Abstract
KU is a heterodimer, consisting of ∼70 and ∼80 kDa subunits (KU70 and KU80, respectively), which is involved in a variety of nuclear functions. We generated tbKU80-deficient trypanosomes to explore the potential role of the tbKU complex in telomere maintenance and transcriptional regulation of variant surface glycoprotein (VSG) genes in Trypanosoma brucei. Using real-time PCR, we demonstrated that the expression of several different VSG genes remains tightly regulated in tbKU80-deficient bloodstream-form cell lines, suggesting that VSG transcription profiles do not change in these cells. Owing to developmental silencing of the VSG Expression Sites (ES), no VSG is transcribed in the insect procyclic stage. With a green fluorescent protein reporter system, we showed that tbKU80-deficient mutants are fully capable of ES silencing after differentiation into procyclic forms. Using T7 RNA polymerase to explore the transcriptional accessibility of ES chromatin in vivo, we demonstrated that tbKU80-deficient bloodstream-form cells were able to generate transcriptionally repressed ES chromatin after differentiation into procyclic cells. Finally, we demonstrated progressive telomere shortening in tbKU80-deficient mutants. The possible function of tbKU80 in telomere maintenance and regulation of telomerase is discussed.
INTRODUCTION
Trypanosoma brucei is a unicellular eukaryotic parasite that causes sleeping sickness in humans and nagana in African cattle. In the mammalian host, the bloodstream form of the parasite is enveloped by a dense surface coat consisting of a single member of the variant surface glycoprotein (VSG) family. The periodic switching of VSGs, a process known as antigenic variation, allows the parasite to escape the host's immune response (1,2). There are several hundred VSG genes encoded at various loci in the parasite's genome (3). However, the expressed VSG is always located at the end of the chromosome adjacent to a telomere in a polycistronic transcription unit known as an Expression Site (ES). Current estimates suggest that there are approximately twenty potential ESs within the genome, of which only one is transcribed at any given time. All the others are transcriptionally repressed. After parasite ingestion by the vector Glossina, the Tsetse, bloodstream forms differentiate into procyclic forms in which all ESs are transcriptionally repressed. Navarro et al. (4) demonstrated that developmental ES repression correlates with a decrease of transcriptional accessibility of chromatin, suggesting that it is mediated by chromatin remodeling. Although there has been considerable progress in understanding the monoallelic VSG transcription (5,6), the molecular mechanisms governing switching and transcriptional repression are still not understood. Since the active ES is always located at a telomere, studies on telomere-binding proteins in trypanosomes might shed light on ES transcriptional regulation. Recently, Conway et al. (7) identified a homologue of the telomeric KU complex in T.brucei. KU is a heterodimer, consisting of two tightly associated polypeptides of ∼80 and ∼70 kDa (KU80 and KU70, respectively). KU proteins have been identified in a variety of organisms, including yeast, flies, humans and mice. They are involved in a variety of nuclear functions, including DNA double strand break repair by the non-homologous end joining pathway (NHEJ), V(D)J recombination of immunoglobulin genes and chromosome integrity (8). Importantly, KU is critical for telomere maintenance. Two KU-dependent phenomena, which have been investigated intensively in Saccharomyces cerevisiae, are of particular interest to us: the transcriptional silencing of genes that are adjacent to telomeres, a phenomenon termed telomere position effect (TPE), and telomere length regulation (TLR) (9–12). In yeast, transcriptionally repressed telomeric chromatin is organized in discrete foci in the nuclear periphery (13–15). These telomere clusters co-localize with pools of silent information regulator proteins (sir2–4), which are essential for TPE (13). Deletion of yeast KU genes disrupts the subnuclear organization of telomeres, redistributes SIR proteins and interferes with TPE, suggesting that KU is important for establishment or maintenance of silent telomeric chromatin (11,14,16,17). Recently, elegant experiments revealed that the localization of telomeres near the periphery is necessary, albeit not sufficient, for silencing and that other KU-independent pathways exist to tether telomeres to the nuclear periphery (18–21). The position-dependent expression of a subtelomeric surface glycoprotein gene has been described in S.cerevisiae recently (22). Expression of the FLO10 gene is variegated in several mutant strains and transcriptional repression of FLO10 is dependent on the subtelomeric location of the gene. Interestingly, both SIR and KU proteins are critical for transcriptional silencing of this locus.
In the last decade, many telomeric proteins that are involved in chromosome stability and TLR have been characterized (23). In S.cerevisiae, the KU complex binds to telomeric repeats in vivo, where it interacts with a variety of proteins and protects telomeres from nucleolytic and recombinational activities (24–26). The importance of KU for TLR was revealed after discovering that yku70 and yku80 mutants possess abnormally short telomeres (9,10). Additional experiments demonstrated that KU interacts with the RNA component of the telomerase complex (27,28), suggesting that KU is involved in the recruitment of telomerase to chromosome ends (29). In mammalian cells, the rate of progressive telomere shortening strongly correlates with the length of a G-rich single stranded overhang (G-overhang), which is found at the end of each telomere throughout the cell cycle (30,31). Interestingly, wild-type S.cerevisiae acquires a detectable G-overhang only during late S-phase. However, KU-deficient yeast cells posses a G-overhang throughout the cell cycle, implicating that KU regulates the G-overhang structure (24,32). Experiments in Schizosaccharomyces pombe contrast significantly with studies done in S.cerevisiae. Although the KU complex is required for maintenance of telomere length in S.pombe, KU is dispensable for transcriptional silencing at telomeres and is located throughout the nucleus rather than concentrated at telomeric repeats (33). A similar role for KU has been proposed in T.brucei, where the deletion of tbKU appeared to affect telomere maintenance but not transcriptional control of telomeric metacyclic ESs in the procyclic (tsetse midgut) stage of the trypanosome life cycle (7). However, the role of tbKU in allelic exclusion and developmental regulation of bloodstream-form ESs has not been addressed and accurate measurements of telomere shortening at silent ESs are needed to pursue the hypotheses concerning telomere structure and maintenance in T.brucei.
In this study, we show that the exclusive expression of the active ES is not affected in tbKU80-deficient bloodstream forms. Furthermore, we demonstrate that tbKU80-deficient cells are fully competent to silence ESs after differentiation into procyclic forms. Finally, we evaluate telomere shortening rates and analyze the G-overhang structure in tbKU80-deficient trypanosomes to define more precisely the role of the tbKU complex in TLR. We demonstrate that the shortening rates in the absence of tbKU80 correlate with the predicted telomere shortening due to the end replication problem.
