Abstract
Lipid accumulation is a pathological feature of every type of kidney injury. Despite this striking histological feature, physiological accumulation of lipids in the kidney is poorly understood. We studied whether the accumulation of lipids in the fasted kidney are derived from lipoproteins or NEFAs. With overnight fasting, kidneys accumulated triglyceride, but had reduced levels of ceramide and glycosphingolipid species. Fasting led to a nearly 5-fold increase in kidney uptake of plasma [14C]oleic acid. Increasing circulating NEFAs using a β adrenergic receptor agonist caused a 15-fold greater accumulation of lipid in the kidney, while mice with reduced NEFAs due to adipose tissue deficiency of adipose triglyceride lipase had reduced triglycerides. Cluster of differentiation (Cd)36 mRNA increased 2-fold, and angiopoietin-like 4 (Angptl4), an LPL inhibitor, increased 10-fold. Fasting-induced kidney lipid accumulation was not affected by inhibition of LPL with poloxamer 407 or by use of mice with induced genetic LPL deletion. Despite the increase in CD36 expression with fasting, genetic loss of CD36 did not alter fatty acid uptake or triglyceride accumulation. Our data demonstrate that fasting-induced triglyceride accumulation in the kidney correlates with the plasma concentrations of NEFAs, but is not due to uptake of lipoprotein lipids and does not involve the fatty acid transporter, CD36.
Keywords: kidney metabolism, lipoprotein lipase, cluster of differentiation 36
The kidney is one of the most energy consuming tissues in the body, with approximately two-thirds of its oxidative substrate provided by fatty acids (1). Excess fatty acid in the cell is esterified to form triglyceride and stored in lipid droplets. While the entire kidney can utilize fatty acids, the cortex and the epithelial tubule cells depend on fatty acid oxidation (FAO) for ATP (2).
The cortex is also the site of large accumulations of neutral lipids during kidney disease and injury (2). Pathways that contribute to lipid accumulation in the kidney are not well-established. Some studies suggest that during obesity, diabetes, and aging, an increase in the SREBPs leads to greater de novo fatty acid synthesis, which drives lipid accumulation and decreases renal function (3–6). Other studies have shown that renal failure causes decreased FAO (7, 8), which could independently increase lipid accumulation.
Multiple pathways are dysregulated in chronic kidney disease (CKD), making this issue difficult to study. On the other hand, neutral lipid accumulation also occurs during fasting (9). In many tissues, such as the heart, muscle, and adipose tissue, LPL and cluster of differentiation (CD)36 are necessary for proper transport of fatty acids into the tissues (10, 11). LPL is synthesized by parenchymal cells and translocates to the endothelial surface, where it hydrolyzes VLDL and chylomicron triglycerides to produce NEFAs. NEFAs can then either diffuse through the membrane in a process called “flip-flop” (12) or transfer across the membrane by a saturable receptor-mediated process (13). CD36 has been postulated to mediate active transport of NEFAs into cells (14). Like the heart, skeletal muscle, and adipose tissue, the kidney robustly expresses both LPL and CD36 (15, 16). However, in an unbiased assessment of renal genes in mice with diabetic kidney disease, Cd36 was reduced, as were other genes associated with renal FAO (17). This suggests that FAO and lipid uptake into the kidney are coordinately regulated.
In the current study, we first determined NEFA uptake into the kidney during fasting and then modulated plasma NEFA levels and assessed whether this altered kidney triglyceride accumulation. We then tested to determine whether triglyceride-rich lipoproteins or NEFAs were the source of kidney triglyceride stores. In addition, we tested to determine how kidney CD36 and LPL were affected by fasting and whether these known moderators of triglyceride metabolism affect fasting-induced lipid accumulation in the kidney.
MATERIALS AND METHODS
Animal studies
We used 10- to 16-week-old male and female C57BL/6 mice, Cd36−/− mice (18), floxed Lpl mice (Lplfl/fl), inducible (i)Lpl−/− mice, floxed Mttp (Mttpfl/fl) and liver-specific Mttp knockout mice (L-Mttp−/−) mice, and adipocyte-specific adipose triglyceride lipase (Atgl) knockout mice (AAKO) (19, 20). Mice were raised on a normal chow diet. Littermates were used as controls for all studies. All procedures were approved by the New York University Langone Medical Center, Washington University, State University of New York Downstate, and University of Pittsburgh Institutional Animal Care and Use Committees. Mice of each genotype were divided into two groups; one group was fasted 16 h overnight and the other was allowed to feed ad libitum for the same time period. All mice were then euthanized with a lethal injection of 100 mg/kg ketamine and 10 mg/kg xylazine. Animals were dissected open and then perfused by cardiac puncture in the heart with 5 ml of PBS until the liver and kidneys blanched. Tissues were dissected out, snap-frozen in liquid nitrogen, and stored at −80°C for further use. Kidneys were also bisected and embedded into Tissue-Tek OCT compound (Sakura) for oil-red O histology.
Poloxamer 407 (P407) was prepared in PBS as previously described (21). Mice were injected intraperitoneally with 1 mg/g bodyweight of P407 and then fasted for 16 h. Control mice were injected with an equivalent volume of PBS.
iLpl−/− animals were generated as described previously (22). Briefly, β-actin-driven tamoxifen-inducible-Cre (Mer/Cre/Mer) transgenic mice were crossed with LPL-flanked loxP sites mice to obtain the β-actin-MerCreMer/Lplfl/fl offspring, designated iLpl−/−. The iLpl−/− mice were given an intraperitoneal injection of 1 mg of 4-hydroxytamoxifen (Sigma) in peanut oil for five consecutive days.
β3-adrenergic receptor agonist, CL 316,243 (Sigma), was dissolved in PBS and injected into C57BL/6 mice at 1 mg/kg at two time points (2:00 PM and 6:00 PM) (23). The mice were allowed to feed ad libitum and euthanized the following morning.
Microsomal triglyceride transfer protein (MTTP) was inhibited using BMS-212122 [MTTP inhibitor (MTTPi)] as previously described (24). Briefly, the MTTPi was diluted in DMSO and given to mice orally at a dose of 1 mg/kg bodyweight for seven consecutive days. Control mice were given an equivalent volume of DMSO. After the final dose, mice were divided into two groups and either fasted overnight for 16 h or allowed access to food ad libitum and euthanized.
Measurement of plasma lipids and glucose
One hundred microliters of blood were drawn from each animal and then centrifuged at 10,600 g on a table top centrifuge for 10 min to obtain plasma. Plasma was used to measure triglycerides and NEFA using the Thermo Scientific Infinity assay (Thermo Scientific) and the Wako NEFA kit, respectively. Glucose was measured from whole blood using a One Touch Ultra 2™ glucometer. Triglyceride-rich lipoproteins (density < 1.006 g/ml) were separated by sequential density ultracentrifugation of plasma in a TLA100 rotor as described in Kako et al. (25).
Lipid extraction and measurement
The lipid extraction protocol was adapted from the Folch method (26) and modified slightly from Trent et al. (10). Briefly, approximately 100 mg of tissue were homogenized in 500 μl of ice-cold PBS using stainless steel beads for 30 s in a bead beater homogenizer. From each sample, 50 μl were removed for protein analysis, and 1.5 ml of 2:1 chloroform:methanol were added to the rest of the homogenate in a glass test tube. Samples were then centrifuged for 10 min at 2,050 g at 4°C. The lower organic phase was separated with a glass Pasteur pipette and blown dry with nitrogen gas. The dried lipid was then dissolved with 500 μl of 2% Triton X-100 in chloroform, further dried, and then dissolved in double distilled water. The sample of tissue lysate put aside was used to assay protein content using Bradford reagent (Bio-Rad) following the manufacturer’s instructions. Using the tissue lipid extract, assays for triglyceride were performed using the previously described assay for plasma lipid. Lipid measurements were normalized to protein content of each sample or milligram of tissue weight.
