Abstract
GTPases of immunity-associated proteins (GIMAPs) are expressed in lymphocytes and regulate survival/death signaling and cell development within the immune system. We found that human GIMAP6 is expressed primarily in T cell lines. By sorting human peripheral blood mononuclear cells and performing quantitative RT-PCR, GIMAP6 was found to be expressed in CD3+ cells. In Jurkat cells that had been knocked down for GIMAP6, treatment with hydrogen peroxide, FasL, or okadaic acid significantly increased cell death/apoptosis. Exogenous expression of GMAP6 protected Huh-7 cells from apoptosis, suggesting that GIMAP6 is an anti-apoptotic protein. Furthermore, knockdown of GIMAP6 not only rendered Jurkat cells sensitive to apoptosis but also accelerated T cell activation under phorbol 12-myristate 13-acetate/ionomycin treatment conditions. Using this experimental system, we also observed a down-regulation of p65 phosphorylation (Ser-536) in GIMAP6 knockdown cells, indicating that GIMAP6 might display anti-apoptotic function through NF-κB activation. The conclusion from the study on cultured T cells was corroborated by the analysis of primary CD3+ T cells, showing that specific knockdown of GIMAP6 led to enhancement of phorbol 12-myristate 13-acetate/ionomycin-mediated activation signals. To characterize the biochemical properties of GIMAP6, we purified the recombinant GIMAP6 to homogeneity and revealed that GIMAP6 had ATPase as well as GTPase activity. We further demonstrated that the hydrolysis activity of GIMAP6 was not essential for its anti-apoptotic function in Huh-7 cells. Combining the expression data, biochemical properties, and cellular features, we conclude that GIMAP6 plays a role in modulating immune function and that it does this by controlling cell death and the activation of T cells.
Keywords: AP1 transcription factor (AP-1), apoptosis, ATPase, GTPase, NF-κB, phorbol ester, T cell activation, immunity
Introduction
GTPases of immunity-associated proteins (GIMAPs),2 also named immune-associated nucleotide-binding proteins (IANs), have been implicated in the regulation of cell survival/death and in the development of lymphomyeloid cells (1, 2). The human GIMAP gene family consists of seven functional members (GIMAP1, GIMAP2, GIMAP4, GIMAP5, GIMAP6, GIMAP7, and GIMAP8) as well as one pseudogene (GIMAP3). These genes are clustered in a 293-kb region of human chromosome 7 (1, 3). Originally these genes were identified in angiosperm plants as being involved in mediating defense responses following bacterial infection (4). Later, GIMAP genes were found to form a novel immunity-associated family that is well conserved among vertebrates (1, 3).
The GIMAP family members are characterized by a common AIG1 domain and the presence of coiled-coil motifs. The AIG1 domain contains a GTP/ATP binding P-loop motif and two functionally undefined motifs: the conserved box and the IAN motif. No GTP or ATP binding has been observed to occur with GIMAP1, but weak GTP binding has been detected with mouse Gimap1 (5). By way of contrast, GIMAP4 binds GDP and GTP specifically and shows GTP hydrolytic activity (6). Recently, an X-ray crystallography study has revealed that GIMAP2 is a monomeric protein and has GDP binding potential (7). On the other hand, GIMAP7 has been shown to be a dimeric protein and to have GTP hydrolysis activity (8). Furthermore, the authors of the latter study have suggested that the GTPase activity associated with the GIMAP family is controlled via homodimerization and/or heterodimerization and that these events may have implications with respect to lymphocyte survival (8).
The function(s) unique to each individual GIMAP gene are not entirely clear. Some GIMAP members have been reported to be involved in cell death/survival. In a transgenic mouse study, deletion of Gimap1 resulted in a severe reduction in peripheral T cell numbers and a profound deficit in mature peripheral B cells (9). The follow-up studies further suggested that Gimap1 is essential for the survival of both activated and naïve peripheral B cells (10) and that Gimap1 is intrinsically critical for maintaining peripheral T cell survival (11). Peripheral T cells from Gimap4-deficient mice have been shown to exhibit a significant delay in cell death when this is induced by serum starvation, γ irradiation, etoposide treatment, or dexamethasone treatment; this suggests that mouse Gimap4 acts as an accelerator of apoptosis in T cells (12). Unlike mouse Gimap4, GIMAP5 and mouse Gimap8 appear to act as apoptosis inhibitors in cell-based assays. GIMAP5 is able to function as an anti-apoptotic effector by protecting Jurkat T cells against okadaic acid and γ radiation (13). Similarly, mouse Gimap8 protects NIH 3T3 and CHO-K1 cells from anisomycin-induced apoptosis (14).
T cell activation and differentiation are important steps in the development of adaptive immunity. Naïve T cells require at least two signals, CD3 and CD28, for activation. Following external stimulation, the secondary messenger 1,2-diacylglycerol and inositol triphosphate are released, and these then activate a complicated series of multiple signaling cascades that orchestrate nuclear factor of activated T cells (NFAT), nuclear factor κB (NF-κB), and activator protein 1 (AP-1) to stimulate IL-2 production. Mammalian GIMAPs are known to be expressed predominantly within lymphomyeloid tissues, indicating a role in lymphocyte functionality and homeostasis (2, 3). Some GIMAPs have been reported to function as key regulators during CD4+ T lymphocytes differentiation. It should be noted that GIMAP1 and GIMAP4 have been found to be differentially regulated during Th1 and Th2 differentiation (15). In 2015, a study reported that GIMAP4 affects T cell differentiation via the regulation of interferon γ secretion (16). Previously, we discovered that the RNA expression of the GIMAP family genes, but not of other neighboring non-GIMAP genes, as detected by a microarray analysis, is uniformly lower in human lung tumor tissue samples (17). This finding was confirmed by quantitative PCR assays using a total of 20 non-small-cell lung carcinoma (NSCLC) samples. We also observed that the expression level of GIMAP genes is higher in adjacent non-tumor tissues compared with lung cancer tumor tissue. This finding indicates that GIMAP genes might be involved in either the pathogenesis of NSCLC or in the immune reaction to NSCLC. Among the GIMAPs, GIMAP6 and GIMAP8 showed the highest normal/tumor expression ratio in lung cancer paired samples (17). Compared with GIMAP8, little is known about GIMAP6 regarding its biological function. In this study, we have focused on GIMAP6 and set out to investigate its function in T cells and to characterize its biochemical properties.
Results
Tissue specificity of GIMAP6 expression
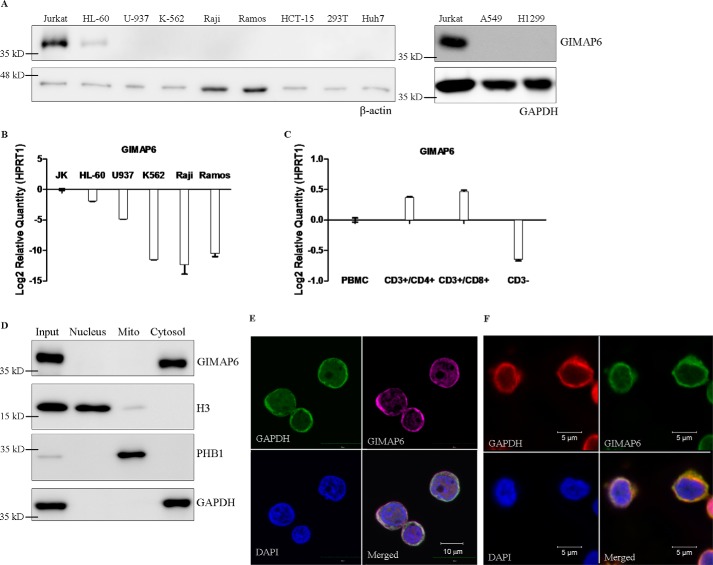
To determine the expression pattern of GIMAP6 in different tissues, we analyzed various cell lines from the hematopoietic system, including T lymphocytes (Jurkat), promyeloblasts (HL-60), monocytes (U-937), lymphoblasts (K-562), B lymphocytes (Raji and Ramos), and various cancer derived cell lines of the colon (HCT-15), kidney (293T), liver (Huh-7), and lungs (A549 and H1299). Immunoblotting and quantitative PCR analysis showed that GIMAP6 was detected primarily in T lymphocytes (Fig. 1, A and B). This contrasts with the expression pattern of mouse Gimap6, which is seen equally in T lymphocytes and B lymphocytes (1). To confirm this finding, we isolated the CD3−, CD3+/CD4+, and CD3+/CD8+ fractions of PBMCs (supplemental Fig. S1) and carried out quantitative PCR assays to determine the expression level of GIMAP6 in these different lymphocyte subsets. As shown in Fig. 1C, both CD3+/CD4+ and CD3+/CD8+ cells showed expression of GIMAP6, whereas CD3− cells were negative. Because CD3− cells within PBMCs are mostly composed of monocytes and B lymphocytes, we concluded that GIMAP6 in humans is predominantly expressed in CD3+ T lymphocytes.
Figure 1.
GIMAP6 expression and protein distribution in T lymphocytes. A, cell lines representing the various different hematopoietic lineages, including T lymphocytes (Jurkat), promyeloblasts (HL-60), monocytes (U-937), lymphoblasts (K-562), and B lymphocytes (Raji and Ramos), as well as cell lines derived from colon (HCT-15), kidney (293T), liver (Huh-7), and lung (A549 and H1299), were analyzed for GIMAP6 protein expression by immunoblot analysis. The amount of protein loaded onto each lane was estimated by anti-β-actin or anti-GAPDH. B, GIMAP6 RNA expression levels in the hematopoietic cell lines were analyzed by quantitative PCR assay. The results were normalized against the expression level of HPRT1. Error bars indicate standard deviation. C, GIMAP6 RNA expression in three subsets of human PBMCs: CD3+/CD4+, CD3+/CD8+, and CD3− cells. These were analyzed by quantitative PCR assay. The PBMCs were obtained from a healthy donor. HPRT1 was used to normalize the dataset. Error bars indicate standard deviation. D and E, cellular distribution of endogenous GIMAP6 in Jurkat T cells. Immunoblot analysis of various protein preparations, including that of the total cell lysate (Input), revealed that endogenous GIMAP6 is restricted to the cytosolic fraction of Jurkat T lymphocyte lysates (D). E, confocal laser-scanning microscopy showed that GIMAP6 (purple, Alexa Fluor 647 goat anti-rabbit IgG) co-localized with the cytosolic marker GAPDH (green, Alexa Fluor 488 goat anti-mouse IgG). DAPI (blue) was used as a nuclear tracking dye. F, cellular distribution of endogenous GIMAP6 in primary CD3+ T cells. Primary CD3+ T cells enriched from PBMCs were used to perform immunofluorescence microscopy, which showed that GIMAP6 (green, Alexa Fluor 488 goat anti-rabbit IgG) co-localized with the cytosolic marker GAPDH (red, Alexa Fluor 594 goat anti-mouse IgG). DAPI (blue) was used as a nuclear tracking dye.
