Abstract
The HO gene in Saccharomyces cerevisiae is regulated by a large and complex promoter that is similar to promoters in higher order eukaryotes. Within this promoter are 10 potential binding sites for the a1-α2 heterodimer, which represses HO and other haploid-specific genes in diploid yeast cells. We have determined that a1-α2 binds to these sites with differing affinity, and that while certain strong-affinity sites are crucial for repression of HO, some of the weak-affinity sites are dispensable. However, these weak-affinity a1-α2-binding sites are strongly conserved in related yeast species and have a role in maintaining repression upon the loss of strong-affinity sites. We found that these weak sites are sufficient for a1-α2 to partially repress HO and recruit the Tup1-Cyc8 (Tup1-Ssn6) co-repressor complex to the HO promoter. We demonstrate that the Swi5 activator protein is not bound to URS1 in diploid cells, suggesting that recruitment of the Tup1-Cyc8 complex by a1-α2 prevents DNA binding by activator proteins resulting in repression of HO.
INTRODUCTION
Regulation of transcription in the yeast Saccharomyces cerevisiae often occurs through relatively simple sets of promoter elements, usually located within a few hundred bases upstream of the transcription start site. In contrast to most yeast promoters, the HO gene is regulated by an atypically large, modular promoter containing an extensive array of binding sites for transcriptional regulators, which is similar to the promoters of higher eukaryotes (Figure 1A) (1–5). Activation of HO transcription proceeds through an ordered recruitment of transcription factors to its promoter, which is initiated by a complex formed by two sequence-specific DNA-binding proteins, Swi5 and Pho2. SWI5 encodes a Zinc-finger protein that is sequestered in the cytoplasm until late anaphase, when it enters the nucleus and activates the transcription of several genes before being rapidly degraded (5–7). The Swi5–Pho2 complex binds to two sites at the distal end of the HO promoter, known as URS1 (Upstream Regulatory Sequence 1, see Figure 1A) (8). Upon binding to URS1, Swi5 recruits both the SWI/SNF chromatin remodeling complex and the Srb/Mediator (SRB/MED) complex to URS1 (6,9). The recruitment of these co-activator complexes to URS1 is followed by the degradation of Swi5 and the stepwise appearance at URS2 of the SAGA complex, Swi4/Swi6 and SRB/MED (6,9). This results in the recruitment of SRB/MED and RNA Polymerase II, in two distinct steps, to the HO TATA box (9,10) and subsequent HO transcription at the G1/S stage of the cell cycle (3). Expression of HO leads to a switch in mating type that is initiated by the HO endonuclease, which cleaves a specific DNA sequence at the MAT locus (11).
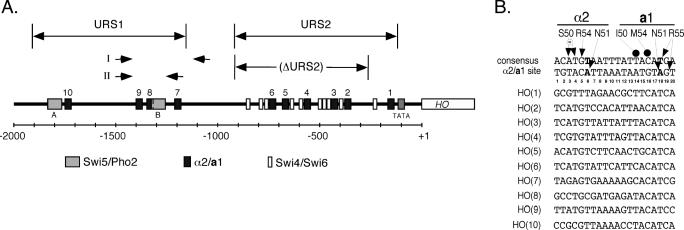
Figure 1.
Position and sequence of a1-α2-binding sites of the HO promoter. (A) A schematic map of the HO promoter is shown, divided into the URS1 and URS2 regions as indicated. The positions of a1-α2 sites (black boxes, numbered from 1 to 10), Swi5/Pho2 sites (gray boxes, designated A and B), and Swi4/Swi6 sites (white boxes) are shown. The scale bar indicates the number of base pairs from the start of the HO ORF. Also shown are PCR primer sets I (Figure 5B) and II (Figure 7), and the URS2 deletion used for strains in Figures 4 and 5. (B) Sequences of the a1-α2-binding sites of the HO promoter (top strand only), aligned with a consensus a1-α2 site [from Jin et al. (32), both top and bottom strands shown]. The major-groove base-specific contacts made by residues of α2 and a1 are shown [from Li et al. (55)], with closed arrows indicating direct hydrogen bonds, a circled ‘w’ indicating water-mediated contacts and closed circles indicating van der Waals contacts. The T6 and T18 bases (in bold) were mutated for mutational analysis of the individual a1-α2 sites within the context of the HO promoter.
In addition to being expressed at a specific point in the cell cycle, HO transcription is restricted to the mother cells from a cell division (4). Expression of HO is repressed in daughter cells by Ash1, whose mRNA is asymmetrically localized to daughter cells during mitosis (12–14). Repression of HO in haploid daughters prevents mating-type switching in these cells. This allows for conjugation with its haploid mother cell that has switched mating type, forming a diploid cell (15). In heterozygous diploid MATa/MATα cells, transcription of HO is repressed by the MATα2 and MATa1 proteins (16). This prevents switching of one of the MAT loci, and the subsequent formation of homozygous diploids (MATa/MATa or MATα/MATα). Such homozygous diploids cannot sporulate, yet are still able to mate with a haploid cell to form a heterozygous triploid cell, which would have problems such as chromosomal non-disjunction during meiosis. Repression of HO in heterozygous diploid cells is thereby an important process by which yeasts maintain three stable cell types: two haploid mating types and a non-mating heterozygous diploid.
The MATα2 and MATa1 proteins (hereafter referred to as α2 and a1, respectively) each contain a homeodomain, a DNA-binding motif that is conserved from yeast to humans (17). α2 and a1 form a heterodimer (a1-α2) that cooperatively binds DNA in a sequence-specific manner (18). The a1-α2 heterodimer represses several haploid-specific genes, including HO, in diploid cells (19). α2 also associates with another cofactor, Mcm1, to form a complex in haploid MATα and diploid cells that represses mating-type a-specific genes, such as STE6 (19). The a1-α2 and α2-Mcm1 complexes repress transcription by the recruitment of the Tup1-Cyc8 (also known as Tup1-Ssn6) co-repressor complex (20,21), which has been shown to negatively regulate several functionally diverse sets of genes in yeast and have conserved homologs in higher order eukaryotes (22). Tup1-Cyc8 has been proposed to repress transcription through two mechanisms: (i) inhibitory interaction with SRB/MED (23–27) and (ii) the creation of a repressive chromatin environment through nucleosome positioning (28,29) and co-recruitment of histone deacetylases (30,31).
Sequence analysis of the HO promoter revealed 10 elements that shared similarity to known a1-α2-binding sites from the MATα1 and STE5 promoters (2). Comparison of these elements to the MATα1 and STE5 a1-α2-binding sites (2), along with mutational analysis of a consensus a1-α2-binding site (32), predicted that some of the sites may be strong-affinity sites, while others may only be weakly bound, if at all. These predictions raise the question of whether these weaker sites are bound by the a1-α2 heterodimer in vivo, and if they have a functional role in the cell. Our results demonstrate that the 10 a1-α2 binding sites of the HO promoter exhibit varying degrees of binding affinity and repression on their own. Some of these weak-affinity a1-α2 sites are strongly conserved among related yeasts and appear to play an auxiliary role to stronger affinity sites within the context of the genomic HO promoter. These findings illustrate how promoter architecture and the use of sites of varying affinities may be important regulators of transcriptional repression in the complex promoters of higher eukaryotes.