MATERIALS AND METHODS
Trypanosome cell lines and plasmid constructions
T.brucei bloodstream forms [strain Lister 427, antigenic type MITat 1.2 and clone 221a (34)] were cultured in HMI-9 at 37°C (35). This strain had been manipulated to generate the bloodstream and procyclic cell lines ‘single marker’ and ‘29-13’, respectively (36). Both cell lines constitutively express T7 RNA polymerase. Bloodstream forms were cultured in HMI-9 containing 2.5 μg/ml G418 (Sigma). Procyclic forms were cultured in SDM-79 (37) containing 15 μg/ml G418 and 25 μg/ml hygromycin (Sigma). Bloodstream forms were differentiated into procyclic forms as described previously (38). The green fluorescent protein (GFP)–tbKU80 fusion protein was expressed from a derivate of the pLew82 vector (39). To generate the expression construct for TY-epitope-tagged tbKU70, pLew111 [pLew82 without luciferase open reading frame (ORF)] was amplified by inverse PCR introducing a XhoI restriction site at the 5′ end and StuI restriction site at the 3′ end and excluding the bleomycin gene. The blasticidin gene was released from pHD309-bsr (40) by digesting with SmaI and XhoI. The two fragments were ligated generating pFlan1. The tbKU70 ORF was amplified by PCR from genomic DNA using primers that introduced a TY-tag (MEVHTNQDPLDA) sequence at the 5′ end. The PCR product was cloned into pFlan1. These T7 promoter driven cassettes were integrated into the rDNA spacer of the single marker cell line. Expression was induced by adding 500 ng/ml of tetracycline to the medium.
The constructs used to generate tbKU80 deletion mutants contained ∼300 bp of 5′ and 3′ tbKU80 non-coding sequences (UTRs) for targeting, separated by drug resistance cassettes. Both 3′-UTRs and 5′-UTRs were amplified by PCR from genomic DNA and cloned into pBluescript II SK+ (Stratagene), generating pCJ10. The hygromycin phosphotransferase ORF, flanked by ∼100 bp of 5′-UTRs and 300 bp of 3′-UTRs from the actin gene, was removed from pHD309 (40) and cloned into PmeI-digested pCJ10. The puromycinN-acetyltransferase ORF, flanked by ∼100 bp of 5′-UTRs and 220 of 3′-UTRs from the actin gene, was released from pHD309. This fragment was treated with DNA polymerase I to generate blunt ends and cloned into PmeI-digested pCJ10. The final construct was cut with NotI to release the targeting cassette for transfections. To generate the GFP reporter constructs, the GFP cassettes were amplified from pCO57 with two different primer pairs. One primer pair included the T7 promoter upstream of the GFP ORF another primer pair excluded the T7 promoter. The two PCR products were each cloned into SpeI/StuI-digested pbRn6 (41). The blasticidin S deaminase ORF was amplified from pHD309-bsd including the 3′-UTRs and 5′-UTRs. The selectable marker was then cloned into the two NheI-digested constructs described above. The final constructs were cut with SacI and XhoI to release the GFP-reporter cassettes for transfection. All transfections were performed as described previously (40).
Immunofluorescence
This protocol was carried out as described previously, with minor modifications (42). Briefly, 1 × 106 bloodstream forms, induced with tetracycline, were washed with trypanosome dilution buffer [TDB, (43)] before fixing in 2% formaldehyde for 10 min at 4°C. The fixed cells were attached onto glass cover slips by centrifugation at 2000 g for 5 min at 4°C and permeabilized with 0.2% NP-40 in phosphate buffered saline (PBS) for 10 min at room temperature. BB2 mouse monoclonal IgG (gift from Keith Gull) and anti-GFP rabbit polyclonal IgG (Molecular Probes) were used to detect TY-tbKU70 or GFP-tagged tbKU80, respectively, and DAPI was used to stain DNA. BB2 and anti-GFP were visualized with tetramethyl rhodamine isothiocyanate-conjugated goat anti-mouse or fluorescein isothiocyanate-conjugated donkey anti-rabbit antibodies (Jackson Laboratories), respectively. Cells were mounted in anti-fade mounting solution (Vectashield, Vecta Laboratories Inc.) and analyzed with a fluorescence microscope (Nikon Optiphot). Representative images were captured using a DeltaVision image restoration microscopy system (Applied Precision). Deconvolution was performed using the softWoRx™ V 3.0 Software.
Immunoprecipitation
This protocol was carried out with minor modifications according the manufacturer (Molecular Probes). Briefly, 1 × 108 bloodstream forms were harvested, washed once with ice-cold TDB and resuspended in 1 ml wash buffer A1 (50 mM Tris, pH 7.5, 150 mM NaCl, 1 mM EDTA and 0.1% NP-40). Cells were sonicated for 30 s using a Micro Ultrasonic Cell Disruptor (Kontes). The lysate was preincubated with 50 μl protein G agarose beads (Sigma) for 1 h at 4°C. The beads were removed by centrifugation and the lysate was transferred to a fresh tube. An aliquot of 10 μg anti-GFP polyclonal rabbit IgG (Molecular Probes) was added and incubated overnight at 4°C. The following day samples were incubated with 30 μl protein G agarose beads (Sigma) for 90 min at 4°C. The beads were then washed for 20 min sequentially with 1 ml buffer A1, A2 (50 mM Tris, pH 7.5, 150 mM NaCl and 0.1% NP-40) and A3 (50 mM Tris, pH 7.5 and 0.1% NP-40). Proteins were eluted from the beads by boiling with 50 μl loading buffer (50 mM Tris–HCl, pH 6.8, 5% 2-mercaptoethanol, 2% SDS, 0.1% bromophenol blue and 10% glycerol), separated by SDS–PAGE electrophoresis, and electroblotted onto a nitrocellulose membrane. tbTY-KU70 and tbGFP-KU80 were analyzed by immunoblotting with monoclonal BB2 anti-TY (gift from Keith Gull) or monoclonal anti-GFP (Roche) antibodies, respectively. Primary antibodies were detected with horseradish-peroxidase-conjugated sheep anti-mouse antibodies (Amersham-Pharmacia).
Real-time-PCR
Total RNA from 1 × 108 bloodstream forms was isolated using RNA Stat-60 (Tel-Test) according to the manufacturer's instructions. An aliquot of 10 μg of total RNA was incubated with RQ1 RNAse-free DNAse (Promega) according to the supplier's recommendations followed by phenol/chloroform/isoamyl alcohol extraction and ethanol precipitation. The precipitated RNA was resolved in 10 μl DEPC-treated water and used for reverse transcriptase reaction using the ProStar First Strand RT–PCR Kit (Stratagene) with the supplied random primers. PCR was performed with an Abi Prism 7900 HT Sequence Detection System (Applied Biosystems) according to the manufacturer's instructions. All values were normalized to an α-tubulin internal control. Post-amplification melting curves were performed to confirm that a single PCR product was produced in each reaction. Genomic DNA was used to generate a standard curve with which the copy numbers of gene products in each sample were calculated. The sequences of primers used in the real-time PCR can be obtained upon request.
Fluorescence cell analysis
Samples of living parasites were analyzed with a FACS Calibur system following the manufacturer's recommendations (Becton Dickinson). Briefly, 1 × 106 bloodstream forms were collected from a log-phase cell culture and GFP-auto-fluorescence was analyzed in medium. The mean fluorescence intensity was determined using CellQuest software (Becton Dickinson).
