Abstract
The cytotoxic T-lymphocyte (CTL) response plays an important role in the control of respiratory syncytial virus (RSV) replication and the establishment of a Th1-CD4+ T cell response against the virus. Despite lacking Major Histocompatibility Complex I (MHC I)-restricted epitopes, the attachment G glycoprotein of RSV enhances CTL activity toward other RSV antigens, and this effect depends on its conserved central region. Here, we report that RSV-G can also improve CTL activity toward antigens from unrelated pathogens such as influenza, and that a mutant form of RSV-G lacking four conserved cysteine residues at positions 173, 176, 182, and 186 fails to enhance CTL responses. Our results indicate that these conserved residues are essential for the wide-spectrum pro-CTL activity displayed by the protein.
Keywords: RSV, G glycoprotein, Influenza, Cellular immunity, Vaccines
Introduction
Respiratory syncytial virus (RSV) is a negative-sense, single-stranded RNA virus of the family Paramyxoviridae that causes lower respiratory tract infections [1]. RSV infects approximately half of the infant population during the first year of life, and more than 90% of infants by the end of their second year [1, 2]. The large majority of pediatric patients undergoing primary RSV infection develop lower respiratory illness, which in severe cases requires hospitalization [1, 2]. In fact, RSV is the major cause of pediatric hospitalizations due to viral respiratory infections in the United States and in the world [1, 2]. Despite its high public health impact, no RSV vaccine has ever been licensed.
Efficient adaptive immune responses to most viral pathogens typically require the activation of anti-viral cytotoxic T lymphocytes (CTL) [3]. Antigen presentation of viral peptides via Major Histocompatibility Complex class I (MHC I) molecules triggers the activation of CD8+ T cells, which produce antiviral cytokines and induce programmed cell death in infected cells [3]. In the case of RSV infections, CD8+ T cells against viral antigens have been reported to play an important role in protective immunity and recovery from infection [4–6]. Activation of RSV-specific CTLs during experimental vaccinations also has been shown to modulate the activity of CD4+ T cells, resulting in Th1 skewing of the T helper response [7–9]. Since Th2 responses against RSV have been associated with aberrant immune reactions, most notably enhanced RSV disease (ERD), it is believed that a safe and effective vaccine against this virus should evoke Th1 responses [7, 8, 10–14]. Thus, CTL activity against RSV plays a critical role during infection directly by eliminating infected cells and indirectly by contributing to the establishment of an appropriate cytokine environment conducive to viral clearance.
In BALB/c mice, the dominant RSV-specific CTL epitope is an H2-Kd-restricted peptide encompassing residues 82–90 of the anti-termination factor M2-1 (M282–90) [15–18]. Approximately 40% of the primary RSV-specific H-2d-restricted CTL response is directed against this peptide [19]. We have recently shown that another viral protein, the attachment glycoprotein (G), is critical for the generation of a robust anti-M282–90 CTL response during RSV infections [20]. In addition, Mei and colleagues have reported that CTL responses elicited by vaccines carrying the M282–90 epitope fused to the Measles F protein were greatly enhanced by co-immunization with a recombinant fragment of G containing the conserved central region of the protein [21, 22]. Interestingly, G lacks H-2d-restricted epitopes in mice and, therefore, cannot elicit a CTL response against itself [7, 9, 15, 23, 24]. Furthermore, no anti-G CTL activity has been reported in humans [17, 24–26]. Therefore, despite its lack of CTL epitopes, G appears to act in trans to boost CTL responses against other RSV antigens by an unknown mechanism [20].
In cells infected with RSV, the glycoprotein G is produced in two forms: a full-length, membrane-anchored polypeptide with a short cytoplasmic domain, and a truncated secreted soluble polypeptide [1, 27]. The sequence of G that is common to both forms of the protein bears two variable mucin-like segments and a conserved central region rich in cysteine residues (G-cysteine-rich region or GCRR) [27]. Within the GCRR, there are four cysteines at positions 173, 176, 182, and 186 that are highly conserved between RSV antigenic subgroups A and B, and which form a cystine noose domain via two disulfide bonds between amino acids 173–186 and 176–182, respectively [1, 23] (Fig. 1). We have recently shown that the pro-CTL effect of G resides within this conserved cystine noose domain [20]. Therefore, we speculated that the conserved cysteine residues within the GCRR are critical to enhance CTL responses. Here, we report that a mutant form of RSV-G lacking cysteines 173, 176, 182, and 186 fails to enhance CTL responses in RSV-infected mice, indicating that these residues are essential for the pro-CTL activity mediated by the glycoprotein. Furthermore, we showed that the CTL-enhancing activity of G can also be elicited against unrelated antigens from influenza virus, indicating that G has broad pro-CTL adjuvant capabilities.
Fig. 1.
