Abstract
Animal-associated microbial communities have important effects on host phenotypes. Individuals within and among species differ in the strains and species of microbes that they harbour, but how natural selection shapes the distribution and abundance of symbionts in natural populations is not well understood. Symbionts can be beneficial in certain environments but also impose costs on their hosts. Consequently, individuals that can or cannot associate with symbionts will be favoured under different ecological circumstances. As a result, we predict that individuals within a species vary in terms of how well they accept and maintain symbionts. In pea aphids, the frequency of endosymbionts varies among host-plant-associated populations (‘biotypes’). We show that aphid genotypes from different biotypes vary in how well they accept and maintain symbionts after horizontal transfer. We find that aphids from biotypes that frequently harbour symbionts are better able to associate with novel symbionts than those from biotypes that less frequently harbour symbionts. Intraspecific variation in the ability of hosts to interact with symbionts is an understudied factor explaining patterns of host–symbiont association.
Keywords: endosymbionts, horizontal transfer, pea aphid, mutualism
1. Introduction
Symbiotic microbes within animals have important effects on their hosts, from increasing resistance against natural enemies to synthesizing nutrients that are missing from host diets [1]. Critical to the study of host–symbiont associations is the observation that individuals within and among species differ in the microbes that they harbour. For example, the vertically transmitted facultative bacterial endosymbionts of insects, which by definition are not essential to host survival or reproduction, are often found at intermediate frequencies within and among populations [2]. Variation in symbiont abundance is in part driven by natural selection acting on differences in the relative fitness of hosts with and without symbionts (a process that has been documented in natural populations of Drosophila neotestacea [3] and Bemisia whiteflies [4]). Symbiont-conferred benefits are balanced by the costs associated with harbouring microbes [5], and this trade-off is expected to affect symbiont population dynamics [6].
Harbouring a symbiont is not a passive process for hosts, which have evolved mechanisms for nurturing symbionts and regulating their growth [7]. Natural selection might act on these traits to favour the spread of host genotypes that can associate with symbionts, or those that cannot, depending on the relative costs and benefits of symbiont association. For natural selection to act on these mechanisms, there must be heritable variation among individuals in their ability to associate with symbionts. This variation has been shown to play a role in horizontal transfers of symbionts between different host species [8,9], which occurs on evolutionary [10] and even ecological timescales [11]. Symbionts have also been shown to move laterally within species [12], but whether there is intraspecific variation among hosts in the ability to associate with symbionts is unknown.
Here we address this issue using the pea aphid (Acyrthosiphon pisum) model system. Aphids harbour an obligate nutritional symbiont and can also host several species of chiefly vertically transmitted facultative symbionts. Aphid symbionts benefit their hosts in a number of ways [13], but harbouring symbionts also incurs costs for aphids [14]. Pea aphids feed on many species of Fabaceae, and form host-plant-associated populations (‘biotypes’) that are to differing extents genetically differentiated [15]. Biotypes vary in the frequency with which individuals harbour facultative symbionts [16]: some biotypes are strongly associated with a particular symbiont species, while other biotypes seldom carry facultative symbionts (table 1, which shows data from [12]). We include genotypes from four aphid biotypes in our study, and examine how well they accept and maintain associations with the globally distributed bacteria Regiella insecticola (which protects aphids against fungal pathogens) and Serratia symbiotica (which increases host tolerance of heat-stress) [13]. Aphids from the Trifolium spp. biotype harbour Regiella at a high frequency but rarely associate with Serratia, and aphids from the Pisum sativum biotype frequently harbour Serratia but rarely associate with Regiella (table 1). By contrast, aphids from the Lathyrus pratensis and Lotus corniculatus biotypes less commonly carry facultative symbionts [12] (table 1). We ask if genotypes vary in their ability to accept and maintain a symbiont infection, and whether this variation reflects the patterns of symbiont distribution found in natural populations.
Table 1.