In vivo NEFA uptake
NEFA uptake was assessed in C57BL/6 mice either fasted 16 h overnight or allowed access to food ad libitum for the same time and in Cd36−/− and Cd36fl/fl mice fasted 16 h overnight. The [1-14C]oleic acid (PerkinElmer Life Sciences) was complexed to 0.6% fatty acid-free BSA (Sigma). Mice were injected intravenously with 1.5 × 106 cpm of [1-14C]oleic acid-BSA and blood was collected at 0.5, 2, and 5 min after injection, after which the mice were euthanized. Plasma was collected as previously described. The body cavity was perfused with 5 ml of PBS by cardiac puncture and tissues were extracted. Tissues were homogenized in 1 ml of PBS and radioactive counts were measured using a LS 6500 multipurpose scintillation counter (Beckman Coulter). For C57BL/6 mice, radioactivity per gram tissue was normalized to average plasma NEFA levels in either the fed or fasted group. For Cd36−/− and Cd36fl/fl mice, radioactivity per gram tissue was normalized to 2 min plasma cpm counts.
Renal gene expression
Total RNA was purified from an approximately 50 mg piece of kidney cortex using TRIzol reagent (Invitrogen) according to the instructions of the manufacturer. cDNA was synthesized using Verso cDNA kit (Thermo Scientific) and quantitative real-time PCR was performed with Power SYBR Green PCR Master Mix (Life Technologies) using a Quant Studio 7 Flex analyzer (Life Technologies). Genes of interest were normalized against 18s rRNA. Primer sequences are listed in supplemental Table S2.
RNA sequencing
Raw sequencing data were received in FASTQ format and mapped against the hg19 human reference genome using Tophat 2.0.9. The resulting BAM alignment files were processed using the HTSeq 0.6.1 python framework and respective hg19 General Transfer Format gene annotation, obtained from the UCSC database. Differentially expressed genes (DEGs) were identified using Bioconductor package DESeq2 (3.2), which analyzes RNA sequencing data based on a negative binomial distribution model. To control for false discovery rate, resulting values were adjusted using the Benjamini and Hochberg method. Ingenuity Pathway Analysis was then performed on the DEG in order to determine the top canonical pathways being altered. Genes with an adjusted p-value <0.05 were determined to be differentially expressed. A heat map was created in Microsoft Excel, normalizing all samples to the fed group and expressing the values as fold change.
Lipidomics
Long-chain fatty acyl-CoAs, acyl carnitines, and ceramides were analyzed by targeted metabolomics as previously described (27–29). Briefly, approximately 20 mg of renal cortex were homogenized and extracted with cold 8:1:1 methanol:chloroform:water containing known amounts of C17:0 acyl-CoA (Sigma-Aldrich,), isotope-labeled carnitines (Cambridge Isotope Laboratories), and C17:0 and C25:0 ceramide (Avanti Polar Lipids) internal standards. An equal volume from each tissue sample was combined to generate a pool sample to monitor analytical variability. Ceramides were extracted following the method of Bligh and Dyer (30) and the organic layer was dried under vacuum and resuspended in 60:40 acetonitrile:isopropanol. As an additional control, a mixture of seven standard ceramide compounds was simultaneously extracted and analyzed. Acyl-CoAs, acyl carnitines, and ceramides were quantified by LC/ESI-MS/MS in the multiple reaction monitoring mode using an Agilent 6410 triple quadrupole MS system equipped with an Agilent 1200 LC system. Concentrations were calculated by ratios of peak areas of samples to known concentrations of internal standards. Data were normalized to tissue weight. All solvents were LC-MS grade (Sigma-Aldrich).
Histology
Frozen sections were cut to 10 μM, air dried, fixed with 4% paraformaldehyde, and washed with double distilled water. Sections were rinsed with 60% isopropanol and stained with freshly prepared oil-red O for 15 min and rinsed again with 60% isopropanol. Sections were then placed in modified Mayer’s hematoxylin for 1 min and washed with distilled water. Slides were then mounted with cover slips using glycerin jelly. Images were taken using a Leica SCN400F whole slide scanner.
Statistics
Data are expressed as mean ± SD. Data were analyzed by the use of unpaired Student’s t-test or two-way ANOVA Tukey’s multiple comparison tests.
RESULTS
FAO increases along with triglyceride accumulation in the kidney after a fast
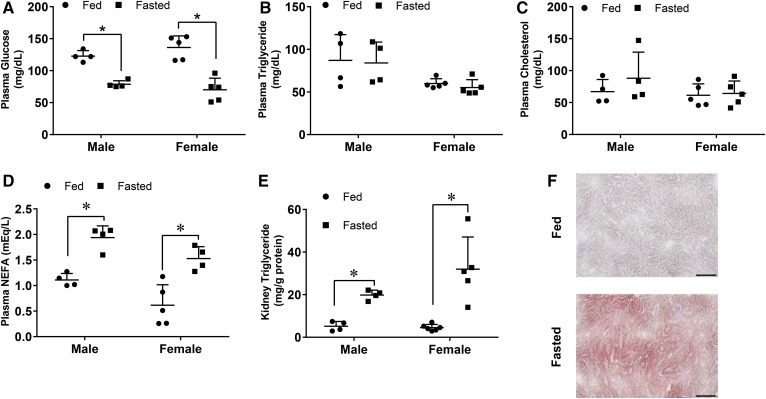
To study lipid accumulation in the kidney, we used an overnight fast as a triglyceride accumulation model (9). After an overnight fast, plasma glucose in both male and female mice decreased by nearly half, while triglycerides and cholesterol were not significantly altered (Fig. 1A–C). NEFAs increased ∼2-fold (Fig. 1D). As in the heart and liver, an overnight fast induced triglyceride accumulation in the kidney; the average triglyceride increase was ∼3-fold in males and 5-fold in females (Fig. 1E). This accumulation occurred primarily in the renal cortex, as shown by oil-red O staining of kidney sections (Fig. 1F).
Fig. 1.
Kidney triglycerides increased in both male and female mice after an overnight fast. A–E: Male and female mice were either fasted or given food ad libitum for 16 h. Blood was drawn to measure glucose and plasma lipids. Lipids were extracted for intracellular triglyceride measurements. A: Plasma glucose. B: Plasma triglycerides. C: Plasma total cholesterol. D: Plasma NEFAs. E: Kidney triglyceride content. F: Kidneys from male mice were formalin fixed and sectioned for oil-red O staining. Scale bar is representative of 100 μM at 20× magnification. N = 4–6/group. *P < 0.05. Results are presented as mean ± SD. *Results compared by unpaired Student’s t-test within each sex group, no significant differences were found between male and female groups using the two-way ANOVA Tukey’s multiple comparison test.