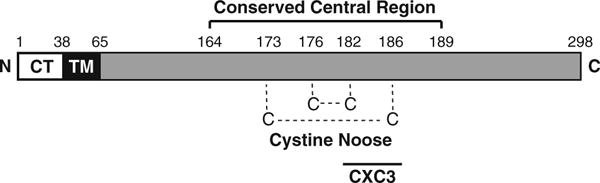
Schematic representation of the membrane-bound form of the RSV G glycoprotein. The cytosolic (CT), transmembrane (TM), and extracellular domains of the protein are depicted by white, black, and gray rectangles, respectively. The sequence of G between amino acids 164 and 189, representing the conserved central region is labeled on top. The disulfide bonds that form the cystine noose domain between conserved cysteines 173 and 186, and between cysteines 176 and 182 are indicated with dotted lines. The CX3C motif between residues 182 and 186 is denoted with a black bar at the bottom. N amino terminus and C carboxy terminus
Materials and methods
DNA vaccines
The pCDNA3.1 plasmid coding for the wild type G gene of RSV (Gwt) was a generous gift from Dr. Ralph Tripp, University of Georgia. To create the quadruple alanine G mutant construct (C4A), we used Gwt as a template in a PCR reaction to generate an intermediate pCDNA3.1 vector carrying the G gene with a deletion in the sequence coding for the GCRR. In the process, we engineered unique EcoRI and BamHI sites flanking this deletion at positions 484 and 563, respectively. Subsequently, annealed primers with compatible EcoRI and BamHI ends, and coding for a mutant central region where cysteines 173, 176, 182, and 186 were replaced by alanines, was introduced by ligation with T4 DNA ligase (Invitrogen, Carsbad, CA). The resulting construct was then propagated in DH5α cells, and transformants were selected for their growth in the presence of ampicillin. Restriction enzyme analysis and DNA sequencing were performed to confirm the correct orientation and reading frame of the inserted gene segment. For immunization purposes, plasmid DNA was purified from DH5α cultures using the Endotoxin-Free Plasmid Mega Preparation Kit (Qiagen, Hildun, Germany). The DNA preparations were quantified by optical density measurements at 260 nm.
Expression and localization of G proteins
To visualize G protein expression at the cell membrane, 293 cells were grown on glass coverslips and transiently transfected with either Gwt, C4A or pCDNA3.1 plasmids using lipofectamine 2000 (Invitrogen) following manufacturer’s instructions. Twenty-four hours after transfection cells were washed with phosphate-buffered saline (PBS) and fixed for 15 min at room temperature with 4% freshly prepared paraformaldehyde in PBS, followed by additional washes with PBS. Immunofluorescence assays were performed by incubating the samples with a goat anti-RSV polyclonal antibody (ViroStat, Portland, ME) at a 1:500 dilution in PBS, followed by anti-goat antibody conjugated to Alexa fluor 568 (Invitrogen) diluted at 1:100 in PBS. Coverslips were then mounted in Vectashield mounting medium (Vector Laboratories, Burlingame, CA) on microscope slides. Samples were analyzed with a Zeiss Axio Imager.Z1 microscope, equipped with 60×/NA 1.40 optics and Apotome apparatus, coupled to a computer driven Zeiss AxioCAM digital camera (MRm), using Zeiss Axio Vision (4.4) software.
Immunoblots
To assess in vivo expression of G proteins encoded by the DNA vaccines, mice were inoculated with 10 μl inoculums containing 5 μg of either Gwt, C4A or pCDNA3.1 plasmids diluted in TE buffer and boosted 3 days later with the same constructs (three animals per plasmid). The day following the booster, lungs were extracted and minced in ice-cold lysis buffer (50 mM Tris–HCl [pH 7.4], 150 mM NaCl, 1% triton x-100, 1% Sodium deoxycholate, 1 mM EDTA, and 0, 1% SDS) in the presence of a protease inhibitor cocktail (Sigma-Aldrich, St. Louis, MO). After 45 min in ice, samples were centrifuged at 10,000×g at 4°C for 10 min, supernatants were collected and samples from each group of mice were pooled together. For each pool, aproximately 20 μg of total lung protein were added loading buffer, incubated at 37°C for 30 min, electrophoresed in NuPage Tris–acetate gels (Invitrogen) and transferred to Immobilon P membranes (Millipore, Billerica, MA). The membranes were then probed with a primary goat anti-RSV polyclonal antibody (ViroStat) followed by bovine anti-goat immunoglobulin coupled to horseradish peroxidase (BD Biosciences, San Jose, CA). Signals were detected using chemiluminescent substrates (Pierce Biotechnology, Rockford, IL).
Mice and immunizations
Four to six week-old female BALB/c mice (The Jackson Laboratory, Bal Harbor, ME) were used for experiments involving RSV infections. Mice were housed under laminar flow hoods in an environmentally controlled specific pathogen-free animal facility. Intranasal (I.N.) infections were performed with 106 plaque forming units (pfu) of either live wild type RSV-A strain, or a recombinant RSV carrying a deletion in the G coding region from residues 172 to 187 within the GCRR (rRSVΔG172–187) [28, 29]. Viruses were propagated in Vero cells and harvested as previously described [30]. DNA vaccines, prepared as 10 μl inoculums containing 5 μg of plasmid diluted in TE buffer, were co-administered with the indicated viruses intranasally at the beginning of each experiment and then given as a booster 3 days later. In all experiments, an empty pCDNA 3.1 plasmid was used as a negative control.
For experiments with influenza virus, 4–6 week-old female C57BL/6 mice were inoculated intranasally with 1.6 × 106 pfu of influenza H1N1 A/New Caledonia/99 strain together with 10 μg of either an empty pCDNA 3.1 plasmid or Gwt. Three days later, I.N. inoculation with the plasmids alone (i.e. booster) was performed. All experimentation was approved by and performed according to guidelines of Vanderbilt University and the National Institutes of Health.
Isolation of pulmonary mononuclear cells (PMCs)
Nine days after the initial co-administration of virus and DNA vaccine, mice were sacrificed and their lungs removed. Lung tissue was rinsed, minced, and digested with 3,500 Dornase U of DNase I (Calbiochem)/ml and 75 U of collagenase (Life-Technologies)/ml at 37°C for 2 h. Digested tissue was then adjusted to 0.01 M EDTA, chilled on ice, and filtered through 100 μm-pore-size nylon monofilament cloth (PGS) to obtain individual cells. The cells were pelleted, resuspended, and subjected to centrifugation in Ficoll-Paque Plus solution (Amersham Pharmacia Biotech) at 400×g and 20°C for 30 min. The PMC interface was collected, washed twice, and resuspended in 5 ml of RPMI 1640 medium (Life Technologies) containing 10% fetal bovine serum (FBS), 100 U of penicillin/ml, and 100 μg of streptomycin sulfate/ml [20].
