Abstract
The notion of an attractor has been widely employed in thinking about the nonlinear dynamics of organisms and biological phenomena as systems and as processes. The notion of a landscape with valleys and mountains encoding multiple attractors, however, has a rigorous foundation only for closed, thermodynamically non-driven, chemical systems, such as a protein. Recent advances in the theory of nonlinear stochastic dynamical systems and its applications to mesoscopic reaction networks, one reaction at a time, have provided a new basis for a landscape of open, driven biochemical reaction systems under sustained chemostat. The theory is equally applicable not only to intracellular dynamics of biochemical regulatory networks within an individual cell but also to tissue dynamics of heterogeneous interacting cell populations. The landscape for an individual cell, applicable to a population of isogenic non-interacting cells under the same environmental conditions, is defined on the counting space of intracellular chemical compositions x = (x1,x2, … ,xN) in a cell, where xℓ is the concentration of the ℓth biochemical species. Equivalently, for heterogeneous cell population dynamics xℓ is the number density of cells of the ℓth cell type. One of the insights derived from the landscape perspective is that the life history of an individual organism, which occurs on the hillsides of a landscape, is nearly deterministic and ‘programmed’, while population-wise an asynchronous non-equilibrium steady state resides mostly in the lowlands of the landscape. We argue that a dynamic ‘blue-sky’ bifurcation, as a representation of Waddington's landscape, is a more robust mechanism for a cell fate decision and subsequent differentiation than the widely pictured pitch-fork bifurcation. We revisit, in terms of the chemostatic driving forces upon active, living matter, the notions of near-equilibrium thermodynamic branches versus far-from-equilibrium states. The emergent landscape perspective permits a quantitative discussion of a wide range of biological phenomena as nonlinear, stochastic dynamics.
Keywords: emergent landscapes, biochemical reaction networks, heterogeneous cell population dynamics, non-equilibrium thermodynamics, phase transition, cell differentiation
1. Introduction
Living organisms are complex; they are collective behaviours of many interacting individuals each with an internal dynamics of its own. At the level of a single cell, the individuals are the major biochemical players, e.g. polymerases, transcription factors, signalling kinases and GTPases, that form an intracellular biochemical reaction network. Single-molecule biophysics has established that individual protein molecules actually have ‘molecular individualism’ [1,2]. However, a population of ‘identical’ protein molecules usually can be accurately characterized statistically, in terms of a few key kinetic parameters: a diffusion constant in an aqueous solution at room temperature, a few rate constants for conformational transitions and a pair of rate constants for the bi-molecular association–dissociation reaction with a substrate.
An analogous kinetic picture exists for a tumour consisting of distinct cell subtypes, even within an isogenic, clonal cell population. Given what is now known as the ‘non-genetic heterogeneity’ of cell phenotypes, the cells are the individuals: phenotypic switching of each individual cell can be thought of as uni-molecular conformational transitions. Cell birth and death processes can be mapped to the synthesis and degradation of a biochemical species, etc. Then at the whole tumour level, there is an interactive cell population dynamics that involves autocrine and paracrine cell-to-cell communication that influences the rates of the above processes, as well as cell migration, predation and prey, etc. Classifying isogenic individual cells according to biomarkers, gene expression patterns and biological functions is one of the most important tasks in this study of non-genetic heterogeneity using single-cell technologies; this is not very different from identifying conformational states of biomacromolecules, as a central task of physical biochemistry using spectroscopies. Indeed, the experiments of extracting the cells in the tail of a population distribution and observing the repopulation kinetics [3] share the same idea as a laser ablation of protein conformations followed by observing relaxation kinetics [4].
The dynamics of both a biochemical reaction network and a heterogeneous cell population can be quantitatively represented in terms of a nonlinear, stochastic dynamical system (NSDS) [5–7]. One of the essential new insights from the NSDS theory is the existence of an emergent, global, non-equilibrium landscape that represents the dynamics. This landscape is not the ‘mechanism’ of the dynamics per se, rather it represents Onsager's thermodynamic force that is responsible for the non-equilibrium kinetic transients in the system.1 This perspective agrees with what K. Pearson [10] once said, ‘[a]ll laws must ultimately be merged into laws of motion’, and fits P. W. Anderson's [11] theory on the emergent phenomenon that ‘each level can require a whole new conceptual structure’.
We make a distinction in the terminologies of ‘non-driven’ and ‘equilibrium’: an equilibrium is a stationary state of a non-driven system which can also undergo time-dependent processes that are progressing towards the equilibrium. The stationary state of a thermodynamically driven system, however, will be a non-equilibrium steady state (NESS) [12,13].
Epistemologically, it is also important to draw a line between the theory of ‘combinatorial landscapes’ [14] and the theory of ‘emergent landscapes’. The former was motivated mainly from optimization problems and the growing usages of the landscape concept in protein science. The existence of a scalar function in these problems is considered as given; it either epitomizes the engineering objective, the quantity to be optimized, or is guaranteed by the equilibrium physics of systems without a thermodynamic driving force. By contrast, the significance of the latter, as a scientific theory, is to establish the existence of a landscape function in the first place, and to demonstrate its relevance, as well as possible computations, for driven stochastic dynamical systems.2
With a given landscape, be it from engineering, equilibrium physics or emergence, the mathematical and computational challenges are the same as those for non-convex problems. In protein science, it usually depends on the biological questions whether one considers certain types of atomic motion in a protein as merely structural fluctuations and others as conformational transitions [22–27]. The distinction can never be made completely clear, especially for proteins with a rugged free energy landscape [4,14]. This is a valuable lesson for current single-cell biology and theories of the emergent landscape.
The mathematical theory of an NSDS reveals a particular method to ‘discover’ the emergent landscape [20,21,28]: in principle, if one can measure the ergodic probability distribution pssV(n1, n2, … ,nN) for the NESS of a dynamical system, where nk is the population size of the kth species and V is the geometrical size of the reaction system, then the landscape
| 1.1 |
in which x = (x1, x2, … ,xN) and xk = nk/V is the number density of the kth subpopulation. Of course, in reality this is not feasible since a biological organism has only finite nk's, and its environment is constantly changing. Still, the mathematical object in (1.1) provides the theory in [29] with a rigorous foundation [20,21,30]. In fact, the act of taking all  echoes a fundamental idea in condensed matter physics [11]: ‘It is only as the [system] is considered to be a many-body system—in what is often called the
echoes a fundamental idea in condensed matter physics [11]: ‘It is only as the [system] is considered to be a many-body system—in what is often called the  limit—that [emergent] behaviour is rigorously definable’.
limit—that [emergent] behaviour is rigorously definable’.
In the present review, we discuss and investigate some key consequences of this landscape perspective of living, biological processes. The presentation is structured as follows. In §2, using a simple enzyme kinetic model as illustration, we introduce the emergent landscape φ(x) in an explicit formula. We show, in particular, that, when the chemical reaction system is situated within an equilibrium environment, this φ(x) is precisely the Gibbs free energy function for a closed chemical system: the partial derivative ∂φ(x)/∂xk then is the chemical potential of the kth species, and the steady state at the global minimum of φ(x) satisfies the detailed balance. We then illustrate that, for the same enzymatic reaction system under a sustained chemostatic chemical potential difference, its φ(x) is a global property of the NESS kinetics, which has a sustained non-zero flux. The local transition rates and the emergent global φ need not to be consistent. Recall that, for non-driven chemical kinetics, the equilibrium energy landscape is related to both equilibrium probability distribution and kinetic transitions between two energy wells; for driven biochemical kinetics, global and local kinetic landscapes are no longer the same [30,31]. There is a breakdown of the detailed balance.
In §3, we study a nonlinear kinetic model in which an increasing chemostatic driving force Δμext leads to a saddle–node (blue-sky) bifurcation. Using a specific reaction scheme known as the Schlögl model [32], the explicit formula for the non-equilibrium, emergent global φ can be again obtained. We then revisit and refine the notion of ‘thermodynamic versus kinetic branches’ of steady states articulated in [33]. The notion of states that are near to or far from equilibrium naturally arises: the former is a continuation of the equilibrium steady state from which the latter is separated by a barrier; a spontaneous catastrophe that causes a transition from the latter to the former is exponentially rare in an autonomous system.
Describing a biological organism, such a bifurcation structure is encoded in a much larger global landscape. In §4, we explore the notion of self-organization from an inanimate near-equilibrium state to a far-from-equilibrium state, not by chance but by necessity [34]. Through an explicit formula, we show that such a system will have an emergent global φ that has remarkable resemblance to C. H. Waddington's epigenetic landscape for cell differentiation [35]. Indeed, we see differentiation in a broader sense as ‘emergence of a new state’, which is precisely what the theory of nonlinear bifurcation seeks to characterize and what Waddington's landscape tries to represent. In physics, phase transition with symmetry breaking is the process that generates new states of matter [11,36]; it should not escape the reader's notice that a saddle–node bifurcation is the manifestation of the same phase transition, realized in a mesoscopic-sized system and viewed at a relatively short time scale [37,38].
The landscape provides a global perspective of a life-history of an autonomous individual organism under a stationary environment. The overall trend is that of a downhill motion on a ‘hillside’, which is nearly deterministic and programmed. The major milestone events are ‘shallow’ local minima on the hillside. In §5, we discuss the dialectical views of, on the one hand, a stochastic NESS that occurs within a landscape basin, which can of course contain many smaller basins; and on the other hand, the ‘downhill flow’. What should be considered as a basin and what should be considered as a hillside, of course, depends on one's biological question. By self-organization, we mean a process with which some form of overall order spontaneously arises from local interactions independent of initial conditions. In the landscape perspective, such behaviour is represented by an attractor and the basin associated with it.
The paper concludes in §6, in which we compare and contrast the NSDS theory with two other prominent schools of thought: the theory of complex systems biology [39] and Eigen–Schuster's theory of replicator dynamics [40]. In addition, we discuss the relation among the emergent landscape perspective, statistical certainty and reconstruction of the sequential steps leading to a rare event, as well as the nature of living matter.