MATERIALS AND METHODS
Plasmids and yeast strains
Heterologous CYC1-lacZ transcription reporter plasmids were made by inserting annealed oligonucleotides bearing an a1-α2 site with 5′-TCGA overhangs into the SalI site of pAV73 (2 μ URA3) (33). pYJ103, a pAV73-based lacZ transcription reporter containing the consensus a1-α2 site, and pJM130, a CEN LEU2 plasmid bearing MATα, have been described previously (32,34).
pG333, bearing the HO open reading frame (ORF) and 4 kb of upstream non-coding sequence (35), was digested with MfeI and SapI, followed by Klenow fill-in and re-ligation to yield pJR067. The 734 bp SacI–BamHI fragment of pJR067 was subcloned into a derivative of pUC19 (which had been digested with HindIII and XbaI, the ends made blunt with Klenow polymerase and re-ligated to yield pJR081), yielding pJR082. Single a1-α2 sites were mutated in pJR067 and pJR082 by site-directed mutagenesis using the QuickChange kit (Stratagene). The mutations introduced at these a1-α2 sites substitute both the T6 and T18 bases (as numbered in Figure 1B) to guanosine, as mutation of either a1 or α2 half-site in this manner has been shown to prevent a1-α2 binding (32). Note that in the case of the HO(8) site, only the T18G mutation was constructed, as position 6 of the HO(8) site is cytosine, a base that has been shown to fully derepress transcription when at the corresponding position in an a1-α2 consensus site (32). Multiple mutations were created by subcloning the SacI/SphI and SphI/BamHI fragments within pJR082-based plasmids, then subcloning the SacI/BamHI fragments from these plasmids into pJR067 and its derivatives. URS2 was deleted in pJR067-based plasmids by digestion with SnaBI and Acc65I, after which the ends were made blunt with Klenow polymerase and re-ligated.
A plasmid-based 13xMyc-tagged version of Tup1 was made by subcloning the TUP1 ORF into pNJ1547, a CEN TRP1 plasmid bearing a 13xMyc-tagged version of SGS1 and a C-terminal multiple cloning site (a gift from Steven Brill) (36). The TUP1 ORF and 750 bp of the TUP1 5′-untranslated region were PCR-amplified using primers containing either XhoI or XmaI sites, then subcloned (in frame) into XhoI/XmaI-digested pNJ1547 (thereby removing SGS1), yielding pJR134.
Yeast strain AJ83 (MATa ura3 his3 leu2 trp1) was used for β-galactosidase assays and has been described previously (37). All other yeast strains are isogenic to W303 (his3 ade2 trp1 ura3 leu2) and its derivatives W1011-3B (MATα HIS3 ade2) and W1346-3C (MATa his3 ADE2).
To avoid differences in HO expression in mother and daughter cells of the strains being assayed, ASH1 was deleted in W1011-3B and W1346-3C by PCR-mediated one-step gene disruption (38) using LEU2, yielding JRY101 and JRY102, respectively. Successful integration of the ash1Δ::LEU2 PCR fragment was confirmed by PCR using primers within the LEU2 and SPE1 (adjacent to the genomic ASH1 locus) ORFs.
For genomic integration of mutant a1-α2 sites into the HO promoter, a 1.5 kb SspI fragment of pRS306, containing URA3, was ligated into NruI/BsaBI-digested pJR067 to create pJR068. A 2.3 kb BsaAI/KpnI fragment from pJR068 was then used to replace HO with URA3 by homologous recombination in JRY101 and JRY102, yielding JRY113 and JRY114, respectively. pJR067 and its derivatives were digested with AvaII to yield 3.7 kb (3.1 kb for URS2 deletions) fragments that were then used to replace hoΔ::URA3 by homologous recombination in both JRY113 and JRY114. A complete list of the strains used in these experiments is available on request.
Diploid strains were constructed by crossing strains of opposite mating type (all either MATα HIS3 or MATa ADE2) on YEPD for 8 h at 30°C, followed by selection for diploids on SD-his-ade. Diploidy (as well as proper transcriptional repression by a1-α2) was confirmed prior to each in vivo assay by the inability of each strain to mate (38).
For the time-course chromatin immunoprecipitation (ChIP) experiments (Figure 7), the MATa strain K8144 (6) was made pseudo-diploid by the integration of MATα at URA3. This was performed by ligating a 6 kb BglI fragment (containing MATα) from pJM130 to a 2.6 kb BglI fragment (containing URA3) of pRS306 (39), yielding pJR120. XcmI-digested pJR120 was then integrated at URA3 of K8144 by standard techniques, yielding JRY200 (MATa, URA3::MATα).
Figure 7.
Swi5 does not bind to the HO promoter in the presence of a1-α2. (A) A culture of JRY200 (MATa, URA3::MATα, GAL1,10-CDC20, Swi5-Myc9) was arrested in mitosis by growth in media lacking galactose, then released by the addition of galactose. Aliquots were taken at the indicated times and crosslinked for ChIP. Cell lysates were divided for ChIP of Swi5-Myc9 and α2. After reversing the crosslinks, purified DNA was used as a template for multiplex PCR using primer set II (see Figure 1A) and primers that amplify the STE6 (as a positive control for α2) and EGT2 (as a positive control for Swi5) promoters. Primer sets that amplify portions of the YDL238c and YDL223c ORFs (the top and bottom primer sets, respectively, in each lane) were used as negative controls. Lane 1, PCR from template of Total Chromatin (TC) sample of DNA purified from a whole cell lysate. Lanes 2–9, PCR from templates of DNA immunoprecipitated using antibodies to c-Myc (Top, for Swi5-Myc9) and α2 (Bottom). (B) A culture of the haploid K8144 (MATa, GAL1,10-CDC20, Swi5-Myc9) strain was arrested in mitosis and processed for ChIP with antibodies to c-Myc as described in (A).
β-galactosidase assays
Strain AJ83 was transformed with pJM130, then with CYC1-lacZ reporter plasmids containing different a1-α2-binding sites. Three independent transformants were cultured in selective media to maintain all transformed plasmids, and liquid β-galactosidase assays were performed as described previously (40).
Electrophoretic mobility shift assays (EMSA)
EMSAs were performed as previously described using purified α2128–210 and a166–126 protein fragments and 32P-labeled PCR products amplified from pAV73-based CYC1-lacZ reporter plasmids (41).
