Abstract
In-frame stop codons normally signal termination during mRNA translation, but they can be read as ‘sense’ (readthrough) depending on their context, comprising the 6 nt preceding and following the stop codon. To identify novel contexts directing readthrough, under-represented 5′ and 3′ stop codon contexts from Saccharomyces cerevisiae were identified by genome-wide survey in silico. In contrast with the nucleotide bias 3′ of the stop codon, codon bias in the two codon positions 5′ of the termination codon showed no correlation with known effects on stop codon readthrough. However, individually, poor 5′ and 3′ context elements were equally as effective in promoting stop codon readthrough in vivo, readthrough which in both cases responded identically to changes in release factor concentration. A novel method analysing specific nucleotide combinations in the 3′ context region revealed positions +1,2,3,5 and +1,2,3,6 after the stop codon were most predictive of termination efficiency. Downstream of yeast open reading frames (ORFs), further in-frame stop codons were significantly over-represented at the +1, +2 and +3 codon positions after the ORF, acting to limit readthrough. Thus selection against stop codon readthrough is a dominant force acting on 3′, but not on 5′, nucleotides, with detectable selection on nucleotides as far downstream as +6 nucleotides. The approaches described can be employed to define potential readthrough contexts for any genome.
INTRODUCTION
Open reading frames (ORFs) within an mRNA are terminated by an in-frame stop codon, which is recognized during translation by the binding of a protein release factor to the ribosomal A site. In eukaryotes, a single release factor, eRF1, recognizes all three stop codons (1). The efficiency of termination is enhanced by a second, GTPase, release factor eRF3 (2,3). The definition of the 3′ boundary of a gene by a stop codon determines the C-terminus of the encoded protein. Using stop codon position to predict the C-terminal amino acid sequence of a protein pre-supposes however that the process of translation termination is both accurate and efficient.
Although release factor recognition is normally efficient, in certain circumstances, a stop codon can be decoded not by the cognate release factor, but instead by a mis-cognate tRNA (4). Examples of tRNAs which decode stop codons include yeast tRNAGUCGln [decodes CAG and UAG, (5)], and tRNAUUGGln [decodes CAA and UAA, (6)]. tRNA-decoding of a stop codon occurs most frequently when the stop codon nucleotide context is unfavourable for release factor recognition. Numerous studies in E.coli, humans and yeast have helped define the elements 5′ and 3′ of the stop codon which either favour, or compromise, termination efficiency. In yeast and E.coli, a strong statistical bias exists in the identity of the nucleotides immediately 3′ of the stop codon (7), a bias increased in sub-groups of highly expressed genes (8). In vivo studies confirm that the identity of the three nucleotides following the stop codon profoundly influence stop recognition by release factors (9–11). In yeast, this zone of influence extends as far as 6 nt downstream of the stop codon (12). However, the relative importance of the six 3′ nucleotides in determining termination efficiency is not clear from this latter study. The nucleotide environment 5′ of the stop codon is also important as a termination efficiency determinant. In E.coli, the combined chemical properties of the last two amino acids in the nascent peptide contribute to stop recognition efficiency, with amino acids involved in α-helix formation favouring efficient termination (13,14). In yeast, the penultimate amino acid in the nascent peptide (encoded by the codon in the ribosomal E-site), together with the identity of the tRNA in the P-site, exert regulatory influence on termination efficiency (15). It is not clear whether such 5′ effects direct selection of the C-terminal amino acid sequence of proteins, nor how the identity of the two codons preceding the stop codon interact with the six 3′ nucleotides in directing termination efficiency.
Inefficient stop codon recognition, or readthrough, is used as part of a programmed gene regulatory strategy by a number of viruses. Expression of the Tobacco Mosaic Virus (TMV) RNA replicase domain requires readthrough of a UAG stop codon (16). TMV readthrough is regulated primarily by the 6 nt downstream (3′) of the stop codon (17). Murine leukaemia virus (MuLV) gag and pol ORFs are separated by an in-frame stop codon which is readthrough (translated) with a defined frequency of 5%, resulting in the synthesis of a gag–pol fusion protein (18). This readthrough is stimulated by a downstream RNA pseudoknot (19). However, it is becoming clear that examples of programmed stop codon readthrough governed by stop codon context are not limited to viruses. By identifying cellular genes where the stop codon terminating the ORF is followed by a significantly long (>200 nt) downstream ORF (dORF), genes in budding yeast and Drosophila have been identified whose expression is also regulated in this way (20–22). Subsequent testing of these stop codons and their surrounding context using reporter gene constructs has confirmed that some are readthrough at significant frequencies (20,22). Interestingly, some of the identified stop codons which are readthrough do not have obviously poor 5′ and 3′ contexts, implying our knowledge of the interplay and nature of 5′ and 3′ context effects is incomplete (20).
Of equal interest, but far more difficult to identify, are cellular stop codon readthrough events where the resulting added peptide tag is very short. The C-termini of proteins can be important determinants of protein targeting, stability and activity, and extending the C-terminus via stop codon readthrough has the potential to markedly alter the properties of the parent protein. For instance, the stop codon of the yeast PDE2 gene, encoding a cAMP phosphodiesterase, is found in a very similar context to the TMV stop codon. It is readthrough with an efficiency of between 2.2 and 8%, depending upon the genetic background, lengthening the Pde2p protein by 20 amino acids (23). This causes physiologically relevant alterations in yeast cellular cAMP levels, modulating stress responses (23). This type of programmed stop codon readthrough potentially makes an important contribution to the spectrum of gene regulatory strategies employed by the cell. It can be used to expand the range of polypeptides encoded by a core set of genes. However, the genome-wide identification of candidate stop codon readthrough events is dependent upon our ability to accurately define what constitutes a leaky stop codon context. Here, we ask whether detailed, genome-wide surveys of stop codon contexts in S.cerevisiae can generate predictive rules for context-driven stop codon readthrough and reveal the selective forces operating on stop codon context. In vivo assays of stop codon recognition are used to characterize potential weak stop codon contexts. The results facilitate attempts to identify novel candidates for programmed stop codon readthrough in silico from eukaryote genome sequences.
MATERIALS AND METHODS
Microbial strains and growth conditions
The S.cerevisiae strain used in this study was 76D694 (leu2-3,112 ura3-52 ade1-14UGA lys2-864UAG his7-1UAA [PSI+]; a gift from Dr S. Liebman, University of Chicago, IL). An isogenic [psi−] variant of strain 76D694, lacking the Sup35p [PSI] prion, was also created for use in this study by two successive platings on solid YPD medium containing 5 mM guanidine hydrochloride. Yeast strains were grown using standard conditions (24) on either YPD complete medium (2% w/v glucose, 2% w/v Bacto-peptone, 1% w/v yeast extract) or synthetic defined minimal medium [SD; 0.67% w/v yeast defined minimal medium without amino acids (Formedium), 2% w/v glucose] supplemented with the appropriate amino acids and bases at 2 mg ml−1. Escherichia coli strain XL1Blue (recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F′ proAB lacIqZΔM15 Tn10(Tetr)] was used throughout for cloning experiments and grown as described previously (25).
Plasmid constructs
DNA manipulation and plasmid construction was carried out according to standard protocols (25). To generate plasmid pJR16, a 3.3 kb XhoI–NotI fragment from plasmid pUKC606 [(3); a gift from M.F. Tuite, University of Kent] containing the entire SUP35 gene and its promoter was cloned into the XbaI site of pRS424 [multicopy-TRP1; (26)]. Plasmid pUKC802 (multicopy URA3 carrying a genomic SUP45 fragment) is described elsewhere (27). To assay readthrough of a stop codon in a given nucleotide context, oligonucleotide cassettes containing stop codons and flanking 5′ and 3′ sequence were cloned into the unique NotI site of the pAC98 vector (single-copy, LEU2; a generous gift from J.-P. Rousset, Université Paris-Sud, France). The pAC98 vector is an almost identical relative of pAC74 (28), and pAC99 (29), differing only in the identity of the unique cloning site at the junction of the reporter genes. pAC98 directs the expression of a β-galactosidase–luciferase fusion protein, and has a unique NotI site at the junction of the two reporter genes. Pairs of oligonucleotides (Table 1) were annealed with the production of NotI-compatible 5′ overhangs, and ligated into NotI-restricted pAC98. Cloning the following pairwise combinations of oligonucleotides into pAC98 in this way produced the vectors listed in brackets; PGf and PGr (pAC98-PG); GPf and GPr (pAC98-GP); PPf and PPr (pAC98-PP); RPS2f and RPS2r (pAC98-RPS2); Star1f and Star1r (pAC98-Star1); Star3f and Star3r (pAC98-Star3); PDE2f and PDE2r (pAC98-PDE2); PDE2-conf and PDE2-conr (pAC98-PDE2CON). Plasmids were sequenced through the cloned region using Big-Dye 3.0 chemistry (Applied Biosystems). Sequencing reactions were run on an Applied Biosystems ABI377 automated sequencer. Plasmids with verified cloned regions were transformed into yeast using a standard lithium acetate-based protocol (30) for reporter gene assay.