2. Emergent landscape of a simple enzymatic reaction in a non-equilibrium steady state
2.1. Cyclic enzymatic reactions as a caricature of open chemical systems
Let us first consider a single enzyme molecule E that catalyses a reversible biochemical reaction between S and P, as shown in figure 1, one molecule at a time [41].
Figure 1.
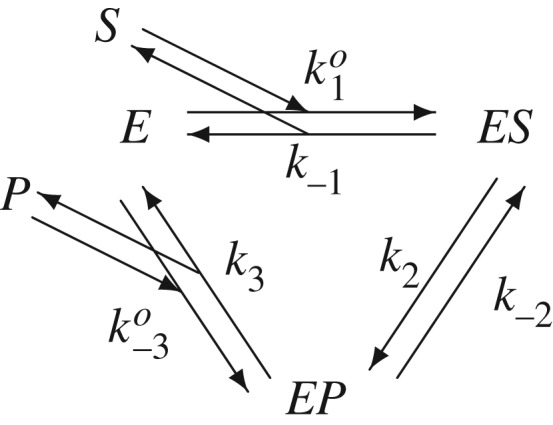
Kinetic scheme for a reversible enzymatic reaction with substrate S and product P. k−1, k2, k−2 and k3 are first-order rate constants; and ko1 and ko−3 are second-order rate constants. The system of biochemical reactions is in equilibrium if and only if the concentrations of S and P, [S] and [P], satisfy k1k2k3 = k−3k−2k−1, in which pseudo-first-order rate constants k1 = ko1[S] and k−3 = ko−3[P].
It is important to note that there are two types of reaction rate constants in figure 1: first-order k−1, k±2, and k3 for unimolecular reactions, and second-order ko1 and ko−3 for bimolecular reactions E + S → ES and E + P → EP, respectively. However, if an open chemical reaction system is sustained under a chemostatic environment, such as biochemical reactions inside a single cell in a Petri dish, then the concentrations of S and P are expected to be sustained externally to the reaction system. In this case, one can lump ko1[S]ext≡k1 and ko−3[P]ext≡k−3. In such a system, let us further assume that each of the six first-order, or pseudo-first-order, reactions is elementary: that is, it has an exponentially distributed waiting time for the enzyme to jump from one state to another. The jump is instantaneous; the time is in the waiting and the mean time is the reciprocal of the first-order, or pseudo-first-order, rate constant of the reaction.
The probability of the enzyme being in any one of the three states at time t, then, follows the chemical master equation [42–45]
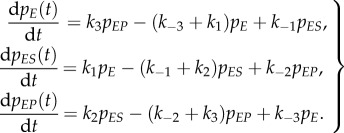 |
2.1 |
With all k's being non-zero, it is easy to show that this set of equations has a unique steady-state probability distribution π = (πE, πES, πEP).
Let us now consider a system that consists of N enzyme molecules in a reaction volume V, each undergoes the reactions in figure 1, among which nE, nES and nEP ≡ N − nE − nES are the molecular numbers in state E, ES and EP in the steady state. nE, nES, nEP still have fluctuations, no matter how large N is, as long as it is not infinite. Then, the steady-state probability
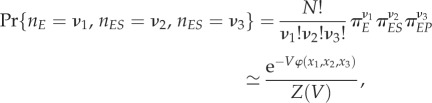 |
2.2 |
in which x1 = nE/V, x2 = nES/V and x3 = nEP/V are the concentrations of E, ES and EP, respectively. And
| 2.3 |
and
| 2.4 |
In the state space of the concentrations of chemical species x = (x1, x2, x3), we call φ(x) in (2.3) a global, kinetic or non-equilibrium landscape. Let us introduce three partial derivatives
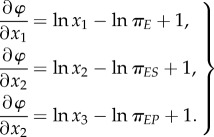 |
2.5 |
We note a very important mathematical result: if the six k's satisfy the detailed balance: πEk1 = πESk−1, πESk2 = πEPk−2 and πEPk3 = πEk−3, then
| 2.6 |
in which k2/k−2 is actually the equilibrium constant between ES and EP. In fact, (2.6) multiplied by kBT is the standard chemical potential difference μES − μEP for the ideal solution of the enzyme molecules. Therefore, φ(x1, x2, x3) is the Gibbs potential function of the equilibrium system, and ∂φ/∂xk then is the chemical potential of the species k.
2.2. Landscape, detailed balance and the Gibbs potential function
The Gibbs function represents the thermodynamics of a spontaneous process in a closed, non-driven chemical reaction system towards its chemical equilibrium. In the above simple example, this is encoded in the detailed balance assumption we made. The mathematical results in equations (2.2)–(2.4), however, are equally valid for driven, open chemical systems with a chemostat. Therefore, they are applicable to the biochemical reaction networks in living cells. In fact, we note that
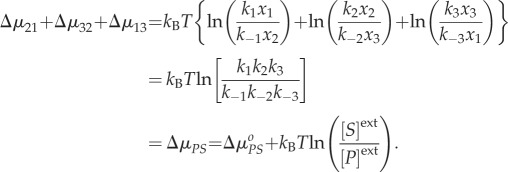 |
2.7 |
In a living cell, almost no biochemical reaction is at its chemical equilibrium. The expression in (2.7) is precisely the amount of chemical potential difference between a substrate molecule S under concentration [S]ext and a product molecule P under concentration [P]ext. If S and P are in a chemical equilibrium, then the ratio of rate constants inside the  , which is the well-known Wegscheider–Lewis cycle condition [46,47].
, which is the well-known Wegscheider–Lewis cycle condition [46,47].
2.3. The emergent global landscape
Therefore, with a sustained, external chemostatic chemical driving force ΔμPS, an open reaction ‘network’ inside a living cell like the one in figure 2 is the rule rather than an exception.
Figure 2.
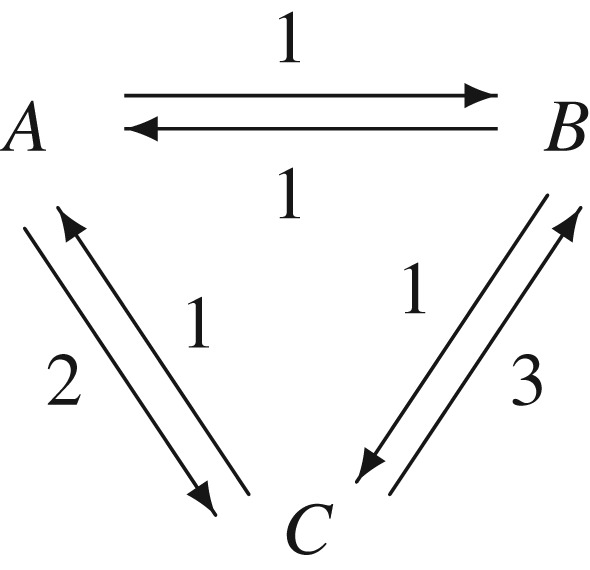
A particular example of the enzymatic reactions in figure 1. Since the k1k2k3 = 1 ≠ k−1k−2k−3 = 6, the system is driven under a chemostatic chemical potential ΔμPS = kBT lnγ, where γ = k1k2k3/(k−1k−2k−3).
In figure 2, we see locally that the probability of  is twice as likely as
is twice as likely as 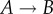 . However, the steady-state probability distribution for the three-state kinetic cycle is
. However, the steady-state probability distribution for the three-state kinetic cycle is
| 2.8 |
Note that this set of distributions has the following, important characteristics:
| 2.9 |
in which kXY is the transition rate constant from state X to state Y . There is a sustained net circular flux going from  , one-quarter of a round per unit time, driven by an external Δμ = − kBT ln 3 < 0.
, one-quarter of a round per unit time, driven by an external Δμ = − kBT ln 3 < 0.
We also note several very important features of this system which has no detailed balance. First, πB/πC = 2 ≠ kCB/kBC = 3. Second, the NESS state probabilities for B and C, πB:πC = 2, are very different from the 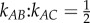 . In other words, the local dynamics among the connected states is completely different from the global, long time probability π = (πA,πB,πC). The steady-state probability π has the ultimate permanence.
. In other words, the local dynamics among the connected states is completely different from the global, long time probability π = (πA,πB,πC). The steady-state probability π has the ultimate permanence.
Just as in the equilibrium chemical thermodynamics, the π, being something ‘invariant’, has a fundamental role to play in the macroscopic dynamics of the simple system in figure 2. Following the steps in equations (2.2)–(2.4), on the space of the concentrations of chemical species x = (xA, xB, xC), we have φ in (2.4) again, but this time as an emergent global landscape.
Consider now the corresponding macroscopic reaction system with concentrations xA(t), xB(t) and xC(t) for the molecules in the three states. Then they follow the deterministic kinetic equation
| 2.10a |
| 2.10b |
| 2.10c |
We have the important mathematical result that
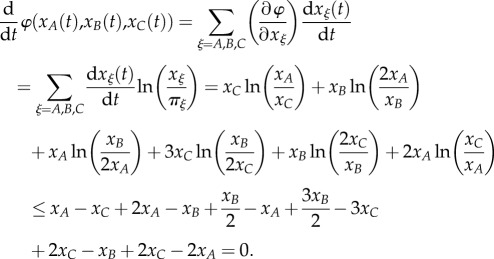 |
That is,
| 2.11 |
All the above x's are non-negative and we used the inequality ln x ≤ x − 1. The emergent φ has a similar property to the Gibbs function for a closed chemical reaction system. Inequality (2.11) is one origin of the organizational power of a global landscape.
2.3.1. Local transition rates and emergent global φ
Fundamentally different from the equilibrium free energy landscape φeq(x), which predicts the equilibrium probability distribution as well as the ratio of the transition rates of a reversible reaction, non-equilibrium theory has a global potential and a local potential, and they are different [30,31]. The global potential is related to the NESS probability distribution, while the local potential is related to the transitions between two adjacent basins of attraction. The relationship between the global and local potentials is very much analogous to the π's in equation (2.8) and the ratios of the rate constants in figure 2. Actually, in this work, the term ‘non-driven chemical system’ is synonymous with ‘chemical system without chemostat’; thus, the pseudo-rate constants satisfy the detailed balance, and the local and global potentials are the same.
