Abstract
PPARα has been known to play a pivotal role in orchestrating lipid, glucose, and amino acid metabolism via transcriptional regulation of its target gene expression during energy deprivation. Recent evidence has also suggested that PPARα is involved in bile acid metabolism, but how PPARα modulates the homeostasis of bile acids during fasting is still not clear. In a mechanistic study aiming to dissect the spectrum of PPARα target genes involved in metabolic response to fasting, we identified a novel mouse gene (herein named mL-STL for mouse liver-sulfotransferase-like) that shared extensive homology with the Sult2a subfamily of a superfamily of cytosolic sulfotransferases, implying its potential function in sulfonation. The mL-STL gene expressed predominantly in liver in fed state, but PPARα was required to sustain its expression during fasting, suggesting a critical role of PPARα in regulating the mL-STL-mediated sulfonation during fasting. Functional studies using recombinant His-tagged mL-STL protein revealed its narrow sulfonating activities toward 7α-hydroxyl primary bile acids, including cholic acid, chenodeoxycholic acid, and α-muricholic acid, and thus suggesting that mL-STL may be the major hepatic bile acid sulfonating enzyme in mice. Together, these studies identified a novel PPARα-dependent gene and uncovered a new role of PPARα as being an essential regulator in bile acid biotransformation via sulfonation during fasting.
Keywords: gene expression, liver, nutrition, nuclear receptors/PPAR, steroid hormone, sulfonation, mouse liver-sulfotransferase-like
PPARα, a nuclear hormone receptor and a ligand-dependent transcription factor, has long been known as a nutritional metabolic sensor in regulating the energy homeostasis during energy deprivation (1). PPARα has been well studied for its critical roles in controlling the expression of its target genes involved in lipid, glucose, and amino acid metabolism during fasting (2, 3). Recent studies have also suggested a role of PPARα in regulating bile acid synthesis (4, 5). However, the overall impact of PPARα on bile acid homeostasis during fasting remains elusive.
Traditionally, bile acids are primarily considered as amphipathic detergent molecules that can facilitate absorption of dietary lipid and fat-soluble vitamins in the intestine as well as modulate cholesterol catabolism in the liver (6). In mammals, two groups of bile acids, namely, the primary and secondary bile acids, are found. Bile acids that are synthesized from cholesterol in the liver are termed primary bile acids, whereas bile acids that are formed by bacterial conversion of primary bile acids in the colon during their enterohepatic cycling are termed secondary bile acids (7). After their biosynthesis from cholesterol, bile acids are N-acyl amidated (conjugated) with the amino acids taurine or glycine in a species-dependent manner. In humans, the principal primary bile acids are cholic acid (CA) and chenodeoxycholic acid (CDCA) and their taurine and glycine amidates, whereas lithocholic acid (LCA) and deoxycholic acid (DCA) are the secondary bile acids (8). In mice, CDCA, once formed, is converted into more hydrophilic muricholic acid (MCA) and most of the bile acids are amidated to taurine (9, 10).
Increasing lines of evidence suggest that bile acids can also function as signaling molecules in controlling glucose, lipid, and energy homeostasis, either via activation of cell surface receptors or modification of gene expression upon binding to nuclear receptors (11–13). Three major bile acid-mediated signaling pathways have been reported. First, bile acids activate the MAPK pathway (14, 15). Second, bile acids are endogenous activators of the G-protein coupled membrane receptor TGR5 (16, 17). Third, bile acids are natural ligands that activate the nuclear hormone receptor farnesoid X receptor (12, 17–19). Through activation of these signaling pathways, bile acids cannot just modulate their own enterohepatic circulation, but also act as a metabolic sensor to regulate lipid, glucose, and energy homeostasis. The intriguing question that remains is under which physiological conditions bile acids exert these endocrine functions. It has been proposed that bile acids exert their signaling functions mainly during feeding and active digestion (11). Under these conditions, bile acids are secreted in the intestine, reabsorbed into the liver through enterohepatic circulation, and then spilled over in the systemic circulation. Through blood circulation, bile acids act as a metabolic signal to the liver, intestine, and other organs that a meal has been taken and that nutrition molecules such as lipids and glucose will become available for digestion and absorption. Thus, bile acid homeostasis has a direct role in maintaining energy balance.
Bile acid homeostasis is tightly coordinated through its synthesis, reabsorption, and removal by hepatic phase II conjugation, such as sulfonation. Sulfonation is an enzymatic reaction carried out by a superfamily of cytosolic sulfotransferases (SULTs), in which a sulfonate (SO3−) group is transferred from the universal donor 3′-phosphoadenosine 5′-phosphosulfate (PAPS) to a hydroxyl, sulfhydryl, or amino group of various exogenous and endogenous compounds, such as bile acids (20). In general, sulfonation has been regarded as a detoxification reaction, leading to more water-soluble products and therefore aiding their excretion in urine or bile. Sulfonation of bile acids can alter the bile acid pool, and thus predict a change to the energy balance. However, the role of bile acid sulfonation in maintaining the bile acid homeostasis, and thus energy balance, has never been explored.
In our long-term efforts to characterize how PPARα directs metabolic changes in response to energy deprivation (21–23), we have identified a PPARα-regulated and 7α-hydroxyl bile acid-preferring sulfonating cytosolic SULT, herein named mL-STL (mouse liver-sulfotransferase-like), during fasting. In this paper, we report the isolation, cloning, expression, and characterization of the novel mouse liver cytosolic SULT, mL-STL. Results from the present study further strengthened the notion that there is a direct engagement of PPARα in the biotransformation of bile acids through sulfonation during energy deprivation.
MATERIALS AND METHODS
Animal studies
Three-month-old male WT and PPARα-null (KO) mice on a SV/129 background were used in all experiments (24). For the starvation experiment, both WT and KO mice were fasted for 6, 12, 24, 48, and 72 h, whereas control mice were fed with a normal mouse chow diet. For 4-chloro-6-(2,3-xylidino)-2-pyrimidinylthioacetic acid (Wy-14,643) treatment, mice were fed with a mouse chow diet (Bio-Serv, Frenchtown, NJ) containing 0.0 or 0.1% (w/w) Wy-14,643 (Chemsyn Laboratories, Lenexa, KS) for 24 h, 1 week, 2 weeks, 6 months, and 11 months. At the end of experiments, mice were decapitated, and tissues were removed for RNA and protein extraction. Animal procedures were approved by The Chinese University of Hong Kong Animal Experimentation Ethics Committee.
Fluorescent differential display
Fluorescent differential display (FDD) was used to compare the mRNA expression patterns between the WT and KO mice under both fed and fasting conditions as described in a previous study (21). Briefly, total RNA from the livers was extracted with TRIzol reagent (Invitrogen, Carlsbad, CA), according to the manufacturer’s instruction. The anchored primer 8 (AP8) and arbitrary primer 1 (ARP1) (supplemental Table S1) were used for the FDD analysis.
Rapid amplification of cDNA ends
The 5′- and 3′-rapid amplification of cDNA ends (RACE) was performed to obtain the full-length cDNA sequence of the mL-STL gene. Two commercial kits, including 5′/3′ RACE (Roche, Mannheim, Germany) and GeneRacer™ (Invitrogen), based on different PCR primer design strategies were used for the analysis. The 5′ and 3′ gene-specific primers (supplemental Table S1) were designed based on the internal sequences of the 910 bp mL-STL FDD cDNA fragment. Total liver RNA from a 3-month-old male WT control and WT fasted mouse was used as RNA templates for the RACE reactions. The RACE product was subcloned into a pCR®II-TOPO® vector (Invitrogen), and the nucleotide sequences of the RACE subclones were determined by DNA sequencing using M13 reverse, M13 forward, and the mL-STL gene-specific sequencing primers (supplemental Table S1).
Construction of His-mL-STL and mL-STL overexpression vectors
Two overexpression vectors were constructed for the study. The mini-pRSETA (mpRSETA) expression vector (Invitrogen), with a 6× His-tag, was used to overexpress the His-tagged mL-STL (His-mL-STL) recombinant fusion proteins in Escherichia coli for SULT enzymatic assays. The modified mpRSET vector (mmpRSET vector), without a 6× His-tag, was used to produce an mL-STL recombinant protein for polyclonal antiserum production. To clone the mL-STL cDNA into the mpRSETA expression vector with a 6× His-tag, the open reading frame (ORF) of mL-STL cDNA (1,042 bp) was PCR amplified from the 1,738 bp 5′-RACE clone 5′#17-Fed(I) by using a pair of forward mL-STL-FP278-Pst I and reverse mL-STL-RP1319-Hind III primers (supplemental Table S1). To clone the mL-STL cDNA into the mmpRSET expression vector without the 6× His-tag, the ORF was PCR amplified by using a pair of forward mL-STL-FP278-Nde I and the same reverse mL-STL-RP1319-Hind III primers (supplemental Table S1). The subclones were confirmed with restriction enzyme mappings, followed by DNA sequencing with the designated sequencing primers (supplemental Table S1). The sequencing results were compared against DNA sequences predicted by Vector NTI Advance 10.
Overexpression of recombinant His-mL-STL and mL-STL proteins
The His-mL-STL and mL-STL expression vectors were transformed into E. coli strain BL21(DE3)pLysS, and protein expression was then induced by using 0.2 mM isopropyl-β-d-1-thiogalactopyranoside (IPTG) for 16 h at 25°C. After protein induction, the bacterial cells were pelleted at 6,160 g in a Beckman JA-14 rotor at 4°C for 15 min and resuspended in their respective lysis buffers. The His-mL-STL bacterial cells were resuspended in 40 ml of lysis buffer A (50 mM Tris-HCl, pH 8, 500 mM NaCl, 30 mM imidazole, 1 mM PMSF, and 0.05% β-mercaptoethanol), while the mL-STL bacterial cells were in lysis buffer B (20 mM sodium phosphate buffer, pH 7.4). The recombinant proteins were released from the bacterial cells by sonication using a Bandelin Sonopuls HD2070 ultrasonic homogenizer (Berlin, Germany). The sonicated cell lysates of His-mL-STL and mL-STL proteins were centrifuged at 149,000 g for 1 h and 20,820 g for 10 min, respectively, at 4°C to obtain the supernatant. After centrifugation, the His-mL-STL supernatant was filtered through a 0.22 μm syringe filter (Millipore, Billerica, MA) and subjected to column purification, whereas the mL-STL inclusion bodies were resuspended in lysis buffer B and used for polyclonal mL-STL antiserum production.
Production of polyclonal mL-STL antiserum
The recombinant mL-STL protein pelleted in inclusion bodies was semipurified by resolving on a preparative SDS-PAGE, according to the standard procedure (25). Four hundred microliters of inclusion bodies were mixed with 100 μl of 5× sample buffer (125 mM Tris-HCl, pH 6.8, 25% glycerol, 5% SDS, 0.05% bromophenol blue, and 14.4 mM β-mercaptoethanol), boiled for 10 min, and resolved in a 12% preparative SDS-PAGE at 60 V for 16 h. After electrophoresis, the gel was stained with 1% Coomassie brilliant blue R dissolved in distilled water. The protein band corresponding to the molecular mass of recombinant mL-STL protein was excised and placed in a 5 ml Wheaton Teflon-glass homogenizer (Wheaton, Millville, NJ). The gel slices were homogenized in PBS (137 mM NaCl, 2.7 mM KCl, 10.1 mM Na2HPO4, and 1.7 mM KH2PO4) by using a Wheaton overhead stirrer until the gel slices were completely homogenized. The homogenized gel lysate was emulsified with complete Freund’s adjuvant at a ratio of 1:1 for injection into 3-month-old New Zealand White rabbits after the preimmune blood was collected from their ear veins. Each rabbit was immunized with ∼100 μg of protein at its neck. Five follow-up booster injections, each with 100 μg of protein in incomplete Freund’s adjuvant, were performed at 1 month intervals, and 40 ml of blood was collected from the ear vein of each rabbit 10–14 days after each boosting. The blood was allowed to coagulate at room temperature for 1 h, followed by two consecutive centrifugations at 2,500 g for 20 min. The serum was stored at −80°C until used for Western blot analysis.
Purification of recombinant His-mL-STL protein
A two-step column chromatographic method using a Bio-Rad NGC™ Discover 10 chromatography system (Bio-Rad, Hercules, CA) was used for purification of recombinant His-mL-STL fusion protein. The supernatant collected from crude cell lysate was subject to two 5 ml nickel affinity HiTrapTM (GE/17-0409-01) chelating high-performance columns coupled in series (GE Healthcare, Piscataway, NJ). The columns were charged with nickel ion and preequilibrated with a binding buffer (50 mM Tris-HCl, pH 8, 500 mM NaCl, and 30 mM imidazole). The samples were then loaded onto the column, and 170 ml of binding buffer was passed through the column at a flow rate of 4 ml/min to remove unbound proteins. Then, the column was washed with 120 ml of 86.4 mM imidazole in a mixed binding and elution buffer (50 mM Tris-HCl, pH 8, 500 mM NaCl, and 500 mM imidazole) to remove nonspecific protein binding. The bound His-mL-STL proteins were eluted from the column with 28 ml of 208.6 mM imidazole in an elution buffer. The 1 ml fractions containing the His-mL-STL proteins were pooled, added up to 2 mM DTT, and concentrated to 3 ml by centrifugation at 2,880 g at 12°C by using an Amicon ultra-15 centrifugal filter (Millipore). The concentrated protein was then centrifuged at 20,820 g for 10 min, filtered through a 0.22 μm syringe filter, and loaded at a flow rate of 1 ml/min onto a HiLoad Superdex 200 prep grade size exclusion column (16 × 600 mm) (GE Healthcare) preequilibrated with a size-exclusion buffer (50 mM Tris-HCl, pH 8, and 150 mM NaCl). The His-mL-STL proteins were eluted with 100 ml of the same buffer, and the eluted fractions containing the His-mL-STL proteins were pooled and stored (within 1 week) at 4°C for SDS-PAGE, Western blot analysis, mass spectrometry analysis, and SULT assays.
Substrate specificity screening of recombinant His-mL-STL protein
The 1-butanol extraction method (26) using a radiolabeled-[35S]PAPS cofactor was used to screen for the His-mL-STL-mediated SULT activity toward the prototype hydroxysteroid SULT2 substrates (27, 28). In brief, the 100 μl incubation mixture comprised 100 mM sodium acetate buffer (pH 5.5) or 100 mM sodium phosphate buffer (pH 6.5 or 7.5), 2.5 mM MgCl2, 20 μg of purified His-mL-STL protein, 10 or 100 μM substrates, and 50 μM PAPS containing 0.015 μCi [35S]PAPS (PerkinElmer, Upplands Väsby, Sweden). The vehicle controls were also run in parallel for detection of background sulfonation. A reaction mixture without the addition of substrate as well as extraction was used for calculating the scintillation counting efficiency. The reaction mixture was incubated at 37°C for 1 h, and then the reaction was terminated by the addition of 400 μl of 1 M ammonium hydroxide, followed by extraction with 1 ml of 1-butanol. The unreacted [35S]PAPS was precipitated, and the [35S]labeled sulfonated product was partitioned in supernatant after centrifugation at 70 g for 10 min. Six hundred microliters of 1-butanol layer was transferred into a scintillation counting vial containing 4 ml of Optiphase HiSafe 3 scintillation cocktail (PerkinElmer). Specific activity was calculated from the liquid scintillation counting by using a Beckman Coulter LS6500 liquid scintillation counter (Beckman Coulter, Fullerton, CA).