Table 1. Oligonucleotides used in this study.
| Name | Sequence (5′–3′) |
|---|---|
| Star1f | GG CCT CCA CAA TAG TTA AAA TAT CAA AAT T |
| Star1r | GG CCA ATT TTG ATA TTT TAA CTA TTG TGG A |
| Star3f | GG CCT CCA CAA TAG TTT TTG TAT CAA AAT T |
| Star3r | GG CCA ATT TTG ATA CAA AAA CTA TTG TGG A |
| RPS2f | GG CCT AGA TTC TAA GCT TGT TGT CTA CAA ATT T |
| RPS2r | GG CCA AAT TTG TAG ACA ACA AGC TTA GAA TCT A |
| PPf | GG CCT GGG CAA TAA CAA GAA TGT CTA CAA ATT T |
| PPr | GG CCA AAT TTG TAG ACA TTC TTG TTA TTG CCC A |
| PGf | GG CCT GGG CAA TAA GCT TGT TGT CTA CAA ATT T |
| PGr | GG CCA AAT TTG TAG ACA ACA AGC TTA TTG CCC A |
| GPf | GG CCT AGA TTC TAA CAA GAA TGT CTA CAA ATT T |
| GPr | GG CCA AAT TTG TAG ACA TTC TTG TTA GAA TCT A |
| PDE2f | GG CCT CCA CAA TAG CAA GAA TAT CAA AAT T |
| PDE2r | GG CCA ATT TTG ATA TTC TTG CTA TTG TGG A |
| PDE2-conf | GG CCT CCA CAA TAG ATC TAC TAT CAA AAT T |
| PDE2-conr | GG CCA ATT TTG ATA GTA GAT CTA TTG TGG A |
Quantification of nonsense suppression efficiency using the β-galactosidase/luciferase dual reporter system
β-galactosidase and luciferase activities in strains transformed with pAC98 vector derivatives were assayed using the Dual-Light kit (Tropix/Applied Biosystems) with the following modifications. Yeast cultures (5 ml) were grown in SD medium lacking leucine until the cell density reached OD600 0.3–0.5. Yeast were harvested and washed by centrifugation (3000 g, 5 min) in Dual-Light lysis buffer. Cells were disrupted by vortexing with glass beads (400 mesh, Sigma) in 200 μl Dual-Light kit lysis buffer to which protease inhibitors had been added at the recommended concentration (Mini-protease tablet, Roche). Aliquots (10 μl) of yeast lysate were assayed for luciferase and β-galactosidase activities as directed by the Dual-Light kit instructions using a Berthold LB9507 luminometer, ensuring enzyme activities lay within previously determined linear response limits of the Dual-Light reaction.
Bioinformatic processing and analysis of stop codon context sequences
The genome sequence of Saccharomyces cerevisiae, divided into 16 separate files, one for each chromosome, was downloaded from the Saccharomyces Genome Database, together with a list of the co-ordinates of all genes [Dolinski, K., Balakrishnan, R., Christie, K. R., Costanzo, M. C., Dwight, S. S., Engel, S. R., Fisk, D. G., Hirschman, J. E., Hong, E. L., Nash, R., Oughtred, R., Theesfeld, C. L., Binkley, G., Lane, C., Schroeder, M., Sethuraman, A., Dong, S., Weng, S., Miyasato, S., Andrada, R., Botstein, D., and Cherry, J. M. ‘Saccharomyces Genome Database’ ftp://ftp.yeastgenome.org/yeast/ (15.1.2003)]. Stop codons and their surrounding nucleotides were extracted from whole chromosome sequence flat files using STOPSCAN, a program written in the language Perl. The program is freely available on request from the authors. Using the co-ordinates for each gene, the program extracts the two codons that precede the stop codon of a given gene, the stop codon itself, and the downstream 6 nt. It searches for the next in-frame stop codon downstream of the ORF-terminating stop codon, and translates the intervening code into peptide sequence. In doing so, data is output from STOPSCAN in a tab-delimited form suitable for loading into a spreadsheet package. Manipulations of the data were performed using standard spreadsheet functions within Microsoft Excel.
Frequency of in-frame stop codons downstream of an ORF
The expected frequency of occurrence of stop codons in the 3′-UTR was calculated using a library of yeast 3′-UTR sequences extracted from the TransTerm database (31). Each entry comprises 100 nt sequence immediately downstream of the stop codon [the average length of a yeast 3′-UTR (32)]. Three thousand such sequences from chromosomes I–VIII were screened for the presence of stop codons in any frame using a sliding 3 nt window, a total screen of 218 000 nucleotide triplets. This analysis was performed using standard spreadsheet functions in Microsoft Excel. This analysis revealed that the probability of encountering TAA, TAG or TGA codons in this downstream region was 0.02711, 0.01327 and 0.01912, respectively, a total probability (p) of 0.0595. This probability was used to calculate a geometric progression probability distribution for encountering the next in-frame stop codon in the 3′-UTR where
probability of encountering tandem stop signal = [p],
probability of encountering a one codon extension is the probability of non-encounter of stop codon followed by encounter of stop codon) = [1 − p]p.
non-encounter of stop codon n times followed by encounter of stop codon = [1 − p]np.
A data mining method for analysing 3′ context data
A new data mining method was employed to analyse the 3′ context nucleotide frequencies (33). This new method directs the learning process of a self-learning algorithm, such as a Kohonen Self-Organizing Map [SOM (34)] by analysing the importance of individual descriptors in the system (in this case each nucleotide position). The data mining method utilizes a type of SOM that trains on the data set, grouping items which share common characteristics. The SOM was implemented using MATLAB with Neural Network Toolbox (Mathworks, Natick, MA). Usually, a SOM contains vectors of numerical values which are compared with data items in a similar format, and are updated using numerical learning rules and compared using numerical distance functions. For example, standard SOMs use the Euclidian distance metric which treats each dimension input in an n-dimensional space as equal and returns a value that gives an indication of the ‘distance’ between two data points described by this n-dimensional space. The SOM utilizes this distance metric to group similar data points together in an automated fashion, and a trained SOM contains a number of vectors of n-numerical values that describe these groupings. The training process therefore begins with a number of random groupings in the SOM, and the distance of these groupings from each individual data input is calculated. The grouping that is closest to the data input is moved closer to the values in the data input by a predefined learning function, which normally allows large movements at the beginning of training and smaller steps thereafter. Training can be stopped after a predefined number of steps or when the error of the overall SOM drops to a suitable value. After training on the available data, the differences between the vectors learnt by the SOM allow the data to be partitioned into separate groups, in which all grouped data items contain similar features, and in particular an examination of what makes the groupings different from each other to be made. This aspect of the process is controlled by a novel algorithm that allows the data mining method to be applied so that only results that are valid in the context of the current problem are returned to the user. The analysis of discrete ‘values’ (nucleotides) in the 3′ context dataset [S.cerevisiae 3′-UTR database (32)] required modifications to be made to the basic SOM algorithm. In extending this technique to analyse nucleotide data, the numerical vectors are replaced by fitness scores for each combination of possible nucleotides. These fitness scores can be updated using a modified set of learning rules. An example of this fitness score is the Levenshtein distance (35), which can be used to compare strings of characters, and gives a numerical similarity measure. After the network is trained, using the Levenshtein distance in place of the Euclidian distance given in the example above, the groupings determined by the SOM were examined and the dominant features for each group identified. In this case, for each dataset comprising combinations of four nucleotides taken from the 6-nt 3′ context, the modified SOM was used to group 4-nt words on the basis of shared common characteristics. This grouping was undertaken with no a priori knowledge of the frequencies of each nucleotide ‘word’, since the SOM is an unsupervised learning algorithm: different groups are generated purely on the basis of shared features which are apparent in the data. After the groupings were formed, the observed and expected frequencies of 4-nt ‘words’ within the group were compared, allowing the significance of a given grouping of contexts to be determined. This automated comparison directed the learning process (33). A fuller description of this approach is the subject of a manuscript in preparation (Williams, I., Stansfield, I. and Starkey, A. J. in preparation).
Analysis of the 6 nt following stop codons in the yeast genome
The population of 6-nt ‘words’ downstream of stop codons, output using the STOPSCAN program, was compared to a control population of 6-nt words derived using a sliding window from a group of 3000 randomly selected, 100-nt, S.cerevisiae 3′-UTRs extracted from the TransTerm database (31).