3. Nonlinear bifurcation and far-from-equilibrium state of a system
In §2, we have illustrated several key non-equilibrium features of an open biochemical reaction network. Nonlinearity with multi-stability is another key characteristic of biochemical reaction networks with feedback regulations [48]. A saddle–node (or blue-sky) bifurcation is one of the widely observed dynamic ‘mechanisms’ for how a system shifts from having a unique steady state to having multiple stable steady states, e.g. attractors.
To illustrate this feature, let us consider the nonlinear reaction kinetics in (3.1). Gene regulatory networks of transcription factors, phosphorylation–dephosphorylation signalling networks with feedback regulation, and many other biochemical processes can be mapped to this simple nonlinear chemical reaction system, as a ‘model of models’ [49,50].
A saddle--node bifurcation with catastrophe is the signature phenomenon of a phase transition on a relatively short time scale. For a nonlinear dynamical system with a parameter λ, it predicts a range of the parameter, the interval (λ1,λ3) in figure 3, at which two stable steady states (i.e. three steady states when a saddle point is included) are possible, depending on the initial condition of the dynamical system. With the presence of fluctuations, the mean value of their probability distribution changes with λ. The sharpness of the transition curve, however, increases with decreasing noise. Then in the limit of zero noise, coexistence can only occur at one, ‘critical’ parameter value [38,48], the λ2 in figure 3. This is what physicists call a first-order phase transition.
Figure 3.
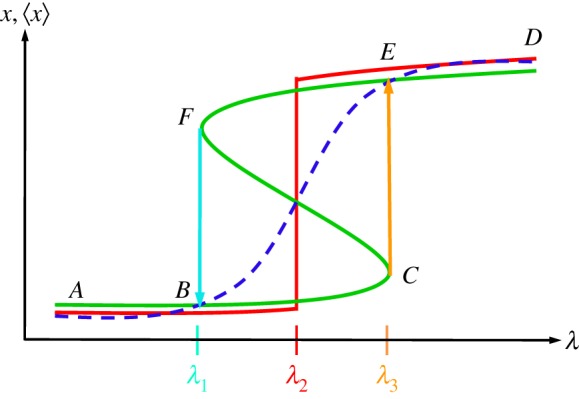
Steady states as functions of a parameter λ in a nonlinear dynamical system that undergoes a saddle–node bifurcation. The bifurcation is also called blue sky since a pair of stable and unstable steady states appears ‘out of the blue’ at F, and at C. The green S-shaped curve shows three steady states when λ is in the range of (λ1, λ3). The middle branch (FC) represents an unstable steady state. For a rapidly decreasing (increasing) λ, the x follows the upper (lower) branch and then the  (
( ) transition to the lower (upper) branch. In the presence of fluctuations and with a slowly changing λ, the average value of x, 〈x〉, changes with λ as shown by the blue dashed curve. The dashed blue curve becomes steeper and steeper with decreasing noise (not shown). In the limit of zero noise, it becomes the red, discontinuous curve, with a critical parameter value at λ2. In the zero-noise limit, the system behaves differently from that predicted by the deterministic dynamics.
) transition to the lower (upper) branch. In the presence of fluctuations and with a slowly changing λ, the average value of x, 〈x〉, changes with λ as shown by the blue dashed curve. The dashed blue curve becomes steeper and steeper with decreasing noise (not shown). In the limit of zero noise, it becomes the red, discontinuous curve, with a critical parameter value at λ2. In the zero-noise limit, the system behaves differently from that predicted by the deterministic dynamics.
3.1. Bistability in a nonlinear reaction system
We shall be particularly interested in how the Δμ from the chemostat induces bistability in an open chemical reaction system [51]. Using the Schlögl model [32,52,53],
| 3.1 |
as an example, we shall establish a connection between the non-equilibrium thermodynamics and nonlinear kinetics.
Let the concentrations for X, A and B be x, a and b, with a and b being sustained by an environment while x changes dynamically. The reaction, thus the x(t), eventually reaches the chemical equilibrium if the environmental concentrations a and b satisfy
| 3.2 |
Note that the equilibrium constant for the ‘overall reaction’
is simply KAB = k+1k+2/k−1k−2. Therefore,
| 3.3 |
which is the chemostatic chemical potential difference from the environment upon the system. If ΔμBA ≠ 0, the reaction reaches a NESS. The nonlinear dynamics of x(t) follows the law of mass action,
| 3.4 |
Introducing non-dimensionalized τ = k+2t and u = (k−1/k+2)1/2x, then
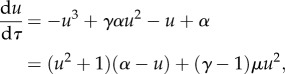 |
in which
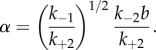 |
When γ = 1, for example, ΔμBA = 0, and the system approaches its unique equilibrium steady state ueq = α. Figure 4 shows chemical steady state(s) u* as a function, or three functions, of the chemical driving force lnγ = ΔμBA/kBT. In a NESS, there is a sustained net transport flux in the reaction system, Jness = k+1ax*2 − k−1x*3 = k2x* − k−2b, from A to B when μA > μA. In the non-dimensionalized variables, u* is related to γ through γ = u*/α + 1/αu* − 1/u*2.
Figure 4.
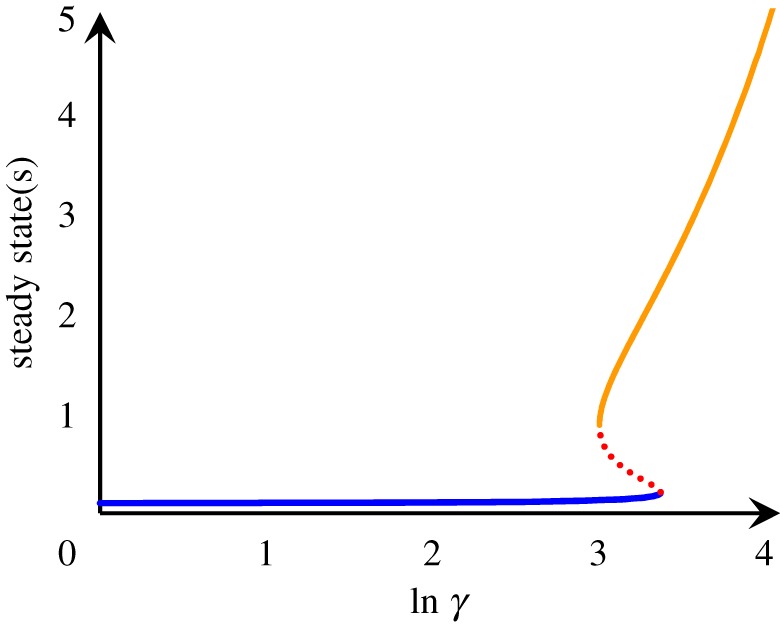
Bifurcation diagram for nonlinear dynamics (3.5) with α = 0.1. The steady state(s) is u* and the steady-state flux is (u* − α). When lnγ = 0, the chemical steady state is unique, which is an equilibrium state with u* = α and zero flux. The blue, lower branch, therefore, is a continuation of the equilibrium state, which we shall call the near-equilibrium branch. A saddle–node bifurcation occurs at lnγ = 2.938, when a pair of stable (orange, upper branch) and unstable (red, dashed branch) states emerge. The orange far-from-equilibrium branch is separated, thus protected, from the near-equilibrium branch by the red dashed line. When lnγ > 3.30, no spontaneous rare event can occur that endangers the far-from-equilibrium state. A distinct state of active matter arises. At a macroscopic scale and in an ergodic limit, the two branches coexist only at a single critical value of γ* = 20.36, lnγ* = 3.014 ∈ (2.938, 3.30).
With increasing γ, the system, driven stronger and stronger, is further and further away from its chemical equilibrium, but the uss represented by the blue branch in figure 4 changes very little. One can use
| 3.6 |
as a measure for characterizing the responsiveness of u* with increasing γ. Equation (3.6) indicates that, when u* ≪ 1, the response is nearly zero; however, if u* ≫ 1, then the response is approximately linear with slope α. When γ > 2.938, a new (orange) stable steady state appears, out of the blue, far from the equilibrium. The near- and far-from-equilibrium steady states are separated by an unstable steady state, represented by the red dashed line. The far-from-equilibrium (orange) steady state is characterized by several fundamental features:
(a) It is robust against internal and external perturbations due to the attractorial structure. In condensed matter physics, ‘robustness’ has been called ‘rigidity’ [11] and ‘protected behaviour’ [37], among many other terms.
(b) Starting from the near-equilibrium branch, reaching the far-from-equilibrium state spontaneously is an exponentially rare event, which takes an exponentially long time. This is another meaning of ‘being far’, in a kinetic sense.
(c) The appearance of the far-from-equilibrium branch is an emergent phenomenon with a dynamic symmetry breaking [36,38].
(d) Finally, but not the least, with a sufficient energy supply γ ≫ 1, the far-from-equilibrium state reaches complete ‘safety’ without the possibility of accidental deterioration to the blue branch: the near-equilibrium branch disappears completely from the state space.
One naturally identifies the far-from-equilibrium orange branch as a ‘new form of matter’: in the macroscopic limit, there is a discontinuous phase transition; the near- and far-from-equilibrium branches only coexist at a critical condition of γ*.
The γ* can be obtained from the emergent landscape of the system (3.1). The landscape can be computed [54,55]:
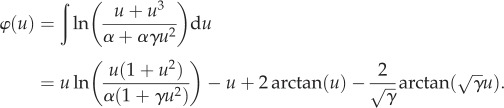 |
3.7 |
It is a Lyapunov function for the solution of the differential equation (3.5), u(t):
| 3.8 |
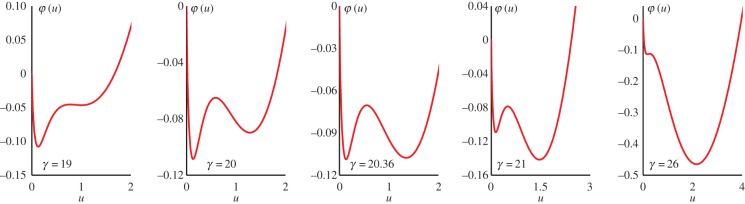
Figure 5 shows the φ(u) for α = 0.1 and several different values of γ. The value of γ* is found to be 20.36.
