Abstract
Recognition of ‘foreign’ DNA by Type I restriction–modification (R-M) enzymes elicits an ATP-dependent switch from methylase to endonuclease activity, which involves DNA translocation by the restriction subunit HsdR. Type I R-M enzymes are composed of three (Hsd) subunits with a stoichiometry of HsdR2:HsdM2:HsdS1 (R2-complex). However, the EcoR124I R-M enzyme can also exist as a cleavage deficient, sub-assembly of HsdR1:HsdM2:HsdS1 (R1-complex). ATPγS was used to trap initial translocation complexes, which were visualized by Atomic Force Microscopy (AFM). In the R1-complex, a small bulge, associated with a shortening in the contour-length of the DNA of 8 nm, was observed. This bulge was found to be sensitive to single-strand DNA nucleases, indicative of non-duplexed DNA. R2-complexes appeared larger in the AFM images and the DNA contour length showed a shortening of ∼11 nm, suggesting that two bulges were formed. Disclosure of the structure of the first stage after the recognition-translocation switch of Type I restriction enzymes forms an important first step in resolving a detailed mechanistic picture of DNA translocation by SF-II DNA translocation motors.
INTRODUCTION
Type I restriction-modification (R-M) enzymes are the most complex of the many R-M systems known [for a recent review see (1)]. They are multisubunit, multifunctional enzymes composed of three separate subunits (HsdR—the restriction/motor subunit, HsdM—the methylation subunit, and HsdS—the DNA binding subunit). The active endonuclease (REase) is composed of all three subunits in a ratio 2:2:1 (HsdR2:HsdM2:HsdS1, or R2-complex). The R2-complex also functions as a DNA methyltransferase (MTase), an ATPase, and as a ‘DNA pulling’ molecular motor (2,3).
HsdM and HsdS alone are sufficient to assemble an independent MTase with a stoichiometry of M2S1 (4). The R-M enzyme EcoR124I can be assembled in vitro from the core MTase by addition of the motor subunit HsdR (5). However, the purified EcoR124I REase exists as an equilibrium mixture of two species—R2M2S1 and R1M2S1 of which only the former is able to cleave DNA (5,6); although the R1-complex is an ATPase and is able to translocate DNA (5,7,8). The R2-complex is relatively unstable and can dissociate into free HsdR subunit and the restriction-deficient R1-complex intermediate, under concentrations expected in vivo (5). This situation assists structural analysis of the EcoR124I R–M enzyme because complexes with only one HsdR subunit present (i.e. R1-complex) can more easily be visualized by AFM.
Unlike other restriction endonucleases, Type I R-M enzymes cut distal to the DNA recognition site to which they bind. DNA cleavage can occur many thousands of base pairs from the recognition site, using a process of DNA movement known as DNA translocation (2). This REase-based motor activity is driven by ATP hydrolysis (3), but unlike other DNA-based motors (e.g. DNA polymerase) it does not involve a linear tracking motion along the DNA; instead the motor remains bound at the recognition site and ‘pulls’ the adjacent DNA toward the bound enzyme (Figure 1). A detailed study of DNA translocation by Type I restriction enzymes has recently been published, based on single molecule analysis using Magnetic Tweezers (8). This intriguing motor activity presents the enzyme with an interesting topological problem at the initiation of translocation—how to ‘grasp’ adjacent DNA and produce the first loop of DNA (9).
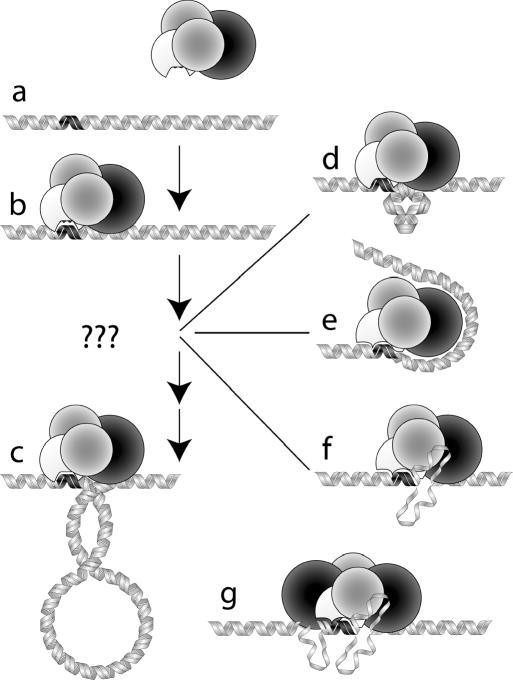
Figure 1.
