Abstract
Homologous recombination (HR) is used in vertebrate somatic cells for essential, RAD51-dependent, repair of DNA double-strand-breaks (DSBs), but inappropriate HR can cause genome instability. A transcriptional transactivation-independent role for p53 in suppressing HR has been established, but is not detected in all HR assays. To address the basis of such exceptions, and the possibility that suppression by p53 may be discriminatory, we have conducted a controlled comparison of the effects of p53 depletion on three different kinds of HR. We show that, within the same cells, p53 depletion promotes both intra-chromosomal HR (ICHR) and extra-chromosomal HR (ECHR), but not homologous DNA integration (gene targeting; GT). This conclusion holds true for both spontaneous and DSB-induced ICHR and GT. We show further that non-conservative ICHR is more susceptible than conservative ICHR to inhibition by p53. These results provide strong evidence that p53 can discriminate between different forms of HR and, despite the fact that GT is used experimentally for gene disruption, is consistent with the possibility that p53 preferentially suppresses genome-destabilizing forms of HR. While the mechanism of suppression by p53 remains unclear, our data suggest that it is independent of mismatch repair and of changes in RAD51 protein levels.
INTRODUCTION
Homology-directed repair (HDR) of DNA uses homologous recombination (HR) for the accurate repair of DNA double strand breaks (DSBs) and requires an intact DNA template homologous to the damaged locus (1). During the S and G2 phases of the cell cycle, templates with perfect homology are available in the form of sister chromatids to repair DSBs generated intrinsically during DNA replication, or by exogenous agents (e.g. ionizing irradiation). In vertebrate cells, such repair is not only genome-stabilizing but also essential, given that deletion of the key HR gene RAD51 is lethal (2,3). Templates with homology that may or may not be perfect are also available throughout the cell cycle, on chromosome homologues or, for repeat sequences, elsewhere in the genome, but their use is genome-destabilizing, leading to loss of heterozygosity or genome rearrangements (4,5). Mechanisms for restricting the amount, or perhaps the type, of HR have therefore evolved. Thus many proteins have been implicated in suppressing HR, including products of the oncogenes bcl-2 (6), bcr-abl (7) or bcl-x(L) (8) and the tumour suppressors MSH2 (9–11) BRCA2 (12–14), BLM and WRN (15,16) and p53 (17).
Stimulation of HR in response to p53 inactivation was first described for an extra-chromosomal HR (ECHR) assay involving replicating SV40 genomes (18), but has also been demonstrated for ECHR assays with non-replicating plasmids (19,20). Intra-chromosomal HR (ICHR) assays are consistently stimulated by p53 inactivation (21–26), and it has been established that this effect depends on parts of the p53 protein distinct from those required for transcriptional transactivation, G1 arrest and apoptosis in response to DNA damage (23–27). The mechanism by which p53 suppresses HR remains uncertain, but many biochemical properties of p53 that may be relevant have been described. These include exonuclease, DNA renaturation and DNA strand-transfer activities, an ability to bind DNA recombination intermediates, and associations with proteins implicated in HR (RAD51, RAD52, RAD54, RPA) or its control (BLM, WRN, BRCA1, BRCA2, MSH2) [see (20) and (28) and references therein]. In the case of RAD51, there is evidence that a direct interaction with p53 is required for suppression of HR in vivo (20). Given that RAD51 levels are elevated in p53-inactive and other immortal cells (29,30), and that RAD51 over-expression promotes various kinds of HR [reviewed in (31)], p53 could conceivably suppress HR by reducing RAD51 expression or stability.
An important question concerning the suppression of HR by p53 is whether it is discriminatory. It is conceivable that p53 globally suppresses HR, leaving sufficient activity to maintain cell viability. Alternatively, p53 may discriminate between genome-stabilizing and genome-destabilizing HR events, suppressing only the latter. Such discrimination offers obvious advantages, and could even be essential, given the lethality of RAD51 disruption. There is evidence (28,32,33) that p53 may, like the mismatch repair (MMR) system (10,11), preferentially suppress HR between imperfectly homologous sequences, and this would certainly represent a discriminatory, genome-stabilizing suppression of HR. The possibility that p53 may discriminate on the basis of variables other than the extent of homology, such as the relative positions of HR substrates, remains to be tested, however. A small number of studies (34–36) describe HR assays that were p53-insensitive, but it remains unclear whether these can be taken as evidence of discriminatory suppression or whether some aspect of the host cells or assay conditions used precluded detection of p53-sensitivity. In one study (35), for example, it was pointed out that the p53-insensitivity of gene targeting (GT; HR between chromosomal and extra-chromosomal DNA) might be attributable to the unusual cytoplasmic location of p53 in the mouse ES cells used. Similarly, further HR assays must be carried out to determine whether all HR in HCT116 (colon carcinoma) cells is p53-insenstive, or whether the reported p53-insensitivity of GT and sister chromatid exchange (SCE) in these cells (34) is indicative of discriminatory suppression. Also, the p53-insensitivity of ECHR in mouse embryo fibroblasts (36) contrasts with its p53-sensitivity in others cell lines (18–20), suggesting that some unusual aspect of the host cells or ECHR assay conditions used, may have precluded detection of p53-sensitivity.
Evidence for discriminatory suppression of HR by p53 therefore requires further experiments in which different kinds of HR assays are compared in the same cell lines. Accordingly, we describe here a comparison of ICHR, ECHR and GT in both human fibrosarcoma (HT1080) and embryonic kidney (HEK293) cell lines and show that GT is insensitive to p53 depletion by RNA interference, whereas ECHR and ICHR are both stimulated. Furthermore, we find that ECHR is p53-sensitive in colon carcinoma (HCT116) cells where GT and SCE were previously shown to be p53-insensitive (34), and that non-conservative ICHR is suppressed more than conservative ICHR. Our data therefore provide strong evidence that suppression of HR by p53 is indeed discriminatory, and is characterized by a remarkable inactivity towards GT. Despite the experimental use of GT for genome disruption, we argue that such discrimination is compatible with the notion that p53 preferentially suppresses genome-destabilizing HR. Finally, because we find no evidence of RAD51 accumulation following p53-depletion, and because ECHR suppression is detectable in MMR-defective HCT116 cells, we conclude that suppression by p53 is MMR-independent and cannot be explained in terms of RAD51 depletion.
MATERIALS AND METHODS
Cells
HT1080 and HEK293 cell lines were from the American Tissue Culture Collection, except for GT experiments in HEK293 where a derivative (HEK293E, Invitrogen) expressing the Epstein Barr virus protein EBNA-1 was used. Conditions of culture were as described previously (37).
Plasmids
pCMV3xnls-I-SceI (38), pCMVβ (BD Biosciences), pCX-EGFP, p451-2 and p429-1 (39) and pHPRThyg (37) have been described. A version of pDRneo (40) was used in which the original XhoI site was changed to a HindIII site. To make pSUPER-p53/neo, the p53 shRNA cassette was removed from pSUPER-p53 (41) as a 247 bp HincII/PstI fragment and cloned into pEGFP-C1 (Invitrogen), which had been cut with PstI, and AseI to remove the EGFP cassette. A similar procedure was used to make pSUPER/neo using the equivalent HincII/PstI fragment from pSUPER (41). The pHPRThyg derivative pHPRThygBcl− was generated by linearization at the unique BclI sites (see Figure 5), followed by end-filling and religation.
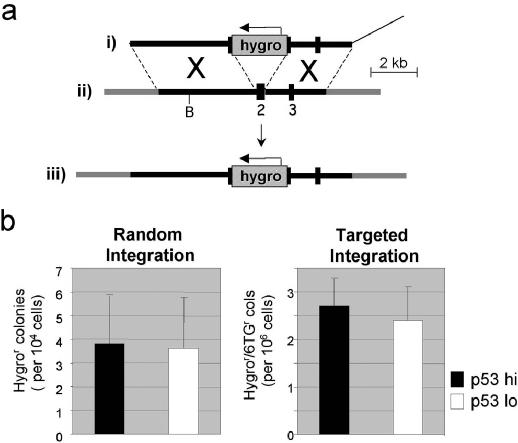
Figure 5.