The barium precipitation method (29, 30) using a radiolabeled-[35S]PAPS cofactor was used to screen for the His-mL-STL-mediated SULT activity toward SULT1 prototype substrates (31). Typically, the 150 μl incubation mixture contained 100 mM sodium acetate buffer (pH 5.5) or 100 mM sodium phosphate buffer (pH 6.5 or 7.5), 2.5 mM MgCl2, 20 μg of purified His-mL-STL protein, 50 or 100 μM substrates, and 50 μM PAPS containing 0.015 μCi [35S]PAPS. The vehicle controls were performed in parallel for detection of background sulfonation, and a reaction mixture without substrate and precipitation was used for calculating the scintillation counting efficiency. The reaction mixture was incubated at 37°C for 1 h, and the reaction was terminated by the addition of 200 μl of 100 mM barium acetate. The unreacted [35S]PAPS was removed by precipitation with 200 μl of 100 mM barium hydroxide and 200 μl of 100 mM zinc sulfate, followed by centrifugation at 11,700 g for 10 min. Five hundred microliters of supernatant were removed and mixed with 4 ml of Optiphase HiSafe 3 scintillation cocktail, and the specific activity was calculated from the liquid scintillation counting.
Subcellular fractionation
Subcellular fractionation of mouse livers was carried out by using the sequential differential ultracentrifugation method. Both WT and KO mice from fed and 72 h-fasted treatment groups were sacrificed by cervical dislocation, and their livers were excised. The dissected livers were minced into small pieces on ice and homogenized by using a Wheaton overhead stirrer at speed 2 in a Wheaton Potter-Elvehjem tissue grinder containing 20 ml of homogenizing buffer (0.1 M sodium phosphate, 0.125 M KCl, 1 mM EDTA, 1 mM PMSF, 1 mM DTT, and 0.25 M sucrose, pH 7.4). The homogenate was then gravity-filtered with two layers of nylon cloth and centrifuged in a Beckman centrifuge by using a JA-20 rotor at 1,000 g for 20 min at 4°C to obtain the nuclear pellet and S1 supernatant. The S1 supernatant was then centrifuged at 3,000 g for 10 min at 4°C to obtain the heavy mitochondrial pellet and S2 supernatant. The S2 supernatant was centrifuged at 12,000 g for 20 min at 4°C to obtain the light mitochondrial pellet and S3 fraction. Finally, the S3 supernatant was centrifuged at 100,000 g for 1 h at 4°C in a Beckman OptimaTM XL-100K ultracentrifuge using a 70 Ti rotor to obtain the microsomal pellet and cytosolic supernatant. All pellets were resuspended in 1 ml of a resuspension buffer (50 mM Tris-HCl, 150 mM NaCl, 1% NP-40, 0.5% Na-deoxycholate, 1 mM PMSF, and 1× proteinase inhibitor cocktail) and stored at −80°C for Western blot analysis. Protein concentration was measured by using the Pierce™ BCA protein assay kit (Invitrogen) according to the manufacturer’s instructions.
SDS-PAGE and Western blot analysis
SDS-PAGE was performed according to Laemmli (25). Different amounts of proteins were electrophoresed in 12% or 14% gels at 50 V for 16 h, after which the proteins on the gel were transferred onto a 0.45 μm BioTrace™ PVDF transfer membrane (Pall Life Sciences, Ann Arbor, MI) at 70 V for 3 h. The membrane was blocked in 4% nonfat dry milk (Nestle, Vevey, Switzerland) in an immunoblotting buffer (50 mM Tris-HCl, pH 7.4, 0.8 M NaCl, and 20 mM CaCl2) at room temperature for 1 h, followed by an overnight incubation in primary antibodies at room temperature on an orbital shaker. The primary antibodies included a mouse monoclonal β-actin (Sigma-Aldrich, St. Louis, MO; catalog no. A-5441, clone AC-15, lot 044K4760) and the in-house-produced fifth bled rabbit polyclonal anti-mouse-mL-STL antiserum. After an overnight incubation, the membrane was washed with distilled water and then incubated with either an alkaline phosphate (AP)-goat anti-mouse IgG (Zymed, San Francisco, CA; catalog no. 81-6522) and/or AP-goat anti-rabbit IgG secondary antibodies (Zymed; catalog no. 65-6122) at room temperature for 2 h with shaking. After washing, the membrane was developed in nitroblue tetrazolium chloride/5-bromo-4-chloro-3-indolyl-phosphate-4-toluidine salt substrates in a detection buffer (100 mM Tris-HCl, pH 9.5, 50 mM MgCl2, and 100 mM NaCl). The reaction was stopped with distilled water, and the signal on the membrane was captured by a scanner (Epson Expression 1600 Pro, Epson, Nagano, Japan) with an Adobe Photoshop 6.0 program (Adobe, San Jose, CA).
Formaldehyde agarose gel electrophoresis and Northern blot analysis
Northern blot analysis was performed as described earlier (21). Briefly, total RNA (20 µg/lane) was electrophoresed in a 1% formaldehyde gel and transferred onto an N+-nylon membrane. The membrane was then hybridized with a digoxigenin (DIG)-labeled cDNA probe by using the 5′-RACE cDNA subclone 5′#9-Fed(I) (supplemental Fig. S3) as a DNA template amplified with a pair of forward mL-STL-FP19 and reverse mL-STL-RP1884 primers (supplemental Table S1) using a PCR DIG-labeling mix.
Statistical analysis
Results are expressed as mean ± SEM. Differences between means were compared by one- or two-way ANOVA with post hoc test for multiple pairwise comparisons using Prism (Version 6; GraphPad Software, Inc., La Jolla, CA). P < 0.05 is considered statistically significant, and P values were indicated categorically: *P < 0.05, **P < 0.01, ***P < 0.001.
RESULTS
A novel mouse mL-STL cDNA fragment was identified by mRNA FDD analysis
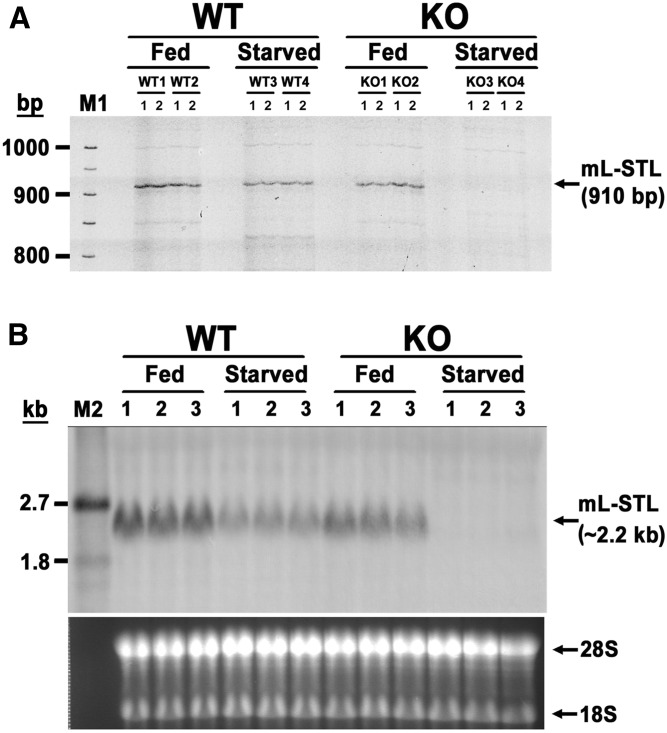
In our long-term goal to uncover novel PPARα target genes and PPARα-mediated signaling pathways in the adaptive response to metabolic perturbation during energy deprivation, we compared the liver mRNA expression profiles between WT and KO mice under fed or 72 h fasted states using the mRNA FDD analysis (21) and isolated a 910 bp partial cDNA fragment (Fig. 1A and supplemental Fig. S1A) , whose DNA sequence showed 99% similarity to the 3′ end of a 2,147 bp mouse Riken cDNA 2810007J24, transcript variant 1 (GenBank accession no. NM_175250) (supplemental Fig. S1B). Because this partial cDNA was obtained from mouse liver and predicted to have SULT activity, we hereby designated it mouse liver-sulfotransferase-like (mL-STL) cDNA. This mL-STL cDNA fragment was constitutively expressed in both WT and KO mice under the fed condition, but it was dramatically downregulated in the PPARα-deficient mice during 72 h fasting (Fig. 1A). This differential expression pattern was further confirmed by using the 910 bp mL-STL cDNA fragment as a probe in the Northern blot analysis (Fig. 1B). A ∼2.2 kb mRNA transcript was detected in the livers of both WT and KO fed mice, but this transcript was differentially reduced in the WT and KO mice during 72 h fasting. Thus, these data strongly suggest that PPARα is required for sustaining the expression of mL-STL mRNA level during fasting. However, the molecular links between PPARα and the mL-STL gene, as well as its physiological role in energy deprivation, require further investigation.
Fig. 1.
A 910 bp partial mL-STL cDNA fragment was identified by FDD analysis. A: FDD analysis. Total liver RNA (0.2 µg/sample) from male WT and PPARα-null (KO) mice (n = 2 per treatment group) either fed with a rodent chow diet (Fed) or fasted for 72 h (Starved) was reverse-transcribed by using a 3′ anchored primer AP8. Subsequent PCRs were performed in duplicate (1 and 2) for each mouse by using a 3′-tetramethylrhodamine (TMR)-labeled oligo(dT) AP8 and a 5′ arbitrary primer ARP1. The TMR-labeled fluorescent PCR products were resolved in a 5.6% denaturing polyacrylamide gel. M1, TMR-labeled molecular weight DNA marker. B: Northern blot analysis. Total RNA (30 µg/lane) from the livers of male WT and KO mice either fed or starved (n = 3 per treatment group) was electrophoretically separated in a 1% formaldehyde agarose gel, transferred onto a nylon membrane, and hybridized with the 910 bp DIG-labeled FDD fragment amplified with a pair of AP8 and ARP1 primers using a PCR DIG-labeling mix. The integrity and uniformity of RNA are indicated by 28S and 18S on the formaldehyde agarose gel. M2, DIG-labeled RNA molecular weight marker I (0.39–6.9 kb).
mL-STL cDNAs exhibit multiple 5′ termini and skipping of nucleotides
As a first step to analyze the mL-STL gene, 5′- and 3′-RACE analyses were performed to obtain the full-length cDNA sequence using total RNA from the WT fed and starved liver samples. A single predominant 5′-RACE amplicon of ∼1,900 bp was obtained from the WT fed liver RNA by using the 5′-RACE primer I, whereas a shorter amplification product of ∼1,400 bp was obtained from both the WT fed and starved liver RNA by using the 5′-RACE primer R (supplemental Fig. S2A, B). Similarly, one major 3′-RACE amplicon of ∼800 bp was obtained from the WT fed liver RNA sample by using the 3′-RACE primer S (supplemental Fig. S2A, B). Both the 5′- and 3′-RACE amplicons were subcloned in a 4 kb pCR®II-TOPO® vector, and the nucleotide sequences in 28 of the 5′-RACE and 12 of the 3′-RACE clones were sequenced, aligned, and compared.
Sequence analysis of the 28 mL-STL 5′-RACE clones revealed that dramatic differences in their 5′ termini as well as skipping of a stretch of nucleotides were noted (supplemental Fig. S3). The longest cDNA transcripts amplified by the 5′-RACE primers R and I were 1,381 [5′#2-Starved(R)] and 1,877 [5′#9-Fed(I)] bp, respectively. In all, 10 different 5′ terminus sequences (L1–L10) were observed from the 28 clones, with the majority (20/28) of them displaying the shorter (L8 and L9) 5′ termini. In addition, a stretch of 95 nucleotides was missing in the middle sequences of two 5′-RACE clones, 5′#22-Fed(I) and 5′#12-Starved(R), suggesting that exon skipping had occurred in these clones. Surprisingly, an extra 8 nucleotides (TTTTTCAG) were also observed in the same 5′-RACE clone, 5′#12-Starved(R), and this might be due to the splicing errors that occurred during the processing of premRNA to mRNA in this clone. These 5′-RACE cDNA sequences were then blasted against the NCBI mouse database, and results (supplemental Fig. S3) indicated that our 5′-RACE mL-STL cDNAs shared the highest sequence homology to mouse Riken cDNA 2810007J24 transcript variant 1 (Riken_v1) (GenBank accession no. NM_175250) and variant 2 (Riken_v2) (GenBank accession no. NM_001199306). Riken_v1 is 2,147 bp long and has the 5′ terminus sequence identical to the 5′-RACE clone 5′#2-Starved(R) with the longest 5′ end sequence. It was of interest to note that the first 96 nucleotides in the 5′ end of Riken_v2 cDNA did not match with the 5′ terminus sequences of all 5′-RACE clones examined, suggesting that this is the alternative splicing variant of exon 1. No skipping of nucleotides was reported in either of the Riken cDNA variants.
Sequence analysis of the 12 mL-STL 3′-RACE clones indicated that only one type of 3′-RACE cDNA, but with different polyadenylation cleavage sites (PACSs) and length of the poly(A) tails, was evident (supplemental Fig. S4). The longest 3′-RACE cDNA obtained was 797 bp [3′#9-Fed(S) and 3′#16-Fed(S)], whereas the shortest ones were 786 bp [3′#13-Fed(S) and 3′#14-Fed(S)]. All of their nucleotide sequences were identical except in their 3′ untranslated regions (UTRs). A single upstream polyadenylation signal (PAS) of the ATTAAA type (32) and four different downstream alternative PACS variants (33, 34) were found from the 12 clones examined. The most abundant PACS variant was designated as WT PACS (33), and eight (67%) clones showed a WT PACS 15 bp downstream of the PAS. In addition, an extra “A” nucleotide was found at the nucleotide positions 1,986 and 1,936 of both Riken cDNAs, but it was absent in all 3′-RACE clones obtained in the present study. No poly(A) tail sequences were reported in the two Riken cDNAs.