ELISPOT assays and granzyme B detection
Nitrocellulose-based 96-well microtiter plates (Millititer HA, Millipore, Bedford, MA) were coated overnight at 4°C with 5 μg/ml of anti-IFN-γ monoclonal antibody (clone R4-6A2, BD Biosciences, Pasadena, CA). Lung mononu-clear cells extracted from infected animals (see above) were incubated for 18 h with target A-20 cells previously loaded with either RSV-M282–90, influenza NP366–374 or influenza PA224–233 peptides, as described [20]. IFN-γ producing cells were detected with biotynilated anti-IFN-γ (clone XMG1.2, BD Biosciences) followed by peroxidase-conjugated avidin and 3-amino-9-ethylcarbazole substrate (Sigma-Aldrich). All assays were performed in triplicates.
Expression of cytolytic granzyme B enzyme by pulmonary CD8+ lymphocytes was quantitated by intracellular staining followed by flow cytometry. Briefly, lung mononuclear cells were counted and incubated with target A-20 cells previously loaded with PA224–233 peptide in the presence of GolgiStop (Invitrogen) for 18 h. After stimulation, lung mononuclear cells were washed twice with PBS containing 2% FBS, treated with Fc Block (BD Biosciences) to block Fc receptors, and stained with FITC-conjugated rat anti-mouse CD8α monoclonal antibody (BD Biosciences). Subsequently, cells were washed twice with PBS, fixed, and permeabilized with Cytofix/Cytoperm Solution (BD Biosciences), followed by staining with phycoerythrin-conjugated anti-granzyme B antibody (eBiosciences; cat# 12-8822). Flow cytometry analysis was performed using a FACS Calibur flow cytometer (BD Biosciences). A total of 30,000 cells were analyzed per sample.
RSV titers in the lungs
Lungs from infected mice were removed aseptically at 4, 7, or 9 days after the initial co-administration of virus and DNA vaccines and ground in 3 ml of Hanks media (Gibco/Invitrogen). Debris was removed by centrifugation at 2,000 rpm for 5 min and supernatants were serially diluted and plated on monolayers of Vero cells. Cells were then overlayed with Opti-MEM (Gibco/Invitrogen) containing 2% fetal calf serum, 0.8% methylcellulose, glutamine, and antibiotics, and incubated for 5 days. Plates were then stained by the immunoperoxidase method as previously described [20, 31] and results expressed as plaque forming units/gram of lung (pfu/g).
Lung histopathology and pneumonia severity score
The lungs of mice were removed 7 days after co-inoculation with rRSVΔG172–187 plus the indicated DNA vaccines, fixed, sectioned, and stained with hematoxylin and eosin (HE) as described elsewhere [20]. A previously described severity scoring system was used by blinded observers to characterize the degrees of pulmonary infiltration [32]. Briefly, the vessels and bronchi were initially scored as “1” when no or few infiltrating cells were present, “2” when focal aggregates of infiltrating cells were present or the structure was cuffed by one definite layer of infiltrating cells, and “3” when two or more definite layers of infiltrating cells with or without focal aggregates were present. Subsequently, pneumonia was categorized as mild (>60% of vessels and bronchi with scores of 1 and 0% with scores of 3), moderate (>30% with scores of 2 and/or >20% with scores of 3), or severe (>20% with scores of 3).
Statistical analysis
Data were analyzed with statistical software (Statview). Comparisons were made using the Kruskal–Wallis test where appropriate.
Results
Delivery of the G glycoprotein via DNA vaccination during RSV infections enhances the CTL response against the virus
The RSV G glycoprotein was recently shown to modulate CTL responses during RSV infections in BALB/c mice, and recombinant viruses either lacking the G gene or carrying a deleted form of G missing the central cystine noose region were defective at eliciting a robust CTL activity [20]. To determine whether inoculation of a DNA vaccine coding for the G glycoprotein of RSV could enhance CTL responses against the virus, we utilized a pCDNA3.1 plasmid carrying a full-length G gene under the control of a CMV-promoter (named Gwt). We co-inoculated BALB/c mice intranasally with this construct and 106 pfu of wild type RSV, and examine IFN-γ production in M282–90-specific lung mononuclear cells by ELISPOT. As shown in Fig. 2a, the number of spots containing IFN-γ-producing, M282–90-specific CTLs was dramatically increased in animals co-inoculated with RSV and Gwt compared to animals infected either with RSV alone or co-inoculated with RSV and a control empty vector (pCDNA 3.1). This result demonstrates that addition of exogenous G via a DNA vaccine can enhance CTL activity during wild type RSV infections.
Fig. 2.