The DNA translocation process for the R1-complex. (a) The black region on the DNA represents the DNA binding (recognition) site of the enzyme, which is represented by the four globular subunits of the R1-complex: HsdS (white), HsdM (grey) and HsdR (black). (b) HsdS is bound to the DNA at the recognition site and HsdR begins to contact adjacent DNA sequences. (c) The motor translocates adjacent DNA through the motor/DNA complex, which remains tightly bound to the recognition sequence. Translocation produces an expanding loop of negatively supercoiled DNA. In the early stages of translocation, a very small loop would require a significant energetic penalty and is thus unlikely. (d) The inherent stiffness of DNA prevents bending over short distances and a bulge of the type shown would require a large amount of energy from protein–DNA interactions. (e) Wrapping of DNA around the HsdR subunit in an attempt to overcome the problem associated with bending the relatively stiff DNA. (f) Alternatively, unwinding of DNA can be used to overcome the problems associated with the persistence length of DNA, resulting in an extrusion or a bubble of ssDNA. (g) R2 initial complexes exhibit a much larger, more intricate structure that is more difficult to analyse with AFM imaging.
The complex molecular motor-function makes these enzymes particularly interesting. The restriction subunit (HsdR) is responsible for this motor activity and contains a series of conserved amino acid motifs (DEAD box motifs) including a Walker-type ATP binding site, which is associated with helicase-like activities (10). They belong to a large superfamily (SF-II) of helicase-like enzymes (11) that also include Type III R-M enzymes, chromatin remodelling factors and a few chimeric enzymes. It has been suggested that chromatin-remodelling factors also make use of DNA translocation, in a similar mode as Type I restriction enzymes, which stresses the significance of a detailed analysis of the translocation process of the SF-II superfamily (11,12).
ATP is an allosteric effector for the REase and is closely involved in the initial switch between methylation activity and translocation activity which leads to DNA cleavage (13). In vivo, the normal function of these enzymes is that of a maintenance methylase—following DNA replication, the enzyme will methylate specific adenines on the newly synthesized strand of the recognition sequence. However, invasion of the bacterial cell by ‘foreign’ DNA (usually bacteriophage DNA) elicits an ATP-dependent switch in behaviour of the Type I R-M enzyme, and the enzyme becomes an endonuclease. To accomplish this switch in activity, the enzyme must read and compare the two adenines of the recognition sequence (one on each strand), and, if they are both unmethylated, the enzyme will undergo a conformational change resulting in motor and endonuclease activity. The switch in function was shown to be an ATP-dependent event by Yuan et al. (13) who ‘trapped’ the initial complex in filter binding studies. The non-hydrolysable ATP-derivative ATPγS can also be used to trap this initial complex by preventing translocation and allows studies of the initiation of translocation.
It has been shown that the REase only ‘grasps’ the DNA in cis (14) suggesting a loop would form adjacent to the DNA-bound REase. However, DNA footprinting studies (15,16) have shown little difference between the footprints of the MTase and that of the REase suggesting the enzyme ‘grasps’ the DNA remote to the sequence recognition site. To initiate translocation, the enzyme would need to produce a loop that is smaller than the enzyme, and far shorter than the persistence length of DNA, making this mechanism very unlikely (Figure 1d). Another possibility is that the DNA initially wraps around the motor subunit, forming multiple DNA-protein interactions along the loop (Figure 1e). This would not have been detected in the DNA footprinting studies mentioned above because of the limited length of the oligoduplexes used (56 bp). Alternatively, the endonuclease may induce a distortion of the usual double-stranded DNA (dsDNA) structure (such as a region of non-duplexed DNA) to allow the enzyme to ‘grasp’ adjacent DNA over short distances (Figure 1f).