Subcellular localization of GIMAP6 protein
To determine the distribution of GIMAP6 protein in different cellular compartments, subcellular fractionation was performed on Jurkat T lymphocytes to separate the nucleus, the cytosol, and the heavy membrane fraction, with the last containing the mitochondria. Specific antibodies (anti-H3, anti-GAPDH, and anti-PHB1) were used as markers to confirm the purity of the fractions from the different compartments. Immunoblot analysis showed that endogenous GIMAP6 was present primarily in the cytosol of Jurkat T lymphocytes (Fig. 1D). This result is consistent with the results obtained by confocal microscopy, which show that GIMAP6 co-localizes with the cytosolic marker GAPDH (Fig. 1E). Consistent with the result obtained from Jurkat cells, immunofluorescence microscopy of primary CD3+ T cells showed that GIMAP6 co-localized with the cytosolic marker GAPDH (Fig. 1F).
GIMAP6 displays an anti-apoptotic function
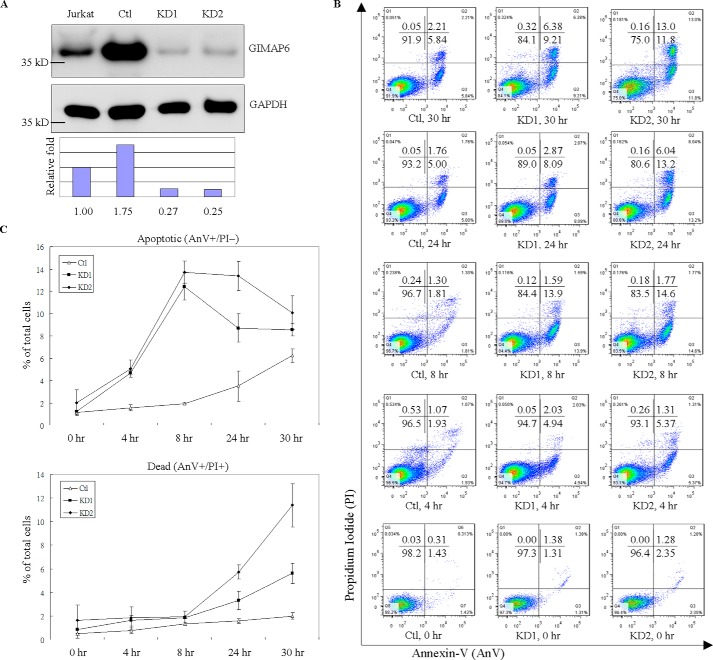
Previous studies have reported that members of the GIMAP family play a role in either cell death or cell survival (1, 12–14, 18). Thus we investigated the possible function of GIMAP6 in the programmed death of T cells by comparing wild-type control cells with GIMAP6-deficient cells. Stable knockdown cells (KD1 and KD2) were generated using anti-GIMAP6 shRNA, whereas control cells (Ctl) were prepared using the pLKO.1 empty vector plasmid. After single colony selection, the relative -fold of GIMAP6 expression was determined by immunoblot analysis (Fig. 2A). To reveal the extent to which cell apoptosis/death was delayed by GIMAP6, an H2O2-mediated, multiple time point apoptosis assay was performed. Various Jurkat-derived cell lines (Ctl, KD1, and KD2) were treated with 100 μm H2O2 for 4, 8, 24, and 30 h to induce cell apoptosis/death (Fig. 2, B and C). The apoptosis signal, phosphatidylserine exposure, was then measured by flow cytometry using a BD Annexin-V apoptosis detection kit. With Annexin-V (AnV)/propidium iodide (PI) staining, GIMAP6 displayed an apparent anti-apoptotic effect, as indicated by the difference between the control and knockdown lines. In these experiments, the apoptotic group (AnV+/PI−) showed a greater increasing trend in knockdown cells compared with control cells after treatment with H2O2 for 4 h, and this difference became more remarkable at 8 h. Compared with knockdown cells, the increase in apoptotic cells (AnV+/PI−) among control cells was postponed until 24 h after treatment with H2O2. In parallel, the cell distribution of knockdown cells, upon being stained with AnV/PI, shifted from AnV+/PI− to AnV+/PI+ after treatment with H2O2 for 24 h. At 30 h, the AnV+/PI− group continued to decrease, whereas the AnV+/PI+ group remained on the increase. This shift in the cell distribution indicates that the cells were progressing from an apoptotic phase to a dead phase. The results also show that GIMAP6 stable knockdown cells are not only more prone to apoptosis than control cells but also that they progress toward the dead phase earlier. Taken together, it can be concluded that GIMAP6 protects cells from apoptosis by delaying apoptosis progress when a H2O2-mediated apoptosis assay is carried out with Jurkat T cells.
Figure 2.
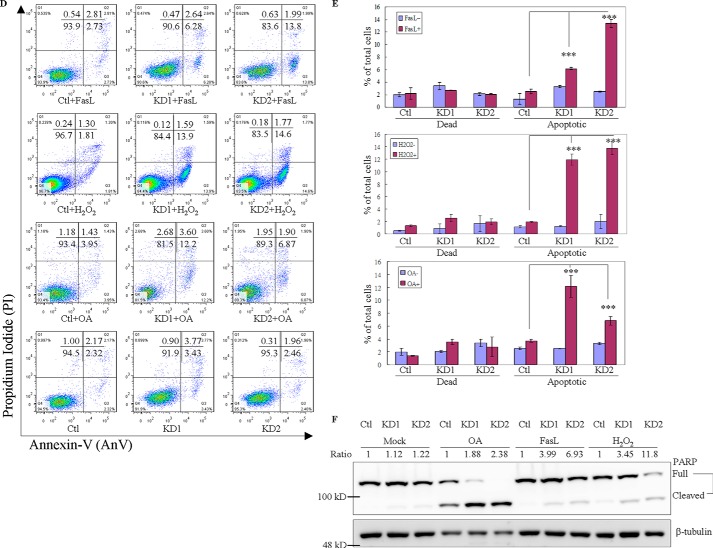
GIMAP6 functions as an anti-apoptotic effector in Jurkat T lymphocytes. A, reduction of GIMAP6 protein in Jurkat T lymphocytes by anti-GIMAP6 shRNA knockdown was confirmed by immunoblot analysis. The Ctl expressed a relatively higher level of GIMAP6 compared with the two knockdown lines (KD1 and KD2) or even compared with the parental Jurkat T cell (Jurkat). The relative -fold expression levels were calculated as GIMAP6/GAPDH ratio, with the relative signal intensity in the Jurkat lane taken as 1. B, knockdown of GIMAP6 accelerated cell apoptosis/death in a H2O2-mediated multiple time point apoptosis assay. Jurkat-derived cell lines (Ctl, KD1, and KD2) were treated with 100 μm H2O2 for 4, 8, 24, and 30 h to induce cell apoptosis/death. Cells were then harvested and stained with Annexin V-FITC/PI, followed by flow cytometry analysis (Fig. 2B). The quantitative results are shown in C. Cells that stained Annexin-V+/PI− were considered to be apoptotic cells, whereas cells that were Annexin-V+/PI+ were considered to be dead cells. The data represent the average of two independent experiments with three triplicate measurements (mean ± S.D.). D, the effects of GIMAP6 on H2O2-mediated, FasL-mediated, and OA-mediated apoptosis. Jurkat-derived cell lines were treated with either 100 μm H2O2 for 8 h, 8 ng/ml FasL for 3 h, or 50 nm OA for 6 h to induce cell apoptosis. Cells were then harvested and stained with Annexin V-FITC/PI, followed by flow cytometry analysis. The quantified results are shown in E. The results represent the average of at least two independent experiments with triplicate measurements (mean ± S.D.; ***, p < 0.001, Student's t test). F, similarly, the various cell lines were treated with apoptosis-inducing agents for 24 h and then analyzed by immunoblot analysis to identify the full-length and cleaved form of PARP. The PARP cleavage ratio was calculated as cleaved/(cleaved + full-length) PARP and normalized against β-tubulin. Relative signal intensity (Ratio) in the Ctl lane for each condition was taken as 1.
To further investigate the anti-apoptotic effect of GIMAP6, a number of apoptosis-inducing agents was used to perform sets of similar experiments with appropriately adjusted incubation times. Specifically, incubation was carried out for 6 h with 50 nm okadaic acid (OA) or for 3 h with 8 ng/ml FasL. These results were compared with 8-h treatment with 100 μm H2O2 (Fig. 2, D and E). Similar to the H2O2-mediated apoptosis assay, GIMAP6 also showed anti-apoptotic effects with OA- and FasL-induced apoptosis. To determine whether this finding is related to downstream activation of apoptosis, an alternative assay was performed. Treated and untreated control cells and GIMAP6 stable knockdown cells were immunoblotted using anti-PARP antibodies. Consistently, after apoptotic induction, the knockdown groups had a higher ratio of the cleaved form of PARP compared with the control group (Fig. 2F). Taken together, these findings indicate that GIMAP6 functions as an anti-apoptotic effector in Jurkat T lymphocytes.
