Abstract
The objective of this descriptive study was to determine Felis domesticus (cat) and Canis familiaris (dog) protein epitopes that bind strongly to selected HLA class II alleles to identify synthetic vaccine candidate epitopes and to identify individuals/populations who are likely to respond to vaccines. FASTA amino acid sequences of experimentally validated allergenic proteins of house cat and dog were identified using International Union of Immunological Societies (IUIS) allergen nomenclature database. NetMHCII 2.2 server was used to determine binding affinities in the form of 1-log 50 k and in nM with commonly found HLA II alleles. Screening of house cat and dog allergenic proteins identified 4 (with 2 isoforms for chain 1 and 3 isoforms for chain 2 for fel d 1) and 6 proteins, respectively. Number of strong binders from each protein against each HLA type was determined as potential candidate for allergen immunotherapy. HLA-DRB1*0101 bound maximum number of epitopes (207 and 275 from house cat and dog, respectively) while HLA-DRB1*0802 bound none. We conclude that HLA specific epitope prediction can help identify synthetic peptide vaccine candidates and predict response as well.
Key Words: Binding affinity, Bioinformatics, Dog, House cat, Human leukocyte antigen
Introduction
Allergic diseases are increasing worldwide and an allergic family history is one of the strongest risk factors for childhood allergy (Lowe et al., 2004 ▶). Some studies (Asher et al., 2006 ▶) strongly suggest that environmental factors also play an important role. Although pets are known to aggravate asthma, allergic rhinitis, and eczema in sensitized individuals (TePas et al., 2006 ▶), controversy remains about whether early life pet exposure is a risk factor or a protective factor in their development. Current guidelines issued in Australia (Prescott and Tang, 2005 ▶), the United States (Expert Panel Report, 2007), and the United Kingdom (Douglas et al., 2008 ▶) and by the Global Initiative for Asthma (Bateman et al., 2008 ▶) all agree there is currently insufficient evidence to provide any recommendations in relation to pet. Sensitization to animal allergens is one of the most important risk factors for developing allergic diseases such as asthma, rhinitis and atopic dermatitis, particularly in occupationally exposed workers. The sensitization phase involves processing and presentation of inhaled aeroallergens to T lymphocytes with activation of Th2 cells. The release of Th2-mediatedcytokines (IL-4, IL-5 and IL-13) leads to IgE synthesis, mast cell degranulation and eosinophilic response (Agarwal, 2009 ▶). T cells recognize this antigen when presented with human leukocyte antigen (HLA) complex on the surface of antigen presenting cells. HLA alleles show differential binding affinities for various epitopes in antigens, and this determines the basis of protective response against foreign antigens by the individual. This differential binding is due to allele specific amino acid composition and thus distinct polarity and stereochemistry of antigen binding pockets on HLA molecule (Tipu and Ahmed, 2014 ▶; Tipu et al., 2014 ▶). As of Jan 2016, 3356 HLA-A, 4179 HLA-B, 2902 HLA-C and 1976 HLA-DR alleles (besides other HLA class I and II loci) have been identified (HLA Nomenclature, 2016 ▶).
Short synthetic peptides are being evaluated for use as vaccines for autoimmune and allergic disorders. Their use allows delivery of epitopes of allergens to T lymphocytes for generation of immune response. These also carry the advantages of low cost, easy purification and good stability, characteristics that are difficult to reproduce in allergen extracts (Moldaver and Larche, 2011 ▶). This “vaccinomics” approach is already being employed in designing bacterial and viral vaccines (Whelan et al., 2013 ▶). Until recently the use of peptides has been restricted to cat allergy, ragweed allergy and bee venom, with varying success (Sirskyj et al., 2011 ▶). For a peptide to be a successful vaccine candidate, it must first bind to HLA molecules in order to be presented to T lymphocytes for immune response generation (Sirskyj et al., 2011 ▶).
In this study we aimed to first identify experimentally validated allergenic proteins from online databases for two species of pets, house cat and dog. Peptides from these allergenic proteins were analyzed for their binding with representative HLA-DRB1 alleles using artificial neural networks method. Since helper T cells response and B cells stimulation for antibody generation require presentation with HLA class II alleles. However, it must be emphasized that ultimately such predictive in silico work needs to be confirmed through in vivo experiments (Sirskyj et al., 2011 ▶).