In this paper, we have used Atomic Force Microscopy (AFM) on single DNA-restriction enzyme complexes to observe the conformational changes associated with the initial switch from recognition to translocation. We have used ATPγS to trap these translocation initiation complexes. Using the R1-complex produced by mixing equimolar ratios of MTase and HsdR, we could observe a small bulge extending from the protein–DNA complex that was only formed when ATPγS was added. This bulge was accompanied by a shortening of the contour length of the DNA molecule. As expected, the bulge was more difficult to delineate in the R2-complex, but additional shortening of the DNA contour length was observed supporting the idea of a two-bulge complex. Thus, we obtained a detailed picture of the important first-step of initial loop formation that may be generic for all superfamily II enzymes that act as DNA-based molecular motors.
MATERIALS AND METHODS
In vitro restriction assays
Cleavage assays of the plasmid pCFD30 and titration of MTase with HsdR were carried out as previously described (6). The concentration of REase was based on the concentration of added MTase, which was determined from the extinction coefficient. Gel retardation assays were as described by Janscák et al. (6).
AFM methods
For translocation experiments, a 2364 bp and a 724 bp DNA fragment with a single s124 recognition site at 175 bp was produced by PCR. HsdR and MTase were mixed at equal molar ratio in 300 mM NaCl, 10 mM Tris–HCl (pH 8), 1 mM EDTA and 50% glycerol. After 5 min the stable R1-complex was subsequently diluted 10 times in 20 mM HEPES (pH 6.5), 5 mM MgCl2 together with 724 bp DNA at a molar ratio of 1:2, and warmed to 37°C.
Muscovite mica was cleaved and pre-treated with poly-lysine before sample deposition. Translocation by the R1-complex was initiated by adding ATP to the reaction mixture, to a final concentration of 0.5 mM. After 10, 30 and 60 s respectively, translocation was quenched by depositing 2.5 μl of the reaction mixture onto the poly-lysine coated mica. The sample was then rinsed with MilliQ water, quickly blown dry in a stream of nitrogen gas and directly imaged with AFM.
For imaging of the initial complex, a 263 bp DNA fragment containing a single recognition site at 126 bp was produced by PCR. The smaller length simplifies measurement of the contour length. ATP was replaced by ATPγS and the mixture was incubated for 30 s.
Samples were imaged with a Nanoscope III Multimode AFM from Digital Instruments (Santa Barbara, USA). The images were acquired in tapping mode in air, with a scan range of 1 μm at 512 × 512 pixels using silicon tips OMCL-AC160TS-W2 purchased from Olympus Optical Co., Ltd (Tokyo, Japan). Image processing and analysis were carried out using IDL (Interactive Data Language, RSI, Boulder, USA) and consisted of a background correction. Contour lengths were measured by manually assigning coarse coordinates that were subsequently improved using an algorithm that looks for the highest features perpendicular to the coarse trace, followed by moderate low-pass filtering of the coordinates. End points were assigned to positions where the DNA trace was reduced to half its height. Average contour lengths were determined by fitting a Gaussian profile to the length distribution histograms.
P1 nuclease digestions
P1 nuclease buffer consisted of 50 mM Tris–HCl (pH 8.0), 10 mM MgCl2, 1 mM DTT and 50 mM NaCl: 10 nM pCDF30 and ATP or ATPγS (final concentration 2 mM) were added. R2 or R1-complex was assembled in the presence of cleavage buffer and ATPγS or ATP, and incubated for 10 min at room temperature. Then, 0.3 U P1 was added and the reaction was incubated for a further 30 min at 37°C. Reactions were terminated by adding 0.5 vol of 150 mM EDTA, 3% (w/v) SDS and 60 μg/ml proteinase K followed by incubation at 37°C for 30 min.
RESULTS
Loop formation during DNA translocation
R1-complex of the EcoR124I R–M enzyme was produced by mixing equimolar ratios of MTase and HsdR (5). Analysis of these complexes, bound to DNA, using the AFM showed a uniform collection of similar-sized complexes, with very few R2-complexes (larger) observed (data not shown). That these complexes were indeed functional R1-complexes was confirmed by demonstrating unidirectional translocation (single loop formation) as shown in Figure 2.
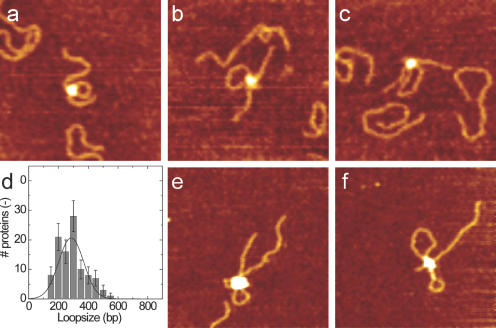
Figure 2.