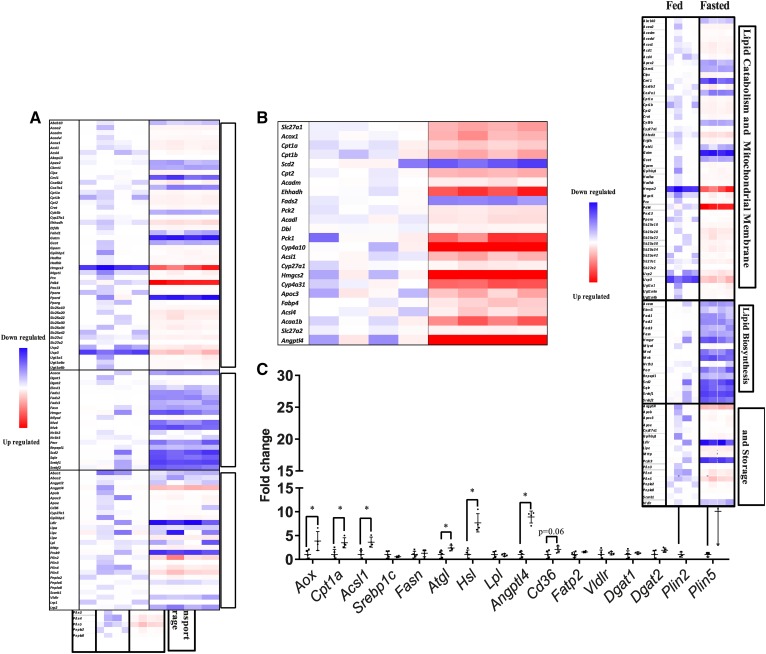
To assess which lipid pathways were changed in kidneys from fasted animals, mRNAs of proteins involved in FAO, de novo lipid synthesis, and lipid transport pathways were analyzed by RNA sequencing of whole kidney from fed and fasted mice. mRNA levels of genes that mediate de novo lipogenesis were decreased, including Fasn, squalene epoxidase (Sqle), acetyl-CoA carboxylase α (Acaca), and sterol regulatory element-binding transcription factor 1 (Srebf1) and 2 (Fig. 2A). However, despite greater lipid accumulation, mRNA levels of genes involved in lipid oxidation and mitochondrial pathways were increased, including those of pyruvate dehydrogenase kinase 4 (Pdk4), alternative oxidase (Aox), carnitine palmitoyltransferase 1a (Cpt1a), and carnitine O-acetyltransferase (Crat) (Fig. 2A). PPARα (Ppara) and Ppard were increased and Pparg remained unchanged (Fig. 2A). Among the more oxidative portions of the kidney, Ppara is the dominantly expressed isoform (31) Ingenuity pathway analysis using the KEGG database showed that PPAR signaling was the most differentially regulated pathway, with 23 associated genes changed (Table 1). A majority of PPAR-regulated genes were also increased after fasting (Fig. 2B). In concordance with the RNA sequencing data, Aox, Cpt1a, and acetyl-CoA synthetase long-chain 1 (Acsl1) were all increased ∼4-fold when assessed by quantitative PCR (Fig. 2C). Genes associated with lipid uptake were also assessed. Cd36 mRNA levels increased almost 2-fold after fasting by quantitative PCR, though it did not quite reach significance. Fatty acid transport protein 2/solute carrier family member 27 member 2 (Fatp2/Slc27A2), a primarily intracellular enzyme that traps NEFAs by esterifying them to CoA (32), was unchanged (Fig. 2C). In addition, the lipoprotein receptors, LDL receptor, LDL receptor related protein (Lrp)1, Lrp2 (megalin), and VLDL receptor (Vldlr), remained unchanged. While the Lpl mRNA level was unchanged, angiopoietin-like 4 (Angptl4), an LPL inhibitor, was increased 9-fold. Lipid droplet-associated protein perilipin (Plin)2 increased 20-fold and Plin5 increased 10-fold. Intracellular lipases needed to utilize triglyceride stored in lipid droplets also increased; hormone sensitive lipase (Hsl/Lipe) mRNA increased 7-fold and adipose triglyceride lipase (Atgl/Pnpla2) 2-fold. Others have reported that, after a fast, both HSL and ATGL activation by phosphorylation increases in the kidney (33). Taken together, this gene expression profile suggests that, despite increased mRNA levels of genes regulating FAO and decreased mRNA levels of genes mediating de novo lipogenesis, the kidney stores excess fatty acids in lipid droplets during fasting. This suggests an excess of fatty acid transport into the kidney.
Fig. 2.
The kidney in the fasted state upregulates lipid oxidation genes and downregulates lipid synthesis genes. RNA sequencing of fed and fasted male kidneys. Data presented as fold change normalized to the fed group. DEGs were identified with Bioconductor package DEseq2 (3.2) and Benjamini and Hochberg’s method was used to control for false discovery rate. A: Canonical genes in pathways related to lipid metabolism and mitochondrial membranes, lipid biosynthesis and lipid transport, and lipolysis. B: Canonical genes in the PPAR pathway. C: Quantitative PCR of selected lipid metabolism genes, normalized to ribosomal 18s. Experiments were performed in male mice, N = 3–5. *P < 0.05. Results are presented as mean ± SD. Results for quantitative PCR were compared by unpaired Student’s t-test.
TABLE 1.
Ten most significantly altered pathways in the kidney between the fed and fasted state
| KEGG Pathway | P | Genes |
| PPAR signaling pathway | 8.36E-08 | SLC27A1, ACOX1, CPT1B, SCD2, CPT2, ACADM, EHHADH, FADS2, PCK2, ACADL, DBI, CPT1A, PCK1, CYP4A10, ACSL1, CYP27A1, HMGCS2, CYP4A31, APOC3, FABP4, ACSL4, ACAA1B, SLC27A2, ANGPTL4 |
| Fatty acid metabolism | 4.71E-06 | ACOX1, CPT1B, ACAA2, ACADM, CPT2, EHHADH, ACADL, HADHA, CPT1A, HADHB, ACADVL, CYP4A10, ACSL1, CYP4A31, ACSL4, ACAA1B |
| Phosphatidylinositol signaling system | 0.030783 | PLCB4, PIK3C2G, PLCG1, INPP5K, PIK3CB, PI4KA, DGKZ, DGKH, DGKI, INPP5D, PLCB1, ITPR1 |
| Biosynthesis of unsaturated fatty acids | 1.99E-05 | PECR, ACOX1, SCD2, FADS1, ACOT2, FADS2, ACOT1, ACAA1B, ACOT4, HADHA, ACOT3 |
| Adipocytokine signaling pathway | 0.034582 | CPT1B, ACSL1, LEPR, IKBKG, ACSL4, PCK2, STAT3, AKT3, CPT1A, AKT2, PCK1 |
| Arginine and proline metabolism | 0.020652 | SAT1, ODC1, GATM, GLUD1, ARG2, MAOB, CKMT1, DAO, NOS3, CKB |
| Glycine, serine, and threonine metabolism | 0.002628 | GLYCTK, GATM, MAOB, PHGDH, GCAT, DAO, PSAT1, CBS, GLDC |
| Amino sugar and nucleotide sugar metabolism | 0.019358 | GALK1, RENBP, GNPDA1, MPI, GM8615, CMAH, UGDH, NAGK, UXS1, PMM1 |
| Valine, leucine and isoleucine degradation | 0.024859 | ACAA2, DBT, ACADM, BCAT2, HMGCS2, EHHADH, ACAA1B, HADHA, HADHB |
| Terpenoid backbone biosynthesis | 3.68E-05 | DHDDS, MVD, HMGCS2, HMGCR, FDPS, MVK, IDI1, PDSS1 |
Ingenuity pathway analysis was performed on the DEGs to determine most canonical pathways being altered, according to the KEGG pathway database. Pathways are listed in order of most significantly different.
Lipidomic changes
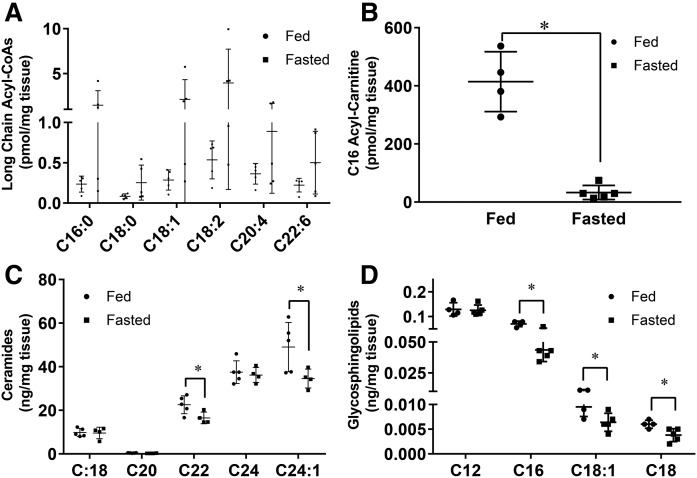
Lipidomics was performed to analyze fatty acid species in the kidney after an overnight fast. There was an overall increase in long-chain acyl-CoAs (Fig. 3A), although significance for changes in individual fatty acids could not be shown due to wide variation in the fasted group. Levels of the long-chain acyl-carnitine, C16, decreased 10-fold (Fig. 3B). This is opposite to what was reported by Koves et al. (34) to happen in skeletal muscles. Additionally, a decline in long-chain ceramides and their derivatives, glycosphingolipids, in the kidney was observed after fasting (Fig. 3C, D). An excess of ceramides is toxic to cells (35, 36). These data suggest that to compensate for increased lipid oxidation, uptake of either triglyceride-rich lipoproteins or NEFAs increases, leading to triglyceride, but not ceramide, accumulation.
Fig. 3.
Ceramides and glycosphingolipids are lowered in the fasted kidney. Lipidomics were performed using LC/ESI-MS/MS on kidneys of mice that were fasted or given food ad libitum for 16 h. A: Long-chain acyl CoAs. B: C16 acyl-carnitine. C: Ceramides. D: Glycosphingolipids. Experiments were performed in male mice, N = 4–5/group. *P < 0.05. Results are presented as mean ± SD. Results compared using unpaired Student’s t-test.
