Abstract
Alternative splicing has recently emerged as a major mechanism of regulation in the human genome, occurring in perhaps 40–60% of human genes. Thus, microarray studies of functional regulation could, in principle, be extended to detect not only the changes in the overall expression of a gene, but also changes in its splicing pattern between different tissues. However, since changes in the total expression of a gene and changes in its alternative splicing can be mixed in complex ways among a set of samples, separating these effects can be difficult, and is essential for their accurate assessment. We present a simple and general approach for distinguishing changes in alternative splicing from changes in expression, based on detecting systematic anti-correlation between the log-ratios of two different samples versus a pool containing both samples. We have tested this analysis method on microarray data for five human tissues, generated using a standard microarray platform and experimental protocols shown previously to be sensitive to alternative splicing. Our automatic analysis was able to detect a wide variety of tissue-specific alternative splicing events, such as exon skipping,mutually exclusive exons, alternative 3′ and alternative 5′ splicing, alternative initiation and alternative termination, all of which were validated by independent reverse-transcriptase PCR experiments, with validation rates of 70–85%. Our analysis method also enables hierarchical clustering of genes and samples by the level of similarity to their alternative splicing patterns, revealing patterns of tissue-specific regulation that are distinct from those obtained by hierarchical clustering of gene expression from the same microarray data. Our data and analysis source code are available from http://www.bioinformatics.ucla.edu/ASAP.
INTRODUCTION
Genome-wide studies of gene expression using DNA microarrays have recently become a powerful tool for identifying new patterns of functional regulation (1). One major challenge of these studies is the volume and complexity of data they produce, which have spawned an entire research field of microarray data analysis. These methods seek to overcome two basic problems common in analysis of these data: first, distinguishing signal versus noise; and second, interpreting their biological meaning, often by translating the quantitative data into a clustering of the samples by the level of similarity of their expression profiles.
A very different approach to the study of functional regulation has been suggested recently by the widespread observation of alternative splicing in the transcripts of human and other species (2–4). Alternative splicing is not a simple quantitative change (e.g. an increase in the amount of mRNA expressed from a gene), but a qualitative change in the structure of the gene product itself (5). It can alter the protein's domain composition (6,7), shift it from a membrane-bound receptor to a soluble, secreted protein (8), or even block its translation (9). Instead of simply changing the amount of a gene's transcript, alternative splicing changes the transcript to a new and different form, which can carry out a different function. Since these are often substantial changes (e.g. addition or removal of an entire exon), the prospect of reliably detecting such qualitative changes on a genome-wide scale, using DNA microarrays or other technologies, is very attractive. Moreover, several groups have demonstrated that microarray-based detection of alternative splicing is possible (10–16).
To make automated discovery of alternative splicing from microarray data broadly practical, several related problems in microarray data analysis must be solved. Most fundamentally, such analysis must distinguish changes in splicing from changes in overall gene expression, since both can be mixed in complex ways in a set of samples, and each can confuse the interpretation of the other. Studies have shown that using many probes for each gene improves the reliability of the resulting gene expression measurement. For example, the well-known Affymetrix GeneChip design typically includes 11 or more pairs of perfect-match and single-base mismatch probes for each gene (17). The dChip analysis software of Li and Wong (18) uses these probe sets to identify the subset of probes that vary in the same way over all the samples, and to discover individual probes or arrays that are outliers, behaving inconsistently versus the consensus of the other probes. From the analysis of all these data, dChip produces a single number (the expression level) for the gene in a given sample, based on the most consistent probe pairs, and effectively ignores the probes that are inconsistent with this value. In contrast, detection of multiple distinct splice forms will require generating not just one but many different expression values per gene (one for each distinct transcript form). This is rendered especially difficult by the fact that we do not necessarily know what distinct forms we need to detect, or even how many. It can be difficult to deconvolute differences in gene expression, alternative splicing and probe sensitivities (e.g. see Figure 1D). These problems are not addressed by existing microarray data analysis, which operates under a simpler set of assumptions. For example, dChip sensibly throws away the probes that do not behave consistently over all the samples. But it is precisely these probes that potentially indicate the presence of alternative splice forms (15,16,19).
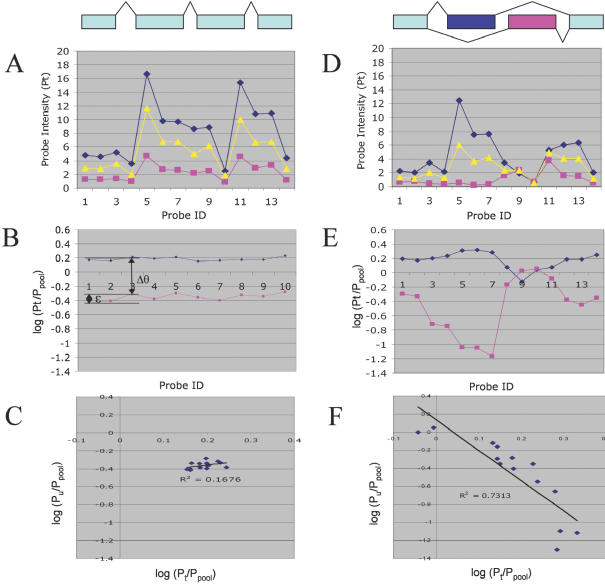
Figure 1.
Distinguishing changes in splicing from changes in total gene expression. Simulated data for multiple probes in a single gene, for two tissues (blue, pink) versus a pooled sample (yellow) representing their average, in the absence (A–C) or presence (D–F) of alternative splicing. (A) In the absence of alternative splicing, the probe intensities for different probes in a gene should show a similar profile in different tissues, reflecting the specific probe sensitivities. (B) Taking the log-ratio of each probe intensity versus its intensity for the pooled sample eliminates the effect of probe sensitivity differences, leaving only the difference in total expression (Δθ) and random probe variation (ɛ). (C) A scatterplot of each probe's log-ratio (versus pool) for tissue t versus tissue u yields a random scatter, whose position reveals their difference in total expression. (D) The introduction of alternative splicing can cause the profiles of the two tissues to differ significantly. (E) Because the pool (yellow in D) is always intermediate between the two tissues, taking the log-ratio versus pool tends to reveal alternative splicing as a ‘mirror image’ pattern. When the proportion of a splice form increases in tissue t relative to tissue u, for probes that are preferentially sensitive to that form (σ > 1), the log-ratio for tissue t will increase, while the log-ratio for tissue u will decrease, and vice versa. (F) This gives rise to a clear pattern of anti-correlation in the scatterplot of the log-ratios.
In this paper, we describe a method for analyzing microarray data that is designed to solve these problems. We present a basic theory for distinguishing changes in alternative splicing from changes in gene expression, and apply this to the detection of statistically significant evidence of tissue-specific alternative splicing from microarray data. We have tested this method on microarray data for five human tissue samples, generated using a standard microarray platform shown previously to be sensitive to alternative splicing. Our analysis method was able to identify strong evidence for a wide variety of tissue-specific alternative splicing events, including exon skip, alternative 5′ and alternative 3′ splicing, alternative promoter usage and alternative termination, which we have tested using independent PCR experiments. Our microarray data and analysis source code are available from http://www.bioinformatics.ucla.edu/ASAP. The microarray data discussed in this paper have been deposited in NCBIs Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) and are accessible through GEO series accession number GSE 1989.
MATERIALS AND METHODS
Distinguishing alternative splicing from gene expression in microarray data
The probe response Ptj for a specific probe j to a specific tissue sample t can be modeled as the product of the gene expression level θt in the tissue sample and the probe sensitivity ϕj for probe j, with additional factors for the baseline signal νj (which microarray analysis software usually seeks to subtract out) and a random error term ɛ (18):
![]()
Taking the ratio of Ptj to the response of the same probe to a sample pool p that contains an equal mixture of two tissue samples:
![]()
In the absence of alternative splicing, plotting the log of this ratio for one tissue t on the x-axis versus that for a second tissue u on the y-axis, for all probes j for a single gene, should yield a random scatter (reflecting the distribution of ɛ; see Figure 1C) whose centroid (x,y) indicates the difference in expression between the two tissues, i.e.