To assemble the full-length consensus mL-STL cDNA variant sequences, the 5′-RACE clone with the longest 5′-RACE sequence [5′#2-Starved(R)] and the 3′-RACE clone with the longest 3′-RACE sequence [3′#9-Fed(S)] were used to compile the “long-form” 2,168 bp mL-STL1 cDNA variant, whereas the 5′-RACE clone 5′#12-Starved(R) with the 95 bp missing fragment and the same 3′-RACE clone [3′#9-Fed(S)] were used to compile the “short-form” 1,927 bp mL-STL2 cDNA variant (Fig. 2A, B). The nucleotide sequences of the long-form mL-STL1 and short-form mL-STL2 transcript variants were subsequently deposited into the NCBI nucleotide database with assigned GenBank accession nos. EU486166 and EU486167, respectively.
Fig. 2.
Full-length mL-STL cDNA has alternative exonic splicing variants and shares high amino acid sequence homology to the SULT2A family. A: Combined mL-STL1 and mL-STL2 cDNA sequences. The sequence of the mL-STL cDNA was deduced from the 5′- and 3′-RACE products. The 5′ and 3′ UTRs are in lowercase, whereas the coding region is in uppercase. The deduced amino acid sequence is presented underneath its corresponding cDNA nucleotide sequences. The numbering of the cDNA sequence is relative to the first TSS. Solid triangles on the 5′ region denote the different putative TSSs, and the numbers of 5′-RACE clones that possess such TSSs are indicated. The 95 bp of exon 5 (shaded) are completely missing in mL-STL2 cDNA. Solid circles on the 3′ region denote the different PACSs, and the numbers of 3′-RACE clones that possess such PACS are indicated. The PAS is double underlined. B: Schematic diagram of the deduced mouse mL-STL genomic structure and its four exonic splicing variants together with the two Riken 2810007J24 variants. Open boxes represent the 5′ and 3′ untranslated sequences, whereas the black boxes represent the amino acid coding sequences. All four mL-STL variants have the same start codon (ATG) located in exon 2. The stop codon (TAA) of mL-STL_v1 and mL-STL_v2 variants is in exon 7, whereas the stop codon (TGA) of mL-STL_v3 and mL-STL_v4 variants is at the junction of exons 4 and 6. C: Dendrogram analysis of the mL-STL1 isoform based on the percentage of amino acid sequence homology to the 18 published mouse SULTs. The figure was produced by using ClustalW (http://www.ebi.ac.uk/clustalw/).
mL-STL gene has multiple alternative exonic splicing variants
To predict the chromosomal localization as well as genomic structure of the mL-STL gene, the long and short forms of mL-STL cDNAs, as well as Riken_v1 and Riken_v2 variants, were blasted with the mouse genomic database. The exon–intron boundaries of the mL-STL gene were then deduced by aligning the long-form mL-STL1 transcript sequence with the predicted corresponding mouse genomic sequence (GenBank accession no. NT_187034). Results indicated that the mL-STL gene was located on the minus strand of chromosome 7 band A1. Similar to Riken_v1 (NM_175250), the long-form mL-STL gene was composed of seven exons separated by six introns, with a total gene size of 35,902 bp (Table 1). The splicing sites of exon–intron boundaries of the long-form mL-STL gene conformed to the standard gt/ag rule (35). An exceptionally long (18,391 bp) intron 1 was predicted in the mL-STL gene. The longest exon was in exon 7 (1,133 bp), whereas the shortest one was in exon 5 (95 bp). Two alternative splicing exon 1 variants, herein named exons 1A (96 bp) and 1B (146 bp), were noted, with the shorter Riken_v2 variant possessing exon 1A, whereas the longer Riken_v1 variant as well as the long-form mL-STL1 transcript contained exon 1B (Fig. 2B).
TABLE 1.
Predicted exon-intron boundaries of the mL-STL gene
| Exon number | Exon size (bp) | Position in cDNAa | 3′-Splice acceptor siteb | 5′-Splice donor siteb | Intron number | Intron size (bp) |
| 1 | 146 | 1–146 | ……TGCTTCAGgttagttc | 1 | 18,391 | |
| 2 | 267 | 147–413 | ttcaacagCAACTTGA…… | ……AAAATCAGgtatgtat | 2 | 2,227 |
| 3 | 209 | 414–622 | tcattcagGAACCCAC…… | ……AGGCAAAGgtcagtgc | 3 | 1,784 |
| 4 | 127 | 623–749 | tattgaagGTGATTTA…… | ……AGGAAAATgtaagtat | 4 | 7,124 |
| 5 | 95 | 750–844 | ctgtacagCATATTTT…… | ……TGAAAAAGgtaaccac | 5 | 2,420 |
| 6 | 169 | 845–1,013 | tccttcagGATACAAG…… | ……GAGAAAAGgtagagac | 6 | 1,809 |
| 7 | 1,133 | 1,014–2,146 | tttttcagGCATCACA…… |
According to the mL-STL cDNA as shown in Fig. 2.
Exonic sequences are shown in uppercase, whereas intronic sequences are shown in lowercase. The invariant nucleotides (ag/gt) are in boldface.
Difference in the length of exon 1B was also noted in 14 of the 5′-RACE clones (supplemental Fig. S3). All mL-STL cDNA transcripts with either a complete or partial exon 1B together with exons 2–7 were designated as mL-STL variant 1 (mL-STL_v1) (Fig. 2B). Alternative splicing of the entire exon 1B was also observed in 12 of the 5′-RACE clones examined, and these variants were hereby designated as mL-STL variant 2 (mL-STL_v2). The ORFs of mL-STL_v1 and mL-STL_v2 were 849 bp, and an adequate Kozak consensus ATG started at nucleotide 278 at exon 2, and thus exon 1 was 5′ UTR. The ORF extended to a TAA stop codon at nucleotide 1,126 at exon 7. Because exon 1 was 5′ UTR, both mL-STL_v1 and mL-STL_v2 encoded the same protein. The translated protein was composed of 282 amino acid residues, and the deduced long-form mL-STL protein had a calculated molecular mass of 33.3 kDa and a theoretical isoelectric point (pI) of 6.56 (http://web.expasy.org/protparam/).
Skipping of an entire 95 bp of exon 5 was found in two short-form mL-STL2 variants. The short-form variant that possessed exons 1–4, 6, and 7 was designated as mL-STL variant 3 (ml-SLT_v3), whereas the other one without exon 1 was designated as mL-STL variant 4 (mL-STL_v4) (Fig. 2B). Because exon 1 was 5′ UTR, these two mL-STL variants encoded the same shorter truncated protein. The ORF was 474 bp, and the adequate Kozak consensus ATG started at the same nucleotide 278 at exon 2 of the long form, but it extended to an early TGA stop codon at nucleotide 751, which spanned between the end nucleotide T of exon 4 and beginning nucleotides GA of exon 6 of the long-form mL-STL gene. As a result of the frame shift of the ORF in these proteins, the truncated protein was composed of 157 amino acid residues and had a predicted molecular mass of 18.6 kDa with a theoretical pI of 7.8.
The mL-STL gene belongs to the Sult2a family of a superfamily of cytosolic SULTs
To determine whether there is any predicted functional domain, in silico analysis of the mL-STL gene sequence was performed (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). Our results indicated that exons 2–7 of the long-form mL-STL gene encoded a complete cytosolic SULT domain, whereas the sulfonation domain was truncated in the short-form protein due to the absence of 95 bp in its cDNA fragment (Fig. 2A).
To classify the mL-STL into the SULT gene families as well as to get some clues regarding the possible substrate candidates of the mL-STL1 protein, its amino acid sequence similarity was compared with 18 other published mouse SULT amino acid sequences by using Dendrogram analysis (Fig. 2C). Our results revealed that mL-STL1 shared the highest amino acid sequence similarity to the SULT2A subfamily, thus suggesting that the mL-STL1 enzyme was a novel Sult2a isoform, which can catalyze the SULT2A prototype substrates. Similar to other known cytosolic SULTs (36), the N-terminal/5′-phosphosulfate binding (5′-PSB) conserved sequence YPKSGTxW, 3′-phosphate binding (3′-PB) motif RNPRDVLVSGY, and C-terminal/3′-PSB conserved sequence RKGxxGDWKNxFT were found in the mL-STL1 identified in the present study (Fig. 2A). In addition, a signature FSSKA motif sequence, which is highly conserved among the SULT2 family (37) in mammals, was also noted (Fig. 2A).
To characterize the shorter mL-STL2 amino acid sequence, both the long mL-STL1 and short mL-STL2 amino acid sequences were aligned and compared (supplemental Fig. S5). Only the N-terminal conserved sequence YPKSGTHW, SULT2 signature motif FSSKA, and 3′-PB sequence RNPRDVLVSGY were retained in the mL-STL2 (mL-STL_v2) isoform. The absence of the C-terminal conserved sequence RKGITGDWKNHFT in the mL-STL2 sequence suggested that the binding to PAPS at the 3′ terminal was handicapped, and thus the mL-STL2-catalyzed sulfonation reaction was likely to be nonfunctional. Therefore, only the mL-STL1 (mL-STL_v1) isoform was further characterized in the present study and is referred to as mL-STL throughout the study.
Native mL-STL protein was subcellularly localized in the cytosol of mouse liver
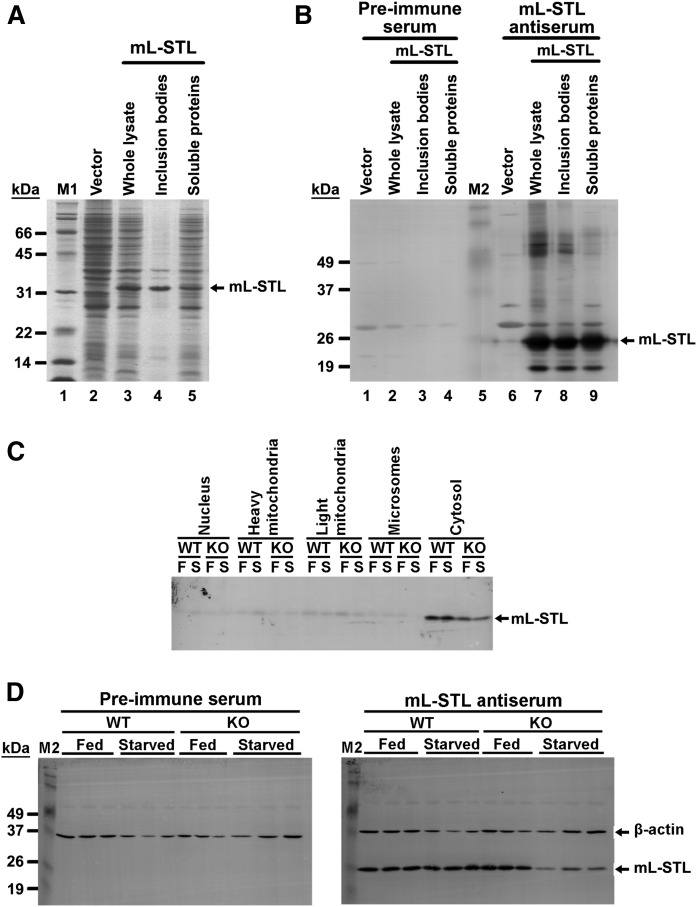
Before studying the subcellular localization of native mL-STL proteins in mouse liver, an mL-STL-specific antiserum was first prepared from immunizing rabbits with a bacterial overexpressed recombinant mL-STL protein. As shown in Fig. 3A, a ∼33 kDa protein, which matched with the expected size of the recombinant mL-STL protein, was detected in whole lysate of E. coli (Fig. 3A, lane 3), compared with the empty vector control (Fig. 3A, lane 2). The recombinant mL-STL proteins were soluble (Fig. 3A, lane 5), but the majority of them were misfolded and present in inclusion bodies (Fig. 3A, lane 4). The identity of recombinant mL-STL protein was confirmed by MALDI-TOF peptide sequencing (supplemental Fig. S6). The mL-STL inclusion bodies were then resolved by a 12% preparative SDS-PAGE, and the protein band corresponding to mL-STL was excised from the SDS-PAGE gel (data not shown) and injected into rabbits for producing the anti-mL-STL antiserum. The antigenicity of the fifth bled polyclonal mL-STL antiserum was confirmed by using Western blot analysis. As shown in Fig. 3B, the mL-STL antiserum detected a major immunoreactive band with an apparent molecular mass of ∼26 kDa (Fig. 3B, lanes 7–9), which was close to the expected size of the mL-STL recombinant protein (33 kDa). The slight size difference might have resulted from the coupling of pink or blue chromophores to the standard proteins in the prestained protein markers, which retarded their electromigration and thus affected the apparent molecular mass relative to the unstained protein standards. The ∼26 kDa immunoreactive band was absent when the mL-STL recombinant protein was incubated with the preimmune serum (Fig. 3B, lanes 2–4), suggesting that the fifth bled antiserum was specific to the mL-STL recombinant protein.
Fig. 3.
An mL-STL-specific polyclonal antiserum confirms the subcellular localization of the native mL-STL protein in liver cytosol. A: SDS-PAGE analysis of recombinant mL-STL protein in E. coli cell lysate. Different fractions of bacterial cell lysate (20 µg/lane) were electrophoresed in a 12% SDS-PAGE gel followed by Coomassie blue staining. M1, Bio-Rad protein standards, broad range (6.5–200 kDa). B: Western blot analysis of antigenicity of the mL-STL antiserum produced in recognizing the recombinant mL-STL protein in E. coli cell lysate. Different fractions of bacterial cell lysate (20 µg/lane) were electrophoresed in a 12% SDS-PAGE, transferred onto a PVDF membrane, and incubated with either a preimmune (1:1,000) or an in-house-produced fifth bled mL-STL antiserum (1:2,000). M2, Invitrogen Benchmark prestained protein ladder (6–180 kDa). C: Western blot analysis of subcellular localization of native mL-STL protein in male mouse liver. Different subcellular protein fractions (5 µg/lane) from the livers of WT and KO mice either fed (F) or fasted for 72 h (S) were separated in a 12% SDS-PAGE gel, transferred onto a PVDF membrane, and incubated with either a preimmune (1:1,000) or an mL-STL antiserum (1:4,000). D: Western blot analysis of antigenicity of the mL-STL antiserum produced in recognizing the native mL-STL protein in liver cytosol. Cytosolic fractions (20 µg/lane) obtained from the livers of male WT and KO mice either fed or starved for 72 h were separated in a 12% SDS-PAGE gel, transferred onto a PVDF membrane, and incubated with either a preimmune (1:1,000) or an mL-STL antiserum (1:4,000). Anti-β-actin antibody (1:3,000) was used in parallel reactions to assess the integrity and uniformity of the cytosolic proteins. M2, Invitrogen Benchmark prestained protein ladder (6–180 kDa).