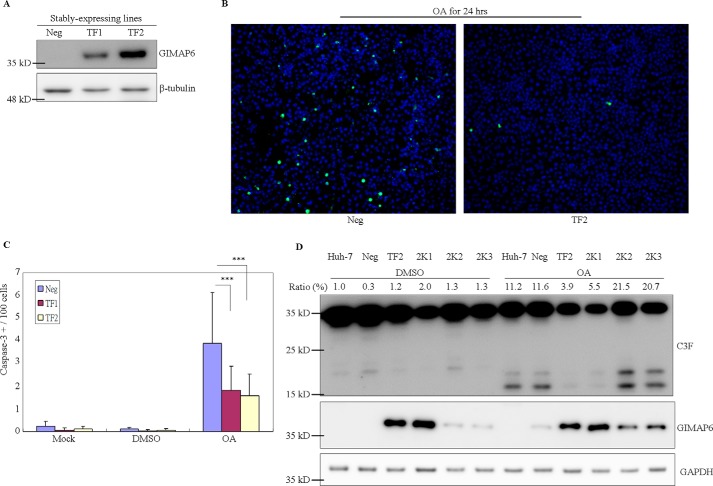
We also used an overexpression approach to demonstrate the anti-apoptotic function of GIMAP6. After stable transfection of the cell line Huh-7, which does not express GIMAP6, single colonies were picked, and the exogenous expression of GIMAP6 was confirmed by immunoblotting (Fig. 3A). Cells expressing no (Neg), a low level (TF1), and a high level (TF2) of GIMAP6 were then incubated with various cell death inducers; namely, 50 nm okadaic acid, 50 μm etoposide, 10 ng/ml FasL, and 200 μm H2O2. The results were compared with control cells (2% DMSO) after 24 h. It was found that Huh-7 cells show a high resistance to etoposide, FasL, and H2O2, and therefore okadaic acid was used as the apoptosis inducer during subsequent experiments (data not shown). An apoptotic marker, the cleaved form of caspase-3, was measured by high-throughput immunofluorescence (Fig. 3B). Under these conditions, protection against okadaic acid-induced apoptosis was observed in cells overexpressing GIMAP6, both TF1 and TF2 (Fig. 3C). To confirm that this anti-apoptotic effect in the presence of okadaic acid was attributed to GIMAP6, we knocked down GIMAP6 using shRNA treatment in Huh-7 cells expressing large amounts of GIMAP6 (TF2). The knockdown was able to partially restore the sensitivity of these cells to the inducing agent. An increase in the cleaved form of caspase-3 was also observed in the knockdown cell lines (2K2 and 2K3) after okadaic acid treatment compared with the high-level GIMAP6-expressing cell lines (TF2 and 2K1) (Fig. 3D). Thus, it can be concluded that GIMAP6 displays an anti-apoptotic effect during these transfection assays and that GIMAP6 is sufficient to protect non-lymphoid Huh-7 cells against okadaic acid-induced apoptosis.
Figure 3.
Exogenous GIMAP6 displays anti-apoptotic effects in Huh-7 stable transfectant cells. A, the GIMAP6 expression level in Huh-7 derivative cells was assessed by immunoblot analysis. After transfection with a recombinant pcDNA3.1(+) vector carrying the GIMAP6 sequence, stably expressing single colonies were picked under G418 selection (500 μg/ml) for 2 weeks. Three drug-resistant clones (Neg, TF1, and TF2) were analyzed for GIMAP6 expression, and these clones were found to show substantial variation in protein levels by immunoblot analysis. B and C, the effect of GIMAP6 on OA-mediated cell apoptosis. B, cells were treated with OA for 24 h, and the level of immunofluorescence staining was analyzed. The cleaved form of caspase-3 (Cell Signaling Technology, 9661S) was used as an apoptotic marker (green), and DAPI (blue) was used as a counterstain as well as a total cell dye. C, the GIMAP6-negative cell line (Neg) was found to have a relatively higher proportion of caspase-3-positive cells compared with the GIMAP6-expressing cell lines (TF1 and TF2). The data represent the average of two independent experiments (mean ± S.D.). ***, p < 0.001, Student's t test. D, GIMAP6 knockdown restored the apoptosis-prone phenotype of the stably expressing cells. The stably expressing cell line (TF2) was transfected with anti-GIMAP6 shRNA to knock down the exogenously expressed GIMAP6. Single colonies (2K1, 2K2, and 2K3) were picked after puromycin selection (2 μg/ml) for 1 week. The result showed that the OA effect on low-GIMAP6-expressing cells (2K2 and 2K3) was restored to the original apoptosis-prone status; this should be compared with the situation when high-GIMAP6-expressing cells (TF2 and 2K1) were subjected to the same treatment procedure. C3F is an antibody that recognizes both the full-length and cleaved form of caspase-3 (GeneTex, GTX110543). The caspase-3 cleavage percentage (Ratio %) was calculated as cleaved/(cleaved + full-length) caspase-3.
GIMAP6 has GTP/ATP hydrolysis activity
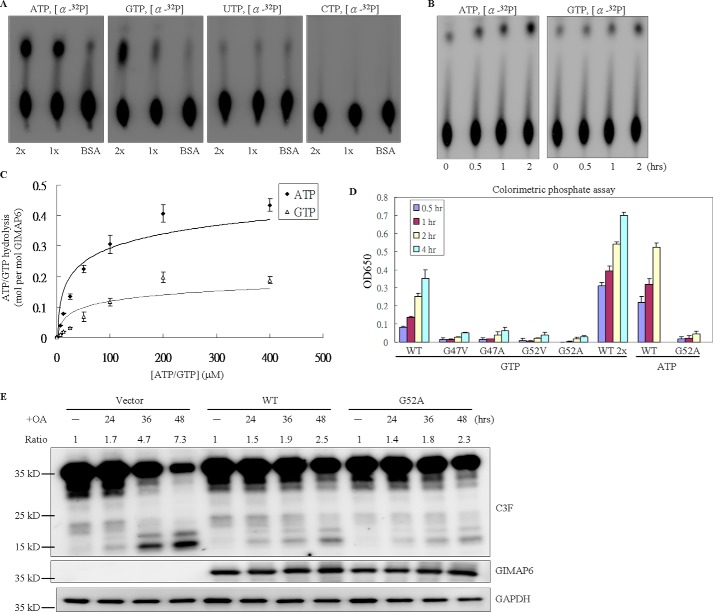
All GIMAP family members are grouped together within the P-loop NTPase superfamily (NCBI Position-specific Scoring Matrix (PSSM) ID 214148) because of the presence of the AIG1 GTP-binding domain (NCBI Conserved Domain Database (CDD) cd01852) (19). A previous study has revealed that some GIMAP family members are able to bind GTP, whereas others have GTP hydrolysis activity (5, 6, 8). Therefore, the three-dimensional structure of GIMAP6 was predicted from the protein sequence using PHYRE, a structure prediction tool. The prediction result showed that GIMAP6 would seem to have a structure very similar to those of GTP-binding proteins with NTP hydrolase activity. Because GTP hydrolysis plays an important role in regulating the activity of GTP-binding proteins, it is important to measure the GTPase activity of GIMAP6. To characterize the biochemical properties of GIMAP6, GTP hydrolysis assays were performed using two different methods, the traditional isotope-labeled NTP hydrolysis assay and the non-radioactive colorimetric phosphate detection assay. The traditional isotope-labeled NTP hydrolysis assay uses α-32P-labeled ATP, GTP, UTP, and CTP as substrates when performing the experiments. The results showed that GIMAP6 exhibits intrinsic GTPase activity and that this activity is dependent on the presence of Mg2+ ions (data not shown). To our surprise, GIMAP6 also showed ATP hydrolysis activity (Fig. 4A). A time course assay of ATP and GTP hydrolysis by GIMAP6 was then carried out to confirm this finding (Fig. 4B). This confirmed that GIMAP6 is the first protein in the GIMAP family to have ATPase activity. Even though such a finding has never been reported previously for the GIMAP family, the coexistence of GTP and ATP hydrolysis activity does occur in some other P-loop NTPase superfamily members (20–22). To further investigate the enzyme kinetics of GIMAP6 and to determine the turnover number and the Michaelis-Menten constant of the enzyme, saturation assays were performed (Fig. 4C). The Kcat was calculated as Kcat = Vmax/[E], and the ratio Kcat/Km was used to monitor enzyme efficiency. We calculated that, for ATP, Kcat was 0.695 (1/min), Km was 104.44 (μm), and Kcat/Km for ATP was 111 (M−1s−1). For GTP, Kcat was 0.102 (1/min), Km was 46.38 (μm), and Kcat/Km for ATP was 36.3 (M−1s−1). Thus, we concluded that the recombinant GIMAP6 had a higher affinity to GTP but showed a higher catalytic rate to ATP.
Figure 4.
Hydrolysis activity of GIMAP6 for ATP and GTP is not associated with the antiapoptotic function. A, the intrinsic hydrolysis activity of the indicated protein was measured in the presence of 2 mm MgCl2 for 2 h at 37 °C by TLC analysis. Specifically, 33 nm of α-32P-labeled ATP, GTP, UTP, and CTP were used separately as substrates, and then hydrolysis activity was measured by TLC. 1x and 2x indicate the relative amounts of GIMAP6 and BSA, used as the control. B, ATP and GTP hydrolysis activity time course. The reaction was incubated at 37 °C. At the indicated time points, a sample of the reaction product (0.5 μl) was removed and stopped by adding 250 μm EDTA, followed by incubation at 95 °C on a heating block for 5 min. C, rate of ATP/GTP hydrolysis by GIMAP6 in relation to the ATP/GTP concentration. To obtain the Kcat data, a saturation curve was obtained by using increasing substrate concentrations under hydrolysis conditions when there was a constant concentration of GIMAP6. Rates of ATP/GTP hydrolysis by GIMAP6 were determined by Lineweaver-Burk plot. D, the GTP/ATP hydrolysis activity of various GIMAP6 mutants (G47V, G47A, G52V, and G52A) was greatly reduced compared with that of the wild-type GIMAP6 protein (WT) using a colorimetric phosphate detection assay. The data represent the average of two independent experiments with triplicate measurements (mean ± S.D.). E, the anti-apoptotic effect of the G52A mutant compared with that of the wild-type protein. A transient transfection assay was conducted using Huh-7 cells and pcDNA3.1(+) (Vector). Apoptosis, which was induced by OA treatment, was measured by immunoblotting against the C3F antibody (which recognizes both the full-length and cleaved form of caspase-3; GeneTex, GTX110543). WT and G52A denote cells transiently expressing wild-type GIMAP6 and G52A mutant protein, respectively. Note that, 36 and 48 h after OA treatment, the expression level of the active-form caspase-3 was reduced to the same level as that of the wild-type GIMAP6 and the G52A mutant. The caspase-3 cleavage was calculated as cleaved/(cleaved + full-length) caspase-3, and the relative signal intensity (Ratio) in the untreated lane (−) for each group was taken as 1.