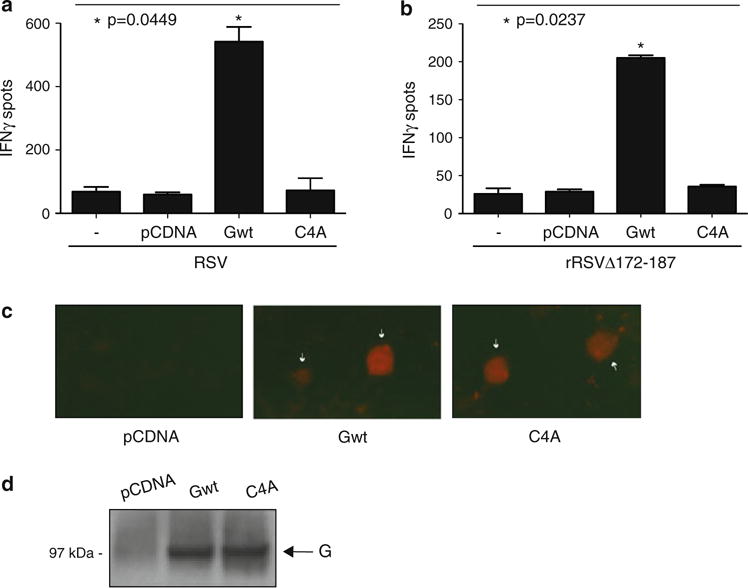
Expression of exogenous G protein during RSV infections enhances CTL responses in a manner dependent on cysteines 173, 176, 182 and 186. a BALB/c mice were co-inoculated intranasally with 106 pfu of WT RSV A (RSV) and 5 μg of one of the following plasmids: pCDNA 3.1 (an empty control plasmid), Gwt (a plasmid carrying a WT G gene), or C4A (a plasmid carrying a mutant form of the G gene with a quadruple alanine substitution at positions 173, 176, 182, and 186). Three days post-inoculation the animals received a booster of 5 μg of the indicated plasmids and 6 days later their lungs were extracted to isolate PMC. A control group of mice (−) was infected intranasally with 106 pfu of RSV in the absence of plasmid and sacrificed at day 9 post-infection for lung extraction and PMC isolation. Isolated cells were tested in an ELISPOT assay using A-20 presenting cells loaded with the M282–90 peptide (see “Materials and methods” section) and the frequency of M282–90-specific IFN-ÓELI-SPOTS was determined (P = 0.0449; Kruskal–Wallis test). b Mice were co-inoculated with 106 pfu of rRSVΔG172–187, a recombinant RSV virus lacking the coding region for residues 172 to 187 of the conserved cystine noose domain of G, and 5 μg of the indicated plasmids (either pCDNA 3.1, Gwt or C4A). Three days later the animals received a booster with the same plasmids and 6 days after the booster, PMCs were isolated and tested by ELISPOT assay. A control group of mice infected intranasally with 106 pfu of rRSVΔG172–187 in the absence of plasmid and sacrificed at day 9 post-infection (−) was included in these experiments. The frequencies of M282–90-specific IFN-γ ELISPOTS in the different experimental groups are shown (P = 0.0237; Kruskal–Wallis test). Data for both panels are presented as the mean ;SEM (error bars) and are representative of three experiments using three mice per infection per experiment. c Detection of membrane-bound G proteins by immunofluorescence. Human 293 cells were transiently transfected with either pCDNA 3.1, Gwt or C4A. Twenty-four hours after transfection cells were fixed with paraformaldehyde (but not permeabilized) and stained with anti-RSV antibody followed by Alexa fluor 568-conjugated secondary antibody. Immunofluorescence signals for each indicated sample are shown. White arrows indicate transfected cells expressing the G glycoprotein. Original magnification 20×. d DNA vaccine-mediated protein expression in lungs of BALB/c mice. Immunoblot analysis of lung extracts from mice inoculated intrana-sally with either pCDNA 3.1, Gwt or C4A was performed using an anti-RSV polyclonal antibody
We then tested whether exogenous addition of Gwt could restore normal CTL responses in animals infected with a recombinant RSV virus lacking the coding region for residues 172 to 187 of the conserved cystine noose domain (rRSVΔG172–187). This deletion mutant RSV has been shown to induce a defective CTL response in BALB/c mice [20]. To this end, we co-inoculated animals intranasally with Gwt and 106 pfu of rRSVΔG172–187 and examine IFN-γ production of M282–90-specific lung mononuclear cells by ELISPOT. As shown in Fig. 2b, the number of spots containing IFN-γ-producing, M282–90-specific CTLs increased approximately five fold in rRSVΔG172–187-infected animals receiving the Gwt plasmid compared to animals receiving the pCDNA3.1 control or infected with the virus alone. This result demonstrates that inoculation of mice with a DNA vaccine carrying the wild type G gene can restore CTL activity to viruses lacking the GCRR.
Mutation of the four conserved cysteine residues within the cystine noose domain abrogates the pro-CTL activity of RSV G
The noose region of G has four highly conserved cysteines at positions 173, 176, 182, and 186, which are largely preserved in RSV strains belonging to antigenic subgroups A and B (Fig. 1) [1, 23]. To test whether these conserved residues are necessary for G-induced CTL activity, we generated a plasmid construct carrying a mutated G gene containing alanine substitutions at positions 173, 176, 182, and 186 (named C4A). Transient transfection experiments in 293 cells showed that C4A was expressed at the cell surface as efficiently as Gwt, indicating that the mutations did not disrupt normal production or trafficking of the protein in mammalian cells (Fig. 2c). Additionally, immunoblot analysis of lung extracts showed that Gwt and C4A were expressed at similar levels in mice intranasally inoculated with these constructs (Fig. 2d).
We then co-inoculated BALB/c mice intranasally with C4A and either wild type RSV or rRSVΔG172–187, and compared the induced CTL responses with those of infected animals receiving either Gwt or pCDNA3.1 control. As shown in Fig. 2a, co-administration of the quadruple alanine mutant G and wild type RSV failed to enhance the cytotoxic response when compared to Gwt supplementation. Similarly, in experiments performed with rRSVΔG172–187, infected animals receiving C4A displayed a significantly reduced CTL activity when compared to those receiving Gwt (Fig. 2b). These results demonstrate that the conserved cysteine residues within the noose domain of G are critical for the pro-CTL activity displayed by the protein.
