Abstract
Errors during the pre-mRNA splicing of metazoan genes can degrade the transmission of genetic information, and have been associated with a variety of human diseases. In order to characterize the mutagenic and pathogenic potential of mis-splicing, we have surveyed and quantified the aberrant splice variants in the human hypoxanthine phosphoribosyl transferase (HPRT) and DNA polymerase β (POLB) in the presence and the absence of the Nonsense Mediated Decay (NMD) pathway, which removes transcripts with premature termination codons. POLB exhibits a high frequency of splice variants (40–60%), whereas the frequency of HPRT splice variants is considerably lower (∼1%). Treatment of cells with emetine to inactivate NMD alters both the spectrum and frequency of splice variants of POLB and HPRT. It is not certain at this point, whether POLB and HPRT splice variants are the result of regulated alternative splicing processes or the result of aberrant splicing, but it appears likely that at least some of the variants are the result of splicing errors. Several mechanisms that may contribute to aberrant splicing are discussed.
INTRODUCTION
The integrity of genetic information is of critical importance to organisms. However, it is not simply the accumulation of mutations in genes that matters; it is becoming increasingly clear that there are serious phenotypic consequences associated with reductions in the fidelity of transmission of genetic information as it flows from DNA. In addition to genetic mutations, errors in the epigenetic processes that affect this fidelity, such as methylation, transcription, RNA processing and translation, can interfere with the steps that transmute genotype into phenotype (1).
In metazoa, an epigenetic process that is crucial in maintaining a normal flow of genetic information is the processing of primary RNA transcripts prior to translation, including the removal of intronic sequences, a process referred to as pre-mRNA splicing (hereafter, splicing). The splicing reaction is carried out by the spliceosome, a collection of five small nuclear RNAs and >145 proteins (2,3). The fidelity of splicing is dependent upon an intricate network of RNA–RNA, RNA–protein and protein–protein interactions, in order to identify the correct splice sites at the exon–intron junction in the context of numerous ‘cryptic’ splice sites that resemble the consensus sequences (4). Moreover, different splice sites must be selected during alternative splicing that plays an important role in regulating the temporal and spatial expression of a large number of genes (5). Indeed the spliceosome has been referred to as the most complex cellular machine ever characterized (3).
Remarkably, little attention has been focused on the error rate of the splicing process and its mutagenic and pathogenic potential. Splice variants are frequently observed in genetic studies but they are assumed to be products of legitimate alternative splicing. However, it has become evident that some splice variants are associated with human pathologies, such as cancer (6–8), Alzheimer's (9,10), amyotrophic lateral sclerosis (11), ataxia telangiectasia (12), cystic fibrosis (13) and other diseases (14,15). The relationship between splice variants and disease or senescence has been reviewed recently (14,16).
Clearly, to fully appreciate the role of splice variants in disease, many questions need to be investigated. What is the frequency of splice variants in normal, healthy cells? Are most splice variants generated by a regulated process or are they generated by splicing errors? What are the molecular mechanisms that contribute to splicing errors? What types of endogenous and exogenous processes can interfere with accurate splicing? What is the genotoxic and pathogenic potential of splice variants?
In order to address these questions, we have previously characterized exon skipping in the hypoxanthine phosphoribosyl transferase (HPRT) gene in primary fibroblasts (17). The study yielded preliminary evidence, which suggests that besides the splice variants observed, additional variants could be generated, but were removed by the Nonsense Mediated Decay (NMD) pathway. NMD detects and removes transcripts with termination codons >50–55 nt upstream from the last exon–exon junction (Premature Termination Codons, PTCs) (18).
In this paper, we report on our studies to estimate the frequency of HPRT and DNA polymerase β (POLB) splice variants in the transformed human lymphoblastoid cell line TK6, the untransformed human lung fibroblast cell line MRC5, and the untransformed human skin fibroblast cell line AG08906 in the presence or absence of the NMD pathway inhibitor emetine (19). Furthermore, we discuss some of the mechanisms that can give rise to splice variants.
METHODS
Cell description, source and growth
The following cells were used in the study: (i) MRC5 cells are untransformed normal fetal lung fibroblasts obtained from the Coriell Cell Repositories (Repository number AG05965). All reagents were obtained from Sigma Canada. The cells were grown in Eagle's minimal essential medium with Hank's BSS, 26 mM HEPES, 10% uninactivated fetal bovine serum, 2 mM l-glutamine at 37°C in 5% CO2. The medium was supplemented with 1 × 10−1 mM hypoxanthine, 4 × 10−4 mM aminopterin and 1.6 × 10−2 mM thymidine to ensure that the cells maintained a functional HPRT gene. (ii) AG08906 cells are untransformed skin fibroblasts isolated in the course of Baltimore Longitudinal Study on Aging. They were obtained from Coriell Cell Repositories and were grown as MRC5. (iii) TK6 lymphoblastoid cells were a gift from Dr Howard Liber. The cells were grown in RPMI 1640 supplemented with 10% heat-inactivated horse serum (Sigma) at 37°C in 5% CO2.
RNA preparation and reverse transcription
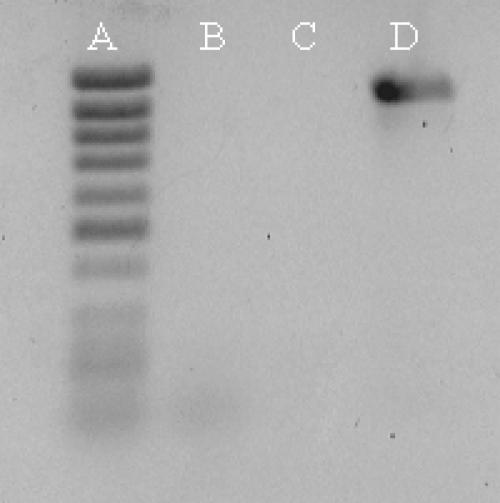
Total RNA was isolated from 5 × 106 cells using the GenElute total RNA miniprep kit (Sigma). HeLa cell RNA was also obtained from GibcoBRL. First strand synthesis from 1 μg of total RNA was performed using the Thermoscript RT–PCR kit (GibcoBRL) and an oligo(dT)22 primer. Control reactions were performed in the absence of reverse transcriptase to exclude the possibility that genomic DNA contamination of our RNA preparations can template amplification products (Figure 1).