The anti-apoptotic function of GIMAP6 is independent of the GTP/ATP hydrolysis activities of the enzyme
To study whether the GTPase/ATPase activity of GIMAP6 is required for the anti-apoptotic function of the enzyme, we generated four mutant enzymes (G47V, G47A, G52V, and G52A) in which GTP/ATP binding by the P-loop motif should be absent. A previous study has shown that site-directed mutagenesis in either of the two glycine residues of the P-loop is able to greatly diminished enzyme activity by reducing the affinity of the enzyme for GTP and/or ATP (20, 23–25). Using these mutants with reduced GTP/ATP binding, it was found that the GTP/ATP hydrolysis activity of GIMAP6, as measured by the non-radioactive colorimetric phosphate assay, had been significantly reduced (Fig. 4D).
To determine the effect of this reduced GTP/ATP hydrolysis activity on the anti-apoptotic function of GIMAP6, the WT and the G52A mutant of GIMAP6 were transiently transfected into Huh-7 cells for 24 h. The cells were then harvested after OA treatment for 24, 36, and 48 h. Compared with cells containing the WT enzyme, no significant difference in the anti-apoptotic effect of GIMAP6 was observed with the G52A mutant. This was determined by measuring the level of active-form caspase-3 present in the cells by immunoblot analysis (Fig. 4E). These findings indicated that, under our assay conditions at least, the anti-apoptotic function of GIMAP6 is independent of the GTP/ATP hydrolysis activity of the enzyme.
Knockdown of GIMAP6 accelerates PMA/ionomycin-induced T cell activation
T cell activation and differentiation play a major role in adaptive immunity. Small GTPases are known to be involved in these specific processes. For example, GIMAP1 and GIMAP 4 have been reported to be differentially regulated during CD4+ T helper differentiation (15). In this study, we have demonstrated that PMA/ionomycin-induced T cell apoptosis and activation is modulated by the level of expression of GIMAP6 (Fig. 5). Ctl and knockdown (KD1 and KD2) cells were treated with PMA/ionomycin for 24 h, and the cells were harvested and stained with Annexin-V-FITC to detect apoptosis (Fig. 5, A and B) and with CD69 to detect T cell activation (Fig. 5, A and C). Additionally, culture medium was collected to allow the measurement of IL-2 secretion (Fig. 5D).
Figure 5.
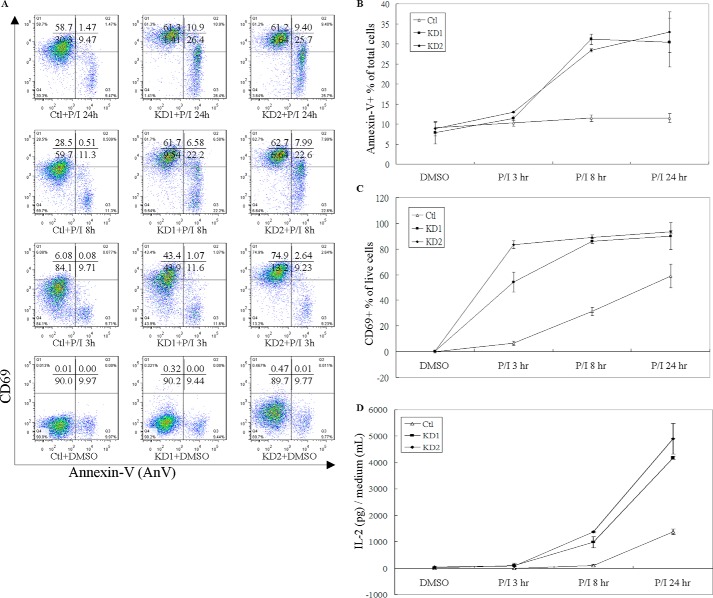
GIMAP6 knockdown accelerates the PMA/ionomycin induction of T cell activation. A, Jurkat-derived cell lines (Ctl, KD1, and KD2) were treated with either PMA/ionomycin or an equal concentration of DMSO to determine the effect of GIMAP6 on T cell activation. To monitor the rate of cell apoptosis/death and T cell activation, PMA/ionomycin-treated cells were harvested at 3, 8, and 24 h and stained for the phosphatidylserine exposure marker Annexin V-FITC and for the T cell activation surface marker CD69-PE. The results were analyzed and quantified by FlowJo 7.6.1. B, the effect of GIMAP6 expression on PMA/ionomycin-induced cell death. All AnV+ cells were taken as apoptotic/dead cells, and the remaining AnV− cells were taken as live cells. C, the effect of GIMAP6 expression on PMA/ionomycin-induced T cell activation. To discriminate activated T cells from inactivated T cells across all live cells, the surface marker CD69 was used to calculate the percentage of CD69+/AnV−. D, the effect of GIMAP6 expression on PMA/ionomycin-induced IL-2 secretion. The data represent the average of at least two independent experiments with triplicate measurements (mean ± S.D.). P/I indicates that the cells were treated with both PMA (10 ng/ml) and ionomycin (1 μg/ml).
Consistent with the finding that GIMAP6 shows an anti-apoptotic function after cells are treated with H2O2, FasL, and OA (Fig. 4), GIMAP6 displayed similar results in terms of a T cell activation assay when T-cell receptor (TCR)-independent stimulation with PMA/ionomycin was carried out. Knockdown (KD1 and KD2) cells showed a higher AnV+ percentage than control cells (Fig. 5A). This difference became apparent after treatment with PMA/ionomycin for 8 h and was sustained until 24 h after treatment. Also agreeing with the above, high GIMAP6-expressing control cells remained resistant to PMA/ionomycin-induced apoptosis/death (Fig. 5B). In parallel, we also measured the T cell activation ratio by detecting the surface marker CD69 (Fig. 5C). After excluding any apoptotic/dead cells (AnV+), we observed that GIMAP6 knockdown cells (KD1 and KD2) showed a relatively high activation ratio (CD69+) among the live cells (AnV−). The effect of GIMAP6 expression on T cell activation became apparent after treatment with PMA/ionomycin for 3 h. At 8 h, nearly 90% of live knockdown cells were activated; that is, they had become CD69+. By way of contrast, the control cells showed only 60% of cells activated after treatment for 24 h. Thus we conclude that GIMAP6 knockdown is able to accelerate T cell activation in this experimental system.
We also determined the level of cytokine IL-2 secretion as a measure of T cell activation (Fig. 5D). IL-2 is a potent growth factor and plays an important role in T cell activation and homeostasis (26). In this experiment, IL-2 secretion by GIMAP6 knockdown cells became detectable after treating the cells with PMA/ionomycin for 8 h. We were only able to observe IL-2 secretion after 24 h in control cells. Moreover, the IL-2 concentration released by the knockdown cells was nearly 4-fold higher than that of the control cells. These results totally agree with the surface marker CD69 staining results; both indicate that knockdown of GIMAP6 accelerates the activation of T cells. Our findings are consistent with previous studies showing that IL-2 acts as a sensitizer and renders T cells susceptible to the induction of apoptosis during T cell activation and proliferation (27–29). This might help to explain why we have observed a high apoptotic/dead ratio in high IL-2-secreting GIMAP6 knockdown cells (Fig. 5, B and D).
GIMAP6 displays anti-apoptotic function in PMA/ionomycin-induced T cell activation through the phosphorylation of NF-κB p65
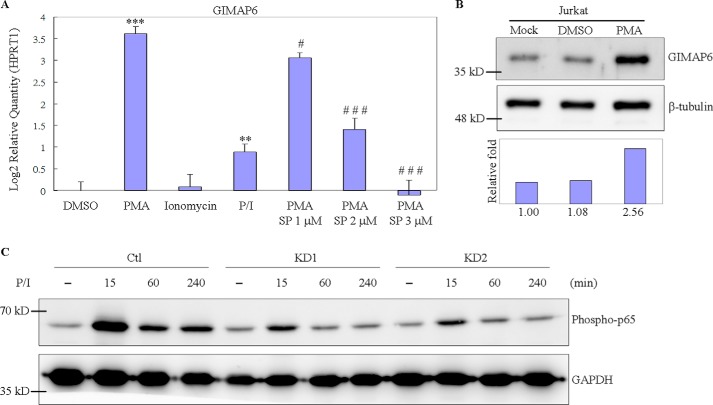
To investigate whether the anti-apoptotic protein GIMAP6 plays a role in T cell functioning, PMA and/or ionomycin were used to induce T cell activation. We found that long-term exposure of Jurkat T cell to PMA greatly induced GIMAP6 expression. After treatment for 24 h, the expression level of GIMAP6 increased 8-fold at the RNA level (Fig. 6A). Consistently, the change in the level of protein was 2.56 times higher after 48 h (Fig. 6B). Previous reports have shown that treating Jurkat T cells with PMA results in an up-regulation of AP-1 and a down-regulation of NF-κB activity (30, 31). We speculated that the induction of GIMAP6 might be controlled via either the protein kinase C/AP-1 or the NF-κB signal transduction cascades. To investigate these possibilities, an inhibitor was used to suppress the induction. When AP-1 and NF-κB activity were suppressed in parallel by treating the cells with 1–3 μm SP100030, there was significant suppression of GIMAP6 at the RNA level (Fig. 6A). We further observed that, when cells were treated with SP100030 alone, there was a slight induction of expression of GIMAP6 rather than a suppression of GIMAP6 expression (data not shown). These findings indicate that SP100030 is not able to suppress the endogenous expression of GIMAP6. SP100030 has been reported to be a potent dual inhibitor of AP-1 and NF-κB and has been used to inhibit cytokine production selectively in T cells (30–32). Because both PMA and SP100030 suppress NF-κB activity and AP-1 activity is absolutely necessary for the induction of GIMAP6, it can be concluded that the induction of GIMAP6 expression by PMA occurs via the AP-1 signaling cascade.
Figure 6.
GIMAP6 expression and NF-κB activation in PMA/ionomycin-treated Jurkat cells. A, up-regulation of GIMAP6 in Jurkat T cells was detected after treatment with PMA and/or ionomycin, and the effect of PMA induction was suppressed by the AP-1 inhibitor SP100030. As a control, Jurkat T cells were treated with an equal concentration of DMSO. RNA was harvested at 24 h for quantitative PCR. The expression levels of GIMAP6 relative to HPRT1 are presented as the log2 -fold. Statistical calculation was determined using Student's t test. Significance relative to the DMSO group is indicated as ***, p < 0.001 and **, p < 0.01. Significance relative to the PMA group is indicated as ###, p < 0.001 and #, p < 0.05. SP indicates SP100030 at the indicated concentration. B, cell lysates were also harvested at 48 h for immunoblot analysis. The relative -fold change was calculated as GIMAP6/β-tubulin, and the intensity in Mock was taken as 1. C, reduction of p65 phosphorylation (Ser-536) under the PMA/ionomycin treatment condition is associated with GIMAP6 knockdown. Three Jurkat-derived cells (Ctl, KD1, and KD2) were harvested at the indicated time points after PMA/ionomycin induction.