Percentage of aphids from different biotypes that are uninfected, harbour Regiella insecticola, or harbour Serratia symbiotica. Data are from [12]. The four rows in bold type indicate the biotypes used in this study.
| biotype | % uninfected (no secondary symbionts) | % harbouring Regiella | % harbouring Serratia | no. aphids sampled |
|---|---|---|---|---|
| Pisum sativum | 5.8% | 5.1% | 81.8% | 137 |
| Trifolium spp. | 15.2% | 53.0% | 11.4% | 132 |
| Lotus corniculatus | 33.3% | 4.4% | 48.9% | 45 |
| Lathyrus pratensis | 40.5% | 4.8% | 9.5% | 42 |
| Lotus pedunculatus | 15.6% | 13.3% | 8.9% | 45 |
| Vicia cracca | 18.5% | 0% | 66.7% | 27 |
| Cytisus scoparius | 14.1% | 0% | 83.1% | 71 |
| Medicago sativa | 15.8% | 23.2% | 19.5% | 241 |
| Onobrychis viciifolia | 31.3% | 31.3% | 18.8% | 16 |
| Medicago lupulina | 18.8% | 4.2% | 45.8% | 48 |
| Melilotus spp. | 18.2% | 36.4% | 31.8% | 22 |
| Ononis spp. | 25.8% | 12.9% | 3.2% | 31 |
| average of all aphids | 17.6% | 18.6% | 36.9% | |
| average across biotypes | 21.1% | 18.9% | 35.8% |
2. Material and methods
We collected the aphids used in the experiment in southern England (table 2). Pea aphids are parthenogenetic in the summer, allowing us to maintain asexual genotypes in the laboratory at 16 L : 8 D and 14°C. We maintained lines on Vicia faba, a host-plant on which all biotypes are able to feed. We used microsatellite markers to confirm that genotypes belonged to the biotypes associated with their collection plant [15], and we screened lines for seven aphid facultative symbionts using PCR [12]. We cleared lines of any facultative symbionts using antibiotics [17], and waited more than 15 generations before the experiment to ensure there were no maternal effects of clearance treatment.
Table 2.
Information on aphid genotypes.
| host genotype | location collected (UK county) | year collected | original symbionts | biotype |
|---|---|---|---|---|
| 602a | Oxfordshire | 2014 | Serratia | Pisum sativum |
| 619 | Oxfordshire | 2014 | Serratia | Pisum sativum |
| 622 | Oxfordshire | 2014 | none | Pisum sativum |
| 126 | Berkshire | 2003 | Regiella | Trifolium spp. |
| 313a | Gloucestershire | 2007 | Regiella | Trifolium spp. |
| 319 | Gloucestershire | 2012 | Regiella | Trifolium spp. |
| 502 | Berkshire | 2003 | none | Lathyrus pratensis |
| 630 | Oxfordshire | 2013 | none | Lathyrus pratensis |
| 671 | Oxfordshire | 2014 | none | Lathyrus pratensis |
| 451 | Berkshire | 2010 | none | Lotus corniculatus |
| 663 | Oxfordshire | 2014 | none | Lotus corniculatus |
| 664 | Oxfordshire | 2014 | none | Lotus corniculatus |
aThe two lines that were the original hosts of the symbiont strains used in this study.
We used established protocols (following reference [9]) to experimentally transfer symbionts between aphids. We injected 0.25 µl of haemolymph from an infected adult donor aphid into the thorax of a one-day-old 1st instar recipient using a capillary needle. Injected aphids (generation 0) were reared on leaves of V. faba in Petri dishes at 14°C until they began reproducing. We collected approximately the tenth offspring of each surviving injected aphid and reared it to adulthood (generation 1), allowed it to reproduce to establish a line, and then extracted its DNA using proteinase K and ethanol precipitation. PCR was used to screen the generation 1 aphids for symbiont acceptance. We included three aphid genotypes from each of the four biotypes (table 2). We injected a total of 478 aphids (108–124 per biotype), half with a single strain of Regiella (strain 313, from an aphid collected in Glouchestershire, UK) and half with a single strain of Serratia (strain 602, collected in Oxfordshire, UK). Note that the aphid genotypes from the Trifolium biotype carried Regiella when originally collected (which were subsequently cleared as described above), two of the three Pisum genotypes harboured Serratia while the third harboured no secondary symbionts, and all of the genotypes from the Lotus and Lathyrus biotypes originally hosted no secondary symbionts (table 2).