GT and RI in HT1080 cells are unaffected by p53 knockdown. (a) The SalI-linearized HPRT GT construct pHPRThyg (i), and part of the X-chromosomal HPRT gene before (ii) and after (iii) the targeted integration of pHPRThyg, are shown. Regions of homology between pHPRThyg and the HPRT gene are shown as thick black lines or numbered boxes (exons), flanking regions of the HPRT gene in grey and vector DNA as a thin line. The Hygro cassette in pHPRThyg is shown as a stippled box. B = BclI. (b) The frequencies of random and targeted integration of pHPRT hyg with (white) or without (black) p53 knockdown are shown. The data represent average values and SD values for 11 electroporations (see Table 1 and Supplementary Material, Table 2).
pPU-I-RO/zeo was made in three steps. First, pBL-Puro/R was made by cloning a 1.3 kb PvuII/BamH1(e/f) fragment, carrying the Puro expression cassette from pPUR (Clontech), into the EcoRV site of pBSKS(+) (Stratagene), with the PvuII end oriented adjacent to the EcoR1 site of pBSKS(+). Second, sites for I-SceI and SacI (annealed oligonucleotides 5′-AATTACCCTGTTATCCCTAAGAGCTCT-3′ and 5′-AGAGCTCTTAGGGATAACAGGGTAATT-3′) were cloned into the MscI site of pBL-PUR/R to make pPU-I-RO. Third, a 1.4 kb NotI/XhoI(e/f) fragment, carrying the Zeo cassette from pfloxZeo (see below), was cloned into the NotI (e/f) site of pPU-I-RO, to make pPU-I-RO/zeo. Three steps were required to make pfloxZeo. First, pBSloxP was made by cloning a loxP site (annealed oligonucleotides 5′-CTAGATAACTTCGTATAATGTATGCTATACGAAGTTATC-3′ and 5′-ACTAGATAACTTCGTATAGCATACATTATACGAAGTTAT-3′) into the SpeI site of pBSKS(+). Second, a 1.3 kb SspI/BamHI fragment, carrying the Zeo cassette from pZeoSV (Invitrogen), was cloned into the EcoRV/BamHI site of pBSloxP to form pBSZeoloxP. Third, MscI and loxP sites (annealed oligonucleotides 5′-TCGAGTGGCCAATAACTTCGTATAATGTATGCTATACGAAGTTATA-3′ and 5′-AGCTTATAACTTCGTATAGCATACATTATACGAAGTTATTGGCCAC-3′) were cloned into the HindIII/XhoI site of pBSZeoloxP to form pfloxZeo, whose loxP site are in the same orientation.
To make p5′ΔPURO, an AspI (e/f)/HindIII fragment from pBL-Puro/R was cloned into EcoRV/HindIII-cut pBSloxPhygro, which was made by cloning the Hygro cassette, as a 1.6 kb AccI(e/f)/BamH1(e/f) fragment from pSV2hygro, into the XbaI(e/f) site of pBSloxP.
Stable transfection
Electroporation was used as described in (37) for gene targeting because lipofection is less efficient in this context (42), and for other stable transfections because it favours single integrations. For HT1080 cells, 10 μg of each plasmid was used per electroporation. For HEK293 cells, 2.5 μg of each plasmid was used per electroporation. Electroporation typically caused some cell death (e.g. 10–20%), but no systematic differences in killing were evident for the various treatments used prior to electroporation. The following concentrations were used for selecting drug-resistant colonies: G418 (200 μg/ml active concentration), Hygromycin (100 μg/ml), zeocin (200 μg/ml) and puromycine (0.8 μg/ml) 6TG (45 μg/ml). pSUPER-p53/neo and pSUPER/neo were delivered by lipofection and G418R clones were screened by western blot: all (6/6) pSUPER/neo transfectants had normal p53 levels whereas 19/36 pSUPER-p53/neo transfectants showed substantial p53 knockdown. DRneo was delivered by electroporation after linearization with BsaI. HygroR colonies were cloned or pooled (>1000 per pool) for further analysis. Clones with a single integration were identified by Southern analysis. pPU-I-RO/zeo was delivered by electroporation after linearization with SpeI and clone 293PU-I-6.2 was one of several a zeocin-resistant clones shown by Southern analysis to have a single integration. Unless stated otherwise, p5′ΔPURO was linearized with BsiWI, and pHPRThyg with SalI, prior to electroporation.
siRNA transfection
RNA oligonucleotide pairs specific for p53 were 5′-CGUACGCGGAAUACUUCGAdTdT-3′ and 5′-UCGAACUAUUCCGCGUACGdTdT-3′, and for luciferase were 5′-UUGCAAUGGAUGAUUUGAUGCdTdT-3′ and 5′-GCAACUAAUCAUCCAUUGCAAdTdT-3′, were obtained pre-annealed from Dharmacon and delivered by oligofectamine (Invitrogen) according to manufacturer's instructions. Cells were plated at 50% confluence in 24-well plates or in large (15 cm diameter) plates. The following day, a mixture containing siRNA (1200 nM, unless stated otherwise) and oligofectamine (3%, v/v) in OptiMEM was prepared according to manufacturer's instructions, and added to cells (100 μl/well, or 2 ml/15 cm plate) with 5 volumes of medium, to give a final siRNA concentration of 200 nM. After 48 h, the siRNA-containing medium was replaced with normal medium; if necessary at this stage, cells were trypsinized and replated at lower dilution to avoid confluence. After a further 24 h, cells were processed for western blots, γ-irradiation or HR assays. Western analyses were always used to confirm p53 knockdown. In many cases, ECHR and GT were carried out on the same batch of siRNA-transfected cells.
Western blots
Immunoblots were as described previously (39). For p53 detection, a primary monoclonal antibody to human p53 l (DAKO, M7001; 1:1000 dilution) and a secondary, horseradish peroxidase-conjugated goat anti-mouse immunoglobulin antibody (DAKO, P0447; 1:1000 dilution) were used.
γ-irradiation
Cells were irradiated with a 137Cs source (CIS BIO IBL 367 irradiator) at a dose rate of 1.85 Gy/min while still attached to the wells of a 6-well plate.
Flow cytometry
Flow cytometric analysis of cells for DNA content or EGFP expression was as described previously (39).
Southern blots
Standard methods were used as described elsewhere. Single integrations of DRneo were identified by BamH1 digests and probed with a neo probe; colonies with only one fragment were chosen. Single pPU-I-RO/zeo integrations were identified by SacI digests probed with a puro probe: colonies with only one fragment in addition to the 3.5 kb fragment were chosen. The neo probe was a 400 bp PCR product from the S2neo cassette, adjacent to the HindIII site. The Puro probe was a 1 kb HindIII fragment from pBL-Puro/R. The hygro probe was a 945 bp AatII/ScaI fragment of the hygromycin open reading frame.
ICHR assays
DRneo-transfected cells were transfected with siRNA (see above) in the absence of hygromycin selection. Cells were then trypsinized, counted, electroporated (3–7 million per electroporation) with pCMV3xnls-I-Sce1 or pCMVβ and replated at ∼3 million cells per 15 cm plate. After a further 24–48 h, selection in G418 was started, and continued for 10–14 days when drug-resistant colonies were counted. To score for conservative ICHR only, selection was continued for another 5–7 days in the presence of both G418 and hygromycin, and the number of surviving colonies counted. Frequencies are expressed as colonies per million cells electroporated.