Telomere blot
This protocol was carried out with minor modifications as described previously (42). Briefly, 5 × 108 bloodstream forms were harvested, washed once in TDB, resuspended in 1 ml TNE (10 mM Tris, pH 7.4, 100 mM NaCl and 10 mM EDTA) and lysed by adding 1 ml TNES (TNE + 1% SDS) supplemented with 100 μg/ml proteinase K (Roche). After 3 h incubation at 37°C, the DNA was extracted with phenol/chloroform, precipitated and resuspended in TNE supplemented with 100 μg/ml RNase A. After incubation for 3 h at 37°C, treatment with proteinase K (100 μg/ml) followed for another hour. DNA was then extracted with phenol/chloroform, isopropanol precipitated, and resuspended in TE (10 mM Tris, pH 7.4 and 10 mM EDTA). DNA was digested overnight with a mixture of AluI, HinfI and RsaI under recommended conditions (New England Biolabs). DNA restriction fragments were separated on an 1% agarose gel and transferred onto a nitrocellulose membrane. Telomere restriction fragments were detected using a probe containing TTAGGG-repeats that was labeled with [α-32P]dCTP as described previously (44).
G-strand overhang-assay
G-strand overhang-assays were performed following a protocol described by Wellinger et al. (32) with minor modifications. DNA was extracted from bloodstream forms and HeLa cells (kindly provided by Dr Bibo Li). Briefly, 2 × 108 cells were harvested, washed twice in ice-cold TDB and resuspended in 1 ml sterile TNE. An aliquot of 1 ml TNES + 100 μg/ml proteinase K was added and incubated overnight at 37°C. After phenol/chloroform extraction, the DNA was precipitated, gently resuspended in 300 μl TNE + 100 μg/ml RNase A and incubated for 3 h at 37°C. After adding 300 μl TNES + proteinase K, the DNA was incubated overnight at 37°C followed by phenol/chloroform extraction as described above. An aliquot of 10–20 μg of gDNA was digested overnight with MboI and AluI (New England Biolabs) and separated on a 0.8% agarose gel. The gel was dried and pre-hybridized in 0.5 M NaPO4, pH 7.2, 1 mM EDTA, pH 8.0, 7% SDS and 1% BSA [Church Mix, (45)] for 2 h at 30°C. An aliquot of 50 ng of oligonucleotides (CCCTAA)4 or (TTAGGG)4 were labeled with 32P-labelled ATP by T4 polynucleotide kinase (New England Biolabs) for 45 min at 37°C. Hybridization was performed overnight at 30°C in Church Mix. After washing three times for 30 min with 4× SSC (60 mM Na3C6O7H5 and 600 mM NaCl) and once for 30 min with 4× SSC and 0.1% SDS at 20°C, the gel was exposed to a phosphorimager screen (Molecular Dynamics) for at least 72 h. Subsequently, the gel was denatured for 30 min in 1.5 M NaCl and 0.5 M NaOH, and neutralized for 15 min in 3 M NaCl and 0.5 M Tris–HCl, pH 7.0, followed by pre-hybridization in Church Mix at 55°C for 1 h and hybridization overnight with the same probes. After washing as described above but at 55°C, the gel was exposed to a phosphorimager screen for 3–5 h.
RESULTS
Expression pattern and co-localization of tbKU80 and tbKU70
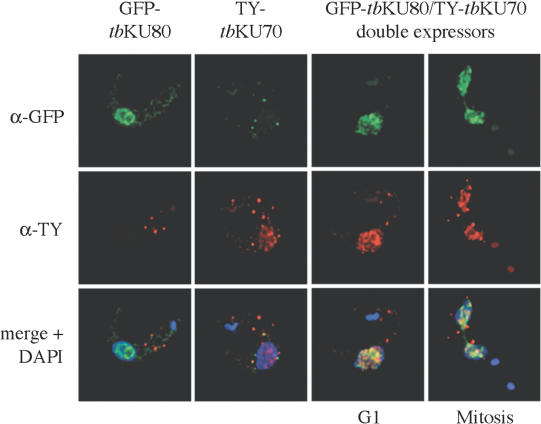
Two homologs of KU proteins, tbKU70 and tbKU80, were recently identified in T.brucei (7). Before proceeding to functional analysis of these proteins, we first wanted to characterize the tbKU complex in trypanosomes. In particular, we were interested in the nuclear distribution of tbKU and the interaction between tbKU80 and tbKU70. In order to avoid the need for tbKU-specific antibodies, we fused GFP and a TY peptide tag to tbKU80 and tbKU70, respectively. By indirect immunofluoresence (IF), both GFP–tbKU80 and TY–tbKU70 were clearly detectable in the nucleus (Figure 1, left panels). The signals in the cytoplasm seem to be unspecific cross reactivity of the antibodies, because they were also detectable in the controls. The distribution of the proteins was not homogeneous, but was manifested as distinguishable foci that were excluded from some areas in the nucleus. Since this distribution was similar to the pattern observed with a telomere probe used in fluorescence in situ hybridization assays (FISH, data not shown), we compared both patterns by combining IF and FISH (see Supplementary Material). Specific co-localization of tbKU and telomeres could not be detected. Partial overlap of GFP–tbKU80 and telomere signals is considered to be coincidental and would mask any telomere-specific signal. Next, we generated a cell line that expressed both GFP–tbKU80 and TY-tagged tbKU70 to investigate if the two proteins form a complex in trypanosomes. Both proteins clearly co-localized in the nucleus of this cell line (Figure 1, right panels). This co-localization was observed in all cells and was not cell cycle dependent, as demonstrated by two examples of cells that are in G1 phase (one nucleus) or mitosis (two nuclei).
Figure 1.
Expression pattern and co-localization of tbKU80 and tbKU70. Deconvolution of single-focal-plane images of bloodstream forms. Indirect IF shows distribution of GFP–tbKU80 (green fluorescence), TY–tbKU70 (red fluorescence) and DAPI-stained nuclei (blue fluorescence). The two left panels show the distribution of tbKU80 and tbKU70 separately in the nucleus of two bloodstream-form cell lines. The two right panels show co-localization of tbKU80 and tbKU70 (merge, yellow fluorescence) in one representative trypanosome of ‘double expressors’ cell line in G1 and one trypanosome during late mitosis.
tbKU80 and tbKU70 interact in vivo
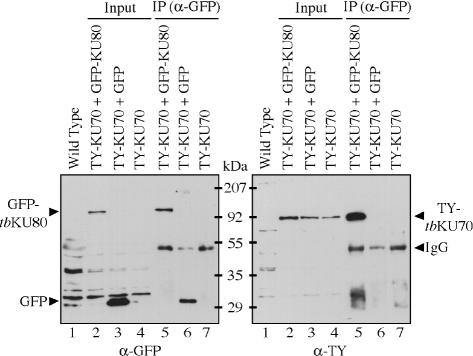
Since co-localization does not prove that two proteins physically interact, we carried out co-immunoprecipitation (co-IP) assays with a cell line that co-expressed TY–tbKU70 and GFP–tbKU80. As controls, we generated two additional cell lines: one expressing TY–tbKU70 and GFP and one only TY–tbKU70. Protein expression levels of all cell lines were analyzed with the corresponding antibodies (Figure 2, input). The parental cell line (single marker) served as negative control (Figure 2, wild type). After IP was performed with a polyclonal antibody directed against GFP, the precipitated proteins were analyzed with a monoclonal α-GFP antibody [Figure 2, left panel, IP (α-GFP)] or a monoclonal α-TY antibody [Figure 2, right panel, IP (α-GFP)]. Only in the cell line expressing both fusion proteins it was possible to co-immunoprecipitate TY–tbKU70 with an antibody specific for GFP–tbKU80 (Figure 2, right panel, lane 5). TY–tbKU70 was not detectable in the control precipitates, suggesting that tbKU exists as a heterodimer in T.brucei similar to the complex described in other organisms.