Materials and Methods
This descriptive study was carried out in Januray 2016 at Armed Forces Institute of Pathology (AFIP), Rawalpindi, using specific databases and tools. Felis domesticus (cat) and Canis familiaris (dog) were screened for identified allergenic proteins using International Union of Immunological Societies (IUIS) allergen nomenclature database (Allergen Nomenclature, 2016 ▶). FASTA format of all these revealed proteins (and their nucleotides) were saved for further analysis. The FASTA sequences for each individual protein were fed into NetMHCII 2.2 server from Technical University of Denmark. This server breaks the amino acid sequence of tested protein into 15 amino acid peptide sequences and predicts binding of each peptide sequence to most commonly found HLA alleles, using artificial neural networks. This server has been reported to provide very good binding results in predicting HLA-DRB1 locus (Wand et al., 2010 ▶). For our study we analyzed only HLA-DRB1 locus. It gives affinity of peptides with HLA alleles in the form of 1-log 50 k and in nM, with higher and lower values indicating stronger binding, respectively (Nielsen et al., 2007 ▶; Nielsen and Lund, 2009 ▶). The affinity of each allergenic protein was tested for binding with HLA-DRB1*01, 03, 04, 07, 08, 09, 11, 13 and 15 with HLA DRB1*0101, 0301, 0401, 0701, 0802, 0901, 1101, 1302, 1501 as representative alleles. From the results, strong binders (peptide sequences binding strongly with respective HLA alleles) were determined with affinity threshold value of less than 50 nM.
Results
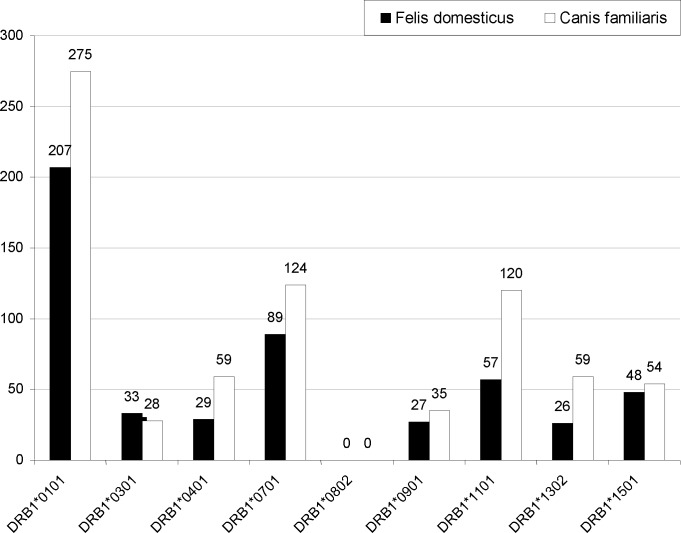
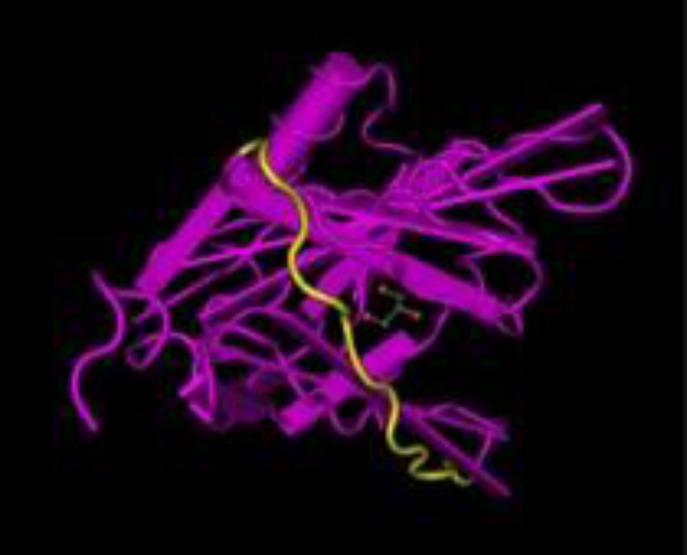
Both the F. domesticus and C. familiaris belong to Animalia chordate of Carnivore order. Screening F. domesticus in IUIS database revealed 4 allergenic proteins (with 2 and 3 isoforms of chain 1 and chain 2, respectively for fel d 1 protein) belonging to different families, for which both nucleotide and peptide sequence is available. Similar screening for C. familiaris revealed 6 allergenic proteins. These proteins along with their biochemical names, GenBank nucleotide ID and Uniprot ID are shown in Table 1. Table 2 summarizes individually, the number of high affinity epitopes of each allergenic protein (mentioned above) against re-presentative HLA alleles and represents results of steps 3 and 4 in “Materials and Methods” section. Figure 1 shows total number of epitopes in all proteins to which our representative HLA alleles show high binding. It is evident that HLA-DRB1*0101 binds maximum number of epitopes from allergenic proteins of both species while HLA-DRB1*0801 binds none. Exact amino acid sequence of allergenic epitopes and their corresponding binding HLA alleles are available upon request. For example, C. familiaris Can f 2 protein has an epitope starting at 80th position, GQCEKVSLTAFKTAT. It binds strongly with HLA-DRB1*0101 with affinity 31.3 nM. Figure 2 shows 3-D structure of protein with this epitope highlighted.