![]()
To take into account multiple splice forms f for a gene, we can designate a separate probe sensitivity ϕfj to each specific form, and express splice form f's quantity (as a fraction of the total transcripts for the gene in tissue t) as ωtf. Then the probe response becomes:
![]()
which reduces to the original expression when the ωtf are constant over the set of tissues. Consider a form f that increases in tissue t relative to the other tissue u (i.e. ωtf > ωuf). Since the pool p is the average of both tissues, ωuf < ωpf < ωtf. Thus, for probes j that are specific to a splice form f, log(Ptj/Ppj) > 0 and log(Puj/Ppj) < 0 (assuming that θt = θu). Moreover, as log(Ptj/Ppj) becomes increasingly positive, log(Puj/Ppj) becomes increasingly negative. Thus, when tissues t and u display markedly different splicing, the plot of their log-ratios relative to the pool should be negatively correlated (Figure 1F). On the other hand, if a tissue u shares a similar increase in splice f as observed in tissue t, its probe responses for this gene should show a positive correlation versus tissue t.
To examine this signal in more detail, consider the simplest case, where only two splice forms f and g are present. We can define the relative sensitivity σj for a probe j as the ratio of its sensitivity for the two forms, σj = ϕfj/ϕgj. Since ωtf + ωtg = 1, we can rewrite the expression for the probe signal as
![]()
We can consider the log-ratio of the probe signal (versus pool) under four different scenarios: when the probe has preferential sensitivity for form f (σj ≫ 1); when the probe does not distinguish f and g (σj = 1); when the probe has preferential sensitivity for g (σj ≈ 0); and intermediate values of σj (e.g. 0.2 < σj < 5). In the first case (σj ≫ 1) we obtain
![]()
Thus, for probes that are specifically sensitive to f, a change in splicing that produces more form f will result in a positive shift in the signal relative to the pool. Examples of such probes include a probe for an exon that is only included in one splice form, or a junctional probe that matches a splice that is included in only one particular splice form. In contrast, for the second case (σj = 1), the sensitivity to alternative splicing ωtf vanishes, and the log-ratio reduces to the original ‘pure gene expression’ expression of Li and Wong (18). Typically, such probes match ‘constitutive’ exons that are included in every splice form of the gene. The third case (σj ≈ 0) is simply the inverse of the first case (where the probe is sensitive to splice form g instead of f), resulting in
![]()
Finally, many probes may have partial specificity; that is, they prefer one splice form, but not strongly. This corresponds to intermediate values of σj that are not too far from σj = 1, e.g. 0.2 < σj < 5. Typically, these are probes that match a splice junction; even if this splice is specific to a single form, the probe may overlap a neighboring constitutive exon enough to have significant sensitivity to other forms containing that exon.
Thus, an important criterion for the detection of alternative splicing is the design of a probe set with wide variation in σj values, including probes that are highly specific to individual forms (with very large or very small σj values), probes with intermediate specificity, and probes to constitutive exons (with σj = 1, essential for an unbiased measure of total gene expression). This variation in σj gives rise to systematic shifts in the individual probe responses that shift them from random scatter (the pure gene expression case, Figure 1A–C) to the strong pattern of anti-correlation indicative of alternative splicing (Figure 1D–F). If these systematic shifts [log(ωtf/ωpf)] are significantly larger than the level of random variations (ɛ), they will be detectible as negative or positive correlations between the log-ratios (relative to pool) of different tissues.
Probe design
We designed probes for 316 human genes (see Supplementary Materials for Table) based on gene structure information from our ASAP database (20). For each gene, we designed probes for individual exons and splice junctions reported by ASAP: exon probes were 40 nt in length; and splice junction probes were 36 nt. Probes for constitutive exons and splice junctions were included in each gene as controls with σj = 1 as described above. To each probe sequence, we added a tail of 20–24 bases of poly(T) (yielding a total probe length of exactly 60 nt) to raise the probe sequence off the surface of the array (14). For each splice junction between two exons, five probe sequences were generated: (i) the last 36 nt of the first exon; (ii) the last 27 nt of the first exon+the first 9 nt of the second exon; (iii) the last 18 nt of the first exon+the first 18 nt of the second exon; (iv) the last 9 nt of the first exon+the first 27 nt of the second exon; and (v) the first 36 nt of the second exon. This design ensures the presence of multiple independent probes spanning the junction and that one of the probes lies entirely within each exon, yet is highly related to the junction probes via substantial overlap. For each probe sequence, we checked for matches to other regions of the human genome sequence using BLAST (21) with an expectation cutoff of 10−10, and also checked for stem–loop sequences that might cause hairpin secondary structures, using the EMBOSS program einverted (22), and a cutoff of at least 6 bp of complementary sequence. However, in many cases it was not possible to avoid such potentially problematic sequences, due to the design constraint of tracking individual exons and splice junctions. For splices identified in ASAP as alternative splicing, we automatically generated probe sequences for both the splice and its neighboring exons. In a subset of genes, we constructed probes for all exons and splices in the gene. The average number of probes per gene in our design was 22. Microarrays based on our probe design were generously fabricated and contributed by Agilent Technologies Inc. (Palo Alto, CA) using their standard 8.4k format.
Tissue samples and RNA extraction
Bone marrow total RNA was ordered from Clontech (Palo Alto, CA). Testes and liver tissue samples were obtained from the UCLA Tissue Bank. Brain and muscle tissue samples were obtained from the University of Maryland Brain and Tissue Bank. Total RNA was extracted from 200–400 mg of frozen tissue using Trizol (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. Total RNA was additionally purified with the RNAeasy kit (Qiagen). All total RNA samples were run on the Agilent 2100 Bioanalyzer (Agilent Technologies Inc.) to check their quality and integrity.
Array experiments
Labeled cDNA for array hybridizations was generated by linear amplification of total RNA, followed by direct labeling of the amplification products. Briefly, RNA samples from five normal tissues were individually amplified using the BD SMART™ mRNA amplification kit (BD Biosciences, Palo Alto, CA). An aliquot of 1.5 μg of total RNA from each normal tissue was used as a substrate for amplification according to the manufacturer's specifications. Each amplified sample was run on the Agilent 2100 Bioanalyzer to check the quality and integrity of the mRNA. Typically, the amplification products migrated as a broad peak, with the majority of transcripts ranging from 500 to 4000 bp. The amplified mRNAs were prepared for hybridization as follows: a pool of all the samples to be hybridized to the splicing arrays was assembled by mixing equal amounts of mRNA from each of the five tissue samples and four glioblastoma (GBM) samples (data not discussed in this study). This comparator pool was used for all hybridizations. For each hybridization, 250 ng of the amplified normal tissue mRNA or GBM mRNA and 250 ng of the pool were labeled with Cyanine 5-dCTP and Cyanine 3-dCTP, respectively (Perkin Elmer, Boston, MA) using the Agilent Fluorescent Direct Label kit (Agilent Technologies Inc.). Dye-swapped labeling reactions were performed in parallel and hybridized to the second array on the slide. Labeling reactions were carried out in accordance with the manufacturer's instructions, with the following modification: instead of the DNA primer provided with the kit, 2 μg of random hexamers (MWG Biotech, High Point, NC) were used to prime the reaction. The entire Cyanine 5 and Cyanine 3 labeling reactions were combined according to the Fluorescent Direct Label kit protocol and were prepared for hybridization to the array using the In situ Hybridization kit Plus (Agilent Technologies Inc.) according to the manufacturer's instructions, with the exception that hybridizations were performed for 36–48 h. Slides were washed and dried according to the manufacturer's specifications and scanned on an Agilent Microarray Scanner (Model G2565BA; Agilent Technologies Inc.).
Scanned images were analyzed using the Agilent Feature Extraction Software (Version A.7.1.1; Agilent Technologies Inc.). Approximately 200 individual probes were excluded owing to saturation or labeling artifacts (e.g. poor correlation of the dye-swap log-ratios). To account for slight variations in sample labeling/loading, processed signals for each individual tissue were rescaled to make the mean fluorescent intensity of each of the four replicate arrays for that tissue equal.