In silico prediction indicated that the mL-STL is a cytoplasmic protein (http://psort.hgc.jp/form2.html) without any signal peptide (http://www.cbs.dtu.dk/services/SignalP/). To confirm its subcellular localization, the mouse livers from both WT and KO under fed and fasting conditions were fractionated into various subcellular fractions by differential ultracentrifugation, followed by Western blot analysis. As expected, a strong immunoreactive band corresponding to the native mL-STL protein was detected in the cytosol of both WT and KO mice of the fed or starved group (Fig. 3C), and this protein expression pattern was in line with the mRNA expression profile (Fig. 1B). Only very weak signals were observed in the nuclei, heavy mitochondria, light mitochondria, and endoplasmic reticulum fractions of these animals, and this might have been due to the cross-contamination of the residual amount of cytosolic proteins trapped in these fractions. To further confirm the specificity of our mL-STL antiserum in recognizing the native mL-STL proteins in liver cytosol, the cytosolic fractions of WT and KO mice during fed and fasting conditions were probed with a preimmune and an mL-STL antisera for comparison by using Western blot analysis. The housekeeping protein β-actin was used as a loading control. As shown in Fig. 3D, an immunoreactive band corresponding to the native mL-STL protein was detected in both WT and KO mice fed a normal mouse diet, but the level of mL-STL protein significantly declined in the KO mice under fasting conditions. This immunoreactive band was absent when the blot was probed with the preimmune serum. Thus, these data confirmed that mL-STL is a cytosolic protein in mouse livers.
mL-STL is a liver-specific and male-dominant cytosolic SULT
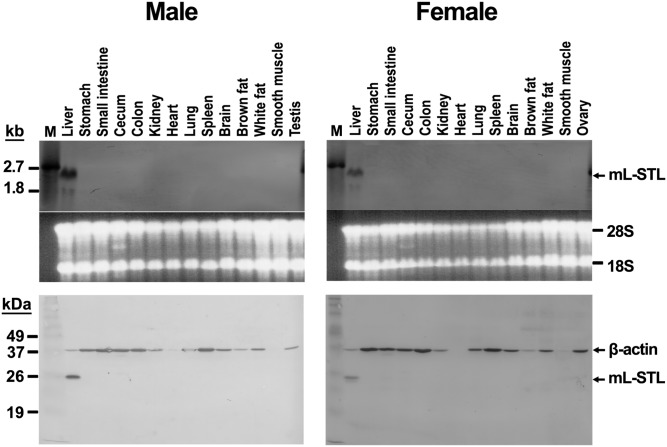
Next, we studied the tissue distribution of mL-STL mRNA and protein in both male and female WT mice using Northern and Western blot analyses, respectively. As shown in Fig. 4 (top), a prominent ∼2.2 kb mL-STL mRNA transcript was constitutively expressed only in the livers of both male and female animals. In addition, a minor ∼1.8 kb mRNA transcript was observed in the liver, but so far we do not know its identity. No detectable expression of mL-STL mRNA was found in other mouse tissues, including the stomach, small intestine, cecum, colon, kidney, heart, lung, spleen, brain, brown fat, white fat, smooth muscle, testis, and ovaries. This result was in agreement with the Western blot analysis (Fig. 4, bottom), in which the mL-STL cytosolic protein was only expressed in the mouse liver of both sexes, suggesting that mL-STL is a liver-specific protein. It was also interesting to note that higher expressions of mL-STL mRNA and protein were noted in males compared with females and that dramatic differences in β-actin protein expression were evident in different mouse tissues.
Fig. 4.
mL-STL is a liver-specific and male-dominant protein. Different tissues excised from WT male and female mice were subject to Northern blot (top) and Western blot (bottom) analyses. For Northern blot analysis, total RNA (20 µg/lane) was electrophoresed in a 1% formaldehyde agarose gel, transferred onto a N+-nylon membrane, and hybridized with a DIG-labeled mL-STL cDNA probe. The integrity and uniformity of the RNA are indicated by 28S and 18S on the formaldehyde agarose gel. M, DIG-labeled RNA molecular mass marker I (0.39–6.9 kb). For Western blot analysis, cytosolic proteins (20 µg/lane) were electrophoresed in a 12% SDS-PAGE gel, transferred onto a PVDF nylon membrane, and incubated with an mL-STL antiserum (1:5,000) and a mouse monoclonal anti-β-actin (1:30,000) antibody. β-actin was used as a loading control. M, Invitrogen Benchmark prestained protein ladder (6–180 kDa). The experiment was repeated with three different batches of animals, and the data shown are representative of Northern (top) and Western (bottom) blots.
mL-STL mRNA and protein express postnatally
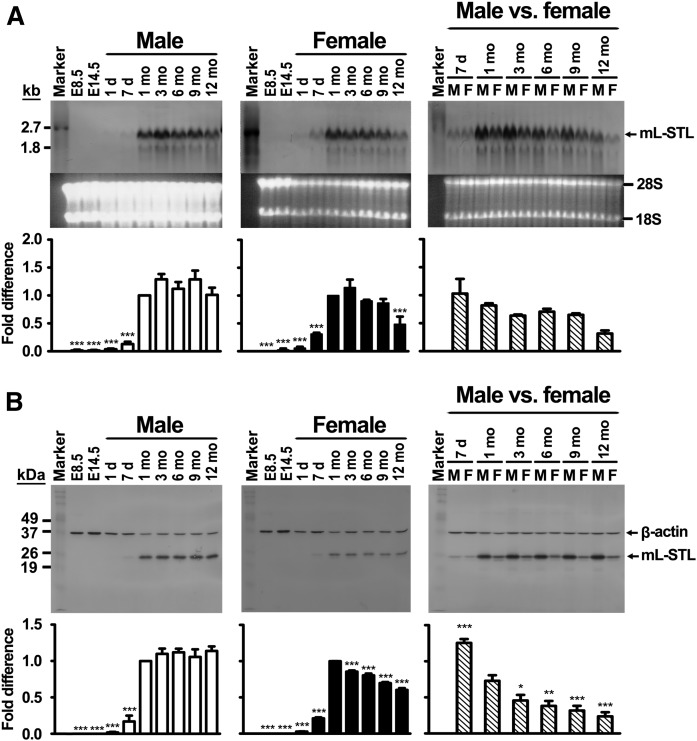
To further probe into the possible biological functions of the mL-STL gene, the ontogenic expression profile of mL-STL mRNA and proteins in postimplantation embryonic stages as well as postnatal hepatic tissues of both male and female mice was performed by using Northern and Western blot analyses, respectively. Our results indicated that no observable expression of mL-STL mRNA (Fig. 5A) was detected in embryonic day 8.5 (E8.5) and E14.5 embryos. Remarkably, expression of mL-STL mRNA was detected right after birth and reached maximally at 1 month in both sexes. After that, the mL-STL expression level was maintained in males, but declined in females, in a time-dependent manner. This result was in agreement with the Western blot analysis (Fig. 5B), in which mL-STL protein was detected in livers postnatally, peaked at 1 month old, and then declined in female, but was maintained in male, animals. In addition, significantly higher levels of mL-STL mRNA (Fig. 5A) and protein (Fig. 5B) expressions were detected in all adult stages of male examined, suggesting that it is a male-dominant Sult2a cytosolic SULT that expressed postnatally.
Fig. 5.
Hepatic mL-STL mRNA and protein express postnatally. Total RNA and cytosolic proteins were extracted from E8.5 and E14.5 embryos as well as from 1 day (1 d), 7 days (7 d), 1 month (1 mo), 3 months (3 mo), 6 months (6 mo), 9 months (9 mo), and 12 months (12 mo) male (M) and female (F) WT mouse livers. A: Northern blot analysis. Total RNA (20 µg/lane) was electrophoresed in a 1% formaldehyde agarose gel. The integrity and uniformity of the RNA are indicated by 28S and 18S on the formaldehyde agarose gel. RNA marker, DIG-labeled RNA molecular mass marker I (0.39–6.9 kb). B: Western blot analysis. Cytosolic proteins (20 µg/lane) were electrophoresed in a 12% SDS-PAGE gel, and the protein blots were incubated with an mL-STL antiserum (1:5,000) and a mouse monoclonal anti-β-actin (1:30,000) antibody. β-actin was used as a loading control. Protein marker, Invitrogen Benchmark prestained protein ladder (6–180 kDa). The experiment was repeated with three different batches of animals, and the results of one representative Northern and Western blot are shown in (A) and (B), respectively. The ImageJ program (http://imagej.nih.gov/ij/) was used to quantify the mL-STL mRNA and protein expression signals on the Northern and Western blots, respectively. The mRNA expression signals on Northern blots are expressed in absolute arbitrary units, whereas the protein expression signals on Western blots are expressed in relative arbitrary units after normalizing against β-actin. Then, the mL-STL mRNA expression (in arbitrary units) and protein expression (in relative arbitrary units) in each time point was normalized against 1 month (which exhibited the highest expression level) and expressed as fold difference in both male and female. Difference in temporal mL-STL expression in each sex was analyzed by one-way ANOVA followed by Dunnett’s post hoc test. For male and female comparison, the mL-STL mRNA expression in Northern blot (A) was quantified into arbitrary units, whereas the mL-STL protein expression in Western blot (B) was quantified in relative arbitrary units after normalization to β-actin. Then, the arbitrary/relative arbitrary unit of the female was normalized against its corresponding male at each time point and expressed as the fold difference. Difference in temporal mL-STL expression in female versus male was compared with 1 month by one-way ANOVA followed by Dunnett’s post hoc test. *** P < 0.001; ** P < 0.01; * P < 0.05.
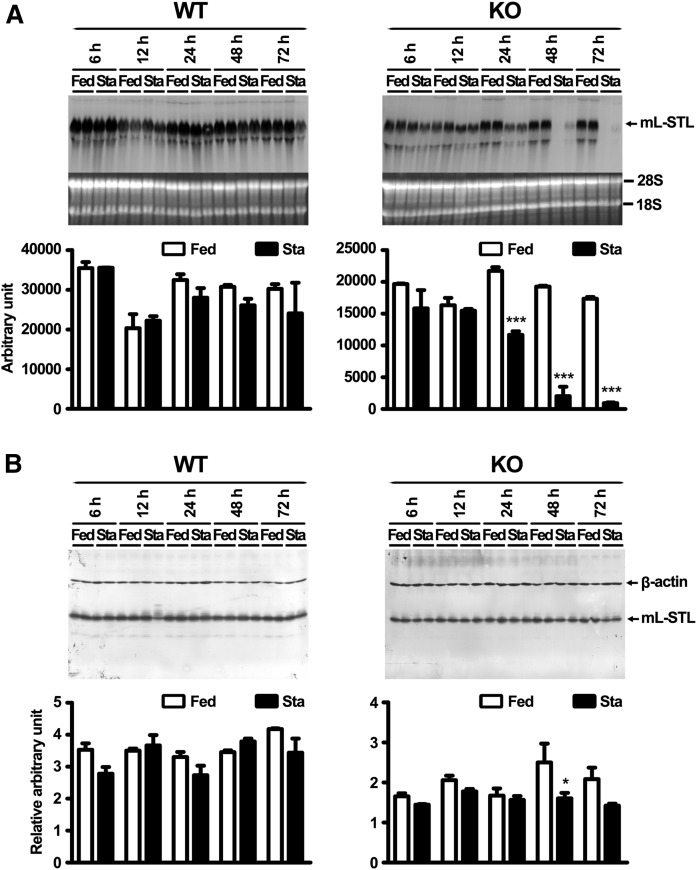
Fasting downregulates mL-STL expression in a time- and PPARα-dependent manner
To further understand the effect of fasting on mL-STL regulation, the temporal expression profile of mL-STL mRNA and protein at 6, 12, 24, 48, and 72 h after fasting was analyzed by using Northern and Western blot analyses. In the KO mice, fasting induced a significant decrease in hepatic expression of mL-STL mRNA transcript at 24 h after food deprivation and continued at 48 and 72 h (Fig. 6A). In contrast, only a slight, but statistically insignificant, suppression of mL-STL mRNA expression was observed at 24, 48, and 72 h in the WT mice during the same time course of treatment. This result was in slight disagreement with the distinct suppression of mL-STL mRNA expression observed in the WT mice at 72 h of fasting (Fig. 1B). This discrepancy might be due to the difference in the length of the two probes that were used for the Northern blot analyses, because one had a shorter, 910 bp probe in Fig. 1B, whereas the other, shown in Fig. 6A, had a longer, 1.8 kb probe. The longer probe had higher efficiency of hybridization to the mL-STL RNA, which led to oversaturation of the color signals on the blot (Fig. 6A) upon overexposure. Similar to the mRNA expression pattern, fasting had a time-dependent inhibition on the mL-STL protein expression in the KO mice, but not significantly in the WT mice. However, the decrease in protein expression was delayed at 48 and 72 h in the KO mice (Fig. 6B). It was also interesting to observe that the reduction of the mL-STL protein level was not as dramatic as the mRNA level in the KO mice.
Fig. 6.
Fasting downregulates mL-STL expression in a time- and PPARα-dependent manner. Northern blot analysis of total RNA (A) and Western blot analysis of cytosolic proteins (B) from the livers of WT and KO mice either fed with a rodent chow diet (Fed) or deprived of food (Sta) for 6, 12, 24, 48, and 72 h are shown. A: Northern blot analysis. Total RNA (30 µg/lane) from each treatment group (n = 2 per treatment group) was subject to 1% formaldehyde-agarose gel. The integrity and uniformity of the RNA are indicated by 28S and 18S on the formaldehyde agarose gel. B: Western blot analysis. Cytosolic proteins (20 µg/lane) from each treatment group (n = 2 per treatment group) were electrophoresed in a 12% SDS-PAGE, and the protein blots were incubated with an mL-STL antiserum (1:4,000) and a mouse monoclonal anti-β-actin (1:30,000) antibody. β-actin was used as a loading control. The ImageJ program (http://imagej.nih.gov/ij/) was used to quantify the mL-STL mRNA and protein expression signals on the Northern and Western blots, respectively. The mRNA expression signals on Northern blots were expressed in absolute arbitrary units, whereas the protein expression signals on Western blots were expressed in relative arbitrary unit after normalizing against β-actin. Difference in temporal mL-STL expression between starved and fed group in each blot was analyzed by two-way ANOVA followed by Bonferroni post hoc test. *** P < 0.001; * P < 0.05. Error bars represent SD.