The NF-κB signal transduction cascade is a key regulator of the life and death of T cells. Loss of NF-κB activation initiates activation-induced cell death (AICD) in T cells (33, 34), which will cause accumulation of reactive oxygen species and suppression of BCL-2 expression, finally leading to the release of BIM and resulting in T cell death through the intrinsic apoptotic pathway (35). In this study, we observed that NF-κB activation is affected by the GIMAP6 expression level in PMA/ionomycin-mediated activation. As shown in Fig. 6C, GIMAP6 knockdown showed a significant decrement of p65 phosphorylation (Ser-536) under the PMA/ionomycin treatment condition compared with control cells. Because loss of NF-κB activation can result in T cell death via apoptosis, the decreased phosphorylation of p65 (Ser-536) in GIMAP6 knockdown cells (KD1 and KD2) might help to explain the high apoptosis/dead ratio observed in the Annexin-V assay (Fig. 5B). Thus, we conclude that GIMAP6 knockdown leads to decreased phosphorylation of p65 (Ser-536) in PMA/ionomycin-stimulated activation, providing a possible mechanism of how GIMAP6 functions as an anti-apoptotic protein.
Activation with PMA/ionomycin is affected by GIMAP6 expression in human primary T cells
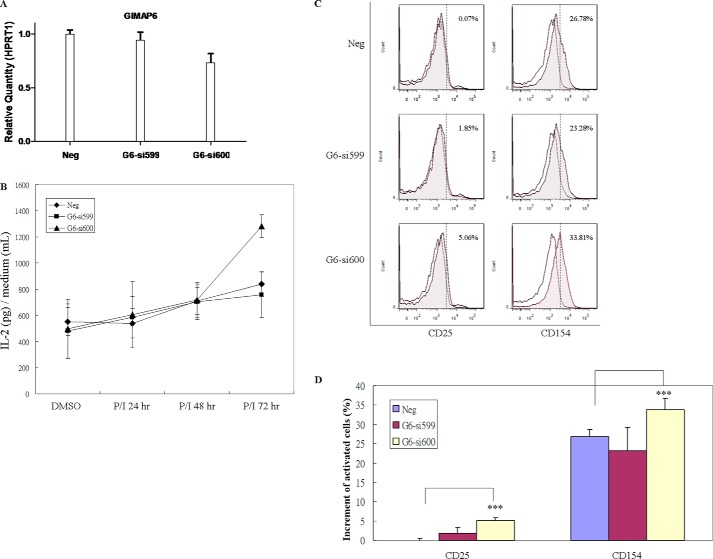
As Jurkat cells are derived through human T cell leukemia/lymphoma virus type I infection and have accumulated multiple genetic mutations that contribute to immortalization (36), we further investigated whether GIMAP6 knockdown had an effect on PMA/ionomycin-induced activation in human primary T cells. To this end, CD3+ T cells were enriched from PBMCs (supplemental Fig. S2), and knockdown of GIMAP6 was performed by transfection of either negative control (Neg) or anti-GIMAP6 siRNAs (G6-si599 and G6-si600) into the cells (supplemental Fig. S3). After 48 h, the knockdown efficiency of GIMAP6 was confirmed by quantitative RT-PCR assay. As shown in Fig. 7A, the knockdown efficiency of the anti-GIMAP6 siRNA G6-si600 and G6-si599 group in CD3+ T cells averaged 26.8% and 6%, respectively. Following the conclusion of the analysis in Jurkat cells (Fig. 5), we hypothesized that a higher level of activation should be observed after knocking down GIMAP6 in primary CD3+ T cells. We then measured PMA/ionomycin-induced IL-2 secretion in these three transfectant groups. As shown in Fig. 7B, no significant difference between these three transfectants was observed at 24 and 48 h after treatment with PMA/ionomycin. However, 72 h after stimulation with PMA/ionomycin, IL-2 production enhancement by 53% was observed in the high knockdown efficiency group (G6-si600). Thus, we concluded that the GIMAP6 expression level had an effect on the increased IL-2 production stimulated by PMA/ionomycin.
Figure 7.
Increased IL-2 secretion and CD25 and CD154 surface expression by GIMAP6 knockdown in primary CD3+ T cells after activation with PMA/ionomycin. A, GIMAP6 RNA expression in primary CD3+ T cells after transfection of siRNA; these were analyzed by quantitative PCR assay. CD3+ T cells were enriched from PBMCs, which were obtained from two healthy donors, and knockdown of GIMAP6 was performed by electroporation with either negative control (Neg) or siRNA oligonucleotides against GIMAP6 (G6-si599 and G6-si600). To monitor the efficiency of GIMAP6 knockdown, transfected cells were harvested at 48 h, and total RNA was extracted for the quantitative RT-PCR assay. HPRT1 was used to normalize the dataset. Error bars indicate standard deviation. B, the effect of GIMAP6 expression on P/I-induced IL-2 secretion. Cells were incubated with either P/I or DMSO (as a control) for 72 h. P/I indicates that the cells were treated with both PMA (10 ng/ml) and ionomycin (1 μg/ml). Media were harvested at the indicated times (24, 48, and 72 h). The data represent the average of two independent experiments with triplicate measurements (mean ± S.D.). C, the effect of GIMAP6 expression on the induction of surface antigens. Transfected primary CD3+ T cells were stimulated with either P/I or an equivalent concentration of DMSO for 72 h. Expression of CD25 and CD154 was analyzed using flow cytometry. The gray histograms show the profiles of P/I-treated cells, whereas the white histograms show those of DMSO-treated cells. The percentage in the top right corner of each panel indicates the increment of positive cells. The results were analyzed and quantified by FlowJo 7.6.1 and are shown in D. The data represent the average of two independent experiments with triplicate measurements (mean ± S.D.). ***, p < 0.001, Student's t test.
To confirm that primary T cell activation is sensitive to the GIMAP6 level, we analyzed the PMA/ionomycin-induced expression of the cell surface antigens CD25 (IL-2 receptor α chain) and CD154 (CD40 ligand). Consistent with the IL-2 production level, the high knockdown efficiency group (G6-si600) showed an elevated level of both antigens 72 h after induction (Fig. 7, C and D). These results demonstrate that down-regulation of GIMAP6 leads to enhanced PMA/ionomycin-stimulated activation in various activation signals, supporting the role of endogenous GIMAP6 as a negative regulator of T cell activation.
Discussion
Across species, the biological functions of the GIMAP family generally seem to be related to immune reactions against internal or external stimuli. Although the structural features of the GIMAP proteins are evolutionarily conserved in vertebrates and angiosperm plants, the biochemical and molecular properties underlying various functions of the GIMAP proteins are not fully characterized (2). In this study, we have demonstrated that GIMAP6 is a T cell-associated cytosolic protein with an anti-apoptotic function and that it plays a role in regulating T cell activation.
Apoptosis plays a critical role in T cell development and function. Dysregulation of apoptosis in the immune system results in autoimmunity, tumorigenesis, or immunodeficiency. We have clearly shown that GIMAP6 knockdown not only made cells apoptosis-prone but also accelerated T cell activation when treated with PMA/ionomycin. One possibility is that the reduction of GIMAP6 facilitates apoptosis and speeds up the removal of cells without appropriate signals, and this eventually accelerates T cell activation. Therefore, we speculate that GIMAP6 can function as an anti-apoptotic protein as well as a blockade to T cell activation. That is to say, physiologically, GIMAP6 in the immune system might serve as a T cell preserver to keep cells alive and retard or prohibit reactivation.
Taking functional and biochemical approaches, we have gained new insights about the molecular mechanisms of GIMAP6 in modulating T cell functions. We used three different apoptosis inducers, H2O2, FasL, and OA, to treat cultured cells. Although all three inducers are able to trigger cell death, they appear to act via a number of signaling pathways. Therefore, we concluded that the anti-apoptotic function of GIMAP6 might involve the intrinsic apoptosis pathway. A previous report has indicated that mouse IAN1 (mouse Gimap4) binds to Bax and IAN4 (mouse Gimap3) and that IAN5 (mouse Gimap5) binds to Bcl-2 and Bcl-xL, all of which are directly involved in the apoptosis pathway (1). In an attempt to gain further insights into the anti-apoptotic mechanism of GIMAP6, an antibody array was used to screen interaction with GIMAP6 and binding to GIMAP6. So far, we have found no evidence to support the direct binding of GIMAP6 to any appropriate candidate protein (data not shown). While this study was in progress, we became aware of a publication by Pascall et al. (37) that reports that GIMAP6 interacts with the Atg8 homologue GABARAPL2 and that it is recruited to autophagosomes. The biological significance of the GIMAP6-GABARAPL2 protein-protein interaction remains unclear, and its relevance to controlling cell death in T cell development and activation warrants further investigation.
The balance between activation and apoptosis plays an important role in maintaining T cell homeostasis. We observed an elevated level of apoptosis and activation in GIMAP6 knockdown cells when treated with PMA/ionomycin. Also, the anti-apoptotic function of GIMAP6 is related to NF-κB activation (Figs. 5B and 6C). These findings are in accordance with the knowledge that restimulation of expanded T cells in the absence of co-stimulatory factors can lead to efficient induction of cell death, known as AICD. NF-κB plays an important role in this process, as loss of NF-κB activity results in T cells death through the intrinsic apoptotic pathway (27, 35). In our study, down-regulation of p65 phosphorylation and increase in apoptosis were observed simultaneously in GIMAP6 knockdown cells after treatment with PMA/ionomycin (Figs. 5B and 6C). This implies that phosphorylation of p65 and its downstream cascade AICD is affected by GIMAP6 expression level.