Significance testing
For a given gene, we calculated the correlation coefficient r for each pair of arrays using the log-ratios (tissue versus pool) of probes for that gene. This produced six correlation coefficients for the possible pairings of the four replicate arrays for each tissue, and 16 correlation coefficients for the possible pairings of replicate arrays between a specific pair of tissues. As a threshold of significant evidence of alternative splicing, we used a mean correlation value of r < −0.5 between a pair of tissues, and a confidence value of P < 0.001. We calculated the P-value using the two-sample Wilcoxon test (in the R software package, http://www.r-project.org) to assess the significance of the difference in r values between the six replicate array pairs for one tissue, versus the r values calculated for the sixteen possible replicate pairs comparing that tissue to a second tissue. As a variation, we also calculated the jackknife for each r value, by removing each probe to find the one whose removal caused the greatest decrease in absolute value of the correlation coefficient.
To check for subtle differences between individual pairs of tissues, we calculated the geometric mean intensity for each probe from the replicate arrays of the two tissues. We used this geometric mean as a ‘computed pool’ value for the two tissues, and calculated log-ratios for a given probe on a given array over its geometric mean value in the computed pool. Using these log-ratios, we calculated correlation r values for a given gene as usual, both for comparing pairs of replicate arrays for a single tissue, and for comparing one array from the first tissue versus one array from the second tissue. We identified evidence for alternative splicing using a threshold of a mean correlation value r < −0.5 for comparisons of the two tissues, and mean correlation values r > 0.5 for comparisons between replicate arrays of each tissue, and P < 0.001 as above. This analysis yielded similar results to the analysis based on the experimental pool composed of the five tissues plus four glioblastomas.
Hierarchical clustering
For each gene, we calculated the average r-coefficient for each array versus a consensus set initially consisting of all arrays (but excluding arrays from the same tissue), using the geometric mean of all five tissues as a pool. We applied an iterative computation for each gene. If the mean r value for a given tissue was below −0.3 (indicating significant negative correlation versus the consensus set), it was removed from the consensus set, and the r values recalculated, until no further changes in the consensus set occurred (typically only one or two cycles). Hierarchical clustering on the final r values was performed using dChip (18). This approach highlights simple splicing changes where the samples divide into just two distinct patterns of splicing (distinguished in this view by negative versus positive r values), but tends to obscure more complex cases where more than two distinct splicing patterns are present (such case are weakly visible as negative r values across several tissues).
We also clustered the same microarray data by gene expression values. We removed probes with coefficient of variance >0.15, calculated the average log-ratio (sample versus pool) for each gene in each sample, and performed hierarchical clustering using dChip (18), on both samples and genes, applying dChip's standardization to both columns and rows.
Real-time PCR validation
The same amplified RNA samples used for fluorescent labeling and microarray hybridization were used as a substrate for reverse transcription reactions and real-time PCR. An aliquot of 20 ng of amplified RNA was used in each reverse transcription reaction. Reactions were primed with random hexamers using the Taqman Reverse Transcription Reagent kit (Applied Biosystems, Foster City, CA) according to the manufacturer's specifications. About 1/20th of each reverse transcription reaction (corresponding to 1 ng of input mRNA) was used in each PCR reaction to assay tissue-specific alternative splicing. PCR reactions were carried out using the SYBR Green PCR Core Reagent kit (Applied Biosystems) according to the manufacturer's specifications with the exception that the reaction volume was reduced to 25 μl. Reactions were incubated in the DNA Engine Opticon2 Continuous Fluorescence Detection System (MJ Research, Waltham, MA). Reactions containing a wide range of titrations of the input reverse transcription products (1, 0.2, 0.02, 0.01 and 0.0033 ng input mRNA) were performed in parallel using primers annealing to the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene sequence. These reactions were used as a standard curve for quantification of the amplification products by the OpticonMonitor2 Analysis Software (version 2.0; MJ Research). Cycling parameters were as follows: 2 min at 50°C, 10 min at 95°C, followed by 35 cycles of 45 s at 95°C, 1 min at 55°C, fluorescence reading and 1 min at 72°C. A final extension of 10 min at 72°C was performed, followed by a melting curve (from 60 to 95°C with a fluorescence reading every 0.2°C) to determine the size distribution of the amplification products. The entire PCR reaction was resolved on a 1.5% agarose gel stained with SYBR Green I (Molecular Probes, Eugene, OR). PCR primers (MWG Biotech) were designed using the freely available Primer3 Software (Whitehead Institute, MIT) (for primer sequences see Supplementary Material).
The cycle number at which the fluorescence signal exceeds the detection threshold (Ct) was used as a basis for comparison with the microarray results. Ct values are inversely proportional to the log of the quantity of starting material being amplified. To derive a consensus measure of Log Total from Ct values for a given sample measured on different days, we calculated a baseline Ct value for each day (by taking the average Ct of a consistent set of samples), and took the average of the differences between the baseline for a given day and the Ct value measured on that day. Each Ct measurement was repeated in at least two experiments. Since the baseline value has no inherent significance, shifts in our Log Total measure have meaning, but its absolute value does not. Moreover, different sets of primers do not amplify with identical kinetics, so comparisons between data from different primer pairs are not meaningful.
cDNA synthesis, PCR and sequencing
The cDNA was synthesized using either oligo(dT)12–18 or random hexamers and Stratascript reverse transcriptase using the StrataScript First-Strand Synthesis System (Stratagene, La Jolla, CA). cDNAs from both reactions were pooled before performing PCR. This was done to increase coverage of the entire transcript.
Gene-specific primers were designed using MIT Primer3 software and synthesized by MWG Biotech. All primers flanked at least one exon–intron junction (to rule out artifacts from genomic DNA contamination), and all had Tm between 55 and 60°C. Primer sequences are available online. GAPDH levels were monitored as a housekeeping control. Touchdown PCR was performed on the MJR PTC-0200 thermal cycler (MJ Research, South San Francisco, CA) using Taq polymerase (Qiagen). Touchdown PCR conditions were as follows: 95°C for 2 min; 10× (95°C for 1 min; 65°C for 1 min with a decrease of 1°C per cycle; and 72°C for 1 min), 30× (95°C for 1 min; 55°C for 1 min; 72°C for 1 min); 72°C for 10 min; hold at 4°C. Reaction products were run on a 2–2.5% agarose gel and visualized by staining with ethidium bromide (Sigma–Aldrich, St Louis, MO). As an internal control for successful PCR, we required that at least one band, corresponding to the known splice form in GenBank, was observed. PCR products were gel purified using a Qiaquick gel purification kit (Qiagen). Sequencing of gel-purified products in both directions was performed by Laragen Inc. (Los Angeles, CA), using gene-specific primers and Amersham MegaBACE 1000 sequencers (Amersham Pharmacia Biotech, Piscataway, NJ). The results confirmed the expected DNA sequences in all cases.