ECHR assays
The previously described ECHR assay was used (39) except that HR DNA substrates were delivered by lipofectamine 2000 and this was preceded by siRNA transfection. Briefly, siRNA-transfected cells (as above) were processed immediately, if in 24-well plates or, if in 15 cm plates, were trypsinized and distributed into wells of 24-well plate at ∼50% confluence and allowed to attach for 4–6 h before proceeding. Cells were transfected with a positive control plasmid (pCX-EGFP), a negative control plasmid (p451-2) or equal weights of ECHR substrates (p451-2 and SalI-linearized p429-1). For each experiment, a single amount of control plasmid was transfected (0.8 or 1 μg per well), whereas varying amounts of ECHR substrates (0.125–2 μg of each) were used. Cells were analysed for EGFP expression by flow cytometry 24 h after the addition of DNA. The % ECHR was calculated as [(% EGFP + cells after p451-2/p429-1 cotransfection) − (% EGFP + cells after p429-1 transfection)] × 100/% EGFP + cells after pCX-EGFP transfection). Values for (% EGFP + cells after p429-1 transfection) were usually zero.
GT and RI assays
The HPRT-GT assay was used as previously described (37) and, in some instances (see Supplementary Material, Table 2), was preceded by siRNA transfection in 15 cm plates (see above). For each electroporation, ∼10–25 million cells were used, 95% of which were selected in hygromycin and 6TG, the remaining 5% selected in hygromycin only. For PURO-GT, which was always preceded by siRNA transfection in 15 cm plates, cells were trypsinized and counted. For each electroporation, 1–5 million cells were co-electroporated with p5′DPURO and pCMV3xnls-I-SceI or pCMVβ β. All or most (95%) of the electroporated cells were replated (∼3 million per 15 cm plate) for selection in puromycine. Remaining cells were replated (∼0.5 million per 9 cm plate) for selection in hygromycin. Selection was started 24–48 h after electroporation and continued for 7–14 days and colonies counted. To score for insertion events, puromycine-resistant colonies were selected for a further 10 days in the presence of both hygromycin and puromycin, and colonies recounted. GT and RI frequencies are expressed as colonies per million cells electroporated.
RESULTS
Transient and stable knockdown of p53 expression in HT1080 and HEK 293 cells
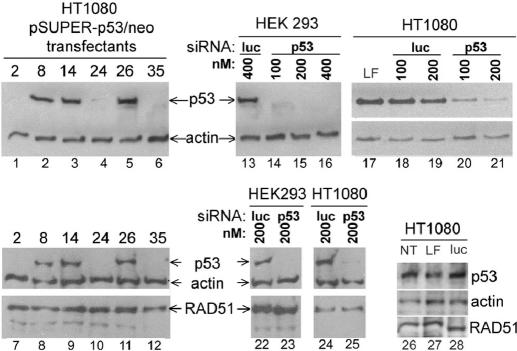
Knockdown of p53 expression by RNA interference (RNAi) was achieved in both HT1080 and HEK293 cells. In HT1080 cells, both transient and stable knockdown were demonstrated. For stable knockdown, cells were transfected to G418-resistance with pSUPER-p53/neo, a modified version of pSUPER-p53 (41), which expresses a short hairpin RNA (shRNA) against p53 mRNA and the neomycin phosphotransferase gene (neo). A plasmid (pSUPER/neo) equivalent to pSUPER-p53/neo but lacking shRNA-encoding DNA, was also used. G418-resistant (G418R) clones were screened by western analyses for p53 expression. Six pSUPER-p53/neo transfectants were chosen for further analysis: clones 2, 24 and 35, which showed substantial p53 knockdown, and clones 8, 14 and 26 in which no knockdown was detectable (Figure 1, lanes 1–6). One pSUPER/neo transfectant (clone 6) was also used as a no-knockdown control in some experiments. Transient knockdown was achieved in both HT1080 and HEK293 cells with short interfering RNA (siRNA) equivalent to the shRNA encoded by pSuper-p53. Knockdown similar to that seen for stable shRNA expression was seen 72 h after siRNA delivery by lipofection in both cell types while control (luciferase) siRNA had no such effect (Figure 1, lanes 13–21). Similar analyses extended to include RAD51 detection showed that, in HT1080 and HEK293 cells at least, RAD51 does not accumulate in response to p53 depletion (Figure 1, lanes 7–12 and 22–25).
Figure 1.
Stable and transient p53 knockdown in HT1080 and HEK293 cells. Immunoblots for p53, RAD51 and actin expression were carried out on HT1080 clones stably transfected with pSUPER derivatives (lanes 1–12), or on HT1080 and HEK293 cells transiently transfected with siRNA specific for p53 or luciferase, as indicated (lanes 13–28). LF, Lipofectamine only, NT, no treatment. In lane 28, luciferase siRNA was used at 200 nM.
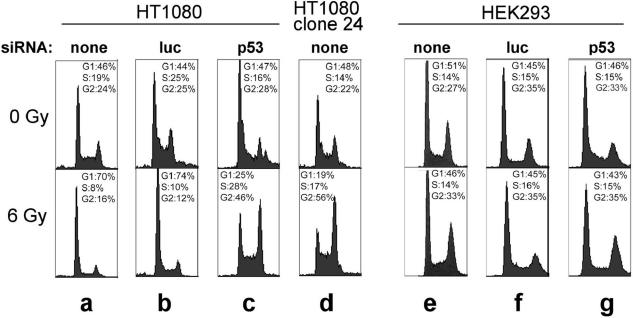
To confirm that the reduction in p53 was sufficient to have physiological consequences, DNA damage-induced cell cycle arrest in G1, a well-known p53-mediated response (43), was tested. Parental HT1080 cells show a typical reduction in S-phase and increase in G1-phase cells 24 h after irradiation, whether or not they are transfected with siRNA against luciferase (Figure 2a and b). This response is maintained in cells transiently transfected with luciferase siRNA, but in cells transiently transfected with p53 siRNA or stably transfected with pSuper-p53 (clone 24), the response is lost and p53- independent G2 accumulation predominates (Figure 2c and d). Consistent with the fact that HEK293 cells constitutively express the adenovirus E1A and E1B genes (44), known inhibitors of transcriptional activation by p53 (45–48), we detected no DNA damage-induced G1 arrest in HEK293 cells (Figure 2e–g). G2 arrest was barely if at all detectable, suggesting that p53-independent G2 arrest is somehow weakened in HEK293 cells. That p53 knockdown was physiologically effective in HEK293 cells was therefore established only in the experiments described below showing HR to be stimulated.
Figure 2.
The effect of p53 knockdown on DNA damage-induced G1 arrest. The DNA content of HT1080 (a–d) and HEK293 (e–g) cells was measured flow cytometrically 24 h after mock irradiation (top) or 6 Gy of γ-irradiation (bottom). Cells were either untreated or transfected with siRNA specific for luciferase or p53, as indicated.
Stimulation of spontaneous and I-SceI-induced ICHR by p53 knockdown
Several measurements of the effect of p53 depletion on ICHR were made (Supplementary Material, Table 1) and representative data are shown in Figure 3. To measure the frequency of ICHR, the plasmid DRneo (40) was introduced into both HT1080 and HEK293 cells and stably transfected cells were selected in hygromycin. Pools of hygromycin-resistant (hygroR) colonies (HT/DRneoP1, 293/DRneoP1 etc.), or individual HT1080 clones shown to carry a single copy of DRneo (clones HT/DRneoC9 and HT/DRneoC22) were analysed. ICHR between the two defective neo cassettes in DRneo generates a functional neo cassette, and this can occur by conservative or non-conservative pathways (Figure 3a). Selection in G418 alone yields clones that have undergone either conservative or non-conservative events, whereas selection in both G418 and hygromycin yields only clones that have undergone conservative events; 24 h before starting these selections, cells were electroporated with a control construct (pCMVβ) or an expression construct for I-SceI (pCMV3xnls-I-SceI), which stimulates HR by making a DSB in the left-hand neo cassette of pDRneo, and 72 h before electroporation, control (luciferase) or experimental (p53) siRNA was introduced into the cells by lipofection. Control experiments (data not shown) established that transfection efficiencies, transcription from the CMV promoter and colony forming ability were not affected by p53 status.
Figure 3.
