Abstract
The stabilization and processing of stalled replication forks is required to maintain genome integrity in all organisms. In an effort to identify novel proteins that might be involved in stabilizing stalled replication forks, Saccharomyces cerevisiae mutant wss1Δ was isolated from a high-throughput screening of ∼5000 deletion strains for genes involved in the response to continuous, low-intensity UV irradiation. Disruption of WSS1 resulted in synergistic increases in UV sensitivity with null mutants of genes involved in recombination (RAD52) and cell cycle control (RAD9 and RAD24). WSS1 was also found to interact genetically with SGS1, TOP3, SRS2 and CTF4, which are involved in recombination, repair of replication forks and the establishment of sister chromatid cohesion. A yeast two-hybrid screen identified a potential physical interaction between Wss1 and both Psy2 and Tof1. Genetic interactions were also detected between PSY2 and TOF1, as well as between each gene and RAD52 and SRS2, and between WSS1 and TOF1. Tof1 is known to be involved in stabilizing stalled replication forks and our data suggest that Wss1 and Psy2 similarly function to stabilize or process stalled or collapsed replication forks.
INTRODUCTION
DNA is a labile molecule (1), and it is thus essential that a cell rapidly recognize DNA damage, coordinate cell cycle progression, and efficiently restore DNA to its native state. The cellular response to DNA damage has been particularly well characterized in Saccharomyces cerevisiae through the isolation of mutants that are hypersensitive to specific DNA damaging agents. From these studies, three groups of functionally related genes have been identified and are referred to as the RAD3, RAD6 and RAD52 epistasis groups. The RAD3 and RAD6 genes encode proteins that function in nucleotide excision repair (NER) and post-replication repair (PRR), respectively. The RAD52 genes encode proteins involved in homology-dependent recombination and double-strand break (DSB) repair.
In addition to facilitating DSB repair, the RAD52 group genes are involved in repairing stalled or collapsed replication forks (2–5). These pathways are based on homologous recombination and require Rad52 to recruit Rad51 to sites of DSBs or collapsed replication forks. Rad51, the S.cerevisiae analog of the bacterial recombination mediator RecA, forms single-stranded DNA–protein filaments, which facilitate the formation of recombination intermediates by catalyzing strand invasion of the 3′-overhang into a homologous duplex. Sgs1, a member of the RecQ family of helicases, forms a complex with Top3 (Sgs1/Top3), and is thought to process Rad51-recombination intermediates to produce non-crossover recombination products (6–8). Sgs1 is also thought to convert recombination intermediates into forms that are required to stabilize (9) and restart stalled replication forks (10,11), and to bind Mec1 (9). Consistent with these activities, Sgs1/Top3 is required for Rad53 activation when cells are damaged in the absence of Rad24, possibly by acting in a pathway that overlaps with Rad9 (12). In addition, Sgs1 binds Rad53 in vitro and in vivo and these proteins colocalize in S-phase specific nuclear foci (12).
In addition to Sgs1, S.cerevisiae has another helicase involved in resolving recombination intermediates, Srs2. Mutations in SRS2 have been found to suppress the sensitivity associated with mutation of RAD6 (13–15), suggesting that Srs2 acts to funnel intermediates from recombinational repair to PRR. Interestingly, suppression of the sensitivity observed in rad6Δ mutants is dependent on the presence of Rad52 (14,16). In addition, Srs2 has been shown to efficiently displace Rad51 from DNA in vitro (17,18). Thus, it has been proposed that Srs2 acts as an anti-recombinase by catalyzing the removal of Rad51 from recombination intermediates and causing their disassembly, which allows for repair by other means (8,17,18).
In addition to these direct repair pathways, cells have evolved mechanisms to stabilize replication forks when they stall, due to the presence of damage or the absence of necessary substrates, such that replication may be restarted when cellular conditions return to normal. Two key proteins in this regard are Mrc1 and Tof1 (and the Tof1 binding partner Csm3), which are thought to be components of the processive replication fork where they help to prevent the uncoupling of the replication proteins from the associated DNA (19–21). The Schizosaccharomyces pombe analog of Tof1, Swi1, is also thought to stabilize collapsed replication forks in a conformation recognized by Mrc1 and checkpoint sensor proteins (22). Once activated, the checkpoint system halts cell cycle progression, inhibits the firing of late origins of replication and induces a transcriptional response that facilitates survival.
Mrc1, Tof1 and Csm3 also appear to play an important role in establishing cohesion between sister chromatids by helping to recruit an alternate RFC complex (the Ctf18–RFC complex) to stalled replication forks (23). The Ctf18–RFC complex can then load PCNA or an alternative clamp loader (24–28), which facilitates loading of polymerase κ (25). The result is the establishment of cohesion between sister chromatids as they emerge from the modified replication fork. Important links between replication and cohesion are mediated by Ctf4 and Ctf8, which are thought to act in association with replication forks and are also required for robust sister chromatid cohesion. Ctf8 is a component of the Ctf18–RFC complex and Ctf4 binds polymerase α and is thought to participate in repair of lagging strand synthesis (29). Neither CTF4 nor CTF8 is essential, unless MRC1, TOF1 or other genes that facilitate the establishment or maintenance of cohesion are absent (29–32). In addition, the ctf4Δ rad52Δ double mutant exhibits severe growth defects (33). These results suggest that sister chromatid cohesion is important for the stabilization and recombinational repair of stalled replication forks. One possibility is that these proteins play a role in establishing cohesion between sister chromatids as they emerge from the replication fork, and in the absence of such juxtaposed sister chromatids, homologs are inappropriately used during recombinational repair if the fork stalls.
Here, we report the identification and initial characterization of WSS1, a gene that when deleted, renders cells sensitive to UVB (310 nm) and UVC (254 nm) irradiation, and results in severe genetic phenotypes when deleted in recombination, cohesion and cell cycle checkpoint deficient backgrounds. We also report the characterization of PSY2 and TOF1, which encode proteins identified by a yeast two-hybrid screen for Wss1 interactors. PSY2 and TOF1 were found to display significant genetic interactions with each other, with several of the same genes identified for WSS1, as well as with WSS1 itself. The data are consistent with a model wherein Wss1, Tof1 and Psy2, function to stabilize or process stalled replication forks.
MATERIALS AND METHODS
Yeast strains used in this study are listed in Table 1. For genome-wide screening, ∼5000 deletion strains of the S.cerevisiae deletion project were pooled as described previously (34) (http://www-sequence.stanford.edu/group/yeast_deletion_project/deletions3.html). UVB irradiations were performed using a UVP-UVXX bench lamp with two 302 nm midrange bulbs (UVP Inc., Upland, CA) filtered by a 2 mm glass pane to yield an overall intensity of 1.04 Js−1 m−2, a dose that results in little delay of doubling time (<1%) in wild-type cells. UVC irradiations were performed using a G8T5 germicidal tube (Ushio America, Cypress, CA). UV fluences were determined using a UVX radiometer with UVX-25 and −31 sensors (Ultraviolet Products). Methyl methane sulfonate (MMS, Aldrich), 5-fluoro-orotic acid (5-FOA, RPI), hydroxyurea (HU, US Biological) and X-gal (Amersham) were also used in this study. Yeast extract/peptone/dextrose (YPD) media and various selective media were prepared as described previously (35). High-throughput screening of the yeast deletion library and microarray analysis were performed as described previously (36).
