Abstract
Recent studies of genome-wide transcriptional regulatory network (TRN) revealed several intriguing structural and dynamic features of gene expression at a system level. Unfortunately, the network under study is often far from complete. A critical question is thus how much the network is incomplete and to what extent this would affect the results of analysis. Here we compare the Escherichia coli TRN built by Shen-Orr et al. (Nature Genet., 31, 64–68) with two TRNs reconstructed from RegulonDB and Ecocyc respectively and present an extended E.coli TRN by integrating information from these databases and literature. The scale of the extended TRN is about twice as large as the previous ones. The new network preserves the multi-layer hierarchical structure which we recently reported but has more layers. More global regulators are inferred. While the feed forward loop (FFL) is confirmed to be highly representative in the network, the distribution of the different types of FFLs is different from that based on the incomplete network. In contrast to the notion of motif aggregation and formation of homologous motif clusters, we found that most FFLs interact and form a giant motif cluster. Furthermore, we show that only a small portion of the genes is solely regulated by only one FFL. Many genes are regulated by two or more interacting FFLs or other more complicated network motifs together with transcriptional factors not belonging to any network motifs, thereby forming complex regulatory circuits. Overall, the extended TRN represents a more solid basis for structural and functional analysis of genome-wide gene regulation in E.coli.
INTRODUCTION
The study of genome-wide transcriptional regulatory network (TRN) has drawn much attention in the last few years because it offers the possibility to better understand the topology and function of gene regulation of cellular responses to environmental changes at a system level (1–12). A prerequisite for this kind of studies is the TRN reconstruction. However, the reconstruction of genome-scale TRN is not an easy task. This is because: (i) we cannot directly obtain regulatory relationships from gene annotation information as in the case of the relationship between gene and metabolic enzyme (13); (ii) computationally predicted relationships between transcription factors and their regulated genes by methods such as binding motif analysis are often not reliable due to the existence of short binding sequence that is not well conserved among organisms (14–16). A combination of genomic information with genome-wide expression data under diverse experimental conditions is often necessary and represents a promising approach (6,8,17–19). Till now, the majority of studies of genome-scale TRN is focused on the two experimentally well-studied model microorganisms: Saccharomyces cerevisiae and Escherichia coli. Information on large-scale TRNs of other organisms is rather limited (14,20). RegulonDB (21) and Ecocyc (22) are the two most prominent databases for E.coli regulatory network that collect information on regulatory relationships through literature study and manual curation to maintain a high-quality data. However, the reconstruction of the E.coli TRN from these databases is not straightforward because the gene–gene regulatory relationships are stored in different files. It is also difficult to compare the data in these two databases since the information is stored in different formats and a different gene nomenclature system is used. Based on RegulonDB and new information from literature, Shen-Orr et al. (10) compiled a list of transcriptional regulatory interactions in E.coli through which one can build the TRN (this network is termed ‘TRN-SO’ in the following parts). This dataset has been used in several recent structural studies of E.coli TRN (10,23,24). These studies have revealed several interesting structural features of TRN. Network motifs, which are considered as the basic building blocks of TRN, have been identified and shown to have important specific functions and implications for the dynamic control of gene regulation (3,10,23,25,26). Dobrin et al. (23) further showed that the two previously identified motif types of TRN (i.e. the feed-forward loop and the bi-fan motifs) aggregate into homologous motif clusters. In our recent study of the E.coli TRN we have revealed a multi-layer hierarchical regulation structure (24). By decomposing this structure, 10 global regulators and 39 functional modules have been identified and shown to have clearly defined biological functions. It is understood that the qualitative significance and quantitative reliability of the results from the structural studies mentioned above may very much depend on the completeness of the network under study. No work has been published so far to address this question.
In an effort to study the gene regulatory network of E.coli with respect to its responses to environmental stress conditions such as iron deficiency and extremely low-pH values, we noticed that several gene regulatory relationships found in literature are not covered in the TRN-SO network (10). This prompted us to examine and extend the regulatory network of E.coli by integrating information from different databases and directly from literature. We then performed a structural and motif analysis of the extended network and compared the results with those previously reported. We not only confirmed certain previous findings such as the multi-layer hierarchical structure, but also found large differences in the number and distribution of network motifs and identified discrepancies in the proposed organization principle of motifs in the network. Overall, our extended TRN of E.coli represents a more solid basis for the structural and functional analysis of genome-wide gene regulation in this microorganism.