ICHR assays. (a) The DRneo substrate is depicted before and after conservative or non-conservative ICHR. Thick arrows represent neo (white) and hygro (stippled) expression cassettes; circles represent their associated promoter regions. Relevant sites for HindIII (H) and fragments it generates are shown. The 3′ neo cassette lacks 5′ sequences including the promoter region and the S2neo cassette is disrupted by insertion of an I-SceI site at the NcoI (N) site. (b–d) The effect of control (black) or p53 (white or grey) siRNA on frequencies of G418R (grey background) or G418R/HygroR (white background) colonies derived from the indicated DRneo-carrying cells, with or without I-SceI expression, as indicated. (d) Southern analysis of three G418R and three G418R/HygroR colonies derived from HT/DRneoP2. Control (C) DNA was 10 pg of DRneo. All DNA was digested with HindIII and probed with the neo cassette probe indicated in (a).
In both pools and individual clones of pDRneo-transfected HT1080 cells, p53 depletion caused a marked stimulation of total (conservative + non-conservative) ICHR, with or without stimulation of HR by I-SceI (Figure 3b; Supplementary Material, Table 1, Expts 1–3). In cultures (HT/DRneoP2 and HT/DRneoP3) that had been extensively passaged in the absence of hygromycin before siRNA treatment, the baseline frequency of G418-resistance was relatively high (Figure 3c; Supplementary Material, Table 1, Expts 4 and 5). Despite this expected accumulation of functional neo genes during passaging, stimulation of G418R colony formation by p53 knockdown and by I-SceI expression were still detected, and the same was true for colonies resistant to both G418 and hygromycin (representing conservative ICHR). The relative frequencies of G418R and G418R HygR colonies suggested that 17–34% of total ICHR was conservative. Interestingly, the fold increase in total ICHR following p53 depletion was consistently higher than that for conservative ICHR. Thus, the average fold increase in the frequency of G418R colonies in response to p53 depletion (with or without I-SceI expression) was 2.6 (SD 0.51, n = 4), which is significantly higher (p = 0.032) than the equivalent figure of 1.6 (SD 0.17, n = 4) for G418R/HygroR colonies. As a control, the average fold increases in the frequency of G418R and G418R HygroR colonies in response to I-SceI expression (with or without p53 depletion) were 2.2 (SD 0.36, n = 4) and 1.9 (SD 0.5, n = 4), respectively, and not significantly different (p = 0.35). Southern analysis of G418R colonies derived from HT/DRneoP2 indicated that 12/12 had undergone non-conservative ICHR, a result consistent with the predominance of non-conservative events, although in larger sample, a proportion (∼25%) of conservative events would have been expected. As expected, all (8/8) G418R HygR colonies analysed had undergone conservative events. Representative Southern analyses are shown in Figure 3e.
Similar results were obtained in HEK293 pools 293/DRneoP1 and 293/DRneoP2 (Figure 3d; Supplementary Material, Table 1, Expts 6 and 7) providing clear evidence that p53 depletion stimulates both spontaneous and I-SceI-induced ICHR. As in HT1080 cells, p53-sensitivity was evident whether selection was for total ICHR or for conservative ICHR events only (Figure 3c), and the fold increases were greater for the former than for the latter (8.6 versus 2.9 and 15.1 versus 4.2). Together these results indicate that, in this ICHR system, non-conservative ICHR is more frequent than conservative ICHR and that, while p53 can suppress both, it preferentially suppresses the former.
ECHR is stimulated by p53 knockdown
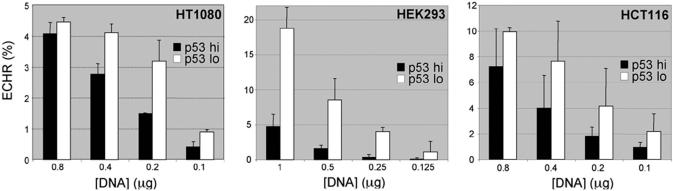
To measure ECHR, we co-transfected two plasmids (p451-2 and p429-1) with different defective EGFP cassettes that can undergo intermolecular HR to form a functional EGFP cassette (39). Cells were transfected with siRNA 72 h before introducing the recombination substrates, and analysed by flow cytometry for EGFP expression 24 h after. The ability of p53 knockdown to stimulate ECHR was evident in both HT1080 and HEK293 cells, though more pronounced in the latter cells (Figure 4). Interestingly, the degree of sensitivity to p53-depletion was dependent on the concentration of DNA used, sensitivity being lower at high concentrations, especially in HT1080 cells. The maximum levels of ECHR were ∼4-fold higher in HEK293 cells than in HT1080 cells (Figure 4). The ability to detect an effect of p53 depletion on ECHR may therefore depend on the HR capacity of the cells used and, where this is low, require the use of low concentrations of DNA substrates. Using the same assay, we also found ECHR to be stimulated by p53 depletion in HCT116 cells (Figure 4).
Figure 4.
ECHR assays. The indicated cells were transfected with siRNA against luciferase (black) or p53 (white) and, 72 h later, with the indicated amount of ECHR substrates, or with a plasmid for constitutive EGFP expression. After a further 24 h, the fraction of EGFP+ve cells was measured and expressed as a percentage of the fraction of EGFP+ve cells generated with the positive control. Measurements were done in duplicate or triplicate and SD values are indicated.
GT and random integration are not affected by p53 knockdown
To assess the effects of p53 knockdown on GT in HT1080 cells, we used a previously described targeting construct (pHPRThyg) for disruption of the HPRT gene (37). Targeted integration of pHPRThyg at the HPRT gene results in cells that are resistant to both 6-thioguanine (6TG) and hygromycin whereas random integration (RI) generates cells that are only hygror (Figure 5a). In a series of electroporations (Table 1), the effects of p53 knockdown, stably, transiently or both, on random and targeted integration frequencies were measured. Transient p53 knockdown was achieved by lipofection of siRNA 72 h prior to the introduction, by electroporation, of the pHPRThyg (methods). Western analyses on samples taken at the time of electroporation confirmed that p53 depletions similar to those shown in Figure 1 were achieved, and Southern analyses confirmed that colonies resistant to both drugs had undergone GT at the HPRT locus (data not shown). The results, whether taken individually or averaged for the three experiments, show that, in contrast to ICHR and ECHR, both GT and RI are insensitive to p53 status (Table 2 and Figure 5b, Supplementary Material).
Table 1. Targeted and random DNA integration are unaffected by p53 knockdown in HT1080.
| Expt | Cellsa | p53 | nb | RI (×104)c | TI (×106)c | TI/RI (%) |
|---|---|---|---|---|---|---|
| A | Clone 26 | hi | 3 | 6.4 ± 2.02 | 3.33 ± 0.57 | 0.53 ± 0.08 |
| Clone 24 | lo | 2 | 7.05 ± 0.07 | 3.45 ± 0.07 | 0.49 ± 0.05 | |
| B | Clones 6, 8 or 14 | hi | 4 | 3.5 ± 0.46 | 2.41 ± 0.02 | 0.67 ± 0.09 |
| Clones 2, 24 or parental HT1080 | lo | 3 | 4.2 ± 2.11 | 2.28 ± 0.83 | 0.59 ± 0.16 | |
| C | Clones 6, 8, 14, or 26 | hi | 4 | 2.1 ± 0.14 | 2.4 ± 0.55 | 1.1 ± 0.28 |
| Clones 2, 24 or 35 | lo | 6 | 2.2 ± 0.46 | 2.1 ± 0.43 | 1.00 ± 0.25 | |
| A–C | hi | 11 | 3.8 ± 2.03 | 2.7 ± 0.58 | 0.81 ± 0.33 | |
| lo | 11 | 3.6 ± 2.16 | 2.4 ± 0.7 | 0.79 ± 0.3 |
aThe indicated clones were grown individually or as combined cultures. In some cases, luciferase or p53 siRNA were used. For full details see Supplementary Material, Table 2.
bNumber of electroporations; 0.9–2.4 × 107 cells per electroporation.
cTargeted integration (TI) and random integration (RI) frequencies (average ± SD).