S1 nuclease assays
RNA was isolated from logarithmically growing yeast cells by hot acid phenol extraction (38). S1 nuclease assays were performed as in (38); briefly, 40 μg of RNA was mixed with 32P-labeled oligonucleotides (2.5 × 105 c.p.m. per sample) corresponding to regions of the HO and ACT1 ORFs (8), denatured at 75°C for 10 min, then allowed to anneal overnight at 55°C. S1 nuclease (AmershamPharmacia Biotech) was added (160 U per sample) and followed by digestion for 1 h at 37°C. Non-digested probe was then recovered by ethanol precipitation and electrophoresed on an 8% polyacrylamide/42% (w/v) urea/TBE gel. Autoradiographs were viewed and quantified using a Storm PhosphorImager and ImageQuant Mac v.2 software.
ChIP assays
ChIP assays of asynchronous cultures (Figures 5B and 6) were performed as described previously (41) using a polyclonal antibody to α2 (a gift from Sandy Johnson) and, for Tup1-Myc13, a monoclonal antibody to the c-Myc epitope (Babco). For ChIPs of Tup1 (Figure 6), cultures were crosslinked in formaldehyde (1% final concentration) for 90 min, which was determined to be necessary to detect Tup1. For the time-course ChIP experiments (Figure 7), cultures of K8144 or JRY200 were grown to an OD600 of 0.4 in YEP media containing 2% raffinose and 2% galactose, then for 4 h in YEP media containing only 2% raffinose to arrest the cells in mitosis as described previously (9). After the addition of 2% galactose, samples were taken at timed intervals and processed for ChIP analysis as described previously (41), except that fixation was stopped by the addition of glycine to 125 mM, followed by incubation at room temperature for 5 min. Cell lysates were then split into two fractions for immunoprecipitations of α2, using a polyclonal antibody to α2 as before, and Swi5-Myc9, using a monoclonal antibody to the c-Myc epitope (Babco). DNA from total chromatin and immunoprecipitation samples was purified and used as a template for multiplex PCR. In general, 1/50th–1/25th of the immunoprecipitated DNA was used as a template in PCRs consisting of 25 cycles. In order to conclusively test for the presence or absence of a signal by a particular sample, higher concentrations of template and/or PCRs consisting of 30 cycles were used. All ChIP results were judged according to conditions that resulted in the absence of amplification by one or two negative control primer sets, as detailed in the results.
Figure 5.
Further in vivo analysis of weak-affinity a1-α2-binding sites. Cultures of the indicated strains were grown to mid-logarithmic phase and divided for S1 nuclease assays and ChIP. (A) A schematic is shown indicating the relative positions of the HO regulatory sites and the region of promoter deleted in the construct used for these experiments. S1 nuclease assays of purified RNA, as described in Figure 4. Below each sample is the normalized HO/ACT1 expression ratio (the average of at least two independent assays) expressed as a percentage of the wild-type MATα strain. (B) ChIP of α2. α2 was immunoprecipitated from formaldehyde-crosslinked cultures using a polyclonal antibody to α2. After reversing the crosslinks, purified DNA was used as a template for multiplex PCR using primer set I (see Figure 1A) and primers that amplify the STE6 promoter (as a positive control for α2 in both haploid and diploid strains) and part of the YDL223c ORF (as a negative control). The first lane (unnumbered) is a PCR from template of Total Chromatin (TC) sample of DNA purified from a whole cell lysate. Lanes numbered 1–11 are PCRs from templates of immunoprecipitated DNA of the same lanes in (A). All strains are ash1Δ::LEU2 and ΔURS2 (as indicated in Figure 1A), except for the wild-type MATα, which does not have URS2 deleted.
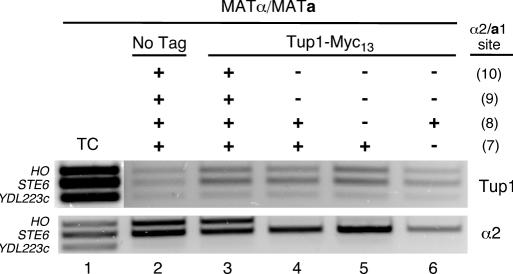
Figure 6.
ChIP of Tup1 at the HO promoter. Diploid cultures of strains bearing either a blank CEN TRP1 plasmid (lane 2) or pJR134 (Tup1-Myc13, lanes 3–6) were grown to mid-logarithmic phase and crosslinked for ChIP analysis. Cell lysates were divided for ChIP of Tup1-Myc13 and α2. After reversing the crosslinks, purified DNA was used as a template for multiplex PCR using primer set I (see Figure 1A) and primers that amplify the STE6 promoter (as a positive control for α2 and Tup1) and part of the YDL223c ORF (as a negative control). Lane 1, PCR from template of Total Chromatin (TC) sample of DNA purified from a whole cell lysate. Lanes 2–6, PCR from templates of DNA immunoprecipitated using antibodies to c-Myc (Top, for Tup1-Myc13) and α2 (Bottom). All strains are ash1Δ::LEU2 and ΔURS2 (as indicated in Figure 1A). The status of each a1-α2 site (shown at the right) is indicated as wild-type (+) or mutant (−), as in Figure 4.
Computational analysis
Genomic sequences from S.cerevisiae, S.bayanus, S.mikatae, S.paradoxus and S.kudriavzevii were obtained from the Saccharomyces Genome Database (www.yeastgenome.org) (42,43). A segmented multiple sequence alignment of the HO promoter region from the five species was performed using the DIALIGN 2 program (44–46). A measure of the significance for the alignment of the sites was then constructed by taking the alignment scores and averaging over each base pair in the site. The background probability of the average alignment scores was calculated by a similar analysis on two other regions of the genome, a region 2000–3000 bp upstream of HO and a region spanning positions 81213–83471 on chromosome IV which is 800 bp from the nearest annotated gene.
RESULTS
Analysis of isolated a1-α2-binding sites of the HO promoter
To assess the relative strengths of each of the 10 predicted a1-α2-binding sites found in the HO promoter, each individual site was cloned between the UASCYC1 and TATA box of a plasmid-based CYC1-lacZ reporter. The level of repression for each reporter was monitored by liquid β-galactosidase assay in a MATa strain transformed with a CEN plasmid bearing MATα (34). As shown in Figure 2A, the relative strength of each isolated a1-α2 site from the HO promoter generally agrees with predictions based on the sequence of each binding site (2,32). Several sites, HO(10), HO(9), HO(6), HO(4) and HO(3), exhibit very strong a1-α2-mediated repression, near the level of a consensus a1-α2 site. The remaining sites demonstrate considerably weaker levels of repression, from nearly non-functional [HO(8) and HO(1)] to moderate strength [HO(7), HO(5) and HO(2)] when compared with a reporter plasmid lacking an a1-α2 site.