METHODS
E.coli TRN from different data sources
We downloaded the databases RegulonDB (version 4.0, http://www.cifn.unam.mx/Computational_Genomics/regulondb/) and Ecocyc (version 8.0, www.ecocyc.org) from the Internet (Ecocyc also got data from RegulonDB; personal communication with Julio Collado-Vides). For the RegulonDB database, we extracted the gene–gene transcriptional regulatory relationships from six files: product_table.dat (relationships between gene and its coded polypeptide product), polyp_prot_link.dat (relationships between polypeptides and proteins), conformation_table.dat (the modified protein conformation), regulatory_interaction.dat (which promoter is regulated by which activated protein), transcription_unit.dat (which transcription unit does an operon belong to) and trans_gene_link.dat (which genes are in which transcription unit). Starting from one gene, we identified all the corresponding genes regulated by it from the information stored in these files, thus making it possible to represent the TRN as a graph in which nodes represent genes and links as transcriptional regulations. Besides these files, we also used the file ‘promoter.dat’ to obtain more regulatory interactions for which the promoters do not belong to any known transcriptional units. In addition, except for sigma 70, all the other sigma factors are regarded as transcription factors and the interactions between them and their regulated genes are also added in the network. In this way, we obtained a network with 1024 genes and 2065 interactions. In contrast, the popularly used TRN-SO network contains only 855 genes and 1330 interactions between the genes (10).
For the database Ecocyc, the gene–gene regulatory relationships were extracted from three files, namely bindrxns.dat (protein–promoter and protein–binding site relationships), transunits.dat (which genes, promoters and binding sites are in a transcriptional unit) and proteins.dat (protein–gene information) respectively. We found that there are many missing links in the database. For example, there are several promoters and binding sites that are regulated by proteins in the ‘bindrxns.dat’ file but not included in the ‘transunits.dat’ file. In this case, we obtained the genes corresponding to the promoters and binding sites directly from the files ‘promoters.dat’ and ‘dnabindsites.dat’. Thus the resulted network from Ecocyc includes 959 genes and 2034 interactions among these genes. The interactions by the alternative sigma factors are also included in this network.
RESULTS AND DISCUSSION
An extended E.coli TRN
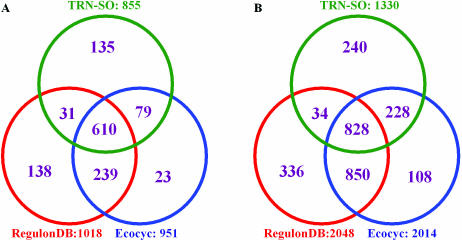
A major problem while comparing the TRN from different data sources is that they often use different gene IDs. RegulonDB assigns a new ECK number for each gene in the E.coli genome which is mainly based on the original genome annotation, the so-called ‘Blattner “b” number system’ (27). However, this annotation has been extensively updated since the genome sequence is completed. A number of genes are deleted or merged and certain new genes are added. A more reliable and up-to-date annotation is the EcoGene database developed by Rudd (28). The EcoGene accession number (EG number) is used in Ecocyc for gene representation. However, there are also a lot of genes in Ecocyc that use a different nomenclature system. For example, many genes have IDs starting with G rather than EG. To avoid this confusion, we mapped all the genes in the regulatory network to genes in the EcoGene database. We found that one gene (b0725, corresponding to ECK120000726 in RegulonDB and G6388 in Ecocyc) has no EG number because it has been removed in the new annotation. There are six pairs of genes that have replicate EG numbers (araH_1,2; gatR_1,2; gntU_1,2; ilvG_1,2; tdcG_1,2; phnE, b4103) because they are merged in the new annotation. By using the consistent gene ID, we compared the three TRNs from different sources for the same organism E.coli K-12. Figure 1 shows the differences in the number of genes and regulatory interactions among the three TRNs reconstructed for E.coli. It should be mentioned that the common genes in the three TRNs shown in Figure 1 make up only half of the total genes, while the common interactions are only about one-third of the total interactions. Therefore integrating information from different resources is important for obtaining a more complete TRN for E.coli. A combined network that includes all the 2624 interactions from the three data sets has been produced. In addition, we further extended this network by adding 23 additional genes and ∼100 regulatory relationships through literature survey. These new regulatory relationships are mainly involved in iron response and acid resistance (29,30). Specifically, a small regulatory RNA, ryhB, and eight genes regulated by it were added in the network (30). We noticed that no small RNA regulated interactions are included in RegulonDB and Ecocyc though many small regulatory RNA have been identified in recent years (31). One possible explanation is that the regulatory mechanisms for most regulatory RNAs are still not clear. Many of them may regulate at posttranscriptional level as antisense RNAs and thus are different from transcription factors that regulate the target genes at transcriptional level.