RESULTS
Efficient translation termination is ensured by positive selection for additional in-frame stop codons downstream of ORFs
Efficient stop codon recognition is important in preventing synthesis of a C-terminally extended polypeptide. The requirement for efficient termination is thought to drive the selection of nucleotide contexts 3′ of the stop codon which are more favourable for stop codon recognition. One way to ensure that proteins are not translated with large C-terminal extensions is to select both for efficient termination contexts, and for additional, in-frame stop codons 3′ of the primary termination codon. E.coli does not have a greater than expected abundance of tandem stop signals (36). Here, this type of analysis was applied to a eukaryote, to examine if yeast exhibits an increased abundance of downstream stop signals not only immediately after the primary stop signal, but also in the immediately following downstream positions.
The length of the dORF 3′ of all yeast ORFs in the genome were determined. This data set allowed the frequency of encounter of the next in-frame stop codon to be determined at each triplet position downstream of the primary stop codon for the entire yeast genome. This observed distribution of downstream stop signals was compared to an expected distribution, calculated from a geometric probability distribution using a p value for encountering a stop codon in 3′-UTR sequences determined as described (Materials and Methods). The significance of any differences between observed and expected distributions was assessed using the Poisson approximation of the binomial distribution (36,37).
The results show clearly that overall, the observed distribution of downstream stop signals closely matches the predicted distribution at most downstream codon positions (Figure 1). However, there is an over-abundance of ‘next in-frame’ stop signals in the +1, +2 and +3 codon positions downstream of the primary stop signal. This indicates the use of secondary ‘safety’ stop signals selected to limit stop codon readthrough effects. The increased abundance of stop codons at the +1, +2 and +3 positions was tested using the Poisson approximation to the binomial distribution, and found to be highly significant at all three positions (<0.1%; not shown). Additional evidence that these ‘safety’ stops are functional comes from the observation that of a collection of 169 highly expressed cytoplasmic ribosomal protein and translation factor genes, 35% had a downstream stop codon in the +1, +2 or +3 codon positions. This was significantly more than expected (19.5%, p < 0.0001, data not shown).
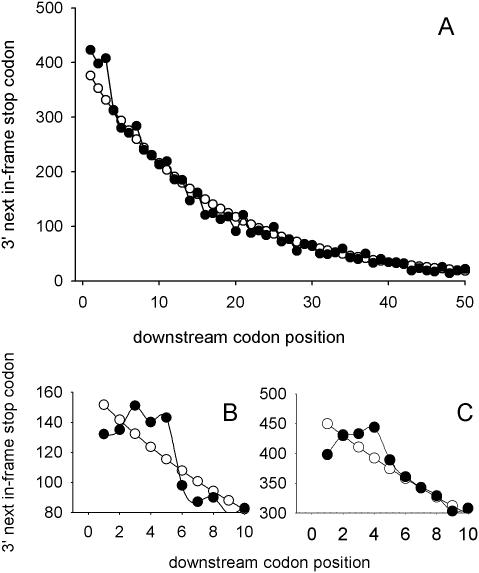
Figure 1.
Three fungal species have a significantly increased abundance of second, in-frame stop codons downstream of ORFs. (A) The lengths of the downstream ORF 3′ of all yeast ORFs was recorded, identifying the position of the next-in-frame stop codon for all genes in S.cerevisiae. The data were separated into bins corresponding to the first 50 codon positions after the ORF stop codon, giving the frequency of ‘next in-frame’ stops for each codon position. These observed frequencies (closed circles) were plotted along with an expected frequency (open circles) derived using a geometric probability distribution and knowledge of the probability of encountering a stop codon in the 3′-UTR (Materials and Methods). The analysis was repeated for all ORFs on chromosome I of S.pombe (B), and the complete Neurospora crassa genome (C).
Saccharomyces cerevisiae is unusual in that its eRF3 release factor can exist in two states, a prion form termed [PSI+] and a wild-type, soluble [psi−] form (38). In the [PSI+] state, eRF3 is inherited in a self-propagating, aggregated form, increasing readthrough of stop codons. The S.cerevisiae genome has thus arguably evolved in a cellular environment characterized by high levels of stop codon readthrough. This may have increased selective pressure for downstream stop codons to limit the effects of readthrough. To control for this possibility, the abundance of downstream stop codons was calculated for two additional fungal species, Neurospora crassa (39) and Schizosaccharomyces pombe (40), using genome sequence (Figure 1B and C). Neither of these fungi is known to exhibit an eRF3 prion phenomenon. Certainly, the S.pombe eRF3 protein lacks the Gln-, Asn- and Pro-rich repeat containing N-terminal sequences thought to be crucial for prion propagation (41). The data shows clearly that these two fungi also have increased abundances of stop codons in the immediate downstream region (codon positions +3, +4 and in S.pombe, +5), again indicative of a selective pressure to limit stop codon readthrough. It was noticeable that both S.pombe and N.crassa exhibit a reduced abundance of stop codons in the +1 codon position (tandem stop signals), and that stop codons in the +2 position were found as often as expected. This may reflect the lack of an eRF3 prion state in these fungi and thus a weaker selective pressure for ‘safety’ stop codons. Alternatively, particular nucleotides 3′ of the primary stop codon may be preferred which consequentially reduces the occurrence of stop codons at the +1 and +2 positions.
Codon usage in the −1 and −2 codon positions preceding the stop codon is not correlated with known effects on translation termination
The ability to predict which stop codons are inefficiently recognized by the termination apparatus depends upon a complete knowledge of the combined influences of the 5′ and 3′ context effects. Current understanding of the 5′ effects in yeast is incomplete. Originally, it was thought that 5′ context effects were contributed by the −2 position codon (the penultimate sense codon) based on the identity of the encoded amino acid, and by the −1 position codon based upon the identity of the decoding, P-site, tRNA (15). However, knowledge of −1 codon effects is restricted to 24 of the 61 sense codons (15). Furthermore, a recent report questioned whether −1 codon effects were tRNA-mediated, instead ascribing the main effect to the two mRNA nucleotides immediately preceding the stop codon, with paired adenines directing the greatest amount of readthrough (42).
In order to address this apparent contradiction, we sought to identify 5′ contexts which showed reduced abundance in the genome, indicative of a readthrough stimulating effect. −1 and −2 position codons were extracted from the database of stop contexts produced by the STOPSCAN program, and these observed frequencies were compared with expected frequencies derived from the codon usage data for S.cerevisiae. A derived frequency ratio was correlated with previously published effects on readthrough efficiency (15). No obvious correlation with readthrough efficiency was detectable at the −2 position (Figure 2A). Although proline codons were under-represented, consistent with their ability to direct readthrough (15), arginine, another amino acid incompatible with efficient termination, was over-represented (Figure 2). The overabundance of this amino acid in the penultimate amino acid position has been previously recognized by studies of C-terminal amino acid bias (43,44). The codon bias analysis was repeated for the −1 position, and as for the −2 position, no inverse correlation was observed between abundance and the ability to cause stop codon readthrough (data not shown). Two codons reported to enhance readthrough at the −1 position, ATA and CAA, were in fact either over-represented or found as frequently as expected, respectively. The most over-represented codon was AAA (Lys), which by virtue of its two adenine residues immediately preceding the stop codon, would be expected to direct high levels of readthrough and therefore be under-represented (42). Again, two previous studies looking at C-terminal peptide sequences in the yeast genome have described a marked overabundance of lysine codons at the C-terminus of yeast proteins (43,44). This bias is likely to arise due to selection at the level of peptide sequence. There appeared to be no consistent selection against those −1 position codons reported to direct readthrough (data not shown).
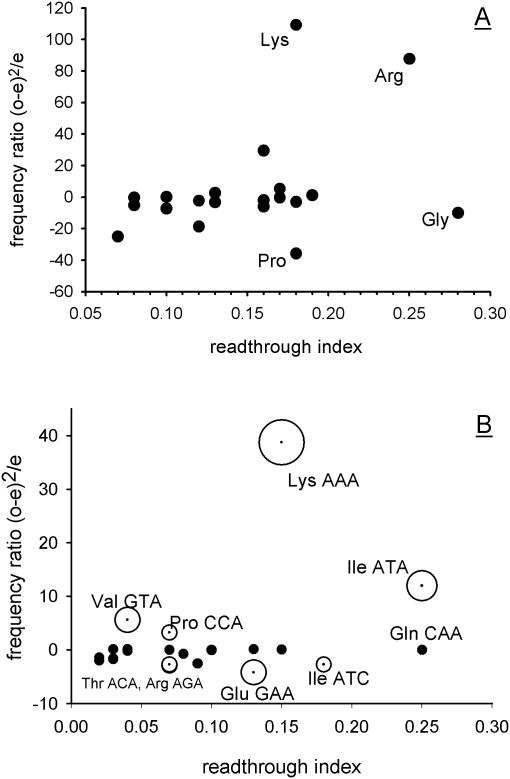
Figure 2.