Table 1. S.cerevisiae strains used in this study.
| Strain | Genotype | Source |
|---|---|---|
| BY4741 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | ATCC |
| BY4742 | MATα his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | ATCC |
| BY4743 | MATa/α his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 met15Δ0/MET15 lys2Δ0/LYS2 ura3Δ0/ura3Δ0 | ATCC |
| orfΔa | BY4741 orfΔ::kanMX4 | Res gen |
| FR007c | BY4742 rad24Δ::HIS3MX6 | (36) |
| FR062c | BY4741 rad52Δ::LEU2 | (36) |
| FR661 | BY4742 wss1Δ::HIS3MX6 | This study |
| FR355c | MATa rad9Δ:: kanMX4 wss1Δ::LEU2 | This study |
| FR172b | MATa rad14Δ::kanMX4 wss1Δ::HIS3MX6 | This study |
| FR191b | MATa rad18Δ::kanMX4 wss1Δ::HIS3MX6 | This study |
| FR244b | MATa wss1Δ::kanMX4 rad24Δ::HIS3MX6 | This study |
| FR097c | BY4741 wss1Δ::kanMX4 rad52Δ::LEU2 | This study |
| FR163b | MATα rad10Δ::kanMX4 wss1Δ::HIS3MX6 | This study |
| FR200c | BY4741 wss1Δ::LEU2 | This study |
| FR221c | BY4741 srs2Δ::kanMX4 wss1Δ::LEU2 | This study |
| FR219c | BY4741 rad5Δ::kanMX4 wss1Δ::LEU2 | This study |
| FR204c | BY4741 rad6Δ::kanMX4 wss1Δ::LEU2 | This study |
| FR216c | BY4741 mms2Δ::kanMX4 wss1Δ::LEU2 | This study |
| FR144b | BY4741 ubc13Δ::kanMX4 wss1Δ::HIS3MX6 | This study |
| FR680c | BY4741 ctf4Δ::kanMX4 wss1Δ::LEU2 | This study |
| FR288c | BY4741 psy2Δ::LEU2 | This study |
| FR292c | BY4741 srs2Δ::kanMX4 psy2Δ::LEU2 | This study |
| FR289c | BY4741 wss1Δ::kanMX4 psy2Δ::LEU2 | This study |
| FR720c | BY4741 psy2Δ::kanMX4 rad52Δ::LEU2 | This study |
| FR319c | BY4741 tof1Δ::LEU2 | This study |
| FR357c | BY4741 srs2Δ::kanMX4 tof1Δ::LEU2 | This study |
| FR323c | BY4741 wss1Δ::kanMX4 tof1Δ::LEU2 | This study |
| FR334c | BY4741 psy2Δ::kanMX4 tof1Δ::LEU2 | This study |
| FR721c | BY4741 tof1Δ::kanMX4 rad52Δ::LEU2 | This study |
| W1588-4C | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 | (59) |
| FR659c | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 wss1Δ::LEU2 | This study |
| U1619-9D | MATa top3::TRP1 pWJ1189 ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 lys2Δ | (59) |
| FR660c | MATa top3::TRP1 pWJ1189 ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 lys2Δ wss1Δ::LEU2 | This study |
aStrains with defined gene deletions orfΔ::kanMX4 were constructed as part of the yeast deletion project.
bDerived from mating and subsequent sporulation of BY4741/2 haploids.
cDerived from one-step disruption by transforming strain BY4741 (or BY4742) orfΔ::kanMX4 with a PCR-amplified, gene targeted deletion cassette.
Competitive growth assays
The homozygous deletion pool was grown in YPD to a density of 2 × 107 cells/ml, representing ∼4000 cells of each strain. The culture was then split to inoculate two 200 ml cultures in 170 × 90 mm crystallization dishes at a density of 1 × 106 cells/ml. The cultures were covered with cellophane to minimize evaporation and maintain sterility. The UVB irradiated culture was grown under continuous exposure for 72 h, diluting every 8 h to maintain log phase growth. The control culture was treated identically, but grown in the dark. Samples of 1 × 108 cells were removed from the logarithmically growing pools at each dilution step. Sample preparation and analysis was performed as described previously (36).
Verification of strain sensitivity
Individual strains identified from the competitive growth assay were grown to mid-log phase and diluted to 1 × 104 cells/ml in 25 ml of YPD, in 80 × 40 mm crystallization dishes, covered with cellophane. Cultures were grown for 17 h, diluted and grown for an additional 17 h. Aliquots were collected at the dilution step and the end of the experiment, and plated onto YPD. Plates were grown for 40 h and photographed.
Complementation assay
Open reading frames (ORFs) were PCR amplified from S288C genomic DNA, isogenic to BY4743, by using primers found in Table 2. In each case, the ORF was amplified along with a ∼500 bp upstream of the start codon. PCR-amplified products were digested with XhoI/SacII and ligated into the low copy, centromeric plasmid pRS416 (37). Wild-type (BY4741) and the WSS1 and PSY2 deletion strains were independently transformed according to published protocols (38), with both pRS416 and pRS416 containing the appropriate gene. Aliquots of mid-log phase cultures were plated on selective media and treated with UVC or MMS.
Table 2. Oligonucleotides used in this study.
| Target gene | Sequence (5′–3′) |
|---|---|
| Cloning into pRS416 | |
| WSS1 | TTGGTTCTCGAGTATCTCCCTTTGCCAATTGG |
| TTGGTTCCGCGGATTTTCGAGTTCTTCGCTGTGG | |
| PSY2 | TTGGTTCTCGAGCGCCAATAAATTCGGCTTTGA |
| TTGGTTCCGCGGTCATCAAGTACTTGCATTCAT | |
| Cloning into pEG202 | |
| WSS1 | TTGGTTGAATTCGGTGGTTCTGGTGGTAAGACAGAAGGAATAAAA |
| TTGGTTCTCGAGTTAAGTGAGATCAATAACTTC | |
| Deletion using pUG73 | |
| WSS1 | CGCATATTTTGAAGATATTCTAAATAAGAGAGATTGATTACAGCTGAAGCTTCGTACGC |
| CATACTTATAATTTTCGAGTTCTTCGCTGTGGACAAGAGAGCATAGGCCACTAGTGGATCTG | |
| PSY2 | CAGAAATTCTAACTGAAAAGTTTAGGATTTACGTATAGTAAGAGTACAGCTGAAGCTTCGTACGC |
| CTTCTTACAACCATGACCGTTGTGCTAGCTTTTTATTCTTCTTTCCGCATAGGCCACTAGTGGATCTG | |
| TOF1 | TAGCTTGTGGGGTTTAGTGTATCTTTAATATAGGAGGGGCACACCAGCTGAAGCTTCGTACGC |
| CTAAAATTACACGTATTAAAGGGATTAATTACTACATATTCATTCGCATAGGCCACTAGTGGATCTG | |
| Confirmation of pUG73 based gene disruption | |
| WSS1-5′ | CTTCCCAGCTTATTTCCC |
| GTTATTCTTCGTCTCTCC | |
| wss1Δ-5′ | CTTCCCAGCTTATTTCCC |
| AGTTATCCTTGGATTTGG | |
| WSS1-3′ | AATCACTTCAAAGTGACC |
| TTTTTTGCAAGACTACCG | |
| wss1Δ-3′ | ATCTCATGGATGATATCC |
| TTTTTTGCAAGACTACCG | |
| PSY2-5′ | GTATCTCTGACTGTCCCC |
| TGCTGGTCGTTGGAGGGC | |
| psy2Δ-5′ | GTATCTCTGACTGTCCCC |
| AGTTATCCTTGGATTTGG | |
| PSY2-3′ | AAAAATTTATATGCGCCGG |
| CCAAGGCATGCAAGAAGG | |
| psy2Δ-3′ | ATCTCATGGATGATATCC |
| CCAAGGCATGCAAGAAGG | |
| TOF1-5′ | ACTTCGGTATTGTAGAGC |
| ATGCTTCTTTAGCTCACC | |
| tof1Δ-5′ | ACTTCGGTATTGTAGAGC |
| AGTTATCCTTGGATTTGG | |
| TOF1-3′ | GAAAGGATGAATAATGGG |
| GGAAAATGAAATTACGGG | |
| tof1Δ-3′ | ATCTCATGGATGATATCC |
| GGAAAATGAAATTACGGG | |
Cell cycle checkpoint assays
DNA cell cycle manipulation, sample preparation and flow cytometry were performed as described previously (39,40). To examine the G1/S checkpoint, cells were arrested in G1 with α-factor (6 mg/ml) for 150 min. Cell were treated with 0.25% MMS during the final 30 min of the arrest and then released. Control cells were arrested but not exposed to MMS. Samples were removed every 15 min, fixed with formaldehyde and examined microscopically. To analyze cell cycle delay at the G2/M transition, log phase cultures were arrested with nocodazole (15 mg/ml) for 150 min. During the final 30 min of the arrest, cells were treated with 0.25% MMS and then released from both MMS and nocodazole. Aliquots were removed every 15 min, fixed with paraformaldehyde and resuspended in buffer (100 mM KPO4 and 1.2 M sorbitol). Nuclei were counterstained with 4′,6′-diamidino-2-phenylindole and cells were visualized with a Nikon E600 microscope. Images were captured with a Coolsnap HQ CCD camera (Roper Scientific) using IPLab software (Scanalytics).