Figure 1.
Comparison of the TRN of E.coli from three different data sources. (A) The difference in gene content and (B) the difference in regulatory interactions.
Our extended TRN altogether includes 1278 genes and 2724 interactions. Compared with the TRN-SO network, the new network contains one-and-a-half times more genes and more than twice the regulatory interactions. Although it is still not a complete network for the transcriptional regulation in E.coli, it provides more reliability for structural and functional analysis at genome level. Comparison of the structure of the new network with that of the previous one can help us to estimate to what extent the incompleteness of the information affects the results of structural analysis.
Multi-layer hierarchical structure of the extended network
To investigate whether the organizational structure of the combined network is in consistence with the previous TRN-SO network, we analyzed the connectivity structure of the obtained network using methods based on graph theory. A component analysis revealed that there are seven two-gene regulatory loops (A regulates B and B also regulates A) in the new network, which is different from the previous finding based on TRN-SO (24). We found that both genes in the same loop are in the same operon and thus are regulated by the same set of transcription factors. This finding also explains why we did not obtain such loops in previous studies while using operons as nodes (24). Certain pairs of the genes in the loops code for different subunits of a transcriptional regulator such as ihfAB, whereas others may code for antagonistic regulators, which regulate almost the same set of genes. For example, marA codes for a transcription activator of the multiple antibiotic resistance locus and marR codes for a transcription repressor of the same genes. The two genes are in the same operon and both regulate the expression of this operon, resulting in a two gene regulatory loop.
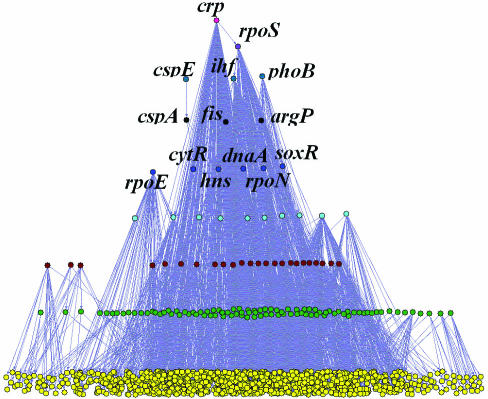
By placing the two genes in a loop at the same layer, we obtained a multi-layer hierarchical structure (Figure 2) similar to that found previously (24). However, nine layers instead of five are in the regulatory hierarchy. This result is not surprising if we consider the fact that the new network includes more interactions among the regulators than the previous one. Among the 14 regulators in the top five layers, six have been identified as global regulators in our previous studies (crp, rpoS, ihf, cspA, hns and rpoN). Four other regulators (phoB, fis, soxR and rpoE) have also been identified as global regulators in three previous papers (5,10,32). DnaA is annotated as ‘initiator protein for DNA synthesis and global transcription regulator’ in Ecogene database (28). Therefore this result supports the conclusion that the top layer regulators tend to be global regulators.
Figure 2.
The multi-layer hierarchical structure of the extended E.coli TRN.
The confirmation of the multi-layer hierarchical structure in the extended TRN strongly implies that it is an underlying structure of the TRN in E.coli. A possible biological explanation for the existence of this hierarchical structure is that the interactions in TRN are between proteins and genes. Only after a regulating gene has been transcribed, translated and eventually further modified by cofactors or other proteins, it can regulate the target gene. A feedback from the regulated gene at transcriptional level may delay the process for the target gene to access a desired expression level in a new environment. Feedback control may be mainly through other interactions (e.g. metabolite and protein interaction) at post-transcriptional level rather than through transcriptional interactions between proteins and genes (24,33). For example, a gene at the bottom layer may code for a metabolic enzyme, the product of which can bind to a regulator which in turn regulates its expression. In this case, the feedback is through metabolite–protein interaction to change the activity of the transcription factor and then to affect the expression of the regulated gene. Therefore, to fully understand the gene expression regulation, an integrated network that includes different interactions is needed.