Symbiont acceptance (determined by PCR amplification) was analysed using generalized linear mixed models with a binomial response variable, after checking for overdispersion, using the lme4 package in R v. 3.2 [18,19]. Symbiont species and aphid biotype were modelled as fixed effects. Genotype and a block effect grouping aphids injected with the same needle were modelled as random effects. The maximal model included the two random effects, symbiont species, biotype, and the interaction between symbiont species and biotype. We first removed the interaction term, followed by the fixed effects (biotype, then symbiont species). We compared the explanatory power of successive models using ANOVA. Post-hoc tests among biotypes were performed using the multcomp package in R v. 3.2 [20].
We then assessed maintenance of Regiella by genotypes from the Trifolium, Lotus and Lathyrus biotypes. Lines that were established from the screened aphid in generation 1 were reared on V. faba until generation 3. We again used approximately the tenth offspring of each aphid to establish both subsequent generations. At generation 3, two adult aphids from each line (which were the great-granddaughters of injected aphids) were frozen, and DNA extraction and PCR were performed as above. Lines were scored as having either retained or lost their symbiont between generations 1 and 3, and these data were analysed as above.
3. Results and discussion
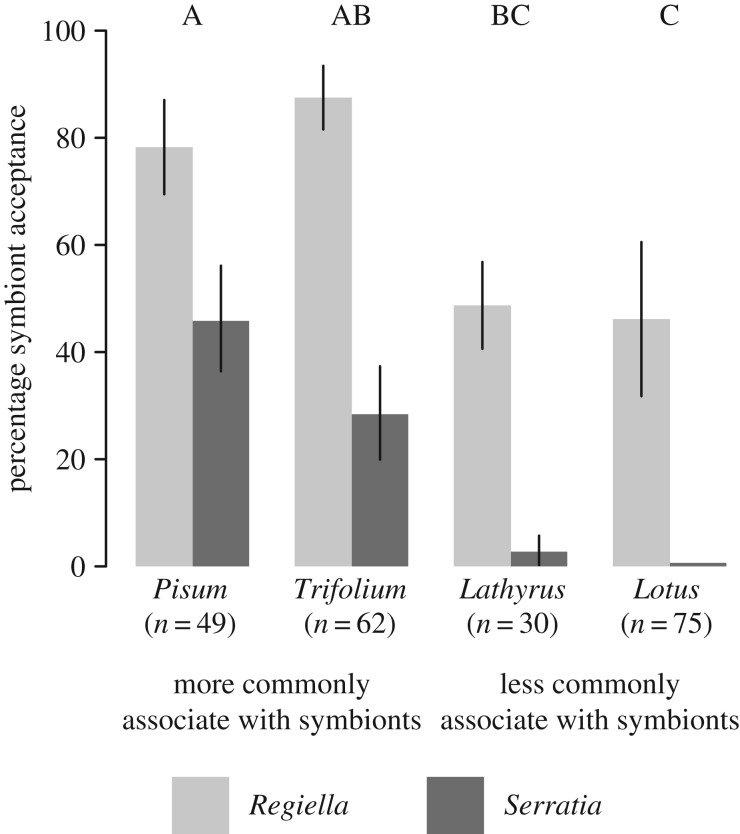
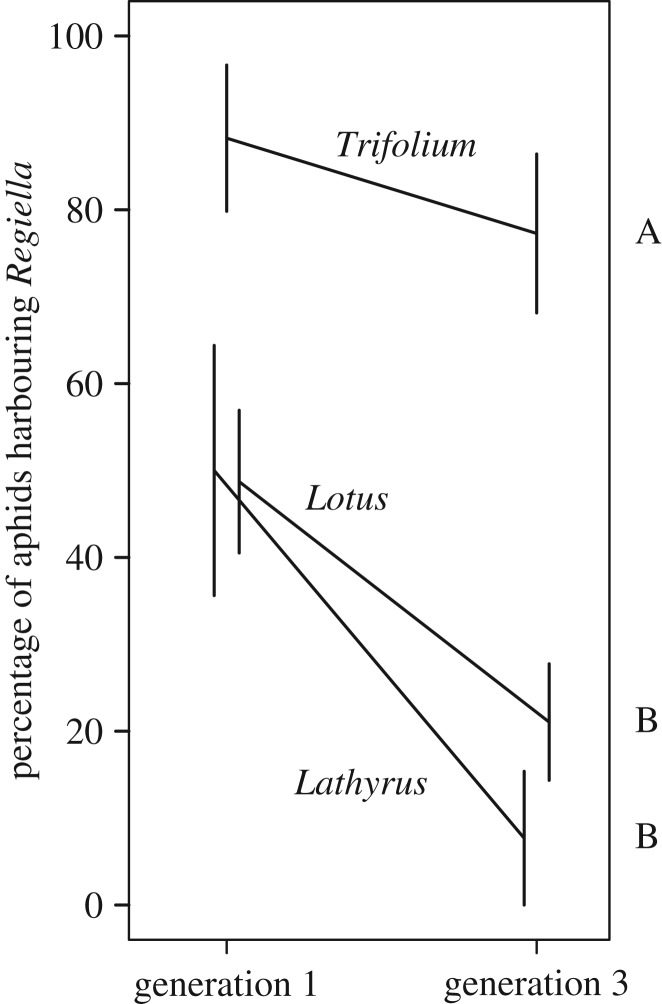
Biotypes differed in how well they accepted symbionts after injection (χ2 = 11.2, 3 d.f, p = 0.01; figure 1), demonstrating that there is intraspecific variation among pea aphids in the ability to associate with symbionts. Overall, Serratia established at a lower rate than Regiella (χ2 = 40.7, 1 d.f, p < 0.0001; figure 1) which could reflect biological differences, but might be an artefact of how well the symbionts survived injection. There was no interaction between symbiont species and aphid biotype (χ2 = 3.90, 3 d.f., p = 0.27). This is surprising given the different phenotypic effects these symbiont species have on their hosts and that they are not found at high frequencies in the same biotypes [16]. One possibility is that the ability to