Wy-14,643 treatment downregulates mL-STL expression in a time- and PPARα-dependent manner
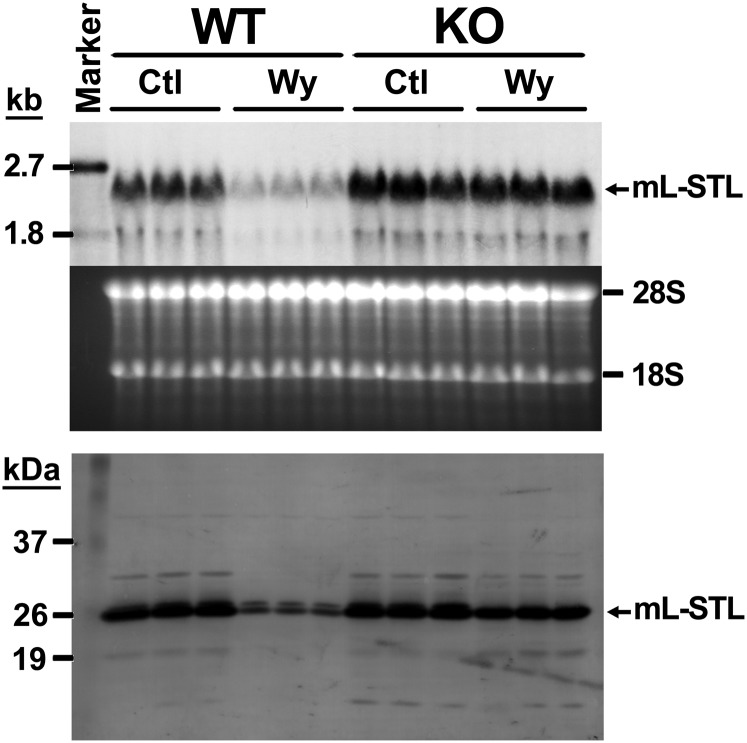
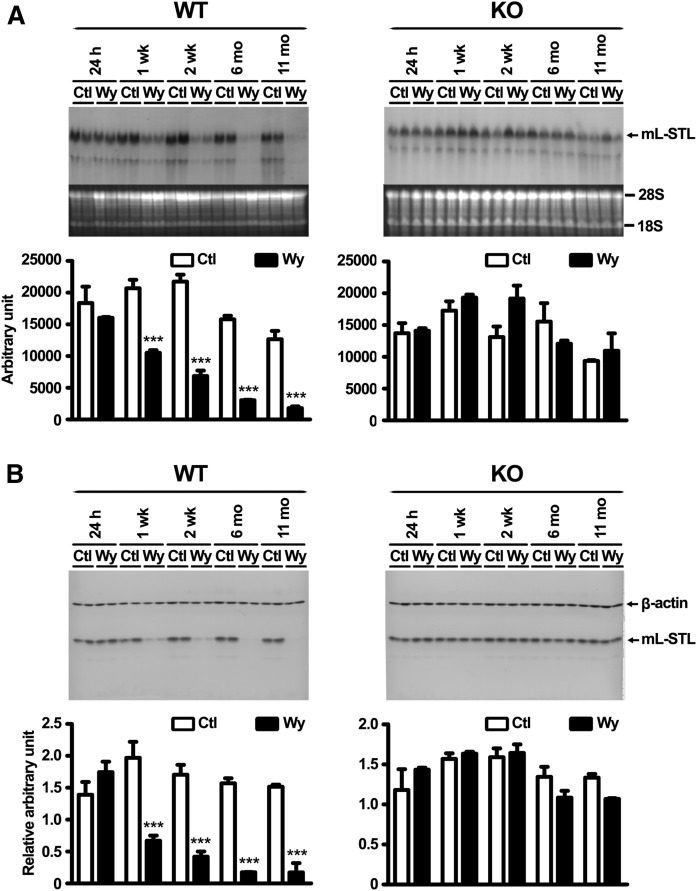
It has been reported that many PPARα-regulated genes are responsive under fasting as well as after treatment with PPARα agonists such as Wy-14,643 (2, 3). To further examine whether the mL-STL gene was also responsive to Wy-14,643, Northern and Western blot analyses were performed in livers from both WT and KO mice after 2 weeks of 0.1% (w/w) Wy-14,643 treatment. Constitutive expression of hepatic mL-STL mRNA (Fig. 7, top) and protein (Fig. 7, bottom) was observed in the livers of both WT and KO mice fed with a control diet, whereas 2 weeks of Wy-14,643 treatment resulted in dramatic suppression of mL-STL mRNA and protein expressions in the WT, but not in KO, mice, suggesting that the downregulation was mediated through the activation of PPARα upon Wy-14,643 treatment. Temporal studies revealed that the effect of Wy-14,643 treatment on the downregulation of mL-STL mRNA (Fig. 8A) and protein (Fig. 8B) was observed as early as after 1 week and peaked at 11 months when compared with their corresponding WT control mice. No such downregulation of the mL-STL protein was observed in the control and Wy-14,643-treated KO mice during the entire treatment period, suggesting that mL-STL is a putative PPARα-regulated gene.
Fig. 7.
Wy-14,643 treatment downregulates mL-STL mRNA and protein expression. Total RNA or cytosolic proteins were extracted from the livers of WT and KO mice either fed with a control rodent diet (Ctl) or a 0.1% Wy-14,643 (w/w) diet (Wy) for 2 weeks and were subject to Northern and Western blot analyses. For Northern blot analysis, total RNA (30 µg/lane) from each treatment group (n = 3 per treatment group) was electrophoresed in a 1% formaldehyde agarose gel, transferred onto a N+-nylon membrane, and hybridized with a DIG-labeled mL-STL cDNA probe. The integrity and uniformity of the RNA are indicated by 28S and 18S on the formaldehyde agarose gel. RNA marker, DIG-labeled RNA molecular mass marker I (0.39–6.9 kb). For Western blot analysis, cytosolic proteins (20 µg/lane) from each treatment group (n = 3 per treatment group) were electrophoresed in a 12% SDS-PAGE gel, transferred to a PVDF nylon membrane, and incubated with an mL-STL antiserum (1:5,000). Protein marker, Invitrogen Benchmark prestained protein ladder (6–180 kDa). Representatives of Northern and Western blots are shown in the top and bottom, respectively.
Fig. 8.
Wy-14,643 treatment downregulates mL-STL expression in a time- and PPARα-dependent manner. Northern blot analysis of total RNA (A) and Western blot analysis of cytosolic proteins (B) from the livers of WT and KO mice either fed with a control rodent diet (Ctl) or a 0.1% Wy-14,643 (w/w) diet (Wy) for 24 h, 1 week (1 wk), 2 weeks (2 wk), 6 months (6 mo), and 11 months (11 mo) are shown. A: Northern blot analysis. Total RNA (30 µg/lane) from each treatment group (n = 2 per treatment group) was subject to 1% formaldehyde-agarose gel. The integrity and uniformity of the RNA are indicated by 18S and 28S on the formaldehyde-agarose gel. B: Western blot analysis. Cytosolic proteins (20 µg/lane) were electrophoresed in a 14% SDS-PAGE gel, and the protein blots were incubated with an mL-STL antiserum (1:4,000) and a mouse monoclonal anti-β-actin (1:30,000) antibody. β-actin was used as a loading control. The ImageJ program (http://imagej.nih.gov/ij/) was used to quantify the mL-STL mRNA and protein expression signals on the Northern and Western blots, respectively. The mRNA expression signals on Northern blots were expressed in absolute arbitrary units, whereas the protein expression signals on Western blots were expressed in relative arbitrary units after normalizing against β-actin. Difference in temporal mL-STL expression between Wy-14,643 treatment and control in each blot was analyzed by two-way ANOVA followed by Bonferroni post hoc test. *** P < 0.001. Error bars represent SD.
Recombinant His-mL-STL protein preferentially catalyzes primary bile acid substrates
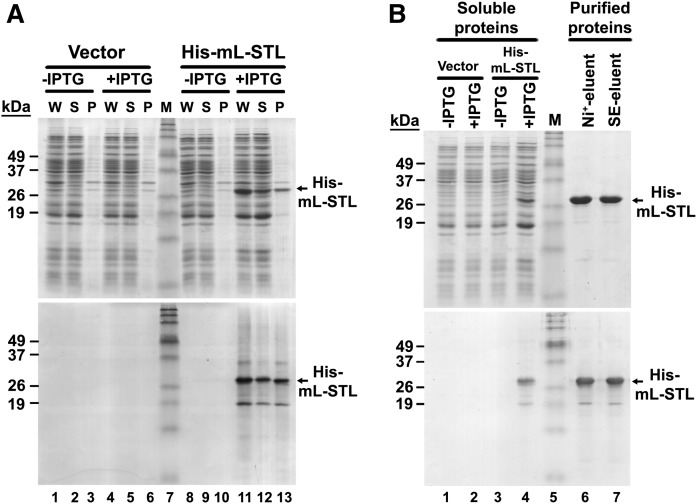
As a first step to study the substrate specificity of mL-STL protein, a His-tag recombinant mL-STL fusion protein (His-mL-STL) was cloned and overexpressed. To overexpress the recombinant His-mL-STL protein, the mpRSETA expression vector was transformed into an E. coli strain BL21(DE3)pLysS, and protein expression was induced with 0.2 mM IPTG at 25°C for 16 h. The recombinant His-mL-STL protein was overexpressed in the whole lysate of E. coli (Fig. 9A, top, lane 11). Half of the expressed protein was soluble (Fig. 9A, top, lane 12), whereas the remaining protein was misfolded to form inclusion bodies (Fig. 9A, top, lane 13). No detectable His-mL-STL protein expression was observed in the E. coli transformed with the His-mL-STL expression vector without IPTG induction (Fig. 9A, top, lanes 8–10). Similarly, no observable expression of the His-mL-STL protein was found in the empty vector control either in the presence (Fig. 9A, top, lanes 4–6) or absence (Fig. 9A, top, lanes 1–3) of IPTG induction. Western blot analysis revealed that the predicted His-mL-STL proteins in the total cell lysate (Fig. 9A, bottom, lane 11), soluble proteins (lane 12), and inclusion bodies (lane 13) were immunoreacted to the polyclonal antiserum raised against the recombinant mL-STL protein, which further confirmed the identity of the His-mL-STL protein.
Fig. 9.
Overexpression and column purification of recombinant His-mL-STL protein. SDS-PAGE and Western blot analysis of bacterial overexpressed (A) and column-purified (B) recombinant His-mL-STL proteins is shown. A: The recombinant His-mL-STL protein was overexpressed in BL21(DE3)pLysS cells with (+IPTG) or without (−IPTG) 0.2 mM IPTG induction. The E. coli cells transformed with the mpRSETA vector served as a negative control (Vector). The whole cell lysate (W) was fractionated into soluble proteins (S) and inclusion bodies (P). The bacterial protein fractions (25 µg/lane) were electrophoresed in a 14% SDS-PAGE gel followed by Coomassie blue staining (top) and Western blot analysis (bottom). B: The His-mL-STL proteins (5 µg/lane) purified from the nickel affinity HiTrap (Ni+-eluent) and size exclusion (SE-eluent) columns were electrophoresed in a 14% SDS-PAGE gel followed by Coomassie blue staining (top) and Western blot analysis (bottom). M, Invitrogen Benchmark prestained protein ladder (6–180 kDa).
Next, the overexpressed His-mL-STL protein was purified from the soluble fraction of bacterial cell lysate by using a two-step column chromatographic process. As shown in Fig. 9B (top, lane 6), the His-mL-STL protein was purified to near-homogeneity after the first nickel-affinity column purification (supplemental Fig. S7A). No noticeable increase in purity (Fig. 9B, top, lane 7) was observed after subsequent size-exclusion column chromatography (supplemental Fig. S7B). The identity of purified proteins was first confirmed by Western blot analysis (Fig. 9B, bottom), followed by MALDI-TOF peptide sequencing analysis (supplemental Fig. S8).
Because the amino acid sequence of the mL-STL gene showed high similarity to the mammalian SULT2A family (Fig. 2C), we first tested whether the column-purified recombinant His-mL-STL protein could catalyze the prototype SULT2 hydroxysteroid substrates, including bile acids, steroid hormones, cholesterol, and corticosterone. It is known that SULT activity is pH-dependent and exhibits substrate inhibition at high concentrations of their preferred substrates (38), presumably through binding at an allosteric site (39). To ensure that the SULT activity was measured at its near-optimal pH, and not within the substrate inhibition concentrations, as a first step, the SULT activity was examined at pH 5.5, 6.5, and 7.5 at a low substrate concentration (10 μM). As shown in Table 2, the His-mL-STL enzyme exhibited a significant pH-dependent decrease in SULT activities as the pH increased from 5.5 to 7.5, suggesting that the His-mL-STL protein has the highest catalytic activity at acidic pH 5.5. Among the 15 groups of compounds studied, the highest SULT activity was observed in three kinds of primary bile acids/salts/conjugated forms, including CA, CDCA, and α-muricholic acid (α-MCA) (Table 2). Low mL-STL-mediated SULT activity was observed with β-muricholic acid (β-MCA), whereas very low activity was observed in ω-muricholic (ω-MCA), ursodeoxycholic acid (UDCA), DCA, LCA, taurolithocholic acid sodium salt, androsterone, dehydroepiandrosterone (DHEA), pregnenolone, cholesterol, 22(S)-hydroxycholesterol, 22(R)-hydroxycholesterol, and corticosterone (Table 2). Next, we studied whether substrate inhibition occurred as the substrate concentration increased from 10 to 100 μM at the optimal pH at 5.5. In general, no significant substrate inhibition was detected in most compounds tested as the substrate concentrations increased from 10 to 100 μM (Table 2).
TABLE 2.