The intra-S phase checkpoint was assayed by resuspending 3 × 107 α-factor arrested cells in 25 ml pre-warmed YPD with and without 0.03% MMS. Aliquots of 2 ml were removed at 10 min intervals and fixed in 70% ethanol overnight at 4°C. Samples were then treated with 0.5 ml RNase [2 mg/ml RNase A (Sigma), 50 mM Tris, pH 8.0] for 2–4 h, 0.2 ml pepsin [5 mg/ml pepsin (Sigma), 4.5 mg/ml concentrated HCl in H2O] for 30 min, and resuspended in 50 mM Tris, pH 7.5. For analysis, cells were diluted into 1 ml of SYTOX Green solution [1 mM SYTOX (Molecular Probes) in 50 mM Tris, pH 7.5] and sonicated. Samples were analyzed by flow cytometry using a FACScalibur (Becton Dickinson Immunocytometry Systems), with an argon laser tuned to 488 nm. The FL1 detector with a standard 530/30 band pass filter was used in the acquisition of SYTOX Green fluorescence.
Rad53 phosphorylation assay
Two aliquots of 1 × 108 log phase cells were collected and resuspended in 10 ml of YPAD with or without 0.2% MMS. Cultures were then incubated for 2 h at 30°C with shaking. Whole cell extracts were prepared as described previously (41), separated on 6.5% SDS–PAGE gels and transferred onto PVDF. Immunostaining was performed with a goat polyclonal antiserum to Rad53 (Santa Cruz Biotechnology Inc.) used at a final dilution of 1:1000 followed by overnight incubation. Horseradish peroxidase-linked secondary anti-goat antibody (Santa Cruz Biotechnology Inc.) was used at a final dilution of 1:5000 with a 60 min incubation period. Chemiluminescent detection was performed with ECL Plus™ (Amersham Pharamacia Biotech).
Yeast two-hybrid
The DupLEX-A yeast two-hybrid system (OriGene Technologies, Inc., Rockville, MD) was employed. YHR134W was PCR amplified from S288C DNA. The 5′ PCR primer contained a BamHI restriction site and a linker sequence encoding Gly-Gly-Ser-Gly-Gly. The 3′ PCR primer contained an XhoI restriction site. The PCR product was digested and ligated into BamHI/XhoI digested pEG202. Screening was done in yeast strain EGY48, using the reporter plasmid pSH18-34 and a genomic library (∼2 × 106 fragments) in pJG4-5 obtained from R. Brent (Molecular Sciences Institute, Berkeley, CA). Proper activation of the LEU2 and lacZ reporter genes (located in EGY48 and pSH18-34, respectively) were used to identify clones with productive binding interactions. Library plasmids that tested positive for interaction were isolated and individually screened by reconstruction of the system in a direct assay. Plasmids testing positive under all criteria were sequenced to identify the interacting protein.
Genetic analysis
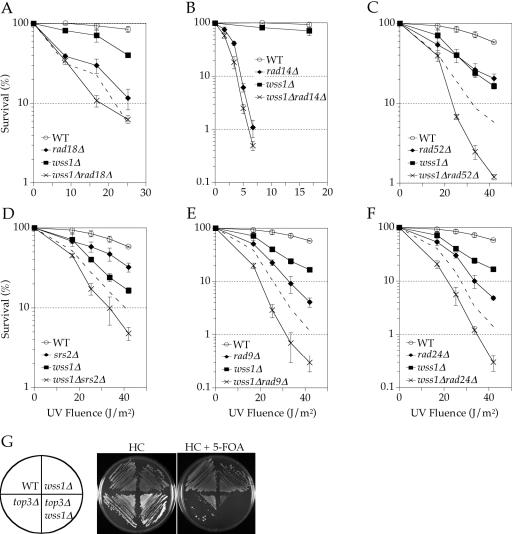
Double mutants of rad14Δ, rad18Δ, rad24Δ and rad52Δ with wss1Δ were constructed as reported previously (35,36,42,43). All additional double mutants were constructed by transformation of a PCR generated DNA deletion cassette specific for WSS1, PSY2 or TOF1. The disruption cassettes were generated by amplification of the LEU2 gene from pUG73 (44), using the gene-specific primers listed in Table 2. Gene disruption was confirmed by colony PCR. Epistasis analyses of the double mutants were conducted using colony survival assays by comparing the UV or MMS sensitivity of the various double mutants with those of the corresponding single mutants. In these assays, cultures were grown to mid-log phase, diluted to ∼6 × 106 cells/ml and serially diluted in 5-fold increments. For UV assays, appropriate dilutions were plated, irradiated, grown for 2 days in the dark and counted. Cell cultures were prepared identically for MMS experiments and spread on agar media with the indicated concentration of MMS. Cells were grown for 3 days and counted. Several independent spores or transformants were analyzed for each double mutant to rule out anomalous phenotypes. The predicted additive interaction viability curves (Figure 2A, C–F) were calculated by multiplying the sensitivity of one single mutant relative to wild type (viability of mutant/viability of wild type) by the measured viability of the other single mutant.
Figure 2.
Genetic epistasis analysis of WSS1. (A–F) Colony survival assays following UVC irradiation of yeast containing wild-type or null alleles of WSS1 in combination with wild-type or null alleles of representatives from known DNA damage repair pathways (see Materials and Methods). In all cases, the WT background is BY4741. The mutant strains are as listed in Table 1. The dashed lines (A and D–F) represent the predicted additive interaction from the sensitivities of the respective single mutants. (G) Simultaneous deletion of WSS1 and TOP3 results in lethality. The background strain for WT, wss1Δ (FR659), top3Δ (U1619-9D) and wss1Δ top3Δ (FR660) is W1588-4C. Strains U1619-9D and FR660 are top3Δ and harbor a plasmid expressing TOP3.
RESULTS
Identification of WSS1
In a competitive fitness assay, a pool of logarithmically growing homozygous diploid deletion strains was subjected to continuous UVB irradiation. A control pool was grown in parallel without UVB exposure. During the experiment, pools were sampled every 8 h, genomic DNA was extracted from the cells, tags corresponding to individual deletion strains were PCR amplified (using conserved flanking sequences) and hybridization intensities from oligonucleotide arrays were collected. The hybridization data were used to calculate the relative growth rate for each deletion mutant in the pool. Mutants judged to be sensitive exhibited growth rates that were at least 20% lower than wild type. Strains that did not exhibit at least two hybridized tag signals at the initial time point that were greater than five times the background signal were not considered for further analysis.
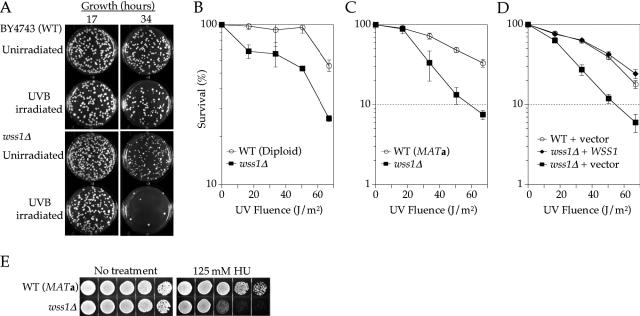
Eight putative UVB-sensitive strains were identified and tested individually to verify the hybridization data (mnn1Δ, wss1Δ, isc1Δ, yor322cΔ, yor235wΔ, clb5Δ, dpb3Δ and ybr277cΔ). Growth rates of the homozygous diploid deletion strains in rich liquid media with and without UVB treatment were compared with that of BY4743. One strain, wss1Δ, was verified as phenotypically more sensitive than wild type to continuous low-dose UVB irradiation (Figure 1A). The eight strains were also tested for sensitivity to acute UVC irradiation, as well as MMS. Deletion of WSS1 resulted in sensitivity to UVC in diploid cells homozygous for the deletion, as well as in MATa haploid cells (Figure 1B and C). During this study it was reported that wss1Δ was isolated in a high-throughput screen for acute UVC and UVB sensitivity (45). The strain clb5Δ was also found to be sensitive to UVC; however, given that this gene encodes a well-studied cyclin, it was not further characterized.
