Abstract
Glioblastoma multiforme (GBM) is the most aggressive of the gliomas, a collection of tumors arising from glia in the central nervous system. Possible associations between the human cytomegalovirus (HCMV) and the JC virus with GBM are now attracting interest. Our present aim was to investigate the prevalence of the two viruses in Iranian patients from Kerman’s cities in the south of Iran. In addition, the expression rates of pp65, large T antigen and p53 proteins were assessed and their relation with GBM evaluated using reverse transcription real time PCR (rReal Time PCR). A total of 199 patients with GBM cancer were enrolled, with mean±SD ages of 50.0±19.5 and 50.7±19.6 years for males and females, respectively. The P53 rate was dramatically low suggesting an aetiological role,. Large T antigen expression was found in JC positive samples, while the PP65 antigen was observed in patients positive for CMV and JC. HCMV products and JC virus with oncogenic potential may induce the development of various tumors including glioblastomas. The JC virus produces an early gene product, T-antigen, which has the ability to associate with and functionally inactivate well-studied tumor suppressor proteins including p53 and pRB.
Keywords: Glioblastoma, HCMV, JC Virus, P53, large T Antigen
Introduction
Glioblastoma multiform (GBM) is the most aggressive primary brain tumor with a median survival of approximately one year, and over 95% of patients surviving less than two years. GBM is the most aggressive of the gliomas which are divided into four grades; unluckily, the most aggressive of these, glioblastoma multiforme (GBM), is also the most common in humans. Several DNA viruses such as Epstein Barr virus (EBV), SV 40, humanpapilloma virus (HPV), JC virus, human cytomegalovirus (HCMV) and hepatitis B have been previously implicated in different cancers (Zhang et al., 2013). HCMV is in Herpesviral order and in β-herpes virus group, CMV does not usually causes clinical disease except in immunocompromised hosts. The DNA of CMV can be detected in colon cancer, malignant glioma, and some prostate cancers have shown in recent studies (dos Santos et al., 2014). HCMV antigen detected in a high percent of malignant tumors including prostate cancer, colorectal carcinoma, skin cancer, and malignant astrocytoma(Ottenhausen et al., 2014). Several CMV proteins interact and inactivate proteins of the Rb family (Retinoblastoma) as well as p53, resulting functions of p53 and Rb were down-regulate (Costa et al., 2013). It is unknown that HCMV plays a role in the pathogenesis of malignant brain tumors, or tumor growth provides an environment supportive for local reactivation and propagation of the virus (Sabbatino et al., 2014). So we chose HCMV to investigate the detection of HCMV proteins and nucleic acids in the tumors of patients with glioblastoma multiform (GBM)same to other researcher (Lucas et al., 2011). JC virus is implicated in different tumors of humans in central nervous system (CNS)(Del Valle et al., 2000). The first human polyomaviruses JC (JC) was discovered in 1971 and it was isolated first from the brain of a patient suffering from progressive multifocal leukoencephalopathy (PML)(Major et al., 1984). Polyomaviruses are a subfamily of non enveloped DNA viruses with icosahedral capsids that contain small, circular, double-stranded DNA genomes(Del Valle et al., 2002; Pina-Oviedo et al., 2006). The DNA of JC virus has been detected in several kinds of human malignancies, such as brain tumors of glial origin, colon carcinoma and medulloblastomas. The T-antigen as an early viral product has the ability to inactivate well-studied tumor suppressor proteins including p53 and pRb(Devireddy et al., 1996; Tognon et al., 1996; Poch et al., 2007). The experimental observations have suggested the presence of the JCV DNA and viral oncogenic protein expression in human brain tumors (Del Valle et al., 2002; Djuzenova et al., 2015). The aim of this study is to determine the presence of CMV and JC DNA and expression level of pp65 mRNA, T antigen and p53 omit expressed on diagnosed GBM tissues in Kerman province southeast of RAN.
