Abstract
Background:
Investigations of methods for detection of mutations have uncovered major weaknesses of direct sequencing and pyrosequencing, with their high costs and low sensitivity in screening for both known and unknown mutations. High resolution melting (HRM) analysis is an alternative tool for the rapid detection of mutations. Here we describe the accuracy of HRM in screening for KRAS and BRAF mutations in metastatic colorectal cancer (mCRCs) samples.
Materials and Methods:
A total of 1000 mCRC patients in Mehr Hospital, Tehran, Iran, from Feb 2008 to May 2012 were examined for KRAS mutations and 242 of them were selected for further assessment of BRAF mutations by HRM analysis. In order to calculate the sensitivity and specificity, HRM results were checked by pyrosequencing as the golden standard and Dxs Therascreen as a further method.
Results:
In the total of 1,000 participants, there were 664 (66.4%) with wild type and 336 (33.6%) with mutant codons 12 and/or 13 of the KRAS gene. Among 242 samples randomly checked for the BRAF gene, all were wild type by HRM. Pyrosequencing and Dxs Therascreen results were in line with those of the HRM. In this regard, the sensitivity and specificity of HRM were evaluated as 100%.
Conclusion:
The findings suggest that the HRM, in comparison with DNA sequencing, is a more appropriate method for precise scanning of KRAS and BRAF mutations. It is also possible to state that HRM may be an attractive technique for the detection of known or unknown somatic mutations in other genes.
Keywords: High-Resolution Melting (HRM), mutation analysis, metastatic colorectal cancer (mCRC)
Introduction
Rapid and reliable genetic screening method of human cancers is one of the interesting fields of research (Simi et al., 2008). There are several diagnostic methods for the investigation of cancerous mutational status of patients such as direct DNA sequencing and pyrosequencing (golden standard), TaqMan array, single-strand conformation analysis, allele-specific PCR (Dxs Therascreen), but they have some defects, including needing manipulation of amplified fragments or being labor intensive and time consuming(Tan et al., 2008; Wooster et al., 2006; Morandi et al., 2012; Packham et al., 2009; Sundstrom et al., 2010; Fransen et al., 2004).
Sanger sequencing, is approved as a gold standard of sequence variation analysis and has high reliability but its disadvantages raised the applicability limitation as a diagnostic tool in routine laboratories and restricted its functionality in research centers, respectively. One of its weak points is the use of several steps by distinct protocols (e.g. PCR, amplicon purification, labelling) that increased the contamination risks (Morandi et al., 2012; Packham et al., 2009; Krypuy et al., 2006). Given that, Sanger sequencing pitfalls such as being time-consuming, labor-intensive, and expensive or being in need of sophisticated devices introduce the suggestion of its gradual replacement by other sensitive, reliable, and efficient methods, (Morandi et al., 2012; Packham et al., 2009; Krypuy et al., 2006).
High resolution melting (HRM) analysis is a high throughput emerging PCR-based method for rapid scanning of hereditary or somatic mutations with high accuracy (Simi et al., 2008; Krypuy et al., 2006; Negru et al., 2014). The screening of mutations by this method is based on the measurement of modifications in the melting profile of duplex, when the DNA is exposed to an increasing temperature. These modifications monitored using the fluorescent dyes, which intercalate double-stranded fragments but no single-stranded fragments (Reed and Wittwer, 2004). Patterns of melting profile influenced by various variants, as a result wild type and homozygote-heterozygote sequences have different melting profile patterns (Boyd et al., 2011).