Figure 1.
POLB amplification from different TK6-derived templates. Lane A shows the size marker. Lane B shows lack of amplification from mock cDNA reaction without RT. Lane C shows lack of amplification from genomic DNA. Lane D shows successful amplification from oligo(dT) primed cDNA template.
Detection and quantification of splice variants by quantitative PCR
The presence of specific POLB and HPRT transcripts with missing exons was probed using PCR primers complementary to the exon junction sequences generated by exon-skipping as described previously (17). The frequency of the splice variants relative to wild-type (WT) transcripts was estimated using quantitative PCR. The PCR conditions were as follows: Eppendorf 1× buffer, 0.15 mM MgCl2, 0.2 mM dNTPs, 1 U Qiagen Taq polymerase and 200 nM of each primer, one of which was radioactively labelled with γ-32P. The template concentration used was one-tenth of the first strand sequence reaction (equivalent to 200 ng of total RNA). Each amplification cycle included a denaturation step (92°C for 30 s), an annealing step (56.5°C for 30 s) and an extension step (74.8°C for 50 s). The PCR was performed in an Eppendorf gradient apparatus. Amplification of the WT transcripts and transcripts with a skipped exon was carried out separately for various number of cycles and the reaction products were separated in a 6% non-denaturing polyacrylamide gel. The amount of product was then quantified using a fluorescent image analyzer (Fuji FL3000G). The data were analyzed to ensure that amplification was within the exponential range, were mathematically transformed in order to be linearized, and the initial template concentration was calculated using linear regression. A cloned copy of the entire HPRT or POLB coding sequence was used as a template in all reactions as a control for non-specific amplification. Each quantification experiment was repeated three times. The York University Core Molecular Biology and DNA Sequencing Facility performed all sequencing.
Detection and quantification of splice variants by transcript cloning and sequencing
Transcript cloning involved the reverse transcription of poly(A) mRNAs to cDNA, the amplification of transcripts using gene specific primers, and the cloning of the amplified products into vectors. Oligo(dT)_ primed cDNA, synthesized from total RNA as described above, was amplified using gene-specific primers. The PCR product was purified using spin columns (microcon-PCR; Millipore) and it was then ligated into the vector pGEM-T Easy kit (Promega). Blue/white colony screening was performed as per vendor's instructions and each white colony was then picked into 100 μl of distilled water and heated to 97°C to lyse. The lysed colony was then used as a template (2 μl) for the amplification of POLB or HPRT sequences. PCR products were then analyzed by electrophoresis and sequencing.
RESULTS AND DISCUSSION
POLB and HPRT as indicators for splice variant diversity
POLB is an excellent indicator locus for studying splice variants for several reasons. POLB is considered a ‘housekeeping’ gene with an even expression profile of ∼6 mRNA molecules per cell throughout the cell cycle (20). POLB is, however, inducible under certain circumstances such as oxidative stress, or caloric restriction (21,22). Multiple POLB splice variants have been observed in many cell types, including cancer cells (23,24). Some of these variant transcripts may be translated into protein, which could then compete with the WT protein as dominant-negative mutants. Indeed, there have been reports that a POLB splice variant missing exon 11, codes for a dominant-negative protein that may be present with elevated frequency in some tumors (24,25). Since POLB is a key enzyme in DNA repair, any perturbations in its expression or function can lead to increased mutation frequency and genomic instability. In order to be able to detect pathology-associated changes in the splicing patterns, it is necessary to survey splice patterns in healthy cells of different lineages. Consequently, we have characterized POLB transcripts in TK6 lymphoblastoid cells and MRC5 fibroblasts, using both transcript cloning and splice variant-specific quantitative PCR.
HPRT also offers several advantages as a genetic marker for the investigation of splicing fidelity. It is located in the X chromosome and therefore, human cells are functionally hemizygotic for the locus. It is also considered a ‘house-keeping’ gene, with an even expression profile, but unlike POLB it has not been shown to be subject to expression induction. Moreover, selection schemes exist for both the presence and absence of functional HPRT alleles, thus, enabling the elimination of cells with DNA mutations that affect splicing. Finally, HPRT has been used extensively as a mutagenesis indicator locus, so there exists a large database of mutations and a great deal of information about splice mutants (26).
POLB splice variants in TK6, MRC5 and AG08906 cells
In TK6 cells, sequence analysis of 87 POLB cloned transcripts revealed that 59.8% were WT, with the remaining 40.1% of the transcripts comprising eight different splice variants, generated by either intron inclusion or exon skipping events (Table 1). Among the transcripts that retained intronic sequences was a splice variant retaining 105 nt of intron 6 (Table 2). This variant has been previously observed and the intronic sequence has been termed exon α (23). This splice variant represented 4.6% of POLB transcripts observed, and is capable of encoding a protein which, however, does not have a polymerase function (23). The most frequently observed splice variant was exon 2 skipping, which represented 13.8% of all transcripts. Exon 11 skipping represented 10.4% of all transcripts and those two variants jointly accounted for over 60% of the observed splice variants.