Material and Methods
Patients
Paraffin embedded blocks samples from patients with GBM were collected during Jun 2001 -May2014, from Neurosurgery referral center in Shahid Bahonar hospitals Kerman province, Iran. In total of 220 specimens were found, but 199 samples were entered to our study and others patients were lost due to inadequate, absence of histological material. Also 100 histopathologic samples were selected from peritumoral normal brain tissue as control group. The present study is based on a retrospective examination of GBM diagnostic biopsy or surgery samples from clinical cases, all original hematoxylin and eosin (H&E) slides and/or H and E recut from tissue blocks were reviewed. In total, samples were screened for CMV DNA, level expression mRNA of pp65, JC DNA, level expression of large T antigen and level of p53 omit expressed. This project was approved by the Neuroscience research center ethics committee of the Kerman University of Medical Sciences.
Deparaffination samples
Paraffinated blocks from the 199 tumor samples were cut in 5-μm sections and eight of these sections were collected in a 1.5ml strile micro-centrifuge tube. Samples were de-waxed in 500 μl xylene, All micro-centrifuge tubes located for 10 min in a 60 °C heated block and centrifuged at 8,000 rpm. Then supernatant was removed. This step was repeated three times. Add 500 μl absolute ethanol, centrifuge at 10,000 rpm for 1 min, the samples were then dried in a 60°C heated block with open lids for 10-20 min for remove residual ethanol.
Tissue digestion
According to samples (biopsy or Paraffinated blocks), 200-400 μl of Tissue Lysis Buffer was added to each tube. Tissue lysis buffer is contain,4 M Urea, 200 mMTris, 20 mMNaCl, 200 mM EDTA; PH=7.4 (25°C). To all tubes added 20-40 μl proteinase K, then Samples were gently vortexes and located for 10 min in a 60°C heated block, and all samples were subsequently incubated at 37°C overnight.
DNA and RNA Extraction
The next day, 200 μl of Binding Buffer, contain [6 M Guanidine- Hcl, 10mM Urea, 10mM Tris-Hcl, 20% Tritonx-100(v/v); PH=4.4(25°C)], was added to each tube with gently vortex. DNA was isolated using a QIAamp DNA Mini kit (Qiagen, Germany) and Total RNA was extracted using RNAeasy mini kit (Qiagen, Germany) according to the manufacturer’s instructions. Extracted DNA and RNA were resuspended in 100μl of pre-warmed Elution buffer and stored at -70°C until use.
Virus Detection using Real time PCR
Real time PCR was carried out using CMV and JC Real Time PCR kit (Inter Lab Servise, Russia) following instruction manual.
rReal Time PCR
For determination expression level of pp65, p53 and largeT- antigen mRNA, reverse transcription real time PCR (rReal Time PCR) was carried out by using the first strand cDNA synthesis kit by Revert AidcDNA synthesis kit (Thermoscientific, USA). Briefly, RNA samples were heated to 65°C for 10 minutes and then chilled on ice. First-strand cDNA reaction mix was added according to the manufacturer’s protocol. One μl of DTT solution, and 1 μl of random hexamer (N)6primer (0.2ug) were then added to the heat-denatured RNA. Samples were mixed properly by pipetting up and down several times and then incubated for 1 hour at 42°C. For Real time PCR, the QuantiTect Probe PCR Kit (Qiagen, Germany) used base on instruction kit. Real time PCR primers and probes were design for pp65, p53 and T antigen mRNA after alignment of these regions between all of them in EBML-EBI and as an internal control, β-Actin were purchased from Metabion company (Germany)(Table 1).
Table 1.