KRAS and BRAF oncogenes are two genes of interest in cancerous cells’ investigation and their mutational status is important to patient’s cancer outcome. In this context, the sensitive and reliable methods compared and analyzed elsewhere (Borras et al., 2011). Common signaling pathways such as KRAS/BRAF/MEK/ERK activated in colorectal cancer (CRC) (Popovici et al., 2012; Vakil et al., 2016). Hydrolysis of guanosine triphosphate (GTP) molecules has a critical role in inactivation of complex KRAS genes produced by signal transduction protein (Pinto et al., 2011; Ji et al., 2007). Mutants of KRAS gene are less sensitive to hydrolysis and this leads to cell proliferation mediated KRAS (Ji et al., 2007; Koochak et al., 2016). In CRC patients, KRAS gene mutations were identified about 30-40% (Andreyev et al., 2001; Andreyev et al., 1998), which KRAS exon 2 mutations are more frequent (almost 90% at codons 12/13 in CRC patients) (Janakiraman et al., 2010; De Roock et al., 2011). KRAS activates the cytosolic protein kinase which encodes by BRAF gene (Roth et al., 2010). BRAF mutations induced cell propagation by MAPKs signaling cascade (Guedes et al., 2013; Allegra et al., 2009). BRAF mutation rates in CRC patients estimates approximately 10% and the most common substitution in BRAF gene is V600E in CRC patients (Minoo et al., 2007; Di Nicolantonio et al., 2008).
Recently, before any specific therapy in CRC patients recommended the screening of KRAS and BRAF mutations as a necessity and due to the presence of some screening methods in this area, the reliable, rapid, and cost-effective diagnostic method is one of research interests. (Pinto et al., 2011). In the current study, our main objective was to validate the performance of HRM technique compared to allele specific PCR (DxS) and Pyrosequencing by samples (n=1000) that originated from colorectal cancer tissues for the detection of KRAS and BRAF mutations in selected subsets.
Material and Methods
Participants
Samples were collected from Iranian mCRC patients who referred to Mehr Hospital, the main referral center for cancerous patients in Tehran, Iran from Feb. 2008 to May 2012 and enrolled in this study. Written consent obtained from each patients and ethical committee of Iran University of Medical Sciences, Tehran, Iran approved ethics of present study. A total of 1,000 mCRC FFPE samples included 427 (42.7%) female by the average age of 55 years and 573 (57.3%) male by the average age of 57 years were enrolled (Table 1).
Table 1.
Demographic Characteristics of 1,000 mCRC Patients
| Demographic and | pathological characteristics | N | % |
|---|---|---|---|
| Sex | Male | 573 | 57.3 |
| Female | 427 | 42.7 | |
| Age (y) | ≤50 | 325 | 32.5 |
| ˃50 | 675 | 67.5 | |
| Tumor differentiation | Well Differentiated | 439 | 43.9 |
| Moderate Differentiated | 384 | 38.4 | |
| Poor differentiated | 164 | 16.4 | |
| Undifferentiated | 13 | 1.3 |
Formalin-fixed paraffin-embedded (FFPE) tissue DNA extraction
FFPE tissues were manually dissected by block trimming from 10 μm thickness collected in a microtube for genetic testing and digested by proteinase K at 56°C for 3 days in a rotating incubator. QIAamp® DNA FFPE Tissue Kit (QIAGEN, Hilden, Germany) used for genomic DNA extraction according to the manufacturer’s protocol. NanoDrop ND-1000® (Thermo Fisher Scientific Inc., Waltham, MA, USA) spectrophotometry used for concentration (ng/ul) and purity (OD 260/280nm) quantification of extracted DNA. Then, the extracted DNA was kept at -20°C before use.
Pyrosequencing
The Qiagen PyroMark KRAS v.2.0 Q96 kit by specific primers used for the amplification of KRAS gene codons 12 and 13. Master mix compounds of 12.5 μl PyroMark PCR Master Mix, 2.5 μl CoralLoad Concentrate, 1 μl PCR Primer KRAS 12/13, 4 μl Water (supplied) and 5 μl sample DNA (2–10 ng concentration) in a total volume of 25 μl. PCR reactions performed on Veriti 96 well Applied BioSystem thermal cycler by the heating program: initial denaturation at 95°C 15 min, 42 cycles of 95°C 20 s (denaturation), 53°C 30 s (annealing), 72°C 20 s (extension) and one step of final extension at 72°C for 5 minutes.