Network motifs and motif organization
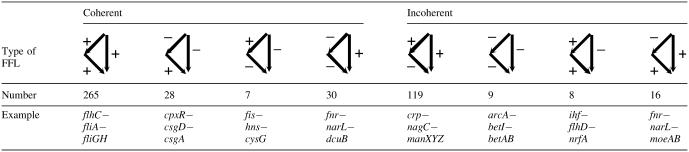
To calculate network motifs in the E.coli TRN, we removed all the loops in the network (including the autoregulatory loops and the two-gene regulatory loops). We then used the program Mfinder developed by Kashtan et al. (34) to generate the motif profiles. In agreement with previous findings, feed-forward loop (FFL) is the only three-node motif (10). There are 712 FFLs in the network, far more than the 42 FFLs found in the TRN-SO network (25). One reason for this large difference is the use of operons rather than genes as nodes in the TRN-SO network. When we use genes as nodes, 162 FFLs are obtained in the TRN-SO network. It is still less than one-fourth of that found in the new network. Regarding the regulatory function (activation or repression), the distribution of the different types of FFLs in the extended E.coli TRN is shown in Table 1 (due to the existence of dual regulation, the sum of the number of the eight types of FFLs are not equal to 712). The first four types are the so-called coherent FFLs in which the direct effect of the up regulator is consistent with its indirect effect through the mid regulator (25). In contrast, the last four types of FFLs are incoherent because the direct effect of the up regulator is contradictive with its indirect effect. The total number of incoherent FFLs is 152, which is only a little less than half of the number of the coherent FFLs (330). This result is inconsistent with the result from the TRN-SO network where only 14 of the 162 FFLs are incoherent (7 of the 42 FFLs are incoherent while using operons as nodes), but very similar to the network of S.cerevisiae (25 of 56 FFLs are incoherent) studied by the same authors (25). Another interesting point is that the first and the fifth FFL types predominate the coherent and incoherent FFLs respectively in both networks of E.coli and S.cerevisiae (25). These results indicate that the distribution and predominance of FFLs in the TRNs of both E.coli and S.cerevisiae have similar patterns.
Table 1. Coherent and incoherent feed forward loops in the E.coli TRN.
We further examined the distribution of different types of FFLs for various regulators. Most of the regulators are found to regulate only one or two types of FFLs. For example, flhDC, lysR, soxS, rob and tdcR mainly regulate type one FFL, while modE regulates type five FFL and cpxR regulates type two FFL. Most of the type four FFLs and the type eight FFLs are regulated by fnr. Mangan et al. (25,35) studied the dynamic behavior of these different types of FFLs. They found that the incoherent FFLs could speed up the responses of the target gene while the coherent FFLs delay the response. The feature of less feedback regulation at transcription level as demonstrated by the multi-layer structure may imply another possible function of incoherent FFLs. By activating a gene and at the same time activating a regulator which represses the target gene, the upper regulators can control the gene expression at a proper level. More in-depth studies may help to examine whether such a mechanism is a general mechanism for gene expression regulation in TRN.
In a recent study, Dobrin et al. (23) showed that network motifs are organized in a hierarchical way in the E.coli regulatory network: first, interacting motifs form motif clusters; motif clusters of different motifs are then connected to make a motif super cluster which is regarded as the backbone of the whole network. However, this concept of network motif organization is not valid for the extended network. We found that 701 of the 712 FFLs are connected to form a giant motif cluster with 435 genes, while the remaining 11 FFLs form four very small clusters. The reason for this large discrepancy is that our new network includes more interactions and thus obtains more motifs that can link the previously disconnected motif clusters together. Therefore, caution should be taken in dealing with results from an incomplete network, especially when drawing conclusions about general organizational principle(s).