Sequence Primer and Probes for Real Time PCR in This Study
| Name | Forward | Reverse | Probe | Location |
|---|---|---|---|---|
| P53 | CAGCATCTTATCCGAGTG | GATGGTGGTACAGTCAGA | CCAACCTCAGGCGGCTCATA | 147-269 |
| Large T Antigen JCV | GCAGCCTATGTATGGTATG | CCTGGAAGTTCCTCTGTC | ACTCTAACCTCCTCTACCTGAGCA | 201-272 |
| PP65 CMV | GCAGAACCAGTGGAAAGA | GCAGAACCAGTGGAAAGA | CGTACTGGTCACCTATCACCTGC | 303-483 |
Statistical analysis
Chi-square test or Fisher’s exact test was conducted using SPSS version 17 for the association between the presence of DNA viruses genome and other characterizes (values P=0.05 were considered statistically significant).
Results
One hundred ninety-nine patients with GBM cancer were selected during Jun 2001- May2014. Paraffinated blocks samples from these patients were selected for evaluation frequency of HCMV, JC virus and evaluation level gene expression of P53, large T antigen and PP65 using Real Time PCR. From total 199 samples, 81(41.3%) samples were female and 118(58.7%) were male. Mean ±SD of age for case group, male and female were 50.0±19.5 and 50.7±19.6 years old, respectively and for control group were 50± 18.5 for male and 51±16.2 years old age for female. Twentyfive samples(25%) were female and 75 samples (75%) were male. Minimum and maximum age in case group for male were 6 and 84 years old and for female were 9 and 86 years old (Table 2).
Table 2.
PCR Results in GBM Patients
| Test | Sex | Result | ||
|---|---|---|---|---|
| Negative(%) | Positive(%) | Negative/Positive % | ||
| JC PCR | Female | 67 (33.7) | 14 (7.0) | 4.7% |
| Male | 107 (53.8) | 11 (5.5) | 9.7% | |
| CMV PCR | Female | 73 (36.7) | 8 (4.0) | 9.12% |
| Male | 106 (53.3) | 12 (6.0) | 8.83% |
In Table 3, Different areas of Kerman province which 199 GBM samples collected was illustrated and the mean age of the patients with PCR test results for JC and CMV was shown. It is important that Rafsanjan city has a separated academic center and sometimes the patients do not go to Kerman medical centers, therefore, the statitistic due to the Rafsanjan may be more than on the table. As it can be seen, the highest frequency of positive CMV PCR is in Kerman city with 12 positive CMV and the second high frequent city for CMV is Jiroft city with 3 positive PCR test. The same results were captured for JC and Kerman city has the high frequency with 15 positive JC PCR and Jiroft is next with 3 positive result and also Sirjan city has shown 3 positive JC PCR and standing beside of Jiroft. It is also observed that from the total patients with GBM, 20 cases have positive CMV PCR and 25 cases are positive for JC PCR. In this study no positive CMV was reported in Kahnoug, Ravar, Zarand, Baft cities and also no positive results for JC PCR was reported in Bardsir, Baft and Bam cities. As it has been shown, Bam city has the highest range of age with the mean of 52.3 ± 17 years and after that Kerman has the mean of 51.4 ± 6 years. The youngest rang of age is related to Kahnoug city with 22±9 year and after that Baft city is standing with the mean 35.1±13 year. Our data also illustrated that there is a significant relationship between tumors and aging. Therefore, by increasing the age, the risk of GBM raises. The results showed that the probability of GBM in men is higher than women in Kerman province(P value<0.05). Geographic map of Kerman province and its different cities are presented in figure 1 and the distribution of patients with GBM is shown. Kerman with 69% is the most abundant and after that Jiroft city with 9% and Sirjan with 8% are abundant. On the other hand Kahnooj, Bardsir and Baft cities with 1% have the lowest frequency for GBM. To catch more information for the role of P53 protein in making tumors, all groups of this study used for P53 detection. For control or normal samples, sidelines of benign tumors were collected. Different rate of P53 expression is shown in figure 2 and it is clear that in control groups the expression of p53 is normal but in case group, the rate of P53 dramatically drops which suggests that the reduction in amount of p53 may involve in making tumors. As it can be seen the expression levels of p53 in positive JC, CMV cases are substantially reduced comparing with negative samples or control groups. Probably, other materials that are produced and released in virus life cycle may lead to reduced p53 in these cases.