The Qiagen PyroMark BRAF kit used for the detection of BRAF mutation according to the manufacturers instructions. Primers for BRAF codon 600 were obtained from the PyroMarkBraf v.2.0 kit. The PyroMark Q96 software was used for sequence analysis.
PCR product confirmation of amplification by agarose gel (Sigma-Aldrich, St. Louis, USA) was recorded. Gel Band Purification Kit (GE Healthcare, Little Chalfont, UK) used for the purification of PCR products. Purified PCR products run on a PyroMark ID Pyrosequencing machine (QIAGEN, Germany) for sequencing reactions and PyroMark Q96 analysis software (QIAGEN, Germany) used for the analysis of electroperograms. All electropherograms were read manually.
Dxs Therascreen
Sequence variation of KRAS oncogene by six aminoacids (Gly > Ala, Asp, Arg, Cys, Ser and Val) on codon 12 and one (Gly > Asp) on codon 13 was investigated using TheraScreen DxS KRAS Mutation Kits KR-21 and KR-22 (QiaGen, Hilden, Germany) according to its recommended internal reaction control and a synthetic control template for the calculation of KRAS mutation degree differences. The LightCycler® Adapt Software v1.1 (Roche Diagnostics, Germany) was used for further analysis. Primers feature were included the 3´ end sequence-specific to mutation and Real-time PCR-Scorpion W primer tags.
TheraScreen K-RAS Mutation Kit version DU001PE used for PCR reactions by LightCyclerW480 II (Roche Applied Science, Penzberg, Germany) according to the instructions. Total reaction volume were 25 μl and thermal cycler protocol were included steps: initial denaturation at 95°C in 4 min, 45 cycle at 95°C in 30 sec., (denaturation) and 60°C in 1 min (annealing). The fluorescence acquisition at the 60°C step (annealing) performed. Assay components included 19.8 μl reaction mix, 0.2 μl Taq, and 5 μl of sample/control by a total volume of 25 μl in each reaction tubes. LightCycler Analysis Software 1.5.0 SP3 program (Roche Applied Science, Penzberg, Germany) used for reports of the Ct and ΔCt values.
High resolution melting (HRM)
HRM method performed for the screening of KRAS mutation. LightCycler® 480 II Real-Time System (Roche Diagnostics, Basel, Switzerland) provided for PCR amplification and HRM. LightCycler® 480 Gene Scanning Software Version 1.5 (Roche diagnostics, Germany) used for data analysis.
PCR reaction performed by the following protocol: forward primer (GCC TGC TGA AAA TGA CTG AA) 1 μl and reverse primer (TAT CGT CAA GGC ACT CTT GC) 1 μl, ddH2O 10 μl, genomic DNA (10 ng/μl) 3 μl, HRM master mix (QIAGEN) Mat. NO. 1057636 20 µl in a final volume of 35 μl. Termocycler program for KRAS gene amplification were as follows: Initial PCR activation step (Pre-amplification) at 95°C for 5 min by ramp rate 4.4°C/s, amplification stage included 55 cycles of denaturation at 95°C for 10 s by ramp rate 4.4°C/s, annealing 63°C 30 s by ramp rate 2.2°C/s, and melting analysis at 63°C for 5 min by the ramp rate of 4.4°C/s, 70°C for 1 s by the ramp rate of 4.4°C/s, 90°C continuous fluorescence data acquisition by the ramp rate of 0.02°C/s and 25 acquisitions per second, then cooling samples after HRM at 40°C 30 s by the ramp rate of 2.2°C/s.