The differences in CTL activity induced by wild type versus the quadruple alanine mutant G proteins do not correlate with differences in pulmonary RSV replication or lung pathology
The level of the CTL response against RSV is typically influenced by the replication titer attained by the virus during infection [1]. Differences in viral load directly affect the amount of antigen being produced and presented, and can also indirectly affect the inflammatory process at the site of replication. Therefore, we tested whether the difference in CTL responses observed in RSV-infected animals co-inoculated with either G wild type or mutant C4A was associated with differences in viral replication levels. To this end, we extracted the lungs of BALB/c mice at 4, 7, or 9 days after co-inoculation with rRSVΔG172–187 plus the different DNA vaccines and determined RSV titers. As shown in Fig. 3a, rRSVΔG172–187 replicated to similar levels in the lungs of infected animals irrespective of the DNA construct co-inoculated at the time of infection (i.e. control, Gwt, or C4A). A similar result was obtained when the lung titers of animals co-inoculated with wild type RSV and either control, Gwt or C4A plasmids was assessed at day 4 post-infection (data not shown). We then assessed lung histopathology in animals co-inoculated with rRSVΔG172–187 and the DNA vaccines. As shown in Fig. 3b, cellular infiltration and tissue damage at day 7 post-infection were comparable in animals receiving pCDNA3.1, Gwt, or C4A. A pneumonia score performed on these lung samples yielded similar values for all animal groups tested (i.e. moderate), further supporting this conclusion. A similar result was observed when wild type RSV was used instead of rRSVΔG172–187 (data not shown). Therefore, the differences in the magnitude of CTL responses observed in animals receiving exogenous Gwt versus mutant C4A cannot be explained by a differential impact of these DNA vaccines on either RSV replication titers or severity of lung damage.
Fig. 3.
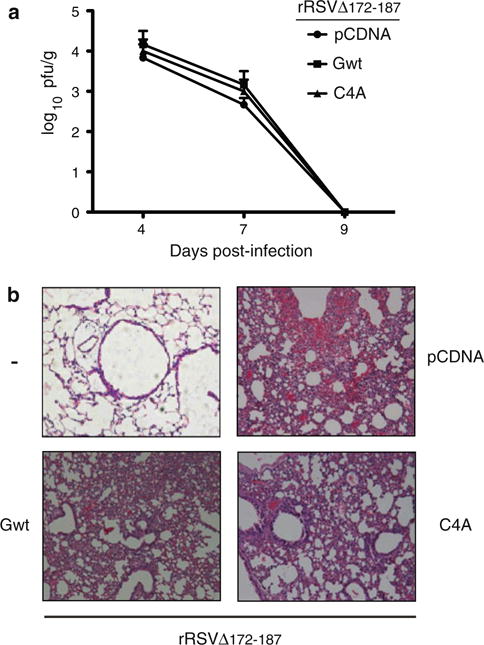
RSV lung titers and pulmonary histopathology. a Mice were co-inoculated intranasally with 106 pfu of rRSVΔG172–187 and 5 μg of the indicated plasmids (i.e. pCDNA 3.1, Gwt, or C4A). Three days post-inoculation the animals received a booster of 5 μg of the indicated plasmids, and were later sacrificed at 1, 4, or 6 days after the booster to obtain lung tissue. RSV titration was performed as described in “Materials and methods” section. RSV titers are shown as log of plaque forming units per gram of lung tissue (pfu/g) and represent the mean ;SEM of three experiments using five mice per infection per experiment. b Pulmonary histopathology in mice during peak inflammation at 7 days post co-inoculation with rRSVΔG172–187 and the indicated plasmids; (−) indicates mocked infected and no plasmid. Lung sections stained with hematoxylin and eosin are shown (original magnification 10×)
The G glycoprotein of RSV enhances CTL activity against non-RSV antigens
The ability of RSV-G to act in trans to boost immune responses against other RSV proteins in the context of an RSV infection suggested that G might have broad-spectrum adjuvant activity. To determine whether the CTL enhancing function of G could also be elicited toward RSV-unrelated antigens and in the absence of an RSV infection, we tested the effect of the glycoprotein in the context of a heterologous infection with influenza virus. Animal models in C57BL/6 mice have shown that two major viral epitopes co-dominate the immune response during primary infection against influenza. These Db-restricted peptides are located in the viral nucleoprotein (NP366–374) and the acidic polymerase (PA224–233) [33, 34]. As shown in Fig. 4a, mice co-immunized with H1N1 and Gwt displayed enhanced CTL activity against both NP366–374 and PA224–233 when compared to mice co-inoculated with influenza virus and pCDNA3.1 control. Increased CTL activity was confirmed by assessing granzyme-B expression in PA224–233-activated CD8+ lung PMCs using flow cytometry (Fig. 4b).
Fig. 4.