Lipid accumulation in the kidney is dependent on serum NEFAs
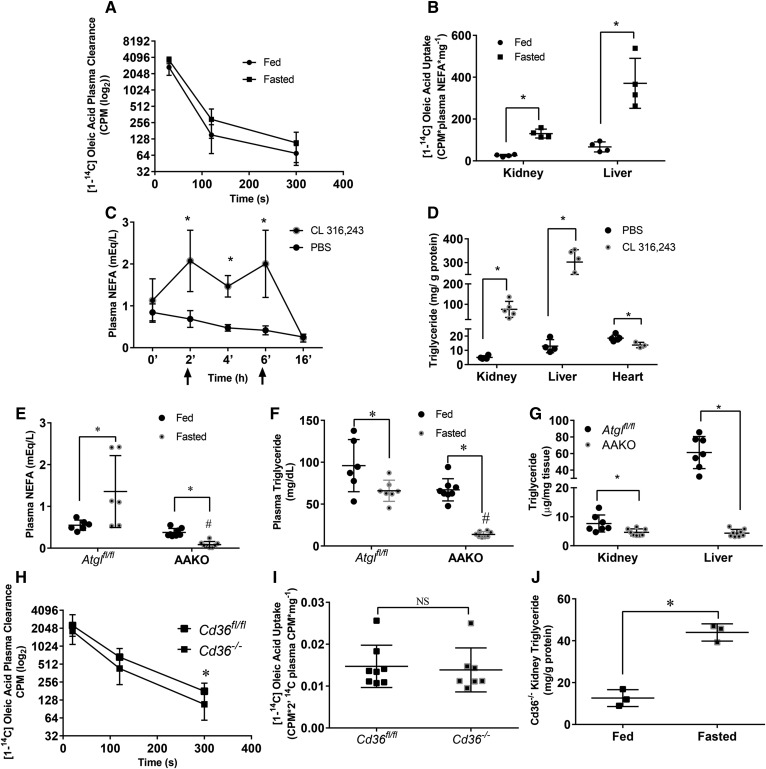
To determine whether fasting increases uptake of albumin-bound NEFAs, we performed an uptake study of [14C]oleic acid in the fed and fasted state. After 5 min, nearly all the [14C]oleic acid tracer was cleared from the circulation (Fig. 4A). The liver took up a majority of fatty acids in both fed and fasted mice and accumulated more NEFAs during fasting. Similarly, kidney uptake of NEFAs was increased ∼4-fold with fasting (Fig. 4B).
Fig. 4.
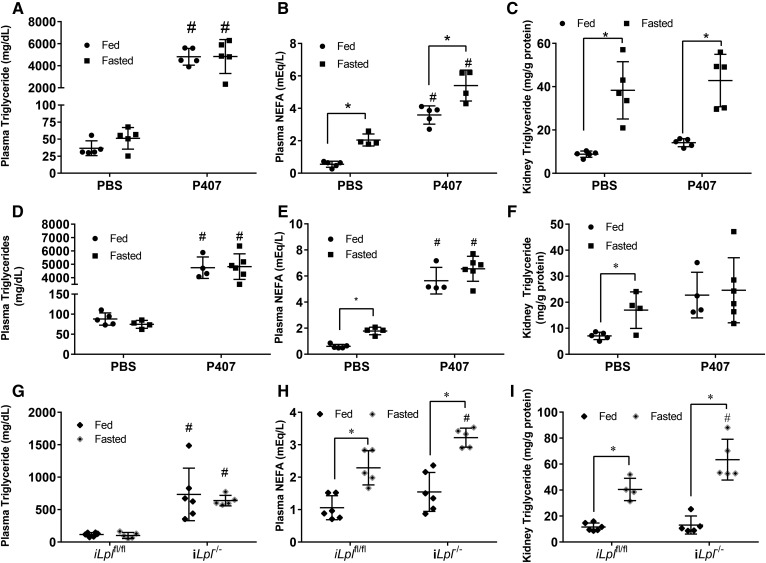
Plasma free fatty acids determine kidney triglyceride content and do not require CD36 for transport. A, B: [1-14C]oleic acid (OA) was injected intravenously into male mice. The presence of radioactive signal was measured in plasma or tissue homogenate using a LS 6500 multipurpose scintillation counter. A: Plasma clearance of [1-14C]OA over time. B: Liver and kidney 14C label 300s after injection. Experiments were performed in male mice, N = 4–5/group. C, D: Male mice were injected with CL 316,243 at 2 and 6 h time points, and blood samples were drawn at indicated points in order to measure NEFA concentration in the blood. Mice were euthanized at the 16 h time point to collect tissues, N = 4–5/group. C: Plasma NEFAs over time after two CL 316,243 injections (indicated by arrows). D: Tissue triglyceride content 16 h after first injection. E–G: Female adipocyte Atgl knockout mice (AAKO) or littermate controls were fasted for 16 h and eithanized, N = 6–8/group. Plasma NEFA (E) and triglyceride (F) of AAKO and control mice in the fed and 16 h fasted state. G: Kidney and liver triglyceride of AAKO and control mice after a 16 h fast. H–J: [1-14C]OA was injected intravenously into male mice. Presence of radioactive signal was measured in plasma or tissue homogenate as mentioned before. H: Plasma clearance of [1-14C]OA over time, N = 8–9/group. I: OA uptake in fasted Cd36fl/fl and Cd36−/− N = 8–9/group. Kidney lipids were extracted from kidneys of Cd36−/− in the fed and fasted state using 2:1 methanol:chloroform. J: Kidney triglyceride in fed and fasted Cd36−/−, N = 3/group.*P < 0.05, #P < 0.05. *Comparison between feeding status using unpaired Student’s t-test. #Comparison between genotype using the two-way ANOVA Tukey’s multiple comparison test. Results are presented as mean ± SD.
We next tested to determine whether altered plasma NEFA levels would lead to parallel changes in kidney triglycerides. To increase NEFAs in the fed state, we used a β adrenergic receptor agonist, CL 316,238. Plasma NEFAs were increased 2-fold over time with two intraperitoneal injections (Fig. 4C). This treatment increased kidney intracellular triglyceride levels ∼15-fold (Fig. 4D); a similar triglyceride increase was found in liver, but not heart. To determine whether reduced circulating fatty acid levels would reduce kidney triglyceride after fasting, we studied mice with an adipocyte-specific deletion of Atgl (mice are denoted AAKO). As had been shown previously (20), AAKO mice have lower circulating NEFAs (Fig. 4E), and unlike floxed control mice (Atglfl/fl littermates), NEFAs decreased with fasting. Plasma triglycerides were also reduced in this model (Fig. 4F). Mirroring the liver, the kidney also had an ∼40% reduction in fasting triglyceride content compared with fasted controls (Fig. 4G). Liver triglyceride accumulation with fasting was reduced in AAKO mice, as was reported previously (20). These data demonstrate that renal triglyceride during fasting is modulated by plasma NEFA levels.
Lipid accumulation in the kidney is not dependent on CD36
We then asked whether NEFA uptake required CD36, a known fatty acid transporter highly expressed in the kidney and increased 2-fold with fasting (Fig. 2C). However, uptake of oleic acid into kidneys from Cd36−/− mice was not decreased (Fig. 4H, I), and kidneys from Cd36−/− mice had the same amount of triglyceride increase with fasting as did wild-type mice (Fig. 4J compared with Fig. 1E). Thus, the kidney does not require CD36 for fasting-induced lipid accumulation.
Triglyceride accumulation in the kidney does not require LPL or circulating lipoproteins
LPL is required for fasting triglyceride accumulation in the heart (10); like in the heart, but not in the liver, LPL is amply expressed in the kidney (15, 37). To assess whether LPL and lipoproteins are necessary for triglyceride accumulation in the kidney, we injected mice with a surfactant compound, P407. P407 blocks the clearance of lipoproteins from the circulation and causes lipemia (38). In our mice, triglyceride increased from ∼50 mg/dl to over 5,000 mg/dl (Fig. 5A). P407 also increased plasma NEFAs in both fed and fasted mice (Fig. 5B), likely due to association with triglyceride-rich lipoproteins (10). P407 did not decrease triglyceride accumulation in the kidney after a fast, but led to a ∼20% increase in triglycerides during the fed state (Fig. 5C). Similar results were found in females (Fig. 5D–F) as in males.
Fig. 5.