Figure 1.
DNA damage sensitivity and complementation of wss1Δ. (A) Sensitivity of homozygous diploid wss1Δ to chronic irradiation with UV-B (see Materials and Methods). All pictures for each respective timepoint are from identical dilutions of the WT and wss1Δ cultures. The strains were WT (Diploid; BY4743) and wss1Δ (from homozygous diploid deletion library). (B–D) Colony survival assays following UVC irradiation of yeast containing wild-type or null alleles of WSS1 (see Materials and Methods). The mutant strains are as listed in Table 1. (D) WT (BY4741) or wss1Δ (from MATa deletion library) were transformed with the plasmid pRS416 (vector) or pRS416 expressing full-length WSS1 regulated by its endogenous promoter (WSS1). (E) Sensitivity of wild-type (BY4741) and wss1Δ to HU. Five-fold serial dilutions of ∼105 cells were plated on YPD agar with or without 125 mM HU. Images were collected three days after plating.
Verification that the increased sensitivity to UVC was caused by the deletion of WSS1 and not an artifact of strain construction was accomplished by complementation with plasmid-borne WSS1 and acute UVC irradiation. We did not investigate complementation of the UVB sensitivity, since selective media is incompatible with the growth assay. The region of the chromosome containing WSS1 and an upstream region containing the endogenous promoter (∼500 bp) were cloned into pRS416. The wss1Δ (MATa haploid) cells transformed with the plasmid containing WSS1 were less sensitive to UVC (as sensitive as the wild-type control) than the single mutant transformed with an empty vector (Figure 1D). We also examined the HU sensitivity of the WSS1 deletion strain. HU is thought to induce replication fork stalling by depleting cellular levels of dNTPs (46). Deletion of WSS1 resulted in cells that grew significantly more slowly in the presence of HU (Figure 1E). The UV and HU sensitivity associated with the deletion of WSS1 implies that its protein product may be important when replication forks stall due to damage or insufficient dNTPs.
To determine if WSS1 plays a role in the damage response that is related to one of the known epistasis groups, it was deleted in strains carrying rad18Δ (PRR), rad14Δ (NER) or rad52Δ (recombination). The wss1Δ rad18Δ double mutant is additively more sensitive to UVC than either single mutant (Figure 2A), implying Wss1 does not function in PRR. The wss1Δ rad14Δ double mutant is only marginally more sensitive than either single mutant (Figure 2B). Given the difficulty in interpreting epistatic relationships when one deletion strain is very sensitive and the other is not, we conclude only that the double mutant is not synergistically more sensitive to UV damage. Based on these results and the additional genetic interactions reported in Table 3, and discussed below, we conclude that WSS1 does not contribute to either PRR or NER. In contrast, the wss1Δ rad52Δ mutant displayed a synergistic increase in sensitivity to UVC irradiation, relative to the corresponding single mutants (Figure 2C). These data suggest that Wss1 and Rad52 play a role in different pathways that act on a common intermediate.
Table 3. Genetic interactions of WSS1.
| Gene deletion | Phenotype with wss1Δa |
|---|---|
| rad9Δ | Synergistic |
| rad14Δ | Not synergisticb |
| rad18Δ | Additive |
| rad24Δ | Synergistic |
| rad52Δ | Synergistic |
| rad5Δ | Additive |
| rad6Δ | Additive |
| rad10Δ | Not synergisticb |
| mms2Δ | Additive |
| ubc13Δ | Additive |
| srs2Δ | Synergistic |
| psy2Δ | Not determinedc |
| tof1Δ | Synergistic |
| ctf4Δ | Synergistic |
| sgs1Δ | Synthetic lethald |
| top3Δ | Synthetic lethal |
Further characterization of Wss1 in the DNA damage response
It was reported previously that wss1Δ is synthetically lethal in an sgs1Δ background (47,48). Based on this interaction, we examined the effect of TOP3 deletion, since at least some aspects of Sgs1 functions are mediated by a complex of Sgs1 and Top3 (6,7). A synthetic growth defect in the absence of damage as well as a synergistic increase in sensitivity to UVC was observed in the wss1Δ top3Δ double mutant, but the mutant was viable (data not shown). To investigate the possibility that suppressor mutations arose in the wss1Δ top3Δ strain, WSS1 was deleted in strains W1588-4C and U1619-9D (the latter of which carries a top3Δ mutation and a URA3-marked TOP3 plasmid). In the top3Δ strain, loss of the complementing plasmid due to counterselection in the presence of 5-FOA yielded colonies that were slow growing, yet viable (Figure 2G). However, counterselection in the wss1Δ top3Δ strain did not give rise to any viable colonies (even after 6 days of growth). Thus, we conclude that suppressor mutation(s) arose during construction of the double mutant strain by gene disruption in a top3Δ background and that without the suppressor mutation(s), simultaneous deletion of WSS1 and TOP3 is lethal. This is consistent with the synthetic lethality reported for wss1Δ and sgs1Δ, and suggests that Wss1 and the Sgs1/Top3 complex may act in different pathways that process a common intermediate.
To define the function of WSS1 further, a genetic interaction with SRS2 was investigated. SRS2 encodes helicase with functions that at least partially overlap with SGS1 in processing recombination intermediates. The wss1Δ srs2Δ double mutant exhibited a mild synergistic sensitivity to UVC treatment (Figure 2D). One interpretation of these data is that Srs2 acts to oppose inappropriate recombination, and that this function becomes more critical in the absence of Wss1. We favor this interpretation because it is also consistent with the hypothesis that Wss1 functions in a pathway that recognizes stalled replication forks that are otherwise recognized and processed by Rad52 and Sgs1/Top3.
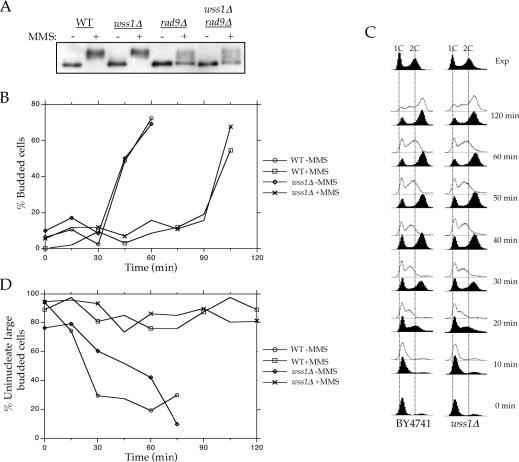
In addition to their roles in recombination, Srs2, Sgs1 and Top3 are thought to play roles in the cell cycle checkpoint response (9,12,49,50). To determine if WSS1 plays a role in the cell cycle checkpoint response, we examined wss1Δ rad9Δ and wss1Δ rad24Δ double mutants. In both cases, we observed a synergistic increase in sensitivity to UVC treatment (Figure 2E and F). To determine if Wss1 plays a direct role in the cell cycle response, we examined Rad53 phosphorylation in the presence and absence of either Rad9 and/or Wss1 (Figure 3A). Deletion of RAD9 resulted in a significant reduction of the amount of phosphorylated Rad53 following MMS damage. In contrast, deletion of WSS1, either in the wild-type or rad9Δ strain, had no effect. Thus, the genetic and biochemical data suggest that Wss1 does not play a direct role in the checkpoint response, but rather in its absence, Rad9 and Rad24-mediated cell cycle arrest becomes more critical.
Figure 3.