Table 1. POLB splice variants in TK6 cells characterized by transcript cloning.
| POLB splice variants | PTC | Putative protein length (amino acids) | Splice variant frequency (%)a | Splice variant frequency relative to WT frequency (%)b | ||
|---|---|---|---|---|---|---|
| Untreated TK6 | Emetine-treated TK6 | Untreated TK6 | Emetine-treated TK6 | |||
| Intron retention | ||||||
| Σ exon α | No | 370 | 4.6 | 4.7 | 7.7 | 9.5 |
| Σ exon β | Yes | 193 | 0 | 11.8 | 0 | 23.8 |
| Σ exons α and β | Yes | 228 | 0 | 2.4 | 0 | 4.8 |
| WT | No | 335 | 59.8 | 49.4 | 100 | 100 |
| Exon skipping | ||||||
| Δ exon 2 | Yes | 26 | 13.8 | 9.4 | 23.1 | 19.0 |
| Δ exons 2 and 3 | Yes | 28 | 0 | 1.2 | 0 | 2.4 |
| Δ exons 2 and 11 | Yes | 26 | 0 | 1.2 | 0 | 2.4 |
| Δ exons 2, 3 and 11 | Yes | 28 | 0 | 4.7 | 0 | 9.5 |
| Δ exons 2, 11–13 | Yes | 26 | 1.1 | 0 | 1.9 | 0 |
| Δ exons 2–13 | No | 51 | 0 | 1.2 | 0 | 2.4 |
| Δ exon 4 | No | 310 | 1.1 | 0 | 1.9 | 0 |
| Δ exons 4 and 5 | Yes | 96 | 0 | 1.2 | 0 | 2.4 |
| Δ exons 4–11 | No | 161 | 2.3 | 0 | 3.8 | 0 |
| Δ exons 4–11 and 13 | No | 93 | 0 | 1.2 | 0 | 2.4 |
| Δ exons 4–13 | No | 115 | 0 | 2.4 | 0 | 4.8 |
| Δ exon 11 | No | 306 | 10.3 | 5.9 | 17.3 | 11.9 |
| Δ exons 11–13 | No | 261 | 2.3 | 2.4 | 3.8 | 4.8 |
| Δ exons 12 and 13 | No | 290 | 4.6 | 0 | 7.7 | 0 |
aThis value is calculated according to the formula frequency = (number of observations of the splice variant/total number of transcripts analyzed) × 100.
bThis value is calculated according to the formula frequency = (number of observations of the splice variant/number of observations of the WT) × 100.
Table 2. Retained intronic sequences in POLB splice variants.
| Name | Sequence (numbering according to GeneBank entry AF491812) |
|---|---|
| Exon α (partial intron 6) | ag-17032-GCT CAC AGC TGG ATT CAT GCC CAG TAA AGG GAC ACC TGA ATG GAA CTG AGT CAC TTT TAG ACT TAA TAT GGG ATG TTA TGA CAA TTC TTA AGT TAA AAA ATG CAG-17136 |
| Exon β (partial intron 9) | ag-23714-ATT CTG CTG TCT ACA TCA ATA CAC CTG AAT AGT TGG ACA GAA AAT TGA AAT CTT TTA ACT AAT TCT AAC TAT GAA GCA CAG TGA AAT AGA AAG TTA GGC TGT AAG AA-23820 |
Similar types of splice variants were identified in the sequence analysis of 76 POLB transcripts in MRC5 cells (Table 3). The frequency of WT transcripts was 44.7%, which is lower than the WT frequency in TK6 cells. With the exception of exon 4 to 10 skip, which was only detected in MRC5, the splice variants observed in MRC5 were also detectable in TK6. The lower frequency of WT transcripts was accompanied by a significantly elevated frequency of exon 2 skip. (Fisher Exact Test, P = 0.0233 for exon 2 skip.) High frequency of exon 2 skip was also observed in the analysis of 56 POLB transcripts in another untransformed fibroblast cell line, AG08906 (Table 4). In AG08906, exon 2 skip accounted for 32.1% of the transcripts. It is not clear at this point whether the higher frequency of exon 2 skip in the untransformed skin and lung fibroblasts relative to TK6 is due to the differences in tissue type or the transformation status.
Table 3. POLB splice variants in MRC5 cells characterized by transcript cloning.
| POLB splice variants | PTC | Putative protein length (amino acids) | Splice variant frequency (%) | Splice variant frequency relative to WT frequency (%) | ||
|---|---|---|---|---|---|---|
| Untreated MRC5 | Emetine-treated MRC5 | Untreated MRC5 | Emetine-treated MRC5 | |||
| Intron retention | ||||||
| Σ exon α | No | 370 | 6.6 | 5.4 | 14.7 | 14.3 |
| Σ exon β | Yes | 193 | 0 | 8.1 | 0 | 21.4 |
| Σ exons α and β | Yes | 228 | 0 | 2.7 | 0 | 4.8 |
| WT | NO | 335 | 44.7 | 37.8 | 100 | 100 |
| Exon skipping | ||||||
| Δ exon 2 | Yes | 26 | 26.3 | 18.9 | 58.8 | 50.0 |
| Δ exons 2, 3 and 11 | Yes | 28 | 0 | 5.4 | 0 | 14.3 |
| Δ exons 2, 11–13 | Yes | 26 | 0 | 5.4 | 0 | 14.3 |
| Δ exon 4 | No | 310 | 3.9 | 2.7 | 8.8 | 7.1 |
| Δ exons 4–10 | No | 190 | 1.3 | 0 | 2.9 | 0 |
| Δ exons 4–13 | No | 115 | 0 | 2.7 | 7.1 | |
| Δ exon 11 | No | 306 | 14.5 | 8.1 | 32.4 | 21.4 |
| Δ exons 11–13 | No | 261 | 2.6 | 2.7 | 5.9 | 7.1 |
Please see notes in Table 1.
Table 4. POLB splice variants in AG08906 cells characterized by transcript cloning.
| POLB splice variants | PTC | Splice variant frequency (%) | Splice variant frequency relative to WT frequency (%) |
|---|---|---|---|
| Intron retention | |||
| Σ exon α | No | 8.9 | 23.8 |
| WT | No | 37.5 | 100 |
| Exon skipping | |||
| Δ exon 2 | Yes | 32.1 | 85.7 |
| Δ exon 4 | No | 7.1 | 19.0 |
| Δ exon 11 | No | 10.7 | 28.6 |
| Δ exons 11–13 | No | 3.6 | 9.5 |
Please see notes in Table 1.
To eliminate the possibility that a DNA mutation was responsible for the skipping of exon 2, we sequenced the genomic sequences of intron 1 and exon 2 of 31 different MRC5, TK6, and, AG08906 isolates. Indeed, there were no genomic DNA mutations detected.