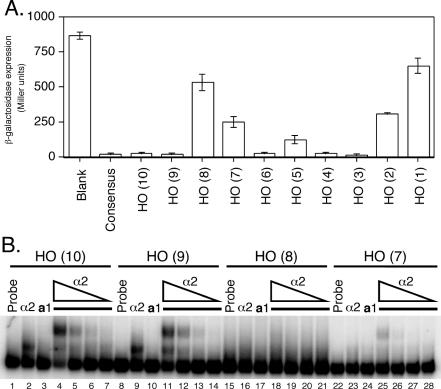
Figure 2.
Analysis of isolated HO a1-α2 sites reveals differential binding affinity. (A) β-galactosidase assay of heterologous CYC1-lacZ reporters with the indicated a1-α2 binding sites (x-axis) inserted between the UAS and TATA box of CYC1. Reporters were assayed in a MATa strain bearing a plasmid-based copy of MATα, with β-galactosidase expression (y-axis) indicated in Miller units (min−1 ml−1). (B) EMSAs using 32P-labeled oligonucleotide probes corresponding to the a1-α2 sites of URS1. Probes were incubated with either no protein (lanes 1, 8, 15 and 22), 8.2 × 10−9 M α2128–210 (lanes 2, 9, 16 and 23), or 2.8 × 10−9 M a166–126 (lanes 3, 10, 17 and 24). For lanes 4–7, 11–14, 18–21 and 25–28, a166–126 was kept at a constant concentration of 2.8 × 10−9 M, while α2128–210 was serially diluted 5-fold from 8.2 × 10−9 M (lanes 4, 11, 18 and 25) to 6.6 × 10−11 M (lanes 7, 14, 21 and 28).
To assess whether the levels of repression correlate with a1-α2 DNA-binding affinity in vitro, the four a1-α2-binding sites from URS1 were used as probes for EMSAs using purified α2 and a1 proteins. The results of these assays (Figure 2B) correlate well with the in vivo repression data, as the a1-α2 heterodimer binds strongly to sites HO(10) and HO(9), while binding weakly to HO(7) and very little to HO(8). This in vitro data thereby confirms the presence of strong a1-α2-binding sites [HO(10) and HO(9)] as well as weak sites [HO(8) and HO(7)] within URS1.
Phylogenetic analysis of HO promoter sequences
Since several of the predicted a1-α2 sites in the HO promoter only weakly repress transcription on their own and are not bound with strong affinity by the complex, this raises the question of whether these sites have a functional role in the context of the HO promoter. One approach to address this question is to determine whether these sites are conserved in related yeast species (42,43,47). Conservation of these sequences would suggest that they have a functional role in the cell.
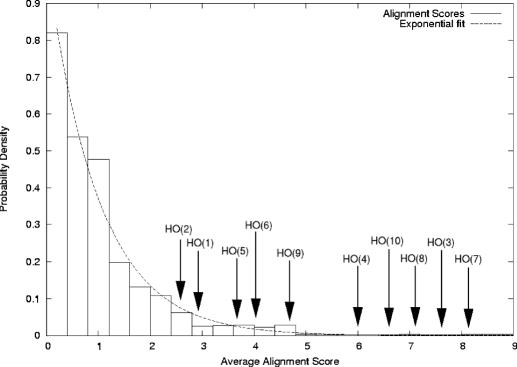
We compared the conservation of the sites in the HO promoter among five sequenced Saccharomyces sensu stricto (S.cerevisae, S.bayanus, S.mikatae, S.paradoxus and S.kudriavzevii) using the DIALIGN 2 multiple sequence alignment program (44). The average alignment scores for these sites were compared against the distribution of scores generated by the same analysis of two other non-ORF regions in the genome. The resulting distribution of scores for both the HO promoter and the other regions fits an exponential function (Figure 3). The average scores of many of the HO sites are in the tail of the distribution and 7 of the 10 a1-α2 sites appeared to be well conserved among the different species. As might be expected, several of the strong-repressor sites, such as HO(3), HO(4) and HO(10), are highly conserved among the different species and have high scores in the analysis. In contrast, HO(1) and HO(2), which were weak repressor sites on their own, showed significantly lower values in the analysis. In addition, the conservation of the HO(5) site, which is also a weak repressor site on its own, is borderline, with a 2.4% significance. Interestingly, the two other weak repressor sites, HO(7) and HO(8), are strongly conserved among the different yeasts. In fact, these sites appear to be more highly conserved than HO(4), HO(6) and HO(9), which are all strong a1-α2 repressor sites. The fact that HO(7) and HO(8) are highly conserved strongly suggests that they have a functional role in regulating the HO promoter, even though they only weakly function on their own.
Figure 3.
Conservation of the a1-α2 sites in related species of yeast. A segmented multiple sequence alignment was performed for the HO promoter region and two control regions from S.cerevisiae, S.bayanus, S.mikatae, S.paradoxus and S.kudriavzevii. The chart shows the background number of occurrences of the average alignment scores for 20 bp segments in a 3000 bp neutral region close to the HO gene. The average alignment scores for the 10 predicted a1-α2 sites are shown. A high average alignment score indicates the site is strongly conserved in the different yeast species.
Analysis of a1-α2-binding sites within the intact HO promoter
The presence of conserved weak-affinity a1-α2 sites within URS1 led to the question of their contribution to repression of HO. We therefore constructed integrated mutant versions of each of the a1-α2 sites of URS1 within the context of the full HO promoter. The mutations introduced at these a1-α2 sites substituted key bases that are absolutely required for DNA binding by α2 and a1 (T6G and T18G mutations, respectively, as numbered in Figure 1B) (32). These mutations disrupt the Asn51-adenine contact demonstrated by all homeodomains (17), and each mutation on its own has been shown to completely abrogate repression by a1-α2 when introduced into a consensus a1-α2 site (32). Mutant sites were integrated into the genome at the HO locus of haploid MATa and MATα strains by homologous recombination. All of the mutant haploid strains expressed HO mRNA at levels comparable to the wild-type haploid strains, indicating that the mutations in the a1-α2 sites did not affect the HO expression (data not shown). Haploid strains were then mated to form diploid strains that were then assayed for the expression of HO mRNA by S1 nuclease digestion.