Codon bias at the −1 and −2 codon positions preceding the stop codon is not predictive of effects on stop codon readthrough. Codon usage at the −1 and −2 codon positions preceding the stop codon was recorded for all S.cerevisiae ORFs. These observed frequencies (o) were compared with expected frequencies (e) derived from a codon usage table for all yeast ORFs. A frequency ratio [(o − e)2/e] was calculated for each of the codons at the −1 position. For the −2 position, the encoded amino acid frequency was recorded (the primary readthrough determinant at this position (15). The frequency ratio was assigned a polarity depending on whether the codon or encoded amino acid was either over-represented (positive polarity) or under-represented (negative polarity) relative to the expected frequency. (A) The −2 position codons were surveyed for all yeast genes, and the polar chi-squared values for each amino acid were plotted against the known effect of amino acid at this position on stop codon readthrough (15). (B) The process was repeated for −1 position codons, surveyed in a sub-set of genes containing other poor −2 position and stop codon context elements (see text for details). The significance level of under- or over-abundance of −1 position codons is indicated by the three sizes of bubbles used [0.001% (largest), 0.01% and 0.1% (smallest)].
It remained possible that the selection against those −1 codons promoting stop codon readthrough might be detectable if a sub-set of stop codons were extracted from the database comprising both (i) a flanking ‘poor’ −2 codon directing high levels of readthrough (15) and (ii) a weakly terminating stop codon/+1 position tetranucleotide (11). Stop codon/+1 nucleotide combinations UAG-C, UGA-G, UGA-C or UAA-C found in the genome survey combined with a −2 codon position encoding any of the readthrough-promoting amino acids Gly, Arg, Cys, Pro or Lys (15), were analysed for a −1 codon position bias. It would be expected that −1 codon positions which are known to direct stop codon readthrough would be under-represented (15,36).
The results showed that frequencies of all −1 codons to which a readthrough effect has previously been ascribed were well within variation produced by sampling error, with the exception of AAA (Lys), ATA (Ile) and GTA (Val), all known to direct readthrough, which paradoxically were significantly over-represented, and GAA (Glu), AGA (Arg), ACA (Thr) and ATC (Ile) which were significantly under-represented (Figure 2B). Thus despite sampling only those −1 codons found combined with known poor contexts at the −2 codon and stop/+1 positions, no consistent inverse correlation between abundance and published readthrough levels was found (15). Second, there was no detectable selection against codons of the type NAA. NAA codons exert negative effects on termination in yeast if placed immediately before the stop codon, and NAA codons are frequently encountered immediately 5′ of many viral stop codon readthrough signals (42,45). While GAA was significantly under-represented, CAA was found as often as would be expected elsewhere in the ORF, and AAA was over-represented.
The over-representation lysine codons at the −1 position is almost certainly due to the previously described amino acid bias at this position. However, the codon bias of lysine codons at the −1 position was markedly skewed. In the yeast genome as a whole, the codon AAA is used only 1.37-fold more frequently than the synonymous codon AAG. However, at the −1 codon position, AAA is used 3.25-fold more often than AAG (not shown). This over-representation of AAA is significant at the 99% level. However, the sub-group of stop codons with AAA in the −1 codon position do not terminate highly expressed genes (for this sub-group of genes analysed, average Codon adaptive index CAI = 0.146, mean codon bias index = 0.07; data not shown). It is therefore not the case that high level expression of this sub-group of genes has driven lysine AAA codon bias at the −1 position. The over-representation of AAA versus AAG at the −1 position is also inconsistent with the reported stimulatory effect of AA dinucleotides preceding the stop codon on readthrough (42), and instead is evidence either of P-site tRNA-specific interactions with the release factor eRF1, or a direct positive effect of −1 codon AAA on termination (15). Taken as a whole, the results indicate the enhancement of termination efficiency is not a dominant factor acting on selection of the −1 or −2 codons.
5′ and 3′ stop codon context effects show similar interactions with the release factor apparatus
The absence of clear selection against poor 5′ contexts indicates that these six nucleotides may only be significant determinants of readthrough in combination with a poor 3′ context. In order to test this hypothesis, poor 5′ (6 nt) and 3′ (6 nt) stop codon contexts were employed singly and in combination to replace the otherwise good 5′ and 3′ context elements of the efficient stop signal terminating the RPS2 ribosomal protein gene (Figure 3A). The 5′ context least favourable to efficient termination was identified from previous work as a glycine codon in the −2 position, combined with a CAA glutamine codon in the −1 position (15). Six nucleotides of immediate 3′ context, derived from the PDE2 gene, were used as a representative poor 3′ context (23).
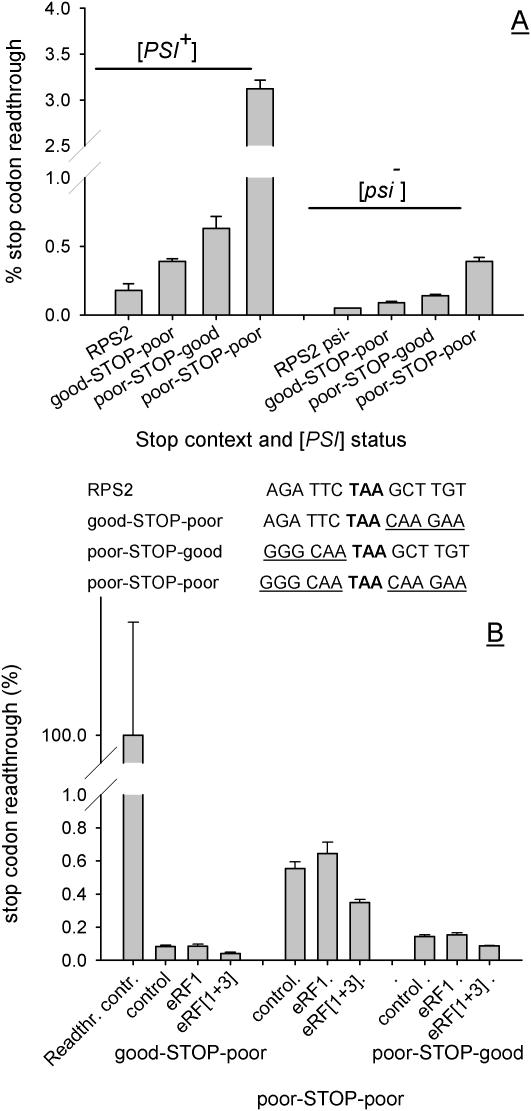
Figure 3.
Stop codon readthrough directed by either poor 5′ or 3′ context is identically responsive to release factor levels. Sequences of 6 nt representing poor 5′ or 3′ stop codon contexts were used singly or in combination to replace the stop codon context elements of the RPS2 gene [hybrid sequences listed in (A)], and cloned into a dicistronic stop codon readthrough vector system (Materials and Methods). (A) Plasmids pAC98-RPS2, pAC98-GP, pAC98-PP and pAC98-PG were transformed into the [PSI+] and [psi−] derivatives of yeast strain 76D694, and levels of stop codon readthrough determined. (B) Levels of stop codon readthrough were measured in the same strains additionally transformed with either the pRS426 and YEp24 vectors (labelled control), pRS426 with the multicopy eRF1 vector pUKC802 (labelled eRF1), or pJR16 and pUKC802 which together direct over-expression of both release factors (labelled eRF[1+3]). For (A and B), the mean of three readthrough determinations is shown (bars represent the SD, n = 3).
The results indicate that poor 5′ and 3′ context elements are both able to direct significant stop codon readthrough when placed in an otherwise good context environment. A poor 5′ context (GGG CAA) produced 0.65% stop codon readthrough in a [PSI+] background when used to replace the RPS2 5′ context elements (Figure 3A). A poor 3′ context (CAAGAA) similarly produced 0.4% readthrough when used to replace the RPS2 3′ context elements. When combined, these two poor context elements synergistically produced 3% readthrough (Figure 3). This observed synergism spanning 12 nt is consistent with previous studies examining a more limited 6 nt flanking the stop codon. The effect was not dependent on the reduced levels of soluble eRF3 found in a [PSI+] genetic background, since similar relative levels of readthrough were directed by all hybrid contexts in a [psi−] genetic background where release factor abundance is wild-type. The discovery that the 5′ context six nucleotides exert effects on readthrough comparable in magnitude to effects exerted by the 3′ six nucleotides is at odds with the observation that only the 3′ context is apparently under selective pressure to optimize termination efficiency.