Table 2. Targeted and random DNA integration are unaffected by p53 knockdown in HEK293.
| Expt | EPa | siRNA | I-SceI | N (×10−6) | RI (hygror) (×104)b | TI (puror) (×106)b | TI (puror/hyror) (×106)b | TI/RI (%) |
|---|---|---|---|---|---|---|---|---|
| A | 1 | luc | − | 5.09 | 5.5 ± 0.61 | 0.13 ± 0.11 | ND | 0.025 ± 0.022 |
| 2 | luc | + | 2.25 | 4.8 ± 0.4 | 67.9 ± 11.1 | ND | 14.5 ± 2.9 | |
| 3 | p53 | − | 3.69 | 6.7 ± 1.1 | 0.18 ± 0.31 | ND | 0.023 ± 0.039 | |
| 4 | p53 | + | 1.29 | 5.5 ± 0.5 | 53.5 ± 8.3 | ND | 9.6 ± 0.89 | |
| B | 1 | luc | + | 4.31 | ND | 36 ± 8.9 | 11.7 ± 1.1 | ND |
| 2 | p53 | + | 4.00 | ND | 23.3 ± 4.0 | 9.5 ± 4.4 | ND |
aElectroporations, done in triplicate.
bRandom integration (RI) and targeted integration (TI) frequencies (average ± SD). TI (puror) measured replacement and insertional targeting, TI (puror/hyror) measured insertional targeting only.
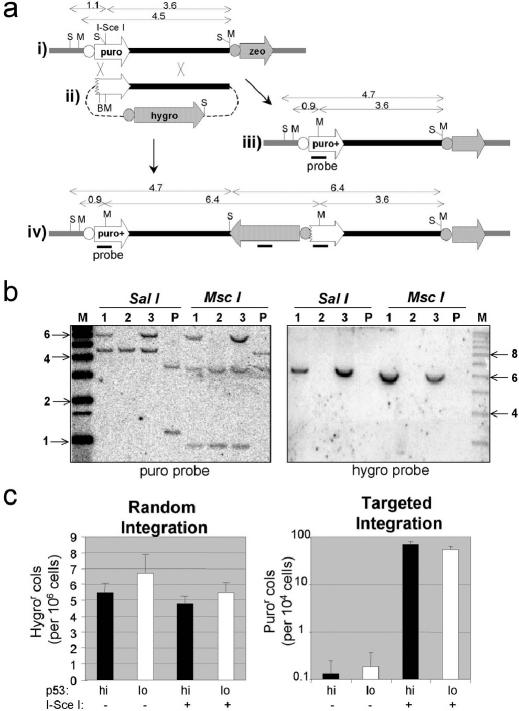
A different system was required to assess GT in HEK293 cells. An HEK293 transfectant (293PU-I-6.2), carrying a single copy of the target construct pPU-I-RO/zeo (Figure 6a, i), was transfected with the targeting construct p5′ΔPURO (Figure 6a, ii). HR between the defective puromycine-resistance cassettes (puro) in the target and targeting constructs regenerates a functional cassette. In principle, this can occur via replacement- or insertion-type GT mechanisms (Figure 6a) and Southern analyses of PuroR clones confirmed that both types of events occur (Figure 6b). The puro cassette in pPU-I-RO/zeo is defective because an I-SceI site has been inserted into the coding sequence. GT can therefore be greatly stimulated by I-SceI-catalysed DSB at the target locus, as has been shown in other GT systems (49,50). Accordingly, the targeting construct was co-electroporated with pCMV-I-SceI or with a control construct (pCMVβ). Transient p53 knockdown was again achieved by siRNA lipofection 72 h prior to electroporation, and western analysis of samples taken at the time of electroporation were used to confirm p53 knockdown (data not shown). The frequency of hygroR colonies (which is overwhelmingly made up of random integrants) was unaffected by p53 depletion, with or without I-SceI expression (Table 2 and Figure 6c). Without I-SceI stimulation, the total (insertion plus replacement) GT frequency, as indicated by the appearance of puroR colonies, was barely measurable, but I-SceI-stimulated total GT was readily measurable and found to be unaffected by p53 knockdown (Table 2, Figure 6c). The frequency of insertional GT, as indicated by the appearance of puroR/hygroR colonies, was approximately one-third of the total GT frequency, and also unaffected by p53 knockdown (Table 2, Expt B). Southern analyses similar to those in Figure 6b confirmed that puroR/hygroR colonies had the structure expected for insertional GT while PuroR colonies represented a mixture of insertion and replacement GT events (data not shown).
Figure 6.
GT and RI in HEK293 cells are unaffected by p53 knockdown. (a) Maps of the target plasmid pPU-I-RO/zeo as integrated in clone 293PUI-6.2 (i), the targeting construct p5′ΔPURO (ii) and the product of replacement (iii) or insertional (iv) GT events, are shown. Thick black and grey lines represent vector (pBSIIKS(+)), and chromosomal DNA flanking pPU-I-RO/zeo, respectively. Puromycine (black), zeocin (white) and hygromycin (grey) resistance cassettes (thick arrows) including their associated promoter regions (circles) are shown. Relevant sites for SacI (S), MscI (M) and BsiW1(B), and DNA fragments generated by them, are shown. (b) Southern analysis of DNA from clone 293PUI-6.2 (P) and three PuroR clones after co-transfection of uncut p5′ΔPURO and pCMV3xnls-I-Sce1. DNA was digested as indicated and probed with puro (left) or hygro (right) probes. (c) The frequencies of random and targeted integration after transfection of BsiWI-linearized p5′ΔPURO into clone 293PUI-6.2, with (white) or without (black) p53 knockdown, or I-SceI expression (as indicated), are shown. The data are from experiment A in Table 2.
Different sensitivities to p53 do not correspond to differences in the degree of DNA mismatch
It is known that HR is inhibited by the mismatch repair (MMR) system if sequence mismatches are generated during HR (10). Thus, for example, GT is made more efficient by inactivation of the MMR system, but only if mismatches exist between the target locus and targeting construct (11,51,52). It has been proposed that p53 inhibits HR in a similar way, perhaps in cooperation with the MMR system (28,32). None of the particular HR assays we have used here is expected to involve single base-pair mismatches, including HPRT-GT (the targeting construct was made with HT1080 genomic DNA). Insertion/deletion loops (IDLs), which can also be recognized by the MMR system, of the following nucleotide lengths could be generated, however: 25 nt (ECHR), 14 nt (ICHR), 27 nt insertion (puro-GT) and 2119 nt (HPRT-GT). By considering the first three of these, which are all of a similar size, it is clear that p53 insensitivity of GT cannot simply be explained in terms of differences in the extent of DNA mismatches. In the case of HPRT-GT, it is possible that the large IDL is recognized less efficiently by the MMR system and that this might explain p53 insensitivity. To test this directly, we introduced a small (4 bp) insertion into the left-hand arm of homology of pHPRThyg to make pHPRTBclI- (see Methods) and repeated the GT, but again found no evidence for a stimulation of GT following p53 knockdown (Table 3).
Table 3. p53 knockdown does not stimulate HPRT-GT involving mismatches.
| Plasmid | N (×10−6) | n | p53 | RI (×104) | TI (×106) | TI/RI (%) |
|---|---|---|---|---|---|---|
| pHPRThygBclI- | 2.5 | 2 | hi | 2.2 ± 0.5 | 4.8 ± 3.4 | 2.5 |
| pHPRThygBclI- | 2.5 | 2 | lo | 2.0 ± 1.7 | 2.2 ± 0.84 | 1.1 |
DISCUSSION
The results described here not only support the proposed transactivation-independent role for p53 in suppressing HR, but also provide first clear evidence that p53 can discriminate between different forms of HR. In particular, GT is shown to be unaffected by p53 depletion while, in the same cells, ICHR and ECHR are stimulated. Such differential p53-sensitivity holds for a variety of assays in each of the three cell lines where it has been analysed: HT1080, HEK293 and HCT116. Thus GT was found to be p53-insensitive whether it was spontaneous or DSB-induced, or whether it occurred by an insertion or replacement type mechanism. Conversely, ICHR was found to be p53-sensitive regardless of whether it was spontaneous or DSB-induced or whether it occurred by a conservative or non-conservative mechanism (though the latter was more sensitive).