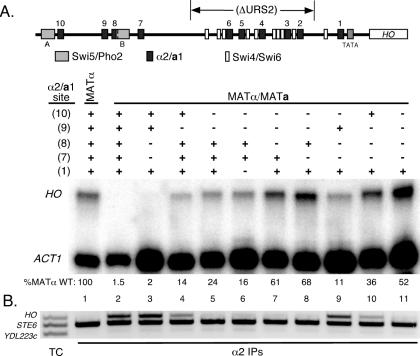
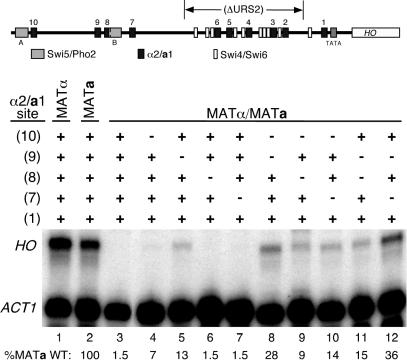
We first examined the effect of mutation of the strong a1-α2 sites HO(10) and HO(9) within the context of the full HO promoter. Mutation of these sites, alone or in combination, did not derepress HO transcription in diploid cells, presumably due to several strong a1-α2 sites within URS2 (data not shown). We therefore constructed the same a1-α2 site mutants in the context of the HO promoter with a large portion of URS2 deleted, as indicated in Figure 1A. This deletion of URS2 leaves intact one Swi4/Swi6 binding site and the HO(1) a1-α2 site, the latter of which was demonstrated to be nearly non-functional on its own (Figure 2A). With URS2 deleted, HO is expressed in haploid strains at the same levels as with the wild-type promoter, and completely repressed in the diploid state (Figure 4) (data not shown). Mutation of either HO(10) or HO(9) in this context caused slight, but significant, derepression of HO transcription in diploid cells. Furthermore, mutation of both sites caused an increase in HO mRNA in comparison with either single mutant. The level of HO expression seen in the HO(10)/HO(9) double mutant was not as high as observed in haploid cells (Figure 4, compare lane 8 with lanes 1 and 2), which suggested that the weaker sites of URS1, HO(8) and HO(7) contribute to HO repression. Mutation of either of these sites on their own caused no observable derepression of HO transcription (Figure 4, lanes 6 and 7), and mutation of both had no further effect (Figure 5A, lane 3). The HO(8) and HO(7) mutations had little effect in combination with the HO(10) mutant (Figure 4, compare lane 4 with lanes 9 and 10), and the HO(8) mutation did not have a significant effect in combination with the HO(9) mutant (Figure 4, compare lane 5 with lane 11). However, the HO(9)/HO(7) double mutation shows a significant increase in the production of HO mRNA, to a level even greater than that of the HO(10)/HO(9) double mutant (Figure 4, compare lane 8 with lane 12).
Figure 4.
In vivo analysis reveals contributions of weak-affinity a1-α2 sites. S1 nuclease assays were performed using 32P-labeled oligonucleotide probes that hybridize to the mRNA of either HO or ACT1, the latter of which was used as a loading control. All strains, both haploid and diploid, are ash1Δ::LEU2 and the URS2 region of HO was deleted as indicated in the schematic map of the HO promoter. The positions of a1-α2 sites (black boxes), Swi5/Pho2 sites (gray boxes, designated A and B), and Swi4/Swi6 sites (white boxes) are shown. The status of each a1-α2 site (shown at the left) is indicated as wild-type (+) or mutant (−), as described in the text. Below each sample is the normalized HO/ACT1 expression ratio (the average of at least two independent assays), expressed as a percentage of the wild-type MATa strain. Wild-type repression of the promoter (lane 3) thereby results in 1.5% the level of HO expression compared with the derepressed promoter (lane 2).
To further assess the contribution of the weak-affinity sites HO(8) and HO(7), we mutated these sites in the context of the HO(10)/HO(9) double mutant. Mutation of either HO(8) or HO(7) shows further derepression of the promoter in this background (Figure 5A, compare lane 5 with lanes 7 and 8). This suggests that the two weak-affinity a1-α2 sites of URS1 can still partially repress HO transcription together, yet are unable to do so on their own. Note that in the HO(10)/HO(9) double mutant background, there is no contribution to repression by the HO(1) site, as mutation of this site did not derepress HO transcription any further (Figure 5A, lane 6).
We also assayed for a1-α2 binding to URS1 in these mutant strains by ChIP, using a polyclonal antibody to α2 and PCR primer set I (Figure 1A), which amplifies a region of the promoter from sites HO(9) to HO(7). As internal controls for these assays, we used primer sets that amplify the α2-Mcm1 site of the STE6 promoter, which is bound by α2 in both haploid and diploid strains, and a portion of the YDL223c ORF, which is 10 kb away from HO and not bound or regulated by α2 in any known manner. As seen in Figure 5B, α2 is present at URS1 in a wild-type diploid strain while being completely absent in a haploid MATα strain. Furthermore, there is roughly a wild-type level of α2 at URS1 when both HO(8) and HO(7) are mutated (Figure 5B, lane 3). Somewhat surprisingly, we did not observe amplification above background (i.e. the negative control band) for the HO primer set in any strain bearing the HO(10)/HO(9) mutation (Figure 5B, lanes 5–8), while observing further derepression with additional mutations to sites HO(8) or HO(7) (Figure 5A, lanes 5–8). This suggests that while HO(8) and HO(7) are still able to partially repress HO with URS2 deleted, the level of in vivo DNA binding by a1-α2 at these sites may be too low for detection by ChIP.
ChIP of Tup1 in a1-α2 site mutant strains
Despite the inability to detect α2 recruitment through sites HO(8) and HO(7), the observation that a1-α2 can partially repress the expression through these sites suggested that the Tup1-Cyc8 complex may still be recruited to URS1 in strains lacking the strong-affinity sites HO(10) and HO(9). Since Tup1-Cyc8 binds to the amino-terminal tails of histones H3 and H4 (48), it is possible that a1-α2 can bind to the weak sites HO(8) and HO(7) at a level that is too low for detection by ChIP, but sufficient to recruit Tup1-Cyc8 to the promoter. We therefore utilized a plasmid-based, Myc13-tagged version of Tup1 to monitor Tup1 binding at URS1 by ChIP. Interestingly, while α2 was not detectable at URS1 when both sites HO(10) and HO(9) are mutated, Tup1 was detectable above the level of a blank control (Figure 6, compare lane 2 with lanes 3–6). Tup1-Myc13 was detected at URS1 when both HO(8) and HO(7) were left intact (Figure 6, lane 4), and when only HO(7) remained (Figure 6, lane 5). A faint band can still be seen at HO when only HO(8) is intact (Figure 6, lane 6), suggesting that Tup1-Cyc8 can still be recruited by a1-α2 weakly binding at this site. Although it is possible that a1-α2 could recruit Tup1-Cyc8 through site HO(1), it should be noted that mutation of this site had no effect in combination with mutations in sites HO(10) and HO(9) (Figure 5, compare lane 5 with lane 6), whereas mutations to HO(8) and HO(7) in this context exhibited derepression (Figure 5, compare lane 5 with lanes 7 and 8).