Genes regulated by interacting network motifs
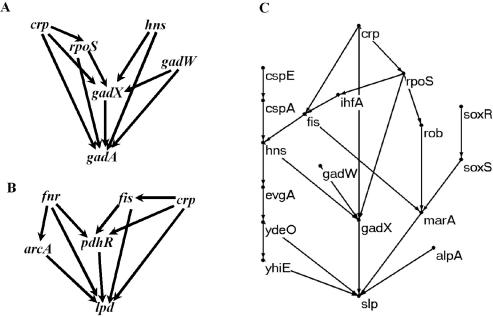
As mentioned above, FFLs are considered to have important functions in controlling the dynamic response of the target gene (25). Therefore it is of interest to check how many genes in the TRN of E.coli are regulated by FFLs and by how many FFLs. We found that there are altogether 400 genes that are regulated by one or more FFLs in the network. Among them, 383 genes are at the bottom layer of the hierarchical structure; they account for only about one-third of the total genes (1121) at the bottom layer. Furthermore, only 56 genes are solely regulated by one FFL. All the other genes are regulated by two or more FFLs or by one FFL together with certain other regulators that do not form a FFL. There are also a few genes (such as cysG, glpAB and nirB) that are regulated by both coherent (delaying the response) and incoherent FFLs (speeding up the response). These results indicate that the previous studies on dynamic behavior of FFLs may be pertinent for only a small part of the genes in the network. It would be interesting to examine the dynamics of target genes that are controlled by several interacting motifs and also by other regulators. Figure 3A and B depict two examples of complex regulatory circuits in which the target gene is regulated by six and five FFLs respectively. In Figure 3A, the target gene gadA codes for glutamate decarboxylase, an important metabolic enzyme in the gamma-aminobutyric acid (GABA) shunt which is important in oxidative stress response of plant cells (36) and bacteria (A.P. Zeng, unpublished data) and is also an important component in the acid resistance system of E.coli (29). In Figure 3B, the target gene lpd codes for lipoamide dehydrogenase which is a component of the pyruvate and 2-oxoglutarate dehydrogenase complexes. Both the pyruvate and 2-oxoglutarate dehydrogenase complexes play key roles in the metabolism of E.coli. lpd also functions as glycine cleavage system L protein. The multiple and important functions of these target genes may explain why they are controlled by several interacting FFLs. In these complex regulatory circuits, the dynamic expression pattern of the target gene will be obviously different from that controlled by one FFL.
Figure 3.
Example of complex regulatory circuits. (A) Gene gadA is regulated by six FFLs; (B) gene lpd is regulated by five FFLs; and (C) gene slp is regulated by 17 regulators.
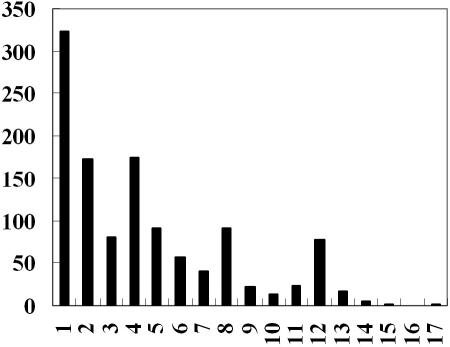
Furthermore, the target gene may also be controlled by regulators not belonging to any FFLs. Figure 4 shows the distribution of the number of regulators that regulate a gene directly or indirectly (the set of these regulators is called the input domain of the target gene in graph theories). A total of 692 genes are regulated by more than two regulators. The most complex regulatory circuit is the one for the gene slp, which codes for an outer membrane lipoprotein induced under carbon starvation and stationary phase (28). It is regulated by 17 regulators as shown in Figure 3C. These regulators participate in cellular responses to various environmental conditions, such as oxidative stress (soxRS), acid stress (gadW, gadX, evgA, ydeO and yhiE), cold shock (cspE, cspA) and multiple antibiotic resistance (marA). This underlies the importance of this gene in stress response. Further studies are required to elucidate the exact function of this gene. Although slp is regulated by only one FFL (ydeO-yhiE-slp), it is in fact also regulated by other FFLs that contain more than three genes. For example, fis-hns-evgA-ydeO-marA-slp forms a six node FFL, while rpoS-rob-marA-gadX-slp makes a five node FFL. One common feature of these FFLs (including the three node FFL) is that one top regulator regulates a target gene through different pathways. Further studies are required to understand the dynamic response of genes under the control of different loops and different regulators. More care should be taken when studying gene expression dynamics in these complex regulatory circuits. It may be completely different from that of a simple three node FFL. Furthermore, only a subset of the regulators (and their regulatory interactions) in the whole network is activated under given environmental conditions as recently shown by Luscombe et al. (2). Therefore, for the complex regulatory circuits shown in Figure 3, it is possible that only one or two motifs are active at a given time. Further studies are required to investigate the dynamic change of the network topology and its effects on gene expression dynamics. Network motif analysis has provided useful information about the gene regulation patterns. It would be interesting to know the dynamic feature and the overall effect of individual motifs if they interact with other motifs within a more complex network and in a dynamically changed environment. The complex regulatory circuits as shown in Figure 3 may be good examples for such studies.