In order to examine this further, the response of the different stop codon contexts to increased release factor abundance was examined. It is known that increasing release factor levels in the cell acts to increase termination efficiency, by out-competing suppressor tRNAs for stop codon recognition (antisuppression). In yeast, previous reports indicate it is necessary to over-express both eRF1 and the GTPase eRF3 to reduce stop codon readthrough (3). In contrast, only eRF1 over-expression is required to generate an antisuppressor effect in mammalian cells (46). Here, expression of either eRF1 alone, or eRF1 and eRF3 together was used to probe the interaction of the release factor apparatus with 5′ and 3′ context elements directing readthrough. The results show, first, that in support of previous findings, over-expression of eRF1 alone has no antisuppressor effect in yeast. Irrespective of the context tested, over-expression of both eRF1 and eRF3 was necessary to reduce readthrough of a stop codon flanked by poor 5′ and 3′ context elements (e.g. poor-STOP-poor; Figure 3B). Second, the results show that the antisuppressor effect of eRF1/eRF3 over-expression was equally effective at limiting readthrough whether directed by poor 5′, or poor 3′ contexts (a 40–50% reduction in each case). The lack of detectable selection against poor 5′ contexts in the genome (Figure 2) is therefore not explained by any differences in the interaction of 5′ context elements with release factor–tRNA competition.
Nucleotide bias in the 6 nt following the stop codon correlates most strongly with the +1,2,3,5 and+1,2,3,6 nucleotide positions
It is known that nucleotide composition immediately downstream of the stop codon is highly biased in all systems so far examined. In yeast, this bias is almost certainly due to a direct effect of the 6 nt following the stop codon on termination efficiency (12). However, this latter study by Namy and colleagues could necessarily only examine a small subset of 22 out of a possible 4096 6-nt stop codon contexts due to the screening method employed. It was also not clear from this study which of the 6 nt 3′ of the stop codon play a dominant role in directing readthrough. A greater understanding of the relative contributions made by each of the 6 nt would enable predictive identification of leaky stop codon contexts on a genome-wide scale.
To identify the relative importance of the downstream context nucleotides, the 6 nt following the stop codons of all Saccharomyces cerevisiae ORFs were extracted, and surveyed for under-represented or missing 6-nt combinations. However, since there are 4096 possible permutations of ATC and G making up a 6-nt ‘word’, the average representation of any given hexamer downstream of the ∼5500 yeast ORFs is <2. Sampling error, rather than selection, thus dictated that many 6-nt words were missing from the ORF-flanking population (data not shown).
Instead, tetramer sequences were analysed from the 3′ contexts of yeast ORF stop codons; for a sequence 4 nt long, there are 256 possible sequence variations. Ignoring bias, each variation should be found ∼23 times within the population of yeast ORFs, increasing the statistical rigour of the analysis. To analyse all six nucleotide positions 3′ of stop codons, the frequencies of tetramer ‘sub-contexts’ were measured. A sub-context was defined as a combination of four nucleotide positions from the six positions following the stop codon. For example, tetramer sub-contexts can be made up from the 6 nt downstream of the stop codon using positions +1234, or +1235, or +1236, or +1346, etc. (where +1 is the nucleotide following the stop codon). In total there are 15 different tetramer sub-contexts.
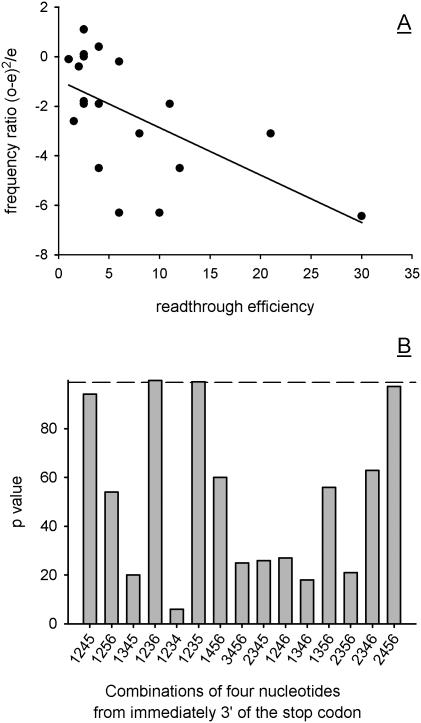
Frequencies of the 256 possible tetrameric sequences occurring downstream of yeast stop codons were recorded for all fifteen sub-contexts. These observed frequencies were compared with expected frequencies derived in the same way from a control sequence population (a sliding 6-nt window sampled 300 000 nt of 3′-UTR sequence derived from the TransTerm database). To correlate abundance with effects on readthrough for all 15 sub-contexts, the relevant sub-context was abstracted from 19 different +6 nucleotide contexts previously reported to direct stop codon readthrough (12). For instance, CAGCTA is a known readthrough-stimulating 3′ context (12). The +1236 subcontext for this sequence is CAGA; its abundance both downstream of stop codons (observed) and in the 3′-UTR population (used to derive an expected f) was compared. For each of 15 sub-context types, the process was repeated with all 19 published readthrough sequences. Derived frequency ratios were plotted against published stop codon readthrough efficiencies, and linear regression used to analyse the level of inverse correlation between abundance and readthrough, producing a p value. Example correlation data is shown for the +1235 combination (Figure 4A). Regression analysis revealed that the abundance of +1235 and +1236 sub-contexts showed a high level of inverse correlation with readthrough efficiency (>99% p value for the regression coefficient; Figure 4B). Nucleotides present in positions +1235 and +1236 of efficient readthrough signals were strongly selected against in the genome in ORF 3′ flanking positions. Combinations of nucleotides at positions +1235 and +1236 following the stop codon are thus the strongest determinants of readthrough. The +4 nucleotide position following the stop codon, in combination with any of the other three positions, was a poor predictor of readthrough efficiency except as part of the +2456 sub-context (p = 98%).
Figure 4.
Identifying key nucleotide components of the 3′ stop codon context. Fifteen different tetramer combinations of nucleotide positions (sub-contexts) from within the six nucleotide positions downstream of the stop codon were tested for an inverse correlation between tetramer sequence usage and known effects on stop codon readthrough. A set of 19 known 3′ contexts known to confer leakiness on the upstream stop codon was used to calibrate the predictive quality of each of the 15 sub-contexts. The frequency of these reference tetrameric sequences in control (3′-UTR) and stop codon flanking populations was compared, producing expected and observed frequencies, respectively. These were used to calculate frequency ratios [(o − e)2/e], which were plotted against known readthrough levels for the 22 reference 3′ contexts. The +1235 subcontext correlation between abundance in the genome and stop codon readthrough efficiency is shown (A). Linear regression analysis was carried out on this type of correlation plot for all 15 sub-context types. The regression p value obtained in each case was plotted in histogram form (B). For reference, the dotted line shows the 99% significance cut-off used.
Having determined which of the 15 combinations of four 3′ nucleotide positions were the best combined predictors of readthrough, those +1235 and +1236 position ‘words’ most under-represented in the genome could be identified (Table 2). As expected, some of these words used CAA in the +123 positions, a key component of the Tobacco Mosaic Virus readthrough signal which is functional in yeast (28), and also of many of the readthrough signals identified by Namy and colleagues (12). However, Some CAA-containing subcontexts identified (+1236 CAAT, CAAC) were not previously known to drive readthrough, although their under-representation suggests they too will direct stop codon suppression. In addition, 4-nt words in the +1236 and +1235 groupings shared common +123 position nucleotides in both under-represented (CAA, CTC, TTT; Table 2A) and over-represented categories (ATC, ATA, AGG, GCC; Table 2A). The TAG and TGA stop codons were also over-represented in the +1236 and +1235 word groupings respectively, accounting partly for the over-abundance of tandem stop signals in S.cerevisiae (Figure 1).
Table 2. Tetranucleotides found 3′ of yeast stop codons with significantly altered abundances.