ChIP of HO regulators following cell cycle arrest
Having established that HO is repressed by a1-α2 binding to sites of varying affinity, we next performed assays to address the mechanism by which HO is repressed by a1-α2 and the Tup1-Cyc8 co-repressor. To elucidate the mode of repression, we performed ChIP experiments to determine whether the Swi5 activator is able to bind to the HO promoter in the presence of a1-α2 and its associated co-repressors. While Swi5 DNA-binding is not detectable at HO in asynchronous cultures, the brief binding of Swi5 to URS1 during the late anaphase stage of the cell cycle can be detected in synchronized haploid cells released from a block in the cell cycle (6). To monitor Swi5 binding by ChIP, we utilized a strain bearing a Myc9-tagged Swi5 that also contains a galactose-inducible CDC20 to arrest the cells in G2 before mitosis (6). We made this MATa strain pseudo-diploid by integrating a copy of MATα at URA3. A culture of this strain was arrested and then aliquots for ChIP analysis were taken at timed intervals following release from the cell cycle block. Microscopic scoring of each time point indicated that the cells were synchronously released from the arrest and proceeded through the cell cycle (data not shown). The ChIP assays showed that the α2 protein bound to both the HO and STE6 promoters through all stages of the assay (Figure 6). In contrast, the Swi5-Myc9 ChIP did not generate a significant band corresponding to the HO promoter for any of the collected time points, suggesting that Swi5-Myc9 was not binding to the promoter. As a positive control for Swi5-Myc9 immunoprecipitation, we also examined the EGT2 promoter, which is activated by Swi5 and is not repressed in diploid cells (49,50). The ChIPs showed that Swi5-Myc9 bound to the EGT2 promoter in a manner that was temporally consistent with previous observations of Swi5 DNA binding in vivo (9), indicating that a Swi5-Myc9 ChIP was detectable in this strain and that the cells were properly synchronized. Immunoprecipitated DNA from time points that demonstrate Swi5-Myc9 binding to the EGT2 promoter were subjected to several different PCR conditions (see Materials and Methods) to examine binding at HO, but no signal above background was obtained (data not shown). We also repeated the assay with the haploid strain (K8144) that was used to make the pseudo-diploid strain (JRY200) and observed Swi5-Myc9 binding at URS1 in a manner consistent with published results (Figure 7B) (6,9). These assays suggest that a1-α2 and its associated co-repressors prevent, or greatly reduce Swi5 binding at URS1.
DISCUSSION
Transcriptional regulation through a complex promoter is an important process by which eukaryotic cells can coordinate several signals to express genes according to various intracellular and extracellular cues (51). The yeast HO promoter is an excellent model for this process, as it coordinates the signals of several sequence-specific DNA-binding proteins to regulate HO transcription according to cell-type and stage in the cell-cycle. HO is repressed in diploid yeast cells by a1-α2 heterodimer, which regulates several haploid-specific genes by binding to specific sequences within their promoters and subsequently recruiting the Tup1-Cyc8 co-repressor complex. Miller et al. (2) identified 10 potential binding sites for the a1-α2 complex within the HO promoter, and we have shown that a1-α2 binds to these sites with widely varying affinities. Our data show that the initial events of HO activation at URS1 are repressed by a1-α2 binding mainly at the strong-affinity sites HO(10) and HO(9). While the weak-affinity sites HO(8) and HO(7) are dispensable for this process, these sites can partially repress HO (with URS2 deleted) in the absence of stronger affinity a1-α2 sites, indicating an auxiliary role for these sites. Both weak sites in URS1 are required for this partial repression (Figure 5), suggesting a level of cooperation between the two sites. We found that either of these weak sites can still recruit the Tup1-Cyc8 corepressor complex to the HO promoter in the absence of detectable occupancy by a1-α2 (Figure 6), suggesting that a1-α2 need only transiently bind to these sites to recruit Tup1-Cyc8. This observation is not unprecedented, as Swi5 binds to URS1 too quickly to be observed in asynchronous cultures, yet sufficiently to recruit the SWI/SNF co-activator complex (6). Our data suggest that while Tup1-Cyc8 is recruited by a1-α2 bound to these weak sites, it may not be recruited at a level sufficient to prevent activation of HO by Swi5 and SWI/SNF. It is interesting to speculate a wider role for a1-α2 in the regulation of genes whose promoters contain weak-affinity sites that deviate from an a1-α2 consensus site (32). Weak-affinity a1-α2 sites that may not be readily identified by conventional computational searches may still serve to partially repress or attenuate the transcription of several genes. Indeed, we have recently demonstrated that the promoters of the PDE1 and MET31 genes appear to have a1-α2 repressor sites that are weakly bound by the complex and may partially repress transcription in vivo (52). If weak-affinity sites were functionally important, then one would expect that they would be conserved in related yeast species. Our finding that HO(7) and HO(8) are more highly conserved than many of the stronger repressor sites supports the model that they have a role in HO regulation. Weak a1-α2 binding sites in other promoters, such as PDE1 and MET31, and even weaker sites in the LSM1 and REX2, are strongly conserved in related yeast species, suggesting these may have also role in fine tuning expression of these genes in diploid cells [R.A.O'F. and A.M.S., unpublished data; (52)].
The placement of weak-affinity a1-α2 sites around Swi5-Pho2 site B (Figure 1A) may also underscore the importance of this element in activation of HO transcription. Mutation of Swi5-Pho2 site B has been shown to cause a more severe defect in HO expression than mutation of site A, indicating a greater contribution to the activation of HO transcription by site B (8). Furthermore, Swi5 and Pho2 bind cooperatively to site B, while only binding in an additive manner to site A (8). Although the a1-α2 HO(7) and HO(8) sites are not absolutely required for HO repression in S.cerevisiae, the conserved placement of these sites around Swi5-Pho2 site B may serve to maintain repression of the promoter in yeast strains, such as S.pastorianus and S.bayanus, lacking one or even both of the stronger sites in URS1 (44). Our data have shown that mutation of either HO(10) or HO(9) weakly derepresses an HO promoter with URS2 deleted. However, the promoter is strongly derepressed when these mutations are made in combination with mutations of the HO(7) and HO(8) sites. The placement of the HO(7) and HO(8) sites on either side of the Swi5/Pho2 activator site may explain why HO(9) is not as highly conserved among the different yeasts as some of the other sites.
While the weak-affinity a1-α2 sites of URS1 serve an auxiliary role in repression of HO in yeast, their presence may give clues to potential functions for such sites in higher order eukaryotes. Many developmental processes feature the formation of gradients of transcriptional regulators that result in boundaries of gene expression and pattern formation (53,54). The placement of weak-affinity repressor binding sites near a strong-affinity site may allow for sustained repression with limiting concentrations of the repressor, whereas genes regulated only by a strong-affinity site may be derepressed. This mechanism could be another level of attenuation of transcriptional regulation, and may further define gene expression boundaries near the end of a gradient of a repressor protein.