For BRAF mutation detection initial PCR activation step (Pre-amplification) at 95°C for 10 min by the ramp rate of 4.4°C/s, amplification stage included 40 cycles of denaturation at 90°C 20 s by the ramp rate of 4.4°C/s, Annealing 67°C 20 s by the ramp rate of 2.2°C/s, and melting analysis at 72°C for 20 s by the ramp rate of 4.4°C/s, 70°C for 1 s by the ramp rate of 4.4°C/s, 90°C continuous fluorescence data acquisition by the ramp rate of 0.02°C/s and 25 acquisitions per second, then cooling samples after HRM at 40°C 30 s by the ramp rate of 2.2°C/s.
Results
Mutational status of samples
A total of 1000 CRC samples were used for screening of KRAS and BRAF by HRM, Pyrosequencing and Dxs Therascreen methods. At first, HRM were performed for all of the subjects, the subsequent results were analyzed, and we have found that there were 33.6% (336/1,000) KRAS mutants and there were not any BRAF mutants. Among 336 KRAS mutants, 286 (85.1%) were codon 12 mutant and 50 (14.9%) were codon 13 mutant (Table 2). As shown in Figure 1, the most common mutation at KRAS codon 12 was Gly12Asp, then Gly12Val, Gly12Ser, Gly12Ala, Gly12Cys and Gly12Arg,. The positive KRAS mutant specimens did not contain any BRAF mutation.
Table 2.
Mutational Status of 1,000 mCRC Patients
| Clinical and pathological characteristics | KRAS wild type, | KRAS | mutant, | N (%) | |
|---|---|---|---|---|---|
| Total | Codon 12 | Codon 13 | |||
| Sex | Male | 402 (60.5%) | 171 (50.9%) | 143 (83.6%) | 28 (16.4%) |
| Female | 262 (39.4%) | 165 (49.1%) | 143 (86.7%) | 22 (13.3%) | |
| Age (y) | ≤50 | 235 (35.4%) | 90 (26.8%) | 74 (82.3%) | 16 (17.7%) |
| ˃50 | 429 (64.6%) | 246 (73.2%) | 212 (86.2%) | 34 (13.8%) | |
| Tumor Differentiation | Well Differentiated | 265 (39.9%) | 173 (51.5%) | 77 (44.5%) | 96 (55.5%) |
| Moderate Differentiated | 279 (42.0%) | 108 (32.1%) | 86 (79.6%) | 22 (20.4%) | |
| Poor differentiated | 109 (16.4%) | 54 (16.1%) | 39 (72.2%) | 15 (27.8%) | |
| Undifferentiated | 11 (1.7%) | 1 (0.3%) | 1 (100.0%) | 0 (0.0%) | |
| Total mCRC | 1,000 patients | 664 (66.4%) | 336 (33.6%) | 286 (85.1%) | 50 (14.9%) |
Figure 1.
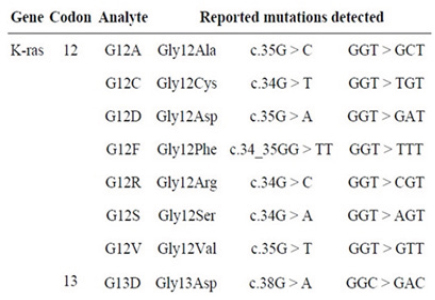
Details of Detected Mutations of KRAS and BRAF
The Pyrosequencing as the golden standard and another Dxs Therascreen method were performed for checking and were compared with HRM results. However, the Pyrosequencing analyses the mutational status the same as HRM (Figure 2) and the Dxs Therascreen results confirmed them, as well. There were no differences in detection of mutational status in KRAS gene in 33.6% of specimens in codon 12 or 13 and BRAF gene in none of them.
Figure 2.