Analysis of the exons skipped in the splice variants revealed a distinct pattern. Most of the splice variants were generated by skipping exons 2, 4 or 11, alone or in combination with other downstream exons. This implies that the majority of splice variants are generated by the use of alternative acceptor splice sites instead of those at introns 1, 3 or 10. Consequently, we investigated whether the strength of these splice sites could account for the observed pattern and the results will be discussed below.
Interestingly, exons 4 and 11 are the only exons that can be skipped without generating a premature termination codon (PTC). In fact, only two of the eight splice variants observed, contain PTCs. Both exceptions involved exon 2 skipping, which generates a frameshift and a PTC after encoding 26 amino acids. This would imply that the exon 2-skip splice variant may evade the NMD pathway by some not yet known mechanism. It has been suggested to us that the transcript may evade detection by translation re-initiation downstream of exon 1 as seen in other genes (C. Valentine, personal communication). The most likely alternative translation initiation site is 463 nt downstream of the legitimate initiation site in exon 8. Re-initiation from this site would result in a truncated protein of 181 amino acids, missing the N-terminus including the 8 kDa domain. Alternatively, there may simply be a read-through of the PTC generated by exon 2 skipping, achieved by codon skipping, frame-shifting or utilization of suppressor tRNA (J. Kranz, personal communication).
Since most of the POLB splice variants detected do not contain PTCs, the question arises whether the observed pattern of splice variants is the result of a regulated process or the effect of the action of the NMD, which removes most splice variants with PTCs. To address this question, we investigated POLB splice variants in TK6 and MRC5 treated with emetine that blocks translation and, therefore, abolishes the translation-dependent NMD pathway.
Sequence analysis of 85 POLB transcripts from emetine-treated TK6 cells, revealed that in the absence of NMD, the spectrum of splice variants was different from that in the untreated cells. The relative frequency of the WT transcript was reduced from 59.1 to 49.4%, and most importantly, new splice variants containing PTCs were now detectable. Two of the new splice variants are involved in intron retention. A splice variant was observed retaining nucleotides 23714–23820 of intron 9 based on the numbering of POLB GenBank entry AF491812. We termed this sequence exon β, according to previous practice. Exon β inclusion is the most frequent (11.8%) splice variant in TK6 with inactivated NMD and this is significantly different inform the NMD proficient TK6 (Fisher Exact Test, P = 0.0016). Exon β was also recovered in a splice variant in combination with exon α. Unlike exon α, exon β inclusion changes the reading frame and introduces a PTC. This would explain why this splice variant is undetectable in NMD proficient cells. As in TK6, analysis of 31 POLB transcripts from emetine-treated MRC5 revealed an increased frequency of splice variants containing PTC (Table 3), indicating that the phenomenon is not tissue specific.
Overall, emetine treatment revealed several splice variants containing PTC, but it does not seem to significantly affect the relative frequency of the most prominent splice variants. In TK6, there were seven splice variants with PTC observed in emetine-treated cells versus two in the untreated cells, both involving exon 2 skip (Tables 1 and 3). This is most likely because emetine inactivates NMD and reveals splice variants normally destroyed. We cannot formally exclude the possibility that emetine treatment affects the splicing pattern of POLB, but we consider this extremely unlikely since the new variants all contain PTC. The spectrum of splice variants without PTC was different in emetine-treated and untreated TK6. Of the nine such variants observed in TK6, only three were found in both (Table 1). However, none of the differences was statistically significant. Considering that most of these splice variants were relatively rare, the different splice patterns of non-PTC containing variants suggests that the diversity of POLB splice variants is even greater than our sampling was able to characterize and thus an exhaustive cataloging of POLB splice variants would require the analysis of a huge number of transcripts. It should be noted that, some of the transcripts observed repeatedly in a study investigating POLB splice variants in bladder cancer study were not detected in our study, even though we characterized many more splice variants (24). The splice variants observed in bladder cancer often contained PTC, and although the authors do not comment on it these observations suggest that NMD defects may play a role in bladder cancer.
As indicated above, it is likely that the spectrum of POLB spice variants is very broad and includes rare transcripts with combinations of multiple exon skipping and intron retention. Consequently, we do not believe that our survey exhaustively detected all POLB splice variants generated even in a single cell type as TK6. In order to ensure that we have detected all POLB splice variants generated by single exon skipping in TK6, we used specific primers and quantitative PCR.
The only POLB variants detected with this method were exon 2 skip, exon 4 skip and exon 11 skip with frequency relative to WT frequency of 19 ± 1%, 2 ± 2%, and 8 ± 1%, respectively, all previously observed with comparable frequencies using transcript cloning. It is important to note that the splice variant frequency calculated using the quantitative PCR method is relative to the WT transcript frequency and not relative to the total number of transcripts as in transcript cloning. If there are several splice variants generated as in POLB the difference is significant. Consequently, the quantitative PCR method yields less information than direct transcript cloning. Nonetheless, as it is clear below, quantitative PCR is very useful in that it can easily detect relatively rare transcripts, whereas direct cloning would require the analysis of thousands of clones for equivalent sensitivity.
HPRT splice variants in TK6 and MRC5 cells
In TK6 cells, sequence analysis of 100 POLB cloned transcripts revealed that 99 were WT and one (1%) was a splice variant missing exons 2 and 3 (Table 5). Similar analysis of 109 transcripts in MRC5 revealed one splice variant missing exon 8 (0.9%). None of the splice variants contained PTCs. It is evident that the frequency of HPRT splice variants is much lower than POLB.
Table 5. Frequency of HPRT splice variants characterized by transcript cloning in TK6.
| Cell type | HPRT splice variants | Splice variant frequency (%) | Splice variant frequency relative to WT frequency (%) | PTC |
|---|---|---|---|---|
| TK6 | WT | 99.0 | 100 | No |
| Δ exons 2 and 3 | 1.0 | 1.0 | No | |
| MRC5 | WT | 99.1 | 100 | No |
| Δ exon 8 | 0.9 | 0.9 | No |
The reduced frequency of HPRT splice variants makes transcript cloning an unproductive methodology for the study of splice variants. Consequently, HPRT splice variants were studied using quantitative PCR and specific primers, taking advantage of the extensive available information about HPRT splice sites, and our previous work in characterizing HPRT splice variants in primary fibroblasts (17).