AFM images of DNA translocation. Typical images of translocation activity by R1-complex after incubation for (a) 10 s, (b) 30 s and (c) 60 s in the presence of ATP and 724 bp DNA fragments, containing a single recognition site at 175 bp. At the position of the protein, which shows up as a white globular feature, one small DNA loop is observed that increases in size with longer incubation times. Scan range, 250 nm; z range, 3 nm. (d) Distribution of the loop size after incubation for 10 s. Translocation distances were fitted with a Gaussian distribution, resulting in an average translocation distance of 2.8 × 102 bp. (e and f) R2 complexes on 2364 bp DNA after incubation with ATP for several minutes. As expected, double loops of DNA are formed. Scan range, 500 nm; z range, 5 nm.
The amount of translocated DNA was determined by tracing the loop size for more than 100 complexes. The resulting distribution for 10 s incubation time is shown in Figure 2d. Though the average loop size increased with longer incubation times, as expected, the apparent translocation velocity is an order of magnitude smaller than previously reported values (7,8). However, though AFM experiments are valuable for qualitative insights into structural changes of DNA–enzyme complexes, quantitative information on the timing can be trusted less due to artefacts related to surface immobilization of the complexes (17,18).
The fixed complexes, imaged by AFM, reveal exciting structural features. Figure 2e and f, shows two examples of R2-complexes after incubation with DNA and ATP. As expected for the fully functional EcoR124I enzyme, two DNA loops are formed. Interestingly, contrary to the R1–DNA complexes, some of the loops formed by R2-complexes show evidence of supercoiling. The loop sizes within single R2-complexes appear to be distinctly different. Assuming the same translocation velocity for both HsdR subunits, this implies that the switch from recognition to translocation occurs independently in both subunits and forms a rate-limiting step for initial DNA translocation [cf. the data presented by Seidel et al. (8)].
Analysis of initial complex formation
DNA translocation by Type I R–M enzymes is halted in the initial stage when ATP is replaced by the non-hydrolysable analogue ATPγS. If either DNA wrapping, or a DNA loop is formed during the initiation stage of translocation, i.e. upon binding of ATPγS, then it should be possible to measure this through a reduction in DNA contour length on a fragment of DNA carrying a single recognition site for the enzyme. A 263 bp fragment of DNA was used to bind the R1-complex in the presence and absence of ATPγS (Figure 3).
Figure 3.
AFM images of initial complex formation of MTase + HsdR(R124I). Two typical AFM images of protein–DNA complexes consisting of R1- (a and b) and R2-complexes (c and d), with and without ATPγS. On the right, the corresponding measured contour length distributions are plotted of free DNA (black) and complexed-DNA (grey). Red lines indicate the fitted mean contour length. Arrows indicate a small extrusion that is only observed when R1-complex is incubated with ATPγS. Analysis shows a decrease in contour length of 8 nm for R1-complexes and 11 nm for R2-complexes incubated in the presence of ATPγS. Scan range, 125 nm; z range, 3 nm.
The contour length of typically 100 DNA molecules with bound protein complexes was obtained in a semi-automatic fashion and compared to the contour length of free DNA molecules in the same sample. In the absence of ATPγS, the contour length of the DNA, in the DNA-protein complexes, was found to be equal to that of free DNA in all experiments. In the presence of ATPγS and HsdR, however, the DNA contour length in R1-complexes was reduced by 8 nm.
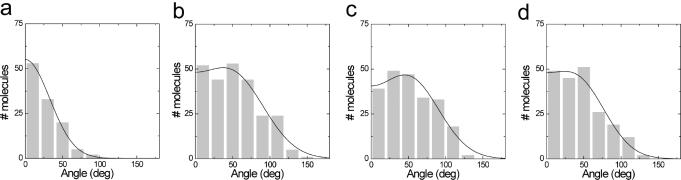
The reduction of contour length is significantly less than would be expected for DNA wrapping around this relatively large protein complex. Furthermore, partial wrapping, as shown in Figure 1e, would induce a significant bend in the trajectory of the bound DNA. In Figure 4, the measured bending angle is plotted for the protein complexes shown in Figure 3. Upon binding of MTase, the DNA is bent by 49°. Binding of R1-complex induces a very similar bend of 51°. For the related EcoKI enzyme, a comparable bending angle of 46° has been reported in the absence of ATP analogues (19). The presence of ATPγS appears to slightly reduce the bend to 40°. Thus, in the absence of ATPγS, R1-complex does not affect the DNA trajectory compared to MTase–DNA complexes, suggesting little HsdR–DNA contact. The relatively small difference in bending angles of 10° that was found when ATPγS was added is not sufficient to account for the measured 8 nm reduction in contour length, ruling out DNA wrapping at the initial stage of translocation.