Acknowledgments
ACKNOWLEDGEMENTS
We thank the members of the Vershon laboratory for plasmid constructs and helpful discussions about the manuscript. We would also like to thank the following for providing materials used for this study: Susan Baxter for α2 and a1 proteins, Steven Brill for pNJ1547, Sandy Johnson for α2 antiserum, Marta Galova of Kim Nasmyth's laboratory for pG333 and yeast strain K8144. This work was supported by the Charles and Joanna Busch Fellowship (Rutgers University) to J.R.M., the Benedict-Michael Graduate Fellowship in Molecular Biology (Rutgers University) to J.R.M. and a grant from the National Institutes of Health (GM49265) to A.K.V.
REFERENCES
- 1.Maxon M.E. and Herskowitz,I. (2001) Ash1p is a site-specific DNA-binding protein that actively represses transcription. Proc. Natl Acad. Sci. USA, 98, 1495–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miller A.M., MacKay,V.L. and Nasmyth,K.A. (1985) Identification and comparison of two sequence elements that confer cell-type specific transcription in yeast. Nature, 314, 598–603. [DOI] [PubMed] [Google Scholar]
- 3.Nasmyth K. (1985) A repetitive DNA sequence that confers cell-cycle START (CDC28)-dependent transcription of the HO gene in yeast. Cell, 42, 225–235. [DOI] [PubMed] [Google Scholar]
- 4.Nasmyth K. (1985) At least 1400 base pairs of 5′-flanking DNA is required for the correct expression of the HO gene in yeast. Cell, 42, 213–223. [DOI] [PubMed] [Google Scholar]
- 5.Stillman D.J., Bankier,A.T., Seddon,A., Groenhout,E.G. and Nasmyth,K.A. (1988) Characterization of a transcription factor involved in mother cell specific transcription of the yeast HO gene. EMBO J., 7, 485–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cosma M.P., Tanaka,T. and Nasmyth,K. (1999) Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle- and developmentally regulated promoter. Cell, 97, 299–311. [DOI] [PubMed] [Google Scholar]
- 7.Nasmyth K., Adolf,G., Lydall,D. and Seddon,A. (1990) The identification of a second cell cycle control on the HO promoter in yeast: cell cycle regulation of SW15 nuclear entry. Cell, 62, 631–647. [DOI] [PubMed] [Google Scholar]
- 8.McBride H.J., Brazas,R.M., Yu,Y., Nasmyth,K. and Stillman,D.J. (1997) Long-range interactions at the HO promoter. Mol. Cell. Biol., 17, 2669–2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bhoite L.T., Yu,Y. and Stillman,D.J. (2001) The Swi5 activator recruits the Mediator complex to the HO promoter without RNA polymerase II. Genes Dev., 15, 2457–2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cosma M.P., Panizza,S. and Nasmyth,K. (2001) Cdk1 triggers association of RNA polymerase to cell cycle promoters only after recruitment of the mediator by SBF. Mol. Cell, 7, 1213–1220. [DOI] [PubMed] [Google Scholar]
- 11.Strathern J.N., Klar,A.J., Hicks,J.B., Abraham,J.A., Ivy,J.M., Nasmyth,K.A. and McGill,C. (1982) Homothallic switching of yeast mating type cassettes is initiated by a double-stranded cut in the MAT locus. Cell, 31, 183–192. [DOI] [PubMed] [Google Scholar]
- 12.Bobola N., Jansen,R.P., Shin,T.H. and Nasmyth,K. (1996) Asymmetric accumulation of Ash1p in postanaphase nuclei depends on a myosin and restricts yeast mating-type switching to mother cells. Cell, 84, 699–709. [DOI] [PubMed] [Google Scholar]
- 13.Jansen R.P., Dowzer,C., Michaelis,C., Galova,M. and Nasmyth,K. (1996) Mother cell-specific HO expression in budding yeast depends on the unconventional myosin myo4p and other cytoplasmic proteins. Cell, 84, 687–697. [DOI] [PubMed] [Google Scholar]
- 14.Sil A. and Herskowitz,I. (1996) Identification of asymmetrically localized determinant, Ash1p, required for lineage-specific transcription of the yeast HO gene. Cell, 84, 711–722. [DOI] [PubMed] [Google Scholar]
- 15.Amon A. (1996) Mother and daughter are doing fine: asymmetric cell division in yeast. Cell, 84, 651–654. [DOI] [PubMed] [Google Scholar]
- 16.Jensen R., Sprague,G.F.,Jr and Herskowitz,I. (1983) Regulation of yeast mating-type interconversion: feedback control of HO gene expression by the mating-type locus. Proc. Natl Acad. Sci. USA, 80, 3035–3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gehring W.J., Affolter,M. and Burglin,T. (1994) Homeodomain proteins. Annu. Rev. Biochem., 63, 487–526. [DOI] [PubMed] [Google Scholar]
- 18.Goutte C. and Johnson,A.D. (1993) Yeast a1 and alpha 2 homeodomain proteins form a DNA-binding activity with properties distinct from those of either protein. J. Mol. Biol., 233, 359–371. [DOI] [PubMed] [Google Scholar]
- 19.Herskowitz I. (1989) A regulatory hierarchy for cell specialization in yeast. Nature, 342, 749–757. [DOI] [PubMed] [Google Scholar]
- 20.Mukai Y., Harashima,S. and Oshima,Y. (1991) AAR1/TUP1 protein, with a structure similar to that of the beta subunit of G proteins, is required for a1-alpha 2 and alpha 2 repression in cell type control of Saccharomyces cerevisiae. Mol. Cell. Biol., 11, 3773–3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keleher C.A., Redd,M.J., Schultz,J., Carlson,M. and Johnson,A.D. (1992) Ssn6-Tup1 is a general repressor of transcription in yeast. Cell, 68, 709–719. [DOI] [PubMed] [Google Scholar]
- 22.Smith R.L. and Johnson,A.D. (2000) Turning genes off by Ssn6-Tup1: a conserved system of transcriptional repression in eukaryotes. Trends Biochem. Sci., 25, 325–330. [DOI] [PubMed] [Google Scholar]
- 23.Kuchin S. and Carlson,M. (1998) Functional relationships of Srb10-Srb11 kinase, carboxy-terminal domain kinase CTDK-I, and transcriptional corepressor Ssn6-Tup1. Mol. Cell. Biol., 18, 1163–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gromoller A. and Lehming,N. (2000) Srb7p is a physical and physiological target of Tup1p. EMBO J., 19, 6845–6852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Papamichos-Chronakis M., Conlan,R.S., Gounalaki,N., Copf,T. and Tzamarias,D. (2000) Hrs1/Med3 is a Cyc8-Tup1 corepressor target in the RNA polymerase II holoenzyme. J. Biol. Chem., 275, 8397–8403. [DOI] [PubMed] [Google Scholar]