An ability to discriminate between different forms of HR may be useful to cells for suppressing genome-destabilizing forms of HR whilst allowing genome-stabilizing forms to proceed. It is interesting to consider how the observed direction of discriminatory suppression may be compatible with such a scheme. The suppression of non-conservative ICHR, which causes deletion, duplications or inversions, is clearly consistent with this scheme. Conservative ICHR, while not leading to such gross rearrangements, is capable of generating mutations [e.g. gene conversion of a normal allele by a pseudogene (17)] and, notwithstanding its value and widespread use as an assay for HDR, must therefore be regarded as genome-stabilizing, and appropriately suppressed by p53. There is no obvious natural correlate for ECHR, but as discussed below, it involves the same mechanism as non-conservative ICHR, and this may explain its sensitivity to p53. GT is widely used in the laboratory for the purpose of gene disruption, and might therefore be regarded as genome-destabilizing, making its lack of suppression by p53 surprising. Under normal circumstances, however, opportunities for GT to occur are extremely rare, compared, for example, to HR between chromosomal repeat sequences. GT therefore probably occurs via an HR pathway that has evolved for another purpose, and we speculate, based on its insensitivity to suppression by p53, that it employs a genome-stabilizing pathway. For example, GT constructs may be able to associate, albeit at low efficiency, with recombination complexes involved in the normal HDR between single-copy sequences on sister chromatids. Unfortunately, there is no direct assay for such genome-stabilizing HDR between non-repeat sequences. The nearest assay is arguably SCE and this also appears to be p53-insensitive (34). Thus a rational physiological basis for the observed discriminatory suppression by p53 can be envisaged, but further work is required to test its validity.
Ultimately, some structural difference at the DNA/chromatin level between GT and ICHR or ECHR must be recognized by p53, directly or indirectly, and the question remains as to what this difference may be. It has been suggested that p53 may suppress HR in concert with the MMR system (28,32). To explain the differential suppression, we have observed in terms of MMR, would require that the ICHR and ECHR assays used here generate heteroduplexes with mismatches while GT assays do not. As already pointed out, however (see results), this is not the case. Furthermore, the HCT116 cell line, in which we observed p53-dependent suppression of ECHR (Figure 4), is MLH1-deficient and therefore defective for MMR (53). These considerations argue against a role for MMR, not only in differential HR suppression by p53, but also in p53-dependent HR suppression generally.
A more likely possibility is that p53 can differentiate between strand invasion (SI) and single-strand annealing (SSA) mechanisms of HR, preferentially suppressing the latter. SI is thought to be RAD51/54-dependent and SSA to be RAD51/54-independent (54). There is evidence that BRCA2 differentially influences these two forms of HR (55), and it is conceivable that p53 works in a related way. This would certainly be consistent with p53-insensitivity of GT that is generally believed to occur by SI, being RAD54-dependent (56) and stimulated by RAD51 over-expression (35,37). It would also be consistent with the p53-sensitivity of most ECHR, which occurs predominately by SSA (57,58). Indeed, in one study of ECHR in HT1080 cells (19), HR between direct repeats was found to be p53-sensitive, whereas HR between inverted repeats (which cannot occur by SSA) was not. Furthermore, the fact that non-conservative ICHR, which most likely occurs by SSA, was more p53-sensitive than conservative ICHR, which cannot occur by a simple SSA mechanism, is again consistent with preferential suppression of SSA. Conservative ICHR was nevertheless clearly p53-sensitive, suggesting that discrimination between SI and SSA cannot fully account for the observed pattern of p53-sensitivies.
It was notable that suppression of HR by p53 was still detectable in HEK293 cells (e.g. Figures 3d and 4) even though these cells express the adenoviral E1A and E1B genes (44) whose products prevent transcriptional transactivation (45,46,48) by binding, at least in the case of E1B, to the N-terminus of p53 (47). We therefore conclude that the transforming activities of E1A/E1B do not include genome destabilization through loss of p53-mediated HR suppression. This observation is also fully consistent with the proposed mechanism for suppression of HR that involves p53 residues quite distinct from those involved in transcriptional transactivation, and probably required for direct interactions with RAD51 (17). Nevertheless, our results do appear to rule out p53-induced RAD51 depletion as the mechanism, because we saw no RAD51 accumulation in response to p53 depletion. Although this contrasts with reports of RAD51 up-regulation in p53-depleted cells (29,30), it is consistent with the fact that RAD51 over-expression stimulates GT (35,37), while p53-depletion does not.
In previous studies where GT was found to be insensitive to p53 status, in HCT116 (34) and ES cells (35), ICHR and ECHR were not analysed, and the possibility that p53-senstivity may depend on the type of HR assays was not considered. Our observation that ECHR is stimulated by p53 depletion in HCT116 cells (Figure 4) argues against the idea of global p53-insensitivity of HR in these cells and in favour of discriminatory suppression. In the case of ES cells, the unusual cytoplasmic localization of p53 appeared to provide a good explanation for p53-insensitivity. Our results suggest, however, that even if p53 had been nuclear, GT may have remained unaffected by p53 depletion. An analysis of the effects of p53 knockouts on ICHR or ECHR in ES cells would therefore be of interest.
In a previous report of differential HR suppression by p53 (36), ECHR appeared to be unaffected to p53 inactivation while, in the same mouse embryo fibroblast (MEF) cells, ICHR was stimulated. On this basis, it was suggested that p53 might suppress only HR involving chromatinized HR substrates. In contrast to that study, our results and those of others (18–20) clearly show ECHR to be p53-sensitive. It is possible that this discrepancy reflects an important difference between the MEF cells used by Willers et al. and the various cell lines used in other studies. Alternatively, given the tendency for p53 sensitivity to be less pronounced at high concentrations of DNA (Figure 4), it is possible that ECHR assays in MEF cells require particularly low DNA concentrations before p53-sensitivity can be detected.
There is evidence that, in addition to suppressing HR, p53 may also suppress non-homologous end-joining (NHEJ) (59,60). We have not studied this directly, but our data (Figures 5 and 6) and those of others (21) indicate that RI, which is thought to occur by a form of NHEJ, is not influenced by p53 status. As there may be more than one NHEJ pathway (31), it is possible that an NHEJ pathway other than that responsible for RI is p53-sensitive. Integration of exogenous DNA, random or targeted, may occur during certain viral infections (61,62) or as a result of apoptosis (63), and can be considered a form of natural mutagenesis. It is therefore interesting to note that, despite its ability to suppress other genome destabilizing events, p53 suppresses neither random nor targeted DNA integration.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to Maria Jasin (pDRneo and pCMV3xnls-I-SceI) and to Reuven Agami (pSUPER and pSUPER-p53) for generously supplying plasmids.