Figure 4.
The distribution of number of regulators (directly or indirectly) for the genes in the E.coli TRN.
Recently, van Nimwegan (37) found by comparative genome analysis that the number of genes in each functional category scales as a power-law of the total number of genes in the genome. Specifically, the exponent for transcriptional regulators is almost two, implying that the number of transcription factors increases much faster than the size of the genome (quadruple transcription factors in a doubled genome). This finding was further verified by Ranea et al. (38) who investigated the distribution of protein superfamilies in 56 different bacterial species. The non-linear scaling observed by these authors could be explained by an increase in complex inter-regulation of transcription factors as illustrated in the examples in Figure 3 and by the multi-layer hierarchical structure shown in Figure 2. The inter-regulation among the transcription factors can lead to a faster growing number of regulators than the number of genes with increase of the genome size.
CONCLUSIONS
We generated a more complete TRN of E.coli by combining information from three different data sources (RegulonDB, Ecocyc and TRN-SO) and literature survey. Only a relatively small part of the regulatory interactions is found to be common in all the three datasets, indicating the importance of data integration for obtaining a more complete and reliable network. Structural analysis of the extended network reveals both the consistency and inconsistency found within results obtained from the network of Shen-Orr et al. recently used in several studies. The new network preserves the multi-layer hierarchical structure but has more layers because of more interactions inside the network. FFLs still remain the only three-node network motif in the network but the number of FFLs increases greatly with the size of the network. The distribution of the different types of FFLs is also different from that derived from the TRN-SO network. Most of the FFLs are connected to form a giant motif cluster instead of forming several small disconnected clusters. Furthermore, only a small portion of the genes is solely regulated by only one FFL. Many of the genes are regulated by two or more interacting FFLs or other more complicated network motifs together with transcriptional factors not belonging to any network motifs. These results underline the importance of having a more complete and reliable network in the study of structure and function of transcriptional regulation of gene expression.
Acknowledgments
ACKNOWLEDGEMENTS
This work was financially supported through the Braunschweig Bioinformatic Competence Center project ‘Intergenomics’ of the Ministry for Education and Research (BMBF), Germany (Grant No. 031U110A) and by the project B6 in the Sonderforschungsbereich 578 der Deutschen Forschungsgemeinschaft (DFG).