SULT2 substrate screen of column-purified recombinant His-mL-STL enzyme using 1-butanol extraction method
| Specific activity (pmol/min/mg), mean ± SEM | ||||||
| Vary pH at 10 μM substrate concentration | Vary substrate concentration at pH 5.5 | |||||
| Compounds | OH Position | pH 5.5 | pH 6.5 | pH 7.5 | 10 μM | 100 μM |
| CA | 3α, 7α, 12α | 578.99 ± 164.72 | 575.18 ± 27.45ns | 217.67 ± 40.86** | 749.53 ± 23.95 | 819.39 ± 111.66ns |
| Sodium cholate | 3α, 7α, 12α | 652.30 ± 89.47 | 566.00 ± 90.29ns | 250.26 ± 32.21*** | 747.25 ± 152.29 | 886.11 ± 115.53ns |
| Taurocholic acid | 3α, 7α, 12α | 720.70 ± 151.04 | 468.24 ± 99.93ns | 222.44 ± 34.56*** | 886.27 ± 134.26 | 1,012.65 ± 229.67ns |
| Sodium taurocholate | 3α, 7α, 12α | 485.07 ± 67.64 | 528.82 ± 34.11ns | 118.50 ± 15.63** | 737.18 ± 16.16 | 633.90 ± 99.2ns |
| Glycocholic acid | 3α, 7α, 12α | 960.14 ± 174.59 | 393.22 ± 33.32*** | 223.14 ± 0.58*** | 1,248.60 ± 142.54 | 987.34 ± 63.79ns |
| Sodium glycocholate | 3α, 7α, 12α | 649.88 ± 111.09 | 467.39 ± 90.63ns | 263.30 ± 35.44*** | 821.63 ± 113.13 | 839.52 ± 146.64ns |
| CDCA | 3α, 7α | 1,287.42 ± 85.87 | 513.88 ± 46.59*** | 198.80 ± 27.30*** | 1,326.47 ± 135.64 | 1,433.67 ± 68.29ns |
| Sodium chenodeoxycholate | 3α, 7α | 1,154.29 ± 146.15 | 755.49 ± 117.35*** | 397.63 ± 10.81*** | 1,121.25 ± 55.34 | 1,563.35 ± 90.81** |
| Sodium taurochenodeoxycholate | 3α, 7α | 1,294.69 ± 142.67 | 934.57 ± 135.95** | 473.98 ± 13.86*** | 1,308.46 ± 106.77 | 1,633.79 ± 70.67ns |
| Sodium glycochenodeoxycholate | 3α, 7α | 1,380.24 ± 90.10 | 618.36 ± 40.76*** | 336.54 ± 30.56*** | 1,474.83 ± 140.74 | 1,451.89 ± 170.04ns |
| α-MCA | 3α, 6β, 7α | 854.42 ± 38.30 | 270.97 ± 61.21*** | 196.44 ± 45.38*** | 629.16 ± 83.76 | 768.81 ± 48.89ns |
| Sodium tauro-α-muricholate | 3α, 6β, 7α | 776.21 ± 4.27 | 433.89 ± 103.21** | 182.89 ± 35.63*** | 722.02 ± 53.87 | 1,084.91 ± 144.82ns |
| β-MCA | 3α, 6β, 7β | 155.85 ± 87.60 | 10.81 ± 4.44ns | 2.61 ± 1.58ns | 76.21 ± 20.56 | 228.34 ± 77.04ns |
| ω-MCA | 3α, 6α, 7β | 30.57 ± 4.04 | −0.65 ± 1.66ns | 2.31 ± 1.09ns | 11.29 ± 2.35 | 35.20 ± 29.32ns |
| UDCA | 3α, 7β | 274.11 ± 159.97 | 1.08 ± 11.58ns | 3.78 ± 3.63ns | 46.97 ± 22.16 | 61.69 ± 31.21ns |
| DCA | 3α, 12α | 13.82 ± 9.27 | −0.76 ± 10.28ns | 6.11 ± 4.27ns | 13.57 ± 12.41 | 7.81 ± 5.23ns |
| Lithocholic acid | 3α | 13.39 ± 3.92 | −4.82 ± 4.87ns | 1.04 ± 2.61ns | 5.86 ± 19.63 | 7.45 ± 5.37ns |
| Sodium taurolithocholate | 3α | 43.97 ± 17.86 | −1.43 ± 2.26ns | 10.95 ± 3.46ns | −2.08 ± 5.63 | 2.86 ± 9.20ns |
| Androsterone | 3α | 34.97 ± 34.49 | 4.36 ± 5.66ns | 0.91 ± 2.34ns | 15.46 ± 8.82 | 22.74 ± 15.58ns |
| DHEA | 3β | 1.60 ± 7.38 | −8.57 ± 2.10ns | −1.40 ± 1.23ns | 0.03 ± 1.99 | −6.08 ± 10.36ns |
| Pregnenolone | 3β | 10.96 ± 4.26 | −9.33 ± 5.07ns | −0.39 ± 2.87ns | 19.53 ± 8.96 | −3.37 ± 1.14ns |
| Cholesterol | 3β | 11.98 ± 7.80 | −2.13 ± 2.87ns | 2.63 ± 3.92ns | 12.11 ± 6.10 | 3.27 ± 6.95ns |
| 22(S)-Hydroxycholesterol | 3β | 18.31 ± 9.12 | 0.23 ± 2.29ns | 0.07 ± 2.14ns | 3.24 ± 3.30 | 11.96 ± 3.98ns |
| 22(R)-Hydroxycholesterol | 3β | 7.13 ± 5.09 | 1.83 ± 2.71ns | 0.66 ± 0.57ns | 4.27 ± 3.41 | 10.28 ± 2.01ns |
| Corticosterone | 11β | 6.78 ± 7.30 | −1.87 ± 2.09ns | 4.48 ± 1.99ns | 7.79 ± 1.17 | 3.94 ± 3.56ns |
Each reaction mixture (100 μl) contained 100 mM sodium acetate buffer (pH 5.5) or 100 mM sodium phosphate buffer (pH 6.5 or 7.5), 2.5 mM MgCl2, 20 μg of purified His-mL-STL protein, 10 or 100 μM substrate, and 50 μM PAPS containing 0.015 μCi [35S]PAPS. The reaction mixture was incubated at 37°C for 1 h, and the reaction was terminated by the addition of ammonium hydroxide. The [35S]product was extracted with 1-butanol, and the specific activity was calculated from the scintillation counting of the [35S]sulfonated product. Data are mean ± SEM of three batches of purified proteins, each performed in duplicate. Differences between compounds and pH at fixed 10 μM substrate concentration as well as differences between compounds and substrate concentrations at pH 5.5 were compared by using two-way ANOVA followed by Bonferroni post hoc test. *** P < 0.001; ** P < 0.01; ns, nonsignificant at P > 0.05 level.
To confirm that the His-mL-STL enzyme activity is specific for SULT2 substrates, other SULT1 signature substrates were also examined at different pH and substrate concentrations. In general, no significant His-mL-STL-mediated SULT activity was detected toward prototype substrates, including phenolic, catecholamines, estrogens, tyrosine/derivatives, thyroid hormones, minoxidil, and 4-phenyl-1, 2, 3, 6-tetrahydropyridine hydrochloride at pH 5.5, 6.5, and 7.5 (Table 3) and at 50 and 100 μM substrate concentrations (Table 3). An exception was with the 2-naphthylamine substrate, in which the highest His-mL-STL enzyme activity was detected at pH 5.5 (Table 3), as well as at 100 μM substrate concentration (Table 3). Thus, our data strongly suggested that the His-mL-STL protein displayed relatively low substrate specificity toward the SULT1 prototype substrates examined.
TABLE 3.
SULT1 substrate screen of column-purified recombinant His-mL-STL enzyme using barium precipitation method
| Specific activity (pmol/min/mg), mean ± SEM | |||||
| Vary pH at 50 μM substrate concentration | Vary substrate concentration at pH 5.5 | ||||
| Compounds | pH 5.5 | pH 6.5 | pH 7.5 | 50 μM | 100 μM |
| Phenols | |||||
| Phenol | 2.28 ± 3.31 | 2.24 ± 0.94ns | 2.71 ± 0.43ns | 3.93 ± 0.77 | 8.12 ± 6.77ns |
| 2-Ethylphenol | 7.59 ± 2.48 | 0.74 ± 2.21ns | 2.28 ± 0.70ns | 2.51 ± 4.64 | 1.31 ± 2.00ns |
| 4-Ethylphenol | 4.36 ± 0.07 | 1.55 ± 2.31ns | 3.17 ± 1.77ns | 1.97 ± 6.81 | 3.18 ± 2.51ns |
| 4-Isoprophylphenol | 5.73 ± 0.27 | 9.19 ± 7.37ns | 3.20 ± 0.05ns | −0.95 ± 0.12 | 3.99 ± 0.08ns |
| 2-Prophylphenol | 7.39 ± 3.36 | 11.04 ± 7.87ns | 0.42 ± 0.42ns | −4.48 ± 0.02 | 0.00 ± 3.40ns |
| 4-Nitrophenol | 4.06 ± 3.55 | 8.17 ± 3.17ns | 5.63 ± 5.11ns | 1.45 ± 2.69 | 2.45 ± 2.23ns |
| 1-Naphthol | 1.21 ± 3.77 | 5.67 ± 3.17ns | 2.28 ± 0.01ns | −1.18 ± 1.48 | 5.50 ± 9.13ns |
| 2-Naphthol | 2.55 ± 0.67 | 8.55 ± 7.18ns | 11.71 ± 9.78ns | 0.51 ± 0.71 | 1.41 ± 6.41ns |
| Acetaminophen | 8.26 ± 0.27 | 8.85 ± 3.41ns | 2.69 ± 1.29ns | 13.07 ± 5.30 | 4.85 ± 9.10ns |
| p-Cresol | 3.36 ± 1.49 | 1.98 ± 1.20ns | 9.26 ± 10.14ns | 12.42 ± 5.06 | 8.53 ± 2.18ns |
| 4-Nitrocatechol | −5.92 ± 4.04 | 3.82 ± 4.50ns | −1.38 ± 0.33ns | −6.45 ± 0.71 | 7.70 ± 1.12ns |
| Resveratrol | 1.27 ± 1.46 | 2.86 ± 6.95ns | 12.62 ± 7.19ns | 3.04 ± 7.54 | −0.62 ± 7.89ns |
| Vanillin | 4.76 ± 1.90 | 8.34 ± 6.30ns | 0.81 ± 1.33ns | 20.60 ± 15.57 | 36.69 ± 28.44ns |
| Catecholamines | |||||
| Dopamine hydrochloride | 0.24 ± 2.80 | 4.21 ± 2.17ns | 0.47 ± 0.82ns | 6.68 ± 5.66 | 2.41 ± 0.37ns |
| Isoprenaline hydrochloride | −11.98 ± 10.27 | −0.91 ± 2.73ns | −5.44 ± 2.46ns | −9.80 ± 5.92 | 3.87 ± 4.55ns |
| Salbutamol | 1.49 ± 2.60 | 6.87 ± 6.42ns | −1.48 ± 4.46ns | 9.04 ± 5.57 | −2.36 ± 3.26ns |
| (±)-Epinephrine | −8.46 ± 11.53 | −5.84 ± 3.34ns | −3.09 ± 0.98ns | −8.76 ± 11.21 | 2.69 ± 2.92ns |
| (−)-Norepinephrine | −13.38 ± 9.11 | −1.62 ± 1.16ns | −7.62 ± 2.71ns | −10.64 ± 10.02 | −2.43 ± 2.34ns |
| Estrogens | |||||
| Estrone | −1.55 ± 2.72 | 0.95 ± 2.91ns | 8.58 ± 8.93ns | −3.54 ± 4.97 | −2.56 ± 1.53ns |
| β-Estradiol | −0.46 ± 0.46 | 1.13 ± 2.05ns | 10.12 ± 12.92ns | 5.77 ± 6.38 | 4.49 ± 1.18ns |
| 17α-Ethynylestradiol | 11.53 ± 0.61 | 10.37 ± 0.99ns | 16.10 ± 9.79ns | 16.02 ± 2.38 | 0.58 ± 7.13ns |
| 2-Hydroxyestradiol | −0.84 ± 1.72 | 3.23 ± 3.13ns | 1.63 ± 1.87ns | 2.05 ± 5.51 | −3.43 ± 0.20ns |
| Amine | |||||
| 2-Naphthylamine | 83.04 ± 2.48 | 37.31 ± 11.30*** | 9.24 ± 7.06*** | 88.95 ± 15.71 | 256.39 ± 101.81*** |
| Tyrosine/derivatives | |||||
| D-Tyrosine | −3.16 ± 2.98 | 2.11 ± 1.43ns | −5.53 ± 0.60ns | 2.14 ± 1.33 | 2.10 ± 5.96ns |
| 3-Nitro-L-tyrosine | −8.89 ± 12.81 | −9.31 ± 9.76ns | 4.32 ± 7.30ns | −10.44 ± 12.48 | 4.80 ± 4.81ns |
| Thyroid hormones | |||||
| 3,5-Diiodo-L-thyronine | −11.57 ± 5.25 | −2.57 ± 1.20ns | 7.31 ± 3.63ns | −3.67 ± 21.24 | 10.37 ± 0.38ns |
| 3,3′,5-Triiodo-L-thyronine | −1.46 ± 8.63 | −1.58 ± 1.60ns | 11.22 ± 10.87ns | 15.94 ± 11.04 | 2.18 ± 1.73ns |
| 3,3′,5′-Triiodo-L-thyronine | −2.05 ± 2.90 | 3.11 ± 0.30ns | 13.02 ± 11.62ns | 8.73 ± 9.55 | −2.09 ± 0.04ns |
| Others | |||||
| Minoxidil | 5.51 ± 5.41 | 0.96 ± 0.06ns | −0.36 ± 4.39ns | 15.13 ± 7.98 | −0.87 ± 7.22ns |
| 4-Phenyl-1,2,3,6-tetrahydropyridine hydrochloride | 1.48 ± 0.46 | 18.30 ± 4.19ns | 8.97 ± 7.15ns | −1.56 ± 2.79 | −1.83 ± 3.19ns |
Each reaction mixture (150 μl) comprised 100 mM sodium acetate buffer (pH 5.5) or 100 mM sodium phosphate buffer (pH 6.5 or 7.5), 2.5 mM MgCl2, 20 μg of purified His-mL-STL protein, 50 or 100 μM substrate, and 50 μM PAPS containing 0.015 μCi [35S]PAPS. The reaction mixture was incubated at 37°C for 1 h, and the reaction was terminated by the addition of barium acetate. The unreacted [35S]PAPS was precipitated by barium hydroxide and zinc sulfate. Specific activity was calculated from the scintillation counting of the [35S]sulfonated product. Data are mean ± SEM of two batches of purified proteins, each performed in duplicate. Differences between compounds and pH at fixed 50 μM substrate concentration as well as differences between compounds and substrate concentrations at pH 5.5 were analyzed by two-way ANOVA followed by Bonferroni post hoc test. *** P < 0.001; ns, nonsignificant at P > 0.05 level.
DISCUSSION
Here, we described the identification, cloning, and characterization of a novel PPARα-dependent mouse liver-specific and male-dominant cytosolic SULT Sult2a, mL-STL, which preferentially catalyzes 7α-hydroxyl primary bile acid substrates in vitro.
Using the PPARα-null (KO) mouse model (24) and FDD approach (21), we identified a new mouse liver cytosolic SULT, herein designated mL-STL, of which its gene expression was under the control of PPARα during the cellular response to fasting as well as under Wy-14,643 treatment, but in an apparently different mechanism. mL-STL mRNA and protein were constitutively expressed in the WT and KO mice under the fed condition, suggesting that the basal expression level of mL-STL was not under the regulation of PPARα. It has been relatively well known that PPARα is activated by exogenous agonist Wy-14,643 as well as endogenous mobilized fatty acids during fasting, resulting in transcriptional upregulation or downregulation of its target genes (2). Consistent with this observation, mL-STL expression was transcriptionally downregulated by Wy-14,643 as well as during fasting in the WT mice. However, the mL-STL expression was also dramatically downregulated in the KO mice during fasting, suggesting that the downregulation of mL-STL expression under the fed-fasted cycle was not limited to the regulation by PPARα, but possibly by the coordinate transcriptional regulation involving other nutrient-sensing nuclear receptor(s). Indeed, recent studies have also revealed the transcriptional coordination of hepatic autophagy-related genes involved in lipid metabolism by farnesoid X receptor and PPARα during the fed-fasted cycles in mouse liver (40). So far, no true PPARα target genes identified have exhibited such a perplexing mode of PPARα regulation during fasting and Wy-14,643 treatment (2). Also, we are uncertain as to whether mL-STL is a direct PPARα target gene. It is known that direct PPARα target genes contain a peroxisome proliferator response element (PPRE) either in its promoter (2) or intronic region (23), which mediates the upregulation or downregulation of its target gene expression during fasting. Identification of a functional PPRE in the mL-STL gene might provide direct evidence that mL-STL is a new PPARα target gene.