Figure 4.
DNA bending by MTase and R1 complexes. Histograms of the distributions of bend angles. Distributions are truncated at 0°, as the direction of bending cannot be determined. Black lines represent fits to truncated Gaussian distributions defined by the average angle α, width σ and an arbitrary normalization factor: (a) DNA α = 0°, σ = 65° ± 1° (mean ± standard error), (b) Mtase + DNA α = 49° ± 3, σ = 86° ± 9°, (c) R1-complex + DNA α = 51° ± 3°, σ = 80° ± 7° and (d) R1-complex + DNA in the presence of ATPγS α = 40° ± 3°, σ = 75° ± 9°.
Careful analysis of the images of R1-complexes in the presence of ATPγS, showed a protrusion at one side of the enzyme (see Figure 3b), which was absent when ATPγS was not added. The height of the newly formed protrusion was about the same as the height of DNA adjacent to the complex, but at this stage, we cannot resolve more detailed structural information. The observed reduction in DNA length, however, is far less than the persistence length of DNA and, therefore, this DNA bulge is likely to exist in a non-B-form structure, e.g. single-stranded DNA (ssDNA).
At a molar ratio of 1:2 MTase/HsdR where it is possible to form both R1- and R2-complexes, the DNA contour length was found to be slightly more reduced, by 11 nm (Figure 3d), and to have a slightly broader distribution. This is to be expected if the observed complexes were to fall into two classes, a group with a reduction in contour length originating from one bulge and another group representing complexes with two bulges. In addition to the change in contour length, the ‘volume’ of the enzymes visualized on the shorter contour length fragments appeared larger than that of the longer contour length fragments (Figure 3) consistent with the presence of R2-complexes. The presence of R2-complexes in this mixture was also confirmed by in vitro DNA cleavage assays (Figure 5, lane 7).
Figure 5.
P1 nuclease digestion of initial complexes. pCFD30 plasmid DNA was used to analyse the effect of P1 nuclease on bulge formation. Lane 1, 1 kb marker; Lanes 2–11, pCFD30; P1, R2-complex, R1-complex, ATPγS and ATP were added as described in the table. P1 digestion in the presence of ATPγS results in increased nicking of covalently closed circular DNA (cccDNA) to produce open circular DNA (ocDNA). R2-complex in the presence of ATP induces DNA cleavage as expected.
On the R2-complexes, no observable protrusions could be resolved. Note however that the larger size of the R2-complexes as depicted in Figure 1g, augments the effect of tip-sample convolution, an effect that is always present in AFM imaging. Because of the significant tip dimensions relative to the size of the molecules, small features in the vicinity of large structures are obscured, although they may well be present. The MTase and R1-complexes are significantly smaller and suffer less from this effect, yielding more detailed data in the vicinity of the enzyme–DNA complex. To account for differences in tip size that may, through convolution, also increase the apparent contour length, we only compared contour lengths from molecules that were imaged within the same sample and with the same tip. Though the width of the distribution of contour lengths is of the same order of magnitude as the observed ATPγS-induced changes in length, the entire distribution of contour lengths is shifted. Thus, the ‘average’ reduction of the contour length can be determined with much higher accuracy than the individual contour lengths, revealing significant changes between incubation with or without ATPγS. Whereas, the apparent contour length of free DNA varied significantly between different preparations and tips, as reflected in the measured contour lengths shown in Figure 3, the measured enzyme-induced ‘change’ in contour length was found to be unaffected and reproducible between different preparations and different tips.
In summary, we observed that the ATPγS-induced switch to the translocation mode of the EcoR124I REase is correlated with the formation of an initial bulge that corresponds to a shortening in DNA length of ∼8 nm. This was indirectly hinted at in R2-complexes but directly visible in R1-complexes.