Figure 5.
Emergent global landscape φ(u) for the reaction kinetic system (3.1), given by (3.7) with α = 0.1 and several values of γ: From left to right: lnγ = 2.044, 3.00, 3.014, 3.045, 3.258. The macroscopic phase transition occurs at γ* = 20.36, when the two local minima have an equal height [38]. The minima (maxima) correspond to the stable (unstable) steady states in figure 4. (Online version in colour.)
3.1.1. Breakdown of ergodicity and symmetry breaking at the mesoscopic scale
Symmetry breaking has always been a phenomenon of time scales. Anderson [11] used progressively larger and larger molecules as an illustration to show that from the possibility of jumping among different ‘attractors’, within a reasonable time scale, to impossible, ‘the symmetry laws have been, not repealed, but broken’. In modern theory of dynamical systems, this is called a breakdown of ergodicity.
A periodic chemical oscillation is a form of temporal symmetry breaking. Systems with detailed balance cannot oscillate. Cellular biochemical reaction networks are full of kinetic cycles. Indeed, as pointed out in [11], ‘Temporal regularity is very commonly observed in living objects’.
3.2. Thermodynamic versus kinetic branches of non-equilibrium state of matter
A piece of inert material under a non-equilibrium condition will have transport fluxes. Even though the flux could be too small to measure, it is nevertheless not at equilibrium. Still, many people would not call such a system ‘active’. Furthermore, when the fluxes are really small, they follow Onsager's linear theory of irreversibility. Is there a qualitative difference between these non-equilibrium systems and systems that are ‘far from equilibrium’?
To distinguish the former from the latter, Nicolis & Prigogine [33] articulated the notion of an ‘equilibrium branch’ in phase space, and the emergence of a ‘kinetic branch’, or dissipative structure, through a transcritical bifurcation, as shown in figure 6. The bifurcation parameter here represents the distance from equilibrium.
Figure 6.
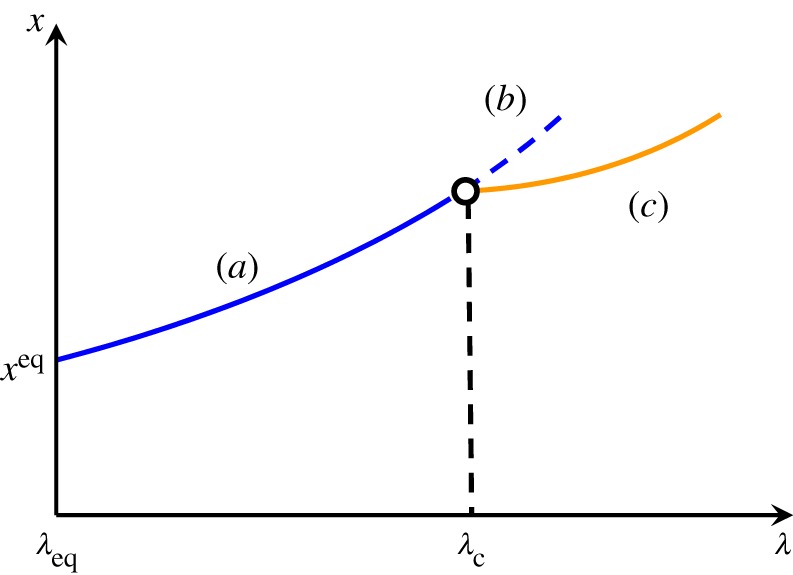
Branching of states as the distance from equilibrium increases: (a) stable part of the thermodynamic branch; (b) this branch becoming unstable; (c) new stable state (dissipative structure) emerging beyond the instability of the thermodynamic branch. (Reproduced from [33].) (Online version in colour.)
A little detour concerning the transcritical bifurcation. Its canonical form is [56]
| 3.9 |
which has an exchange of stabilities between the two branches of steady states, x1 = 0 and x2 = λ − λc, at λ = λc; it is always the lower branch that is stable. However, the transcritical bifurcation is imperfect [56]. For example, let λc = 0 and consider
| 3.10 |
with ε > 0. In this case, the two branches are
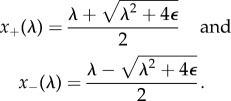 |
3.11 |
They never cross. Bifurcation behaviour that cannot persist under an ε-perturbation, such as the systems in (3.9) and (3.10), is called imperfect. Thus, in biological terms, the bifurcation phenomenon is not robust.
We should not build a scientific theory about dissipative structure based on such a mathematical concept. Rather, we shall refine the notions of ‘thermodynamic branch’ versus ‘kinetic branch’, or, alternatively, inanimate near-equilibrium branch versus far-from-equilibrium branch [5] in terms of the saddle–node bifurcation discussed in §3.1 and depicted in figure 4. The saddle–node bifurcation, together with the catastrophe phenomenon, is robust [38].
We suggest the term inanimate branch as the non-equilibrium continuation (γ > 1) of the thermodynamic equilibrium to describe the branch of steady state that passes through the equilibrium ueq = α when γ = 1. Then the far-from-equilibrium branch has an ‘energy barrier’, the red dashed line, that separates itself from the inanimate branch. The notions of near and far from in this sense are separated by an insurmountable barrier in the macroscopic limit. They are qualitatively different.
4. Physics of complexity meets Waddington
With the concept of near- versus far-from-equilibrium states of a system established, one naturally asks whether and how a chemical reaction system under a suitable external chemostat can spontaneously organize itself into a far-from-equilibrium state, not by chance but by necessity [34]. More precisely, how an autonomous chemical reaction system starts in a near-equilibrium state and reaches a driven steady state that is far from equilibrium. As we have discussed earlier, the notion of self-organization precisely reflects an independency from the initial situations: different systems all reach the same final state; the entire self-organizing process, thus, is a slow kinetic transient very much like the living process of organism development.
In other words, we envision that the γ-axis in figure 4 actually represents a slowly varying dynamic variable. With such a multi-scale evolving dynamics, a system with any initial condition, originating at lnγ = 2, will rapidly settle into the blue, near-equilibrium branch, but ultimately be in a far-from-equilibrium state on the orange branch when lnγ = 4.
4.1. Self-organization and differentiation
In terms of the landscape, one of the simplest realizations of such a multi-scale self-organizing reaction kinetic system is to ‘embed’ a saddle–node bifurcation structure, such as the one in figure 4, into an autonomous dynamics, with the bifurcation parameter being a slowly changing dynamic variable. In other words, we ‘stitch’ the one-dimensional landscapes in figure 5 into a single two-dimensional landscape with the γ as the second dimension, which contains the interval (18.9, 27.1) and goes downward with increasing γ.
Let us consider the following kinetics scheme:
| 4.1 |
in which now a chemical species Y serves as a catalyst for the reaction A + 2X → 3X. The kinetic system, according to the law of mass action, then is
 |
4.2 |
If the rate constant k−3 ≪ k± 2, then one can simply use the value y as a bifurcation parameter for the fast dynamics of x(t), while the y itself changes with time very slowly y(t) = y* + [y(0) − y*]e−k+3t, where y* = k−3c/k+3, and γy(0) < 18.9 and γy* > 27.1.
Again, to obtain the emergent global landscape, one needs to compute the NESS probability distribution according to the NSDS in terms of the chemical master equation [42–45]. When there is a separation of time scales, the steady-state probability distribution pssXY(ℓ, n) can actually be obtained in two separated steps. First, one solves the steady-state distribution for nX with nY = n fixed. This yields the so-called conditional probability pssX | Y(ℓ | n). Then one notices that the kinetics of Y is actually independent of X in (4.2). Thus, one can solve the steady-state distribution for nY, pssY(n). It can be shown that, when putting them together, pssXY(ℓ, n) = pssX|Y(ℓ|n)pssY(n). pssY(ℓ) is called the marginal probability of nY. Then one obtains a landscape
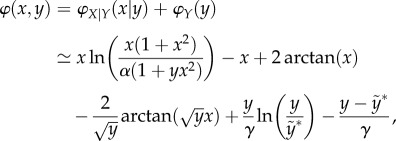 |
in which γ = k+1k+2a/k−1k−2b and 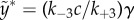 .
.
With value α = 0.1, γ = 30 and 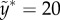 , figure 7 shows the landscape with a narrow trough along x = 0 for y < 15, and a turning point facing an ‘open field with downhill’, somewhere near y = 20.
, figure 7 shows the landscape with a narrow trough along x = 0 for y < 15, and a turning point facing an ‘open field with downhill’, somewhere near y = 20.
Figure 7.
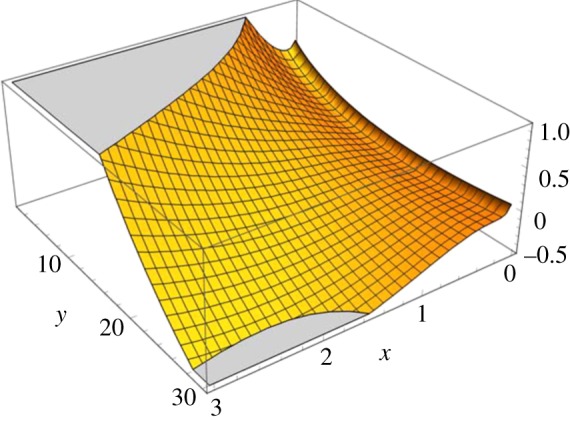
Global, emergent kinetic landscape for the open chemical reaction system in (4.1) according to (4.3). For small y, the trough along x = 0 should be identified as the ‘near-equilibrium’ thermodynamic branch; the ‘lower land’ near x = 3, y = 30 is a far-from-equilibrium state. One can see a curve like that in figure 4, and the set of cross-sections like figure 5, embedded in this landscape, which captures the self-organization into the far-from-equilibrium state.
This example illustrates that one can certainly design a kinetics with a landscape that leads a system from its beginning state near an equilibrium to a final state far from equilibrium, or, more generally, from a simple system to a more complex one. It requires no stretch of the imagination to think that Nature has adopted such a self-organizing mechanism in the process of adaptive evolution; and that thus biological organisms have co-opted such a ‘program’ in their genomes.