Notably, GIMAP6 is up-regulated in T cells activated by PMA/ionomycin (Fig. 6A), whereas GIMAP6 inhibits T cell activation (Fig. 5). This is expected to lead to an elevated reactivation threshold for activated T cells. It may be envisioned that increased levels of GIMAP6 confer the survival of activated T cells by enhancing resistance to both restimulation and activation-induced cell death. Therefore, GIMAP6 may contribute to T cell immunity through regulating T cell viability during microbe infections and autoimmune diseases. Further studies will help establish the exact physiological role of GIMAP6.
In terms of the biochemical properties of the protein, recombinant GIMAP6 has both GTP and ATP hydrolysis activity. Interestingly, ATPase activity has never been reported previously for any GIMAP protein. ATP usually functions either as a signal molecule for protein phosphorylation via protein kinases (38) or as an energy source for the pumps involved in transporting substances inside/outside of cells (39, 40). It also powers the molecular motors of muscle cells during contraction (41). By way of contrast, GTP can act as a molecular switch, and it is a well known part of signal transduction regulation (42). Even though this function is not common, some proteins that contain the P-loop motif, such as nsP2 of Semliki Forest virus, 2C of poliovirus, p206 of turnip yellow mosaic virus (20, 21, 43), elongation factor 3 of yeast, FtsZ of Escherichia coli, and Rab14 of Bombyx mori (silkworm), have been reported as possessing both ATP and GTP hydrolysis activity (22, 44, 45). All of these proteins, including GIMAP6, contain a consensus sequence of GXXXXGKS/T within the phosphate-binding loop (P-loop) (46, 47). This is the signature of the P-loop containing the nucleotide triphosphate hydrolase superfamily, and it is used during the structural classification of proteins (48, 49).
The elution pattern of GIMAP6 indicates that it is present in cells as a multimer (supplemental Fig. S4). The nature of protein interactions in the GIMAP6 complex remains unclear, and it is reasonable to speculate that such multiprotein interaction(s) might have structural, functional, or regulatory implications (50). As GIMAP6 has been shown to exhibit GTP/ATP hydrolysis activity and has been observed to oligomerize, it is possible to connect these two properties based on previous reports. Many P-loop proteins, including the GTPases atToc33 and IIGP1 and the GIMAP family member GIMAP7 (8, 51, 52), have been shown previously to function as oligomers inside the cell for the purpose of hydrolysis activity. Therefore, further study is required to identify the interaction domain of GIMAP6 and to confirm whether oligomerization is required for the GTP/ATP hydrolysis activity of GIMAP6.
Finally, it appears that GIMAPs, by controlling cell survival and death, are able to modulate the proportion of differentiated cell lineages in the hematopoietic system. Different cell lineages express different levels of GIMAP genes. In addition to the Jurkat cell line, in this study, HL-60 is the only other cell line expressing a detectable level of GIMAP6 (Fig. 1, A and B). HL-60 is a promyelocytic cell line that is derived from peripheral blood leukocytes with a myeloblast morphology; it is often used for studying the molecular mechanism of myeloid differentiation (53–55). The significance of GIMAP6 expression in HL-60 cells remains unclear. Based on the above, we propose that, by turning on and off the GIMAP genes that have pro-apoptotic or anti-apoptotic functions, there is a dynamic regulation of the cell numbers of the various components of the hematopoietic system that make up the immune reaction, and these changes in cell numbers are important during development and disease. Thus, our findings regarding the functional and biochemical properties of GIMAP6 warrant further study to advance the knowledge of how GIMAP proteins maintain lymphocyte homeostasis.
Experimental procedures
Purification of recombinant GIMAP6 protein
The full-length human GIMAP6 (GIMAP6, NM_024711) coding sequence was amplified by PCR using primers containing NdeI and XhoI cutting sites and a commercial cDNA clone (MHS1010-98051901, MCLAB) as a template. The amplified DNA was then cloned using the NdeI and XhoI sites into the vector pET29 (Novagen), which resulted in a coding sequence that contained a six-histidine tag at the C terminus. This plasmid was then transformed into E. coli BL21 (DE3) and expressed in the presence of 1 mm isopropyl 1-thio-β-d-galactopyranoside in LB medium. Purification was performed using a HisTrap HP column (GE Healthcare), which was followed by size exclusion chromatography.
Gel filtration assay
Recombinant GIMAP6 was subjected to size exclusion separation on a Superose 12 10/300 GL prepacked column connected to an AKTA-explorer system (GE Healthcare), which was controlled by the UNICORN software. After automatic sample injection, protein separation was performed using elution buffer (100 mm Tris-HCl and 200 mm NaCl (pH 7.4) at a flow velocity of 0.5 ml/per minute, and 1-ml fractions were collected.
Cells and cell culture
Human cell lines, 293T and Huh-7, were maintained in DMEM (Gibco), whereas the Jurkat cell line was maintained in RPMI 1640 medium (Hyclone). All media were supplemented with 10% fetal bovine serum containing 1% penicillin, 1% streptomycin, and 1% glutamine (10378-016, Gibco).
Plasmid preparation and transfection
The previously described GIMAP6 cDNA clone was amplified and cloned into the pcDNA3.1(+) vector (Invitrogen) using EcoRI and XhoI cutting sites. The expression construct was then transformed into E. coli DH5α and grown in LB medium. Plasmids were prepared using a PureLink plasmid midiprep kit (K2100-05, Life Technologies) and then introduced into the various cell lines using transfection reagent (Turbofect R0531, Fermentas) according to the protocol of the manufacturer.
Immunoblotting
Protein samples were separated by electrophoresis using an appropriate percentage SDS-PAGE gel and transferred onto a polyvinylidene difluoride membrane (Millipore). The membrane was blocked with blocking buffer (5% skimmed milk in TBS-T buffer (10 mm Tris, 150 mm NaCl (pH 7.6), and 0.05% Tween 20)) for 1 h at room temperature. The primary antibody was appropriately diluted into the blocking buffer, and the membrane was then incubated with the mixture overnight at 4 °C. Next the membrane was washed with TBS-T buffer. Finally, diluted horseradish peroxidase-conjugated secondary antibody corresponding to the primary antibody was added to the buffer, and the mixture was incubated for 1 h at room temperature. After washing with TBS-T buffer, the membrane was developed using ECL substrate (WBKLS0500, Millipore) and photographed using an ImageQuantTM LAS 4000 (GE Healthcare).
RNA extraction, RT-PCR, and quantitative RT-PCR
Total RNA samples were prepared using TRIzol reagent (Invitrogen) from the indicated cells, and reverse transcription was carried out using 100 ng of total RNA according to the instructions of the manufacturer (Superscript® III, Life Technologies). Quantitative PCR was performed using TaqMan probe-based gene expression analysis in 48-well plates on a StepOne real-time system (Applied Biosystems). The mRNA levels of the target genes in the samples were normalized against HPRT1 (4326321E, Applied Biosystems). The probe ID for the indicated gene is GIMAP6 Hs00226776_m1.
Generation of anti-GIMAP6 antibodies
To characterize GIMAP6 at the protein level, rabbit polyclonal antibodies were raised against recombinant GIMAP6. Rabbits were inoculated with 500 μg of GIMAP6-His fusion protein in 2 ml of PBS mixed with 2 ml of complete Freund adjuvant. Five boosts of 200 μg of protein in 2 ml of PBS plus 2 ml of incomplete Freund adjuvant were administrated over 3 months. All antisera from the rabbit blood used in this study stained only a single protein band with high intensity at the expected molecular size (35 kDa) in immunoblots against GIMAP6-transfected 293T and Huh-7 cells, secondary Jurkat T cells, and human PBMC cell lysates.
Immunofluorescence microscopy
Jurkat or primary CD3+ T cells were collected by centrifugation and washed three times with PBS. After resuspension in PBS, the cells were then cross-linked onto poly-l-lysine-coated coverslides at 4 °C for 30 min and fixed with paraformaldehyde. Following penetration with ice-cold methanol and blocking with 0.5% BSA in PBS for 30 min at room temperature, the cells were incubated with a 1:500 dilution of rabbit anti-GIMAP6 and mouse anti-GAPDH antibody for 1 h at room temperature. After three washes with PBS, the secondary antibody was added at a 1:500 dilution. Alexa Fluor 647 or 488 goat anti-rabbit IgG (Life Technologies) and Alexa Fluor 488 or 594 goat anti-mouse IgG (Life Technologies) was used. Confocal laser-scanning microscopy was performed using an Olympus FV10i or Zeiss LSM 700.
Nucleotide triphosphatase assays
Nucleotide triphosphatase activity was assessed by either thin-layer chromatography (TLC) or colorimetric phosphate detection. To determine the hydrolysis activities for GIMAP6, α-32P-labeled ATP, GTP, UTP, and CTP (NEG503H, NEG506H, NEG507H, and NEG508H, respectively) were used as substrates. Time course experiments were carried out in a 20-μl reaction mixture containing 50 mm Tris (pH 7.9), 100 mm NaCl, 2.5 mm MgCl2, and 2.43 μm wild-type GIMAP6 containing 33 nm α-32P of either ATP or GTP (1 μCi) (PerkinElmer Life Sciences) and a specific concentration of ATP or GTP. The reaction was incubated at 37 °C. For TLC, 2 μl of the α-32P-labeled reaction was removed at various time points and mixed with 2 μl of solution containing 0.2% SDS and 5 mm EDTA. Samples were incubated at 70 °C for 2 min to stop the reaction. A fraction of the stopped reaction (0.5 μl) from each time point was spotted on a polyethyleneimine cellulose TLC plate (Merck), and the plate was developed in 1 m formic acid and 0.5 m LiCl. The plate was then dried and exposed to a PhosphorImager for quantitative analysis. The rate of ATP/GTP hydrolysis at each nucleotide concentration was calculated and plotted.
To determine the hydrolysis activities of the G47V, G47A, G52V, and G52A mutants, colorimetric phosphate detection was performed by adding 100 μl of malachite green reagent to the non-radioactive reaction mixture and incubated for 30 min at room temperature to develop a green color. A buffer blank was used for background subtraction, and the absorbance at 650 nm was measured to calculate the ATP/GTP hydrolysis rate by interpolation onto a phosphate standard curve.
Detection of apoptosis by FACS analysis
Apoptosis was measured using an apoptosis detection kit (Annexin V-FITC and PI) (BD Biosciences) according to the instructions of the manufacturer. Cells were analyzed on a flow cytometer (BD Biosciences, Canto), and we manually compensated for crossover of FITC fluorescence into the PI detection window. The analysis was completed using FlowJo 7.6.1.