Effect of single-strand-specific nucleases on the DNA bulge
The shortening in contour length of only 8 nm suggests that a distinct change in DNA structure is produced during the switch to translocation. The most straightforward explanation of this structure is that it contains ssDNA, as this would allow a bulge of such a short length, given the long (53 nm) persistence length of dsDNA. To investigate this possibility, we examined the effect of single-strand-specific nucleases on the DNA present in the initial complex, in the presence and absence of ATPγS.
Figure 5 shows that addition of ATPγS and P1 nuclease to R1- or R2-complex results in nicking of covalently closed circular DNA (cccDNA) to produce open circular DNA (ocDNA). The yield of ocDNA was never 100%, and did not increase with increasing P1 (lane 10), or EcoR124I (data not shown) concentration, suggesting significant steric hindrance to the attack at the bulge by P1 nuclease. This is supported by the observation that the larger enzyme Mung Bean Nuclease did not cleave the bulge at all (data not shown). Experiments using S1 nuclease supported the observations with P1 nuclease and showed increasing HsdR/ATPγS-dependent nicking activity with increasing S1 nuclease concentration (data not shown). However, experiments using S1 nuclease proved to be more difficult to reproduce and interpret due to significant non-specific activity and incompatibility of the buffer conditions for S1 nuclease and EcoR124I.
The absence of full cleavage by the single-strand nucleases in the presence of R1-complex and ATPγS is interesting since the existence of melted DNA in the bulge would be expected to lead to cleavage of both strands of the DNA—in other words linear DNA should be produced. The observed nicking by both S1 and P1 nucleases suggests that the REase protects one strand of the DNA present in the complex from the nucleases and that the observed protrusion is ssDNA from the complementary strand, which is accessible to the nucleases as drawn schematically in Figure 1f. The absence of significant amounts of linear DNA points at a high stability of the stalled complex during the incubation with both S1 and P1 nuclease.
When ATP replaced ATPγS, as shown in Figure 5 (lane 7), cleavage of the plasmid into linear DNA by R2-complex was observed, confirming full endonuclease activity of the native enzyme in the present conditions. Interestingly however, stalled R1- and R2-complexes result in similar amounts of ocDNA as a result of P1 nuclease activity. The assay cannot distinguish between single or multiple nicks that both result in ocDNA, unless the nicks are on opposite strands and close enough to result in linear DNA. Alternatively, steric hindrance by a single P1 nuclease may prevent nicking of a second bulge. Though we cannot confirm the presence of two bulges, the very similar nicking results point at similar melted DNA structures present in both stalled R1 and R2 complexes.
DISCUSSION
The EcoR124I R-M enzyme is unusual in that it readily dissociates from the REase complex (which has a stoichiometry of HsdR2:HsdM2:HsdS1 or R2-complex) into a R1-complex and free HsdR. For AFM studies, this provides an unique opportunity to visualize structures produced by the R1-complex without the obscuring effect of a large complex consisting of MTase and two HsdR subunits. Indeed, by trapping initial complexes of the R1-complex (produced by the addition of ATPγS), we observed a new structural feature in these stalled complexes, namely a bulge in the DNA.
By quantifying the contour length of stalled initial complexes, we found an overall reduction in contour length of ∼8 nm for R1-complexes and 11 nm for samples that contain a mixture of R1- and R2-complexes. This reduction in contour length was associated with a visible ‘bulge’ in the DNA adjacent to the enzyme in the R1-complex. Such a bulge can be accounted for by a region of ssDNA, which could easily bend to take up the observed structure, with one strand of the bulge present in the enzyme complex and protected from cleavage by ssDNA-specific nucleases by the presence of the REase, but with the other strand extruding from the enzyme (Figure 1f).
A reduction in contour length of ∼8 nm corresponds to ∼24 bp of DNA being separated, which requires a significant amount of energy, of the order of 25 kT. Favourable DNA-protein interactions that require close contact between one strand of DNA and the HsdR subunit are likely to complement the energy gained by ATP binding for this structural transition.
The current results can now explain why no ‘extended’ footprint was observed by Mernagh et al. (16) when comparing the DNA footprint of MTase and REase—the bulge formed by the initial complex is ATP-dependent and no ATP analogues were used in the footprinting studies.