REFERENCES
- 1.Herrgard M.J. and Palsson,B.O. (2004) Flagellar biosynthesis in silico: building quantitative models of regulatory networks. Cell, 117, 689–690. [DOI] [PubMed] [Google Scholar]
- 2.Luscombe N.M., Babu,M.M., Yu,H., Snyder,M., Teichmann,S.A. and Gerstein,M. (2004) Genomic analysis of regulatory network dynamics reveals large topological changes. Nature, 431, 308–312. [DOI] [PubMed] [Google Scholar]
- 3.Milo R., Shen-Orr,S., Itzkovitz,S., Kashtan,N., Chklovskii,D. and Alon,U. (2002) Network motifs: simple building blocks of complex networks. Science, 298, 824–827. [DOI] [PubMed] [Google Scholar]
- 4.Babu M.M., Luscombe,N.M., Aravind,L., Gerstein,M. and Teichmann,S.A. (2004) Structure and evolution of transcriptional regulatory networks. Curr. Opin. Struct. Biol., 14, 283–291. [DOI] [PubMed] [Google Scholar]
- 5.Martinez-Antonio A. and Collado-Vides,J. (2003) Identifying global regulators in transcriptional regulatory networks in bacteria. Curr. Opin. Microbiol., 6, 482–489. [DOI] [PubMed] [Google Scholar]
- 6.Herrgard M.J., Covert,M.W. and Palsson,B.O. (2003) Reconciling gene expression data with known genome-scale regulatory network structures. Genome Res., 13, 2423–2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bar-Joseph Z., Gerber,G.K., Lee,T.I., Rinaldi,N.J., Yoo,J.Y., Robert,F., Gordon,D.B., Fraenkel,E., Jaakkola,T.S., Young,R.A. et al. (2003) Computational discovery of gene modules and regulatory networks. Nat. Biotechnol., 21, 1337–1342. [DOI] [PubMed] [Google Scholar]
- 8.Gutierrez-Rios R.M., Rosenblueth,D.A., Loza,J.A., Huerta,A.M., Glasner,J.D., Blattner,F.R. and Collado-Vides,J. (2003) Regulatory network of Escherichia coli: consistency between literature knowledge and microarray profiles. Genome Res., 13, 2435–2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guelzim N., Bottani,S., Bourgine,P. and Kepes,F. (2002) Topological and causal structure of the yeast transcriptional regulatory network. Nature Genet., 31, 60–63. [DOI] [PubMed] [Google Scholar]
- 10.Shen-Orr S.S., Milo,R., Mangan,S. and Alon,U. (2002) Network motifs in the transcriptional regulation network of Escherichia coli. Nature Genet., 31, 64–68. [DOI] [PubMed] [Google Scholar]
- 11.Lee T.I., Rinaldi,N.J., Robert,F., Odom,D.T., Bar-Joseph,Z., Gerber,G.K., Hannett,N.M., Harbison,C.T., Thompson,C.M., Simon,I. et al. (2002) Transcriptional regulatory networks in Saccharomyces cerevisiae. Science, 298, 799–804. [DOI] [PubMed] [Google Scholar]
- 12.Kao K.C., Yang,Y.L., Boscolo,R., Sabatti,C., Roychowdhury,V. and Liao,J.C. (2004) Transcriptome-based determination of multiple transcription regulator activities in Escherichia coli by using network component analysis. Proc. Natl Acad. Sci. USA, 101, 641–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma H.W. and Zeng,A.P. (2003) Reconstruction of metabolic networks from genome data and analysis of their global structure for various organisms. Bioinformatics, 19, 270–277. [DOI] [PubMed] [Google Scholar]
- 14.Herrgard M.J., Covert,M.W. and Palsson,B.O. (2004) Reconstruction of microbial transcriptional regulatory networks. Curr. Opin. Biotechnol., 15, 70–77. [DOI] [PubMed] [Google Scholar]
- 15.Alkema W.B., Lenhard,B. and Wasserman,W.W. (2004) Regulog analysis: detection of conserved regulatory networks across bacteria: application to Staphylococcus aureus. Genome Res., 14, 1362–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu H., Luscombe,N.M., Lu,H.X., Zhu,X., Xia,Y., Han,J.D., Bertin,N., Chung,S., Vidal,M. and Gerstein,M. (2004) Annotation transfer between genomes: protein–protein interologs and protein–DNA regulogs. Genome Res., 14, 1107–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ihmels J., Friedlander,G., Bergmann,S., Sarig,O., Ziv,Y. and Barkai,N. (2002) Revealing modular organization in the yeast transcriptional network. Nature Genet., 31, 370–377. [DOI] [PubMed] [Google Scholar]
- 18.Segal E., Shapira,M., Regev,A., Pe'er,D., Botstein,D., Koller,D. and Friedman,N. (2003) Module networks: identifying regulatory modules and their condition-specific regulators from gene expression data. Nature Genet., 34, 166–176. [DOI] [PubMed] [Google Scholar]