Table 3.
Distribution GBM and PCR Results in Kerman Province
| Location | Num | Age | CMV PCR | JC PCR | ||
|---|---|---|---|---|---|---|
| (Mean±SD) | Negative | Positive | Negative | Positive | ||
| Bam | 6 | 52.3 ± 17.0 | 4 | 2 | 6 | 0 |
| Baft | 3 | 35.1 ± 13.0 | 3 | 0 | 3 | 0 |
| Bardsir | 3 | 49.0 ± 25.0 | 2 | 1 | 3 | 0 |
| Jiroft | 18 | 47.3 ± 16.0 | 15 | 3 | 15 | 3 |
| Kerman | 137 | 51.4 ± 6.0 | 125 | 12 | 122 | 15 |
| Kahnoug | 2 | 22.0 ± 9.0 | 2 | 0 | 1 | 1 |
| Ravar | 4 | 46.0 ± 19.0 | 4 | 0 | 3 | 1 |
| Sirjan | 16 | 44.7 ± 19.0 | 15 | 1 | 13 | 3 |
| Zarand | 5 | 50.8 ± 13.0 | 5 | 0 | 4 | 1 |
| Rafsanjan | 5 | 46.1 ± 25.0 | 4 | 1 | 4 | 1 |
| Total | 199 | 44.5 ± 16.2 | 179 (89.9%) | 20 (10.1%) | 174 (87.4%) | 25 (12.6%) |
Figure 1.
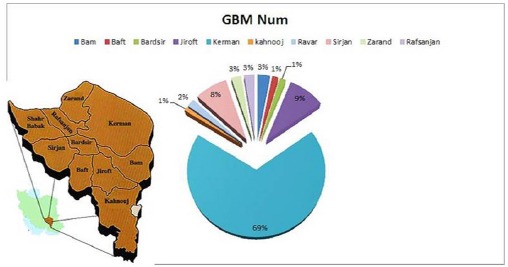
Distribution GBM in Different Towns of Kerman Province
Figure 2.
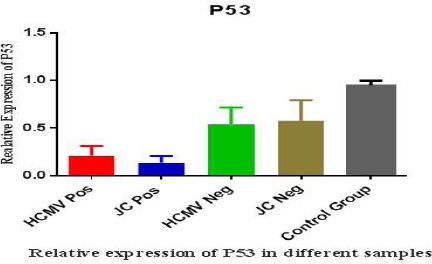
Relative P53 Expression in GBM and Normal Samples
Large T antigen expression is shown in figure 3 and expected that Large T antigen is produced in JC positive samples (green column). In 3 positive cases and 1 negative for HCMV, Large T antigen expression detected and also in one negative JC sample we could find Large T antigen expression. Interestingly, in none of them JC genome was not detected. This is may be load of DNA virus in sample is very low. In figure 4, PP65 antigen expression in the different groups studied and seen the expression rate of PP65 antigen in the positive CMV patients is in the highest level (red column) but as it is shown in one positive JC sample and 2 negative CMV cases some levels of expression were observed. Statistical analysis of results showed that there is a significant correlation between p53 and tumor genesis. Our data defined that there is no significant relationship between Large T antigen expression and presence of JC virus. So it is hard to say that JC viruses have a role in GBM tumors genesis. Also, no relation between the expression levels of pp65 and CMV infection was found in GBM tumors production and it cannot be said that CMV virus is involved in GBM tumors genesis.
Figure 3.
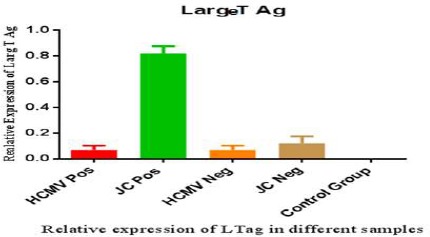
Relative Large T Antigen Expression in GBM and Normal Samples
Figure 4.