Table 1.
Allergenic proteins identified in IUIS database
| S No. | Allergenic protein | Biochemical name | GenBank ID | Uniprot ID |
|---|---|---|---|---|
| Felis domesticus | ||||
| 1 | Fel d 1 | Uteroglobin | M74952 (chain 1), M77341 (chain 2) | P30438 (chain 1), P30440 (chain 2) |
| 2 | Fel d 2 | Serum albumin | X84842 | P49064 |
| 3 | Fel d 3 | Cystatin | AF238996 | Q8WNR9 |
| 4 | Fel d 4 | Lipocalin | AY497902 | Q5VFH6 |
| Canis familiaris | ||||
| 5 | Can f 1 | Lipocalin | AF027177 | O18873 |
| 6 | Can f 2 | Lipocalin | AF027178 | O18874 |
| 7 | Can f 3 | Serum albumin | AB090854 | P49822 |
| 8 | Can f 4 | Lipocalin | GU132996 | D7PBH4 |
| 9 | Can f 5 | Arginine esterase | Y00751 | P09582 |
| 10 | Can f 6 | Lipocalin | HE653774 | H2B3G5 |
Table 2.
Number of strong binders in each allergenic protein against representative HLA allele
| Allergenic protein | HLA II allele |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| 0101 | 0301 | 0401 | 0701 | 0802 | 0901 | 1101 | 1302 | 1501 | |
| Felis domesticus | |||||||||
| Fel d 1 chain 1 | |||||||||
| Isoform 1 | 21 | 1 | 0 | 6 | 0 | 3 | 0 | 4 | 7 |
| Isoform 2 | 14 | 1 | 0 | 6 | 0 | 3 | 0 | 4 | 7 |
| Fel d 1 chain 2 | |||||||||
| Isoform 1 | 18 | 6 | 4 | 8 | 0 | 0 | 0 | 2 | 0 |
| Isoform 2 | 19 | 6 | 4 | 8 | 0 | 0 | 0 | 2 | 0 |
| Isoform 3 | 19 | 6 | 4 | 8 | 0 | 0 | 0 | 2 | 0 |
| Fel d 2 | 74 | 4 | 12 | 38 | 0 | 16 | 43 | 0 | 22 |
| Fel d 3 | 19 | 4 | 0 | 6 | 0 | 0 | 7 | 5 | 12 |
| Fel d 4 | 23 | 5 | 5 | 9 | 0 | 5 | 7 | 7 | 0 |
| Total epitopes identified | 207 | 33 | 29 | 89 | 0 | 27 | 57 | 26 | 48 |
| Canis familiaris | |||||||||
| Can f 1 | 41 | 0 | 8 | 11 | 0 | 0 | 19 | 10 | 0 |
| Can f 2 | 41 | 0 | 20 | 14 | 0 | 8 | 7 | 2 | 16 |
| Can f 3 | 89 | 8 | 10 | 42 | 0 | 17 | 57 | 0 | 15 |
| Can f 4 | 43 | 8 | 12 | 22 | 0 | 1 | 16 | 20 | 12 |
| Can f 5 | 41 | 0 | 0 | 25 | 0 | 5 | 19 | 8 | 0 |
| Can f 6 | 20 | 12 | 9 | 10 | 0 | 4 | 2 | 19 | 11 |
| Total epitopes identified | 275 | 28 | 59 | 124 | 0 | 35 | 120 | 59 | 54 |
Fig. 1.