Figure 2.
Co-IP of GFP–tbKU80 and TY–tbKU70. IPs were performed with an antibody specific for GFP with whole cell lysates of cells co-expressing TY–tbKU70 and GFP–tbKU80 and control cells expressing TY–tbKU70 and GFP or only tbKU70. Whole cell lysates (lanes 2–4, input) or immunoprecipitates (lanes 5–7, IP) were separated by SDS–PAGE and transferred onto a nitrocellulose membrane. Western blots were incubated with an α-GFP antibody (left panel) or an α-TY antibody (right panel). Wild-type cells (lane 1) served as a control for antibody specificity.
Transcriptional silencing of ESs is not affected in tbKU80-deficient trypanosomes
To proceed with functional analysis of tbKU, we replaced both alleles of the tbKU80 gene with selectable markers to generate a tbKU80-deficient cell line. The correct integration of the marker genes was confirmed by Southern blotting (data not shown). Consistent with previous publications, tbKU80 knockout mutants were viable and showed no developmental or growth phenotype [data not shown, (7)]. Only one VSG is normally expressed in bloodstream-form trypanosomes. To examine if tbKU80 is involved in this monoallelic expression of VSG genes, we compared the expression levels of a variety of known VSG genes in wild-type and tbKU80-deficient trypanosomes. Real-time PCR was performed with specific primer pairs to detect transcripts of VSG 221 (single copy gene, active ES), VSG 118 (single copy gene, inactive ES), VSG B (one telomeric and two internal copies), VSG bR2 (two telomeric and one internal copies) or VSG VO2 (one telomeric and one internal copy) with cDNA from wild-type and ΔtbKU80 parasites. As expected, mRNA of the active VSG221 was highly abundant in both cell lines (Table 1). In contrast, transcripts of other tested VSG genes were not detectable at all or are very close to the threshold which might represent unspecific amplification (Table 1). These results suggest that tbKU80-deficient cells are still able to regulate VSG transcription tightly and that tbKU is not involved in ES regulation in bloodstream-form trypanosomes.
Table 1. VSG expression profiles in wild-type and ΔtbKU80 cells.
| Wild-type copy numbers | ΔtbKU80 copy numbers | |
|---|---|---|
| VSG 221 | 1.8 × 108 | 1.4 × 108 |
| VSG 118 | nd | nd |
| VSG B | nd | nd |
| VSG bR2 | 179 | 325 |
| VSG VO2 | 118 | 454 |
Values represent calculated copy numbers of gene products in each sample (nd, not detectable). Results were normalized to α-tubulin.
tbKU80-deficient cells are proficient in establishing developmentally silenced chromatin
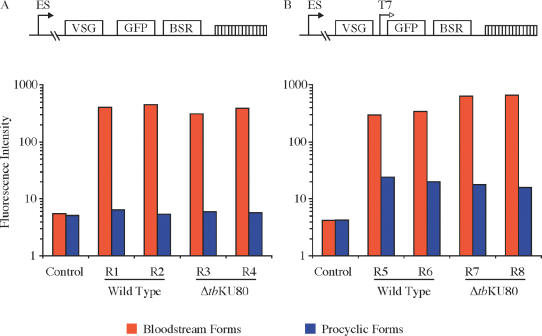
Several groups have reported in detail that KU plays a central role in the establishment of transcriptionally repressed telomeric chromatin in yeast. After differentiation of bloodstream forms to the insect procyclic forms in vitro, transcription from the active ES promoter is turned off. This developmental silencing is promoter specific and depends on the telomeric location of the ES (46,47). Navarro et al. (4) used a T7 RNA polymerase-driven reporter system to study the transcriptional accessibility of the ES. In contrast to ESs in bloodstream forms, the T7 promoter was repressed along the entire ES in procyclic parasites, suggesting that the ES chromatin was remodeled to a less accessible, compact chromatin upon differentiation. We employed a similar assay to evaluate whether tbKU80-deficient cells are still able to establish developmentally silenced chromatin. A GFP reporter cassette driven by a T7 promoter was introduced immediately downstream of the active VSG in bloodstream forms (Figure 3B). In addition, we introduced a GFP cassette without a T7 promoter to assess the transcriptional activity of the endogenous ES promoter (Figure 3A). To compare the developmental ES regulation of wild-type trypanosomes with that of ΔtbKU80 parasites, GFP expression levels were monitored by FACS analysis in bloodstream forms (red bars) and in procyclic forms two weeks after differentiation (blue bars). The results of two representative clones from each reporter cell line are shown. As expected, wild-type cells efficiently down regulated the ES promoter (Figure 3A). GFP expression was reduced to a level comparable to control cells lacking the GFP reporter cassette. A similar effect was observed in ΔtbKU80 cells, suggesting that ES-promoter silencing is not influenced by tbKU80. To test whether chromatin remodeling is affected by the deletion of tbKU80, we differentiated parasites in which the GFP cassette was transcribed by the heterologous T7 polymerase. Upon differentiation, both wild-type cells and ΔtbKU80 mutants were able to reduce reporter activity to ∼10% of bloodstream form activity (Figure 3B) as described previously (4). Only a slight difference, which was within the range of clonal and experimental variation, was observed between wild-type and tbKU80-deficient parasites, suggesting that any effect of tbKU depletion was very minor. These data suggest that the KU complex is not involved in down regulation of the ES promoter in procyclic forms and that tbKU80-deficient cells are fully capable of establishing developmentally silenced chromatin in trypanosomes.
Figure 3.
Developmental silencing of ESs in ΔtbKU80 trypanosomes. (A) Schematic representation of integrated reporter cassette. VSG 221 gene, GFP-reporter (GFP) and selectable marker (bsr) are indicated as boxes. Endogenous ES promoter (ES) is shown as arrow. Telomeres are represented by a hatched box. The GFP-reporter cassette was introduced into the active ES of two independent tbKU80-deficient cell lines (ΔtbKU80/R3 and ΔtbKU80/R4) and wild-type trypanosomes (Wild Type/R1 and Wild Type/R2). To analyze developmental silencing of ES promoter GFP expression was monitored by FACS analysis in bloodstream forms (red bars) and two weeks post-differentiation into procyclic-form trypanosomes (blue bars). Wild-type cells without GFP reporter cassette served as a negative control to evaluate background fluorescence intensity (control). (B) Schematic representation as in (A). Additional T7 promoter is shown as an open arrow. The T7 promoter driven GFP-reporter was introduced into the ESs of tbKU80-deficient parasites generating cell lines ΔtbKU80/R7 and ΔtbKU80/R8 and wild-type cells generating Wild Type/R5 and Wild Type/R6. To analyze chromatin access of the T7 polymerase as a marker for chromatin remodeling, GFP expression was monitored as described above.