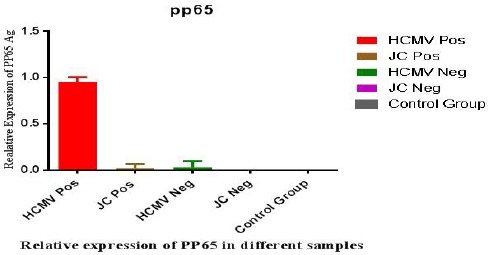
Relative PP65 Antigen Expression in GBM and Normal Samples
Discussion
The role of human cytomegalovirus (HCMV) and JC virus in the pathogenicity of glial tumours is under question. HCMV can products and also transactivate other oncogenic viruses that are associated with malignant gliomas such as JC virus, and may promote oncogenesis. Furthermore, JCV has an oncogenic potential and induces development of various tumors including medulloblastomas and glioblastomas(Lucas et al., 2011; Ghazi et al., 2012; Knight et al., 2013). The JCV is producing early gene product, T-antigen, which has the ability to associate with and functionally inactivate well-studied tumor suppressor proteins including p53 and pRB. GBM tumors are very important and different studies have been done to answer the question of being a relation between viruses like CMV and JC with their proteins such as PP65 and Large T antigen, and GBM tumors (Lucas et al., 2011; Nair et al., 2014). In 2013, Kenneth Alibek in Kazakhstan showed the presence of human cytomegalovirus (HCMV) in brain tumours, namely glioblastoma multiforme (GBM) and medulloblastomas by using highly sensitive detection techniques such as immunohistochemical detection and PCR amplification and HCMV nucleic acids and CMV proteins pp65 were found in GBM tumors (Alibek et al., 2013). In our study, CMV genome was detected in 20 GBM patients out of 199 patients. However, PP65 protein expression detected in CMV positive and beside this, PP65 expression was seen in JC positive and CMV negative patients. In 2011 Charles S. Cobbs, indicates that HCMV could facilitate glioma progression. Although, our data clarify the potential role of HCMV and JC in glioma pathogenesis, GBM in negative JC or negative CMV virus without PP65 were reported (Cobbs, 2011). In another study by Michael E. Scheurer in 2008 in USA, using sensitive immunohistochemical and in situ hybridization methods in glioma samples and detected HCMV antigen and DNA in all cases of glioblastoma which is not the same as our data (Scheurer et al., 2008). Also they did not search for PP65 protein but in our research, we found PP65, in positive and negative CMV PCR samples and positive JC samples. In USA in 2011, Steven Lehrer showed that 80% of patients with newly diagnosed glioblastoma have no detectable cytomegalovirus DNA which is near to our data (Lehrer et al., 2012). We found negative evidence of an association between HCMV or JC virus infection and GBM in this study. In addition, we showed dramatic differences in the expression of PP65 and Large T antigen in these tumors which are irrelevant with tumor production. Based on our data, P53 expression is directly linked with CMV and JC infection and in our positive CMV, JC samples the rate of P53 dropped and helps to progression tumors. In a study by Del Valle in 2001, they have examined 85 clinical specimens with GBM for their possible association with JCV. Gene amplification techniques used to recognize the JCV DNA sequence, and demonstrated the presence of the viral sequence in 49 (69%) of 71 samples and JCV T-antigen in the nuclei of tumor cells in 28 (32.9%) of 85 tested samples detected. In our study, 25 out of 199 samples were positive for JC virus and Larg T antigen in both CMV and JC were detected. In other studies investigated the presence of human polyomaviruses JC virus genome and the expression of the viral Oncoproteine T-antigen in patient with MS(Multiple Sclorosis) and a glioblastoma. They reported the expression of T-antigen, but not p53. In our survey, P53 was detected in both positive CMV and JC patients and decreased comparing to negative patients or control groups and beside the presence of PP65 in positive CMV, we could report some expression in negative CMV and positive JC samples (Major et al., 1984; Lucas et al., 2011; Matlaf et al., 2013; Price et al., 2013). We have got more positive GBM in elderly cases and also reported that the presence and possibility of GBM in men is higher than women. Although researches have a strict causal relationship between HCMV and JC infection and GBM tumors, but future studies need to focus on determining the role of HCMV and JC as a glioma-initiating event. Moreover, finding different proteins with varied functions and different types of CMV and JC can be proposed.