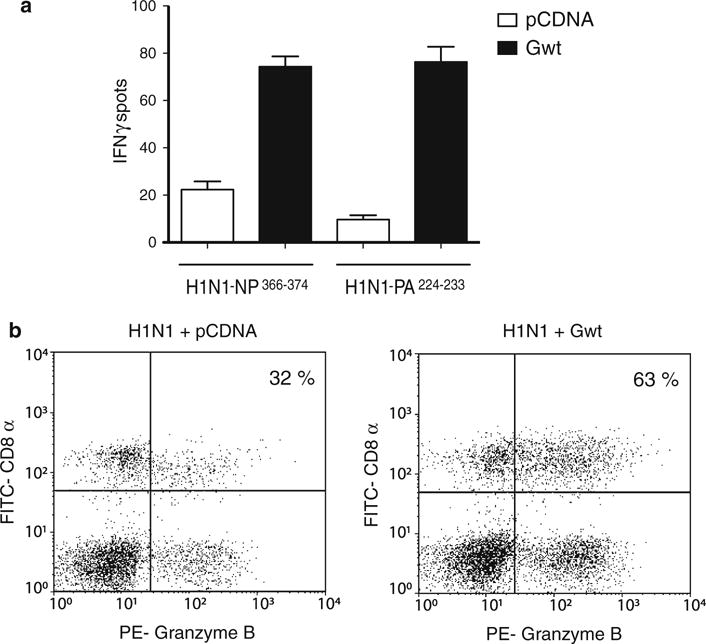
RSV-G enhances CTL activity against flu antigens during H1N1 infections. a C57BL/6 mice were co-inoculated intranasally with 106 pfu of H1N1 and 10 μg of either pCDNA 3.1 or Gwt. Three days post-inoculation, the animals received a booster of 10 μg of the indicated plasmids and 6 days later their lungs were extracted to isolate PMC. Isolated cells were tested in an ELISPOT assay using A-20 presenting cells loaded with either influenza NP366–374 or PA224–233 peptides (see “Materials and methods” section) and the frequency of IFN-©ELISPOTS was determined. Assays were performed in triplicates. b Expression of granzyme B by PA224–233 peptide-specific CD8+ pulmonary CTLs. Lung mononuclear cells from mice co-inoculated intranasally with 106 pfu of H1N1 and 10 μg of either pCDNA 3.1 (left panel) or Gwt (right panel) were activated by A-20 presenting cells previously loaded with influenza PA224–233 peptides. Cells were then fixed and doubly labeled with FITC-conjugated anti-CD8α antibody and phycoerythrin (PE)-conjugated anti-granzyme B antibody. Flow cytometry analysis was performed on a total of 30,000 cells per sample
Discussion
This study demonstrates that (i) the RSV-G glycoprotein can act in trans to enhance CTL responses not only against other RSV antigens but also against peptides from unrelated pathogens such as influenza, and (ii) that highly conserved residues within the cystine noose domain of G, cysteines 173, 176, 182, and 186, are critical for the pro-CTL activity mediated by this viral molecule. While exogenous addition of a DNA vaccine expressing wild type G was able to significantly enhance CTL activity against RSV and influenza antigens, a DNA vaccine coding for a quadruple cysteine to alanine mutant of G was severely defective when tested in the context of RSV infections. The differences in CTL activity elicited by the wild type and mutant G proteins were not associated with changes in either RSV lung titers or severity of lung damage in infected animals.
Cellular immunity mediated by CD8+ T cells plays a main role in the control of RSV replication in the lungs. Increased viral titers early during infection and delayed pulmonary clearance have been reported in CD8+ T cell-depleted BALB/c mice infected with RSV [6]. Conversely, experimental vaccines based exclusively on CTL epitopes have been shown to provide early protection against RSV [15]. In humans, severe RSV lower respiratory tract illness is characterized by the absence of pulmonary CTL responses [35]. Importantly, CTL activity against the virus not only plays a role in eliminating RSV-infected cells but also can affect the overall immune response through the secretion of soluble factors such as IFN-γ [36]. Some of these factors appear to modulate the activity of Th1 CD4+ T cells against RSV in various experimental paradigms [7–9]. Therefore, the capacity of G to boost the CTL response during RSV infections despite lacking MHC class I-restricted epitopes should be considered in the design of future RSV vaccines.
How does G enhance CTL activity against other antigens is not known. Proper activation of the inflammatory response during viral infections is important for effective antigen presentation and consequent activation of T cells. G has been shown to downregulate inflammation via TLR4 interaction at early time points during RSV infection in a GCRR-dependent fashion [31]. Importantly, studies with infants bearing TLR4 loss-of-function single nucleotide polymorphisms have established an epidemiological association between these alleles and increased severity of RSV disease [37]. It is possible that modulation of TLR4 signaling by the GCRR in dendritic cells and macrophages could contribute to reduced T-cell activation by affecting cytokine production and/or antigen presentation [31].
The sequence of G between cysteines 182 and 186 contains a CX3C fractalkine chemokine domain (Fig. 1), which has been shown to bind to the CX3C receptor 1, induce leukocyte chemotaxis, and modulate inflammation and dendritic cell activation [38–40]. The fact that G has been shown to act in vivo as a fractalkine antagonist thereby inhibiting fractalkine-mediated responses [41] suggests that this function of G might be important for its pro-CTL activity. Interestingly, Harcourt et al. have reported that G through its CX3C motif reduces antiviral T cell responses mediated by a subpopulation of CX3CR1+ cells in the spleen [41]. We generated a plasmid coding for a mutant form of G containing a double alanine replacement at cysteines 182 and 186 (i.e. disrupting the fractal-kine motif) and tested it in our assay with total pulmonary cells; this construct was as defective in eliciting CTL activity as the quadruple mutant C4A (data not shown). However, the double 182/186 alanine substitution is not only predicted to disrupt the CX3C motif but also the structure of the noose domain, and, therefore, we cannot distinguish between the individual contribution of each element (i.e. CX3C fractalkine-like domain vs. the entire noose domain). Future experiments using plasmids coding for recombinant versions of G carrying only the fractalkine motif should help elucidate the individual role of this domain in the CTL enhancing activity in the lungs.
In addition to the above-mentioned domains, the cystine noose sequence of G bears homology to the C2 region of the IV subdomain of tumor necrosis factor receptor 1 (TNFR1), a motif that has been implicated in TNF-α binding [42]. It is possible that G through its noose region could affect TNF signaling, consequently modulating TNF-induced apoptosis in either antigen presenting cells or CD8+ T cells, and increasing CTL responses. Finally, the cystine noose domain plays a critical role in the ability of G to induce the production of interleukin (IL)-10, a cytokine involved in the down-regulation of CTL responses in various models [31, 43].