DNA damage cell cycle checkpoint analysis for wss1Δ. Experiments were performed as described in Materials and Methods. (A) Phosphorylation of Rad53 in response to MMS. Mobility of Rad53 was examined after asynchronous cultures were grown in the presence or absence of MMS. Protein extracts were separated by SDS–PAGE and blotted with antibodies specific for Rad53. (B) Assay for G1 DNA damage checkpoint. Briefly, cells were synchronized with α-factor in G1 and treated with MMS (+MMS) or without MMS (−MMS). At the indicated times after release from α-factor, the percentage of budded cells was scored under microscopy. (C) Assay for S-phase DNA damage checkpoint. Cells were synchronized with α-factor in G1 and released in either the presence or absence of MMS. Aliquots of cells were collected at the indicated times after release from α-factor and examined for DNA content by flow cytometry. The dotted lines indicate the DNA content of 1C and 2C unsynchronized cells. The top panel represents exponentially (Exp) growing cells and are included as a reference. (D) Assay for G2/M DNA damage checkpoint. Logarithmically growing cells were arrested with nocodazole and treated or not with MMS. At the indicated times after release of MMS-treated (+MMS) and untreated (−MMS) cultures from nocodazole, the percentage of uninucleate large budded cells was scored using DIC and flourescence microscopy.
We also examined the integrity of the G1-, S- and G2/M-phase damage checkpoints in cells lacking WSS1. Following MMS treatment during α-factor arrest, wild-type and wss1Δ cells showed similar delays before progressing through the G1/S transition, as judged by budding morphology (Figure 3B). The S-phase DNA damage checkpoint was analyzed by monitoring the DNA content of cells after their release from G1 block. When treated with MMS following release from α-factor arrest, wss1Δ and wild-type cells showed virtually identical transitions through S phase (Figure 3C). The G2/M-phase checkpoint was examined by monitoring mitotic division following DNA damage (Figure 3D). When cell cultures were released from nocodazole arrest after MMS treatment, both wild-type and wss1Δ cells entered mitosis at similar rates. Thus, in an otherwise wild-type cell, Wss1 does not appears to be required for the G1-, S- and G2/M-phase damage checkpoints following MMS treatment.
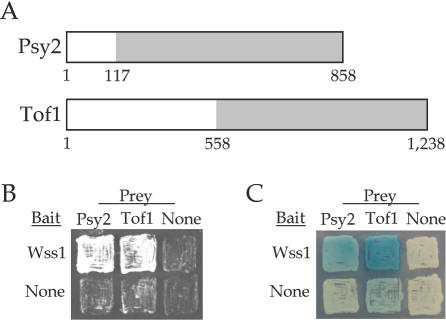
Interaction of Wss1 with Psy2 and Tof1
A yeast two-hybrid screen was performed to identify other proteins that may physically interact with Wss1. The entire WSS1 gene was cloned into the bait vector pEG202 and screened against a S.cerevisiae genomic DNA library of ∼2 × 106 fragments. About 140 colonies growing on selective media were isolated and screened to remove false positives. Library plasmids from the potential positive clones were recovered and sequenced. From 30 individual clones, 11 unique proteins were predicted to interact with Wss1 (Table 4).
Table 4. Wss1 yeast two-hybrid binding partners.
| Gene | Number of copies | Descriptiona |
|---|---|---|
| TyA | 14 | Retrotransposon element |
| OSH3 | 3 | Oxysterol binding protein |
| NOB1 | 2 | Component of 26S proteasome |
| RPN3 | 2 | Proteasome regulation |
| LCB1 | 2 | Sphingolipid biosynthesis |
| TOF1 | 1 | S-phase DNA damage checkpoint |
| FAB1 | 1 | PIP kinase |
| PSY2 | 1 | Interacts with Rad53 and Cdc6 |
| YOR227W | 1 | Uncharacterized |
| GSY1 | 1 | Glycogen metabolism |
| YDR221W | 1 | Uncharacterized |
| FAS1 | 1 | Fatty acid biosynthesis |
aAs obtained from SGD (www.yeastgenome.org).
The majority of the fragments (14 out of 30) identified encoded TyA, a gag protein for the Ty retrotransposon. The strains nob1Δ and rpn3Δ are inviable and were also not further characterized. As indicated in Table 4, a single fragment was obtained for TOF1 and for PSY2. To confirm these interactions, direct yeast two-hybrid analyses were performed with the WSS1 bait plasmid and the recovered TOF1 and PSY2 library plasmids. In each case, a strong binding interaction was verified using the LEU2 reporter gene (Figure 4B). A productive interaction for Psy2 was also verified using the lacZ reporter (Figure 4C). Expression of the Tof1 fragment resulted in a slight autoactivation of lacZ; however, the signal intensity was significantly greater in the presence of Wss1-LexA fusion, also confirming the interaction (Figure 4C). However, initial attempts to immunoprecipitate the corresponding complexes with myc13-tagged Wss1, GST-tagged Tof1 and GST-tagged Psy2 failed. This implies that the physical interactions may depend on the protein concentrations or the cellular environment associated with the two-hybrid experiment, or at least that the complex is not stable under the conditions required for its immunoprecipitation. Nonetheless, the genetic data described below, implies that there is a functional interaction between Wss1, Psy2 and Tof1.
Figure 4.
Wss1 binding interactions. (A) Schematic representation of Psy2 and Tof1 protein sequences. Regions recovered form yeast two-hybrid screen are shaded in gray. (B and C) Direct yeast two-hybrid assays. In all cases, both bait and prey constructs are expressed. (B) Growth on Leu− medium indicates a binding interaction. (C) Growth in the presence of X-gal results in blue color in the presence of a productive binding interaction.
Analysis of tof1Δ and psy2Δ
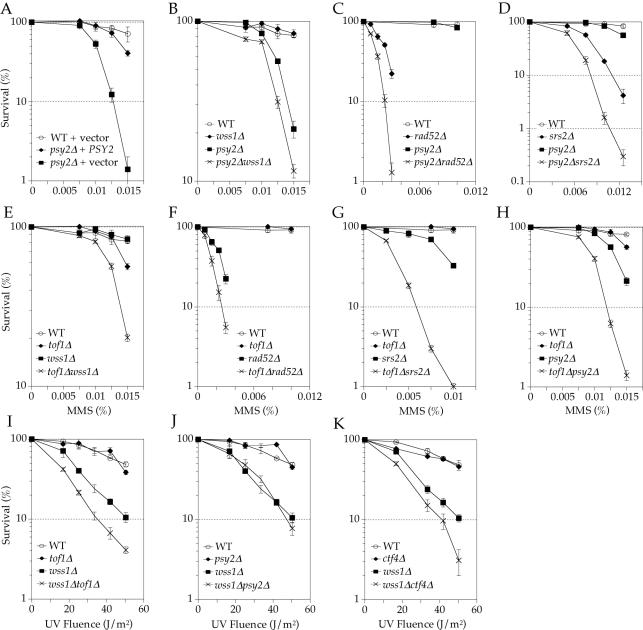
The tof1Δ and psy2Δ strains are both sensitive to MMS (19,36). However, since psy2Δ has not been previously characterized, its sensitivity was first confirmed by complementation (Figure 5A). Double mutants of tof1Δ and psy2Δ were then constructed with wss1Δ, rad52Δ and srs2Δ, as well as with each other. Analysis of MMS sensitivities demonstrated that tof1Δ and psy2Δ are synergistic with wss1Δ, rad52Δ and srs2Δ (Figure 5B–G). Additionally, the tof1Δ psy2Δ double mutant is synergistically more sensitive to MMS (Figure 5H). Sensitivity to UVC treatment also showed that wss1Δ and tof1Δ are synergistic; however, the interaction between psy2Δ and wss1Δ under these conditions could not be determined due to the insensitivity of psy2Δ to UV irradiation (Figure 5I and J). During the course of this work, a synthetic lethal interaction was reported for double mutant spore progeny of a heterozygous tof1Δ srs2Δ mutant (48,51). These genetic interactions suggest that Wss1, Psy2 and Tof1 have at least some functional overlap and they also suggest that the Wss1–Tof1 and Wss1–Psy2 interactions detected in the two-hybrid assay may be functionally significant.
Figure 5.