| Analysis of the six nucleotides 3′ of the stop codon | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A: 4-Position sub-context analysis | B: Data mining analysis | ||||||||||||
| Under-represented 3′ sequences | Fold-reduced abundance | Significance(%) | Over-represented 3′ sequences | Fold-increased abundance | Significance(%) | Under-represented 3′ sequences | Fold-reduced abundance | Significance (%) | |||||
| CAA . . A | 2.27 | 0.0002 | ATC . . C | 2.36 | 1 × 10−5 | C | . | A | . | . | C | 1.92 | 0.001 |
| TTT . . T | 1.45 | 0.013 | ATC . . T | 2.10 | 8 × 10−6 | C | A | A | . | . | . | 1.87 | 0.001 |
| AAG . . G | 1.86 | 0.07 | AGG . . A | 2.11 | 5.4 × 10−5 | C | . | . | C | . | A | 1.77 | 0.005 |
| CAA . . C | 2.22 | 0.18 | TCA . . T | 1.72 | 0.005 | C | . | A | A | . | . | 1.75 | 0.001 |
| CAA . . T | 1.70 | 0.2 | GCC . . C | 2.21 | 0.05 | C | . | . | . | A | A | 1.53 | 0.005 |
| CTC . . T | 2.37 | 0.23 | ATA . . A | 1.44 | 0.01 | C | . | . | . | A | T | 1.53 | 0.005 |
| CAT . . T | 1.67 | 0.27 | GCT . . C | 2.01 | 0.07 | . | . | G | A | . | G | 1.48 | 0.02 |
| CAA . A . | 2.08 | 0.001 | TAG . . T | 1.6 | 0.15 | C | T | . | C | . | . | 1.48 | 0.1 |
| TTT . T . | 1.54 | 0.002 | ATC . A . | 166 | 1.5 × 10−22 | . | A | . | A | . | G | 1.3 | 0.05 |
| CCG . T . | 10.4 | 0.032 | ACA . A . | 15.4 | 0.007 | C | . | A | C | . | . | 1.3 | 0.63 |
| CTC . A . | 2.58 | 0.087 | AGG . A . | 12.3 | 0.04 | . | A | . | T | . | A | 1.2 | 0.47 |
| GAT . A . | 1.84 | 0.1 | GCC . A . | 11.1 | 0.1 | T | T | C | . | . | . | 1.3 | 0.13 |
| CAA . T . | 1.75 | 0.2 | TGA . A . | 10.2 | 0.06 | ||||||||
| CAA . G . | 1.93 | 0.23 | ATA . A . | 9.8 | 0.04 | ||||||||
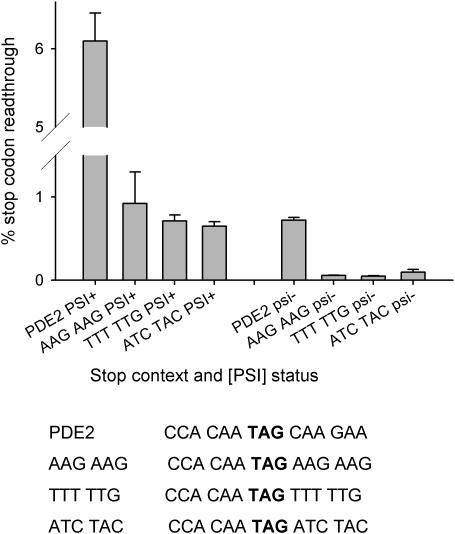
Some novel combinations of 3′ flanking nucleotides emerged from this analysis which were significantly under-represented, predictive of a poor termination context, but which were not previously known readthrough enhancers, such as TTTxTx from the +1235 grouping, and AAGxxG from the +1236 grouping. To test their effect on readthrough, 6-nt words containing these 4-nt groupings were used to substitute the PDE2 gene stop codon 3′ context in the pAC98 readthrough vector system. In each case, the undefined nucleotides (e.g. +4 and +5 in AAGxxG) were selected by identifying a specific AAGxxG context which was most under-represented in the yeast genome. The readthrough level of these novel contexts was compared to that of an over-represented 3′ context (ATCTAC). There was no significant difference in readthrough level between these three sequences, indicating that the under-abundance of some contexts cannot be explained by their negative effect on efficiency of stop codon recognition (Figure 5).
Figure 5.
Testing potential readthrough contexts identified through analysis of genome-wide 3′ contexts. Sequences of 6 nt identified in the 3′ context genome screen which represent potential poor 3′ stop codon contexts were tested for their ability to direct stop codon readthrough. The nucleotide hexamers AAGAAG and TTTTTG were used to replace the corresponding 3′ context element of the PDE2 stop codon environment placed in the pAC98 readthrough test system (Materials and Methods). The PDE2 3′ context was used as a positive control, and an over-represented 3′ sequence (ATCTAC) as a negative control. Levels of stop codon readthrough were determined in both [PSI+] and [psi−] derivatives of yeast strain 76D694. The mean of three readthrough determinations is shown (bars represent the SD, n = 3).
A data mining method to identify poor 3′ contexts de novo
The previous identification of weak 3′ contexts was based on the under-representation of whole groups of 4-nt words, or sub-contexts, downstream of the stop codon. The technique relies on a training set of readthrough data to identify significant sub-contexts. A method which could rapidly identify under-represented combinations of nucleotides from the 3′ stop flanking region, without reference to a training data set of stop codon readthrough data would be of use in probing termination efficiency context effects in novel genomes.
To address this issue, a new data-mining method, utilizing the Kohonen Self-Organizing Map (SOM) was employed. The SOM is a type of neural network which trains on a data set, grouping items which share common characteristics (Materials and Methods). The learning process was guided using an automated comparison of the observed versus expected frequency ratio for each 4-nt word within the groupings generated. The use of the data mining method considerably speeds up the process of identifying groups of potential readthrough contexts on the basis of under-representation.
Expected frequencies were measured from the complete 3′-UTR data set (Materials and Methods). Groupings where >70% of such ratios were <1 (indicating selection against that nucleotide combination) were considered significant and were retained, and the common characteristic for that group recorded. The results from this analysis show clearly that it is possible to identify weak stop codon contexts simply on the basis of careful analysis of the frequencies of different nucleotide combinations using this new method (Table 2B). Patterns identified include CAA at positions +1, 2 and 3 (C1A2A3). This nucleotide context is found in the TMV readthrough signal, which functions in yeast. C1A2A3 was identified three times during the analysis of the 15 sub-context types, and was found as part of 5 of the 8 most leaky stop signals identified previously in vivo (12). C1A3A4 was also identified as a poor context. Although in the previous section, the fourth nucleotide position was shown to be the least important determinant of readthrough (Figure 4B), the data mining approach shows A4 can be important, but crucially, only in combination with C1A3. Comparing this in silico approach to in vivo data, C1A3A4 is found as part of 3 of the 8 most ‘leaky’ 3′ contexts identified in a recent yeast screen, verifying the power of the data mining technique. The utility of this approach is also corroborated by the identification, using the data mining technique, of the patterns C1C4A6, A2A4G6, C1A5T1 and A2T4A6, all of which are components of the ‘leaky’ 3′ contexts previously reported in yeast (12).
DISCUSSION
Stop codons act as the 3′ marker of an ORF, the point at which translation should halt. However, there are an increasing number of known examples of stop codon readthrough, both viral and cellular (18,23). At least in the case of the yeast PDE2 gene and the Drosophila headcase gene, this readthrough has physiological consequences for the cell (22,23,47). Comparison of a range of phenotypes exhibited by different [PSI+] and [psi−] yeast strains has also indicated that yeast physiology can be substantially affected by increasing levels of readthrough (48). There may thus be a population of stop codons in yeast that are leaky, and which when readthrough, generateC-terminally extended proteins with diverse effects on physiology. It seems likely that just as the use of different translation initiation sites at the 5′ end of an ORF allows a cell to express a variety of proteins from a single mRNA, similar mechanisms of variable efficiency of stop codon recognition can allow the expression of proteins with differing C-termini. An ability to predict leaky stop codon contexts for any genome would therefore be extremely useful in identifying proteins with heterogeneous C-termini.
In this study, a detailed survey was made of stop codon context in the yeast genome to identify the types of bias which exist both 3′ and 5′ of the stop codon, with the broad aim of developing predictive techniques which identify ‘tight’, and ‘leaky’ stop codon nucleotide environments for any genome. The work showed clearly that, as previously reported, bias within the 3′ context was strongly correlated with stop codon recognition efficiency. However, 5′ context, both at the −1 and −2 codon positions before the stop codon, was subject to bias which did not correlate with known effects on stop codon recognition. Neither could a bias correlating with termination efficiency be revealed at the −1 codon position by sampling sub-sets of stop codons containing other known poor context elements (Figure 2). It seems as if selection for given amino acids at the C-terminus of proteins may mask any selection operating at the level of termination efficiency, at least in yeast. The C-termini of proteins are often the site of important targeting sequences, such as the endoplasmic reticulum retention and the vacuolar targeting signals. The over-abundance of di-basic pairs of amino acids such as Arg–Lys in the last two positions of proteins may arise because of their ability in this position to interact with phospholipid (44).
This 5′ context analysis did confirm that lysine codons are highly over-represented at the −1 position, as previously reported (43,44). However, there was a marked AAA lysine codon bias at the −1 position inconsistent with the known readthrough stimulating effects of AA dinucleotides before the stop codon. This bias was found within a group of genes with otherwise low codon bias. This −1 position codon bias may be indicative of differential effects of the two lysyl-tRNAs on eRF1 termination efficiency, conflicting with a recent report which found no evidence of selection for −1 codons on the basis of tRNA identity (42). Further work will be necessary to investigate this effect. Strikingly, there was no detectable selection against the use of CAA or ATA codons at the −1 position, despite the known readthrough enhancing effects of these codons at this position.