Whereas enzymes belonging to helicase super family type I (SFI) all appear to be unwinding DNA during translocation, the situation for the SF II enzymes, to which EcoR124I belongs, is far less clear (20). The mechanism through which initiation occurs is still unknown (9). The question arises whether the bulge of ssDNA that we observed is formed as a consequence of DNA translocation in a loop, i.e. by buckling of highly bent DNA. In this case, the observed bulge formation would only transiently be present in the complex and it would only be relevant for those family members that produce a loop. In our experiments, where ATP hydrolysis is blocked, only a single step can be made. The regular step size of EcoR1241 is currently not known, but estimates range from one to a few bp (21). In recent Magnetic-Tweezers-based experiments in our laboratory (R. Seidel, personal communication), an upper limit to the step size of about 3 bp was found. Thus, the large step that we observed in initial complexes is most likely not representative of a single step that is regularly repeated during subsequent translocation, but rather a distinct conformational change specific for initiation of translocation. The absence of full DNA cleavage in the P1 nuclease assay points at a strong interaction of the enzyme with one of the strands of the bulge, protecting it from cleavage. In the case of simple buckling of the DNA, both strands would be subject to digestion, which was not observed. It is therefore likely that the bulge represents a conformation of the initial complex where one strand of DNA is exposed to digestion by single-strand-specific nucleases, whereas the other strand is protected.
The strand separation that we report in initial complexes may be a prerequisite for DNA translocation, in a similar fashion as polymerases and helicases, that track ssDNA during translocation. In this case, the complementary bulge of ssDNA would propagate along the complex during translocation. Subsequently, DNA can reanneal again behind the translocation complex. Though the intermediate steps in the mechanism of helicase activity are currently heavily debated (22), all current mechanisms require strand separation. The observed formation of a small DNA protrusion in the initial translocation complex of HsdR is in accordance with such mechanisms. A mechanism similar to DNA helicases would also form a direct link to the observed induction of supercoiling (23) both ahead and behind the moving subunit. However, it should be noted that strand separation is not required for the creation of distinct topological domains. Future studies should reveal whether initial bulge formation is generic for the first step in translocation of all superfamily II enzymes that act as DNA-based molecular motors. Disclosure of the structure of the first stage after the recognition-translocation switch of Type I restriction enzymes, as reported in this study, forms an important first step in resolving a detailed mechanistic picture of DNA translocation by SF-II DNA translocation motors that are involved in diverse processes like DNA strand separation, DNA cleavage and chromatin remodelling.
Acknowledgments
ACKNOWLEDGEMENTS
We would like to acknowledge the financial support of the European Commission through the Mol Switch project (IST-2001-38036), an EMBO Short-Term Fellowship to K.F. and a NWO Pioneer Grant to C.D.
REFERENCES
- 1.Murray N.E. (2000) Type I restriction systems: sophisticated molecular machines (a legacy of Bertani and Weigle). Microbiol. Mol. Biol. Rev., 64, 412–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yuan R., Hamilton,D.L. and Burckhardt,J. (1980) DNA translocation by the restriction enzyme from E.coli K. Cell, 20, 237–244. [DOI] [PubMed] [Google Scholar]
- 3.Endlich B. and Linn,S. (1985) The DNA restriction endonuclease of Escherichia coli B. I. Studies of the DNA translocation and the ATPase activities. J. Biol. Chem., 260, 5720–5728. [PubMed] [Google Scholar]