Number of high affinity epitopes of Felis domesticus and Canis familiaris binding to representative HLA alleles
Fig. 2.
3-D structure of can f 1 allergenic protein, showing epitope GQCEKVSLTAFKTAT highlighted in yellow (adopted from PDB ID 3L4R, modified in NCBI Cn3-D structures
Discussion
Antigen presentation to T cells by HLA molecules is the key step towards the development of an antigen specific immune response. Specific HLA alleles influence specific IgE responses to airborne allergens (Park et al., 2012 ▶). Determining this genetic association can help identify individuals at risk of developing different disorders (Tipu et al., 2011 ▶). Determining the binding of a peptide to HLA alleles aids designing synthetic peptide vaccines. This bioinformatics approach is known as “reverse vaccinology” and has arisen because conventional experimental approaches are extremely laborious, expensive and time consuming. “reverse vaccinology” involves computational methods to identify all potential candidate immunogens from genome of a pathogen/allergen. Once appropriate vaccine candidates have been identified, genes of interest can be cloned to produce corresponding protein. Subsequent in vivo and in vitro testing can further validate potential use in specific population (Tomar and De, 2010 ▶).
In total we were able to identify 516 (from 4 allergenic proteins) and 754 (from 6 allergenic proteins) 15mer peptide sequences from F. domesticus and C. familiaris, respectively, that showed strong binding to HLA-DRB1 alleles. These epitopes are candidates for wet laboratory testing of synthetic peptides for allergen immunotherapy. Reduction of nasal symptoms after treatment with synthetic peptides from cat proteins has recently been reported (Hafner et al., 2013 ▶). Worm et al. (2011) ▶ have used seven experimentally validated peptides of varying lengths from fel d 1 protein for designing peptide vaccine. All of the peptides (from fel d 1 only) they have tested have been found to be strong binders by us, although length of peptide may vary by few amino acid residues. Oseroff et al. (2012) ▶ have validated in vitro 133 different allergens including those from house cat and dogs. They found several of the 15mer peptide sequences from fel d 1, fel d 2, fel d 3 and can f 3 that showed T lymphocyte stimulation in vitro. All of these peptides have also been found as strong HLA binders in our results.
We also found that HLA-DRB1*0101 could bind the maximum number of epitopes and DRB1*0802 is not able to bind any epitope in either species. All other alleles showed variable binding affinity as depicted in Table 2. Its implication lies in designing the individualized immunotherapy as peptide vaccines according to HLA types dominant in a population. In fact, peptide vaccines restricted to HLA types for different types of cancer are already in place (Berlin et al., 2014 ▶; Sonpavde et al., 2014 ▶). HLA restricted epitopes have also been found useful in peanut allergy subjects (Prickett et al., 2013 ▶). Similarly Immonen et al. (2005) ▶ have determined seven epitopes from can f 1 protein suitable for allergen immunotherapy according to seven commonly found HLA types. Our study was limited by the fact that we tested the peptides in silico only. However, these results identified specific peptides according to HLA types for individual allergenic proteins for in vitro and then in vivo experiments.
This article highlights the integration of “immunoinformatics” and “vaccinomics” for deter-mining candidate peptide sequences from experimentally validated allergenic proteins of two common pets. These computationally determined epitopes will provide the basis for in vitro and then in vivo experiments as potential vaccine candidates. Our results also show that individuals are likely to respond to these epitopes according to their HLA types and provide a base for tailored immunotherapy.
References
- Agarwal, R. Allergic bronchopulmonary aspergillosis. Chest. 2009;135:805–826. doi: 10.1378/chest.08-2586. [DOI] [PubMed] [Google Scholar]
- Allergen Nomenclature: IUIS Allergen Nomenclature Sub-Committee [online] 2016. Available at http://www.allergen.org/.