LPL is not required for triglyceride accumulation in the kidney. A–C: Male mice were injected with P407 or PBS equivalent and fasted or given food ad libitum for 16 h (top panel). Plasma triglycerides (A) and plasma NEFAs (B). C: Kidney triglyceride content. N = 4–5/group. D–F: P407 repeated in females (middle panel) with same conditions as males. G–I: Tamoxifen inducible LPL floxed (iLplfl/fl) and knockout (iLpl−/−) male mice were fasted or given ad libitum access to food for 16 h. Blood was drawn for measurement of plasma lipids and kidney lipids were extracted and measured (bottom panel). Plasma triglycerides (G) and NEFAs (H). I: Triglyceride content of kidneys. N = 4–6/group. *P < 0.05, #P < 0.05. *Comparison between feeding status using unpaired Student’s t-test. #Comparison between genotype or treatment using the two-way ANOVA Tukey’s multiple comparison test.
To further explore the role of LPL, we studied mice with a tamoxifen-inducible deletion of LPL (iLpl−/−) (22) and assessed the changes in renal triglyceride accumulation before and after an overnight fast and compared them to floxed littermate (Lplfl/fl) controls also injected with tamoxifen. Two weeks after the final tamoxifen injections, plasma triglycerides increased more than 20-fold in both fed and fasted iLpl−/− mice (Fig. 5G). Plasma NEFAs slightly increased as well in iLpl−/− mice after fasting (Fig. 5H). Triglycerides in the kidney did not decrease, but rather increased by 25% after fasting, compared with the fasting iLplfl/fl group (Fig. 5I). This might be due to the increased fatty acids in iLpl−/− mice after fasting.
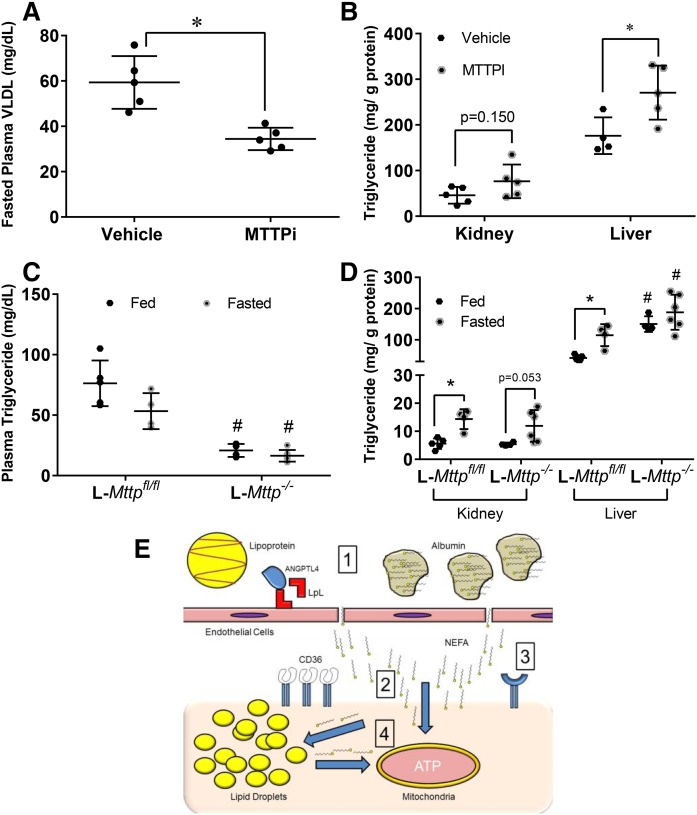
We further explored whether lipoproteins are the source of fatty acids in the kidney by blocking MTTP, a key regulator of lipoprotein formation in the liver and small intestines (39). After a 16 h fast, BMS-212122, a MTTPi, decreased plasma and VLDL triglycerides by approximately 40% (Fig. 6A). MTTPi treatment increased liver triglycerides (Fig. 6B), but unexpectedly, the kidney triglyceride level trended toward an increase (Fig. 6B). It has been previously shown that the kidney can synthesize and secrete lipoproteins (9).
Fig. 6.
Triglyceride-rich lipoproteins are not a significant source of triglyceride in the kidney. A, B: Whole body blockade of MTTP was achieved using BMS-212122 (MTTPi, N = 4–5/group) given orally for 7 days or vehicle (DMSO) equivalent to mice. Data obtained after a 16 h fast. A: VLDL triglyceride levels obtained from ultracentrifugation of plasma of mice treated with MTTPi or vehicle. B: Kidney and liver triglyceride content of fasted mice treated with either MTTPi vehicle. C, D: Liver-specific blockade of MTTP was achieved with a liver-specific knockout of MTTP (L-Mttp−/−, N = 4–6/group). C: Plasma triglycerides of L-Mttp−/− mice versus floxed littermate controls. D: Kidney and liver triglyceride content of L-Mttp−/− mice versus littermate controls. E: Summary figure of research data: 1) During fasting Angptl4 inhibits LPL, so that triglycerides on lipoproteins cannot be hydrolyzed, thus fatty acid-loaded albumin is the primary exogenous source of fatty acids for the kidney. 2) Though CD36 is amply expressed on the kidney, it is not required for the transport of fatty acids into the kidney during fasting, which might involve nonreceptor diffusion across the member. 3) Though CD36 does not appear to act as a major transporter, it does not discount that other transporters may exist. 4) Once inside the cell, fatty acid is shuttled to the mitochondria where it can be oxidized for ATP. If the rate of fatty acid flux into the cells is far greater than the cells capacity for oxidization, excess fatty acids are stored in lipid droplets. Stored fatty acids can also be released from the lipid droplets and oxidized. Experiments were performed in male mice. *P < 0.05, #P < 0.05. *Comparison between feeding status using unpaired Student’s t-test. #Comparison between genotype using the two-way ANOVA Tukey’s multiple comparison test.
To test whether some of the effects of plasma triglyceride reduction were masked by intra-renal MTTP inhibition, we fasted mice with a liver-specific knockdown of Mttp (L-Mttp−/−) (40) for 16 h. Plasma triglycerides were decreased several fold (Fig. 6C), while liver triglycerides in the L-Mttp−/− mice were increased. Fasting kidney triglyceride levels were slightly affected by loss of Mttp in the liver (Fig. 6D). These results suggest that circulating lipoproteins and LPL play a minimal role, if any, in lipid accumulation in the kidney during a fast.
DISCUSSION
Lipid accumulates in the kidney cortex during diseased and nondiseased states. The mechanism(s) by which these lipids accumulate is unclear. In addition, little information is available to determine whether lipid accumulations are pathological or physiological. In this study, we evaluated the physiologic regulation of kidney lipids during fasting and explored the roles of lipid uptake processes that are known to affect triglyceride storage in other tissues. Unlike muscles, kidney lipid storage is not dependent on either LPL or CD36 and correlates with plasma NEFAs. Further data indicated that kidney triglyceride accumulation was dependent on NEFAs from the plasma. Using [14C]oleic acid, we showed that the kidney indeed takes up more NEFAs during a fast (Fig. 4B). We further showed that increasing serum NEFA levels in the fed state with CL 315,343, a β3-adrenergic receptor agonist, markedly increased kidney triglyceride accumulation (Fig. 4C, D). At the same time, heart triglycerides were reduced with CL 315,343 treatment; these data are consistent with our previous studies indicating that circulating triglycerides and not NEFAs are the source of heart triglyceride (10). Conversely, lowering circulating NEFAs using AAKO mice (Fig. 5F) led to reduced kidney triglyceride (Fig. 5G). Our results confirm studies that measured arterial versus venous blood and showed that the kidney takes up more fatty acid during fasting (41).
Fasting-induced lipid accumulation was associated with increased expression of genes that modulated fatty acid metabolism and triglyceride storage and not with greater accumulation of ceramides. In many tissues, dysfunction is associated with the accumulation of potentially toxic lipids, such as ceramides. The nonpathological storage of NEFAs as triglyceride was suggested by an increase in the lipid droplet proteins, Plin2 and Plin5 (Fig. 2A), indicating a capacity for safely storing excess triglyceride. Following fasting, our lipidomic analysis showed a trend to an increase in fatty acyl CoAs, a decrease in C16 acyl-carnitine, and a decrease in long-chain ceramides and glycosphingolipids. An increase in skeletal muscle acyl-carnitines was associated with incomplete FAO in high-fat diet-fed animals (42). Therefore, the decrease in renal acyl-carnitines in the fasted state could result from more complete β-oxidation. An increase in ceramides is associated with cellular apoptosis, as well as insulin resistance (35, 36, 43). There is a dearth of information on the role of ceramides in kidney injury and some of what is reported is conflicting. Sas et al. (28) reported that in a diabetic obesity model of kidney injury, long-chain ceramides and glycosphingolipids are reduced. Others have reported an increase in glycosphingolipids, a product of ceramides, associated with diabetic nephropathy and mesangial expansion (44).