accept symbionts is based on general mechanisms that are not specific at the level of symbiont species, though because the two symbiont species we worked with are both in the family Enterobacteriaceae experiments with more distantly related aphid bacterial endosymbionts would be required to confirm this. We also measured whether genotypes from three biotypes maintain a Regiella infection over the course of several generations. There were again significant differences across biotypes (χ2 = 14.2, 2 d.f., p < 0.001; figure 2) with the Trifolium biotype showing a higher retention frequency than Lathyrus or Lotus.
Figure 1.
Symbiont acceptance. Bars show the percentage of aphids from each biotype (indicated along the x-axis) that accepted symbiont transfer (generation 1). Error bars show standard error. Aphids injected with Regiella or Serratia are shown in light and dark grey, respectively. Statistical significance groups among the biotypes are indicated at the top of the figure. Sample sizes, reflecting the number of aphids that survived after injection, are shown along the x-axis.
Figure 2.
Symbiont maintenance. The frequency of Regiella infection is shown on the y-axis at generations 1 and 3 after transfer. Each line represents an aphid biotype as indicated. Bars show standard error. Significance groups are indicated to the right of the figure.
The biotypes that were successful at accepting and maintaining symbionts (Trifolium and Pisum; significance groups, figures 1 and 2) frequently associate with symbionts in natural populations ([12], shown in table 1). By contrast, the Lathyrus and Lotus biotypes (which are not closely related phylogenetically [21]) have a lower frequency of symbiont carriage, and were poor at accepting and maintaining symbionts in our experiments (figures 1 and 2). Together, the pattern of symbiont association we uncovered reflects the distribution of symbionts in natural aphid populations. We caution, however, that we have only tested three genotypes from each biotype. A more definitive understanding of aphid–symbiont associations across biotypes will require testing additional genotypes with a variety of symbiont backgrounds (originally collected with and without facultative symbionts before antibiotic curing).