4.2. The biochemistry of a γ-driven bifurcation
In a recent experimental investigation, the role of cellular phosphorylation potential, e.g. the γ for ATP hydrolysis, on cell (division) cycle progression has been carefully studied [57]. Cyclin-dependent kinase 1 (Cdk1), also known as cell division cycle protein 2 homologue (Cdc2), is a highly conserved kinase in cell cycle regulation (figure 8). In fission yeast Schizosaccharomyces pombe and in humans, it is encoded by the cdc2 gene, and in the budding yeast Saccharomyces cerevisiae, by the cdc28 gene. The Cdk1 kinase together with its protein substrate and a cyclin form a tertiary complex within which phosphorylation can occur. Phosphorylation of the various protein substrates leads to cell cycle progression.
Figure 8.
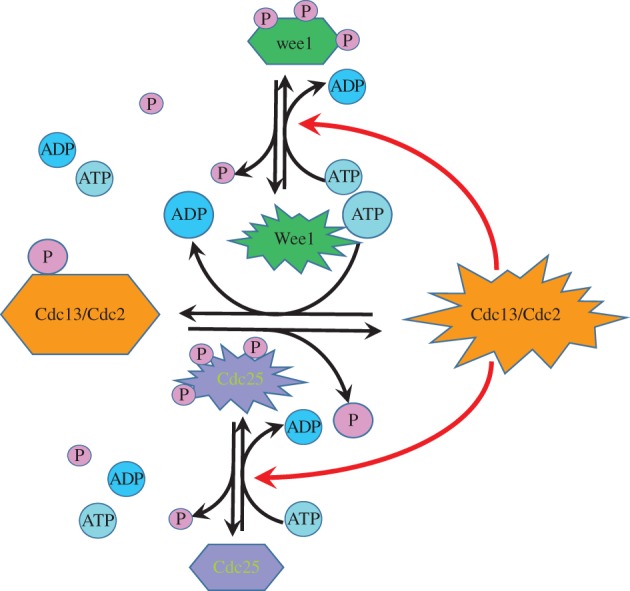
The transition from G2 phase to M phase in a fission yeast and human cell cycle progression is controlled by three types of PdPCs, with an intricate interplay of kinase and phosphatase activations. Such a regulatory network will not function when the free energy associated with ATP hydrolysis, a necessary ingredient for each and every PdPC, is low. This has been experimentally demonstrated recently in the nucleoplasmic extract from fission yeast Schizosaccharomyces pombe [57]. (Reproduced from [57].)
The kinase activity of the Cdc2/Cdc13 complex in fission yeast, where the Cdc protein 13 is a B-type cyclin, is itself regulated through a phosphorylation–dephosphorylation cycle (PdPC), with the kinase Wee1 and the phosphatase Cdc25: the dephosphorylation activates the kinase. The enzymatic activities of both Wee1 and Cdc25 themselves are regulated by two respective types of PdPCs; and there are feedback controls: the active Cdc12/Cdc13 is actually the kinase for both Wee1 and CdC25 phosphorylations!
The transition from G2 phase to M phase in a yeast cell cycle is considered to follow the dynamics of a bistable system. The γ-driven saddle–node bifurcation with a bifurcation diagram is remarkably similar to our figure 4 and has been experimentally observed in the nucleoplasmic extract of fission yeast S. pombe [57].
Finally, the widely employed notion of a cellular decision-making ‘check point’ can in fact be mapped to the concept of the transition state of a chemical reaction. It is located at the saddle point of a landscape, the crossing of which is when a transition from one basin of attraction to another occurs.
The biochemical network and its regulation involved in the yeast cell cycle is one of the best understood cellular systems in terms of biochemical kinetics [58]. We expect that the theory of NSDS and the notion of a landscape will also provide insights into and aid the understanding of many other complex biological processes.
5. Irreversible living process on a landscape
One of the fundamental insights afforded by non-equilibrium thermodynamics of NSDS is the replacing of the celebrated entropy balance equation [33,59,60] dS[x]/dt = σ(x) − hd/T by the free energy balance equation [5,20]:
| 5.1 |
in which φ(x) is the emergent global landscape of the NSDS, cmf(x) ≥ 0 is the rate of the chemostatic energy input into the system, as a chemical motive force, and σ ≥ 0 is the rate of total entropy production. Furthermore, we know that dφ/dt≤0 always. Therefore, the total entropy production rate, σ = ( −dφ/dt) + cmf, is the sum of contributions from two different non-equilibrium processes: dφ/dt represents the dissipation associated with transient kinetic processes and cmf(x) is the entropy production rate associated with the chemostat. Although not precise, the former can be viewed as an ‘internal irreversibility’ and the latter as arising from non-equilibrium coupling between the system and its environment.
In our previous work, attention has been paid mostly to the NESS behaviour of stochastic chemical reaction systems [12,13,48]. We shall now shift our attention to individual cells.
5.1. Major biological events occur on the ‘hillside’ of a landscape
Two clonal cells with identical genomes can exhibit very different phenotypes, best epitomized by the various cell types in the body of multicellular organisms. To a biologist, this is something that needs to be ‘explained’ since the tacit expectation is that the same set of genes would dictate the same cellular behaviour(s) and function(s). However, to a chemist who studies single-molecule conformational transitions, and knowing that there is only a single copy of DNA in a cell, it is the clonal cells' very similar behaviour that requires an explanation. This is a matter of perspectives; and it can be illustrated cogently in terms of the emergent global landscape. Figure 7 has a remarkable resemblance to C. H. Waddington's epigenetic landscape for cell differentiation [35,61]. The emergent global landscape φ rooted in biochemical network dynamics transforms the biological metaphor into a physicochemically based quantitative description [62–64].
First, biology focuses on major ‘life events’ in a living system, while biochemistry usually focuses on a certain part of the intracellular molecular reaction network. The former is a ‘higher-level’ coarse-grained view of a living system. In terms of a landscape then, for a biologist one should consider a global topography and neglect all the minor roughness. In this perspective, a single cell undergoes cell division to become two daughter cells is non-stationary behaviour which is a sequence of 'downhill' events [65]. All interesting biological processes, in a biologist's perspective, are non-stationary; therefore, all major biological events occur on the ‘hillsides’ of a landscape.
What we learned from the landscape theory is that on a hillside, at first-order approximation, the dynamics of different individuals with the same initial condition actually do follow the same deterministic equation as illustrated in §3.1. The stochasticity is mostly reflected in the timing of various events; and the time for an event to occur is more heterogeneous on a more rugged landscape. Differentiation, therefore, indeed can be represented as a Waddington's landscape [62], such as figure 9.
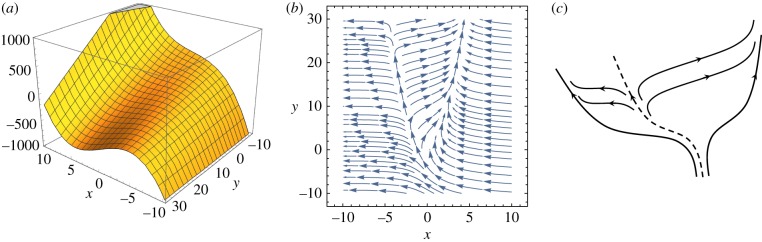
Figure 9.
(a) Landscape U(x,y) = x3 − 2xy − 20y. Consider the motion in the y-direction, which is much slower than that in the x-direction. Then for y near −10, the x is monotonically falling to the minimum at x = −10. However, for y > 0, a local minimum appears at 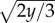 , which is separated from x = −10 by a maximum at
, which is separated from x = −10 by a maximum at  . For y = 30, the minimum and maximum are located at
. For y = 30, the minimum and maximum are located at 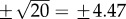 , respectively. (b) The vector field associated with the U(x, y) is:
, respectively. (b) The vector field associated with the U(x, y) is: 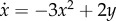 ,
, 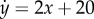 . (c) A schematic for the ‘dynamic flow’ in (b). We suggest that this type of ‘blue-sky’ bifurcation structure is more robust than the widely acknowledged ‘pitch-fork’ bifurcation pictured by C. H. Waddington. On the other hand, with the presence of noise, the two structures are nearly indistinguishable.
. (c) A schematic for the ‘dynamic flow’ in (b). We suggest that this type of ‘blue-sky’ bifurcation structure is more robust than the widely acknowledged ‘pitch-fork’ bifurcation pictured by C. H. Waddington. On the other hand, with the presence of noise, the two structures are nearly indistinguishable.
Continuing this thinking, a path that crosses many consecutive barriers of approximately equal heights can be considered as a path along a flat landscape. Only the one saddle that has a significantly higher barrier, if it exists, needs to be considered. Repeating this approximation significantly reduces the ruggedness of a landscape, ultimately transforming it into a landscape with a ‘smooth’ hillside.
Biochemical studies of intracellular processes, on the other hand, focus either on the kinetics of an individual reaction, which has very little relevance to the ‘global’ cell behaviour, or on the steady state of a biochemical reaction network. Therefore, the very nature of the biochemical studies already puts the focus on an ‘attractor’ of a landscape. A phenotypic switching then is associated with the transition between two attractors. From the hillside perspective mentioned above, this is a very detailed, reductionistic view. On the other hand, transient kinetic studies of a regulatory network without stochasticity do not encompass the overall landscape. Therefore, such studies, although valuable with respect to quantitative details, are not capable of representing the global ‘flow on a hillside of a rugged landscape’.
5.1.1. A need for single-cell kinetic studies
How can we experimentally obtain the global topography of the landscape of a cell? One needs to carry out time-dependent kinetic studies at the level of an entire single cell. Melnykov et al. [66] have developed a fast relaxation approach to single-cell biochemical kinetics based on light-induced transcriptional perturbations. If expanded to all relevant state-space dimensions by multiplexing, which is in principle possible, these methods could have the potential to open up a new vista to quantifying the emergent landscape of biochemical reaction networks. One expects to witness a growing research on whole-cell relaxation kinetic studies parallel to that of chemical and enzymatic reaction kinetic studies in the 1970s using rapid relaxation methods [67–69].