To achieve a global perspective on the changes in kidney lipid metabolism during fasting, we performed RNA sequencing of the kidney. Several enzymes associated with increased ceramide production were reduced. mRNA levels of sphingomyelin phosphodiesterase 1 (Smpd1) which is responsible for hydrolyzing sphingolipids to form ceramides in the fasted kidney, were reduced 2-fold and serine palmitoyltransferase subunits 1 and 2 (Sptlc1 and Sptlc2) that control de novo synthesis of ceramide were unchanged (supplemental Table S1).
FAO appeared to increase with fasting, and de novo synthesis did not appear to increase. Though this was not measured directly, our indirect data strongly suggest that this is the case. mRNA levels of genes in the lipid synthesis pathway were downregulated, while genes in the lipid oxidation pathway, mitochondrial electron transport, and fatty acid transport were upregulated (Fig. 2A). The PPAR pathway was the most differentially regulated pathway among those in the KEGG database, which have been shown to regulate FAO genes (Table 1) (45). These data suggest that, along with greater triglyceride storage, the kidney accumulates an excess supply of substrate for storage while catabolizing more lipids for energy. During fasting, many tissues burn fats rather than glucose and, like the kidney, some of these tissues also store lipids, perhaps as a local energy supply to use in times of greater energy requirements (46). The gene expression profile in the kidney implies greater FAO during fasting. This differs from the heart, which also accumulates triglyceride during fasting, but does not show this same pattern of increased FAO genes (10).
The increase in FAO during fasting is contrary to what is reported in kidney injury. FAO-associated genes are downregulated in the kidneys of mice with folic acid-induced nephropathy, as well as in humans with CKD (7). However, during early diabetic kidney disease, when estimated glomerular filtration rate is not significantly decreased, β-oxidation is increased (28, 29). Therefore, fasting and early diabetes may reflect a need for greater energy requirements by intact tubular cells in the kidney, while CKD illustrates an example of pathologic injury and lipid accumulation due to reduced lipid oxidation.
We sought to determine the transporter required for renal NEFA uptake. A logical transporter was CD36; our quantitative PCR showed an increased trend of Cd36 mRNA levels in the kidney during the fasted state (Fig. 2C). CD36 is a long-chain fatty acid transporter that is important for the uptake of fatty acids in various tissues (47). Surprisingly, [14C]oleic acid uptake in fasted Cd36−/− mouse kidneys was the same as in controls (Fig. 4I). Triglyceride content in the kidney also tripled after a fast in Cd36−/− mice (Fig. 4J). Overall triglyceride levels in the fasted Cd36−/− mouse kidneys were higher when compared with fasted wild-type mice (Fig. 1E). This is likely due to elevated NEFA in the plasma due to the defect in peripheral NEFA uptake by the heart and skeletal muscle (48). Our data demonstrate that CD36 is not necessary for fatty acid uptake in kidneys from fasted mice and, at first glance, appear to contradict the studies of Kang et al. (7), who showed that overexpression of Cd36 in the proximal tubule increases triglyceride accumulation in the kidney. While forced overexpression of Cd36 will lead to accumulation of triglyceride in the kidney, we show that it is not necessary for fatty acid transport or triglyceride accumulation in the kidney. It is possible that other fatty acid transporters, such as those from the FATP family (49, 50), are responsible for uptake in the kidney. However, these FATPs are predominantly intracellular and, rather than being transmembrane transporters, are thought to trap intracellular fatty acids by acyl-CoA synthetase actions (32). Another possible route of NEFA uptake into the kidney is via uptake of albumin. However, the very rapid turnover of NEFAs compared with that of albumin makes this hypothesis unlikely. Moreover, mRNA levels of purported albumin transporters, such as Lrp2, were decreased according to our RNA sequencing (Fig. 2A). Alternatively, fatty acids may enter the kidney via passive diffusion through the membranes, which can occur when fatty acid levels are high (51).
We tested to determine whether fasting-induced renal triglycerides were derived from circulating lipoproteins and whether kidney triglyceride accumulation requires LPL hydrolysis of triglyceride to release NEFAs. Blocking or deleting LPL did not decrease triglyceride accumulation in the kidney after a fast. Although LPL is abundant in the kidney, during the fasted state, increased expression of Angptl4 may inhibit LPL actions (Fig. 2A). Angptl4 is thought to dissociate the LPL dimer, thus inhibiting its enzymatic action (52). Our data are consistent with those of Ruge et al. (53) who reported that kidney LPL activity is highest in the fed state and lowest in the fasted state of mice; we now show that this is likely due to Angptl4. Because LPL loss did not play a role in lipid accumulation in the kidney in either the fed or fasted state, the role of LPL in the kidney is unclear.
In support of our finding that LPL does not play a major role in lipid accumulation, we found that circulating triglyceride levels did not correlate with the intracellular lipid content of the kidney. We lowered circulating triglycerides two ways: pharmacologically with an MTTPi (Fig. 6A) and genetically by specifically knocking out Mttp in the liver (Fig. 6C). The kidney, like the liver, is able to form and release lipoproteins (9). This is highlighted by our data showing that the kidney retains triglycerides when treated with MTTPi, with a trend to increased kidney triglyceride content after a fast (Fig. 6B). When we lowered circulating lipoprotein levels by deleting Mttp in the liver, plasma triglyceride levels were markedly reduced, but triglyceride content in the kidney remained mostly unchanged, although some of the fasted L-Mttp−/− mice had reduced triglycerides (Fig. 6D). Because some NEFAs associate with lipoproteins, this reduction in kidney triglyceride may be due to fewer circulating lipoproteins carrying NEFAs. These MTTP inhibition studies showed that the kidney does not largely rely on exogenous lipoproteins for triglycerides.
In conclusion, we have found that the kidney primarily takes up NEFAs rather than lipoproteins, and high serum levels of NEFAs cause greater lipid accumulation in the kidney. Figure 6E summarizes the findings from our study. The kidney, despite having high levels of expression of Cd36 and Lpl, does not require either of these proteins for fatty acid uptake during fasting. During fasting, LPL actions in the kidney are likely inhibited by increased Angptl4. Taken together, our data show that the kidney behaves similarly to the liver with respect to certain aspects of lipid metabolism (e.g., lipid uptake does not involve LPL or CD36). Both the liver and the kidney receive abundant cardiac output of blood, are intensely metabolically active, and might not depend on a high affinity localized fatty acid uptake pathway. Rather, a lower affinity, but higher capacity, fatty acid uptake process may be sufficient to extract fatty acids from the circulation. This high volume of blood supply to the kidney may also be why NEFAs on albumin are sufficient for lipid accumulation in the kidney. Comparably, less perfused tissues, such as skeletal muscle and adipose tissue, would require the large amounts of fatty acids, in the form of triglyceride, and also the actions of a high affinity NEFA transporter, such as CD36. It should be noted that the molecular events required for NEFA uptake by the liver are incompletely characterized and might require a number of known or unique transporters (54). The fatty acids are either shuttled to the mitochondria for oxidation or to lipid droplets for storage in the form of triglyceride. These stored triglycerides can also be hydrolyzed by HSL and ATGL to release fatty acids for oxidation. Little in the way of kidney lipid metabolism has been studied despite lipid accumulation being present in nearly all forms of kidney injury. Our data add to our understanding of lipid metabolism in the nondiseased kidney.
Supplementary Material
Acknowledgments
The authors would like to acknowledge New York University Langone Medical Center Office of Collaborative Science for their technical services, supported in part by the Cancer Center Support Grant P30CA016087, at the Laura and Isaac Perlmutter Cancer Center. The authors also acknowledge the editing assistance of Stephanie Chiang.