RESULTS
Detection of alternative splicing as a qualitative change in microarray data
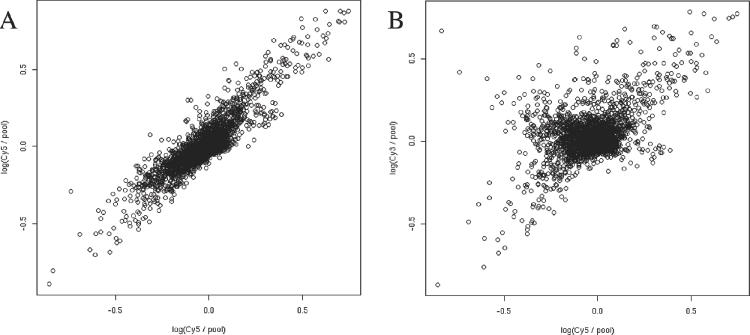
When the probes for a given gene show substantial variation in their intensity profiles over the set of samples, is this noise, or signal? Answering this question is essential for reliable detection of alternative splicing. To address this question in a simple way, we used a pooling strategy in which each sample was compared with a pool composed of an equal mixture of all samples, using two-color labeling (Cy3 versus Cy5) on each array. Each sample was measured on four separate arrays, two using Cy3 for the sample (versus Cy5 for the pool), and two with the dyes reversed. Each probe's fluorescence intensity for the sample was divided by that for the pool, and expressed as a log-ratio value, to remove the effect of systematic differences in probe sensitivity. Comparison of log-ratio values between replicate arrays showed good reproducibility, even between dye-swaps (Figure 2). Overall, 90% of the probes had a coefficient of variation of ≤0.15. It should be emphasized that our analysis method automatically takes into account the higher level of noise typically present in comparisons of arrays with reversed labeling (i.e. dye-swaps), because it assesses the statistical significance of a comparison between two tissues (e.g. Figure 3B) versus the level of variation seen among dye-swapped replicates of an individual tissue (Figure 3D–F).
Figure 2.
Reproducibility of microarray replicate and dye-swap experiments. Comparisons of raw microarray data for muscle versus pool (log-ratio) in (A) replicate arrays both loaded with muscle (labeled as Cy5) versus pool (Cy3); and (B) dye-swapped replicates. The x-axis: muscle (Cy5) versus pool (Cy3); y-axis: muscle (Cy3) versus pool (Cy5).
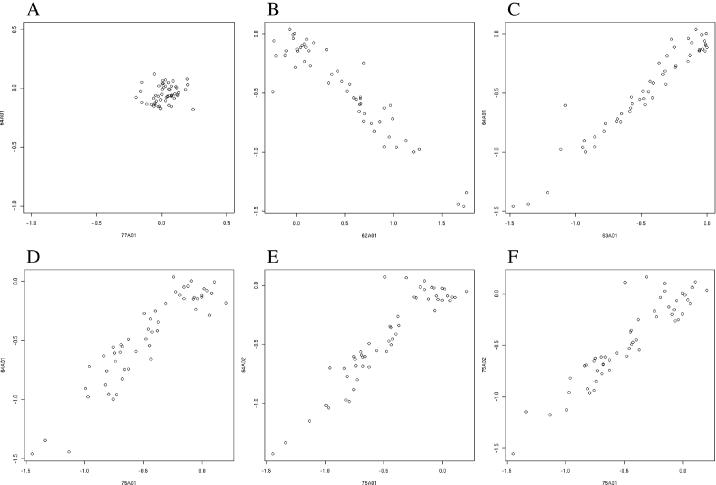
Figure 3.
Alternative splicing signals in the probe data for TPM1. (A) Log-ratio for bone marrow/pool (77A01) versus log-ratio for brain/pool (64A01) for probes in the gene TPM1. The random scatter indicates no differences in splicing between these two tissues. (B) Log-ratio for muscle/pool (77A01) versus log-ratio for brain/pool (64A01). The evident anti-correlation indicates a strong difference in splicing between these tissues. (C) Log-ratio for testes/(brain + muscle pool) (63A01) versus log-ratio for brain/(brain + muscle pool) (64A01). The positive correlation between these two tissues indicates that they share a splicing pattern that is different from the other tissue in the pool (muscle). (D–F) Scatter plots for the four replicate arrays of brain (64A01, 64A02, 75A01 and 75A02) against each other. Arrays 64A01, 64A02 and 75A02 are each shown compared versus array 75A01. These data show that the level of random variation between different measurements of the same sample (in this case, brain, measured as a log-ratio versus brain + muscle pool) is well below the level of systematic changes observed in a comparison versus a tissue with altered splicing (compare with B).
Normalization using the sample/pool ratio enables a simple way for distinguishing alternative splicing (Figure 1). In the default case where the variation among probes reflects random noise instead of a consistent alternative splicing signal, a scatter plot of the normalized values for the probes in a single gene from one tissue versus a second tissue will display random variation (Figure 1C). In contrast, if the gene has two alternative splice forms, one that is found in the first tissue and the other in the remaining tissues, this will give rise to systematic variations in the normalized probe signals (Figure 1D). Since the pool represents an average of all the samples, this produces an unusual and characteristic pattern of variation. Probes for exons and splices that are found only in the first splice form should show a positive log-ratio in the first tissue (versus pool), while probes for exons and splices that are found only in the second splice form should show a negative log-ratio in the first tissue (versus pool). In contrast, in the other tissues, probes that are found only in the first splice form should show a negative log-ratio, while probes that are found only in the second splice form should show a positive log-ratio. In other words, because the pool represents a midpoint between the two splice forms, the normalized signals from the two tissues should be a mirror-image of each other—where one goes up, the other should go down, and vice versa (Figure 1E). This is revealed as a negative correlation between the log-ratios for the two tissues (Figure 1F). In contrast, in the presence of alternative splicing, two samples with the same splice form composition should show the same systematic variations (versus pool), revealed by a strongly positive correlation between the log-ratios for the two tissues. The statistical significance of a given negative correlation pattern between two tissues can be calculated versus the distribution of correlation values between replicate arrays of each tissue versus itself (which should be positive). For example, this analysis detected strong evidence of muscle-specific alternative splicing in tropomyosin 1 (TPM1) (Figure 3). It should be emphasized that a change in overall expression of the gene will not produce such a pattern of correlation (see detailed explanation in Materials and Methods).
Hierarchical clustering of alternative splicing patterns
To test our method on a large dataset, we analyzed data from a set of 19 microarrays representing five human tissues (brain, muscle, liver, bone marrow and testes). Each tissue was represented by four replicate arrays, two labeled with the tissue-Cy3, pool-Cy5, and two labeled with tissue-Cy5, pool-Cy3 (see Materials and Methods). Out of the twenty arrays, one was excluded due to poor quality hybridization. Out of 316 human genes in the array, our analysis identified evidence (P < 0.001) of tissue-specific alternative splicing in 106 genes (listed in Supplementary Materials).
Our analysis provides a simple way to cluster genes and samples according to their alternative splicing patterns (instead of gene expression, as is normally done for microarray data). Hierarchical clustering of the r-values for 316 genes and the 19 microarrays revealed clear clusters, both among genes and among microarray samples (Figure 4A). Clustering of the microarray samples perfectly matched their division into the five tissues. Clustering of the genes showed conspicuous groupings such as muscle-specific alternative splicing (e.g. TPM1, tropomyosin 3 and myosin regulatory light-chain), liver-specific alternative splicing [e.g. fibrinogen γ, apolipoprotein C-II (APOC2) and dolichyl-phosphate mannosyltransferase polypeptide 2], bone marrow-specific alternative splicing [e.g. vascular endothelial growth factor and angiotensin 1 converting enzyme 1 (ACE1)], brain-specific alternative splicing (e.g. tryptophanyl t-RNA synthase and proteolipid protein 1) and testes-specific alternative splicing [e.g. myeloid leukemia factor 1 (MLF1) and DDXL nuclear RNA helicase]. In addition, several clusters showed a shared alternative splicing pattern in two tissues, for example liver and bone marrow (e.g. heat shock 90 kDa protein 1β), or brain and testes (e.g. trafficking protein particle complex 3).