6. Discussion
It is apt to recall a statement in [11]: ‘We recognize that the [cell] is, after all, not macroscopic; it is merely approaching macroscopic behaviour. … Starting with the fundamental laws and a computer, we would have to do two impossible things—solve a problem with infinitely many bodies, and then apply the result to a finite system—before we synthesized this behaviour’. The NSDS theory presented in this work, together with the global, emergent landscape perspective, conceptually organizes the two impossible steps. It shows that Waddington's landscape metaphor has a mathematical, chemical kinetic foundation [62,70].
An NSDS, as a model, integrates some of the essential thoughts of the Brussels school's non-equilibrium thermodynamics and the Stuttgart school's synergetics and symmetry-breaking as the mechanism for generating complexity which originated from the phase transition lore in condensed matter physics [5,38]. It also shares a great deal of concepts, ideas and mathematical techniques with two other bodies of work, one on constructive biology [39] and another on the quasi-species model [71]. We particularly notice the eloquent statement that ‘[o]ur knowledge of physical and chemical systems is, in a final analysis, based on models derived from repeatable experiments’ [71], which firmly puts ‘models’ as the source of knowledge.
6.1. The theory of complex systems biology
There are ample agreements between the NSDS theory and the complex systems biology approach [39]. With a few types of elements (e.g. very low heterogeneity among individuals) and a few simple rules for interactions, complex behaviour can arise. The objective of the latter school is not so much to search for ‘self-organization’ but for ‘emergent universality’. This is further defined as follows [72]:
The approach that should be taken will be constructive in nature. We combine several basic processes, and construct a class of models, and to find universal logic underlying therein. With this logic, biological systems are classified into some universality classes. [An] organism, then, [is] understood as one representative for a universal class, to which the ‘life as it could be’ also belongs.
…
Note the approaches for complex and complicated systems should be distinguished. Since the latter are essentially understood as a combination of simple processes, what should be done here is to search for minimal sets of local processes that can fit real data. On the other hand, for complex systems [( … )], such an approach is not effective. One has to search for a general logic why such a complex system is of necessity and universal.
The first paragraph does not give a clear distinction between self-organization and emergent universality, per se. The real difference resides in the ‘why such … is of necessity and universal’. This is the ultimate question for biological science [73]. The answer to this question can be precisely represented in terms of a ‘landscape’, which quantifies ‘plausibility’. Indeed, the logic of the theory of probability is to consider ‘all possible outcomes and their probabilities’ [74]: the question of necessity is nothing but an overwhelming probability of 1; and the issue of universality, such as theories of thermodynamics and phase transition, has been most cogently represented as limit theorems in probability [75–77]. In a nutshell, the ‘constructive’ nature of a theory for an individual can be reduced (or should be expanded?) into ‘understanding complex systems ensemble and their stochastic dynamics’, as statistical laws. We hasten to add that one very successful mathematical approach to even purely deterministic complex dynamics has been their statistical characterizations [78,79].
There are possible correspondences between the key notions in complex systems biology and NSDS: isologous diversification  symmetry breaking and bifurcation; dynamic consolidation
symmetry breaking and bifurcation; dynamic consolidation  attractor and multi-stability; itinerancy
attractor and multi-stability; itinerancy  emergent inter-basin Markov jumps; and minority control
emergent inter-basin Markov jumps; and minority control  stochasticity is dictated by low copy numbers.
stochasticity is dictated by low copy numbers.
6.2. The theory of replicator dynamics
The number of emergent, discrete phenotypes of a biochemical network, with sufficient robustness, should not be an overwhelmingly large number. The reader is referred to an earlier work in this vein in protein science [80]. Indeed, even though combinations of nucleotide mutations in DNA have an astronomically large number of possibilities, if taking functional protein three-dimensional folds and biochemical network dynamics into consideration, the relevant possible outcomes of mutations should also be limited, e.g. the genotype to phenotype map has a great deal of degeneracy. If we assume that the set of all possible discrete phenotypes 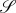 , e.g. attractors, is finite, then at a coarse-grained level the dynamics of subpopulations within an organism can be represented as
, e.g. attractors, is finite, then at a coarse-grained level the dynamics of subpopulations within an organism can be represented as
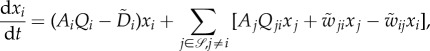 |
6.1 |
in which Ai and 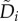 are the per capita birth and death rates of the ith subpopulation, 0 ≤ Qi ≤ 1 represents the proportion that has an exact reproduction, and 0 ≤ Qij ≤ 1 represents ‘erroneous’ reproduction giving birth to a jth individual:
are the per capita birth and death rates of the ith subpopulation, 0 ≤ Qi ≤ 1 represents the proportion that has an exact reproduction, and 0 ≤ Qij ≤ 1 represents ‘erroneous’ reproduction giving birth to a jth individual:
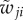 characterizes the rates for phenotype switching from j to i. This dynamical model, originally proposed to account for mutational dynamics, can be equally applicable to epigenetic switchings among attractors.
characterizes the rates for phenotype switching from j to i. This dynamical model, originally proposed to account for mutational dynamics, can be equally applicable to epigenetic switchings among attractors.
Equation (6.1) can be re-written as the standard from of replicator dynamics [40],
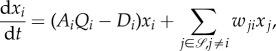 |
6.2a |
with
| 6.2b |
In (6.2), the per capita death rate Di now contains the phenotype transitions from the ith subpopulation to all the other subpopulations j ≠ i. Similarly, the ‘mutation rate’ wji includes all the asymmetric divisions with ‘erroneous’ replication as well as phenotypic transitions. Differentiating these different effects from dynamics requires high-precision single-cell measurements.
Equation (6.1) can also be re-written in a third form:
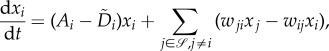 |
6.3 |
in which Ai and 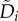 are the per capita birth and death rate irrespective of reproduction errors, and wij contains both the effects of all the asymmetric divisions and phenotypic transitions. This equation was the starting point of our earlier studies [81,82]. On the population level, one cannot distinguish between asymmetric division, e.g. equation (6.1), and symmetric division followed by a phenotypic transition, as in equation (6.3).
are the per capita birth and death rate irrespective of reproduction errors, and wij contains both the effects of all the asymmetric divisions and phenotypic transitions. This equation was the starting point of our earlier studies [81,82]. On the population level, one cannot distinguish between asymmetric division, e.g. equation (6.1), and symmetric division followed by a phenotypic transition, as in equation (6.3).
When stochasticity is introduced into a replicator dynamics, it becomes an NSDS [83,84]. Within the framework of replicator dynamics, Eigen and Schuster and their co-workers have developed the concept of quasi-species [40,71,85], a selection of one distribution, among the subpopulations, against all other distributions. In this context, they also discussed the notions of robustness and punctuated equilibrium [86,87].
6.3. Landscape and statistical certainty
Giving a sustained stationary environment and believing that ‘dynamics’ is the ultimate underlying description [10] of any phenomenon, sequence of events, and functions if any, a landscape can be obtained in two thought experiments with two extreme scenarios. (i) A single, individual system can be followed in an infinitely long time. By infinitely long, we mean it is longer than all the broken symmetries, and reaches an ultimate ergodicity. For a single protein, such as T4 lysozyme at 12°C, this time scale could be already more than 108 s (≃3 years) [88]. For a single cell, this time scale could easily be already longer than the age of the universe [28]. (ii) Alternatively, one has a large population of identical, independent individuals. Macroscopic protein chemistry simply took advantage of large Avogadro number: a micromolar concentration in a 2.5 ml cuvette has 1015 molecules. This reduces 108 s to 10−7 s, if one had single-molecule detection sensitivity, e.g. observing the transition of one out of the 1015. The rates are expected to be Arrhenius like, with inverse temperature replaced by the size of the population.
One does not need to have a full landscape; most of the time only a relatively small portion of it is relevant. This can be explored in a much shorter time scale. Current cellular studies based on cytometry with single-cell sensitivity [89] that measure millions of cells in a population at a single time point may approximate ergodicity for a given extracellular condition [90], and repeating such snapshot measurements under slowly changing conditions captures the hillside dynamics.
An emergent landscape is not the ‘cause’ of a biological dynamics. It is a summary of all its ‘potential transient behaviour’ in statistical terms. Here, we emphasize two insights from the mathematical theory of probability. First, statistical certainty concerning a population of individuals gives very little predictive power on anyone in the population; there is an uncomfortable dualism between ‘statistical truth’ and individual reality. Biochemical kinetics in a test tube has a deterministic description but single enzyme molecules fluctuate with ‘individualism’. Second, a stochastic dynamics with small fluctuations can undergo movements that have large deviations away from its average behaviour. Interestingly, while such movements are rare, when its occurrence is observed, the sequential events leading to the rare outcome are nearly deterministic, since any other possible sequence of events is much less probable, relatively speaking. In other words, retrospective reconstruction of the history of a rare event that occurred can be made with almost 100% confidence in the context of NSDS.
6.4. On living matters
What is living matter? In the classical physics of inanimate matters, different macroscopic states are best understood and characterized through phase transition. There is no doubt that bistability and saddle–node bifurcation with catastrophe are signatures of a macroscopic, first-order phase transition in a biochemical reaction network system. The theoretical result in figure 4 and the recent experimental studies on fission yeast [57] have clearly shown that living matter can be understood as an attractor state emerging through bifurcation with increasing external driving force lnγ, moving further and further away from the equilibrium state where γ = 1. As pointed out recently in [91], ‘[a]nother way to reconceptualize the problem [of the origin of life] is to consider life's emergence as a phase transition that manifests as a sudden change in how chemistry can process and use information and free energy’. In fact, Cronin and Walker [91] said succinctly that ‘[u]nderstanding this phase transition requires new approaches to non-equilibrium physics that hold promise for explaining the origin of structure at multiple hierarchical scales’. The non-equilibrium, global emergent landscape, as both a metaphor and an analytical device, is one development in answering that call.
Acknowledgements
We thank many colleagues for helpful discussions, particularly Elliot Elson, Stuart Kauffman, Vito Quaranta, Thea Tlsty, Yue Wang and X.-J. Cai Zhang.