- Asher, MI, Montefort, S, Björkstén, B, Lai, CK, Strachan, DP, Weiland, SK, Williams, H, ISAAC Phase Three Study Group. Worldwide time trends in the prevalence of symptoms of asthma, allergic rhino-conjunctivitis, and eczema in childhood: ISAAC phases one and three repeat multicountry cross-sectional surveys. Lancet. 2006;368:733–743. doi: 10.1016/S0140-6736(06)69283-0. [DOI] [PubMed] [Google Scholar]
- Bateman, ED, Hurd, SS, Barnes, PJ. Global strategy for asthma management and prevention: GINA executive summary. Europ. Resp. J. 2008;31:143–178. doi: 10.1183/09031936.00138707. [DOI] [PubMed] [Google Scholar]
- Berlin, C, Kowalewski, DJ, Schuster, H, Mirza, N, Walz, S, Handel, M, Schmid-Horch, B, Salih, HR, Kanz, L, Rammensee, HG, Stevanovic, S, Stickel, JS. Mapping the HLA ligandome landscape of acute myeloid leukemia: a targeted approach toward peptide-based immunotherapy. Leukemia. 2014;30:1003–1004. doi: 10.1038/leu.2016.1. [DOI] [PubMed] [Google Scholar]
- Douglas, G, Higgins, B, Barnes, N. British guideline on the management of asthma: a national clinical guideline. Thorax. 2008;63:iv1–iv121. doi: 10.1136/thx.2008.097741. [DOI] [PubMed] [Google Scholar]
- Hafner, R, Couroux, P, Armstrong, K, Patel, D, Larche, M, Haumann, B. Two year persistent treatment effect in reducing nasal symptoms of cat allergy after 4 doses of Cat-PAD, the first in a new class of synthetic peptide immune-regulatory epitopes. Clin. Transl. Allergy. 2013;S3:O7. [Google Scholar]
- HLA Nomenclature: HLA Alleles Numbers [online] 2016. Available at http://hla.alleles.org/nomenclature/stats.html.
- Immonen, A, Farci, S, Taivainen, A, Partanen, J, Pouvelle-Moratille, S, Narvanen, A, Kinnunen, T, Saarelainen, S, Rytkonen-Nissinen, M, Maillere, B, Virtanen, T. T cell epitope-containing peptide of the major dog allergen Can f 1 as candidates for allergen immunotherapy. J. Immunol. 2005;175:3614–3620. doi: 10.4049/jimmunol.175.6.3614. [DOI] [PubMed] [Google Scholar]
- Lowe, L, Custovic, A, Woodcock, A. Childhood asthma. Curr. Allergy Asthm. R. 2004;4:159–165. doi: 10.1007/s11882-004-0062-9. [DOI] [PubMed] [Google Scholar]
- Moldaver, D, Larche, M. Immunotherapy with peptides. Allergy. 2011;66:784–791. doi: 10.1111/j.1398-9995.2011.02610.x. [DOI] [PubMed] [Google Scholar]
- National Asthma Education, Prevention Program. Expert Panel Report 3 (EPR-3): guidelines for the diagnosis and management of asthma-summary report 2007. J. Allergy Clin. Immunol. 2007;120:S94–S138. doi: 10.1016/j.jaci.2007.09.043. [DOI] [PubMed] [Google Scholar]
- Nielsen, M, Lund, O. NN-align An artificial neural network-based alignment algorithm for MHC class II peptide binding prediction. BMC Bioinformatics. 2009;10:296–306. doi: 10.1186/1471-2105-10-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen, M, Lundegaard, C, Lund, O. Prediction of MHC class II binding affinity using SMM-align, a novel stabilization matrix alignment method. BMC Bioinformatics. 2007;8:238–250. doi: 10.1186/1471-2105-8-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oseroff, C, Sidney, J, Vita, R, Tripple, V, McKinney, DM, Southwood, S, Brodie, TM, Sallusto, F, Grey, H, Alam, R, Brodie, D, Greenbaum, JA, Kolla, R, Peters, B, Sette, A. T cell responses to known allergen proteins are differently polarized and account for a variable fraction of total response to allergen extracts. J. Immunol. 