- 19.Yu H., Luscombe,N.M., Qian,J. and Gerstein,M. (2003) Genomic analysis of gene expression relationships in transcriptional regulatory networks. Trends Genet., 19, 422–427. [DOI] [PubMed] [Google Scholar]
- 20.Matys V., Fricke,E., Geffers,R., Gossling,E., Haubrock,M., Hehl,R., Hornischer,K., Karas,D., Kel,A.E., Kel-Margoulis,O.V. et al. (2003) TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res., 31, 374–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Salgado H., Gama-Castro,S., Martinez-Antonio,A., Diaz-Peredo,E., Sanchez-Solano,F., Peralta-Gil,M., Garcia-Alonso,D., Jimenez-Jacinto,V., Santos-Zavaleta,A., Bonavides-Martinez,C. et al. (2004) RegulonDB (version 4.0): transcriptional regulation, operon organization and growth conditions in Escherichia coli K-12. Nucleic Acids Res., 32, D303–D306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karp P.D., Riley,M., Saier,M., Paulsen,I.T., Collado-Vides,J., Paley,S.M., Pellegrini-Toole,A., Bonavides,C. and Gama-Castro,S. (2002) The EcoCyc Database. Nucleic Acids Res., 30, 56–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dobrin R., Beg,Q.K., Barabasi,A.L. and Oltvai,Z.N. (2004) Aggregation of topological motifs in the Escherichia coli transcriptional regulatory network. BMC Bioinformatics, 5, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma H.W., Buer,J. and Zeng,A.P. (2004) Hierarchical structure and modules in the Escherichia coli transcriptional regulatory network revealed by a new top-down approach. BMC Bioinformatics, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mangan S. and Alon,U. (2003) Structure and function of the feed-forward loop network motif. Proc. Natl Acad. Sci. USA, 100, 11980–11985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wolf D.M. and Arkin,A.P. (2003) Motifs, modules and games in bacteria. Curr. Opin. Microbiol., 6, 125–134. [DOI] [PubMed] [Google Scholar]
- 27.Blattner F.R., Plunkett,G.,III, Bloch,C.A., Perna,N.T., Burland,V., Riley,M., Collado-Vides,J., Glasner,J.D., Rode,C.K., Mayhew,G.F. et al. (1997) The complete genome sequence of Escherichia coli K-12. Science, 277, 1453–1474. [DOI] [PubMed] [Google Scholar]
- 28.Rudd K.E. (2000) EcoGene: a genome sequence database for Escherichia coli K-12. Nucleic Acids Res., 28, 60–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Masuda N. and Church,G.M. (2003) Regulatory network of acid resistance genes in Escherichia coli. Mol. Microbiol., 48, 699–712. [DOI] [PubMed] [Google Scholar]
- 30.Masse E. and Gottesman,S. (2002) A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc. Natl Acad. Sci. USA, 99, 4620–4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gottesman S. (2004) The small RNA regulators of Escherichia coli: roles and mechanisms. Annu. Rev. Microbiol., 58, 303–28., 303–328. [DOI] [PubMed] [Google Scholar]
- 32.Madan B.M. and Teichmann,S.A. (2003) Evolution of transcription factors and the gene regulatory network in Escherichia coli. Nucleic Acids Res., 31, 1234–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wall M.E., Hlavacek,W.S. and Savageau,M.A. (2004) Design of gene circuits: lessons from bacteria. Nature Rev. Genet., 5, 34–42. [DOI] [PubMed] [Google Scholar]
- 34.Kashtan N., Itzkovitz,S., Milo,R. and Alon,U. (2004) Efficient sampling algorithm for estimating subgraph concentrations and detecting network motifs. Bioinformatics, 20, 1746–1758. [DOI] [PubMed] [Google Scholar]
- 35.Mangan S., Zaslaver,A. and Alon,U. (2003) The coherent feed forward loop serves as a sign-sensitive delay element in transcription networks. J. Mol. Biol., 334, 197–204. [DOI] [PubMed] [Google Scholar]
- 36.Bouche N. and Fromm,H. (2004) GABA in plants: just a metabolite? Trends Plant Sci., 9, 110–115. [DOI] [PubMed] [Google Scholar]
- 37.van Nimwegen E. (2003) Scaling laws in the functional content of genomes. Trends Genet., 19, 479–484. [DOI] [PubMed] [Google Scholar]
- 38.Ranea J.A., Buchan,D.W., Thornton,J.M. and Orengo,C.A. (2004) Evolution of protein superfamilies and bacterial genome size. J. Mol. Biol., 336, 871–887. [DOI] [PubMed] [Google Scholar]