Endnotes
The notion of a potential function in a non-equilibrium system has long been associated with the logarithm of its stationary probability distribution [15–17]. A reformulation of the standard Ito process based on a potential function has also been suggested as the dynamic foundation for Darwin's theory [18]. The NSDS theory, while sharing with the others many of the same ideas and mathematics, is rooted in a discrete description of population kinetics. The theory proves that the landscape is an emergent property of a mesoscopic dynamics, e.g. equation (1.1) [19–21].
Authors' contributions
All authors contributed equally to this work.
Funding
This work is supported by NIH grant no. R01GM109964.
References
- 1.de Gennes PG. 1997. Molecular individualism. Science 276, 1999–2000. ( 10.1126/science.276.5321.1999) [DOI] [PubMed] [Google Scholar]
- 2.Moerner WE. 2015. Single-molecule spectroscopy, imaging, and photocontrol: foundations for super-resolution microscopy (Nobel lecture). Angew. Chem. Int. Ed. 54, 8067–093. ( 10.1002/anie.201501949) [DOI] [PubMed] [Google Scholar]
- 3.Chang HH, Hemberg M, Barahona M, Ingber DE, Huang S. 2008. Transcriptome-wide noise controls lineage choice in mammalian progenitor cells. Nature 453, 544–547. ( 10.1038/nature06965) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frauenfelder H, Sligar S, Wolynes PG. 1991. The energy landscapes and motions of proteins. Science 254, 1598–1603. ( 10.1126/science.1749933) [DOI] [PubMed] [Google Scholar]
- 5.Qian H. 2016. Nonlinear stochastic dynamics of complex systems, I: a chemical reaction kinetic perspective with mesoscopic nonequilibrium thermodynamics. (http://arxiv.org/abs/1605.08070).
- 6.Thompson LF, Qian H. 2016. Nonlinear stochastic dynamics of complex systems, II: potential of entropic force in Markov systems with nonequilibrium steady state, generalized Gibbs function and criticality. (http://arxiv.org/abs/1605.08070).
- 7.Saakian DB, Qian H. 2016. Nonlinear stochastic dynamics of complex systems, III: noneqilibrium thermodynamics of self-replication kinetics. (http://arxiv.org/abs/1606.02391).
- 8.Qian H. 2006. Open-system nonequilibrium steady-state: statistical thermodynamics, fluctuations and chemical oscillations (feature article). J. Phys. Chem. B 110, 15063–15074. ( 10.1021/jp061858z) [DOI] [PubMed] [Google Scholar]
- 9.Wang J, Xu L, Wang E. 2008. Potential landscape and flux framework of nonequilibrium networks: robustness, dissipation, and coherence of biochemical oscillations. Proc. Natl Acad. Sci. USA 105, 12 271–12 276. ( 10.1073/pnas.0800579105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pearson K. 1943. The grammar of science. London, UK: J. M. Dent & Sons. [Google Scholar]
- 11.Anderson PW. 1972. More is different: broken symmetry and the nature of the hierarchical structure of science. Science 177, 393–396. ( 10.1126/science.177.4047.393) [DOI] [PubMed] [Google Scholar]
- 12.Zhang X-J, Qian H, Qian M. 2012. Stochastic theory of nonequilibrium steady states and its applications (Part I). Phys. Rep. 510, 1–86. ( 10.1016/j.physrep.2011.09.002) [DOI] [Google Scholar]
- 13.Ge H, Qian M, Qian H. 2012. Stochastic theory of nonequilibrium steady states (Part II): applications in chemical biophysics. Phys. Rep. 510, 87–118. ( 10.1016/j.physrep.2011.09.001) [DOI] [Google Scholar]
- 14.Reidys CM, Stadler PF. 2002. Combinatorial landscapes. SIAM Rev. 44, 3–54. ( 10.1137/S0036144501395952) [DOI] [Google Scholar]
- 15.Graham R, Haken H. 1971. Generalized thermodynamic potential for Markoff systems in detailed balance and far from thermal equilibrium. Z. Phys. 243, 289–302. ( 10.1007/BF01394858) [DOI] [Google Scholar]
- 16.Kubo R, Matsuo K, Kitahara K. 1973. Fluctuation and relaxation of macrovariables. J. Stat. Phys. 9, 51–96. ( 10.1007/BF01016797) [DOI] [Google Scholar]
- 17.Nicolis G, Lefever R. 1977. Comment on the kinetic potential and the Maxwell construction in non-equilibrium chemical phase transitions. Phys. Lett. A 62, 469–471. ( 10.1016/0375-9601(77)90069-X) [DOI] [Google Scholar]
- 18.Ao P. 2005. Laws in Darwinian evolutionary theory. Phys. Life Rev. 2, 117–156. ( 10.1016/j.plrev.2005.03.002) [DOI] [Google Scholar]
- 19.Ge H, Qian H. 2010. The physical origins of entropy production, free energy dissipation and their mathematical representations. Phys. Rev. E 81, 051133 ( 10.1103/PhysRevE.81.051133) [DOI] [PubMed] [Google Scholar]
- 20.Ge H, Qian H. 2016. Mesoscopic kinetic basis of macroscopic chemical thermodynamics: a mathematical theory. Phys. Rev. E 94, 052150 ( 10.1103/PhysRevE.94.052150) [DOI] [PubMed] [Google Scholar]
- 21.Ge H, Qian H. 2017. Mathematical formalism of nonequilibrium thermodynamics for nonlinear chemical reaction systems with general rate law. J. Stat. Phys. 166, 190–209. ( 10.1007/s10955-016-1678-6) [DOI] [Google Scholar]
- 22.Qian H. 2002. From discrete protein kinetics to continuous Brownian dynamics: a new perspective. Protein Sci. 11, 1–5. ( 10.1110/ps.ps.18902) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schulz GE, Schirmer RH. 1979. Principles of protein structure. New York, NY: Springer. [Google Scholar]
- 24.Price NC, Stevens L. 1981. Fundamentals of enzymology. Oxford, UK: Oxford University Press. [Google Scholar]
- 25.Fersht A. 1985. Enzyme structure and mechanism, 2nd edn New York, NY: W. H. Freeman. [Google Scholar]
- 26.Welch GR. (ed.). 1986. The fluctuating enzyme. New York, NY: John Wiley & Sons. [Google Scholar]
- 27.Creighton TE. (ed.). 1992. Protein folding. New York, NY: W. H. Freeman. [Google Scholar]
- 28.Qian H, Ge H. 2012. Mesoscopic biochemical basis of isogenetic inheritance and canalization: stochasticity, nonlinearity, and emergent landscape. Mol. Cell. Biomech. 9, 1–30. [PubMed] [Google Scholar]
- 29.Huang S, Eichler G, Bar-Yam Y, Ingber DE. 2005. Cell fates as high-dimensional attractor states of a complex gene regulatory network. Phys. Rev. Lett. 94, 128701 ( 10.1103/PhysRevLett.94.128701) [DOI] [PubMed] [Google Scholar]
- 30.Freidlin MI, Wentzell AD. 1984. Random perturbations of dynamical systems. New York, NY: Spinger. [Google Scholar]
- 31.Ge H, Qian H. 2012. Landscapes of non-gradient dynamics without detailed balance: stable limit cycles and multiple attractors. Chaos 22, 023140 ( 10.1063/1.4729137) [DOI] [PubMed] [Google Scholar]
- 32.Schlögl F. 1972. Chemical reaction models for nonequilibrium phase transition. Z. Phys. 253, 147–161. ( 10.1007/BF01379769) [DOI] [Google Scholar]
- 33.Nicolis G, Prigogine I. 1977. Self-organization in nonequilibrium systems: from dissipative structures to order through fluctuations. New York, NY: Wiley Interscience. [Google Scholar]
- 34.Monod J. 1972. Chance and necessity: an essay on the natural philosophy of modern biology. New York, NY: Vintage Books. [Google Scholar]
- 35.Waddington CH. 1953. The epigenetics of birds. New York, NY: Cambridge University Press. [Google Scholar]
- 36.Hopfield JJ. 1994. Physics, computation, and why biology looks so different. J. Theor. Biol. 171, 53–60. ( 10.1006/jtbi.1994.1211) [DOI] [Google Scholar]
- 37.Laughlin RB, Pines D, Schmalian J, Stojkovicí BP, Wolynes PG. 2000. The middle way. Proc. Natl Acad. Sci. USA 97, 32–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qian H, Ao P, Tu Y, Wang J. 2016. A framework towards understanding mesoscopic phenomena: emergent unpredictability, symmetry breaking and dynamics across scales. Chem. Phys. Lett. 665, 153–161. ( 10.1073/pnas.97.1.32) [DOI] [Google Scholar]
- 39.Kaneko K. 2006. Life: an introduction to complex systems biology. New York, NY: Springer. [Google Scholar]
- 40.Eigen M, Schuster P. 1979. Hypercycle: a principle of natural self-organization. Berlin, Germany: Springer. [DOI] [PubMed] [Google Scholar]
- 41.Lu HP, Xun L, Xie XS. 1998. Single molecule enzymatic dynamics. Science 282, 1877–1882. ( 10.1126/science.282.5395.1877) [DOI] [PubMed] [Google Scholar]
- 42.Delbrück M. 1940. Statistical fluctuations in autocatalytic reactions. J. Chem. Phys. 8, 120–124. ( 10.1063/1.1750549) [DOI] [Google Scholar]
- 43.Kurtz TG. 1972. The relationship between stochastic and deterministic models for chemical reactions. J. Chem. Phys. 57, 2976–2978. ( 10.1063/1.1678692) [DOI] [Google Scholar]
- 44.Liang J, Qian H. 2010. Computational cellular dynamics based on the chemical master equation: a challenge for understanding complexity (survey). J. Comp. Sci. Tech. 25, 154–168. ( 10.1007/s11390-010-9312-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Érdi P, Lente G. 2014. Stochastic chemical kinetics: theory and (mostly) systems biological applications. New York, NY: Springer. [Google Scholar]
- 46.Wegscheider R. 1902. Uber simultane gleichgewichte und die beziehungen zwischen thermodynamik und reaktionskinetik homogener systeme. Z. Phys. Chem. 39, 257–303. [Google Scholar]
- 47.Lewis GN. 1925. A new principle of equilibrium. Proc. Natl Acad. Sci. USA 11, 179–183. ( 10.1073/pnas.11.3.179) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Qian H. 2012. Cooperativity in cellular biochemical processes: noise-enhanced sensitivity, fluctuating enzyme, bistability with nonlinear feedback, and other mechanisms for sigmoidal responses. Annu. Rev. Biophys. 41, 179–204. ( 10.1146/annurev-biophys-050511-102240) [DOI] [PubMed] [Google Scholar]
- 49.Qian H, Bishop LM. 2010. The chemical master equation approach to nonequilibrium steady-state of open biochemical systems: linear single-molecule enzyme kinetics and nonlinear biochemical reaction networks (review). Int. J. Mol. Sci. 11, 3472–3500. ( 10.3390/ijms11093472) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bishop LM, Qian H. 2010. Stochastic bistability and bifurcation in a mesoscopic signaling system with autocatalytic kinase. Biophys. J. 98, 1–11. ( 10.1016/j.bpj.2009.09.055) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Qian H, Reluga TC. 2005. Nonequilibrium thermodynamics and nonlinear kinetics in a cellular signaling switch. Phys. Rev. Lett. 94, 028101 ( 10.1103/PhysRevLett.94.028101) [DOI] [PubMed] [Google Scholar]
- 52.Ross J. 2008. Thermodynamics and fluctuations far from equilibrium. New York, NY: Springer. [Google Scholar]
- 53.Vellela M, Qian H. 2009. Stochastic dynamics and nonequilibrium thermodynamics of a bistable chemical system: the Schlögl model revisited. J. R. Soc. Interface 6, 925–940. ( 10.1098/rsif.2008.0476) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ge H, Qian H. 2009. Thermodynamic limit of a nonequilibrium steady-state: Maxwell-type construction for a bistable biochemical system. Phys. Rev. Lett. 103, 148103 ( 10.1103/PhysRevLett.103.148103) [DOI] [PubMed] [Google Scholar]
- 55.Ge H, Qian H. 2011. Nonequilibrium phase transition in mesoscopic biochemical systems: from stochastic to nonlinear dynamics and beyond. J. R. Soc. Interface 8, 107–116. ( 10.1098/rsif.2010.0202) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Strogatz SH. 1994. Nonlinear dynamics and chaos. Reading, MA: Perseus Books. [Google Scholar]
- 57.Wang T, Zhao J, Ouyang Q, Qian H, Fu YV, Li F. 2016. Phosphorylation energy and nonlinear kinetics as key determinants for G2/M transition in fission yeast cell cycle. (http://arxiv.org/abs/1605.08070).