2012;189:1800–1811. doi: 10.4049/jimmunol.1200850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, H, Ahn, K, Park, MH, Lee, SI. The HLA-DRB1 polymorphism is associated with atopic dermatitis, but not egg allergy in Korean children. Allergy Asthma Immunol. Res. 2012;4:143–149. doi: 10.4168/aair.2012.4.3.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott, SL, Tang, MLK. The Australasian Society of Clinical Immunology and Allergy position statement: summary of allergy prevention in children. Med. J. Aust. 2005;182:464–467. doi: 10.5694/j.1326-5377.2005.tb06787.x. [DOI] [PubMed] [Google Scholar]
- Prickett, SR, Voskamp, AL, Phan, T, Dacumos-Hill, A, Mannering, SI, Rolland, JM, O’Hehir, RE. Ara h 1 CD4+ T cell epitope-based peptides: candidates for a peanut allergy therapeutic. Clin. Exp. Allergy. 2013;43:684–697. doi: 10.1111/cea.12113. doi: 10.1111/cea.12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirskyj, D, Diaz-Mitoma, F, Golshani, A, Kumar, A, Azizi, A. Innovative bioinformatics approaches for developing peptide based vaccines against hypervariable viruses. Immunol. Cell Biol. 2011;89:81–89. doi: 10.1038/icb.2010.65. [DOI] [PubMed] [Google Scholar]
- Sonpavde, G, Wang, M, Peterson, LE, Wang, HY, Joe, T, Mims, MP, Kadmon, D, Ittmann, MM, Wheeler, TM, Gee, AP, Wang, RF, Hayes, TG. HLA-restricted NY-ESO-1 peptide immunotherapy for metastatic castration resistant prostate cancer. Invest. New Drugs. 2014;32:235–242. doi: 10.1007/s10637-013-9960-9. doi: 10.1007/s10637-013-9960-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TePas, EC, Litonjua, AA, Celedón, JC, Sredl, D, Gold, DR. Sensitization to aeroallergens and airway hyper responsiveness at 7 years of age. Chest. 2006;129:1500–1508. doi: 10.1378/chest.129.6.1500. [DOI] [PubMed] [Google Scholar]
- Tipu, HN, Ahmed, D. Differential binding of Aspergillus fumigatus allergenic proteins to human leuko-cyte antigen (HLA) alleles in dry lab: potential for immunotherapy. Cell. Biol.: Res. Ther. 2014;3(1) doi: 10.4172/ 2324-9293.1000111. [Google Scholar]
- Tipu, HN, Ahmed, TA, Bashir, MM. Human leukocyte antigen class II susceptibility conferring alleles among non-insulin dependent diabetes mellitus patients. J. Coll. Physicians Surg. Pak. 2011;21:26–29. [PubMed] [Google Scholar]
- Tipu, HN, Babar, ME, Hussain, T. Binding affinities of dengue virus envelope glycoprotein residues with human leukocyte antigen alleles: dry lab candidates for synthetic vaccines. Life Sci. J. 2014;11:552–557. [Google Scholar]
- Tomar, N, De, RK. Immunoinformatics: an integrated scenario. Immunology. 2010;131:153–168. doi: 10.1111/j.1365-2567.2010.03330.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wand, P, Sidney, J, Kim, Y, Sette, A, Lund, O, Nielsen, M. Peptide binding predictions for HLA DR, DP and DQ molecules. BMC Bioinformatics. 2010;11:568–580. doi: 10.1186/1471-2105-11-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whelan, FJ, Yap, NVL, Surette, MG, Golding, GB, Bowdish, DME. A guide to bioinformatics for immunologists. Front. Immunol. 2013;4:1–16. doi: 10.3389/fimmu.2013.00416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worm, M, Lee, HH, Kleine-Tebbe, J, Hafner, RP, Laidler, P, Healey, D, Buhot, C, Verheof, A, Maillere, B, Kay, AB, Larche, M. Development and preliminary clinical evaluation of a peptide immunotherapy vaccine for cat allergy. J. Allergy Clin. Immunol. 2011;127:89–97. doi: 10.1016/j.jaci.2010.11.029. [DOI] [PubMed] [Google Scholar]