HPRT has been frequently used in mutagenesis studies and there exists a large mutation database (26). Analysis of the database enabled the detection of six cryptic splice sites, which are used by the spliceosome when the legitimate sites are damaged by mutations (26). Use of the cryptic sites results in transcripts with partial intron inclusions, or partial exon skipping (Table 6). Exploiting this information, we investigated whether some of these splice sites are also used spontaneously, in the absence of mutations. Specifically, we investigated the presence and relative frequency of eight exon skipping events, three partial exon skipping events and two intron inclusion events for a total of 13 HPRT splice variants (Tables 6 and 7). Of the 13 HPRT splice variants investigated, we detected five: a combined exon 2 and 3 skipping, exon 4 skipping, exon 7 skipping, exon 8 skipping and partial exon 8 skipping (Table 7). None of these transcripts contained a PTC. The partial skipping of exon 8 appears to be generated by the use of an illegitimate splice acceptor site. The site has been previously shown to be used when the legitimate site or nearby sequences have been affected by mutation. This, however, is the first report of the site being used spontaneously. The frequency of the five HPRT splice variants detected is listed in Table 7. Exon 8 was the most frequently affected exon; fully or partially skipped, it had a frequency relative to the WT transcript of 1.1%.
Table 6. HPRT cryptic splice sites.
| Legitimate splice site | Cryptic splice site | Splice variant | |
|---|---|---|---|
| Cryptic donor splice sites | |||
| Intron | |||
| 1* | GTG/g1704tgagc | cag1752/gtggcg | Inclusion of 1–49 of intron 1 |
| 5* | GAA/g31635taagt | Aag31701/gtaagc | Inclusion of 1–67 of intron 5 |
| Cryptic acceptor splice sites | |||
| Exon | |||
| 2 | tttcag14779/A28TT | TAG32/T | Skip 28–32 of exon 2 |
| 4* | aactag27890/A319AT | CAG327/T | Skip 319–327 of exon 4 |
| 8* | ttttag40032/T533TG | CAG553/A | Skip 533–553 of exon 8 |
| 9* | ttatag41453/C610AT | TAG626/T | Skip 610–626 of exon 9 |
Data adapted from (26). Lowercase indicates intron sequences, uppercase indicates exon sequences, and/refers to an intron/exon boundary. Numbers in italic refer to cDNA numbering (base 1 is the A in the AUG initiation codon). All other numbers refer to genomic numbering (26).
*Indicates cryptic sites investigated in this study.
Table 7. HPRT splice variants characterized in MRC5 by quantitative PCR.
| Splice variant | PTC | Splice variant frequency relative to WT frequency in MRC5 (%) | Splice variant frequency relative to WT frequency in emetine-treated MRC5 (%) |
|---|---|---|---|
| Complete exon skipping | |||
| 2 | Yes | 0 | 0 |
| 3 | Yes | 0 | 0.2 ± 0.3 |
| Exons 2 and 3 | No | 0.4 ± 0.2 | 0.6 ± 0.4 |
| 4 | No | 0.2 ± 0.3 | 0.1 ± 0.3 |
| 5 | No | 0 | 0 |
| 6 | Yes | 0 | 0.4 ± 0.5 |
| 7 | No | 0.1 ± 0.3 | 0.1 ± 0.3 |
| 8 | No | 0.8 ± 0.2 | 0.7 ± 0.3 |
| Partial exon skipping | |||
| 4 | No | 0 | 0 |
| 8 | No | 0.3 ± 0.5 | 0.3 ± 0.4 |
| 9 | Yes | 0 | 0 |
| Partial intron inclusion | |||
| 1 | Yes | N/Da | N/Da |
| 5 | Yes | 0 | 0.6 ± 0.3 |
| Complex event | |||
| Intron 5 and exon 6 | Yes | 0 | 0.1 ± 0.3 |
aUnable to determine.
In HPRT, as in POLB, it appears that several splice variants are eliminated by NMD. In emetine-treated MRC5, the spectrum of aberrant transcripts was dramatically different. We detected nine splice variants (Table 7), including four containing PTC and not seen in untreated MRC5. The frequency of the splice variants without PTC was not affected by emetine treatment. There was an unexpected transcript with a complex splicing pattern among the new splice variants, which has not been reported before. This variant, which we detected using the primers designed to detect intron 5 inclusion, contains the first 2 bases of intron 5 and is missing the first 52 bases of exon 6. It has a junction sequence of -gtAG- where gt is the putative intronic sequence and AG the putative exonic sequence. However, the intron 5 nucleotides 1–4 are gtaa and the exon 6 nucleotides 50–54 are GGCAG. Consequently, it is not possible to assign the A to the intron or the exon unambiguously.
Mechanisms generating splice variants
The data presented above, in addition to many reports in the literature, indicate that splice variant production is a widespread phenomenon. For example, NF1 is reported to produce over 46 splice variants (27). There is also a strong evidence that NMD can detect and remove splice variants containing PTC and may be crucial in preventing the cell's translation machinery from being overloaded with unproductive and even deleterious transcripts. Splice variants represent ∼40% of all POLB transcripts and ∼1% of HPRT transcripts in both TK6 and MRC5. The frequency increases to ∼50% for POLB in emetine-treated cells and to ∼2% for HPRT in TK6. As discussed above, since our survey was not exhaustive, it is likely that the diversity and frequency of splice variants may be even higher.