Footnotes
Abbreviations:
- AAKO
- Adipose-Atgl knock out
- Acsl1
- acetyl-CoA synthetase long-chain 1
- Angptl4
- angiopoietin-like 4
- ATGL
- adipose triglyceride lipase
- CD
- cluster of differentiation
- CKD
- chronic kidney disease
- Cpt1a
- carnitine palmitoyltransferase 1
- DEG
- differentially expressed gene
- FAO
- fatty acid oxidation
- FATP
- fatty acid transport protein
- HSL
- hormone sensitive lipase
- iLpl−/−
- inducible-Lpl−/−
- L-Mttp−/−
- liver-specific knockdown of microsomal triglyceride transfer protein
- Lrp
- LDL receptor related protein
- MTTP
- microsomal triglyceride transfer protein
- MTTPi
- microsomal triglyceride transfer protein inhibitor
- P407
- poloxamer 407
- Plin
- perilipin
This work was supported by National Institutes of Health Grants HL45095 and HL73029 (I.J.G.); DK094292, DK089503, DK082841, DK081943, and DK097153 (S.P.); 2UL1TR000433 (K.M.S.); R56DK046900 (M.M.H.); DK090166 (E.E.K.); DK087635, DK105821, and DK108220 (K.S.); and DK33301 (N.A.A.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. All the authors have declared no competing interests.
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Nieth H., and Schollmeyer P.. 1966. Substrate-utilization of the human kidney. Nature. 209: 1244–1245. [DOI] [PubMed] [Google Scholar]
- 2.Bobulescu I. A. 2010. Renal lipid metabolism and lipotoxicity. Curr. Opin. Nephrol. Hypertens. 19: 393–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang Z., Jiang T., Li J., Proctor G., McManaman J. L., Lucia S., Chua S., and Levi M.. 2005. Regulation of renal lipid metabolism, lipid accumulation, and glomerulosclerosis in FVBdb/db mice with type 2 diabetes. Diabetes. 54: 2328–2335. [DOI] [PubMed] [Google Scholar]
- 4.Jiang T., Wang Z., Proctor G., Moskowitz S., Liebman S. E., Rogers T., Lucia M. S., Li J., and Levi M.. 2005. Diet-induced obesity in C57BL/6J mice causes increased renal lipid accumulation and glomerulosclerosis via a sterol regulatory element-binding protein-1c-dependent pathway. J. Biol. Chem. 280: 32317–32325. [DOI] [PubMed] [Google Scholar]
- 5.Jiang T., Liebman S. E., Lucia M. S., Li J., and Levi M.. 2005. Role of altered renal lipid metabolism and the sterol regulatory element binding proteins in the pathogenesis of age-related renal disease. Kidney Int. 68: 2608–2620. [DOI] [PubMed] [Google Scholar]
- 6.Sun L., Halaihel N., Zhang W., Rogers T., and Levi M.. 2002. Role of sterol regulatory element-binding protein 1 in regulation of renal lipid metabolism and glomerulosclerosis in diabetes mellitus. J. Biol. Chem. 277: 18919–18927. [DOI] [PubMed] [Google Scholar]
- 7.Kang H. M., Ahn S. H., Choi P., Ko Y. A., Han S. H., Chinga F., Park A. S., Tao J., Sharma K., Pullman J., et al. . 2015. Defective fatty acid oxidation in renal tubular epithelial cells has a key role in kidney fibrosis development. Nat. Med. 21: 37–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Simon N., and Hertig A.. 2015. Alteration of fatty acid oxidation in tubular epithelial cells: from acute kidney injury to renal fibrogenesis. Front. Med. (Lausanne). 2: 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krzystanek M., Pedersen T. X., Bartels E. D., Kjaehr J., Straarup E. M., and Nielsen L. B.. 2010. Expression of apolipoprotein B in the kidney attenuates renal lipid accumulation. J. Biol. Chem. 285: 10583–10590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Trent C. M., Yu S., Hu Y., Skoller N., Huggins L. A., Homma S., and Goldberg I. J.. 2014. Lipoprotein lipase activity is required for cardiac lipid droplet production. J. Lipid Res. 55: 645–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goldberg I. J., Trent C. M., and Schulze P. C.. 2012. Lipid metabolism and toxicity in the heart. Cell Metab. 15: 805–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hamilton J. A. 1998. Fatty acid transport: difficult or easy? J. Lipid Res. 39: 467–481. [PubMed] [Google Scholar]
- 13.Schwenk R. W., Holloway G. P., Luiken J. J., Bonen A., and Glatz J. F.. 2010. Fatty acid transport across the cell membrane: regulation by fatty acid transporters. Prostaglandins Leukot. Essent. Fatty Acids. 82: 149–154. [DOI] [PubMed] [Google Scholar]
- 14.Harmon C. M., and Abumrad N. A.. 1993. Binding of sulfosuccinimidyl fatty acids to adipocyte membrane proteins: isolation and amino-terminal sequence of an 88-kD protein implicated in transport of long-chain fatty acids. J. Membr. Biol. 133: 43–49. [DOI] [PubMed] [Google Scholar]
- 15.Goldberg I. J., Soprano D. R., Wyatt M. L., Vanni T. M., Kirchgessner T. G., and Schotz M. C.. 1989. Localization of lipoprotein lipase mRNA in selected rat tissues. J. Lipid Res. 30: 1569–1577. [PubMed] [Google Scholar]
- 16.Susztak K., Ciccone E., McCue P., Sharma K., and Bottinger E. P.. 2005. Multiple metabolic hits converge on CD36 as novel mediator of tubular epithelial apoptosis in diabetic nephropathy. PLoS Med. 2: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Susztak K., Bottinger E., Novetsky A., Liang D., Zhu Y., Ciccone E., Wu D., Dunn S., McCue P., and Sharma K.. 2004. Molecular profiling of diabetic mouse kidney reveals novel genes linked to glomerular disease. Diabetes. 53: 784–794. [DOI] [PubMed] [Google Scholar]
- 18.Febbraio M., Abumrad N. A., Hajjar D. P., Sharma K., Cheng W., Pearce S. F., and Silverstein R. L.. 1999. A null mutation in murine CD36 reveals an important role in fatty acid and lipoprotein metabolism. J. Biol. Chem. 274: 19055–19062. [DOI] [PubMed] [Google Scholar]
- 19.Sitnick M. T., Basantani M. K., Cai L., Schoiswohl G., Yazbeck C. F., Distefano G., Ritov V., DeLany J. P., Schreiber R., Stolz D. B., et al. . 2013. Skeletal muscle triacylglycerol hydrolysis does not influence metabolic complications of obesity. Diabetes. 62: 3350–3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schoiswohl G., Stefanovic-Racic M., Menke M. N., Wills R. C., Surlow B. A., Basantani M. K., Sitnick M. T., Cai L., Yazbeck C. F., Stolz D. B., et al. . 2015. Impact of reduced ATGL-mediated adipocyte lipolysis on obesity-associated insulin resistance and inflammation in male mice. Endocrinology. 156: 3610–3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Millar J. S., Cromley D. A., McCoy M. G., Rader D. J., and Billheimer J. T.. 2005. Determining hepatic triglyceride production in mice: comparison of poloxamer 407 with Triton WR-1339. J. Lipid Res. 46: 2023–2028. [DOI] [PubMed] [Google Scholar]
- 22.Noh H. L., Okajima K., Molkentin J. D., Homma S., and Goldberg I. J.. 2006. Acute lipoprotein lipase deletion in adult mice leads to dyslipidemia and cardiac dysfunction. Am. J. Physiol. Endocrinol. Metab. 291: E755–E760. [Erratum. 2007. Am. J. Physiol. Endocrinol. Metab. 292: E367.] [DOI] [PubMed] [Google Scholar]
- 23.Kosteli A., Sugaru E., Haemmerle G., Martin J. F., Lei J., Zechner R., and Ferrante A. W. Jr. 2010. Weight loss and lipolysis promote a dynamic immune response in murine adipose tissue. J. Clin. Invest. 120: 3466–3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Josekutty J., Iqbal J., Iwawaki T., Kohno K., and Hussain M. M.. 2013. Microsomal triglyceride transfer protein inhibition induces endoplasmic reticulum stress and increases gene transcription via Ire1alpha/cJun to enhance plasma ALT/AST. J. Biol. Chem. 288: 14372–14383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kako Y., Huang L. S., Yang J., Katopodis T., Ramakrishnan R., and Goldberg I. J.. 1999. Streptozotocin-induced diabetes in human apolipoprotein B transgenic mice. Effects on lipoproteins and atherosclerosis. J. Lipid Res. 40: 2185–2194. [PubMed] [Google Scholar]