- 4.Taylor I., Patel,J., Firman,K. and Kneale,G.G. (1992) Purification and biochemical characterisation of the EcoR124 modification methylase. Nucleic Acids Res., 20, 179–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Janscák P., Dryden,D. and Firman,K. (1998) Analysis of the subunit assembly of the type IC restriction-modification enzyme EcoR124I. Nucleic Acids Res., 26, 4439–4445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Janscák P., Abadjieva,A. and Firman,K. (1996) The type I restriction endonuclease R.EcoR124I: over-production and biochemical properties. J. Mol. Biol., 257, 977–991. [DOI] [PubMed] [Google Scholar]
- 7.Firman K. and Szczelkun,M. (2000) Measuring motion on DNA by the type I restriction endonuclease EcoR124I using triplex dissociation. EMBO J., 19, 2094–2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Seidel R., van Noort,J., van der Scheer,C., Bloom,J.G.P., Dekker,N.H., Dutta,C.F., Blundell,A., Robinson,T., Firman,K. and Dekker,C. (2004) Real-time observation of DNA translocation by the type I restriction-modification enzyme EcoR124I. Nature Struct. Mol. Biol., 11, 838–843. [DOI] [PubMed] [Google Scholar]
- 9.Dryden D.T.F. (2004) Reeling in the bases. Nature Struct. Mol. Biol., 11, 804–806. [DOI] [PubMed] [Google Scholar]
- 10.Gorbalenya A.E. and Koonin,E.V. (1991) Endonuclease (R) subunits of type I and type III restriction-modification enzymes contain a helicase-like domain. FEBS Letts., 291, 277–281. [DOI] [PubMed] [Google Scholar]
- 11.Flaus A. and Owen-Hughes,T. (2001) Mechanisms for ATP-dependent chromatin remodelling. Curr. Opin. Genet. Dev., 11, 148–154. [DOI] [PubMed] [Google Scholar]
- 12.Mahdi A.A., Briggs,G.S., Sharples,G.J., Wen,Q. and Lloyd,R.G. (2003) A model for dsDNA translocation revealed by a structural motif common to RecG and Mfd proteins. EMBO J., 22, 724–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yuan R., Bickle,T.A., Ebbers,W. and Brack,C. (1975) Multiple steps in DNA recognition by restriction endonuclease from E. coli K. Nature, 256, 556–560. [DOI] [PubMed] [Google Scholar]
- 14.Szczelkun M.D., Dillingham,M.S., Janscák,P., Firman,K. and Halford,S.E. (1996) Repercussions of DNA tracking by the type IC restriction endonuclease EcoR124I on linear, circular and catenated substrates. EMBO J., 15, 6335–6347. [PMC free article] [PubMed] [Google Scholar]
- 15.Mernagh D.R. and Kneale,G.G. (1996) High resolution footprinting of a type I methyltransferase reveals a large structural distortion within the DNA recognition site. Nucleic Acids Res., 24, 4853–4858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mernagh D.R., Janscák,P., Firman,K. and Kneale,G.G. (1998) Protein–protein and protein–DNA interactions in the Type I restriction endonuclease R.EcoR124I. Biol. Chem., 379, 497–503. [DOI] [PubMed] [Google Scholar]
- 17.Kasas S., Thomson,N.H., Smith,B.L., Hansma,H.G., Zhu,X., Guthold,M., Bustamante,C., Kool,E.T., Kashlev,M. and Hansma,P.K. (1997) Escherichia coli RNA polymerase activity observed using atomic force microscopy. Biochemistry, 36, 461–468. [DOI] [PubMed] [Google Scholar]
- 18.van Noort S.J.T., van der Werf,K.O., Eker,A.P.M., Wyman,C., de Grooth,B.G., van Hulst,N.F. and Greve,J. (1998) Direct visualization of dynamic protein–DNA interactions with a dedicated atomic force microscope. Biophys. J., 74, 2840–2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Walkinshaw M.D., Taylor,P., Sturrock,S.S., Atanasiu,C., Berge,T., Henderson,R.M., Edwardson,J.M. and Dryden,D.T.F. (2002) Structure of Ocr from bacteriophage T7, a protein that mimics B-form DNA. Mol. Cell, 9, 187–194. [DOI] [PubMed] [Google Scholar]
- 20.Halford S.E., Welsh,A.J. and Szczelkun,M.D. (2004) Enzyme-mediated DNA looping. Annu. Rev. Biophys. Biomol. Struct., 33, 1–24. [DOI] [PubMed] [Google Scholar]
- 21.Szczelkun M.D. (2002) Kinetic models of translocation, head-on collision, and DNA cleavage by type I restriction endonucleases. Biochemistry, 41, 2067–2074. [DOI] [PubMed] [Google Scholar]
- 22.Velankar S.S., Soultanas,P., Dillingham,M.S., Subramanya,H.S. and Wigley,D.B. (1999) Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism. Cell, 97, 75–84. [DOI] [PubMed] [Google Scholar]
- 23.Janscak P. and Bickle,T.A. (2000) DNA supercoiling during ATP-dependent DNA translocation by the Type I restriction enzyme EcoAI. J. Mol. Biol., 295, 1089–1099. [DOI] [PubMed] [Google Scholar]