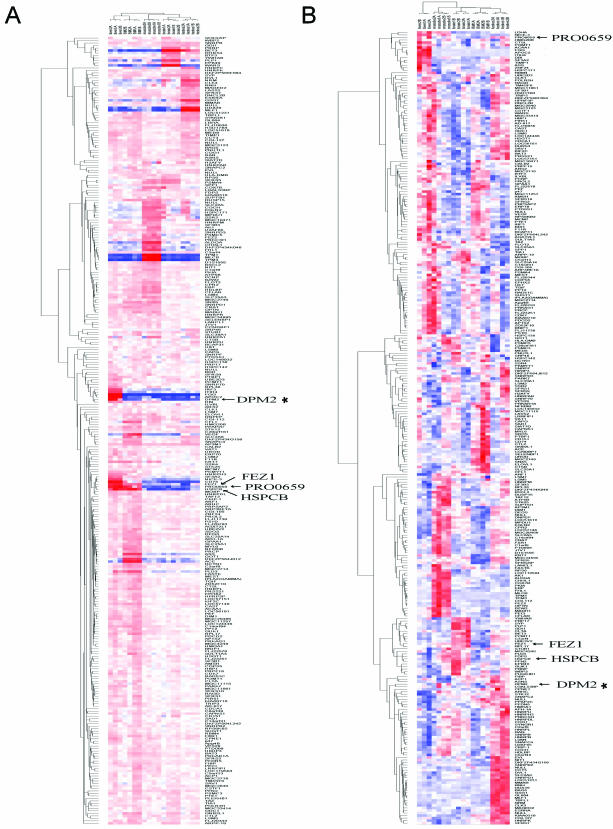
Figure 4.
Hierarchical clustering of alternative splicing for five human tissues. (A) For each array, we computed its mean correlation coefficient r versus arrays from other tissues (see Materials and Methods), and displayed the results using dChip (r = −1, red, representing a divergent splicing pattern; r = +1, blue, representing a consensus splicing pattern) with hierarchical clustering on both the array samples and genes. The simplest tissue-specific alternative splicing examples partition into just two distinct splicing patterns (red versus blue), but more complex alternative splicing patterns are revealed mainly by regions of anti-correlation (red), due to the limitations of this color mapping representation (see Materials and Methods). (B) Gene expression clustering of the same microarray data, using dChip's hierarchical clustering of expression values for each gene, clustering both the samples and genes (see Materials and Methods) (see Supplementary Material for high resolution image).
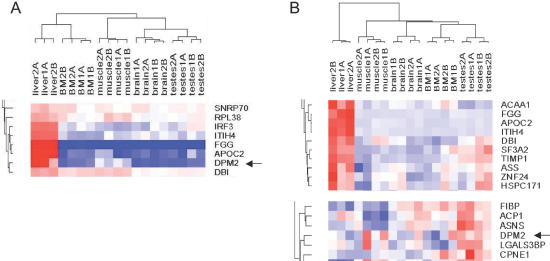
Hierarchical clustering of alternative splicing yields a gene clustering that is distinct from that produced by gene expression clustering of the same microarray data (Figure 4B; full cluster data in Supplementary Material). First, many genes that show up-regulated expression in a tissue actually appear to have a large increase in only one splice form, yielding a strong tissue-specific alternative splicing signal in that tissue. For example, among the approximately 10 genes whose expression is strongly up-regulated in a liver-specific manner, four [ITIH4, fibrinogen gamma (FGG), APOC2 and IRF3] were identified by the alternative splicing analysis as showing strong up-regulation of just one splice form, while other splice forms remained constant (Figure 5; see detailed analysis of APOC2, below). A similar pattern was seen in other tissues (for a detailed example in testes, see analysis of MLF1 below). Second, some genes that show no clear tissue specificity by gene expression, have strongly tissue-specific alternative splicing. For example, DPM2 was clustered by alternative splicing into the liver-specific group, but its gene expression profile displays no such tissue specificity (Figure 5B). Third, alternative splicing reveals some gene clusters that are not present in the gene expression profile. For example, clustering by alternative splicing identified a cluster of genes (FEZ1, PRO0659 and HSPCB; see Figure 4A) with similar alternative splicing in liver and bone marrow. This cluster was not observed in the expression clustering, which scattered these genes to very different clusters in the expression tree (Figure 4B).
Figure 5.
Comparison of alternative splicing clusters versus gene expression clusters. (A) A detailed view of the alternative splicing clusters, showing a cluster of genes with liver-specific alternative splicing. (B) A detailed view of the gene expression clustering, showing the cluster of genes that are up-regulated specifically in liver (above), and the separate cluster containing DPM2 (below).
Validation of alternative splicing signals versus multiple probes of gene structure
Our preceding analysis is a purely statistical procedure that makes no use of actual information about gene structure. Thus, its results can be validated independently by comparing the sets of individual probes that produce correlation or anti-correlation signals with the known gene structure and possible splice patterns. Specifically, we have designed our probe set for each gene to include multiple probes for each possible alternative splice event. Thus, the subset of probes that give a statistically significant alternative splicing signal in our previous analysis (i.e. log-ratio anti-correlation), should match a specific group of probes designed to detect a particular alternative splice event. For example, to detect insertion of an alternatively spliced exon, we designed a probe to that individual exon; a set of five splice probes stepping across the exon–exon junction entering this exon; a similar set of five splice probes stepping across the exon–exon junction exiting this exon; and similar sets of exon and splice probes for the exons flanking this exon (for details see Materials and Methods). Inclusion of this alternatively spliced exon should cause the whole group of splice1 + exon + splice2 probes to be identified by our statistical analysis (which knows nothing about what the individual probes are) as causing anti-correlation, with the surrounding exons and splices identified as constant.
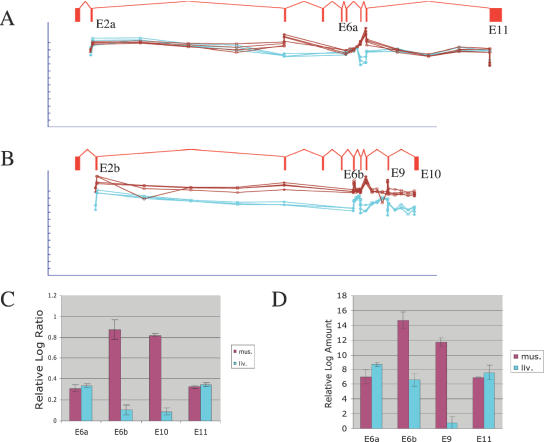
For example, our microarray analysis detected muscle-specific alternative splicing of TPM1 (Figure 6). These data showed strong shifts in the proportions of different splice forms in muscle, relative to other tissues. At the 5′ end of the gene, the data showed mutually exclusive exon usage of exons 2a and b. Both isoforms were observed at approximately equal levels in brain, bone marrow and other tissues, but in muscle the exon 2b isoform was up-regulated. A similar mutually exclusive exon pair was observed at exon 6a and b; the latter was up-regulated in muscle. At the 3′ end, again two splice forms were observed: one including exons 9 and 10 and another replacing these with exon 11 as a 3′ terminal exon. These two forms were observed at the same level in brain, bone marrow and other tissues, but in muscle, the latter form was strongly up-regulated (Figure 6C). Independent real-time PCR experiments showed qualitatively the same result (Figure 6D). Specifically, exon 6a and 11 showed the same level in muscle versus liver, whereas exon 6b and 9 were present at much higher levels in muscle than in liver. Our results are consistent with TPM1 isoforms reported previously in muscle (23), although our data additionally indicate mutually exclusive usage of exons 6a and b as described above.
Figure 6.
Detection of mutually exclusive exons and alternative termination in TPM1. (A) Raw microarray probe intensities for exons and splice junctions contained in a non-muscle splice form of TPM1, for four replicate arrays for muscle (brown) and three replicate arrays for liver (cyan). Each probe is shown immediately beneath the exon or splice junction it matches; the gene structure for a non-muscle transcript inferred from ESTs is shown. Three alternative splicing events are indicated: mutually exclusive exons 2a versus 2b; mutually exclusive exons 6a versus 6b; and alternative termination (exon 11 versus exons 9 and 10). Probe intensity is shown on a log scale; the tick marks represent 2-fold steps. The fact the plots closely overlap indicates that this isoform is expressed at the same level in both tissues. Only the constitutive exons (3, 7 and 8) included in the muscle-specific isoform shown in (B) show up-regulation. (B) Raw microarray probe intensities for exons and splice junctions contained in a muscle-specific splice form of TPM1, displayed as in (A). All the probes for this isoform had stronger fluorescent intensity in muscle, although there are variations in probe sensitivity. (C) Mean log-ratio values for exon probes in exons 6a, 6b, 10 and 11, measured from the muscle or liver microarray experiments. (D) Mean log amount values measured in independent real-time PCR experiments amplifying exons 6a, 6b, 9 and 11, derived from the Ct value (see Materials and Methods).