- 26.Wahi M. and Johnson,A.D. (1995) Identification of genes required for alpha 2 repression in Saccharomyces cerevisiae. Genetics, 140, 79–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zaman Z., Ansari,A.Z., Koh,S.S., Young,R. and Ptashne,M. (2001) Interaction of a transcriptional repressor with the RNA polymerase II holoenzyme plays a crucial role in repression. Proc. Natl Acad. Sci. USA, 98, 2550–2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shimizu M., Roth,S.Y., Szent-Gyorgyi,C. and Simpson,R.T. (1991) Nucleosomes are positioned with base pair precision adjacent to the alpha 2 operator in Saccharomyces cerevisiae. EMBO J., 10, 3033–3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kastaniotis A.J., Mennella,T.A., Konrad,C., Torres,A.M. and Zitomer,R.S. (2000) Roles of transcription factor Mot3 and chromatin in repression of the hypoxic gene ANB1 in yeast. Mol. Cell. Biol., 20, 7088–7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu J., Suka,N., Carlson,M. and Grunstein,M. (2001) TUP1 utilizes histone H3/H2B-specific HDA1 deacetylase to repress gene activity in yeast. Mol. Cell, 7, 117–126. [DOI] [PubMed] [Google Scholar]
- 31.Watson A.D., Edmondson,D.G., Bone,J.R., Mukai,Y., Yu,Y., Du,W., Stillman,D.J. and Roth,S.Y. (2000) Ssn6-tup1 interacts with class I histone deacetylases required for repression. Genes Dev., 14, 2737–2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jin Y., Zhong,H. and Vershon,A.K. (1999) The yeast a1 and alpha2 homeodomain proteins do not contribute equally to heterodimeric DNA binding. Mol. Cell. Biol., 19, 585–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vershon A.K., Hollingsworth,N.M. and Johnson,A.D. (1992) Meiotic induction of the yeast HOP1 gene is controlled by positive and negative regulatory sites. Mol. Cell. Biol., 12, 3706–3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mead J., Zhong,H., Acton,T.B. and Vershon,A.K. (1996) The yeast alpha2 and Mcm1 proteins interact through a region similar to a motif found in homeodomain proteins of higher eukaryotes. Mol. Cell. Biol., 16, 2135–2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tebb G., Moll,T., Dowzer,C. and Nasmyth,K. (1993) SWI5 instability may be necessary but is not sufficient for asymmetric HO expression in yeast. Genes Dev., 7, 517–528. [DOI] [PubMed] [Google Scholar]
- 36.Mullen J.R., Kaliraman,V. and Brill,S.J. (2000) Bipartite structure of the SGS1 DNA helicase in Saccharomyces cerevisiae. Genetics, 154, 1101–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Siliciano P.G. and Tatchell,K. (1984) Transcription and regulatory signals at the mating type locus in yeast. Cell, 37, 969–978. [DOI] [PubMed] [Google Scholar]
- 38.Ausubel F.M., Brent,R., Kingston,R.E., Moore,D.D., Seidman,J.G., Smith,J.A. and Struhl,K. (1998) Current Protocols in Molecular Biology. John Wiley and Sons, Inc, New York, NY. [Google Scholar]
- 39.Sikorski R.S. and Hieter,P. (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics, 122, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Keleher C.A., Goutte,C. and Johnson,A.D. (1988) The yeast cell-type-specific repressor alpha 2 acts cooperatively with a non-cell-type-specific protein. Cell, 53, 927–936. [DOI] [PubMed] [Google Scholar]
- 41.Hart B., Mathias,J.R., Ott,D., McNaughton,L., Anderson,J.S., Vershon,A.K. and Baxter,S.M. (2002) Engineered improvements in DNA-binding function of the MATa1 homeodomain reveal structural changes involved in combinatorial control. J. Mol. Biol., 316, 247–256. [DOI] [PubMed] [Google Scholar]
- 42.Cliften P., Sudarsanam,P., Desikan,A., Fulton,L., Fulton,B., Majors,J., Waterston,R., Cohen,B.A. and Johnston,M. (2003) Finding functional features in Saccharomyces genomes by phylogenetic footprinting. Science, 301, 71–76. [DOI] [PubMed] [Google Scholar]
- 43.Kellis M., Patterson,N., Endrizzi,M., Birren,B. and Lander,E.S. (2003) Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature, 423, 241–254. [DOI] [PubMed] [Google Scholar]
- 44.Morgenstern B. (1999) DIALIGN 2: improvement of the segment-to-segment approach to multiple sequence alignment. Bioinformatics, 15, 211–218. [DOI] [PubMed] [Google Scholar]
- 45.Morgenstern B., Dress,A. and Werner,T. (1996) Multiple DNA and protein sequence alignment based on segment-to-segment comparison. Proc. Natl Acad. Sci. USA, 93, 12098–12103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Morgenstern B., Frech,K., Dress,A. and Werner,T. (1998) DIALIGN: finding local similarities by multiple sequence alignment. Bioinformatics, 14, 290–294. [DOI] [PubMed] [Google Scholar]
- 47.Tamai Y., Tanaka,K., Umemoto,N., Tomizuka,K. and Kaneko,Y. (2000) Diversity of the HO gene encoding an endonuclease for mating-type conversion in the bottom fermenting yeast Saccharomyces pastorianus. Yeast, 16, 1335–1343. [DOI] [PubMed] [Google Scholar]
- 48.Edmondson D.G., Smith,M.M. and Roth,S.Y. (1996) Repression domain of the yeast global repressor Tup1 interacts directly with histones H3 and H4. Genes Dev., 10, 1247–1259. [DOI] [PubMed] [Google Scholar]
- 49.Galitski T., Saldanha,A.J., Styles,C.A., Lander,E.S. and Fink,G.R. (1999) Ploidy regulation of gene expression. Science, 285, 251–254. [DOI] [PubMed] [Google Scholar]
- 50.Kovacech B., Nasmyth,K. and Schuster,T. (1996) EGT2 gene transcription is induced predominantly by Swi5 in early G1. Mol. Cell. Biol., 16, 3264–3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Levine M. and Tjian,R. (2003) Transcription regulation and animal diversity. Nature, 424, 147–151. [DOI] [PubMed] [Google Scholar]
- 52.Nagaraj V.H., O'Flanagan,R.A., Bruning,A.R., Mathias,J.R., Vershon,A.K. and Sengupta,A.M. (2004) Combined analysis of expression data and transcription factor binding sites in the yeast genome. BMC Genomics, 5, 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Small S., Blair,A. and Levine,M. (1992) Regulation of even-skipped stripe 2 in the Drosophila embryo. EMBO J., 11, 4047–4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stanojevic D., Small,S. and Levine,M. (1991) Regulation of a segmentation stripe by overlapping activators and repressors in the Drosophila embryo. Science, 254, 1385–1387. [DOI] [PubMed] [Google Scholar]
- 55.Li T., Jin,Y., Vershon,A.K. and Wolberger,C. (1998) Crystal structure of the MATa1/MATalpha2 homeodomain heterodimer in complex with DNA containing an A-tract. Nucleic Acids Res., 26, 5707–5718. [DOI] [PMC free article] [PubMed] [Google Scholar]