It is not certain at this point whether POLB and HPRT splice variants are the result of regulated alternative splicing processes or the result of aberrant splicing. It is tempting to speculate that these splice variants serve some function in the cell, but clearly most of these splice variants cannot code for a polymerase function (POLB) or a functional transferase (HPRT). Perhaps RNA of the splice variants has a regulatory role. At this point we are faced with two equally unlikely conclusions: either the multiple splice variants, some of which contain PTC and will be eliminated quickly by NMD, serve a cellular function, or the cell can tolerate a splicing process, which generates frequent splicing errors. The choice between the two alternatives is far from clear. What possible function can tens of low frequency splice variants can have? On the other hand if most POLB and HPRT splice variants are indeed the result of aberrant splicing, then several questions arise. How can the cell tolerate such high frequency of aberrant splice variants as that seen in POLB? Could aberrant splicing itself have an adaptive value, perhaps by facilitating the evolution of new genes? Moreover, what mis-splicing mechanisms can account for the differences in splice variant frequency among different genes and also explain why some exons, such as exon 8 of HPRT or exon 2 of POLB appear more vulnerable to mis-splicing than other exons?
Even though the question of the function of splice variants is not yet settled, it appears likely that at least some variants are the result of aberrant splicing. How then could these aberrant splice variants be generated? The hypothesis that transcripts with skipped exons are the result of an active mechanism, which detects premature stop codons and directs the skipping of the exon containing the stop codon sequence (28) is not supported by our data. In our opinion, there are four mechanisms capable of generating aberrant splice variants: (i) stochastic spliceosome errors in splice site recognition. (ii) Errors by the machinery regulating alternative splicing. (iii) Misincorporation errors by RNA polymerase II (POL II) during transcription. (iv) Splice site selection errors due to transcriptional pausing at DNA lesions. We discuss each of them in turn.
First, spliceosome error is the most obvious mechanism for mis-splicing and perhaps the most difficult to evaluate. Splice site selection by the spliceosome is a very complex process that is not fully understood. Nevertheless, as a first approximation, it can be argued that if mis-splicing relates predominantly to errors by the spliceosome in discriminating between legitimate and illegitimate splice sites, then the relative strength of the splice consensus sequences is of paramount importance, with the most errors occurring at the weakest sites. As stated above, we have investigated this hypothesis by examining the strength of the splice sites involved in the skipped exons in HPRT and POLB in our data. Exon skipping involves the illegitimate use of an alternative upstream 3′ acceptor splice site. The strength of the acceptor sites has been evaluated using an information-theory based analysis model (4,29) that evaluates the information content of the splice sites and expresses it in bits (Ri). According to this model, the mean Ri value for splice acceptor sites (28 nt) in humans is 9.35, representing the average amount of information required for splicing. The information content of POLB and HPRT exons has been estimated previously (24,26). Interestingly, exon 8, one of the most frequently skipped exons in the HPRT has a particularly low-scoring acceptor site Ri of 2.8, the weakest acceptor site in HPRT. The alternatively used acceptor in the complete skipping has a higher score, Ri = 8.5, even the cryptic acceptor used in the partial skipping has an Ri of 3.4. In POLB, exon 2 skipping by itself also involves a stronger acceptor splice site. The exon 2 acceptor site has an Ri of 11.1, whereas the exon 3 acceptor has an Ri of 15.6. Other exon skipping events, however, do not fit this pattern. For example, in the skipping of exon 3 in HPRT, the acceptor used has an Ri of 10.0 but the legitimate acceptor has an Ri of 11.3. In the skipping of exon 11 in POLB, the acceptor site used has an Ri of 3.0, but the legitimate acceptor has an Ri of 6.6. On the other hand, mutation spectra using the HPRT as indicator target have demonstrated that DNA lesions are not distributed evenly across the gene, but often cluster on specific exons such as exon 3 and exon 8 (30). Further insights into the strength of splicing signals could be gained by estimating the strength of exonic enhancers. Exonic enhancers have been studied in HPRT, but unfortunately no data about their relative strength is available (31). According to this analysis, the strength of the splice sites influences splicing fidelity but is not sufficient to explain and predict all of the observed aberrant splice variants.
Second, a role for the alternative splicing machinery in aberrant splicing is supported by several lines of evidence. In the adenovirus E1A pre-mRNA splicing reporter system in mouse and green monkey cells, it was shown that cellular exposure to osmotic stress resulted in abnormal splicing patterns in genes by influencing the sub-cellular distribution of heterogeneous nuclear ribonucleoprotein A1, a splicing factor that can regulate alternative splicing in vitro and in vivo by antagonizing the serine–arginine family of proteins (32). A variety of aberrant transcripts were detectable in the affected tissues but not in other tissues. Similarly, in sporadic Alzheimer's disease an aberrant splicing variant of Presenillin-2 (PS2), missing exon 5, was shown to be preferentially expressed and it was suggested that the transcript is a diagnostic feature of the disease (10,33). It was translated to protein, and led to an increased production of Aβ protein. Intriguingly, it has been recently reported that exon 5 skipping in PS2 can be induced by hypoxia (9). The effect was mediated by the hypoxia-induced activation of a trans-acting factor, HMGA1a, which binds the PS2 transcript at exon 5 (9). This is a very significant observation since hypoxia is frequently a feature in tumors.
Third, misincorporation errors by POL II during transcription, which destroy the splice sites, may also result in aberrant splicing. It is difficult to calculate directly how large a mutagenic target the regulatory splice sequences represent, because their consensus is quite loosely conserved and many mutations are tolerated. Therefore, the effective size of the splice sequences as mutagenic targets by POL II errors is likely to be smaller than the actual size of the consensus sequences. Moreover, it is quite difficult to estimate the correct size of the exonic splicing enhancers. Consequently, we have attempted to estimate the effective size of splice sequences according to mutagenic studies. Since 15% of DNA mutations of the HPRT gene lead to mis-splicing (26,29), and the HPRT coding sequence is 657 bp long, we have estimated that the effective size of the sequences affecting splicing is 100 bp (assuming that each base has an equal probability of mutating). This analysis indicates that splice consensus sequences represent a smaller mutagenic target than it would be calculated by simply adding their length. We have estimated that the reported RNA polymerase II misincorporation rate of ∼2.8 × 10−4 (34) is sufficient to account for the observed frequency of HPRT splice variants, but not the frequency of POLB splice variants.