Knockdown of GIMAP6 in human Jurkat T lymphocytes
Human Jurkat T lymphocytes were maintained in RPMI 1640 medium with 10% FBS. The cells were transfected with anti-GIMAP6 shRNA plasmids (TRCN0000151921, TRCN0000152080, TRCN0000152413, TRCN0000154009, and TRCN0000156658) or a control plasmid (pLKO.1.nullT) (provided by Academia Sinica, Taiwan) using the Neon transfection system (Invitrogen) in accordance with the recommended protocol. After limiting dilution followed by selection with 4 μg/ml puromycin (Sigma) for 2 weeks, cloning of single cells was achieved. These cells were then amplified for further study.
Overexpression of GIMAP6 in the HEK293T and Huh-7 cell lines
HEK293T and Huh-7 cells were cultured separately in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum. Transfection of the indicated expression plasmids separately into these cell lines was performed using TurboFect (Fermentas, R0531) according to the instructions of the manufacturer. The transfected cells were then cultured under the indicated conditions for 24 or 48 h. A stably expressing single colony was picked after G418 selection (500 μg/ml) for 2 weeks.
Peripheral blood mononuclear cell subset cell sorting
Peripheral blood mononuclear cells (PBMCs) were collected via Ficoll-Paque Plus (GE Healthcare, 18-1152-69) centrifugation from a fresh blood donor sample according to the instructions of the manufacturer. Cell sorting was accomplished by CD3-PE (BD Biosciences, 555333), CD4-FITC (BD Biosciences, 555346), and CD8-allophycocyanin (APC) (BD Biosciences, 555369) triple staining. Three subsets of cells, CD3+/CD4+, CD3+/CD8+, and CD3−, were collected after flow cytometry cell sorting. RNA was extracted from each subset immediately for use in the subsequent experiments.
Cellular fractionation
Subcellular fractionation was carried out on cultured Jurkat T lymphocytes using the ProteoJET cytoplasmic and nuclear protein extraction kit (Fermentas, K0311). A heavy membrane fraction that contained the mitochondria, a nucleus fraction, and a cytosolic fraction were obtained, and these were analyzed by immunoblotting using the indicated antibodies. For this analysis, histone H3, GAPDH, and PHB1 were used as controls for the nucleus, cytosol, and heavy membrane fractions, respectively.
Cell content screening
Cells were seeded on 24-well plates overnight using 1 × 104 cells/well. After treatment with the indicated reagents/drugs for 24 h, the cells were fixed on plates, and apoptosis detection was performed using rabbit anti-cleaved caspase-3 (Cell Signaling Technology) as the primary antibody and Alexa Flour 488 goat anti-rabbit IgG (Invitrogen) as the secondary antibody. With DAPI counterstaining, 16 images were taken (ImageXpress Micro XLS system) for each well, and the data were analyzed using MetaXpress software (Molecular Devices).
Jurkat T cell activation
Cells were seeded on 24-well plates overnight using 2 × 105 cells/well. After treatment with either PMA/ionomycin or an equivalent concentration of DMSO, cells were harvested at 3, 8, and 24 h and then stained with the phosphatidylserine exposure marker Annexin V-FITC (BD Biosciences) and the T cell activation surface marker CD69-PE (Biolegend, 310906). The results were analyzed and quantified using FlowJo 7.6.1.
IL-2 ELISA
In the Jurkat T cell activation experiments, supernatants of the media were collected from each well after centrifugation and stored at −80 °C until the relevant assays were carried out. IL-2 was determined using a human IL-2 ELISA kit (Biolegend, 431801).
Knockdown of GIMAP6 on human primary CD3+ T cells
After PBMC collection from healthy donors, CD3+ T cells were further enriched using a human CD3 T cell negative selection kit (Biolegend, 480022). The purity of the CD3+ population was examined by flow cytometry with anti-CD3-PE (BD Biosciences, 555333) antibody. The cells were then transfected with anti-GIMAP6 siRNA (Ambion, s54599, s54600) or a negative control (AM4611) using the Neon transfection system (Invitrogen) in accordance with the recommended protocol. The transfection efficiency was monitored by transfecting Cy3-labeled negative control siRNA (AM4621). After transfection for 48 h, the transfection efficiency was checked on fluorescence microscopy. Then cells were either harvested for total RNA extraction or for further experiments.
Primary CD3+ T cell activation
Previously described GIMAP6 knockdown or negative control CD3+ cells were seeded on 24-well plates using 1 × 106 cells/well. After treatment with either PMA/ionomycin or an equivalent concentration of DMSO, cells were harvested at 72 h and then stained with the T cell activation surface marker CD25-APC (Biolegend, 302609) and CD154-Alexa Flour 488 (Biolegend, 310815). The results were analyzed and quantified using FlowJo 7.6.1.
Supplementary Material
Acknowledgments
We thank Dr. Ming-Zong Lai and Dr. I-Cheng Ho for critical reading of this paper and Dr. Ralph Kirby for editing the manuscript. We thank Hui-Ying Weng for donating the blood samples for the study and the staff of the VGH Genome Research Center at National Yang-Ming University for conducting Sanger sequencing.
This work was supported by Intramural Grant MG-105-PP-01 of the National Health Research Institutes, Taiwan, and two extramural grants from Taiwan's Ministry of Health and Welfare (106-0324-01-10-05 and Advance Medical Plan 106-0324-01-10-07). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains supplemental Figs. S1–S4.
- GIMAP
- GTPase of immunity-associated proteins
- IAN
- immune-associated nucleotide-binding protein
- NSCLC
- non-small-cell lung cancer
- PBMC
- peripheral blood mononuclear cell
- Ctl
- control
- AnV
- Annexin-V
- PI
- propidium iodide
- OA
- okadaic acid
- PMA
- phorbol 12-myristate 13-acetate
- AICD
- activation-induced cell death
- P-loop
- phosphate-binding loop
- TLC
- thin-layer chromatography
- PE
- phycoerythrin
- P/I
- phorbol 12-myristate 13-acetate/ionomycin
- PARP
- poly(ADP-ribose) polymerase.
References
- 1. Nitta T., Nasreen M., Seike T., Goji A., Ohigashi I., Miyazaki T., Ohta T., Kanno M., and Takahama Y. (2006) IAN family critically regulates survival and development of T lymphocytes. PLoS Biol. 4, e103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Nitta T., and Takahama Y. (2007) The lymphocyte guard-IANs: regulation of lymphocyte survival by IAN/GIMAP family proteins. Trends Immunol. 28, 58–65 [DOI] [PubMed] [Google Scholar]
- 3. Krücken J., Schroetel R. M., Müller I. U., Saïdani N., Marinovski P., Benten W. P., Stamm O., and Wunderlich F. (2004) Comparative analysis of the human gimap gene cluster encoding a novel GTPase family. Gene 341, 291–304 [DOI] [PubMed] [Google Scholar]
- 4. Reuber T. L., and Ausubel F. M. (1996) Isolation of Arabidopsis genes that differentiate between resistance responses mediated by the RPS2 and RPM1 disease resistance genes. Plant Cell 8, 241–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Stamm O., Krücken J., Schmitt-Wrede H. P., Benten W. P., and Wunderlich F. (2002) Human ortholog to mouse gene imap38 encoding an ER-localizable G-protein belongs to a gene family clustered on chromosome 7q32–36. Gene 282, 159–167 [DOI] [PubMed] [Google Scholar]
- 6. Cambot M., Aresta S., Kahn-Perlès B., de Gunzburg J., and Roméo P. H. (2002) Human immune associated nucleotide 1: a member of a new guanosine triphosphatase family expressed in resting T and B cells. Blood 99, 3293–3301 [DOI] [PubMed] [Google Scholar]
- 7. Schwefel D., Fröhlich C., Eichhorst J., Wiesner B., Behlke J., Aravind L., and Daumke O. (2010) Structural basis of oligomerization in septin-like GTPase of immunity-associated protein 2 (GIMAP2). Proc. Natl. Acad. Sci. U.S.A. 107, 20299–20304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Schwefel D., Arasu B. S., Marino S. F., Lamprecht B., Köchert K., Rosenbaum E., Eichhorst J., Wiesner B., Behlke J., Rocks O., Mathas S., and Daumke O. (2013) Structural insights into the mechanism of GTPase activation in the GIMAP family. Structure 21, 550–559 [DOI] [PubMed] [Google Scholar]
- 9. Saunders A., Webb L. M., Janas M. L., Hutchings A., Pascall J., Carter C., Pugh N., Morgan G., Turner M., and Butcher G. W. (2010) Putative GTPase GIMAP1 is critical for the development of mature B and T lymphocytes. Blood 115, 3249–3257 [DOI] [PubMed] [Google Scholar]
- 10. Webb L. M., Datta P., Bell S. E., Kitamura D., Turner M., and Butcher G. W. (2016) GIMAP1 is essential for the survival of naive and activated B cells in vivo. J. Immunol. 196, 207–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Datta P., Webb L. M., Avdo I., Pascall J., and Butcher G. W. (2017) Survival of mature T cells in the periphery is intrinsically dependent on GIMAP1 in mice. Eur. J. Immunol. 47, 84–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Schnell S., Démollière C., van den Berk P., and Jacobs H. (2006) Gimap4 accelerates T-cell death. Blood 108, 591–599 [DOI] [PubMed] [Google Scholar]
- 13. Sandal T., Aumo L., Hedin L., Gjertsen B. T., and Døskeland S. O. (2003) Irod/Ian5: an inhibitor of γ-radiation- and okadaic acid-induced apoptosis. Mol. Biol. Cell 14, 3292–3304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Krücken J., Epe M., Benten W. P., Falkenroth N., and Wunderlich F. (2005) Malaria-suppressible expression of the anti-apoptotic triple GTPase mGIMAP8. J. Cell Biochem. 96, 339–348 [DOI] [PubMed] [Google Scholar]