tbKU80-deficient trypanosomes exhibit modest telomere shortening
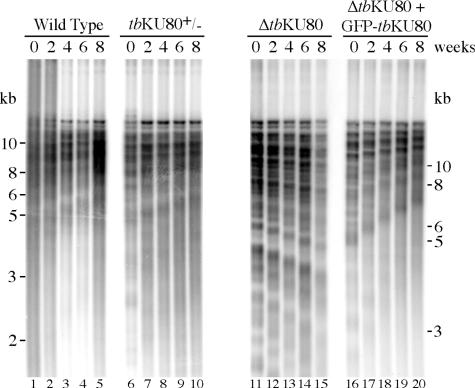
A previous study suggested that tbKU80-deficient mutants exhibit shortened or broken telomeres after extensive propagation, but the precise shortening rate was not determined (7). However, this is important to reveal the function of KU in telomerase recruitment and regulation. To evaluate the telomere shortening rates in tbKU80-deficient trypanosomes, we monitored telomere length in wild-type cells, a tbKU80 single knockout cell line (tbKU80+/−), and a tbKU80-deficient cell line (ΔtbKU80) for eight weeks. To be certain that any phenotype was owing to the loss of tbKU80, a cell line in which GFP–tbKU80 was reintroduced into the ΔtbKU80 mutant was also monitored. Genomic DNA was isolated at two-week intervals and digested with frequently cutting restriction enzymes that do not cut within the repetitive telomeric DNA. This preparation of bulk telomeres was analyzed by Southern blot with a telomere-specific probe. As described previously (48,49), wild-type cells displayed a continual elongation of telomeres (Figure 4, lanes 1–5). Similarly, the deletion of one tbKU80 allele had no apparent effect on telomere growth (lanes 6–10). Strikingly, ΔtbKU80 parasites exhibit progressive telomere shortening (lanes 11–15). A clear reduction of telomere length was visible after only two weeks (lane 12). Overall telomere length was reduced by 0.8–1.0 kb after eight weeks. Eight weeks represent ∼210 generations, so this translates into a telomere shortening rate of ∼4 bp per cell division. Interestingly, identical telomere shortening rates were observed in telomerase-deficient trypanosomes (Oliver Dreesen, unpublished data). Furthermore, we could completely recover the telomere growth defect by reintroduction of GFP–tbKU80 (lanes 16–20), suggesting that this phenotype was directly caused by the loss of tbKU80. Although we cannot exclude a role in end protection, our data suggest that tbKU80 is likely to be important for telomerase activity. Unfortunately, our attempts to make a cell line lacking both telomerase and KU80 were unsuccessful.
Figure 4.
Telomere shortening in ΔtbKU80 trypanosomes. Genomic DNA from wild-type cells, tbKU80 single allele knockout cells (tbKU80+/−), tbKU80-deficient cells (ΔtbKU80) and from a tbKU80-deficient cell line, which expressed an ectopic copy of GFP–tbKU80 (ΔtbKU80 + GFP–tbKU80) was prepared every other week for a period of 8 weeks. The DNA was digested with AluI, HinfI and RsaI, and separated by agarose gel electrophoresis, Southern-blotted and probed with a radiolabeled telomeric (TTAGGG)27 probe. Distinguishable bands of ΔtbKU80 telomeric DNA were used to estimate telomere shortening rates during 8 weeks.
G-overhang structure in ΔtbKU80 trypanosomes
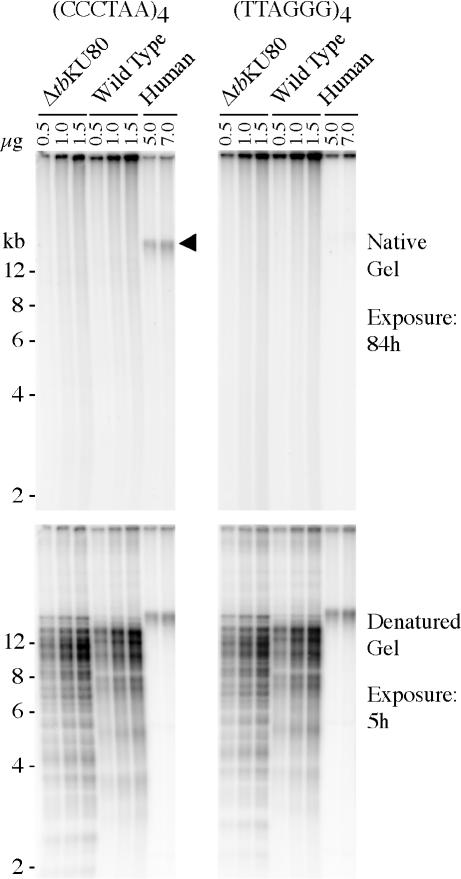
The rate of telomere shortening is dependent on the length of single-stranded G-overhang with which telomeres end (50). To examine the role of tbKU80 in telomere length control, we compared the G-overhang structure in wild-type and tbKU80-deficient trypanosomes. Furthermore, we wanted to know if the loss of tbKU80 would alter G-overhang structure, as has been described in yeast (24). For this purpose, DNA was isolated, digested with enzymes that do not cut repetitive telomeric DNA and separated on agarose gels. The dried native gels were hybridized with a radiolabeled (CCCTAA)4 oligonucleotide probe to detect G-overhangs. Human DNA served as a positive control and G-overhangs were clearly detectable in this sample (Figure 5, upper left panel, arrow head). A (TTAGGG)4 probe, which should not produce a signal, was used to confirm that the DNA was still in its native condition (Figure 5, upper right panel). However, no overhang signal was detectable in DNA derived from either wild-type or tbKU80-deficient trypanosomes with the (CCCTAA)4 probe, despite loading at least 10-fold more trypanosome telomeric DNA compared to the HeLa cell sample (Figure 4, left panels) as is evident after denaturation, when the gels were re-hybridized with the same probes to confirm equal loading (Figure 5, lower panels). The weak signals in the upper panels in lanes with 1 and 1.5 μg DNA are unspecific, as they appear with both probes. As it is not possible to detect overhangs shorter than ∼30 nt with this assay, we therefore conclude that trypanosomes have short overhangs and that, unlike in yeast, tbKU80 has no apparent influence on overhang structure in T.brucei.
Figure 5.
G-overhang structure in ΔtbKU80 trypanosomes. DNA was extracted from tbKU80-deficient cells (ΔtbKU80), wild-type cells (wild type) and from HeLa cells (human), and digested with MboI and AluI restriction enzymes. Different amounts of DNA, as indicated in each lane, were separated by gel electrophoresis and analyzed with radiolabeled probes specific for G-overhangs [(CCCTAA)4, left panels] or C-overhangs [(TTAGGG)4, right panels]. To confirm equal loading the gels were denatured and re-hybridized with the same probes as above (lower panels). Different exposure times of the gels on a phosphorimager (Molecular Dynamics) are indicated on the right.