4. Conclusion
Recent work has shown that insect endosymbionts, which are primarily transmitted vertically, are also transmitted horizontally on ecological timescales [11] both within [12] and among [10] species. These studies suggest that microbial symbionts represent a ‘horizontal gene pool’ of useful adaptations that can be sampled by hosts resulting in adaptation through symbiosis. Our experiments show that this view needs to be more nuanced: host genotype has the potential to influence the distribution of symbionts within populations by determining the probability of acquiring novel infections.
The advantages of possessing a symbiont depend on the costs of carriage, the benefits the symbiont confers in different circumstances, and the probability that the aphid will encounter those circumstances in the field. But these variables only partly explain symbiont distribution. In addition, we need to know the likelihood that an aphid is exposed to a new symbiont and the probability the transfer will be successful. Our results help understand this second process, and allow for the possibility that natural selection might act on the mechanisms underlying host–symbiont associations. Further progress will require more information about frequency and mode of horizontal transfer of endosymbionts, and would be especially aided by additional study of the molecular mechanisms underlying insect–symbiont associations.
Acknowledgements
Ciara Mann provided valuable assistance with the experiments. Julia Ferrari kindly provided aphid genotypes. Two anonymous reviewers provided valuable feedback.
Data accessibility
Data are available from the Dryad digital repository. http://dx.doi.org/10.5061/dryad.k50h1 [22].
Authors' contributions
B.J.P. carried out the study and analysed the data. All authors conceived of and designed the study, drafted the manuscript, gave final approval of the version to be published, and agreed to be accountable for all aspects of the work.
Competing interests
The authors declare no competing interests.
Funding
This research was supported by NERC grant no. NE/K004972/1 to H.C.J.G. and by NSF postdoctoral fellowship DBI1306387 to B.J.P.
References
- 1.Douglas AE. 2015. Multiorganismal insects: diversity and function of resident microorganisms. Annu. Rev. Entomol. 60, 17–34. ( 10.1146/annurev-ento-010814-020822) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ferrari J, Vavre F. 2011. Bacterial symbionts in insects or the story of communities affecting communities. Phil. Trans. R. Soc. B 366, 1389–1400. ( 10.1098/rstb.2010.0226) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jaenike J, Unckless R, Cockburn SN, Boelio LM, Perlman SJ. 2010. Adaptation via symbiosis: recent spread of a Drosophila defensive symbiont. Science 329, 212–215. ( 10.1126/science.1188235) [DOI] [PubMed] [Google Scholar]
- 4.Himler AG, et al. 2011. Rapid spread of a bacterial symbiont in an invasive whitefly is driven by fitness benefits and female bias. Science 332, 254–256. ( 10.1126/science.1199410) [DOI] [PubMed] [Google Scholar]
- 5.Chrostek E, Marialva MSP, Esteves SS, Weinert LA, Martinez J, Jiggins FM, Teixeira L. 2013. Wolbachia variants induce differential protection to viruses in Drosophila melanogaster: a phenotypic and phylogenomic analysis. PLoS Genet. 9, e1003896 ( 10.1371/journal.pgen.1003896) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jaenike J. 2012. Population genetics of beneficial heritable symbionts. Trends Ecol. Evol. 27, 227–233. ( 10.1016/j.tree.2011.10.005) [DOI] [PubMed] [Google Scholar]
- 7.Login FH, Balmand S, Vallier A, Vincent-Monegat C, Vigneron A, Weiss-Gayet M, Rochat D, Heddi A. 2011. Antimicrobial peptides keep insect endosymbionts under control. Science 334, 362–365. ( 10.1126/science.1209728) [DOI] [PubMed] [Google Scholar]