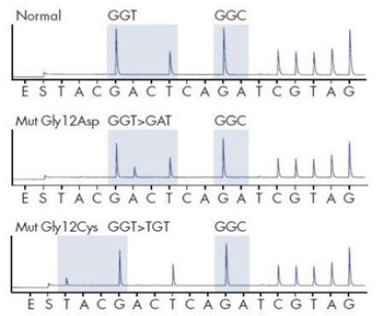
KRAS Analysis by Pyrosequencing. (Top Electropherogram) Wild Type (Normal) - (12Gly-GGT, 13Gly-GGC), (Middle and Bottom Electropherogram) Mutant-KRAS (12Asp-GAT, 12Cys-TGT)
Reliability and Sensitivity of HRM
1,000 samples were screened to evaluate the reliability and sensitivity of HRM assay. HRM results were compared with pyrosequencing and Dxs Therascreen method. Subsequently, the specificity and sensitivity of HRM were assessed as high in detection of 336 KRAS positive mutant samples and 664 wild types compared with two other methods. BRAF mutation was negative in all 242 KRAS mutant samples. Examples of HRM and melting profiles obtained from tissues carrying mutations of KRAS are shown in Figure 3. In all cases, the HRM is approved by the results of pyrosequencing and Dxs Therascreen. Consequently, the specificity and sensitivity of HRM analysis were 100% (Figure 4).
Figure 3.
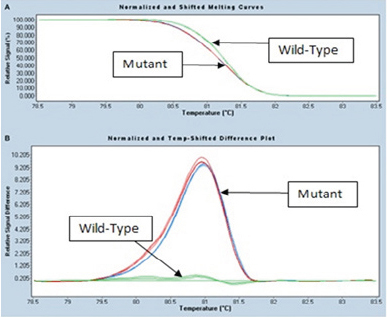
HRM Analysis Melting Profiles Obtained from Tissues Carrying Mutant KRAS
Figure 4.
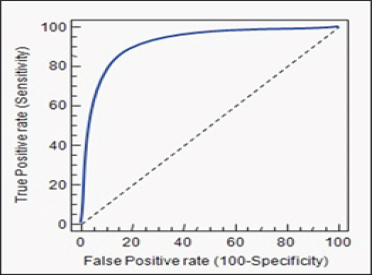
HRM Specificity and Sensitivity Diagram
Discussion
Reliable and practicable screening of mutations in known genes would be requested in future clinical practice(Katsanis and Katsanis, 2013). Hence, there is noticeably an essential need for rapid and low-cost detection of mutations with high accuracy. Though DNA sequencing is recognized as the golden standard, it is less applicable in clinical screening of genes. This limitation can be explained by high costs and low sensitivity of sequencing (Krypuy et al., 2006). The main purpose of current study was to develop a sensitive test, which makes the rapid detection of hot-spot mutations of KRAS, and BRAF oncogenes possible in mCRC. HRM has been established for the detection of mutation from various samples, such as frozen tumor samples and archived specimens in the form of formalin fixed paraffin-embedded and methanol-fixed tissues (Chen et al., 2014). This method provides a reliable and inexpensive test to carry out mutational screenings of tumor specimens in the clinical practice and diagnostic lab (Li et al., 2012; Montgomery et al., 2010). A meta-analysis indicated that the overall values of the sensitivity and specificity of HRM were 0.99 (95% CI 5 0.75–0.82) and 0.99(95% CI 5 0.94–0.98), respectively (Chen et al., 2014). In our study, HRM confirms the results of pyrosequencing and Dxs Therascreen in all formalin fixed paraffin-embedded samples and these finding shown 100% sensitivity and specificity of HRM. The results obtained here suggest that the HRM, in comparison with DNA sequencing, is a more applicable technique for detection of the BRAF and KRAS mutations. It is also worth mentioning that one of the few disadvantages of HRM is that it cannot easily distinguish how many mutation are present, or where within the amplified fragment the mutation is positioned. For this, sequencing of the PCR product is essential (Chateigner-Boutin and Small, 2007).