REFERENCES
- 1.van Gent D.C., Hoeijmakers,J.H. and Kanaar,R. (2001) Chromosomal stability and the DNA double-stranded break connection. Nature Rev. Genet., 2, 196–206. [DOI] [PubMed] [Google Scholar]
- 2.Sonoda E., Sasaki,M.S., Buerstedde,J.M., Bezzubova,O., Shinohara,A., Ogawa,H., Takata,M., Yamaguchi-Iwai,Y. and Takeda,S. (1998) Rad51-deficient vertebrate cells accumulate chromosomal breaks prior to cell death. EMBO J., 17, 598–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lim D.S. and Hasty,P. (1996) A mutation in mouse rad51 results in an early embryonic lethal that is suppressed by a mutation in p53. Mol. Cell. Biol., 16, 7133–7143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Porter A.C.G. (2002) Homologous recombination. In Sons,J.W. (ed.), Wiley Encyclopedia of Molecular Medicine. John Wiley & Sons, Inc., NY, Vol. 3, pp. 1662–1666. [Google Scholar]
- 5.Inoue K. and Lupski,J.R. (2002) Molecular mechanisms for genomic disorders. Annu. Rev. Genomics Hum. Genet., 3, 199–242. [DOI] [PubMed] [Google Scholar]
- 6.Saintigny Y., Dumay,A., Lambert,S. and Lopez,B.S. (2001) A novel role for the Bcl-2 protein family: specific suppression of the RAD51 recombination pathway. EMBO J., 20, 2596–2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Slupianek A., Schmutte,C., Tombline,G., Nieborowska-Skorska,M., Hoser,G., Nowicki,M.O., Pierce,A.J., Fishel,R. and Skorski,T. (2001) BCR/ABL regulates mammalian RecA homologs, resulting in drug resistance. Mol. Cell, 8, 795–806. [DOI] [PubMed] [Google Scholar]
- 8.Wiese C., Pierce,A.J., Gauny,S.S., Jasin,M. and Kronenberg,A. (2002) Gene conversion is strongly induced in human cells by double-strand breaks and is modulated by the expression of BCL-x(L). Cancer Res., 62, 1279–1283. [PubMed] [Google Scholar]
- 9.Elliott B. and Jasin,M. (2001) Repair of double-strand breaks by homologous recombination in mismatch repair-defective mammalian cells. Mol. Cell. Biol., 21, 2671–2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Evans E. and Alani,E. (2000) Roles for mismatch repair factors in regulating genetic recombination. Mol. Cell. Biol., 20, 7839–7844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Wind N., Dekker,M., Berns,A., Radman,M. and te Riele,H. (1995) Inactivation of the mouse Msh2 gene results in mismatch repair deficiency, methylation tolerance, hyperrecombination, and predisposition to cancer. Cell, 82, 321–330. [DOI] [PubMed] [Google Scholar]
- 12.Powell S.N., Willers,H. and Xia,F. (2002) BRCA2 keeps Rad51 in line. High-fidelity homologous recombination prevents breast and ovarian cancer? Mol. Cell, 10, 1262–1263. [DOI] [PubMed] [Google Scholar]
- 13.Jasin M. (2002) Homologous repair of DNA damage and tumorigenesis: the BRCA connection. Oncogene, 21, 8981–8993. [DOI] [PubMed] [Google Scholar]
- 14.Tutt A. and Ashworth,A. (2002) The relationship between the roles of BRCA genes in DNA repair and cancer predisposition. Trends Mol. Med., 8, 571–576. [DOI] [PubMed] [Google Scholar]
- 15.Yang Q., Zhang,R., Wang,X.W., Spillare,E.A., Linke,S.P., Subramanian,D., Griffith,J.D., Li,J.L., Hickson,I.D., Shen,J.C. et al. (2002) The processing of Holliday junctions by BLM and WRN helicases is regulated by p53. J. Biol. Chem., 277, 31980–31987. [DOI] [PubMed] [Google Scholar]
- 16.Traverso G., Bettegowda,C., Kraus,J., Speicher,M.R., Kinzler,K.W., Vogelstein,B. and Lengauer,C. (2003) Hyper-recombination and genetic instability in BLM-deficient epithelial cells. Cancer Res., 63, 8578–8581. [PubMed] [Google Scholar]
- 17.Bertrand P., Saintigny,Y. and Lopez,B.S. (2004) p53's double life: transactivation-independent repression of homologous recombination. Trends Genet., 20, 235–243. [DOI] [PubMed] [Google Scholar]
- 18.Wiesmuller L., Cammenga,J. and Deppert,W.W. (1996) In vivo assay of p53 function in homologous recombination between simian virus 40 chromosomes. J. Virol., 70, 737–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Slebos R.J. and Taylor,J.A. (2001) A novel host cell reactivation assay to assess homologous recombination capacity in human cancer cell lines. Biochem. Biophys. Res. Commun., 281, 212–219. [DOI] [PubMed] [Google Scholar]
- 20.Linke S.P., Sengupta,S., Khabie,N., Jeffries,B.A., Buchhop,S., Miska,S., Henning,W., Pedeux,R., Wang,X.W., Hofseth,L.J. et al. (2003) p53 interacts with hRAD51 and hRAD54, and directly modulates homologous recombination. Cancer Res., 63, 2596–2605. [PubMed] [Google Scholar]
- 21.Mekeel K.L., Tang,W., Kachnic,L.A., Luo,C.M., DeFrank,J.S. and Powell,S.N. (1997) Inactivation of p53 results in high rates of homologous recombination. Oncogene, 14, 1847–1857. [DOI] [PubMed] [Google Scholar]
- 22.Bertrand P., Rouillard,D., Boulet,A., Levalois,C., Soussi,T. and Lopez,B.S. (1997) Increase of spontaneous intrachromosomal homologous recombination in mammalian cells expressing a mutant p53 protein. Oncogene, 14, 1117–1122. [DOI] [PubMed] [Google Scholar]
- 23.Saintigny Y., Rouillard,D., Chaput,B., Soussi,T. and Lopez,B.S. (1999) Mutant p53 proteins stimulate spontaneous and radiation-induced intrachromosomal homologous recombination independently of the alteration of the transactivation activity and of the G1 checkpoint. Oncogene, 18, 3553–3563. [DOI] [PubMed] [Google Scholar]
- 24.Dudenhoffer C., Kurth,M., Janus,F., Deppert,W. and Wiesmuller,L. (1999) Dissociation of the recombination control and the sequence-specific transactivation function of P53. Oncogene, 18, 5773–5784. [DOI] [PubMed] [Google Scholar]
- 25.Willers H., McCarthy,E.E., Wu,B., Wunsch,H., Tang,W., Taghian,D.G., Xia,F. and Powell,S.N. (2000) Dissociation of p53-mediated suppression of homologous recombination from G1/S cell cycle checkpoint control. Oncogene, 19, 632–639. [DOI] [PubMed] [Google Scholar]
- 26.Lu X., Lozano,G. and Donehower,L.A. (2003) Activities of wildtype and mutant p53 in suppression of homologous recombination as measured by a retroviral vector system. Mutat. Res., 522, 69–83. [DOI] [PubMed] [Google Scholar]
- 27.Saintigny Y. and Lopez,B.S. (2002) Homologous recombination induced by replication inhibition, is stimulated by expression of mutant p53. Oncogene, 21, 488–492. [DOI] [PubMed] [Google Scholar]
- 28.Zink D., Mayr,C., Janz,C. and Wiesmuller,L. (2002) Association of p53 and MSH2 with recombinative repair complexes during S phase. Oncogene, 21, 4788–4800. [DOI] [PubMed] [Google Scholar]
- 29.Kumari A., Schultz,N. and Helleday,T. (2004) p53 protects from replication-associated DNA double-strand breaks in mammalian cells. Oncogene, 23, 2324–2329. [DOI] [PubMed] [Google Scholar]
- 30.Xia S.J., Shammas,M.A. and Shmookler Reis,R.J. (1997) Elevated recombination in immortal human cells is mediated by HsRAD51 recombinase. Mol. Cell. Biol., 17, 7151–7158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vasquez K.M., Marburger,K., Intody,Z. and Wilson,J.H. (2001) Manipulating the mammalian genome by homologous recombination. Proc. Natl Acad. Sci. USA, 98, 8403–8410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dudenhoffer C., Rohaly,G., Will,K., Deppert,W. and Wiesmuller,L. (1998) Specific mismatch recognition in heteroduplex intermediates by p53 suggests a role in fidelity control of homologous recombination. Mol. Cell. Biol., 18, 5332–5342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Susse S., Janz,C., Janus,F., Deppert,W. and Wiesmuller,L. (2000) Role of heteroduplex joints in the functional interactions between human Rad51 and wild-type p53. Oncogene, 19, 4500–4512. [DOI] [PubMed] [Google Scholar]