Taken together, the ability of G to modulate several aspects of the inflammatory response, antigen presentation, and CD8+ T cell activation may account for its pro-CTL activity. Regardless of its precise mechanism of action, the effects of the central region of G on the overall cellular immune response against RSV and influenza CTL epitopes described here should be considered in the design of novel immunization strategies against respiratory pathogens. Our results indicate that G-mediated enhancement of CTL responses requires a combination of some or all of the conserved cysteines of the cystine noose domain to be effective. Whether a short peptide encompassing this domain of G is sufficient to elicit CTL responses and could be used as an adjuvant to supplement future vaccines should be investigated.
Acknowledgments
This research was supported by a Pilot Grant from The Linda and Timothy O’Neill Institute at Georgetown University (PMI), AI-054952 (FPP) and Thrasher Research Fund Early Career Award, and Fogarty International Center International Clinical Research Fellows Program at Vanderbilt (R24 TW007988) (GAM). ACM and PA are recipients of doctoral and MFD of post-doctoral awards from CONICET, Argentina.
Contributor Information
Guillermina A. Melendi, INFANT Fundacion, Buenos Aires, ArgentinaDepartment of Pediatrics, Vanderbilt University, Nashville, TN, USA
Dowd Bridget, Department of Human Science, Georgetown University Medical Center, 3700 Reservoir Road, STM 253, Washington, DC 20057, USA.
Ana C. Monsalvo, INFANT Fundacion, Buenos Aires, Argentina
Federico F. Laham, INFANT Fundacion, Buenos Aires, Argentina
Patricio Acosta, INFANT Fundacion, Buenos Aires, Argentina.
Maria Florencia Delgado, INFANT Fundacion, Buenos Aires, Argentina.
Fernando P. Polack, INFANT Fundacion, Buenos Aires, ArgentinaDepartment of Pediatrics, Vanderbilt University, Nashville, TN, USA
Pablo M. Irusta, INFANT Fundacion, Buenos Aires, ArgentinaDepartment of Human Science, Georgetown University Medical Center, 3700 Reservoir Road, STM 253, Washington, DC 20057, USA
References
- 1.Collins P, Chanock RM, Murphy BR. In: Fields Virology. 4. Knipe DM, Howley PM, editors. Lippincott/The Williams & Wilkins Co; Philadelphia: 2001. pp. 1443–1486. [Google Scholar]
- 2.Hall CB. N Engl J Med. 2001;344:1917–1928. doi: 10.1056/NEJM200106213442507. [DOI] [PubMed] [Google Scholar]
- 3.Russell JH, Ley TJ. Annu Rev Immunol. 2002;20:323–370. doi: 10.1146/annurev.immunol.20.100201.131730. [DOI] [PubMed] [Google Scholar]
- 4.Cannon MJ, Openshaw PJ, Askonas BA. J Exp Med. 1988;168:1163–1168. doi: 10.1084/jem.168.3.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cannon MJ, Stott EJ, Taylor G, Askonas BA. Immunology. 1987;62:133–138. [PMC free article] [PubMed] [Google Scholar]
- 6.Graham BS, Bunton LA, Wright PF, Karzon DT. J Clin Investig. 1991;88:1026–1033. doi: 10.1172/JCI115362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hussell T, Baldwin CJ, O’Garra A, Openshaw PJ. Eur J Immunol. 1997;27:3341–3349. doi: 10.1002/eji.1830271233. [DOI] [PubMed] [Google Scholar]
- 8.Sparer TE, Matthews S, Hussell T, Rae AJ, Garcia-Barreno B, Melero JA, Openshaw PJ. J Exp Med. 1998;187:1921–1926. doi: 10.1084/jem.187.11.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Srikiatkhachorn A, Braciale TJ. J Exp Med. 1997;186:421–432. doi: 10.1084/jem.186.3.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Connors M, Kulkarni AB, Firestone CY, Holmes KL, Morse HC, 3rd, Sotnikov AV, Murphy BR. J Virol. 1992;66:7444–7451. doi: 10.1128/jvi.66.12.7444-7451.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Graham BS, Henderson GS, Tang YW, Lu X, Neuzil KM, Colley DG. J Immunol. 1993;151:2032–2040. [PubMed] [Google Scholar]
- 12.Srikiatkhachorn A, Chang W, Braciale TJ. J Virol. 1999;73:6590–6597. doi: 10.1128/jvi.73.8.6590-6597.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tang YW, Graham BS. J Clin Investig. 1994;94:1953–1958. doi: 10.1172/JCI117546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Delgado MF, Coviello S, Monsalvo AC, Melendi GA, Hernandez JZ, Batalle JP, Diaz L, Trento A, Chang HY, Mitzner W, Ravetch J, Melero JA, Irusta PM, Polack FP. Nat Med. 2009;15:34–41. doi: 10.1038/nm.1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Connors M, Kulkarni AB, Collins PL, Firestone CY, Holmes KL, Morse HC, Murphy BR., III J Virol. 1992;66:1277–1281. doi: 10.1128/jvi.66.2.1277-1281.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kulkarni AB, Collins PL, Bacik I, Yewdell JW, Benn-ink JR, Crowe JE, Murphy BR., Jr J Virol. 1995;69:1261–1264. doi: 10.1128/jvi.69.2.1261-1264.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nicholas JA, Rubino KL, Levely ME, Adams EG, Collins PL. J Virol. 1990;64:4232–4241. doi: 10.1128/jvi.64.9.4232-4241.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Openshaw PJ, Anderson K, Wertz GW, Askonas BA. J Virol. 1990;64:1683–1689. doi: 10.1128/jvi.64.4.1683-1689.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chang J, Braciale TJ. Nat Med. 2002;8:54–60. doi: 10.1038/nm0102-54. [DOI] [PubMed] [Google Scholar]