The inability to detect significant 5′ bias correlating with termination efficiency is not caused by lesser effects of 5′ context elements on stop codon readthrough. 5′ elements were slightly more efficient at driving readthrough, in both [PSI+] and a wild-type [psi−] background (Figure 3). Readthrough directed by poor 5′ or 3′ elements could be reduced to the same extent by over-expressing the eRF1 and eRF3 release factors. This demonstrates that 5′ contexts can be as effective as 3′ contexts in interfering with eRF competition for the stop codon, and provides no explanation as to why 5′ contexts should not be subject to selection on the basis of termination effects. Using different assay methods to those previously employed, the results also confirmed that in yeast, over-expression of both release factors is required to produce antisuppression, and that such antisuppression operates in the same way against 5′ and 3′ context elements (Figure 3). The assay method used here, employing a dicistronic reporter, eliminates the possibility of artefacts based on mRNA stability. To explain this result, it has been proposed that eRF3 is naturally in excess in the yeast cell. Over-expression of eRF1 should therefore move to restore eRF1 and eRF3 molar equivalence, causing antisuppression: this was not observed.Conversely, if it is proposed that eRF1 is in excess over eRF3 in the cell, and ectopic expression further increases eRF1 levels, the results clearly indicate that eRF1 alone is unable to compete against suppressor tRNAs. It seems likely therefore that at least in yeast, both eRF1 and eRF3 are together required to generate anti-suppressive action, and that the requirements for antisuppression are therefore different in yeast and mammalian cells.
The study revealed that the strong selection against poor 3′ contexts is helped in fungi by the highly significant selection of second in-frame stop codons downstream of the ORF. These codons are over-represented at the +1, +2 and +3 codon positions. While it cannot be excluded that their occurrence is the fortuitous consequence of the selection for U, G and A rich sequences in the 3′-UTR at this position, it seems likely that they act as ‘safety’ stops, limiting stop codon readthrough by providing a second chance for the translation terminationapparatus. Evolution appears to favour the selection of safety stop codons in addition to an effective and ‘leak-proof’ 3′ stop codon context.
What is the nature of 3′ context effect in yeast? In this study, bias factors have been carefully analysed by genome-wide surveys to identify under-represented sequences downstream of the stop codon. By doing so, it was possible to tease apart the components of the 3′ context which interact and combine to regulate stop codon recognition. By using a training set of known readthrough signals, combinations of 4 nt within the +6 nt region were identified which together determine termination efficiency. These were found to be principally the +1235 and +1236 groupings. The +4 nt position was largely irrelevant, and sequence abundance did not correlate well with readthrough efficiency for all but one of the nine sub-contexts in which it is found. The rules emerging from the genome-wide survey agreed very well with the consensus poor 3′ context CA(A/G)N(UCG)A elucidated by Namy and colleagues on the basis of their 19 readthrough signals. Whether or not all the under-represented contexts identified actually drive stop codon readthrough remains to be tested. However, analysis of the peptide tags encoded by ORFs downstream of poor stop contexts has revealed that as a group, their amino acid composition is significantly more similar to that of the global yeast proteome than that of the peptide tags encoded by dORFs downstream of over-represented, good, contexts (data not shown). This suggests at least some of the dORFs 3′ of weak stop codons may be expressed.
However, while identifying PDE2-like contexts as under-represented, the survey also found several other over- and under-represented groupings downstream of yeast ORFs, including some novel sequences. The under-represented sequences did not direct significant readthrough in a PDE2 test-bed context. The findings indicate that multiple selection forces may operate on the 3′ context, only one of which is termination efficiency. These forces could derive from requirements to optimize contacts between mRNA and ribosomal proteins or to eliminate mRNA secondary structures [known components of readthrough motifs, (4)]. Some contexts might interfere with the rapid juxtaposition of eRF1 domain 1, responsible for stop recognition, and the GGQ motif responsible for triggering peptidyl-release. However, it is known that termination efficiency is defined by both Km value for the stop codon, as well as the Kcat value for the reaction (49). Contexts which impose a kinetic penalty on termination thus also confer a fidelity defect. Alternatively, 3′ context may affect ribosome release. It has been previously suggested that the stop codon context of the GCN4 uORFs 1 and 4 might have differing effects on ribosome release rates from the mRNA, influencing the resumption of scanning by ribosomes following termination (50). In fact, the 3′ context TTTxxT (Figure 5) is part of one of the identified contexts supporting uORF1 resumed scanning function (15), implying it could contribute to slowed ribosome release.
The use of the new data mining method utilizing the Self-organizing map allowed the identification of under-represented groupings of nucleotides de novo, without reference to a training set. This approach thus allows weak context identification for organisms where nothing is known about the 3′ context preferences of the release factor apparatus. This approach was successful in identifying the known weak 3′ context C1A2A3, as well as other triplet nucleotide combinations identified in a recent screen for 3′ contexts directing readthrough, and illustrates the potential of this approach, particularly for identifying inter-dependencies of nucleotides in defining a poor context e.g. A4 combined with C1A3 (Table 2). The analysis of under-represented groupings of nucleotides 3′ of the stop codon shows clearly that the signature of selection against 3′ nucleotide combinations known to drive readthrough is clearly detectable.
The data presented has highlighted the difficulty of identifying weak stop codon contexts in silico. 5′ context determinants in yeast are still the subject of some debate, and the survey of codon bias at the −1 and −2 codon positions presented here has shown that selective forces cannot readily be used to resolve the issue. New techniques presented here are capable of identifying known poor 3′ determinants on the basis of selection against those sequences in ORF-flanking regions. However, despite their reduced abundance, apparently poor contexts are used in yeast, and the stop codons within that context are readthrough at significant frequency (23). The potential for even short peptide tags to alter the cellular localization or activity of a protein is great. Many short peptide tags are known which will target proteins to the membrane using, for instance, a myristoyl group or alter retention in the endoplasmic reticulum. Addition of even a single amino acid could complete a partial targeting signal already present at the C-terminus of a protein. All protein extensions altering targeting could potentially act as genetically dominant, gain of function events. Thus, despite the generally low efficiency of stop codon readthrough events, such events may well have a phenotypic consequence for the cell. The elucidation of the full gamut of context effects regulating stop codon readthrough is thus essential for a full understanding of cellbiology.
Acknowledgments
ACKNOWLEDGEMENTS
Dr S. Liebman (University of Chicago, IL) and Prof. M. F. Tuite (University of Kent, UK) are thanked for gifts of strains and plasmids. This work was supported by grants to I.S. from the Biotechnology and Biological Sciences Research Council, and the Wellcome Trust.
REFERENCES
- 1.Frolova L., Le Goff,X., Rasmussen,H.H., Cheperegin,S., Drugeon,G., Kress,M., Arman,I., Haenni,A.L., Celis,J.E., Philippe,M. et al. (1994) A highly conserved eukaryotic protein family possessing properties of polypeptide-chain release factor. Nature, 372, 701–703. [DOI] [PubMed] [Google Scholar]
- 2.Zhouravleva G., Frolova,L., Le Goff,X., Leguellec,R., Ingevechtomov,S., Kisselev,L. and Philippe,M. (1995) Termination of translation in eukaryotes is governed by 2 interacting polypeptide-chain release factors, eRF1 and eRF3. EMBO J., 14, 4065–4072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stansfield I., Jones,K.M., Kushnirov,V.V., Dagkesamanskaya,A.R., Poznyakovski,A.I., Paushkin,S.V., Nierras,C.R., Cox,B.S., Teravanesyan,M.D. and Tuite,M.F. (1995) The products of the sup45 (erf1) and sup35 genes interact to mediate translation termination in Saccharomyces cerevisiae. EMBO J., 14, 4365–4373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bertram G., Innes,S., Minella,O., Richardson,J. and Stansfield,I. (2001) Endless possibilities: translation termination and stop codon recognition. Microbiology, 147, 255–269. [DOI] [PubMed] [Google Scholar]