Within several investigations, various factors had been testified that might affect the HRM accuracy, although the accuracy of HRM is remarkable (Wittwer, 2009). Takano et al. showed that sensitivity increased, when fresh tumor samples were used for HRM, a possible explanation for this event is degeneration of DNA throughout sampling, or the maintenance of the archived samples (Takano et al., 2007). Wittwer et al., (2009) reported that sensitivity and specificity of HRM for fragments smaller than 400 bp is 100% and they also indicated that sensitivity increased when melt profile contains just one or two melt domains not more. Another study showed that, when the amplicon lengths were between 400 and 1 kb, sensitivity and specificity decreased to 96.1% and 99.4%, respectively (Montgomery et al., 2010). It has been demonstrated that GC content fragments might affect the results of HRM and shown that lower GC content is associated with false negative results (Mader et al., 2008; Taylor, 2009). The finding of this study supports most of the previous investigation results regarding effectible factors on HRM sensitivity and specificity. Our samples almost were high GC content and short length but not fresh and were formalin fixed paraffin-embedded tissues. However, there again the maintenance conditions of archived samples in our lab was appropriate and not seems that DNA might have been degenerated.
Collectively, current study verified the high sensitivity and specificity of HRM and our results seem to highlight the significance of HRM for precise scanning of KRAS mutations and BRAF screening. It is also possible to state that HRM may be a more attractive technique for the detection of known or unknown somatic mutations of other genes, in comparison with other methods. However, further research is required on this topic. In countries like Iran which costs of test affect the treatment, low cost of accurate HRM assay is the best choice.
Acknowledgments
The authors widely acknowledged the patients and their families for participation in this project. This work was supported by Iran University of Medical Sciences.
Conflict of interest
The authors have no conflicts of interests.
References
- Andreyev HJ, Norman AR, Cunningham D, et al. Kirsten ras mutations in patients with colorectal cancer: the ‘RASCAL II’ study. Br J Cancer. 2001;85:692–6. doi: 10.1054/bjoc.2001.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreyev HJ, Norman AR, Cunningham D, Oates JR, Clarke PA. Kirsten ras mutations in patients with colorectal cancer: the multicenter “RASCAL” study. J Natl Cancer Inst. 1998;90:675–84. doi: 10.1093/jnci/90.9.675. [DOI] [PubMed] [Google Scholar]
- Allegra CJ, Jessup JM, Somerfield MR, et al. American Society of Clinical Oncology provisional clinical opinion: testing for KRAS gene mutations in patients with metastatic colorectal carcinoma to predict response to anti-epidermal growth factor receptor monoclonal antibody therapy. J Clin Oncol. 2009;27:2091–6. doi: 10.1200/JCO.2009.21.9170. [DOI] [PubMed] [Google Scholar]
- Borras E, Jurado I, Hernan I, et al. Clinical pharmacogenomic testing of KRAS, BRAF and EGFR mutations by high resolution melting analysis and ultra-deep pyrosequencing. BMC Cancer. 2011;11:406. doi: 10.1186/1471-2407-11-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd EM, Bench AJ, van ‘t Veer MB, et al. High resolution melting analysis for detection of BRAF exon 15 mutations in hairy cell leukaemia and other lymphoid malignancies. Br J Haematol. 2011;155:609–12. doi: 10.1111/j.1365-2141.2011.08868.x. [DOI] [PubMed] [Google Scholar]