In our study, we found the relation between age and sex in GBM patients. We have reported that the presence and possibility of GBM in men is higher than women. The role of human cytomegalovirus (HCMV) and JC virus in the pathogenicity of glial tumours was unknown reported. The JCV is producing early gene product, T-antigen, which has the ability to associate with and functionally inactivate well-studied tumor suppressor proteins including p53 and pRB.
Competing interest
The authors declare that they have no competing interests.
Acknowledgments
The authors of this project are grateful to Kerman Virology Laboratory in Besat clinic staff and their cooperation in collecting samples, and so thanks for Research Center for Tropical and Infectious Disease, Kerman University of Medical Sciences, Kerman, Iran ethics committee of the Kerman University of Medical Sciences in approve this project.
References
- Alibek K, Kakpenova A, Baiken Y. Role of infectious agents in the carcinogenesis of brain and head and neck cancers. Infect Agent Cancer. 2013;8:7. doi: 10.1186/1750-9378-8-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobbs CS. Evolving evidence implicates cytomegalovirus as a promoter of malignant glioma pathogenesis. Herpesviridae. 2011;2:10. doi: 10.1186/2042-4280-2-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa B, Bendinelli S, Gabelloni P, et al. Human glioblastoma multiforme: p53 reactivation by a novel MDM2 inhibitor. PLoS One. 2013;8:e72281. doi: 10.1371/journal.pone.0072281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Valle L, Azizi SA, Krynska B, et al. Reactivation of human neurotropic JC virus expressing oncogenic protein in a recurrent glioblastoma multiforme. Ann Neurol. 2000;48:932–6. [PubMed] [Google Scholar]
- Del Valle L, Delbue S, Gordon J, et al. Expression of JC virus T-antigen in a patient with MS and glioblastoma multiforme. Neurology. 2002;58:895–900. doi: 10.1212/wnl.58.6.895. [DOI] [PubMed] [Google Scholar]
- Devireddy LR, Kumar KU, Pater MM, et al. Evidence for a mechanism of demyelination by human JC virus: negative transcriptional regulation of RNA and protein levels from myelin basic protein gene by large tumor antigen in human glioblastoma cells. J Med Virol. 1996;49:205–11. doi: 10.1002/(SICI)1096-9071(199607)49:3<205::AID-JMV8>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Djuzenova CS, Fiedler V, Memmel S, et al. Actin cytoskeleton organization, cell surface modification and invasion rate of 5 glioblastoma cell lines differing in PTEN and p53 status. Exp Cell Res. 2015;330:346–57. doi: 10.1016/j.yexcr.2014.08.013. [DOI] [PubMed] [Google Scholar]
- Dos Santos CJ, Stangherlin LM, Figueiredo EG, et al. High prevalence of HCMV and viral load in tumor tissues and peripheral blood of glioblastoma multiforme patients. J Med Virol. 2014;86:1953–61. doi: 10.1002/jmv.23820. [DOI] [PubMed] [Google Scholar]
- Ghazi A, Ashoori A, Hanley PJ, et al. Generation of polyclonal CMV-specific T cells for the adoptive immunotherapy of glioblastoma. J Immunother. 2012;35:159–68. doi: 10.1097/CJI.0b013e318247642f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight A, Arnouk H, Britt W, et al. CMV-independent lysis of glioblastoma by ex vivo expanded/activated Vdelta1+gammadelta T cells. PLoS One. 2013;8:e68729. doi: 10.1371/journal.pone.0068729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehrer S, Green S, Ramanathan L, et al. No consistent relationship of glioblastoma incidence and cytomegalovirus seropositivity in whites, blacks, and Hispanics. Anticancer Res. 2012;32:1113–5. [PubMed] [Google Scholar]