- 26.Folch J., Lees M., and Sloane Stanley G. H.. 1957. A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 226: 497–509. [PubMed] [Google Scholar]
- 27.Han C. Y., Umemoto T., Omer M., Den Hartigh L. J., Chiba T., LeBoeuf R., Buller C. L., Sweet I. R., Pennathur S., Abel E. D., et al. . 2012. NADPH oxidase-derived reactive oxygen species increases expression of monocyte chemotactic factor genes in cultured adipocytes. J. Biol. Chem. 287: 10379–10393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sas K. M., Nair V., Byun J., Kayampilly P., Zhang H., Saha J., Brosius F. C. III, Kretzler M., and Pennathur S.. 2015. Targeted lipidomic and transcriptomic analysis identifies dysregulated renal ceramide metabolism in a mouse model of diabetic kidney disease. J. Proteomics Bioinform. 14 (Suppl.): 002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sas K. M., Kayampilly P., Byun J., Nair V., Hinder L. M., Hur J., Zhang H., Lin C., Qi N. R., Michailidis G., et al. . 2016. Tissue-specific metabolic reprogramming drives nutrient flux in diabetic complications. JCI Insight. 1: e86976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bligh E. G., and Dyer W. J.. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37: 911–917. [DOI] [PubMed] [Google Scholar]
- 31.Ruan X., Zheng F., and Guan Y.. 2008. PPARs and the kidney in metabolic syndrome. Am. J. Physiol. Renal Physiol. 294: F1032–F1047. [DOI] [PubMed] [Google Scholar]
- 32.Coe N. R., Smith A. J., Frohnert B. I., Watkins P. A., and Bernlohr D. A.. 1999. The fatty acid transport protein (FATP1) is a very long chain acyl-CoA synthetase. J. Biol. Chem. 274: 36300–36304. [DOI] [PubMed] [Google Scholar]
- 33.Marvyn P. M., Bradley R. M., Button E. B., Mardian E. B., and Duncan R. E.. 2015. Fasting upregulates adipose triglyceride lipase and hormone-sensitive lipase levels and phosphorylation in mouse kidney. Biochem. Cell Biol. 93: 262–267. [DOI] [PubMed] [Google Scholar]
- 34.Koves T. R., Ussher J. R., Noland R. C., Slentz D., Mosedale M., Ilkayeva O., Bain J., Stevens R., Dyck J. R., Newgard C. B., et al. . 2008. Mitochondrial overload and incomplete fatty acid oxidation contribute to skeletal muscle insulin resistance. Cell Metab. 7: 45–56. [DOI] [PubMed] [Google Scholar]
- 35.Dbaibo G. S., El-Assaad W., Krikorian A., Liu B., Diab K., Idriss N. Z., El-Sabban M., Driscoll T. A., Perry D. K., and Hannun Y. A.. 2001. Ceramide generation by two distinct pathways in tumor necrosis factor alpha-induced cell death. FEBS Lett. 503: 7–12. [DOI] [PubMed] [Google Scholar]
- 36.Rotolo J. A., Zhang J., Donepudi M., Lee H., Fuks Z., and Kolesnick R.. 2005. Caspase-dependent and -independent activation of acid sphingomyelinase signaling. J. Biol. Chem. 280: 26425–26434. [DOI] [PubMed] [Google Scholar]
- 37.Merkel M., Eckel R. H., and Goldberg I. J.. 2002. Lipoprotein lipase: genetics, lipid uptake, and regulation. J. Lipid Res. 43: 1997–2006. [DOI] [PubMed] [Google Scholar]
- 38.Han S., Akiyama T. E., Previs S. F., Herath K., Roddy T. P., Jensen K. K., Guan H. P., Murphy B. A., McNamara L. A., Shen X., et al. . 2013. Effects of small interfering RNA-mediated hepatic glucagon receptor inhibition on lipid metabolism in db/db mice. J. Lipid Res. 54: 2615–2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hussain M. M., Shi J., and Dreizen P.. 2003. Microsomal triglyceride transfer protein and its role in apoB-lipoprotein assembly. J. Lipid Res. 44: 22–32. [DOI] [PubMed] [Google Scholar]
- 40.Raabe M., Veniant M. M., Sullivan M. A., Zlot C. H., Bjorkegren J., Nielsen L. B., Wong J. S., Hamilton R. L., and Young S. G.. 1999. Analysis of the role of microsomal triglyceride transfer protein in the liver of tissue-specific knockout mice. J. Clin. Invest. 103: 1287–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Elhamri M., Martin M., Ferrier B., and Baverel G.. 1993. Substrate uptake and utilization by the kidney of fed and starved rats in vivo. Ren. Physiol. Biochem. 16: 311–324. [DOI] [PubMed] [Google Scholar]
- 42.Koves T. R., Li P., An J., Akimoto T., Slentz D., Ilkayeva O., Dohm G. L., Yan Z., Newgard C. B., and Muoio D. M.. 2005. Peroxisome proliferator-activated receptor-gamma co-activator 1alpha-mediated metabolic remodeling of skeletal myocytes mimics exercise training and reverses lipid-induced mitochondrial inefficiency. J. Biol. Chem. 280: 33588–33598. [DOI] [PubMed] [Google Scholar]
- 43.Yang G., Badeanlou L., Bielawski J., Roberts A. J., Hannun Y. A., and Samad F.. 2009. Central role of ceramide biosynthesis in body weight regulation, energy metabolism, and the metabolic syndrome. Am. J. Physiol. Endocrinol. Metab. 297: E211–E224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Subathra M., Korrapati M., Howell L. A., Arthur J. M., Shayman J. A., Schnellmann R. G., and Siskind L. J.. 2015. Kidney glycosphingolipids are elevated early in diabetic nephropathy and mediate hypertrophy of mesangial cells. Am. J. Physiol. Renal Physiol. 309: F204–F215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Poulsen L., Siersbaek M., and Mandrup S.. 2012. PPARs: fatty acid sensors controlling metabolism. Semin. Cell Dev. Biol. 23: 631–639. [DOI] [PubMed] [Google Scholar]
- 46.Egan B., and Zierath J. R.. 2013. Exercise metabolism and the molecular regulation of skeletal muscle adaptation. Cell Metab. 17: 162–184. [DOI] [PubMed] [Google Scholar]
- 47.Goldberg I. J., Eckel R. H., and Abumrad N. A.. 2009. Regulation of fatty acid uptake into tissues: lipoprotein lipase- and CD36-mediated pathways. J. Lipid Res. 50 (Suppl.): S86–S90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Coburn C. T., Knapp F. F. Jr., Febbraio M., Beets A. L., Silverstein R. L., and Abumrad N. A.. 2000. Defective uptake and utilization of long chain fatty acids in muscle and adipose tissues of CD36 knockout mice. J. Biol. Chem. 275: 32523–32529. [DOI] [PubMed] [Google Scholar]
- 49.Falcon A., Doege H., Fluitt A., Tsang B., Watson N., Kay M. A., and Stahl A.. 2010. FATP2 is a hepatic fatty acid transporter and peroxisomal very long-chain acyl-CoA synthetase. Am. J. Physiol. Endocrinol. Metab. 299: E384–E393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kazantzis M., and Stahl A.. 2012. Fatty acid transport proteins, implications in physiology and disease. Biochim. Biophys. Acta. 1821: 852–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hamilton J. A. 1999. Transport of fatty acids across membranes by the diffusion mechanism. Prostaglandins Leukot. Essent. Fatty Acids. 60: 291–297. [DOI] [PubMed] [Google Scholar]
- 52.Sukonina V., Lookene A., Olivecrona T., and Olivecrona G.. 2006. Angiopoietin-like protein 4 converts lipoprotein lipase to inactive monomers and modulates lipase activity in adipose tissue. Proc. Natl. Acad. Sci. USA. 103: 17450–17455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ruge T., Neuger L., Sukonina V., Wu G., Barath S., Gupta J., Frankel B., Christophersen B., Nordstoga K., Olivecrona T., et al. . 2004. Lipoprotein lipase in the kidney: activity varies widely among animal species. Am. J. Physiol. Renal Physiol. 287: F1131–F1139. [DOI] [PubMed] [Google Scholar]
- 54.Mashek D. G. 2013. Hepatic fatty acid trafficking: multiple forks in the road. Adv. Nutr. 4: 697–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.