DISCUSSION
In this study we investigated the role of T.brucei tbKU80 in transcriptional regulation of VSG genes, developmental silencing of ES and telomere maintenance. In order to characterize the KU complex in T.brucei, we first analyzed the localization and distribution pattern of KU in trypanosomes and the interaction between tbKU80 and tbKU70. The localization of tbKU80 is not uniform throughout the nucleus, as it is in some mammalian cells and in S.pombe (33,51). Rather it resembles the distribution of KU in S.cerevisiae, where the KU complex and telomere-repeat-binding protein Rap1p co-localize at telomeric clusters (52). We showed that both tbKU80 and tbKU70 form punctate patterns throughout the nucleus and that they co-localize. With co-IP experiments, we demonstrate that these structures most likely represent heterodimers. In yeast, KU seems to be tightly associated with telomere function and structure. IP experiments showed that telomeric DNA can be co-immunoprecipitated with a tagged yKu80 protein (24). Furthermore, Laroche et al. (16) showed that mutation of KU genes disrupts the subnuclear organization of telomeres in the periphery. Although tbKU distribution patterns are similar to those observed by telomere FISH, the partial co-localization of GFP–tbKU80 and telomeres that we observed by combining IF with FISH might be coincidental. Furthermore, neither the size nor the distribution of telomere clusters was altered in ΔtbKU80 trypanosomes and telomeric repeats could not be co-immunoprecipitated with GFP–tbKU80 (data not shown). These data suggest that tbKU80 does not play a critical role in the nuclear organization of telomere clusters in trypanosomes, and that interactions between tbKU80 and telomeres might be transient or weak. However, firm conclusions about the role of tbKU80 in ES regulation and telomeric silencing cannot be derived from these observations. Work by several groups revealed that redundant telomere-anchoring pathways exist in other cells, and that localization of telomeres to the nuclear periphery is separable from transcriptional repression (16,17,21). Furthermore, recent reports suggest that KU proteins are involved in transcriptional regulation of individual genes and that KU80 is associated with RNA polymerase II elongation sites (53). Since there is a strong evidence that inactive ESs are repressed by attenuation of transcription elongation rather than inhibition of transcription initiation at the ES promoter (5), we analyzed ES transcriptional regulation in ΔtbKU80 trypanosomes.
Two different phenomena were investigated: the exclusive expression of one ES in bloodstream forms and the developmental silencing of the ES after differentiation into procyclic forms. We could not detect any differences in the VSG expression profiles of wild-type cells compared with that of ΔtbKU80 cells. This is consistent with the model of ES transcriptional regulation proposed by Navarro and Gull (6). This model advocates that a unique location, the Expression Site Body (ESB), is responsible for allelic exclusion by sequestering factors that are essential for full ES transcription. The lack of these factors rather than active repression (for example by the KU complex) might be the cause of transcriptional attenuation of inactive ESs in bloodstream forms. However, repression of inactive ESs by unidentified factors cannot be excluded, based on our data, since KU-independent repression is known to occur at fission yeast telomeres and at budding yeast telomeres in strains lacking RIF1 or RIF2 (17,33).
Trypanosomes encounter a different situation after differentiation from bloodstream to procyclic forms, where all ESs are transcriptionally repressed. Analysis of ES localization by FISH with probes specific for telomeres and subtelomeric ES-specific 50 bp repeats suggests that the majority of ESs are in the nuclear periphery (J.Lowell, unpublished data). Furthermore, Navarro et al. (4) demonstrated that developmental ES-silencing correlated with a decrease in transcriptional accessibility of chromatin to an abundantly expressed single-unit T7 RNA polymerase. Transcription from chromosomally integrated T7 promoters is repressed along the entire ES after differentiation into procyclic forms, but not at chromosome-internal loci. In contrast, a T7 promoter integrated into an inactive ES in bloodstream forms is fully accessible, indicating that distinct mechanisms control ES regulation in bloodstream and procyclic forms and that ES repression upon differentiation is mediated by remodeling chromatin to yield a structure that is no longer permissive for transcription. We used the same experimental approach to elucidate the role of tbKU80 in developmental silencing of ES. As shown with GFP reporter cassettes driven by endogenous ES promoter or T7 promoter, ΔtbKU80 trypanosomes are fully capable of silencing their ES promoter after differentiation. In addition, there is no detectable difference in T7 promoter accessibility between wild-type and ΔtbKU80 parasites. Hence, it seems that tbKU80 does not play a significant role in developmental ES regulation and chromatin remodeling. Whether the exclusive telomeric location of ESs is important for developmental transcriptional regulation of the endogenous ES promoter is still not known and is intensively discussed. The spread of transcriptionally repressed heterochromatin from telomeres into subtelomeric regions has been described in other organisms. Since telomeric repression of different promoters has been reported in T.brucei (4,41), it is reasonable to believe that telomeres influence adjacent chromatin structure in trypanosomes. However, it is uncertain whether telomere-induced repression can spread to the ES promoters, which can be as far as 50 kb upstream of the telomere. In order to understand telomeric repression and its role in ES regulation, it is essential to analyze chromatin modifications and modifying enzymes in trypanosomes.
Substantially more is known about trypanosome telomere structure. T.brucei telomeres consist of 10–20 kb duplex TTAGGG repeats that end in T-loop structures (54). In contrast to mammalian or yeast cells, telomeres grow constantly in trypanosomes and, interestingly, the telomere of the active ES is susceptible to large truncations. The KU complex is important for accurate telomere maintenance in many organisms, and, in general, KU deletion leads to loss of telomeric repeats. Telomere shortening reflects the inherent dilemma of replicating linear chromosomes: the end replication problem. In yeast, the removal of short RNA primers and possibly exonuclease activity creates short G-overhangs at chromosome ends. In the absence of telomerase, these G-overhangs cannot be processed, which leads to the loss of 4–6 bp of telomeric DNA at each round of replication. The telomere shortening in ΔtbKU80 trypanosomes, ∼4 bp per cell division, is comparable with the shortening rates of telomerase-deficient parasites. This indicates that tbKU80 is essential for full telomerase activity. Interestingly, Conway et al. (7) reported that the length of very short telomeres did not change over 150 generations in tbKU-deficient trypanosomes. This observation suggests that although tbKU seems to be essential for recruitment or activation of telomerase to long telomeres, it may be dispensable at very short telomeres, where a different unidentified mechanism may recruit or activate telomerase.
One of the parameters that determines telomere shortening rates is the length of the single-stranded G-rich overhang at the end of telomeres. In human cells, for example, shortening rates of ∼50–100 bp per cell division are proportional to the size of the G-overhang (31). In S.cerevisiae, longer overhangs can only be transiently observed in late S-phase (32). During most of the cell cycle, any G-overhangs are short, which is consistent with the low telomere shortening rate in telomerase-deficient cells. KU-deficient yeast cells, however, display longer overhangs throughout the cell cycle, and this might be responsible for dramatically shortened telomeres (24). In contrast to yeast, the telomere shortening in ΔtbKU80 trypanosomes is modest. To understand the apparently different mechanism of telomere maintenance in T.brucei compared with yeast, it was essential to investigate G-overhang structure in wild-type and ΔtbKU80 trypanosomes. Surprisingly, we could not detect G-overhangs either in wild-type nor in ΔtbKU80 cells, which implies that overhangs are short and hence undetectable in our assay. However, the presence of transient longer G-overhangs during S-phase, as described in yeast, cannot be excluded. These findings explain the low shortening rates, assuming that telomerase function is abolished in ΔtbKU80 parasites and that shortening rates are proportional to short G-overhangs. Although this is the most consistent explanation in the light of our data, we cannot exclude the possibility that tbKU80 plays a role in chromosome end protection.
tbKU80 clearly does not play a role in well-characterized functions of the KU complex in other organism, such as DNA damage repair or telomeric silencing [(7) and data not shown]. We propose that the function of tbKU is more comparable to that of KU in S.pombe, where it is involved in telomere maintenance but not in TPE. However, tbKU80 seems to be essential to recruit or activate telomerase, because the deletion of tbKU80 abolished telomerase function, at least at long telomeres.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Keith Gull and colleagues for gifts of antibodies. We thank all members of the Cross lab for useful suggestions, and Joanna Lowell and Sandra Hake for carefully reading the manuscript. We are also grateful to the staff of the Flow Cytometry Resource Center and the Bio-Imaging Resource Center of The Rockefeller University for help and support. This work was supported by the National Institutes of Health (grant number AI50614) and by the Deutsche Forschungsgemeinschaft (grant number JA 1013/1-1).