Detection of exon skipping, alternative 5′ and 3′ splicing, alternative promoter and alternative termination
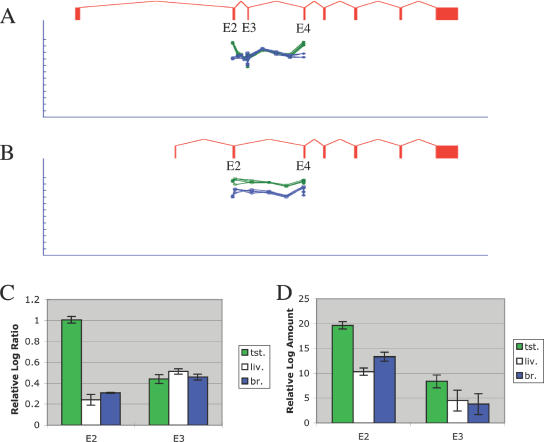
We also observed single exon skip events. For example, correlation analysis of MLF1 detected a testes-specific exon skip (Figure 7). The MLF1 probe data for testes diverged from all of the other eight tissues (r = −0.88, P < 10−4). Alternative splicing of exon 3 was observed. In all tissues, the splice form containing exon 3 appeared to be expressed at a similar level. In contrast, the splice form skipping exon 3 was up-regulated specifically in testes. The hybridization signal for exon 3 remained unchanged, while the signals for exons 2 and 4 increased in testes. Similarly, the splice probes for the junctions between exons 2–3 and 3–4 remained constant, while the splice probe for the junction between exons 2 and 4 (skipping exon 3) increased. These data provide clear evidence of testes-specific regulation of exon skipping in MLF1. Real-time PCR experiments showed a similar overall profile, with strong up-regulation of the exon skip form in testes (Figure 7D), but also showed a weaker increase in the exon 3 including form. No reports of MLF1 alternative splicing were found in PubMed.
Figure 7.
Detection of exon skipping in MLF1. (A) Raw microarray probe intensities for probes testing inclusion of exon 3 in MLF1, for four replicate arrays for testes (green) and four replicate arrays for brain (blue). Each probe is shown immediately beneath the exon or splice junction it matches. Probe intensity is shown on a log-scale; the tick marks represent 2-fold steps. (B) Raw microarray probe intensities for probes testing skipping of exon 3. (C) Mean log-ratio values for exon probes in exons 2 and 3, measured from the testes, liver or brain microarray experiments. (D) Mean log amount values measured in independent real-time PCR experiments amplifying exons 2 and 3, derived from the Ct value (see Materials and Methods).
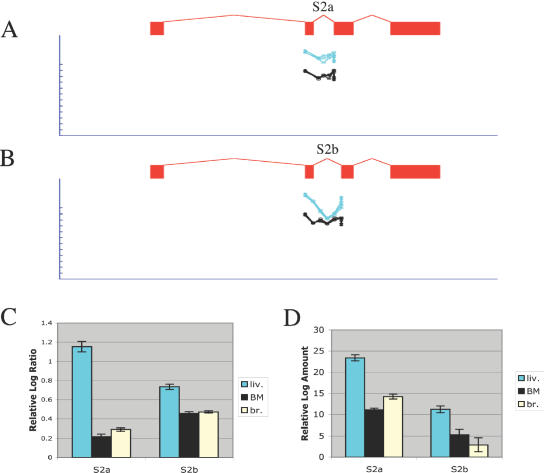
We were also able to detect more subtle changes such as alternative 3′ and alternative 5′ splicing, in which only a single splice is altered, by choosing a different splice site in the same exon. For example, in APOC2, correlation analysis detected an alternative splicing shift in liver (r = −0.80, P = 10−4), which corresponded to alternative usage of two 3′ splice acceptor sites in exon 3 (Figure 8). APOC2 was expressed at higher levels in liver than the other tissues we tested, but this shift was not equal for the two splice forms using these distinct splice acceptor sites. The splice form using the first splice acceptor site in exon 3 was up-regulated much more than the second splice form using the second splice acceptor site in exon 3 (Figure 8C). Independent real-time PCR experiments confirmed this result (Figure 8D). Detection of this alternative splicing shift depended entirely on the splice junction probes: while the probes for exons 2 and 3 and the first of their two splice junctions showed constant intensity, the probes for their second splice junction showed a highly position-specific reduction in intensity in liver. The three central probes (closely centered on this junction, and thus less likely to cross-hybridize to either exon 2 alone or exon 3 alone) showed the strongest drop in signal, while the two probes immediately adjacent to the splice (on either side) closely matched the signals for the respective exons. Alternative splicing of the mouse APOC2 gene in liver has been reported previously, specifically exon skipping of mouse exons 1B and C (24). We also detected alternative 5′ splicing in a wide variety of genes. For example, in ACE1, correlation analysis detected bone marrow-specific alternative splicing of exon 1 (r = −0.65, P < 10−4). While probes in exons 1 and 2 showed no significant shift, probes to the two possible splice junctions showed strikingly different responses in bone marrow compared with other tissues. While the five probes spanning the junction from the first splice donor site in exon 1 to exon 2 were constant in signal in all tissues, including bone marrow, the five probes spanning the junction from the second splice donor site in exon 1 showed a significant increase specifically in bone marrow (see Supplementary Material).
Figure 8.
Detection of alternative 3′ splicing in APOC2. (A) Raw microarray probe intensities for the first splice junction (S2a) between exons 2 and 3, for three replicate arrays for liver (cyan) and four replicate arrays for bone marrow (black). Each probe is shown immediately beneath the exon or splice junction it matches; the gene structure for a muscle-specific transcript inferred from ESTs is shown. Probe intensity is shown on a log-scale; the tick marks represent 2-fold steps. (B) Raw microarray probe intensities for the second splice junction (S2b) between exons 2 and 3. (C) Mean log-ratio values for splice probes for S2a and S2b, measured from the liver, bone marrow or brain microarray experiments. (D) Mean log amount values measured in independent real-time PCR experiments amplifying splice forms S2a and S2b, derived from the Ct value (see Materials and Methods).
Correlation analysis detected other types of isoforms, including alternative initiation and alternative termination. For example, in FGG, we observed a strong anti-correlation between probe values in liver versus other tissues (r = −0.88, P = 10−4), due to a large increase in usage of exon 1B in liver, while the isoform containing exon 1A remained constant (see Supplementary Material).
PCR validation of exon skipping, alternative 5′ and 3′ splicing, alternative promoter and alternative termination
To validate our results further, we performed RT–PCR on a sample of 20 of these genes, including representatives of all the different types of alternative splicing described above (see Supplementary Material). Of these cases, seven were exon skip events, which are easiest to test, since a single pair of primers directed to the exons flanking the exon skip will detect both the exon skip form and the exon inclusion form as products of different lengths. This gives an internal control for each primer pair, specifically, checking that it amplifies the known splice form. Of the seven exon skip cases, five were validated by PCR and sequencing, a 70% validation rate. The remaining test cases involved alternative 3′ splicing, alternative 5′ splicing, alternative initiation and alternative termination, which require a distinct pair of primers for each of the two splice forms. Of the 26 primer pairs for this group, 7 failed to amplify. In the six cases where both primer pairs worked, all six validated the alternative splicing pattern by PCR and sequencing of the products. In every case, DNA sequencing of the PCR products exactly matched the sequences of the expected alternative splice forms. In total, ∼85% (5/7 of the exon skips, plus 6/6 of the other types) of the alternative splicing events identified by the microarray analysis were validated by independent RT–PCR experiments. The total PCR primer failure rate was 21% (7/33), perhaps due to the narrow constraints of primer design (each primer was constrained to an individual exon, rather than being selected from anywhere in the gene).