Two isoforms hereby named mL-STL1 and mL-STL2 were identified from the RACE assays in which the 95 bp of the entire exon 5 was missing in the mL-STL2 cDNA compared with the mL-STL1 isoform, suggesting that they are alternative splicing variants. This splicing phenomenon is not uncommon in the SULT family. It has been reported that a human SULT1C2 gene is an alternative splicing variant of SULT1C1 due to the lack of a 95 bp exonic sequence located between exons 3 and 4 of the gene (41). Because only 8% of the 5′-RACE clones belonged to the mL-STL2 isoform, mL-STL1 appeared to be the major isoform. Both isoforms possessed multiple 5′ termini of different lengths, thus implying the presence of putative multiple transcription start sites (TSSs) in the 5′ end. Multiple TSSs are not uncommon in the SULT family because no canonical TATA motif was located near any of the transcription initiation sites identified in SULTs during the RACE experiments (41). In silico analysis of the mL-STL cDNA sequence revealed that exons 2–7 of the mL-STL1 gene encode a complete cytosolic SULT domain, whereas only a truncated one was presented in the mL-STL2 gene, thus suggesting that only mL-STL1 is a functional SULT.
SULTs in mammalian cells constitute a group of enzymes that catalyze the transfer of a sulfuryl group of cofactor PAPS to the amino or hydroxyl group of substrates (42). SULTs have been divided into several gene families based on the similarity of their amino acid sequences and catalytic properties in mammals (43). Members of each family share at least 45% amino acid sequence similarity, whereas members of the subfamily are 60% or more identical in amino acid sequence. Based on this criterion, our mL-STL1 isoform was found to share the highest amino acid similarity to the SULT2A family and possesses all conserved SULT2A signature sequences (36), and thus it is a new Sult2a isoform. During the preparation of this manuscript, the Riken cDNA 2810007J24, which shares 99% similarity to our mL-STL cDNA sequence, was given an official name of Sult2a8 with predicted DHEA-preferring activity (Gene ID 76971), and, therefore, our mL-STL can be officially renamed as the Sult2a8 isoform. The SULT2A family is composed of several subfamilies, and the isoforms of each subfamily have been characterized in a variety of species. Four SULT2A isoforms (SULT2A1, 2A2, 2A3, and 2A4) have been characterized in rats (43), as well as two isoforms (Sult2a1 and Sult2a2) in mice (44). Seven other Sult2a genes clustering on chromosome 7 have also been examined in mice, but their substrate specificities have not been addressed (45). In humans, only one SULT2A1 isoform has been identified and characterized (46).
Dramatic differences in tissue distribution exist in SULT families. SULT1 family members exhibit the widest tissue distribution of the SULT subfamily. Human SULT1A1 is expressed in the adrenal gland, brain, breast, endometrium, intestine, jejunum, kidney, lung, placenta, and platelets (47). Similarly, rat SULT1A1 is also ubiquitously expressed in a wide range of tissues, with the highest level of expression in the liver (48). In the SULT2 family, the majority of SULT2 enzymes was found in the liver, whereas the extrahepatic expression was extremely limited. In mice, Sult2a1/2 mRNA was found abundantly in female, but not in male, livers, and low levels of Sult2a1/2 mRNA were detected in the brain, gonads, placenta, and uterus (44). In rats, SULT2A1 mRNA was mainly detectable in liver and at significantly lower levels in adrenal (48). In human, SULT2A1 is predominantly expressed in the liver, whereas a very low level of expression was found in the adrenal and small intestine (46, 47). Consistent with the narrow range of tissue distribution of mammalian SULT2A enzymes, our mouse mL-STL mRNA transcript and proteins were only detected in the livers of both males and female animals, suggesting that it is a unique liver-specific Sult2a8 isoform.
Sexual dimorphic expression of SULT is not uncommon in rodents. Mouse Sult1a1, Sult1d1, and Sult1e1 liver mRNA is higher in females than in males (49), whereas Sult1c1 is male-dominant (44). Rat SULT1A1 mRNA expression is approximately twofold higher in males as in females, whereas SULT1C1 is approximately 10-fold higher in males than in females (50). Sexual dimorphism of Sult2a1 is conserved in rodent species, but does not exist in humans. Hepatic mouse Sult2a1/2 expression was 100-fold higher in females than in males (51). Rat liver Sult2a1/2 enzyme activities toward hydroxysteroid, including DHEA and bile acid substrates, are also female-predominant (52). No gender difference in catalyzing DHEA was detected in humans (53). Thus, gender-divergent SULTs in rodents are mostly female-dominant. In the present study, the hepatic mL-STL mRNA and protein expressions in females were lower than in males across the lifespan, suggesting that it is a male-dominant Sult2a. As far as we know, this is the first report of a male-predominant Sult2a in mice. It is known that sexual dimorphism of SULT expression is regulated by sex hormone and/or growth hormone and is isoform specific (50). The female dominance of Sult1a1, Sult1d1, and Sult2a1/2 mRNA expression is attributed to suppressive effects of androgens and male growth hormone secretion pattern in mice (51). In contrast, the male predominance of rat Sult1c1 mRNA expression and enzyme activity is due to stimulatory effects of androgens and male growth hormone secretion pattern in rats (54). In mice, the male-predominant Sult1c1 mRNA expression is only due to the stimulatory effects of androgen, but not the growth hormone (51). Future studies are required to dissect the mechanistic basis of gender differences of mL-STL expression in mouse liver.
Considerable variations in ontogenic expression of SULT were reported. Hepatic mRNA expression of mouse Sult1a1 and 1d1 increases gradually from birth until 22 days and then declines thereafter (44). Mouse Sult1c1 mRNA expression is the highest before birth and decreases thereafter (44). Sult3a1 mRNA expression is low in fetal mouse liver and remains low until 1 month old, when the level increases in females, but not in males (44). Sult2a1/2 mRNA level expresses after birth and reaches a peak at 15–22 days of age and then is barely detectable in male adult mice (44). No age or gender differences in hepatic SULT activity toward DHEA exist in humans (53). Our results here showed that mL-STL was detected only after birth, increased gradually and reached a peak at 30 days, and then declined gradually in females, whereas the level in males was maintained, suggesting that the level of mL-STL is regulated by sex-dependent growth hormone and/or male sex hormone. Ontogenic gender-divergent expressions are also reported for P450s, UDP-glucuronosyltransferases, glutathione S-transferase, and xenobiotic transporters, as well as transcription factors (49). Further studies are required to elucidate the ontogenic sexual dimorphic mechanism of hepatic mL-STL expression between males and females.
The high amino acid sequence similarity of the mL-STL gene to the mammalian SULT2A family provides some hints as to the possible substrate candidates of the mL-STL enzyme identified in the present study. Various types of hydroxysteroids, including DHEA (46, 55), pregnenolone, and bile acids (26) have been reported as substrates for the mammalian SULT2A subfamily. Surprisingly, our mouse recombinant His-mL-STL enzyme exhibited no sulfonating activity toward DHEA, the predicted/hypothetical DHEA-preferring function of Sult2a8. Instead, His-mL-STL exhibited the highest sulfonation activity toward three kinds of primary bile acids/salt/conjugated forms, including CA, CDCA, and α-MCA. All of these bile acid substrates possess several OH targets for sulfonation, but the preferential target for His-mL-STL remains to be clarified.
To gain insight into the putative OH target sulfonated by our His-mL-STL protein in vitro, the positions of OH groups in the potent (CA, CDCA, and α-MCA) and nonpotent catalytic substrates were compared. CA (tri-OH at 3α, 7α, and 12α), CDCA (di-OH at 3α and 7α), and α-MCA (tri-OH at 3α, 6β, and 7α) all possess 3α-OH and 7α-OH, and thus they are potential sulfonation targets of His-mL-STL. However, when the 7α-OH was replaced by a 7β-OH as in β-MCA (tri-OH at 3α, 6β, and 7β) and ω-MCA (tri-OH at 3α, 6α, and 7β), only very weak sulfonating activity was retained, despite the presence of 3α-OH in these compounds, suggesting that only 7α-OH is the key catalytic target of the His-mL-STL protein. Our hypothesis was further substantiated by the lack of His-mL-STL-mediated activity in hydroxysteroid substrates lacking a 7α-OH as in UDCA, DCA, LCA, androsterone, DHEA, pregnenolone, cholesterol, 22(S)-hydroxycholesterol, 22(R)-hydroxycholesterol, and corticosterone. However, direct evidence demonstrating the mL-STL-catalyzed sulfonation of 7α-OH in CA, CDCA, and α-MCA in vivo is still lacking.
The OH position at which bile acids are sulfonated was also studied in humans and rodents. In humans, the primary bile acids are CA and CDCA. Monosulfonation of these bile acids at the 3α-OH position is predominant because only 3-sulfates, and no polysulfates, were detected in human urine (56, 57). In rat hepatocytes, DCA and CDCA were only sulfonated at 3α-OH position to form its corresponding 3-sulfates (58, 59). In mice, the major bile acids are CA, α-MCA, β-MCA, and small amount of CDCA (59). Significant amounts (∼80%) of fecal CA and CDCA were sulfonated at 7α-OH because only 7-sulfates, and no 3-sulfates or 12-sulfates, were detected in mouse feces (60). Furthermore, the sulfonation of CA and CDCA at 7α-OH is male-predominant in the mouse liver (61). Strikingly, both α-MCA and β-MCA were nearly exclusively in the nonsulfate form in mouse feces (60). Thus, it can be concluded that bile acid-sulfates are exclusively present in the monosulfate form in humans and rodents and that sulfonation at 3α-OH is predominant in humans and rats, but at 7α-OH in mice.
Human SULT2A1 was found to play a significant and unique role in the 3α-OH sulfonation, particularly to LCA and tauroLCA (55), but the major Sult2a isoform(s) responsible for catalyzing the sulfonation of 7α-OH bile acid substrates in mice is still not clear. Because bile acid sulfonation is carried out in the liver and our mL-STL was a liver-specific and male-dominant enzyme that preferentially sulfonated 7α-OH of CA, CDCA, and α-MCA in vitro, it is tempting to hypothesize that the male-predominant CA-7-sulfates and CDCA-7-sulfates detected in mouse fecal bile acid in the earlier studies (60, 61) were presumably due to the 7α-OH sulfonation carried out by our newly identified mL-STL enzyme. However, our assumption could only explain why β-MCA was not sulfonated because it possesses a 7β-OH instead of a 7α-OH. The question as to why the α-MCA that has a 7α-OH was not sulfonated by the putative mL-STL enzyme in vivo cannot be answered at present. The lack of mL-STL homologs in humans, as revealed by BLAST search, further supported the fact that 7α-OH bile acid sulfonation is absent in humans and that the mL-STL (Sult2a8) may be the major hepatic bile acid sulfonating enzyme in mice. Currently, an X-ray crystallographic approach has been undertaken to confirm the substrate binding specificity and catalytic mechanism of mL-STL enzyme toward the 7α-OH bile acid substrates.
Although we have shown that mL-STL can sulfonate several primary bile acids in vitro, the physiological significance of PPARα in regulating the mL-STL-mediated sulfonation of bile acids during fasting in an animal model remains to be answered. It is known that during fasting, increased bile acid secretion and reabsorption was detected in mouse enterohepatic circulation and hepatocyte (62). The increased bile acids in hepatocytes act as signaling molecules to activate various nuclear bile acid receptors, such as the farnesoid X receptor, to maintain the bile acid homeostasis and thus energy homeostasis during energy metabolism (18). Given the fact that PPARα is a nutritional sensor (1), and the mL-STL is a PPARα-dependent bile acid-specific sulfonating enzyme during fasting, we hypothesize that the PPARα-mediated mL-STL sulfonation pathway can function as a switch on regulating the bile acid signal concentrations and thus in turn maintaining the energy homeostasis during the adaptive response to fasting. The predominant high hepatic expression of the mL-STL proteins is consistent with the fact that liver is the major site for bile acid metabolism and the mL-STL protein can sulfonate the bile acids in liver, which in turn regulates the bile acid homeostasis and energy metabolism during fasting. Further studies are required to confirm our hypothesis in the mL-STL-null mouse model to be created in our laboratory.
Supplementary Material
Acknowledgments
The authors thank Prof. K. B. Wong of The Chinese University of Hong Kong for providing the modified mpRSET vector for the current study.
Footnotes
Abbreviations:
- AP8
- anchored primer 8
- ARP1
- arbitrary primer 1
- CA
- cholic acid
- CDCA
- chenodeoxycholic acid
- DCA
- deoxycholic acid
- DHEA
- dehydroepiandrosterone
- DIG
- digoxigenin
- FDD
- fluorescent differential display
- IPTG
- isopropyl-β-d-1-thiogalactopyranoside
- KO
- PPARα-null mice
- LCA
- lithocholic acid
- α-MCA
- α-muricholic acid
- β-MCA
- β-muricholic acid
- ω-MCA
- ω-muricholic acid
- mL-STL
- mouse liver-sulfotransferase-like
- mpRSETA
- mini-pRSETA
- ORF
- open reading frame
- PACS
- polyadenylation cleavage site
- PAPS
- 3′-phosphoadenosine 5′-phosphosulfate
- PAS
- polyadenylation signal
- RACE
- rapid amplification of cDNA ends
- SO3−
- sulfonate
- SULT
- sulfotransferase
- TSS
- transcription start site
- UDCA
- ursodeoxycholic acid
- UTR
- untranslated region
- Wy-14
- 643, 4-chloro-6-(2,3-xylidino)-2-pyrimidinylthioacetic acid
The work described in this paper was partially supported by Research Grants Council, University Grants Committee (Hong Kong Special Administrative Region, China) Project No. CUHK 14109414 and Direct Grant Project Code 3132806.