Finally, additional mis-splicing may result from transcriptional pausing in front of DNA lesions. Indeed, it has been shown that transcription and splicing are intimately coordinated and that pausing or even slow processivity by POL II, affects splicing fidelity (35). A link between splicing fidelity and POL II fidelity and processivity is particularly interesting, because it would imply a link between DNA repair and splicing fidelity. Therefore, increased levels of DNA lesions or a reduced rate of repair would result in increased frequency of aberrant transcripts, either because of mutagenic by-pass or pausing by POL II while repair is effected. Indirectly supporting such a relationship are reports that human cells, which are missing functional p53, exhibit a higher frequency of aberrant transcripts (36,37). Intriguingly, it has also been demonstrated that p53 is required for transcription-coupled excision repair (TCR) and therefore, p53-deficient cell lines would be relatively slow in removing DNA lesions from the transcribed strand (37).
In conclusion, it appears that splicing generates a large number of splice variants whose function is not yet known and it is likely that some of them are the result of mis-splicing. As discussed above, several different mechanisms may contribute to mis-splicing. This has important implications in evaluating the role of splice variants in disease, as well as implications in our attempts to understand the observed differential frequency of the various splice variants. However, no single mechanism can explain as yet, all the splice variants observed, but we believe that estimating the relative contribution of the various processes that affect splicing fidelity will be crucial in evaluating the role of splicing in disease and aging.
REFERENCES
- 1.Melki J. and Clark,S. (2002) DNA methylation changes in leukaemia. Semin. Cancer Biol., 12, 347. [DOI] [PubMed] [Google Scholar]
- 2.Hartmuth K., Urlaub,H., Vornlocher,H.P., Will,C.L., Gentzel,M., Wilm,M. and Luhrmann,R. (2002) Protein composition of human prespliceosomes isolated by a tobramycin affinity-selection method. Proc. Natl Acad. Sci. USA, 99, 16719–16724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou Z., Licklider,L.J., Gygi,S.P. and Reed,R. (2002) Comprehensive proteomic analysis of the human spliceosome. Nature, 419, 182–185. [DOI] [PubMed] [Google Scholar]
- 4.Stephens R.M. and Schneider,T.D. (1992) Features of spliceosome evolution and function inferred from an analysis of the information at human splice sites. J. Mol. Biol., 228, 1124–1136. [DOI] [PubMed] [Google Scholar]
- 5.Zhu J., Shendure,J., Mitra,R.D. and Church,G.M. (2003) Single molecule profiling of alternative pre-mRNA splicing. Science, 301, 836–838. [DOI] [PubMed] [Google Scholar]
- 6.Silva J.M., Dominguez,G., Gonzalez-Sancho,J.M., Garcia,J.M., Silva,J., Garcia-Andrade,C., Navarro,A., Munoz,A. and Bonilla,F. (2002) Expression of thyroid hormone receptor/erbA genes is altered in human breast cancer. Oncogene, 21, 4307–4316. [DOI] [PubMed] [Google Scholar]
- 7.Oh Y., Proctor,M.L., Fan,Y.H., Su,L.K., Hong,W.K., Fong,K.M., Sekido,Y.S., Gazdar,A.F., Minna,J.D. and Mao,L. (1998) TSG101 is not mutated in lung cancer but a shortened transcript is frequently expressed in small cell lung cancer. Oncogene, 17, 1141–1148. [DOI] [PubMed] [Google Scholar]
- 8.Carney M.E., Maxwell,G.L., Lancaster,J.M., Gumbs,C., Marks,J., Berchuck,A. and Futreal,P.A. (1998) Aberrant splicing of the TSG101 tumor suppressor gene in human breast and ovarian cancers. J. Soc. Gynaecol. Investig., 5, 281–285. [DOI] [PubMed] [Google Scholar]
- 9.Manabe T., Katayama,T., Sato,N., Gomi,F., Hitomi,J., Yanagita,T., Kudo,T., Honda,A., Mori,Y., Matsuzaki,S. et al. (2003) Induced HMGA1a expression causes aberrant splicing of Presenilin-2 pre-mRNA in sporadic Alzheimer's disease. Cell Death Differ., 10, 698–708. [DOI] [PubMed] [Google Scholar]
- 10.Manabe T., Katayama,T., Sato,N., Kudo,T., Matsuzaki,S., Imaizumi,K. and Tohyama,M. (2002) The cytosolic inclusion bodies that consist of splice variants that lack exon 5 of the presenilin-2 gene differ obviously from Hirano bodies observed in the brain from sporadic cases of Alzheimer's disease patients. Neurosci. Lett., 328, 198–200. [DOI] [PubMed] [Google Scholar]
- 11.Munch C., Ebstein,M., Seefried,U., Zhu,B., Stamm,S., Landwehrmeyer,G.B., Ludolph,A.C., Schwalenstocker,B. and Meyer,T. (2002) Alternative splicing of the 5′-sequences of the mouse EAAT2 glutamate transporter and expression in a transgenic model for amyotrophic lateral sclerosis. J. Neurochem., 82, 594–603. [DOI] [PubMed] [Google Scholar]
- 12.Ejima Y., Yang,L. and Sasaki,M.S. (2000) Aberrant splicing of the ATM gene associated with shortening of the intronic mononucleotide tract in human colon tumor cell lines: a novel mutation target of microsatellite instability. Int. J. Cancer, 86, 262–268. [DOI] [PubMed] [Google Scholar]
- 13.Ramalho A.S., Beck,S., Penque,D., Gonska,T., Seydewitz,H.H., Mall,M. and Amaral,M.D. (2003) Transcript analysis of the cystic fibrosis splicing mutation 1525-1G>A shows use of multiple alternative splicing sites and suggests a putative role of exonic splicing enhancers. J. Med. Genet., 40, e88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Faustino N.A. and Cooper,T.A. (2003) Pre-mRNA splicing and human disease. Genes Dev., 17, 419–437. [DOI] [PubMed] [Google Scholar]