DISCUSSION
We have developed a simple method for distinguishing alternative splicing from changes in gene expression, which could be applied to many types of microarray data. This method also provides a way for clustering genes and samples according to their alternative splicing patterns, producing a regulatory picture that is distinct from hierarchical clustering of the same microarray data according to gene expression. To test these methods, we have applied them to a small experimental dataset generated using a microarray platform and amplification methods shown previously to be sensitive to tissue-specific changes in alternative splicing (14,16). However, we have not analyzed any aspects of the biological questions or interest of this experimental dataset, which, for reasons of space, will be presented elsewhere.
Alternative splicing represents a qualitative change in a gene's expression—production of a different form of the gene product, bearing a different combination of functional elements in its sequence. Thus, it is useful to develop high-throughput methods for detecting such qualitative changes on a genome-wide basis. Fundamentally, alternative splicing provides a very different source of information for tracking regulatory events, monitoring cellular differentiation and classifying different tissues, than has been considered by traditional gene expression analysis. For example, a gene product may be switched from one splice form to another splice form with very different functional properties, while leaving the total amount of gene product unchanged. Although current microarray analysis would be unlikely to detect such a regulatory event, our qualitative analysis could make such changes clear, on a genome-wide scale.
It is also possible that alternative splicing can cause confusion or misinterpretation in existing gene expression analyses. For example, if a gene has two major splice forms that are regulated quite differently, the gene's expression can only be represented accurately by two distinct numbers (one value for the expression level of each form). Seeking to extract a single number as the ‘expression level’ of the gene, may actually be inappropriate in this case. At a minimum, this measurement is likely to be confounded by apparently inconsistent behaviors of different probes for the gene. Indeed, gene expression analyses such as the dChip software (18) are likely to systematically exclude as unreliable, any microarray probes that reveal strong patterns of alternative splicing, since these will diverge from the consistent expression profile across samples observed in the majority of probes for the gene. Thus, one benefit of our qualitative analysis approach is recognition that some probes are not simply unreliable, but contain additional information about the gene's regulation that was not captured by its total expression level.
To avoid such problems, it is essential to distinguish alternative splicing from gene expression within microarray data analysis. Our method provides a simple and general way to do this, but with a number of caveats. First of all, our approach should be considered a discovery method for detecting possible alternative splicing, rather than a validation method, which ensures that a given result is definitely due to alternative splicing. A number of other effects might give rise to systematic variation within the probes for a single gene. For example, if a subset of the probes cross-hybridize to transcripts from a paralogous gene, changes in expression of that paralogous gene would produce the kind of systematic variation (anti-correlation of tissue log-ratios) that our method detects. The nature of alternative splicing probe design makes it difficult to exclude such cross-hybridization entirely. Since alternative splice detection requires probes that match specific exons and splice junctions, probe selection is tightly constrained, and it is often not possible to completely avoid sequences that have a match somewhere else in the human genomic sequence. To weigh the evidence for true alternative splicing versus cross-hybridization to other genes, detailed consideration of the specific gene structure and likely splice forms for the gene in question, its paralogs, and other factors are required, which our method does not take into account. Second, we consider our method to be a qualitative analysis (identification of the presence or absence of changes in splicing), which we consider to be useful in its own right, rather than a quantitative method. Many additional kinds of statistical analysis would be required for such a method. Moreover, alternative splicing arrays have required different amplification protocols than those ordinarily used for expression arrays, because of the necessity of coverage across the full length of the gene (including the 5′ end) (14). There are many questions about the quantitative accuracy and reproducibility of the amplification protocol, which need to be addressed more fully as a prerequisite to reliable quantitative analysis. For example, the amplification method used in this study [and also in previous work (16)] yields substantial quantitative differences versus measurements made from total RNA (25) (http://www.bdbiosciences.com/clontech/archive/OCT03UPD/Smart-mRNA.shtml), although it does provide greatly improved coverage over the full length of transcripts (14).
Despite these technical challenges, there is now broadly reproducible evidence that alternative splicing can be detected using microarrays. Hu et al. (10) used standard Affymetrix array designs to search for evidence of alternative splicing in 1600 rat genes, by performing hybridizations with 10 normal tissue samples. A total of 268 genes (17%) showed signs of alternative splicing, and RT–PCR validation indicated that about half of these represented genuine alternative splice events. Other studies have focused on individual genes with known alternative splicing patterns, to demonstrate that microarray technology can detect these events. Clark et al. (11) used a cDNA spotted array to demonstrate successful detection of experimentally induced intron-retention in a number of Saccharomyces cerevisiae genes containing introns. Yeakley et al. (13) described detection of alternative splicing in six human genes using a fiber-optic microarray platform. Wang et al. (15) reported analysis of quantification of distinct splice forms of two human genes (CD44 and TPM2), using the well-known Affymetrix microarray platform. Castle et al. (14) reported studies of two genes (RB1 and ANXA7), examining in great detail the experimental factors determining probe response as a function of distance from an exon junction, position with the gene and so on. For example, they have analyzed in detail the effect of probe length on accurate detection of both exons and splice junctions. Kampa et al. (19) used Affymetrix microarrays to look for novel transcripts, and provided evidence that most human genes show evidence of more than one distinct isoform (26). By far, the largest microarray study was performed recently by Johnson et al. (16) using exon–exon junction probes to detect exon skipping. This study included probes for over 10 000 human genes and examined 52 distinct tissue samples. For genes in which alternative splice forms had not been previously reported by expressed sequence tag (EST) studies, about half were reported to show microarray evidence of exon-skipping. Validation by RT–PCR suggested that 45% of these positive candidates were genuinely alternatively spliced, indicating new discovery of alternative splicing in a large number of genes (estimated 798 in this study alone). Our work has used a similar microarray platform (Agilent microarrays), but has examined a variety of different types of alternative splicing including exon skipping, alternative 3′ and alternative 5′ splice site usage, alternative initiation and alternative termination.
In comparison with these extensive experimental studies, relatively little has been published on bioinformatics methodology for general detection and analysis of alternative splicing from microarray data. Wang et al. (15) describe a detailed method for quantitating distinct splice forms of a gene, and tested it both on a mixture of two isoforms, and a mixture of three isoforms. This method was designed for quantification of well-known isoforms, as the authors emphasized: ‘This algorithm is intended for splice variant typing, not discovery’. Johnson et al. (16) apparently analyzed their microarray data by fitting the probe intensities to a model of probe sensitivity, based on a single value representing total expression of the gene, and then identifying probes with strong ‘residuals’, indicating a poor fit to this model (16). Both the Wang et al. and Johnson et al. methods are based on constructing a sophisticated model of probe sensitivity and comparing this model to the actual probe behaviors. Our approach is somewhat different. It compares the behavior of probes for a gene in one tissue versus their behavior in other tissues (instead of to a model), and adopts a simpler method of detection designed for discovery of novel alternative splicing. The use of normalization versus tissue-averaged ‘pool’ intensities largely removes probe sensitivity and total gene expression from consideration, enabling our analysis to focus on distinguishing three qualitatively different cases: uncorrelated, random scatter (no evidence of alternative splicing); anti-correlation (the two samples differ in splicing); and correlation (the two sample have the same splicing, compared with other tissues that have different splicing). Computation of this correlation factor for all possible pairs of replicate arrays allows direct assessment of its statistical significance. This simple approach works for ab initio discovery of a wide variety of types of alternative splicing (not just exon skipping), and could be applied to many kinds of microarray designs and data.