REFERENCES
- 1.Barry J.D. and McCulloch,R. (2001) Advances in Parasitology. Academic Press Ltd, London, Vol. 49, pp. 1–70. [DOI] [PubMed] [Google Scholar]
- 2.Cross G.A.M. (1996) Antigenic variation in trypanosomes: secrets surface slowly. Bioessays, 18, 283–291. [DOI] [PubMed] [Google Scholar]
- 3.van der Ploeg L.H.T., Valerio,D., de Lange,T., Bernards,A., Borst,P. and Grosveld,F.G. (1982) An analysis of cosmid clones of nuclear DNA from Trypanosoma brucei shows that the genes for variant surface glycoproteins are clustered in the genome. Nucleic Acids Res., 10, 5905–5923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Navarro M., Cross,G.A.M. and Wirtz,E. (1999) Trypanosoma brucei variant surface glycoprotein regulation involves coupled activation/inactivation and chromatin remodeling of expression sites. EMBO J., 18, 2265–2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vanhamme L., Poelvoorde,P., Pays,A., Tebabi,P., Xong,H.V. and Pays,E. (2000) Differential RNA elongation controls the variant surface glycoprotein gene expression sites of Trypanosoma brucei. Mol. Biol., 36, 328–340. [DOI] [PubMed] [Google Scholar]
- 6.Navarro M. and Gull,K. (2001) A pol I transcriptional body associated with VSG mono-allelic expression in Trypanosoma brucei. Nature, 414, 759–763. [DOI] [PubMed] [Google Scholar]
- 7.Conway C., McCulloch,R., Ginger,M.L., Robinson,N.P., Browitt,A. and Barry,J.D. (2002) Ku is important for telomere maintenance, but not for differential expression of telomeric VSG genes, in African trypanosomes. J. Biol. Chem., 277, 21269–21277. [DOI] [PubMed] [Google Scholar]
- 8.Downs J.A. and Jackson,S.P. (2004) A means to a DNA end: the many roles of Ku. Nature Rev. Mol. Cell Biol., 5, 367–378. [DOI] [PubMed] [Google Scholar]
- 9.Boulton S.J. and Jackson,S.P. (1996) Identification of a Saccharomyces cerevisiae KU80 homologue: roles in DNA double strand break rejoining and in telomeric maintenance. Nucleic Acids Res., 24, 4639–4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Porter S.E., Greenwell,P.W., Ritchie,K.B. and Petes,T.D. (1996) The DNA-binding protein Hdf1p (a putative Ku homologue) is required for maintaining normal telomere length in Saccharomyces cerevisiae. Nucleic Acids Res., 24, 582–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boulton S.J. and Jackson,S.P. (1998) Components of the Ku-dependent non-homologous end-joining pathway are involved in telomeric length maintenance and telomeric silencing. EMBO J., 17, 1819–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tham W.H. and Zakian,V.A. (2002) Transcriptional silencing at Saccharomyces telomeres: implications for other organisms. Oncogene, 21, 512–521. [DOI] [PubMed] [Google Scholar]
- 13.Gotta M., Laroche,T., Formenton,A., Maillet,L., Scherthan,H. and Gasser,S.M. (1996) The clustering of telomeres and colocalization with Rap1, Sir3, and Sir4 proteins in wild-type Saccharomyces cerevisiae. J. Cell Biol., 134, 1349–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feuerbach F., Galy,V., Trelles-Sticken,E., Fromont-Racine,M., Jacquier,A., Gilson,E., Olivo-Marin,J.C., Scherthan,H. and Nehrbass,U. (2002) Nuclear architecture and spatial positioning help establish transcriptional states of telomeres in yeast. Nature Cell Biol., 4, 214–221. [DOI] [PubMed] [Google Scholar]
- 15.Galy V., Olivo-Marin,J.C., Scherthan,H., Doye,V., Rascalou,N. and Nehrbass,U. (2000) Nuclear pore complexes in the organization of silent telomeric chromatin. Nature, 403, 108–112. [DOI] [PubMed] [Google Scholar]
- 16.Laroche T., Martin,S.G., Gotta,M., Gorham,H.C., Pryde,F.E., Louis,E.J. and Gasser,S.M. (1998) Mutation of yeast Ku genes disrupts the subnuclear organization of telomeres. Curr. Biol., 8, 653–656. [DOI] [PubMed] [Google Scholar]
- 17.Mishra K. and Shore,D. (1999) Yeast Ku protein plays a direct role in telomeric silencing and counteracts inhibition by rif proteins. Curr. Biol., 9, 1123–1126. [DOI] [PubMed] [Google Scholar]
- 18.Tham W.H., Wyithe,J.S., Ko Ferrigno,P., Silver,P.A. and Zakian,V.A. (2001) Localization of yeast telomeres to the nuclear periphery is separable from transcriptional repression and telomere stability functions. Mol. Cell, 8, 189–199. [DOI] [PubMed] [Google Scholar]
- 19.Hediger F., Neumann,F.R., Van Houwe,G., Dubrana,K. and Gasser,S.M. (2002) Live imaging of telomeres: yKu and Sir proteins define redundant telomere-anchoring pathways in yeast. Curr. Biol., 12, 2076–2089. [DOI] [PubMed] [Google Scholar]
- 20.Taddei A., Hediger,F., Neumann,F.R., Bauer,C. and Gasser,S.M. (2004) Separation of silencing from perinuclear anchoring functions in yeast Ku80, Sir4 and Esc1 proteins. EMBO J., 23, 1301–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taddei A. and Gasser,S.M. (2004) Multiple pathways for telomere tethering: functional implications of subnuclear position for heterochromatin formation. Biochem. Biophys. Acta, 1677, 120–128. [DOI] [PubMed] [Google Scholar]
- 22.Halme A., Bumgarner,S., Styles,C. and Fink,G.R. (2004) Genetic and epigenetic regulation of the FLO gene family generates cell-surface variation in yeast. Cell, 116, 405–415. [DOI] [PubMed] [Google Scholar]
- 23.McEachern M.J., Krauskopf,A. and Blackburn,E.H. (2000) Telomeres and their control. Ann. Rev. Gen., 34, 331–358. [DOI] [PubMed] [Google Scholar]
- 24.Gravel S., Larrivee,M., Labrecque,P. and Wellinger,R.J. (1998) Yeast Ku as a regulator of chromosomal DNA end structure. Science, 280, 741–744. [DOI] [PubMed] [Google Scholar]
- 25.Polotnianka R.M., Li,J. and Lustig,A.J. (1998) The yeast Ku heterodimer is essential for protection of the telomere against nucleolytic and recombinational activities. Curr. Biol., 8, 831–834. [DOI] [PubMed] [Google Scholar]
- 26.Dubrana K., Perrod,S. and Gasser,S.M. (2001) Turning telomeres off and on. Curr. Opin. Cell Biol., 13, 281–289. [DOI] [PubMed] [Google Scholar]