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Kersten S. 2014. Integrated physiology and systems biology of PPARα. Mol. Metab. 3: 354–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mandard S., Müller M., and Kersten S.. 2004. Peroxisome proliferator-activated receptor α target genes. Cell. Mol. Life Sci. 61: 393–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rakhshandehroo M., Knoch B., Müller M., and Kersten S.. 2010. Peroxisome proliferator-activated receptor alpha target genes. PPAR Res. 2010: 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hunt M. C., Yang Y. Z., Eggertsen G., Carneheim C. M., Gåfvels M., Einarsson C., and Alexson S. E. H.. 2000. The peroxisome proliferator-activated receptor α (PPARα) regulates bile acid biosynthesis. J. Biol. Chem. 275: 28947–28953. [DOI] [PubMed] [Google Scholar]
- 5.Li F., Patterson A. D., Krausz K. W., Tanaka N., and Gonzalez F. J.. 2012. Metabolomics reveals an essential role for peroxisome proliferator-activated receptor α in bile acid homeostasis. J. Lipid Res. 53: 1625–1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Russell D. W. 2009. Fifty years of advances in bile acid synthesis and metabolism. J. Lipid Res. 50(Suppl.): S120–S125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hofmann A. F., and Hagey L. R.. 2008. Bile acids: chemistry, pathochemistry, biology, pathobiology, and therapeutics. Cell. Mol. Life Sci. 65: 2461–2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hylemon P. B., Zhou H., Pandak W. M., Ren S., Gil G., and Dent P.. 2009. Bile acids as regulatory molecules. J. Lipid Res. 50: 1509–1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Aguiar Vallim T. Q., Tarling E. J., and Edwards P. A.. 2013. Pleiotropic roles of bile acids in metabolism. Cell Metab. 17: 657–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li T., and Chiang J. Y.. 2013. Nuclear receptors in bile acid metabolism. Drug Metab. Rev. 45: 145–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Houten S. M., Watanabe M., and Auwerx J.. 2006. Endocrine functions of bile acids. EMBO J. 25: 1419–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou H., and Hylemon P. B.. 2014. Bile acids are nutrient signaling hormones. Steroids. 86: 62–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qi Y., Jiang C., Cheng J., Krausz K. W., Li T., Ferrell J. M., Gonzalez F. J., and Chiang J. Y.. 2015. Bile acid signaling in lipid metabolism: metabolomic and lipidomic analysis of lipid and bile acid markers linked to anti-obesity and anti-diabetes in mice. Biochim. Biophys. Acta. 1851: 19–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gupta S., Stravitz R. T., Dent P., and Hylemon P. B.. 2001. Down-regulation of cholesterol 7α-hydroxylase (CYP7A1) gene expression by bile acids in primary rat hepatocytes is mediated by the c-Jun N-terminal kinase pathway. J. Biol. Chem. 276: 15816–15822. [DOI] [PubMed] [Google Scholar]
- 15.Qiao L., Han S. I., Fang Y. W., Park J. S., Gupta S., Gilfor D., Amorino G., Valerie K., Sealy L., Engelhardt J. F., et al. 2003. Bile acid regulation of C/EBPβ, CREB, and c-Jun function, via the extracellular signal-regulated kinase and c-Jun NH2-terminal kinase pathways, modulates the apoptotic response of hepatocytes. Mol. Cell. Biol. 23: 3052–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maruyama T., Miyamoto Y., Nakamura T., Tamai Y., Okada H., Sugiyama E., Nakamura T., Itadani H., and Tanaka K.. 2002. Identification of membrane-type receptor for bile acids (M-BAR). Biochem. Biophys. Res. Commun. 298: 714–719. [DOI] [PubMed] [Google Scholar]
- 17.Li T., and Chiang J. Y.. 2014. Bile acid signaling in metabolic disease and drug therapy. Pharmacol. Rev. 66: 948–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lefebvre P., Cariou B., Lien F., Kuipers F., and Staels B.. 2009. Role of bile acids and bile acid receptors in metabolic regulation. Physiol. Rev. 89: 147–191. [DOI] [PubMed] [Google Scholar]
- 19.Chiang J. Y. L. 2013. Bile acid metabolism and signaling. Compr. Physiol. 3: 1191–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Strott C. A. 2002. Sulfonation and molecular action. Endocr. Rev. 23: 703–732. [DOI] [PubMed] [Google Scholar]
- 21.Lee S. S. T., Tian L., Lee W. S., and Cheung W. T.. 2002. Application of fluorescent differential display and peroxisome proliferator-activated receptor (PPAR)α-null mice to analyze PPAR target genes. Methods Enzymol. 357: 214–240. [DOI] [PubMed] [Google Scholar]
- 22.Lee S. S. T., Chan W. Y., Lo C. K. C., Wan D. C. C., Tsang D. S. C., and Cheung W. T.. 2004. Requirement of PPARα in maintaining phospholipid and triacylglycerol homeostasis during energy deprivation. J. Lipid Res. 45: 2025–2037. [DOI] [PubMed] [Google Scholar]
- 23.Sun Y., Ng L., Lam W., Lo C. K. C., Chan P. T., Yuen Y. L., Wong P. F., Tsang D. S. C., Cheung W. T., and Lee S. S. T.. 2008. Identification and characterization of a novel mouse peroxisome proliferator-activated receptor α-regulated and starvation-induced gene, Ppsig. Int. J. Biochem. Cell Biol. 40: 1775–1791. [DOI] [PubMed] [Google Scholar]
- 24.Lee S. S. T., Pineau T., Drago J., Lee E. J., Owens J. W., Kroetz D. L., Fernandez-Salguero P. M., Westphal H., and Gonzalez F. J.. 1995. Targeted disruption of the α isoform of the peroxisome proliferator-activated receptor gene in mice results in abolishment of the pleiotropic effects of peroxisome proliferators. Mol. Cell. Biol. 15: 3012–3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laemmli U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 227: 680–685. [DOI] [PubMed] [Google Scholar]
- 26.Barnes S., Buchina E. S., King R. J., McBurnett T., and Taylor K. B.. 1989. Bile acid sulfotransferase I from rat liver sulfates bile acids and 3-hydroxy steroids: purification, N-terminal amino acid sequence, and kinetic properties. J. Lipid Res. 30: 529–540. [PubMed] [Google Scholar]
- 27.Geese W. J., and Raftogianis R. B.. 2001. Biochemical characterization and tissue distribution of human SULT2B1. Biochem. Biophys. Res. Commun. 288: 280–289. [DOI] [PubMed] [Google Scholar]
- 28.Huang J., Bathena S. P., Tong J., Roth M., Hagenbuch B., and Alnouti Y.. 2010. Kinetic analysis of bile acid sulfation by stably expressed human sulfotransferase 2A1 (SULT2A1). Xenobiotica. 40: 184–194. [DOI] [PubMed] [Google Scholar]
- 29.Foldes A., and Meek J. L.. 1973. Rat brain phenolsulfotransferase: partial purification and some properties. Biochim. Biophys. Acta. 327: 365–374. [DOI] [PubMed] [Google Scholar]
- 30.Stanley E. L., Hume R., and Coughtrie M. W. H.. 2005. Expression profiling of human fetal cytosolic sulfotransferases involved in steroid and thyroid hormone metabolism and in detoxification. Mol. Cell. Endocrinol. 240: 32–42. [DOI] [PubMed] [Google Scholar]
- 31.Allali-Hassani A., Pan P. W., Dombrovski L., Najmanovich R., Tempel W., Dong A., Loppnau P., Martin F., Thornton J., Edwards A. M., et al. 2007. Structural and chemical profiling of the human cytosolic sulfotransferases. PLoS Biol. 5: e97 [Erratum. 2007. PLoS Biol. 5: e165.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tian B., Hu J., Zhang H., and Lutz C. S.. 2005. A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res. 33: 201–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pauws E., van Kampen A. H. C., van de Graaf S. A. R., de Vijlder J. J. M., and Ris-Stalpers C.. 2001. Heterogeneity in polyadenylation cleavage sites in mammalian mRNA sequences: implications for SAGE analysis. Nucleic Acids Res. 29: 1690–1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Edwalds-Gilbert G., Veraldi K. L., and Milcarek C.. 1997. Alternative poly(A) site selection in complex transcription units: means to an end? Nucleic Acids Res. 25: 2547–2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hsieh S. J., Lin C. Y., Liu N. H., Chow W. Y., and Tang C. Y.. 2006. GeneAlign: a coding exon prediction tool based on phylogenetical comparisons. Nucleic Acids Res. 34: W280–W284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weinshilboum R. M., Otterness D. M., Aksoy I. A., Wood T. C., Her C. T., and Raftogianis R. B.. 1997. Sulfation and sulfotransferases 1: sulfotransferase molecular biology: cDNAs and genes. FASEB J. 11: 3–14. [PubMed] [Google Scholar]
- 37.Her C., Wood T. C., Eichler E. E., Mohrenweiser H. W., Ramagli L. S., Siciliano M. J., and Weinshilboum R. M.. 1998. Human hydroxysteroid sulfotransferase SULT2B1: two enzymes encoded by a single chromosome 19 gene. Genomics. 53: 284–295. [DOI] [PubMed] [Google Scholar]
- 38.Gulcan H. O., and Duffel M. W.. 2011. Substrate inhibition in human hydroxysteroid sulfotransferase SULT2A1: studies on the formation of catalytically non-productive enzyme complexes. Arch. Biochem. Biophys. 507: 232–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang H., Varlamova O., Vargas F. M., Falany C. N., and Leyh T. S.. 1998. Sulfuryl transfer: the catalytic mechanism of human estrogen sulfotransferase. J. Biol. Chem. 273: 10888–10892. [Erratum, 1998. J. Biol. Chem. 273: 17296.] [DOI] [PubMed] [Google Scholar]
- 40.Lee J. M. 2016. Transcriptional coordination of hepatic autophagy by nutrient-sensing nuclear receptor PPARalpha and FXR. Ann. Pediatr. Endocrinol. Metab. 21: 193–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Freimuth R. R., Raftogianis R. B., Wood T. C., Moon E., Kim U. J., Xu J. P., Siciliano M. J., and Weinshilboum R. M.. 2000. Human sulfotransferases SULT1C1 and SULT1C2: cDNA characterization, gene cloning, and chromosomal localization. Genomics. 65: 157–165. [DOI] [PubMed] [Google Scholar]
- 42.Weinshilboum R., and Otterness D.. 1994. Sulfotransferase enzymes. In Conjugation-Deconjugation Reactions in Drug Metabolism and Toxicity. Kauffman F. C. and Bock K. W., editors. Springer-Verlag, Berlin: 45–78. [Google Scholar]
- 43.Blanchard R. L., Freimuth R. R., Buck J., Weinshilboum R. M., and Coughtrie M. W.. 2004. A proposed nomenclature system for the cytosolic sulfotransferase (SULT) superfamily. Pharmacogenetics. 14: 199–211. [DOI] [PubMed] [Google Scholar]
- 44.Alnouti Y., and Klaassen C. D.. 2006. Tissue distribution and ontogeny of sulfotransferase enzymes in mice. Toxicol. Sci. 93: 242–255. [DOI] [PubMed] [Google Scholar]
- 45.Kocarek T. A., Duanmu Z., Fang H. L., and Runge-Morris M.. 2008. Age- and sex-dependent expression of multiple murine hepatic hydroxysteroid sulfotransferase (SULT2A) genes. Biochem. Pharmacol. 76: 1036–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Otterness D. M., Wieben E. D., Wood T. C., Watson W. G., Madden B. J., McCormick D. J., and Weinshilboum R. M.. 1992. Human liver dehydroepiandrosterone sulfotransferase: molecular cloning and expression of cDNA. Mol. Pharmacol. 41: 865–872. [PubMed] [Google Scholar]
- 47.Gamage N., Barnett A., Hempel N., Duggleby R. G., Windmill K. F., Martin J. L., and McManus M. E.. 2006. Human sulfotransferases and their role in chemical metabolism. Toxicol. Sci. 90: 5–22. [DOI] [PubMed] [Google Scholar]
- 48.Dunn R. T. II, and Klaassen C. D.. 1998. Tissue-specific expression of rat sulfotransferase messenger RNAs. Drug Metab. Dispos. 26: 598–604. [PubMed] [Google Scholar]
- 49.Fu Z. D., Csanaky I. L., and Klaassen C. D.. 2012. Effects of aging on mRNA profiles for drug-metabolizing enzymes and transporters in livers of male and female mice. Drug Metab. Dispos. 40: 1216–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Klaassen C. D., Liu L., and Dunn R. T. II. 1998. Regulation of sulfotransferase mRNA expression in male and female rats of various ages. Chem. Biol. Interact. 109: 299–313. [DOI] [PubMed] [Google Scholar]
- 51.Alnouti Y., and Klaassen C. D.. 2011. Mechanisms of gender-specific regulation of mouse sulfotransferases (Sults). Xenobiotica. 41: 187–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Singer S. S., Giera D., Johnson J., and Sylvester S.. 1976. Enzymatic sulfation of steroids: I. The enzymatic basis for the sex difference in cortisol sulfation by rat liver preparations. Endocrinology. 98: 963–974. [DOI] [PubMed] [Google Scholar]
- 53.Aksoy I. A., Sochorova V., and Weinshilboum R. M.. 1993. Human liver dehydroepiandrosterone sulfotransferase: nature and extent of individual variation. Clin. Pharmacol. Ther. 54: 498–506. [DOI] [PubMed] [Google Scholar]
- 54.Liu L., and Klaassen C. D.. 1996. Ontogeny and hormonal basis of male-dominant rat hepatic sulfotransferases. Mol. Pharmacol. 50: 565–572. [PubMed] [Google Scholar]
- 55.Radominska A., Comer K. A., Zimniak P., Falany J., Iscan M., and Falany C. N.. 1990. Human liver steroid sulphotransferase sulphates bile acids. Biochem. J. 272: 597–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Almé B., Bremmelgaard A., Sjövall J., and Thomassen P.. 1977. Analysis of metabolic profiles of bile acids in urine using a lipophilic anion exchanger and computerized gas-liquid chromatorgaphy-mass spectrometry. J. Lipid Res. 18: 339–362. [PubMed] [Google Scholar]
- 57.Hedenborg G., and Norman A.. 1984. The nature of urinary bile acid conjugates in patients with extrahepatic cholestasis. Scand. J. Clin. Lab. Invest. 44: 725–733. [DOI] [PubMed] [Google Scholar]
- 58.Lambiotte M., and Thierry N.. 1980. Hydroxylation, sulfation, and conjugation of bile acids in rat hepatoma and hepatocyte cultures under the influence of glucocorticoids. J. Biol. Chem. 255: 11324–11331. [PubMed] [Google Scholar]
- 59.Alnouti Y. 2009. Bile acid sulfation: a pathway of bile acid elimination and detoxification. Toxicol. Sci. 108: 225–246. [DOI] [PubMed] [Google Scholar]
- 60.Eyssen H. J., Parmentier G. G., and Mertens J. A.. 1976. Sulfated bile acids in germ-free and conventional mice. Eur. J. Biochem. 66: 507–514. [DOI] [PubMed] [Google Scholar]
- 61.Zhang Y., and Klaassen C. D.. 2010. Effects of feeding bile acids and a bile acid sequestrant on hepatic bile acid composition in mice. J. Lipid Res. 51: 3230–3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sokolović M., Sokolović A., van Roomen C. P., Gruber A., Ottenhoff R., Scheij S., Hakvoort T. B., Lamers W. H., and Groen A. K.. 2010. Unexpected effects of fasting on murine lipid homeostasis-transcriptomic and lipid profiling. J. Hepatol. 52: 737–744. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.