One important foundation for the detection of complex phenomena such as alternative splicing is high-quality hybridization data displaying good specificity, reproducibility and signal-to-noise. Our data validate previous reports of the advantages of the Agilent array platform, which makes longer probe sequences possible (36–40 nt in this study). We wish to emphasize that all the data presented in this paper are raw microarray hybridization intensities directly reflecting the quality of the experimental data. Each data point shown in our figures is the signal from a single spot (on a single microarray), in contrast with common practices such as averaging up to four replicate arrays to suppress noise, or using data from up to 40 hybridization spots per array to obtain a single expression signal. The reproducibility of our data across four replicate arrays (with dye-swaps) indicates a good level of signal-to-noise, taking into account both variation between arrays and variation between different experiments and labeling. The reproducibility of our data for each gene across many different tissues shows that this level of signal-to-noise is also well above the level of variation between different experiments and samples. The use of longer probe sequences [60 nt total, including a specific probe sequence of 36–40 nt on top of a base of 20–24 bases of poly(T) to raise the probe sequence off the surface of the array] appears to work well for clear, reproducible detection of alternative splicing. The Agilent array platform's reproducibility (comparing a single spot between replicate arrays) and consistency (comparing the absolute intensities of many probes for an individual gene) provide a good foundation for detecting alternative splicing. The development of amplification and labeling methods that give robust coverage over the full length of each gene (as opposed to just the 3′ end) has also been crucial to reliable detection of tissue-specific alternative splicing (14,16).
Our approach has many deficiencies that need to be filled. For example, in this paper, we have de-emphasized quantification in favor of qualitative analysis, as a way of stressing the distinct character of alternative splicing when compared with increases or decreases in total gene expression. However, the next stage of analysis requires accurate estimation of the amounts of each distinct splice form. This is clearly more challenging than accurate estimation of the total amount of mRNA for a gene. Based on identification of the individual sets of probes that distinguish different splice forms, it is possible to measure the amounts of each splice form. For example, Wang et al. (15) have described a matrix-based method for estimating the amounts of distinct transcript forms given a set of individual probes that distinguish them.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We wish to thank Drs M. Kronick and J. Collins for helpful comments on this work, and Agilent for kindly providing microarrays. This work has been supported by NIH grant MH65166, DOE grant DEFG0387ER60615, The David and Lucille Packard Foundation and the Henry Singleton Brain Tumor Foundation. Microarray work was supported through the NINDS/NIMH Microarray Consortium site at UCLA.
REFERENCES
- 1.Butte A. (2002) The use and analysis of microarray data. Nature Rev. Drug Discov., 1, 951–960. [DOI] [PubMed] [Google Scholar]
- 2.Mironov A.A., Fickett,J.W. and Gelfand,M.S. (1999) Frequent alternative splicing of human genes. Genome Res., 9, 1288–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brett D., Hanke,J., Lehmann,G., Haase,S., Delbruck,S., Krueger,S., Reich,J. and Bork,P. (2000) EST comparison indicates 38% of human mRNAs contain possible alternative splice forms. FEBS Lett., 474, 83–86. [DOI] [PubMed] [Google Scholar]
- 4.Modrek B. and Lee,C. (2002) A genomic view of alternative splicing. Nature Genet., 30, 13–19. [DOI] [PubMed] [Google Scholar]
- 5.Maniatis T. and Tanis,B. (2002) Alternative pre-mRNA splicing and proteome expansion in metazoans. Nature, 418, 236–243. [DOI] [PubMed] [Google Scholar]
- 6.Kriventseva E.V., Koch,I., Apweiler,R., Vingron,M., Bork,P., Gelfand,M.S. and Sunyaev,S. (2003) Increase of functional diversity by alternative splicing. Trends Genet., 19, 124–128. [DOI] [PubMed] [Google Scholar]
- 7.Resch A., Xing,Y., Modrek,B., Gorlick,M., Riley,R. and Lee,C. (2004) Assessing the impact of alternative splicing on domain interactions in the human proteome. J. Proteome Res., 3, 76–83. [DOI] [PubMed] [Google Scholar]
- 8.Xing Y., Xu,Q. and Lee,C. (2003) Widespread production of novel soluble protein isoforms by alternative splicing removal of transmembrane anchoring domains. FEBS Lett., 555, 572–578. [DOI] [PubMed] [Google Scholar]
- 9.Lewis B.P., Green,R.E. and Brenner,S.E. (2003) Evidence for the widespread coupling of alternative splicing and nonsense-mediated mRNA decay in humans. Proc. Natl Acad. Sci. USA, 100, 189–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu G.K., Madore,S.J., Moldover,B., Jatkoe,T., Balaban,D., Thomas,J. and Wang,Y. (2001) Predicting splice variant from DNA chip expression data. Genome Res., 11, 1237–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clark T.A., Sugnet,C.W. and Ares,M.J. (2002) Genomewide analysis of mRNA processing in yeast using splicing-specific microarrays. Science, 296, 907–910. [DOI] [PubMed] [Google Scholar]
- 12.Kochiwa H., Suzuki,R., Washio,T., Saito,R., Bono,H., Carninci,P., Okazaki,Y., Miki,R., Hayashizaki,Y. and Tomita,M. (2002) Inferring alternative splicing patterns in mouse from a full-length cDNA library and microarray data. Genome Res., 12, 1286–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yeakley J.M., Fan,J.B., Doucet,D., Luo,L., Wickham,E., Ye,Z., Chee,M.S. and Fu,X.D. (2002) Profiling alternative splicing on fiber-optic arrays. Nat. Biotechnol., 20, 353–358. [DOI] [PubMed] [Google Scholar]
- 14.Castle J., Garrett-Engele,P., Armour,C.D., Duenwald,S.J., Loerch,P.M., Meyer,M.R., Schadt,E.E., Stoughton,R., Parrish,M.L., Shoemaker,D.D. et al. (2003) Optimization of oligonucleotide arrays and RNA amplification protocols for analysis of transcript structure and alternative splicing. Genome Biol., 4, R66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang H., Hubbell,E., Hu,J.S., Mei,G., Cline,M., Lu,G., Clark,T., Siani-Rose,M.A., Ares,M., Kulp,D.C. et al. (2003) Gene structure-based splice variant deconvolution using a microarray platform. Bioinformatics, 19 (Suppl. 1), i315–i322. [DOI] [PubMed] [Google Scholar]
- 16.Johnson J.M., Castle,J., Garrett-Engele,P., Kan,Z., Loerch,P.M., Armour,C.D., Santos,R., Schadt,E.E., Stoughton,R. and Shoemaker,D.D. (2003) Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science, 302, 2141–2144. [DOI] [PubMed] [Google Scholar]
- 17.Lipshutz R.J., Fodor,S.P., Gingeras,T.R. and Lockhart,D.J. (1999) High density synthetic oligonucleotide arrays. Nature Genet., 21, 20–24. [DOI] [PubMed] [Google Scholar]
- 18.Li C. and Wong,W.H. (2001) Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc. Natl Acad. Sci. USA, 98, 31–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kampa D., Cheng,J., Kapranov,P., Yamanaka,M., Brubaker,S., Cawley,S., Drenkow,J., Piccolboni,A., Bekiranov,S., Helt,G. et al. (2004) Novel RNAs identified from an in-depth analysis of the transcriptome of human chromosomes 21 and 22. Genome Res., 14, 331–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee C., Atanelov,L., Modrek,B. and Xing,Y. (2003) ASAP: the alternative splicing annotation project. Nucleic Acids Res., 31, 101–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Altschul S.F., Gish,W., Miller,W., Myers,E.W. and Lipman,D.J. (1990) Basic local alignment search tool. J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 22.Rice P., Longden,I. and Bleasby,A. (2000) EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet., 16, 276–277. [DOI] [PubMed] [Google Scholar]
- 23.Perry S.V. (2001) Vertebrate tropomyosin: distribution, properties and function. J. Muscle Res. Cell Motil., 22, 5–49. [DOI] [PubMed] [Google Scholar]
- 24.Hoffer M.J., van Eck,M.M., Havekes,L.M., Hofker,M.H. and Frants,R.R. (1993) Structure and expression of the mouse apolipoprotein C2 gene. Genomics, 17, 45–51. [DOI] [PubMed] [Google Scholar]
- 25.BDBiosciences (2003) BD SMART mRNA amplification kit. Clontechniques, XVIII, No. 4, 8–9. [Google Scholar]
- 26.Kapranov P., Cawley,S.E., Drenkow,J., Bekiranov,S., Strausberg,R.L., Fodor,S.P. and Gingeras,T.R. (2002) Large-scale transcriptional activity in chromosomes 21 and 22. Science, 296, 916–919. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.