- 27.Stellwagen A.E., Haimberger,Z.W., Veatch,J.R. and Gottschling,D.E. (2003) Ku interacts with telomerase RNA to promote telomere addition at native and broken chromosome ends. Genes Dev., 17, 2384–2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peterson S.E., Stellwagen,A.E., Diede,S.J., Singer,M.S., Haimberger,Z.W., Johnson,C.O., Tzoneva,M. and Gottschling,D.E. (2001) The function of a stem–loop in telomerase RNA is linked to the DNA repair protein Ku. Nature Genet., 27, 64–67. [DOI] [PubMed] [Google Scholar]
- 29.Taggart A.K., Teng,S.C. and Zakian,V.A. (2002) Est1p as a cell cycle-regulated activator of telomere-bound telomerase. Science, 297, 1023–1026. [DOI] [PubMed] [Google Scholar]
- 30.Makarov V.L., Hirose,Y. and Langmore,J.P. (1997) Long G tails at both ends of human chromosomes suggest a C strand degradation mechanism for telomere shortening. Cell, 88, 657–666. [DOI] [PubMed] [Google Scholar]
- 31.Huffman K.E., Levene,S.D., Tesmer,V.M., Shay,J.W. and Wright,W.E. (2000) Telomere shortening is proportional to the size of the G-rich telomeric 3′-overhang. J. Biol. Chem., 275, 19719–19722. [DOI] [PubMed] [Google Scholar]
- 32.Wellinger R.J., Wolf,A.J. and Zakian,V.A. (1993) Saccharomyces telomeres acquire single-strand TG1-3 tails late in S-phase. Cell, 72, 51–60. [DOI] [PubMed] [Google Scholar]
- 33.Manolis K.G., Nimmo,E.R., Hartsuiker,E., Carr,A.M., Jeggo,P.A. and Allshire,R.C. (2001) Novel functional requirements for non-homologous DNA end joining in Schizosaccharomyces pombe. EMBO J., 20, 210–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Doyle J.J., Hirumi,H., Hirumi,K., Lupton,E.N. and Cross,G.A.M. (1980) Antigenic variation in clones of animal-infective Trypanosoma brucei derived and maintained in vitro. Parasitology, 80, 359–369. [DOI] [PubMed] [Google Scholar]
- 35.Hirumi H. and Hirumi,K. (1989) Continuous cultivation of Trypanosoma brucei bloodstream-forms in a medium containing a low concentration of serum protein without feeder cell layers. J. Parasitol., 75, 985–989. [PubMed] [Google Scholar]
- 36.Wirtz E., Leal,S., Ochatt,C. and Cross,G.A.M. (1999) A tightly regulated inducible expression system for dominant negative approaches in Trypanosoma brucei. Mol. Biochem. Parasitol., 99, 89–101. [DOI] [PubMed] [Google Scholar]
- 37.Brun R. and Schonenberger,M. (1979) Cultivation and in vitro cloning of procyclic culture forms of Trypanosoma brucei in a semi-defined medium. Acta Trop., 36, 289–292. [PubMed] [Google Scholar]
- 38.Overath P., Czichos,J. and Haas,C. (1986) The effect of citrate/cis-aconitate on oxidative metabolism during transformation of Trypanosoma brucei. Eur. J. Biochem., 160, 175–182. [DOI] [PubMed] [Google Scholar]
- 39.Wirtz E., Hoek,M. and Cross,G.A.M. (1998) Regulated processive transcription of chromatin by T7 RNA polymerase in Trypanosoma brucei. Nucleic Acids Res., 26, 4626–4634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wirtz E., Hartmann,C. and Clayton,C. (1994) Gene expression mediated by bacteriophage T3 and T7 RNA polymerases in transgenic trypanosomes. Nucleic Acids Res., 22, 3887–3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Horn D. and Cross,G.A.M. (1997) Position-dependent and promoter-specific regulation of gene expression in Trypanosoma brucei. EMBO J., 16, 7422–7431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Munoz-Jordan J.L. and Cross,G.A.M. (2001) Telomere shortening and cell cycle arrest in Trypanosoma brucei expressing human telomeric repeat factor TRF1. Mol. Biochem. Parasitol., 114, 169–181. [DOI] [PubMed] [Google Scholar]
- 43.Cross G.A.M. (1975) Identification, purification and properties of clone-specific glycoprotein antigens constituting the surface coat of Trypanosoma brucei. Parasitology, 71, 393–417. [DOI] [PubMed] [Google Scholar]
- 44.de Lange T. (1992) Human telomeres are attached to the nuclear matrix. EMBO J., 11, 717–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Church G.M. and Gilbert,W. (1984) Genomic sequencing. Proc. Natl Acad. Sci. USA, 81, 1991–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rudenko G., Blundell,P.A., Taylor,M.C., Kieft,R. and Borst,P. (1994) VSG gene expression site control in insect form Trypanosoma brucei. EMBO J., 13, 5470–5482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Horn D. and Cross,G.A.M. (1995) A developmentally regulated position effect at a telomeric locus in Trypanosoma brucei. Cell, 83, 555–561. [DOI] [PubMed] [Google Scholar]
- 48.Pays E., Laurent,M., Delinte,K., van Meirvenne,N. and Steinert,M. (1983) Differential size variations between transcriptionally active and inactive telomeres of Trypanosoma brucei. Nucleic Acids Res., 11, 8137–8147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bernards A., Michels,P.A.M., Lincke,C.R. and Borst,P. (1983) Growth of chromosome ends in multiplying trypanosomes. Nature, 303, 592–597. [DOI] [PubMed] [Google Scholar]
- 50.Chakhparonian M. and Wellinger,R.J. (2003) Telomere maintenance and DNA replication: how closely are these two connected? Trends Genet., 19, 439–446. [DOI] [PubMed] [Google Scholar]
- 51.Bertinato J., Schild-Poulter,C. and Hache,R.J.G. (2001) Nuclear localization of Ku antigen is promoted independently by basic motifs in the Ku70 and Ku80 subunits. J. Cell Science, 114, 89–99. [DOI] [PubMed] [Google Scholar]
- 52.Martin S.G., Laroche,T., Suka,N., Grunstein,M. and Gasser,S.M. (1999) Relocalization of telomeric Ku and Sir proteins in response to DNA strand breaks in yeast. Cell, 97, 621–633. [DOI] [PubMed] [Google Scholar]
- 53.Mo X. and Dynan,W.S. (2002) Subnuclear localization of Ku protein: functional association with RNA polymerase II elongation sites. Mol. Cell Biol., 22, 8088–8099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Munoz-Jordan J., Cross,G.A.M., de Lange,T. and Griffith,J.D. (2001) T-loops at trypanosome telomeres. EMBO J., 20, 579–588. [DOI] [PMC free article] [PubMed] [Google Scholar]