes A., Klammt J., and Pfäffle R. (2015) Novel heterozygous IGF1R mutation in two brothers with developing impaired glucose tolerance. J. Pediatr. Endocrinol. Metab. 28, 217–225 [DOI] [PubMed] [Google Scholar]
- 45. Prontera P., Micale L., Verrotti A., Napolioni V., Stangoni G., and Merla G. (2015) A New Homozygous IGF1R variant defines a clinically recognizable incomplete dominant form of SHORT syndrome. Hum. Mutat. 36, 1043–1047 [DOI] [PubMed] [Google Scholar]
- 46. Domené H. M., Hwa V., Jasper H. G., and Rosenfeld R. G. (2011) Acid-labile subunit (ALS) deficiency. Best Pract. Res. Clin. Endocrinol. Metab. 25, 101–113 [DOI] [PubMed] [Google Scholar]
- 47. Poukoulidou T., Kowalczyk J., Metherell L., De Schepper J., and Maes M. (2014) A novel homozygous mutation of the IGFALS gene in a female adolescent: indirect evidence for a contributing role of the circulating IGF-I pool in the pubertal growth spurt. Horm. Res. Paediatr. 81, 422–427 [DOI] [PubMed] [Google Scholar]
- 48. Killian J. K., Oka Y., Jang H. S., Fu X., Waterland R. A., Sohda T., Sakaguchi S., and Jirtle R. L. (2001) Mannose 6-phosphate/insulin-like growth factor 2 receptor (M6P/IGF2R) variants in American and Japanese populations. Hum. Mutat. 18, 25–31 [DOI] [PubMed] [Google Scholar]
- 49. De Souza A. T., Hankins G. R., Washington M. K., Orton T. C., and Jirtle R. L. (1995) M6P/IGF2R gene is mutated in human hepatocellular carcinomas with loss of heterozygosity. Nat. Genet. 11, 447–449 [DOI] [PubMed] [Google Scholar]
- 50. Acuna-Hidalgo R., Veltman J. A., and Hoischen A. (2016) New insights into the generation and role of de novo mutations in health and disease. Genome Biol. 17, 241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Katsanis N. (2016) The continuum of causality in human genetic disorders. Genome Biol. 17, 233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Quintana-Murci L. (2016) Understanding rare and common diseases in the context of human evolution. Genome Biol. 17, 225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lek M., Karczewski K. J., Minikel E. V., Samocha K. E., Banks E., Fennell T., O'Donnell-Luria A. H., Ware J. S., Hill A. J., Cummings B. B., Tukiainen T., Birnbaum D. P., Kosmicki J. A., Duncan L. E., et al. (2016) Analysis of protein-coding genetic variation in 60,706 humans. Nature 536, 285–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ruderfer D. M., Hamamsy T., Lek M., Karczewski K. J., Kavanagh D., Samocha K. E., Exome Aggregation Consortium, Daly M. J., MacArthur D. G., Fromer M., and Purcell S. M. (2016) Patterns of genic intolerance of rare copy number variation in 59,898 human exomes. Nat. Genet. 48, 1107–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zou J., Valiant G., Valiant P., Karczewski K., Chan S. O., Samocha K., Lek M., Sunyaev S., Daly M., and MacArthur D. G. (2016) Quantifying unobserved protein-coding variants in human populations provides a roadmap for large-scale sequencing projects. Nat. Commun. 7, 13293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kruger R. P. (2016) Knockout! Knockout! Who's not there. Cell 167, 289–291 [DOI] [PubMed] [Google Scholar]
- 57. Bahcall O. G. (2016) Genetic variation: ExAC boosts clinical variant interpretation in rare diseases. Nat. Rev. Genet. 17, 584. [DOI] [PubMed] [Google Scholar]
- 58. Duguay S. J., Milewski W. M., Young B. D., Nakayama K., and Steiner D. F. (1997) Processing of wild-type and mutant proinsulin-like growth factor-IA by subtilisin-related proprotein convertases. J. Biol. Chem. 272, 6663–6670 [DOI] [PubMed] [Google Scholar]
- 59. Duguay S. J., Jin Y., Stein J., Duguay A. N., Gardner P., and Steiner D. F. (1998) Post-translational processing of the insulin-like growth factor-2 precursor: analysis of O-glycosylation and endoproteolysis. J. Biol. Chem. 273, 18443–18451 [DOI] [PubMed] [Google Scholar]
- 60. Bikle D. D., Tahimic C., Chang W., Wang Y., Philippou A., and Barton E. R. (2015) Role of IGF-I signaling in muscle bone interactions. Bone 80, 79–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Livingstone C. (2013) IGF2 and cancer. Endocr. Relat. Cancer 20, R321–R339 [DOI] [PubMed] [Google Scholar]
- 62. Brzozowski A. M., Dodson E. J., Dodson G. G., Murshudov G. N., Verma C., Turkenburg J. P., de Bree F. M., and Dauter Z. (2002) Structural origins of the functional divergence of human insulin-like growth factor-I and insulin. Biochemistry 41, 9389–9397 [DOI] [PubMed] [Google Scholar]
- 63. Suh Y., Atzmon G., Cho M. O., Hwang D., Liu B., Leahy D. J., Barzilai N., and Cohen P. (2008) Functionally significant insulin-like growth factor I receptor mutations in centenarians. Proc. Natl. Acad. Sci. U.S.A. 105, 3438–3442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Baron J., Sävendahl L., De Luca F., Dauber A., Phillip M., Wit J. M., and Nilsson O. (2015) Short and tall stature: a new paradigm emerges. Nat. Rev. Endocrinol. 11, 735–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Berryman D. E., Christiansen J. S., Johannsson G., Thorner M. O., and Kopchick J. J. (2008) Role of the GH/IGF-1 axis in lifespan and healthspan: lessons from animal models. Growth Horm. IGF Res. 18, 455–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Jones E. R., Gonzalez-Fortes G., Connell S., Siska V., Eriksson A., Martiniano R., McLaughlin R. L., Gallego Llorente M., Cassidy L. M., Gamba C., Meshveliani T., Bar-Yosef O., Müller W., Belfer-Cohen A., Matskevich Z., et al. (2015) Upper Palaeolithic genomes reveal deep roots of modern Eurasians. Nat. Commun. 6, 8912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Vattathil S., and Akey J. M. (2015) Small amounts of archaic admixture provide big insights into human history. Cell 163, 281–284 [DOI] [PubMed] [Google Scholar]
- 68. Karczewski K. J., Weisburd B., Thomas B., Solomonson M., Ruderfer D. M., Kavanagh D., Hamamsy T., Lek M., Samocha K. E., Cummings B. B., Birnbaum D., The Exome Aggregation Consortium, Daly M. J., and MacArthur D. G. (2017) The ExAC browser: displaying reference data information from over 60 000 exomes. Nucleic Acids Res. 45, D840–D845 [DOI] [PMC free article] [PubMed] [Google Scholar]