- Chateigner-Boutin AL, Small I. A rapid high-throughput method for the detection and quantification of RNA editing based on high-resolution melting of amplicons. Nucleic Acids Res. 2007;35:e114. doi: 10.1093/nar/gkm640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Wang YY, Chuai ZR, et al. High-resolution melting analysis for accurate detection of BRAF mutations: a systematic review and meta-analysis. Sci Rep. 2014;4:4168. doi: 10.1038/srep04168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Roock W, De Vriendt V, Normanno N, Ciardiello F, Tejpar S. KRAS, BRAF, PIK3CA, and PTEN mutations: implications for targeted therapies in metastatic colorectal cancer. Lancet Oncol. 2011;12:594–603. doi: 10.1016/S1470-2045(10)70209-6. [DOI] [PubMed] [Google Scholar]
- Di Nicolantonio F, Martini M, Molinari F, et al. Wild-type BRAF is required for response to panitumumab or cetuximab in metastatic colorectal cancer. J Clin Oncol. 2008;26:5705–12. doi: 10.1200/JCO.2008.18.0786. [DOI] [PubMed] [Google Scholar]
- Fransen K, Klintenas M, Osterstrom A, et al. Mutation analysis of the BRAF, ARAF and RAF-1 genes in human colorectal adenocarcinomas. Carcinogenesis. 2004;25:527–33. doi: 10.1093/carcin/bgh049. [DOI] [PubMed] [Google Scholar]
- Guedes JG, Veiga I, Rocha P, et al. High resolution melting analysis of KRAS, BRAF and PIK3CA in KRAS exon 2 wild-type metastatic colorectal cancer. BMC Cancer. 2013;13:169. doi: 10.1186/1471-2407-13-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janakiraman M, Vakiani E, Zeng Z, et al. Genomic and biological characterization of exon 4 KRAS mutations in human cancer. Cancer Res. 2010;70:5901–11. doi: 10.1158/0008-5472.CAN-10-0192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji H, Wang Z, Perera SA, et al. Mutations in BRAF and KRAS converge on activation of the mitogen-activated protein kinase pathway in lung cancer mouse models. Cancer Res. 2007;67:4933–9. doi: 10.1158/0008-5472.CAN-06-4592. [DOI] [PubMed] [Google Scholar]
- Katsanis SH, Katsanis N. Molecular genetic testing and the future of clinical genomics. Nat Rev Genet. 2013;14:415–26. doi: 10.1038/nrg3493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koochak A, Rakhshani N, Karbalaie Niya MH, et al. Mutation Analysis of KRAS and BRAF Genes in Metastatic Colorectal Cancer: a First Large Scale Study from Iran. Asian Pac J Cancer Prev. 2016;17:603–8. doi: 10.7314/apjcp.2016.17.2.603. [DOI] [PubMed] [Google Scholar]
- Krypuy M, Newnham GM, Thomas DM, Conron M, Dobrovic A. High resolution melting analysis for the rapid and sensitive detection of mutations in clinical samples: KRAS codon 12 and 13 mutations in non-small cell lung cancer. BMC Cancer. 2006;6:295. doi: 10.1186/1471-2407-6-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li BS, Wang XY, Xu AG, et al. High-resolution melting assay (HRMA) is a simple and sensitive stool-based DNA Test for the detection of mutations in colorectal neoplasms. Clin Colorectal Cancer. 2012;11:280–90. doi: 10.1016/j.clcc.2012.04.001. [DOI] [PubMed] [Google Scholar]
- Mader E, Lukas B, Novak J. A strategy to setup codominant microsatellite analysis for high-resolution-melting-curve-analysis (HRM) BMC Genet. 2008;9:69. doi: 10.1186/1471-2156-9-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minoo P, Moyer MP, Jass JR. Role of BRAF-V600E in the serrated pathway of colorectal tumourigenesis. J Pathol. 2007;212:124–33. doi: 10.1002/path.2160. [DOI] [PubMed] [Google Scholar]
- Montgomery JL, Sanford LN, Wittwer CT. High-resolution DNA melting analysis in clinical research and diagnostics. Expert Rev Mol Diagn. 2010;10:219–40. doi: 10.1586/erm.09.84. [DOI] [PubMed] [Google Scholar]