Genetic epistasis analysis of PSY2 and TOF1. Colony survival assays of yeast containing wild-type or null alleles of PSY2 and/or TOF1 in combination with wild-type or null alleles of representatives from known DNA damage repair pathways. Yeasts were either plated on YPD-agar media containing the indicated concentration of MMS (A–H) or UVC irradiated post-plating (I–K) (see Materials and Methods). The mutant strains (all in BY4741 background) are as listed in Table 1.
Genetic interaction of WSS1 with CTF4
Ctf4 is thought to be associated with replication forks and to help establish sister chromatid cohesion after their replication (32). Because CTF4 appears to play an essential but redundant function in establishing cohesion, synthetic interactions between ctf4Δ and other gene mutations have been used to identify proteins involved in establishing or maintaining cohesion (29,31,32). Using this approach, it has already been suggested that Tof1 plays a role in establishing sister chromatid cohesion during replication (23,30,31). To determine if Wss1 plays a similar role, we constructed a wss1Δ ctf4Δ double mutant and found that it was synergistically sensitive to UVC damage (Figure 5K). This result supports the hypothesis that Wss1 recognizes or processes stalled replication forks in a manner that has some functional redundancy with Ctf4.
DISCUSSION
A critical component of the DNA damage response is the maintenance or stabilization of replication forks that have stalled in response to damaged DNA or depleted dNTPs, and must be eventually restarted. Mrc1, Tof1 and Csm3 have already been found to help build stable pausing complexes. These proteins are associated with the replication fork (23) and facilitate checkpoint activation (19,22,52–54) and the establishment of cohesion between sister chromatids, such that they are readily available for recombination repair of the stalled fork (23). While these proteins are thought to eventually recruit the Ctf18–RFC-like complex and polymerse κ to sites where the fork stalls, other proteins are likely to be required.
As part of an effort to identify additional proteins involved in the stabilization of stalled forks, we performed a genome-wide screen in S.cerevisiae for genes whose deletion render yeast cells sensitive to low-intensity, chronic UVB radiation. The screen was performed under permissive conditions that allowed DNA synthesis in the presence of damaged DNA without causing cell cycle arrest or cell death. WSS1 was isolated from this screen, and encodes a 30 kDa protein of unknown function. Wss1 shares 47% similarity and 28% identity to S.pombe, Spcc1442.07, which is thought to be a zinc protease, but Wss1 does not appear to contain the protein motifs commonly found in zinc proteases. Deletion of WSS1 was found to render cells sensitive to chronic UVB, acute UVC irradiation and HU. During the course of these studies, wss1Δ was also identified in a genome-wide screen for UV sensitive strains (45), and reported to be synthetically lethal with sgs1Δ, based on a synthetic genetic array analysis (47,48). WSS1 was originally identified in a screen for high copy suppressors of a temperature-sensitive mutant allele of SMT3 (smt3-331) (55), which encodes a ubiquitin-like protein (SUMO in higher eukaryotes) that is post-translationally conjugated to target proteins and is required for normal chromosome segregation (55,56). The smt3-331 mutants accumulate large-budded cells with sister chromatids separated by short spindles (55). While WSS1 was found to suppress the growth defects associated with smt3-331 mutants, it did not suppress the large-budded phenotype, suggesting that it did not suppress the aberrant chromosome segregation.
We identified Tof1 and Psy2 as binding partners for Wss1 in a yeast two-hybrid screen. The physical interaction between Wss1 and Tof1 suggests that they may perform related functions. To examine the functional relatedness of Wss1, Tof1 and Psy2, we examined the damage sensitivity of the three double mutants, wss1Δ tof1Δ, wss1Δ psy2Δ and psy2Δ tof1Δ. All three were synergistically more sensitivite to MMS than the corresponding single mutants, implying that the three proteins play similar or overlapping roles and recognize a common substrate. This hypothesis is strengthened by the observation that TOF1 deletion increases UV sensitivity only in a wss1Δ background. Because Tof1 is known to function in the stabilization of stalled replication forks (20), we suggest that both Wss1 and Psy2 also function to stabilize or process stalled or collapsed replication forks.
Proteins that stabilize replication forks, for example Mrc1 and Tof1, are often also involved in the checkpoint response to DNA damage, but Wss1 does not appear to play a direct role in cell-cycle wide checkpoint activation. While the wss1Δ rad9Δ and wss1Δ rad24Δ double mutants were synergistically sensitive to UV damage, relative to the single mutants, we found that Rad53 activation in response to MMS was not reduced by the deletion of WSS1 either in a wild-type or rad9Δ background. In addition, the G1-, S- and G2/M-phase damage checkpoints all remained intact following MMS treatment in the absence of Wss1. These results imply that in the absence of Wss1, cells show a greater reliance on checkpoint activation by Rad9 and Rad24.
Based on the idea that Wss1, Psy2 and Tof1 may stabilize or process stalled or collapsed replication forks, we examined the effect of crippling the recombination repair pathways by deleting RAD52. We found that the wss1Δ rad52Δ double mutant is significantly more sensitive to UV damage than predicted by the sensitivities of the two single mutants. We also found that TOF1 and PSY2 show similar genetic interactions with RAD52 in response to MMS damage. Previously, wss1Δ was shown to be synthetically lethal when combined with sgs1Δ, and we have extended these results by showing that it is also synthetic lethal with top3Δ. We interpret these data to mean that Wss1, Tof1 and Psy2 act at an early stage after forks stall and that in their absence, collapsed or broken forks result, which must be processed by Rad52 and Sgs1/Top3.
Similar to Sgs1, Srs2 also plays an important role in processing stalled replication forks. Srs2 destabilizes Rad51 filaments within recombination intermediates and thus acts as an anti-recombinase to help abort aberrant recombination intermediates to allow for repair by other means (17,18). For example, the synthetic lethality observed between mrc1Δ and srs2Δ was suggested to result from unrestrained recombination involving collapsed forks with unestablished cohesion between sister chromatids (23). We find that wss1Δ srs2Δ is synergistically more sensitive to UV irradiation, relative to the wss1Δ and srs2Δ single mutants. Similarly, we observe synergistic increase in MMS sensitivity for tof1Δ srs2Δ and psy2Δ srs2Δ. Thus, we suggest that in the absence of Wss1, Psy2 or Tof1, Srs2 is required to restrain or control recombination.
The data are consistent with a model in which Wss1, Psy2 and Tof1 cooperate, perhaps along with Csm3 and Mrc1, to stabilize replication forks when they stall due to damaged DNA or insufficient dNTP levels. Given the different sensitivities of the corresponding deletion mutants (i.e. wss1Δ is sensitive to UV- but not to MMS-mediated DNA damage, while tof1Δ and psy2Δ are sensitive only to MMS), these proteins may stabilize replication forks specifically in response to different forms of DNA damage. This is consistent with the damage response roles already proposed for Mrc1, Tof1 and Csm3. One model postulates that Mrc1, Tof1 and Csm3 recruit the Ctf18–RFC-like complex to stalled replication forks (23), which then loads PCNA or an alternative clamp loader (24). Polymerase κ in turn replaces the RFC-like complex and ultimately establishes cohesion during replication (24–28). It is presently not clear how Psy2 contributes to stable pausing complexes, but its potential interactions with Cdc6 and Rad53 suggest a cell cycle checkpoint role similar to that already proposed for Mrc1 and Tof1 (57,58).
The precise role of Wss1 in stabilizing stalled replication forks is not yet clear. However, several lines of evidence point to a function intimately related to the establishment of sister chromatid cohesion and recombinational fork repair. First, we find that WSS1 interacts genetically with CTF4, whose protein product is thought to couple DNA synthesis and sister chromatid cohesion. Second, our genetic data suggest that at least some functions of Wss1 overlap with those of Tof1, which is thought to help establish sister chromatid cohesion at stalled replication forks. Finally, overexpression of Wss1 suppresses the growth defect of SMT3 mutants, apparently without suppressing the actual cohesion defects (55). One possibility that is consistent with the data is that Wss1 helps to funnel stalled forks into a pathway that is parallel to cohesion-mediated recombinational repair. This hypothesis is consistent with the increased importance of Rad52, Sgs1/Top3 and Ctf4 in the absence of Wss1, as well as with the ability of Wss1 overexpression to facilitate survival of strain with cohesion defects. The repair pathway favored by Wss1 might also be facilitated by the anti-recombinase activity of Srs2, explaining the increased dependence on Srs2 in the absence of Wss1.