- Lucas KG, Bao L, Bruggeman R, et al. The detection of CMV pp65 and IE1 in glioblastoma multiforme. J Neurooncol. 2011;103:231–8. doi: 10.1007/s11060-010-0383-6. [DOI] [PubMed] [Google Scholar]
- Major EO, Mourrain P, Cummins C. JC virus-induced owl monkey glioblastoma cells in culture: biological properties associated with the viral early gene product. Virol J. 1984;136:359–67. doi: 10.1016/0042-6822(84)90172-7. [DOI] [PubMed] [Google Scholar]
- Matlaf LA, Harkins LE, Bezrookove V, et al. Cytomegalovirus pp71 protein is expressed in human glioblastoma and promotes pro-angiogenic signaling by activation of stem cell factor. PLoS One. 2013;8:e68176. doi: 10.1371/journal.pone.0068176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair SK, De Leon G, Boczkowski D, et al. Recognition and killing of autologous, primary glioblastoma tumor cells by human cytomegalovirus pp65-specific cytotoxic T cells. Clin Cancer Res. 2014;20:2684–94. doi: 10.1158/1078-0432.CCR-13-3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottenhausen M, Bodhinayake I, Schaefer PM, et al. VIGAS and beyond: the impact of HCMV-infection and its treatment in glioblastoma. J Neurosurg. 2014;74:23–4. doi: 10.1227/01.neu.0000445337.42703.0b. [DOI] [PubMed] [Google Scholar]
- Pina-Oviedo S, De Leon-Bojorge B, Cuesta-Mejias T, et al. Glioblastoma multiforme with small cell neuronal-like component: association with human neurotropic JC virus. Acta Neuropathol. 2006;111:388–96. doi: 10.1007/s00401-006-0050-3. [DOI] [PubMed] [Google Scholar]
- Poch E, Minambres R, Mocholi E, et al. RhoE interferes with Rb inactivation and regulates the proliferation and survival of the U87 human glioblastoma cell line. Exp Cell Res. 2007;313:719–31. doi: 10.1016/j.yexcr.2006.11.006. [DOI] [PubMed] [Google Scholar]
- Price RL, Song J, Bingmer K, et al. Cytomegalovirus contributes to glioblastoma in the context of tumor suppressor mutations. Cancer Res. 2013;73:3441–50. doi: 10.1158/0008-5472.CAN-12-3846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabbatino F, Fusciello C, Somma D, et al. Effect of p53 activity on the sensitivity of human glioblastoma cells to PARP-1 inhibitor in combination with topoisomerase I inhibitor or radiation. Cytometry A. 2014;85:953–61. doi: 10.1002/cyto.a.22563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheurer ME, Bondy ML, Aldape KD, et al. Detection of human cytomegalovirus in different histological types of gliomas. Acta Neuropathol. 2008;116:79–86. doi: 10.1007/s00401-008-0359-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tognon M, Casalone R, Martini F, et al. Large T antigen coding sequences of two DNA tumor viruses, BK and SV40, and nonrandom chromosome changes in two glioblastoma cell lines. Cancer Genet Cytogenet. 1996;90:17–23. doi: 10.1016/0165-4608(96)00067-2. [DOI] [PubMed] [Google Scholar]
- Zhang W, Fulci G, Wakimoto H, et al. Combination of oncolytic herpes simplex viruses armed with angiostatin and IL-12 enhances antitumor efficacy in human glioblastoma models. Neoplasia. 2013;15:591–9. doi: 10.1593/neo.13158. [DOI] [PMC free article] [PubMed] [Google Scholar]