- 5.Edelman I. and Culbertson,M.R. (1991) Exceptional codon recognition by the glutamine transfer-RNAs in Saccharomyces cerevisiae. EMBO J., 10, 1481–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pure G.A., Robinson,G.W., Naumovski,L. and Friedberg,E.C. (1985) Partial suppression of an ochre mutation in Saccharomyces cerevisiae by multicopy plasmids containing a normal yeast transfer RNA-Gln gene. J. Mol. Biol., 183, 31–42. [DOI] [PubMed] [Google Scholar]
- 7.Brown C.M., Stockwell,P.A., Trotman,C.N. and Tate,W.P. (1990) The signal for the termination of protein synthesis in prokaryotes. Nucleic Acids Res., 18, 2079–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown C.M., Stockwell,P.A., Trotman,C.N.A. and Tate,W.P. (1990) Sequence-analysis suggests that tetra-nucleotides signal the termination of protein-synthesis in eukaryotes. Nucleic Acids Res., 18, 6339–6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Poole E.S., Brown,C.M. and Tate,W.P. (1995) The identity of the base following the stop codon determines the efficiency of in-vivo translational termination in Escherichia coli. EMBO J., 14, 151–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tate W.P., Poole,E.S., Horsfield,J.A., Mannering,S.A., Brown,C.M., Moffat,J.G., Dalphin,M.E., Mccaughan,K.K., Major,L.L. and Wilson,D.N. (1995) Translational termination efficiency in both bacteria and mammals is regulated by the base following the stop codon. Biochem.Cell Biol., 73, 1095–1103. [DOI] [PubMed] [Google Scholar]
- 11.Bonetti B., Fu,L.W., Moon,J. and Bedwell,D.M. (1995) The efficiency of translation termination is determined by a synergistic interplay between upstream and downstream sequences in Saccharomyces cerevisiae. J. Mol. Biol., 251, 334–345. [DOI] [PubMed] [Google Scholar]
- 12.Namy O., Hatin,I. and Rousset,J.P. (2001) Impact of the six nucleotides downstream of the stop codon on translation termination. EMBO Rep., 2, 787–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mottaguitabar S., Bjornsson,A. and Isaksson,L.A. (1994) The 2nd to last amino-acid in the nascent peptide as a codon context determinant. EMBO J., 13, 249–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bjornsson A., Mottaguitabar,S. and Isaksson,L.A. (1996) Structure of the C-terminal end of the nascent peptide influences translation termination. EMBO J., 15, 1696–1704. [PMC free article] [PubMed] [Google Scholar]
- 15.Mottaguitabar S., Tuite,M.F. and Isaksson,L.A. (1998) The influence of 5′ codon context on translation termination in Saccharomyces cerevisiae. Eur. J. Biochem., 257, 249–254. [DOI] [PubMed] [Google Scholar]
- 16.Pelham H.R. (1978) Leaky UAG termination codon in tobacco mosaic virus RNA. Nature, 272, 469–471. [DOI] [PubMed] [Google Scholar]
- 17.Skuzeski J.M., Nichols,L.M., Gesteland,R.F. and Atkins,J.F. (1991) The signal for a leaky UAG stop codon in several plant viruses includes the two downstream codons. J. Mol. Biol., 218, 365–373. [DOI] [PubMed] [Google Scholar]
- 18.Yoshinaka Y., Katoh,I., Copeland,T.D. and Oroszlan,S. (1985) Murine leukemia virus protease is encoded by the gag-pol gene and is synthesized through suppression of an amber termination codon. Proc. Natl Acad. Sci. USA, 82, 1618–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wills N.M., Gesteland,R.F. and Atkins,J.F. (1991) Evidence that a downstream pseudoknot is required for translational read-through of the Moloney murine leukemia virus gag stop codon. Proc. Natl Acad. Sci. USA, 88, 6991–6995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Namy O., Duchateau-Nguyen,G., Hatin,I., Hermann-Le Denmat,S., Termier,M. and Rousset,J.P. (2003) Identification of stop codon readthrough genes in Saccharomyces cerevisiae. Nucleic Acids Res., 31, 2289–2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robinson D.N. and Cooley,L. (1997) Examination of the function of two kelch proteins generated by stop codon suppression. Development, 124, 1405–1417. [DOI] [PubMed] [Google Scholar]
- 22.Steneberg P. and Samakovlis,C. (2001) A novel stop codon readthrough mechanism produces functional Headcase protein in Drosophila trachea. EMBO Rep., 2, 593–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Namy O., Duchateau-Nguyen,G. and Rousset,J.P. (2002) Translational readthrough of the PDE2 stop codon modulates cAMP levels in Saccharomyces cerevisiae. Mol. Microbiol., 43, 641–652. [DOI] [PubMed] [Google Scholar]
- 24.Sherman F. (1991) Getting started with yeast. Methods Enzymol., 194,3–21. [DOI] [PubMed] [Google Scholar]
- 25.Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York. [Google Scholar]
- 26.Christianson T.W., Sikorski,R.S., Dante,M., Shero,J.H. and Hieter,P. (1992) Multifunctional yeast high-copy-number shuttle vectors. Gene, 110, 119–122. [DOI] [PubMed] [Google Scholar]
- 27.Stansfield I., Grant,C.M., Akhmaloka and Tuite,M.F. (1992) Ribosomal association of the yeast SAL4 (SUP45) gene-product—implications for its role in translation fidelity and termination. Mol. Microbiol., 6,3469–3478. [DOI] [PubMed] [Google Scholar]
- 28.Stahl G., Bidou,L., Rousset,J.P. and Cassan,M. (1995) Versatile vectors to study recoding: conservation of rules between yeast and mammalian cells. Nucleic Acids Res., 23, 1557–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bidou L., Stahl,G., Hatin,I., Namy,O., Rousset,J.P. and Farabaugh,P.J. (2000) Nonsense-mediated decay mutants do not affect programmed −1 frameshifting. RNA, 6, 952–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gietz R.D. and Woods,R.A. (2002) Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol., 350, 87–96. [DOI] [PubMed] [Google Scholar]
- 31.Jacobs G.H., Rackham,O., Stockwell,P.A., Tate,W. and Brown,C.M. (2002) Transterm: a database of mRNAs and translational control elements. Nucleic Acids Res., 30, 310–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Frischmeyer P.A., van Hoof,A., O'Donnell,K., Guerrerio,A.L., Parker,R. and Dietz,H.C. (2002) An mRNA surveillance mechanism that eliminates transcripts lacking termination codons. Science, 295, 2258–2261. [DOI] [PubMed] [Google Scholar]
- 33.Starkey A.J. (2004) Controlled Selection of Inputs. PCT/GB02/00160.
- 34.Kohonen T. (2001) Self-Organising Maps. Springer-Verlag, Berlin, Germany. [Google Scholar]
- 35.Levenshtein V.I. (1966) Binary Codes Capable of Correcting Deletions, Insertions and Reversals. Soviet Phisics Doklady, 10, 707. [Google Scholar]
- 36.Major L.L., Edgar,T.D., Yee,Y.P., Isaksson,L.A. and Tate,W.P.(2002) Tandem termination signals: myth or reality? FEBS Lett., 514,84–89. [DOI] [PubMed] [Google Scholar]
- 37.Sheskin D.J. (2004) Handbook of Parametric and Non-parametric Statistical Procedures. Chapman and Hall/CRC Boca Raton, FL. [Google Scholar]
- 38.Tuite M.F. and Lindquist,S.L. (1996) Maintenance and inheritance of yeast prions. Trends Genet., 12, 467–471. [DOI] [PubMed] [Google Scholar]
- 39.Galagan J.E., Calvo,S.E., Borkovich,K.A., Selker,E.U., Read,N.D., Jaffe,D., FitzHugh,W., Ma,L.J., Smirnov,S., Purcell,S. et al. (2003) The genome sequence of the filamentous fungus Neurospora crassa. Nature, 422, 859–868. [DOI] [PubMed] [Google Scholar]
- 40.Wood V., Gwilliam,R., Rajandream,M.A., Lyne,M., Lyne,R., Stewart,A., Sgouros,J., Peat,N., Hayles,J., Baker,S. et al. (2002) The genome sequence of Schizosaccharomyces pombe. Nature, 415,871–880. [DOI] [PubMed] [Google Scholar]
- 41.Parham S.N., Resende,C.G. and Tuite,M.F. (2001) Oligopeptide repeats in the yeast protein Sup35p stabilize intermolecular prion interactions. EMBO J., 20, 2111–2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tork S., Hatin,I., Rousset,J.P. and Fabret,C. (2004) The major 5′ determinant in stop codon read-through involves two adjacent adenines. Nucleic Acids Res., 32, 415–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gatto G.J. Jr. and Berg,J.M. (2003) Nonrandom tripeptide sequence distributions at protein carboxyl termini. Genome Res., 13, 617–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scheglmann D., Werner,K., Eiselt,G. and Klinger,R. (2002) Role of paired basic residues of protein C-termini in phospholipid binding. Protein Eng., 15, 521–528. [DOI] [PubMed] [Google Scholar]
- 45.Harrell L., Melcher,U. and Atkins,J.F. (2002) Predominance of six different hexanucleotide recoding signals 3′ of read-through stop codons. Nucleic Acids Res., 30, 2011–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Le G., X, Philippe,M. and Jean-Jean,O. (1997) Overexpression of human release factor 1 alone has an antisuppressor effect in human cells. Mol. Cell. Biol., 17, 3164–3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Steneberg P., Englund,C., Kronhamn,J., Weaver,T.A. and Samakovlis,C. (1998) Translational readthrough in the hdc mRNA generates a novel branching inhibitor in the drosophila trachea. Genes Dev., 12, 956–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.True H.L. and Lindquist,S.L. (2000) A yeast prion provides a mechanism for genetic variation and phenotypic diversity. Nature, 407, 477–483. [DOI] [PubMed] [Google Scholar]
- 49.Freistroffer D.V., Kwiatkowski,M., Buckingham,R.H. and Ehrenberg,M. (2000) The accuracy of codon recognition by polypeptide release factors. Proc. Natl Acad. Sci. USA, 97, 2046–2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Grant C.M. and Hinnebusch,A.G. (1994) Effect of sequence context at stop codons on efficiency of reinitiation in GCN4 translational control. Mol. Cell. Biol., 14, 606–618. [DOI] [PMC free article] [PubMed] [Google Scholar]