- 15.Buchner D.A., Trudeau,M. and Meisler,M.H. (2003) SCNM1, a putative RNA splicing factor that modifies disease severity in mice. Science, 301, 967–969. [DOI] [PubMed] [Google Scholar]
- 16.Meshorer E. and Soreq,H. (2002) Pre-mRNA splicing modulations in senescence. Aging Cell, 1, 10–16. [DOI] [PubMed] [Google Scholar]
- 17.Skandalis A., Ninniss,P.J., McCormac,D. and Newton,L. (2002) Spontaneous frequency of exon skipping in the human HPRT gene. Mutat. Res., 501, 37–44. [DOI] [PubMed] [Google Scholar]
- 18.Lewis B.P., Green,R.E. and Brenner,S.E. (2003) Evidence for the widespread coupling of alternative splicing and nonsense-mediated mRNA decay in humans. Proc. Natl Acad. Sci. USA, 100, 189–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carter M.S., Doskow,J., Morris,P., Li,S., Nhim,R.P., Sandstedt,S. and Wilkinson,M.F. (1995) A regulatory mechanism that detects premature nonsense codons in T-cell receptor transcripts in vivo is reversed by protein synthesis inhibitors in vitro. J. Biol. Chem., 270, 28995–29003. [DOI] [PubMed] [Google Scholar]
- 20.Zmudzka B.Z., Fornace,A., Collins,J. and Wilson,S.H. (1988) Characterization of DNA polymerase beta mRNA: cell-cycle and growth response in cultured human cells. Nucleic Acids Res., 16, 9587–9596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cabelof D.C., Yanamadala,S., Raffoul,J.J., Guo,Z., Soofi,A. and Heydari,A.R. (2003) Caloric restriction promotes genomic stability by induction of base excision repair and reversal of its age-related decline. DNA Repair (Amsterdam), 2, 295–307. [DOI] [PubMed] [Google Scholar]
- 22.Cabelof D.C., Raffoul,J.J., Yanamadala,S., Guo,Z. and Heydari,A.R. (2002) Induction of DNA polymerase beta-dependent base excision repair in response to oxidative stress in vivo. Carcinogenesis, 23, 1419–1425. [DOI] [PubMed] [Google Scholar]
- 23.Chyan Y.J., Strauss,P.R., Wood,T.G. and Wilson,S.H. (1996) Identification of novel mRNA isoforms for human DNA polymerase beta. DNA Cell. Biol., 15, 653–659. [DOI] [PubMed] [Google Scholar]
- 24.Thompson T.E., Rogan,P.K., Risinger,J.I. and Taylor,J.A. (2002) Splice variants but not mutations of DNA polymerase beta are common in bladder cancer. Cancer Res., 62, 3251–3256. [PubMed] [Google Scholar]
- 25.Bhattacharyya N., Chen,H.C., Comhair,S., Erzurum,S.C. and Banerjee,S. (1999) Variant forms of DNA polymerase beta in primary lung carcinomas. DNA Cell. Biol., 18, 549–554. [DOI] [PubMed] [Google Scholar]
- 26.O'Neill J.P., Rogan,P.K., Cariello,N. and Nicklas,J.A. (1998) Mutations that alter RNA splicing of the human HPRT gene: a review of the spectrum. Mutat. Res., 411, 179–214. [DOI] [PubMed] [Google Scholar]
- 27.Vandenbroucke I., Callens,T., De Paepe,A. and Messiaen,L. (2002) Complex splicing pattern generates great diversity in human NF1 transcripts. BMC Genomics, 3, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miriami E., Motro,U., Sperling,J. and Sperling,R. (2002) Conservation of an open-reading frame as an element affecting 5′ splice site selection. J. Struct. Biol., 140, 116–122. [DOI] [PubMed] [Google Scholar]
- 29.Rogan P.K., Faux,B.M. and Schneider,T.D. (1998) Information analysis of human splice site mutations. Hum. Mutat., 12, 153–171. [DOI] [PubMed] [Google Scholar]
- 30.Skandalis A., da Cruz,A.D., Curry,J., Nohturfft,A., Curado,M.P. and Glickman,B.W. (1997) Molecular analysis of T-lymphocyte HPRT- mutations in individuals exposed to ionizing radiation in Goiania, Brazil. Environ. Mol. Mutagen., 29, 107–116. [PubMed] [Google Scholar]
- 31.Fairbrother W.G., Yeh,R.F., Sharp,P.A. and Burge,C.B. (2002) Predictive identification of exonic splicing enhancers in human genes. Science, 297, 1007–1013. [DOI] [PubMed] [Google Scholar]
- 32.van der Houven van Oordt W., Diaz-Meco,M.T., Lozano,J., Krainer,A.R., Moscat,J. and Caceres,J.F. (2000) The MKK(3/6)-p38-signaling cascade alters the subcellular distribution of hnRNP A1 and modulates alternative splicing regulation. J. Cell. Biol., 149, 307–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sato N., Imaizumi,K., Manabe,T., Taniguchi,M., Hitomi,J., Katayama,T., Yoneda,T., Morihara,T., Yasuda,Y., Takagi,T. et al. (2001) Increased production of b-amyloid and vulnerability to ER stress by an aberrant spliced form of presenilin-2. J. Biol. Chem., 276, 2108–2114. [DOI] [PubMed] [Google Scholar]
- 34.Shaw R.J., Bonawitz,N.D. and Reines,D. (2002) Use of an in vivo reporter assay to test for transcriptional and translational fidelity in yeast. J. Biol. Chem., 277, 24420–24426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Caceres J.F. and Kornblihtt,A.R. (2002) Alternative splicing: multiple control mechanisms and involvement in human disease. Trends Genet., 18, 186–193. [DOI] [PubMed] [Google Scholar]
- 36.Moyret-Lalle C., Duriez,C., Van Kerckhove,J., Gilbert,C., Wang,Q. and Puisieux,A. (2001) p53 induction prevents accumulation of aberrant transcripts in cancer cells. Cancer Res., 61, 486–488. [PubMed] [Google Scholar]
- 37.Leger C. and Drobetsky,E.A. (2002) Modulation of the DNA damage response in UV-exposed human lymphoblastoid cells through genetic-versus functional-inactivation of the p53 tumor suppressor. Carcinogenesis, 23, 1631–1640. [DOI] [PubMed] [Google Scholar]