- 58.Verdugo A, Vinod PK, Tyson JJ, Novak B. 2013. Molecular mechanisms creating bi-stable switches at cell cycle transitions. Open Biol. 3, 120179 ( 10.1098/rsob.120179) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.de Groot SR, Mazur P. 1984. Nonequilibrium thermodynamics. New York, NY: Dover. [Google Scholar]
- 60.Qian H, Kjelstrup S, Kolomeisky AB, Bedeaux D. 2016. Entropy production in mesoscopic stochastic thermodynamics: nonequilibrium kinetic cycles driven by chemical potentials, temperatures, and mechanical forces (topical review). J. Phys. Condens. Matter. 28, 153004 ( 10.1088/0953-8984/28/15/153004) [DOI] [PubMed] [Google Scholar]
- 61.Ferrell JE. 2012. Bistability, bifurcations, and Waddington's epigenetic landscape. Curr. Biol. 22, R458–R466. ( 10.1016/j.cub.2012.03.045) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang J, Xu L, Wang E, Huang S. 2010. The potential landscape of genetic circuits imposes the arrow of time in stem cell differentiation. Biophys. J. 99, 29–39. ( 10.1016/j.bpj.2010.03.058) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang J, Zhang K, Xu L, Wang E. 2011. Quantifying the Waddington landscape and biological paths for development and differentiation. Proc. Natl Acad. Sci. USA 108, 8257–8262. ( 10.1073/pnas.1017017108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhou P, Li T. 2016. Construction of the landscape for multi-stable systems: potential landscape, quasi-potential, A-type integral and beyond. J. Chem. Phys. 144, 094109 ( 10.1063/1.4943096) [DOI] [PubMed] [Google Scholar]
- 65.Li F, Long T, Lu Y, Ouyang Q, Tang C. 2004. The yeast cell-cycle network is robustly designed. Proc. Natl Acad. Sci. USA 101, 4781–4786. ( 10.1073/pnas.0305937101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Melnykov AV, Jiang Y, Elson EL. 2014. Stochasticity in cellular response to light-induced transcriptional perturbations. Biophys. J. 106(Supplement 1), 373a ( 10.1016/j.bpj.2013.11.2113) [DOI] [Google Scholar]
- 67.Eigen M. 1972. Immeasurably fast reactions. In Nobel lectures in chemistry 1963–1970, pp. 170–203. Amsterdam, The Netherlands: Elsevier. [Google Scholar]
- 68.Gutfreund H. 1976. Kinetics: the grammar of enzymology. FEBS Lett. 62(Suppl), E13–E19. ( 10.1016/0014-5793(76)80849-6) [DOI] [PubMed] [Google Scholar]
- 69.Frieden C. 2008. A lifetime of kinetics. J. Biol. Chem. 283, 19873–19878. ( 10.1074/jbc.X800003200) [DOI] [PubMed] [Google Scholar]
- 70.Manin YI. 2007. Mathematics as metaphore: selected essays of Yuri I. Manin. Providence, RI: AMS Press. [Google Scholar]
- 71.Eigen M, McCaskill J, Schuster P. 1988. Molecular quasi-species. J. Phys. Chem. 92, 6881–6891. ( 10.1021/j100335a010) [DOI] [Google Scholar]
- 72.Kaneko K. 1998. Life as complex systems: viewpoint from intra-inter dynamics. Complexity 3, 53–60. (doi:10.1002/(SICI)1099-0526(199807/08)3:6<53::AID-CPLX7>3.0.CO;2-9) [Google Scholar]
- 73.Kirschner MW, Gerhart JC. 2006. The plausibility of life: resolving Darwin's dilemma. New Haven, CT: Yale Univ. Press. [Google Scholar]
- 74.Jaynes ET. 2003. Probability theory: the logic of science. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 75.Touchette H. 2009. The large deviation approach to statistical mechanics. Phys. Rep. 478, 1–69. ( 10.1016/j.physrep.2009.05.002) [DOI] [Google Scholar]
- 76.Sinai YaG. 1976. Self-similar probability distribution. Theory Probab. Appl. 21, 64–80. ( 10.1137/1121005) [DOI] [Google Scholar]
- 77.Jona-Lasinio G. 2001. Renormalization group and probability theory. Phys. Rep. 352, 439–458. ( 10.1016/S0370-1573(01)00042-4) [DOI] [Google Scholar]
- 78.Eckmann J-P. 1985. Ergodic theory of chaos and strange attractors. Rev. Mod. Phys. 57, 617–656. ( 10.1103/RevModPhys.57.617) [DOI] [Google Scholar]
- 79.Lasota A, Mackey MC. 1994. Chaos, fractals, and noise: stochastic aspects of dynamics. New York, NY: Springer. [Google Scholar]
- 80.Li H, Helling R, Tang C, Wingreen N. 1996. Emergence of preferred structures in a simple model of protein folding. Science 273, 666–669. ( 10.1126/science.273.5275.666) [DOI] [PubMed] [Google Scholar]
- 81.Zhou JX, Pisco AO, Qian H, Huang S. 2014. Nonequilibrium population dynamics of phenotype conversion of cancer cells. PLoS ONE 9, e110714 ( 10.1371/journal.pone.0110714) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Qian H. 2014. Fitness and entropy production in a cell population dynamics with epigenetic phenotype switching. Quant. Biol. 2, 47–53. ( 10.1007/s40484-014-0028-4) [DOI] [Google Scholar]
- 83.Rodriguez-Vargas AM, Schuster P. 1984. The dynamics of catalytic hypercycles—a stochastic simulation. In Stochastic phenomena and chaotic behavior in complex systems (ed. Schuster P.), pp. 208–219. Berlin, Germany: Springer. [Google Scholar]
- 84.Hofbauer J, Sigmund K. 1998. Evolutionary games and population dynamics. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 85.Nowak MA. 1992. What is a quasispecies? Trends Ecol. Evol. 7, 118–121. ( 10.1016/0169-5347(92)90145-2) [DOI] [PubMed] [Google Scholar]
- 86.Fontana W, Schuster P. 1998. Continuity in evolution: on the nature of transitions. Science 280, 1451–1455. ( 10.1126/science.280.5368.1451) [DOI] [PubMed] [Google Scholar]
- 87.Bak P, Sneppen K. 1993. Punctuated equilibrium and criticality in a simple model of evolution. Phys. Rev. Lett. 71, 4083–4086. ( 10.1103/PhysRevLett.71.4083) [DOI] [PubMed] [Google Scholar]
- 88.Chen B-L, Baase WA, Schellman JA. 1992. Folding kinetics of T4 lysozyme and nine mutants at 12°C. Biochemistry 31, 1464–1476. ( 10.1021/bi00120a025) [DOI] [PubMed] [Google Scholar]
- 89.Li Q, Wennborg A, Aurell E, Dekel E, Zou J-Z, Xu Y, Huang S, Ernberg I. 2016. Dynamics inside the cancer cell attractor reveal cell heterogeneity, limits of stability, and escape. Proc. Natl Acad. Sci. USA 113, 2672–2677. ( 10.1073/pnas.1519210113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kafri R, Levy J, Ginzberg MB, Oh S, Lahav G, Kirschner MW. 2013. Dynamics extracted from fixed cells reveal feedback linking cell growth to cell cycle. Nature 494, 480–483. ( 10.1038/nature11897) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cronin L, Walker SI. 2016. Beyond prebiotic chemistry: what dynamic network properties allow the emergence of life? Science 352, 1174–1175. ( 10.1126/science.aaf6310) [DOI] [PubMed] [Google Scholar]