We suggest that Wss1, Psy2 and Tof1, along with Mrc1 and Csm3, function to stabilize or repair stalled or collapsed replication forks and facilitate the resumption of processive DNA synthesis. This may be accomplished by helping to coordinate the various facets of the response, including delaying cell cycle progression, establishing cohesion for recombinational repair or other forms of repair, such as translesion synthesis. Further genetic characterization of these genes and biochemical characterization of their protein products is required to refine this proposed model, and help in elucidating the manner in which the different facets of the damage response are coordinated.
Acknowledgments
ACKNOWLEDGEMENTS
We thank R. Rothstein for providing strains and we gratefully acknowledge the NIH for providing funding (GM068569 to F.E.R.).
REFERENCES
- 1.Nickoloff J.A. and Hoekstra,M.F. (1998) DNA damage and repair. In Nickoloff,J.A. (ed.), Contemporary Cancer Research. Humana Press, Totowa, NJ, Vol. II, pp. 1–32. [Google Scholar]
- 2.Davis A.P. and Symington,L.S. (2004) RAD51-dependent break-induced replication in yeast. Mol. Cell. Biol., 24, 2344–2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sung P., Krejci,L., Komen,S.V. and Sehorn,M.G. (2003) Rad51 recombinase and recombination mediators. J. Biol. Chem., 278, 42729–42732. [DOI] [PubMed] [Google Scholar]
- 4.Pâques F. and Haber,J.E. (1999) Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev., 63, 349–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Symington L.S. (2002) Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol. Mol. Biol. Rev., 66, 630–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mullen J.R., Kaliraman,V. and Brill,S.J. (2000) Bipartite structure of the SGS1 DNA helicase in Saccharomyces cerevisiae. Genetics, 154, 1101–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bennett R.J., Noirot-Gros,M.F. and Wang,J.C. (2000) Interaction between yeast sgs1 helicase and DNA topoisomerase III. J. Biol. Chem., 275, 26898–26905. [DOI] [PubMed] [Google Scholar]
- 8.Ira G., Malkolva,A., Liberi,G., Foiani,M. and Haber,J.E. (2003) Srs2 and Sgs1-Top3 suppress crossovers during double-strand break repair in yeast. Cell, 115, 401–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cobb J.A., Bjergbaek,L., Shimada,K., Frei,C. and Gasser,S.M. (2003) DNA polymerase stabilization at stalled replication forks requires Mec1 and the RecQ helicase Sgs1. EMBO J., 22, 4325–4336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mullen J.R., Kaliraman,V., Ibrahim,S.S. and Brill,S.J. (2001) Requirement for three novel protein complexes in the absence of the Sgs1 DNA helicase in Saccharomyces cerevisiae. Genetics, 157, 103–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fabre F., Chan,A., Heyer,W.D. and Gangloff,S. (2002) Alternate pathways involving Sgs1/Top3, Mus81/Mus81, and Srs2 prevent formation of toxic recombination intermediates from single-stranded gaps created by DNA replication. Proc. Natl Acad. Sci. USA, 99, 16887–16892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frei C. and Gasser,S.M. (2000) The yeast Sgs1p helicase acts upstream of Rad53p in the DNA replication checkpoint and colocalizes with Rad53p in S-phase-specific foci. Genes Dev., 14, 81–96. [PMC free article] [PubMed] [Google Scholar]
- 13.Lawrence C.W. and Christensen,R.B. (1979) Metabolic suppressors of trimethoprim and ultraviolet light sensitivities of Saccharomyces cerevisiae rad6 mutants. J. Bacteriol., 139, 866–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schiestl R.H., Prakash,S. and Prakash,L. (1990) The Srs2 suppressor of Rad6 mutations of Saccharomyces cerevisiae acts by channeling DNA lesions into the Rad52 DNA-repair pathway. Genetics, 124, 817–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Friedl A.A., Liefshitz,B., Steinlauf,R. and Kupiec,M. (2001) Deletion of the SRS2 gene suppresses elevated recombination and DNA damage sensitivity in rad5 and rad18 mutants of Saccharomyces cerevisiae. Mutat. Res., 486, 137–146. [DOI] [PubMed] [Google Scholar]
- 16.Nguyen M.M. and Livingston,D.M. (1997) The effect of a suppressed rad52 mutation on the suppression of rad6 by srs2. Yeast, 13, 1059–1064. [DOI] [PubMed] [Google Scholar]
- 17.Veaute X., Jeusset,J., Soustelle,C., Kowalczykowski,S.C., Le Cam,E. and Fabre,F. (2003) The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature, 423, 309–312. [DOI] [PubMed] [Google Scholar]
- 18.Krejci L., Van Komen,S., Li,Y., Villemain,J., Reddy,M.S., Klein,H., Ellenberger,T. and Sung,P. (2003) DNA helicase Srs2 disrupts the RAd51 presynaptic filament. Nature, 423, 305–309. [DOI] [PubMed] [Google Scholar]
- 19.Foss E.J. (2001) Toflp regulates DNA damage responses during S phase in Saccharomyces cerevisiae. Genetics, 157, 567–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Katou Y., Kanoh,Y., Bando,M., Noguchi,H., Tanaka,H., Ashikari,T., Sugimoto,K. and Shirahige,K. (2003) S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex. Nature, 424, 1078–1083. [DOI] [PubMed] [Google Scholar]
- 21.Osborn A.J. and Elledge,S.J. (2003) Mrc1 is a replication fork component whose phosphorylation in response to DNA replication stress activates Rad53. Genes Dev., 17, 1755–1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Noguchi E., Noguchi,C., Du,L.L. and Russell,P. (2003) Swi1 prevents replication fork collapse and controls checkpoint kinase Cds1. Mol. Cell. Biol., 23, 7861–7874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu H., Boone,C. and Klein,H.L. (2004) Mrc1 is required for sister chromatid cohesion to aid in recombination repair of spontaneous damage. Mol. Cell. Biol., 24, 7082–7090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mayer M.L., Gygi,S.P., Aebersold,R. and Hieter,P. (2001) Identification of RFC(Ctf18p, Ctf8p, Dcc1p): an alternative RFC complex required for sister chromatid cohesion in S.cerevisiae. Mol. Cell, 7, 959–970. [DOI] [PubMed] [Google Scholar]
- 25.Wang Z., Castano,I.B., Panas,A.D.L., Adams,C. and Christman,M.F. (2000) Pol k: a DNA polymerase required for sister chromatid cohesion. Science, 289, 774–779. [DOI] [PubMed] [Google Scholar]
- 26.Uhlmann F. and Nasmyth,K. (1998) Cohesion between sister chromatids must be established during DNA replication. Curr. Biol., 8, 1095–1101. [DOI] [PubMed] [Google Scholar]