- 34.Bunz F., Fauth,C., Speicher,M.R., Dutriaux,A., Sedivy,J.M., Kinzler,K.W., Vogelstein,B. and Lengauer,C. (2002) Targeted inactivation of p53 in human cells does not result in aneuploidy. Cancer Res., 62, 1129–1133. [PubMed] [Google Scholar]
- 35.Dominguez-Bendala J., Priddle,H., Clarke,A. and McWhir,J. (2003) Elevated expression of exogenous Rad51 leads to identical increases in gene-targeting frequency in murine embryonic stem (ES) cells with both functional and dysfunctional p53 genes. Exp. Cell Res., 286, 298–307. [DOI] [PubMed] [Google Scholar]
- 36.Willers H., McCarthy,E.E., Hubbe,P., Dahm-Daphi,J. and Powell,S.N. (2001) Homologous recombination in extrachromosomal plasmid substrates is not suppressed by p53. Carcinogenesis, 22, 1757–1763. [DOI] [PubMed] [Google Scholar]
- 37.Yáñez R.J. and Porter,A.C.G. (1999) Gene targeting is enhanced in human cells overexpressing hRAD51. Gene Ther., 6, 1282–1290. [DOI] [PubMed] [Google Scholar]
- 38.Johnson R.D., Liu,N. and Jasin,M. (1999) Mammalian XRCC2 promotes the repair of DNA double-strand breaks by homologous recombination. Nature, 401, 397–399. [DOI] [PubMed] [Google Scholar]
- 39.Yáñez R.J. and Porter,A.C.G. (2002) Differential effects of Rad52p overexpression on gene targeting and extrachromosomal homologous recombination in a human cell line. Nucleic Acids Res., 30, 740–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liang F., Romanienko,P.J., Weaver,D.T., Jeggo,P.A. and Jasin,M. (1996) Chromosomal double-strand break repair in Ku80-deficient cells. Proc. Natl Acad. Sci. USA, 93, 8929–8933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brummelkamp T.R., Bernards,R. and Agami,R. (2002) A system for stable expression of short interfering RNAs in mammalian cells. Science, 296, 550–553. [DOI] [PubMed] [Google Scholar]
- 42.Yáñez R.J. and Porter,A.C.G. (1999) Influence of DNA delivery method on gene targeting frequencies in human cells. Somat. Cell Mol. Genet., 25, 27–31. [DOI] [PubMed] [Google Scholar]
- 43.Fei P. and El-Deiry,W.S. (2003) P53 and radiation responses. Oncogene, 22, 5774–5783. [DOI] [PubMed] [Google Scholar]
- 44.Nevins J.R. (1982) Induction of the synthesis of a 70,000 dalton mammalian heat shock protein by the adenovirus E1A gene product. Cell, 29, 913–919. [DOI] [PubMed] [Google Scholar]
- 45.Steegenga W.T., Van Laar,T., Shvarts,A., Terleth,C., Van der Eb,A.J. and Jochemsen,A.G. (1995) Distinct modulation of p53 activity in transcription and cell-cycle regulation by the large (54 kDa) and small (21 kDa) adenovirus E1B proteins. Virology, 212, 543–554. [DOI] [PubMed] [Google Scholar]
- 46.Steegenga W.T., van Laar,T., Riteco,N., Mandarino,A., Shvarts,A., van der Eb,A.J. and Jochemsen,A.G. (1996) Adenovirus E1A proteins inhibit activation of transcription by p53. Mol. Cell. Biol., 16, 2101–2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lin J., Chen,J., Elenbaas,B. and Levine,A.J. (1994) Several hydrophobic amino acids in the p53 amino-terminal domain are required for transcriptional activation, binding to mdm-2 and the adenovirus 5 E1B 55-kD protein. Genes Dev., 8, 1235–1246. [DOI] [PubMed] [Google Scholar]
- 48.Yew P.R. and Berk,A.J. (1992) Inhibition of p53 transactivation required for transformation by adenovirus early 1B protein. Nature, 357, 82–85. [DOI] [PubMed] [Google Scholar]
- 49.Jasin M. (1996) Genetic manipulation of genomes with rare-cutting endonucleases. Trends Genet., 12, 224–228. [DOI] [PubMed] [Google Scholar]
- 50.Miller D.G., Petek,L.M. and Russell,D.W. (2003) Human gene targeting by adeno-associated virus vectors is enhanced by DNA double-strand breaks. Mol. Cell. Biol., 23, 3550–3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.te Riele H., Maandag,E.R. and Berns,A. (1992) Highly efficient gene targeting in embryonic stem cells through homologous recombination with isogenic DNA constructs. Proc. Natl Acad. Sci. USA, 89, 5128–5132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Deng C. and Capecchi,M.R. (1992) Reexamination of gene targeting frequency as a function of the extent of homology between the targeting vector and the target locus. Mol. Cell. Biol., 12, 3365–3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Boyer J.C., Umar,A., Risinger,J.I., Lipford,J.R., Kane,M., Yin,S., Barrett,J.C., Kolodner,R.D. and Kunkel,T.A. (1995) Microsatellite instability, mismatch repair deficiency, and genetic defects in human cancer cell lines. Cancer Res., 55, 6063–6070. [PubMed] [Google Scholar]
- 54.Karran P. (2000) DNA double strand break repair in mammalian cells. Curr. Opin. Genet. Dev., 10, 144–150. [DOI] [PubMed] [Google Scholar]
- 55.Tutt A., Bertwistle,D., Valentine,J., Gabriel,A., Swift,S., Ross,G., Griffin,C., Thacker,J. and Ashworth,A. (2001) Mutation in Brca2 stimulates error-prone homology-directed repair of DNA double-strand breaks occurring between repeated sequences. EMBO J., 20, 4704–4716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Essers J., Hendriks,R.W., Swagemakers,S.M., Troelstra,C., de Wit,J., Bootsma,D., Hoeijmakers,J.H. and Kanaar,R. (1997) Disruption of mouse RAD54 reduces ionizing radiation resistance and homologous recombination. Cell, 89, 195–204. [DOI] [PubMed] [Google Scholar]
- 57.Seidman M.M. (1987) Intermolecular homologous recombination between transfected sequences in mammalian cells is primarily nonconservative. Mol. Cell. Biol., 7, 3561–3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lin F.L., Sperle,K. and Sternberg,N. (1984) Model for homologous recombination during transfer of DNA into mouse L cells: role for DNA ends in the recombination process. Mol. Cell. Biol., 4, 1020–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bill C.A., Yu,Y., Miselis,N.R., Little,J.B. and Nickoloff,J.A. (1997) A role for p53 in DNA end rejoining by human cell extracts. Mutat. Res., 385, 21–29. [DOI] [PubMed] [Google Scholar]
- 60.Okorokov A.L., Warnock,L. and Milner,J. (2002) Effect of wild-type, S15D and R175H p53 proteins on DNA end joining in vitro: potential mechanism of DNA double-strand break repair modulation. Carcinogenesis, 23, 549–557. [DOI] [PubMed] [Google Scholar]
- 61.Hirata R., Chamberlain,J., Dong,R. and Russell,D.W. (2002) Targeted transgene insertion into human chromosomes by adeno-associated virus vectors. Nat. Biotechnol., 20, 735–738. [DOI] [PubMed] [Google Scholar]
- 62.Wurtele H., Little,K.C. and Chartrand,P. (2003) Illegitimate DNA integration in mammalian cells. Gene Ther., 10, 1791–1799. [DOI] [PubMed] [Google Scholar]
- 63.Bergsmedh A., Szeles,A., Henriksson,M., Bratt,A., Folkman,M.J., Spetz,A.L. and Holmgren,L. (2001) Horizontal transfer of oncogenes by uptake of apoptotic bodies. Proc. Natl Acad. Sci. USA, 98, 6407–6411. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.