- 15. Filén J. J., Filén S., Moulder R., Tuomela S., Ahlfors H., West A., Kouvonen P., Kantola S., Björkman M., Katajamaa M., Rasool O., Nyman T. A., and Lahesmaa R. (2009) Quantitative proteomics reveals GIMAP family proteins 1 and 4 to be differentially regulated during human T helper cell differentiation. Mol. Cell Proteomics 8, 32–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Heinonen M. T., Kanduri K., Lähdesmäki H. J., Lahesmaa R., and Henttinen T. A. (2015) Tubulin- and actin-associating GIMAP4 is required for IFN-γ secretion during Th cell differentiation. Immunol. Cell Biol. 93, 158–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shiao Y. M., Chang Y. H., Liu Y. M., Li J. C., Su J. S., Liu K. J., Liu Y. F., Lin M. W., and Tsai S. F. (2008) Dysregulation of GIMAP genes in non-small cell lung cancer. Lung Cancer 62, 287–294 [DOI] [PubMed] [Google Scholar]
- 18. Zenz T., Roessner A., Thomas A., Fröhling S., Döhner H., Calabretta B., and Dahéron L. (2004) hIan5: the human ortholog to the rat Ian4/Iddm1/lyp is a new member of the Ian family that is overexpressed in B-cell lymphoid malignancies. Genes Immun. 5, 109–116 [DOI] [PubMed] [Google Scholar]
- 19. Weiss Y., Forêt S., Hayward D. C., Ainsworth T., King R., Ball E. E., and Miller D. J. (2013) The acute transcriptional response of the coral Acropora millepora to immune challenge: expression of GiMAP/IAN genes links the innate immune responses of corals with those of mammals and plants. BMC Genomics 14, 400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rikkonen M., Peränen J., and Kääriäinen L. (1994) ATPase and GTPase activities associated with Semliki Forest virus nonstructural protein nsP2. J. Virol. 68, 5804–5810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rodríguez P. L., and Carrasco L. (1993) Poliovirus protein 2C has ATPase and GTPase activities. J. Biol. Chem. 268, 8105–8110 [PubMed] [Google Scholar]
- 22. Uritani M., and Miyazaki M. (1988) Characterization of the ATPase and GTPase activities of elongation factor 3 (EF-3) purified from yeasts. J. Biochem. 103, 522–530 [DOI] [PubMed] [Google Scholar]
- 23. Deyrup A. T., Krishnan S., Cockburn B. N., and Schwartz N. B. (1998) Deletion and site-directed mutagenesis of the ATP-binding motif (P-loop) in the bifunctional murine ATP-sulfurylase/adenosine 5′-phosphosulfate kinase enzyme. J. Biol. Chem. 273, 9450–9456 [DOI] [PubMed] [Google Scholar]
- 24. Saraste M., Sibbald P. R., and Wittinghofer A. (1990) The P-loop: a common motif in ATP- and GTP-binding proteins. Trends Biochem. Sci. 15, 430–434 [DOI] [PubMed] [Google Scholar]
- 25. Krengel U., Schlichting I., Scherer A., Schumann R., Frech M., John J., Kabsch W., Pai E. F., and Wittinghofer A. (1990) Three-dimensional structures of H-ras p21 mutants: molecular basis for their inability to function as signal switch molecules. Cell 62, 539–548 [DOI] [PubMed] [Google Scholar]
- 26. Boyman O., and Sprent J. (2012) The role of interleukin-2 during homeostasis and activation of the immune system. Nat. Rev. Immunol. 12, 180–190 [DOI] [PubMed] [Google Scholar]
- 27. Krammer P. H., Arnold R., and Lavrik I. N. (2007) Life and death in peripheral T cells. Nat. Rev. Immunol. 7, 532–542 [DOI] [PubMed] [Google Scholar]
- 28. Schmitz I., Krueger A., Baumann S., Schulze-Bergkamen H., Krammer P. H., and Kirchhoff S. (2003) An IL-2-dependent switch between CD95 signaling pathways sensitizes primary human T cells toward CD95-mediated activation-induced cell death. J. Immunol. 171, 2930–2936 [DOI] [PubMed] [Google Scholar]
- 29. Lenardo M. J. (1991) Interleukin-2 programs mouse α β T lymphocytes for apoptosis. Nature 353, 858–861 [DOI] [PubMed] [Google Scholar]
- 30. Khalaf H., Jass J., and Olsson P. E. (2010) Differential cytokine regulation by NF-κB and AP-1 in Jurkat T-cells. BMC Immunol. 11, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ye N., Ding Y., Wild C., Shen Q., and Zhou J. (2014) Small molecule inhibitors targeting activator protein 1 (AP-1). J. Med. Chem. 57, 6930–6948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Moore-Carrasco R., Busquets S., Figueras M., Palanki M., López-Soriano F. J., and Argilés J. M. (2009) Both AP-1 and NF-κB seem to be involved in tumour growth in an experimental rat hepatoma. Anticancer Res. 29, 1315–1317 [PubMed] [Google Scholar]
- 33. Brenner D., Golks A., Kiefer F., Krammer P. H., and Arnold R. (2005) Activation or suppression of NFκB by HPK1 determines sensitivity to activation-induced cell death. EMBO J. 24, 4279–4290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Perkins N. D. (2006) Post-translational modifications regulating the activity and function of the nuclear factor κ B pathway. Oncogene 25, 6717–6730 [DOI] [PubMed] [Google Scholar]
- 35. Hildeman D. A., Mitchell T., Aronow B., Wojciechowski S., Kappler J., and Marrack P. (2003) Control of Bcl-2 expression by reactive oxygen species. Proc. Natl. Acad. Sci. U.S.A. 100, 15035–15040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sharma S., Grandvaux N., Mamane Y., Genin P., Azimi N., Waldmann T., and Hiscott J. (2002) Regulation of IFN regulatory factor 4 expression in human T cell leukemia virus-I-transformed T cells. J. Immunol. 169, 3120–3130 [DOI] [PubMed] [Google Scholar]
- 37. Pascall J. C., Rotondo S., Mukadam A. S., Oxley D., Webster J., Walker S. A., Piron J., Carter C., Ktistakis N. T., and Butcher G. W. (2013) The immune system GTPase GIMAP6 interacts with the Atg8 homologue GABARAPL2 and is recruited to autophagosomes. PLoS ONE 8, e77782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hunter T. (1987) A thousand and one protein kinases. Cell 50, 823–829 [DOI] [PubMed] [Google Scholar]
- 39. Kühlbrandt W. (2004) Biology, structure and mechanism of P-type ATPases. Nat. Rev. Mol. Cell Biol. 5, 282–295 [DOI] [PubMed] [Google Scholar]
- 40. Kühlbrandt W., Auer M., and Scarborough G. A. (1998) Structure of the P-type ATPases. Curr. Opin. Struct. Biol. 8, 510–516 [DOI] [PubMed] [Google Scholar]
- 41. Rayment I., Rypniewski W. R., Schmidt-Bäse K., Smith R., Tomchick D. R., Benning M. M., Winkelmann D. A., Wesenberg G., and Holden H. M. (1993) Three-dimensional structure of myosin subfragment-1: a molecular motor. Science 261, 50–58 [DOI] [PubMed] [Google Scholar]
- 42. Strick D. J., and Elferink L. A. (2005) Rab15 effector protein: a novel protein for receptor recycling from the endocytic recycling compartment. Mol. Biol. Cell 16, 5699–5709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kadaré G., David C., and Haenni A. L. (1996) ATPase, GTPase, and RNA binding activities associated with the 206-kilodalton protein of turnip yellow mosaic virus. J. Virol. 70, 8169–8174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Uno T., Moriwaki T., Isoyama Y., Uno Y., Kanamaru K., Yamagata H., Nakamura M., and Takagi M. (2010) Rab14 from Bombyx mori (Lepidoptera: Bombycidae) shows ATPase activity. Biol. Lett. 6, 379–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. RayChaudhuri D., and Park J. T. (1994) A point mutation converts Escherichia coli FtsZ septation GTPase to an ATPase. J. Biol. Chem. 269, 22941–22944 [PubMed] [Google Scholar]
- 46. Satishchandran C., Hickman Y. N., and Markham G. D. (1992) Characterization of the phosphorylated enzyme intermediate formed in the adenosine 5′-phosphosulfate kinase reaction. Biochemistry 31, 11684–11688 [DOI] [PubMed] [Google Scholar]
- 47. Walker J. E., Saraste M., Runswick M. J., and Gay N. J. (1982) Distantly related sequences in the α- and β-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1, 945–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gough J., Karplus K., Hughey R., and Chothia C. (2001) Assignment of homology to genome sequences using a library of hidden Markov models that represent all proteins of known structure. J. Mol. Biol. 313, 903–919 [DOI] [PubMed] [Google Scholar]
- 49. Wilson D., Madera M., Vogel C., Chothia C., and Gough J. (2007) The SUPERFAMILY database in 2007: families and functions. Nucleic Acids Res. 35, D308–D313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Park J., Lappe M., and Teichmann S. A. (2001) Mapping protein family interactions: intramolecular and intermolecular protein family interaction repertoires in the PDB and yeast. J. Mol. Biol. 307, 929–938 [DOI] [PubMed] [Google Scholar]
- 51. Yeh Y. H., Kesavulu M. M., Li H. M., Wu S. Z., Sun Y. J., Konozy E. H., and Hsiao C. D. (2007) Dimerization is important for the GTPase activity of chloroplast translocon components atToc33 and psToc159. J. Biol. Chem. 282, 13845–13853 [DOI] [PubMed] [Google Scholar]
- 52. Uthaiah R. C., Praefcke G. J., Howard J. C., and Herrmann C. (2003) IIGP1, an interferon-gamma-inducible 47-kDa GTPase of the mouse, showing cooperative enzymatic activity and GTP-dependent multimerization. J. Biol. Chem. 278, 29336–29343 [DOI] [PubMed] [Google Scholar]
- 53. Sugimoto K., Yamada K., Egashira M., Yazaki Y., Hirai H., Kikuchi A., and Oshimi K. (1998) Temporal and spatial distribution of DNA topoisomerase II alters during proliferation, differentiation, and apoptosis in HL-60 cells. Blood 91, 1407–1417 [PubMed] [Google Scholar]
- 54. Breitman T. R., Collins S. J., and Keene B. R. (1980) Replacement of serum by insulin and transferrin supports growth and differentiation of the human promyelocytic cell line, HL-60. Exp. Cell Res. 126, 494–498 [DOI] [PubMed] [Google Scholar]
- 55. Gallagher R., Collins S., Trujillo J., McCredie K., Ahearn M., Tsai S., Metzgar R., Aulakh G., Ting R., Ruscetti F., and Gallo R. (1979) Characterization of the continuous, differentiating myeloid cell line (HL-60) from a patient with acute promyelocytic leukemia. Blood 54, 713–733 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.