- Morandi L, de Biase D, Visani M, et al. Allele specific locked nucleic acid quantitative PCR (ASLNAqPCR): an accurate and cost-effective assay to diagnose and quantify KRAS and BRAF mutation. PloS One. 2012;7:e36084. doi: 10.1371/journal.pone.0036084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negru S, Papadopoulou E, Apessos A, et al. KRAS, NRAS and BRAF mutations in Greek and Romanian patients with colorectal cancer: a cohort study. BMJ. 2014;4:e004652. doi: 10.1136/bmjopen-2013-004652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packham D, Ward RL, Ap Lin V, Hawkins NJ, Hitchins MP. Implementation of novel pyrosequencing assays to screen for common mutations of BRAF and KRAS in a cohort of sporadic colorectal cancers. Diagn Mol Pathol. 2009;18:62–71. doi: 10.1097/PDM.0b013e318182af52. [DOI] [PubMed] [Google Scholar]
- Pinto P, Rocha P, Veiga I, et al. Comparison of methodologies for KRAS mutation detection in metastatic colorectal cancer. Cancer Genet. 2011;204:439–46. doi: 10.1016/j.cancergen.2011.07.003. [DOI] [PubMed] [Google Scholar]
- Popovici V, Budinska E, Tejpar S, et al. Identification of a poor-prognosis BRAF-mutant-like population of patients with colon cancer. J Clin Oncol. 2012;30:1288–95. doi: 10.1200/JCO.2011.39.5814. [DOI] [PubMed] [Google Scholar]
- Reed GH, Wittwer CT. Sensitivity and specificity of single-nucleotide polymorphism scanning by high-resolution melting analysis. Clin Chem. 2004;50:1748–54. doi: 10.1373/clinchem.2003.029751. [DOI] [PubMed] [Google Scholar]
- Roth AD, Tejpar S, Delorenzi M, et al. Prognostic role of KRAS and BRAF in stage II and III resected colon cancer: results of the translational study on the PETACC-3, EORTC 40993, SAKK 60-00 trial. J Clin Oncol. 2010;28:466–74. doi: 10.1200/JCO.2009.23.3452. [DOI] [PubMed] [Google Scholar]
- Simi L, Pratesi N, Vignoli M, et al. High-resolution melting analysis for rapid detection of KRAS, BRAF, and PIK3CA gene mutations in colorectal cancer. Am J Clin Pathol. 2008;130:247–53. doi: 10.1309/LWDY1AXHXUULNVHQ. [DOI] [PubMed] [Google Scholar]
- Sundstrom M, Edlund K, Lindell M, et al. KRAS analysis in colorectal carcinoma: analytical aspects of Pyrosequencing and allele-specific PCR in clinical practice. BMC Cancer. 2004;10:660. doi: 10.1186/1471-2407-10-660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano T, Ohe Y, Tsuta K, et al. Epidermal growth factor receptor mutation detection using high-resolution melting analysis predicts outcomes in patients with advanced non small cell lung cancer treated with gefitinib. Clin Cancer Res. 2007;13:5385–90. doi: 10.1158/1078-0432.CCR-07-0627. [DOI] [PubMed] [Google Scholar]
- Tan YH, Liu Y, Eu KW, et al. Detection of BRAF V600E mutation by pyrosequencing. Pathology. 2008;40:295–8. doi: 10.1080/00313020801911512. [DOI] [PubMed] [Google Scholar]
- Taylor CF. Mutation scanning using high-resolution melting. Biochem Soc Trans. 2009;37:433–7. doi: 10.1042/BST0370433. [DOI] [PubMed] [Google Scholar]
- Wittwer CT. High-resolution DNA melting analysis: advancements and limitations. Hum Mutat. 2009;30:857–9. doi: 10.1002/humu.20951. [DOI] [PubMed] [Google Scholar]
- Wooster R, Futreal AP, Stratton MR. Sequencing analysis of BRAF mutations in human cancers. Methods Enzymol. 2006;407:218–24. doi: 10.1016/S0076-6879(05)07018-7. [DOI] [PubMed] [Google Scholar]
- Vakil L, Najafipour R, Rakhshani N, et al. Investigation of FIH-1 and SOCS3 expression in KRAS mutant and wild-type patients with colorectal cancer. Tumour Biol. 2016;37:8841–8. doi: 10.1007/s13277-015-4723-1. [DOI] [PubMed] [Google Scholar]