- 27.Skibbens R.V., Corson,L.B., Koshland,D. and Heiter,P. (1999) Ctf7p is essential for sister chromatid cohesion and links mitotic chromosome structure to the DNA replication machinery. Genes Dev., 13, 307–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Toth A., Ciosk,R., Uhlmann,F., Galova,M., Schleiffer,A. and Nasmyth,K. (1999) Yeast cohesion complex requires a conserved protein, Eco1p(Ctfy7), to establish cohesion between sister chromatids during DNA replication. Genes Dev., 13, 320–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Formosa T. and Nittis,T. (1999) Dna2 mutants reveal interactions with DNA polymerase a and Ctf4, a Pol a accessory factor, and show that dull Dna2 helicase activity is not essential for growth. Genetics, 151, 1459–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Warren C.D., Eckley,D.M., Lee,M.S., Hanna,J.S., Hughes,A., Peyser,B., Jie,C.F., Irizarry,R. and Spencer,F.A. (2004) S-phase checkpoint genes safeguard high-fidelity sister chromatid cohesion. Mol. Biol. Cell, 15, 1724–1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mayer M.L., Pot,I., Chang,M., Xu,H., Aneliunas,V., Kwok,T., Newitt,R., Aebersold,R., Boone,C., Brown,G.W. and Hieter,P. (2004) Identifiction of protein complexes required for efficient sister chromatid cohesion. Mol. Biol. Cell, 15, 1736–1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hanna J.S., Kroll,E.S., Lundblad,V. and Spencer,F.A. (2001) Saccharomyces cerevisiae CTF18 and CTF4 are required for sister chromatid cohesion. Mol. Cell. Biol., 21, 3144–3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kouprina N., Kroll,E., Bannikov,V., Bliskovsky,V., Gizatullin,R., Kirillov,A., Shestopalov,B., Zakharyev,V., Heiter,P. and Spencer,F. (1992) CTF4 (CHL15) mutants exhibit defective DNA metabolism in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol., 12, 5736–5747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Winzeler E.A. Shoemaker,D.D., Astromoff,A., Liang,H., Anderson,K., Andre,B., Bangham,R., Benito,R., Boeke,J.D., Bussey,H. et al. (1999) Functional characterization of the S.cerevisiae genome by gene deletion and parallel analysis. Science, 285, 901–906. [DOI] [PubMed] [Google Scholar]
- 35.Sherman F., Fink,G.R. and Hicks,J. (1983) Methods in Yeast Genetics. CSHL Press, Cold Spring Harbor, NY. [Google Scholar]
- 36.Hanway D.H., Chin,J.K., Xia,G., Oshiro,G., Winzeler,E.A. and Romesberg,F.E. (2002) Previously uncharacterized genes in the UV- and MMS-induced DNA damage response pathways in yeast. Proc. Natl Acad. Sci. USA, 99, 10605–10610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sikorski R.S. and Hieter,P. (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics, 122, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Agatep R., Kirkpatrick,R.D., Parchaliuk,D.L., Woods,R.A. and Gietz,R.D. (1998) Transformation of Saccharomyces cerevisiae by the lithium acetate/single-stranded carrier DNA/polyethylene glycol protocol. Tech. Tips Online, 1, P01525. [Google Scholar]
- 39.Haase S.B. and Reed,S.I. (2002) Improved flow cytometric analysis of the budding yeast cell cycle. Cell cycle, 1, 132–136. [PubMed] [Google Scholar]
- 40.Naiki T., Kondo,T., Nakada,D., Matsumoto,K. and Sugimoto,K. (2001) Chl12 (Ctf18) forms a novel replication factor C-related complex and functions redundantly with Rad24 in the DNA replication checkpoint pathway. Mol. Cell. Biol., 21, 5838–5845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bonifacino J.S., Dell'Angelica,E.C. and Springer,T.A. (1999) Affinity purification. In Coligan,J.E., Dunn,B.M., Speicher,D.W. and Wingfield,P.T. (eds) Current Protocols in Protein Science. John Wiley & Sons, Inc., Hoboken, NJ, Vol. II, pp. 9.8.1–9.8.20. [Google Scholar]
- 42.Wach A., Brachat,A., Alberti-Segui,C., Rebischung,C. and Philippsen,P. (1997) Heterologous HIS3 marker and GFP reporter modules for PCR-targeting in Saccharomyces cerevisiae. Yeast, 13, 1065–1075. [DOI] [PubMed] [Google Scholar]
- 43.Sprague G. (1991) Assay of yeast mating reaction. Methods Enzymol., 194, 77–93. [DOI] [PubMed] [Google Scholar]
- 44.Gueldener U., Heinisch,J., Koehler,G.J., Voss,D. and Hegemann,J.H. (2002) A second set of loxP marker cassettes for Cre-mediated multiple gene knockouts in budding yeast. Nucleic Acids Res., 30, e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Birrell G.W., Giaever,G., Chu,A.M., Davis,R.W. and Brown,J.M. (2001) A genome-wide screen in Saccharomyces cerevisiae for genes affecting UV radiation sensitivity. Proc. Natl Acad. Sci. USA, 98, 12608–12613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Futcher B. (1994) Analysis of the cell cycle in Saccharomyces cerevisiae. In Fantes,P. and Brooks,R. (eds) The Cell Cycle. A Practical Approach. IRL Press, New York, NY, pp. 69–92. [Google Scholar]
- 47.Tong A.H., Evangelista,M., Parsons,A.B., Xu,H., Bader,G.D., Page,N., Robinson,M., Raghibizadeh,S., Hogue,C.W., Bussey,H., Andrews,B., Tyers,M. and Boone,C. (2001) Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science, 294, 2364–2368. [DOI] [PubMed] [Google Scholar]
- 48.Ooi S.L., Shoemaker,D.D. and Boeke,J.D. (2003) DNA helicase gene interaction network defined using synthetic lethality analyzed by microarray. Nature Genet., 35, 277–286. [DOI] [PubMed] [Google Scholar]
- 49.Liberi G., Chiolo,I., Pellicioli,A., Lopes,M., Plevani,P., Muiz-Falconi,M. and Foiani,M. (2000) Srs2 DNA helicase is involved in checkpoint response and its regulation requires a functional Mec-1 dependent pathway and Cdk1 activity. EMBO J., 19, 5027–5038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chakraverty R.K., Kearsey,J.M., Oakley,T.J., Grenon,M., de la Torres Ruiz,M.-A., Lowdes,N.F. and Hickson,I.D. (2001) Topoisomerase III acts upstream of Rad53p in the S-phase DNA damage checkpoint. Mol. Cell. Biol., 21, 7150–7162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tong A.H.Y., Lesage,G., Bader,G.D., Ding,H.M., Xu,H., Xin,X.F., Young,J., Berriz,G.F., Brost,R.L., Chang,M. et al. (2004) Global mapping of the yeast genetic interaction network. Science, 303, 808–813. [DOI] [PubMed] [Google Scholar]
- 52.Alcasabas A.A., Osborn,A.J., Bachant,J., Hu,F.H., Werler,P.J.H., Bousset,K., Furuya,K., Diffley,J.F.X., Carr,A.M. and Elledge,S.J. (2001) Mrc1 transduces signals of DNA replication stress to activate Rad53. Nature Cell. Biol., 3, 958–965. [DOI] [PubMed] [Google Scholar]
- 53.Tanaka K. and Russell,P. (2001) Mrc1 channels the DNA replication arrest signal to checkpoint kinase Cds1. Nature Cell. Biol., 3, 966–972. [DOI] [PubMed] [Google Scholar]
- 54.Nyberg K.A., Michelson,R.J., Putnam,C.W. and Weinert,T.A. (2002) Toward maintaining the genome: DNA damage and replication checkpoints. Annu. Rev. Genet., 36, 617–656. [DOI] [PubMed] [Google Scholar]
- 55.Biggins S., Bhalla,N., Chang,A., Smith,D.L. and Murray,A.W. (2001) Genes involved in sister chromatid separation and segregation in the budding yeast Saccharomyces cerevisiae. Genetics, 159, 453–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Saitoh H., Pu,R.T. and Dasso,M. (1997) SUMO-1: wrestling with a new ubiquitin-related modifier. Trends Biochem. Sci., 22, 374–376. [DOI] [PubMed] [Google Scholar]
- 57.Zavec P.B., Comino,A., Watt,P. and Komel,R. (2000) Interaction trap experiment with CDC6. Pflugers Arch., 439, R94–R96. [PubMed] [Google Scholar]
- 58.Uetz P., Giot,L., Cagney,G., Mansfield,T.A. and Judson,R.S. (2000) A comprehensive analysis of protein–protein interactions in Saccharomyces cerevisiae. Nature, 403, 601–603. [DOI] [PubMed] [Google Scholar]
- 59.Shor E., Gangloff.S., Wagner,M., Weinstein,J., Price,G. and Rothstein,R. (2002) Mutation in homologous recombination genes rescue top3 slow growth in Saccharomyces cerevisiae. Genetics, 162, 647–662. [DOI] [PMC free article] [PubMed] [Google Scholar]