- 8.Haselkorn TS, Cockburn SN, Hamilton PT, Perlman SJ, Jaenike J. 2013. Infectious adaptation: potential host range of a defensive endosymbiont in Drosophila. Evolution 67, 934–945. ( 10.1111/evo.12020) [DOI] [PubMed] [Google Scholar]
- 9.Łukasik P, Guo H, van Asch M, Henry LM, Godfray HCJ, Ferrari J. 2015. Horizontal transfer of facultative endosymbionts is limited by host relatedness. Evolution 69, 2757–2766. ( 10.1111/evo.12767) [DOI] [PubMed] [Google Scholar]
- 10.Haselkorn TS, Markow TA, Moran NA. 2009. Multiple introductions of the Spiroplasma bacterial endosymbiont into Drosophila. Mol. Ecol. 18, 1294–1305. ( 10.1111/j.1365-294X.2009.04085.x) [DOI] [PubMed] [Google Scholar]
- 11.Duron O, Wilkes TE, Hurst GDD. 2010. Interspecific transmission of a male-killing bacterium on an ecological timescale. Ecol. Lett. 13, 1139–1148. ( 10.1111/j.1461-0248.2010.01502.x) [DOI] [PubMed] [Google Scholar]
- 12.Henry LM, Peccoud J, Simon J-C, Hadfield JD, Maiden MJC, Ferrari J, Godfray HCJ. 2013. Horizontally transmitted symbionts and host colonization of ecological niches. Curr. Biol. 23, 1713–1717. ( 10.1016/j.cub.2013.07.029) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McLean AHC, Parker BJ, Hrček J, Henry LM, Godfray HCJ. 2016. Insect symbionts in food webs. Phil. Trans. R. Soc. B 371, 20150325 ( 10.1098/rstb.2015.0325) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hrček J, McLean AHC, Godfray HCJ. 2016. Symbionts modify interactions between insects and natural enemies in the field. J. Anim. Ecol. 85, 1423–1646. ( 10.1111/1365-2656.12586) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peccoud J, Ollivier A, Plantegenest M, Simon J-C. 2009. A continuum of genetic divergence from sympatric host races to species in the pea aphid complex. Proc. Natl Acad. Sci. USA 106, 7495–7500. ( 10.1073/pnas.0811117106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferrari J, West JA, Via S, Godfray HCJ. 2012. Population genetic structure and secondary symbionts in host-associated populations of the pea aphid complex. Evolution 66, 375–390. ( 10.1111/j.1558-5646.2011.01436.x) [DOI] [PubMed] [Google Scholar]
- 17.McLean AHC, van Asch M, Ferrari J, Godfray HCJ. 2011. Effects of bacterial secondary symbionts on host plant use in pea aphids. Proc. R. Soc. B 278, 760–766. ( 10.1098/rspb.2010.1654) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bates D, Mächler M, Bolker B, Walker S. 2015. Fitting linear mixed-effects models using lme4. J. Stat. Softw. 67, 1–48. ( 10.18637/jss.v067.i01) [DOI] [Google Scholar]
- 19.R Core Development Team. 2015. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. http://www.R-project.org.
- 20.Hothorn T, Bretz F, Westfall P. 2008. Simultaneous inference in general parametric models. Biom. J. 50, 346–363. ( 10.1002/bimj.200810425) [DOI] [PubMed] [Google Scholar]
- 21.Peccoud J, Simon J-C, McLaughlin HJ, Moran NA. 2009. Post-Pleistocene radiation of the pea aphid complex revealed by rapidly evolving endosymbionts. Proc. Natl Acad. Sci. USA 106, 16 315–16 320. ( 10.1073/pnas.0905129106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parker BJ, McLean A, Hrcek J, Gerardo N, Godfray HCJ. 2017. Data from: Establishment and maintenance of aphid endosymbionts after horizontal transfer is dependent on host biotype. Dryad Digital Repository. 10.5061/dryad.k50h1. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Parker BJ, McLean A, Hrcek J, Gerardo N, Godfray HCJ. 2017. Data from: Establishment and maintenance of aphid endosymbionts after horizontal transfer is dependent on host biotype. Dryad Digital Repository. 10.5061/dryad.k50h1. [DOI] [PMC free article] [PubMed]
Data Availability Statement
Data are available from the Dryad digital repository. http://dx.doi.org/10.5061/dryad.k50h1 [22].