- 20.Bukreyev A, Serra ME, Laham FR, Melendi GA, Klee-berger SR, Collins PL, Polack FP. J Virol. 2006;80:5854–5861. doi: 10.1128/JVI.02671-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fan CF, Mei XG. Vaccine. 2005;23:4453–4461. doi: 10.1016/j.vaccine.2005.03.044. [DOI] [PubMed] [Google Scholar]
- 22.Zeng RH, Gong W, Fan CF, Wang YF, Mei XG. Vaccine. 2006;24:941–947. doi: 10.1016/j.vaccine.2005.08.064. [DOI] [PubMed] [Google Scholar]
- 23.Martinez I, Dopazo J, Melero JA. J Gen Virol. 1997;78:2419–2429. doi: 10.1099/0022-1317-78-10-2419. [DOI] [PubMed] [Google Scholar]
- 24.Heidema J, de Bree GJ, De Graaff PM, van Maren WW, Hoogerhout P, Out TA, Kimpen JL, van Bleek GM. J Gen Virol. 2004;85:2365–2374. doi: 10.1099/vir.0.80131-0. [DOI] [PubMed] [Google Scholar]
- 25.Rock MT, Crowe JE., Jr Immunology. 2003;108:474–480. doi: 10.1046/j.1365-2567.2003.01619.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Venter M, Rock M, Puren AJ, Tiemessen CT, Crowe JE., Jr J Virol. 2003;77:7319–7329. doi: 10.1128/JVI.77.13.7319-7329.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Melero JA, Garcia-Barreno B, Martinez I, Pringle CR, Cane PA. J Gen Virol. 1997;78:2411–2418. doi: 10.1099/0022-1317-78-10-2411. [DOI] [PubMed] [Google Scholar]
- 28.Teng MN, Collins PL. J Virol. 2002;76:6164–6171. doi: 10.1128/JVI.76.12.6164-6171.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Teng MN, Whitehead SS, Collins PL. Virology. 2001;289:283–296. doi: 10.1006/viro.2001.1138. [DOI] [PubMed] [Google Scholar]
- 30.Trudel M, Nadon F, Seguin C, Binz H. Virology. 1991;185:749–757. doi: 10.1016/0042-6822(91)90546-n. [DOI] [PubMed] [Google Scholar]
- 31.Polack FP, Irusta PM, Hoffman SJ, Schiatti MP, Melendi GA, Delgado MF, Laham FR, Thumar B, Hendry RM, Melero JA, Karron RA, Collins PL, Kleeberger SR. Proc Natl Acad Sci USA. 2005;102:8996–9001. doi: 10.1073/pnas.0409478102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Melendi GA, Hoffman SJ, Karron RA, Irusta PM, Laham FR, Humbles A, Schofield B, Pan CH, Rabold R, Thumar B, Thumar A, Gerard NP, Mitzner W, Barnum SR, Gerard C, Kleeberger SR, Polack FP. J Virol. 2007;81:991–999. doi: 10.1128/JVI.01783-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Belz GT, Xie W, Altman JD, Doherty PC. J Virol. 2000;74:3486–3493. doi: 10.1128/jvi.74.8.3486-3493.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Belz GT, Xie W, Doherty PC. J Immunol. 2001;166:4627–4633. doi: 10.4049/jimmunol.166.7.4627. [DOI] [PubMed] [Google Scholar]
- 35.Welliver TP, Garofalo RP, Hosakote Y, Hintz KH, Avendano L, Sanchez K, Velozo L, Jafri H, Chavez-Bueno S, Ogra PL, McKinney L, Reed JL, Welliver RC., Sr J Infect Dis. 2007;195:1126–1136. doi: 10.1086/512615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ostler T, Davidson W, Ehl S. Eur J Immunol. 2002;32:2117–2123. doi: 10.1002/1521-4141(200208)32:8<2117::AID-IMMU2117>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 37.Tal G, Mandelberg A, Dalal I, Cesar K, Somekh E, Tal A, Oron A, Itskovich S, Ballin A, Houri S, Beigelman A, Lider O, Rechavi G, Amariglio N. J Infect Dis. 2004;189:2057–2063. doi: 10.1086/420830. [DOI] [PubMed] [Google Scholar]
- 38.Tripp RA, Jones LP, Haynes LM, Zheng H, Murphy PM, Anderson LJ. Nat Immunol. 2001;2:732–738. doi: 10.1038/90675. [DOI] [PubMed] [Google Scholar]
- 39.Guo J, Zhang M, Wang B, Yuan Z, Guo Z, Chen T, Yu Y, Qin Z, Cao X. Int J Cancer. 2003;103:212–220. doi: 10.1002/ijc.10816. [DOI] [PubMed] [Google Scholar]
- 40.Niess JH, Brand S, Gu X, Landsman L, Jung S, McCormick BA, Vyas JM, Boes M, Ploegh HL, Fox JG, Littman DR, Reinecker HC. Science. 2005;307:254–258. doi: 10.1126/science.1102901. [DOI] [PubMed] [Google Scholar]
- 41.Harcourt J, Alvarez R, Jones LP, Henderson C, Anderson LJ, Tripp RA. J Immunol. 2006;176:1600–1608. doi: 10.4049/jimmunol.176.3.1600. [DOI] [PubMed] [Google Scholar]
- 42.Langedijk JP, de Groot BL, Berendsen HJ, van Oirschot JT. Virology. 1998;243:293–302. doi: 10.1006/viro.1998.9066. [DOI] [PubMed] [Google Scholar]
- 43.Kurte M, Lopez M, Aguirre A, Escobar A, Aguillon JC, Charo J, Larsen CG, Kiessling R, Salazar-Onfray F. J Immunol. 2004;173:1731–1737. doi: 10.4049/jimmunol.173.3.1731. [DOI] [PubMed] [Google Scholar]


