Abstract
Background:
Around 95% of the world’s population are infected with the Epstein-Barr virus (EBV), which can persist latent in B lymphocytes and epithelial cells life-long. EBV has been linked with lymphoid and epithelial cancers and persistence of EBV infection in lymphoid or epithelial cells may result in virus-associated B-cell tumors or nasopharyngeal carcinomas (NPC). This study was conducted to determine the frequency of EBV DNA in nasopharyngeal carcinoma tissue of Iranian patients.
Materials and methods:
A total of 50 blocks of formalin-fixed paraffin-embedded tissue of NPCs from 38 (76 %) male and 12 (24%) female patients were collected from archives of Ahvaz hospitals. Sections were cut at 5 µm and DNA was extracted for detection of EBV DNA and EBV typing by mested PCR. DNA sequencing was performed to confirm PCR results. The distribution of EBV DNA was compared among WHO histological subtypes of NPC.
Results:
Some 3 female and 11 (22%) male NPC samples showed positive for EBV DNA type 1, 2/14(22.2%)WHO histological type II and 12/41(29.3%) WHO histological type III.
Conclusions:
The frequency of EBV DNA among NPCs in Iranian patients was found to be 28%, EBV type I predominating. Both WHO histological type II and III NPC subtypes demonstrated approximately the same detection prevalence.
Keywords: Epstein-Bar (EBV), nasopharyngeal carcinoma (NCP), nested PCR
Introduction
EBV is a widespread and infects about 95% of the world population. EBV can remain latent in B lymphocytes and epithelial cells and may implicate in pathogenesis of various lymphoid and epithelial malignancies, such as Burkitt’s lymphoma, Hodgkin’s lymphoma, nasopharyngeal carcinoma (NPC), gastric carcinomas T cell lymphomas, post-transplant lymphoproliferative disease (Fish et al; 2014; Bieging et al., 2009; Young et al., 2004 Sharma et al., 2011; Pathmanathan et al., 1995). Epstein - Barr virus (EBV) is a member of family Herpesviridae and subfamily Gamma Herpesvirus. EBV genome is a double-stranded DNA with 184 Kilobase pairs(Kbp). Based on DNA sequence divergence in the EBNA-2 and EBNA -3 regions, the EBVs have been classified into type I and Type II (Aitken et al.,1994). Type 1 is prevalent more in china while type 2 are common in Africa and New Guinea (Sharma et al.,2011; Khabir et al.,2005). EBV is transmitted via saliva, kissing, sharing food and drinks with someone who has infectious mononucleosis. EBV has two infectivity cycles latency, and lytic (Sharma et al.,2011; Mirzamani et al., 2006; Markus et al.,2012). EBV DNA can persist in episomal form in the memory B-cells, and establishes latent infection by expression of EBV-nuclear antigen 1 (EBNA1) gene (Dawson et al., 2012).
Based on different EBV-associated malignancies several forms of EBV latency genes are expressed (Temple et al., 2014; Tsao et al., 2012). In some epithelial tumors such as NPC EBV express latent membrane proteins, LMP1, LMP2A and LMP2B (Pathmanathan et al., 1995; Parikhit et al., 2016). LMP-1 is a major oncogene which activates NF-κB, JNK and p38 pathways and is critical for EBV-mediated B-cell transformation and induces Epithelial-Mesenchymal Transition (EMT) in tumor cells of NPC (Mancao et al., 2005; Izumi et al., 1999; Borthakur et al.,2016;). In epithelial cells, LMP2A is phosphorylated by csk and LMP2A constitutively activates PI3 kinase and the Akt kinase in the absence of integrin signaling. This partial substitution of integrin signaling by LMP2A leads to cell transformation and anchorage-independent growth of the HaCat keratinocyte cell line (Scholle F et al;Allen et al., 2005; Chen et al., 2005; Iwakiri 2013). LMP-2B impedes LMP-2A function and increases lytic activation of EBV(Rechsteiner et al., 2008; Rovedo et al., 2007). EBNA1 is the only protein expressed in all EBV-associated tumors and is critical for maintenance, replication and transcription of the EBV genome in latency (Wang et al., 2002). NPC is a unique type of head and neck squamous cell carcinoma (HNSCC), highly metastatic disease and has an uncommon malignancy in most populations of the world, with an incidence rate of one case per 100, 000 person-year (Wang et al., 2013). The incidence rate of NPC in China have been reported 2.8/100 000 in men and 1.9/100,000 in women per year (Su-Mei et al., 2011). The incidence rate of NPC have been reported 0.33 per 100000 person-year in Iran(26 Safavi-Naini et al., 2015). Several sensitive methods including in situ hybridization (ISH), Southern blotting and PCR (Tsuchiya et al., 2002), RNA-ISH (RISH), EBERs detection (IARC., 1997) and PCR have been used for detection of EBV genome in cancer tissue (Rocío et al., 2006; Bao-Lin et al., 2012). Currently Quantification of EBV DNA load have been used in early detection of EBV in patients with nasopharyngeal cancer(Xiao-Hui et al., 2015). Since the detection of EBV has not been studied in patients with NCP in Ahvaz city, thus this study was conducted to determine the frequency of EBVDNA type1 and 2 in patients samples with nasopharyngeal carcinoma Ahvaz city hospitals. Ahvaz city is capital of Khozestan province with 2 million population located in the south west region of Iran.
Materials and Methods
50 blocks formalin-fixed paraffin-embedded tissue of patients with NPC were collected from archive of Ahvaz city hospitals during 2005 to 2015. The diagnostic accuracy of nasopharyngeal carcinoma (NPC) was confirmed by a pathologist. The sections of 5 μm thickness were prepared from each sample.
Deparaffinized tissue samples
Deparaffinization was done with xylene and ethanol (Germany, Merk). Initially, all the fragments specimens were placed in Microtubes; then, xylene was added and kept at 45C° for 15 min followed by centrifuge at 14,000rpm. This stage was repeated again. Then, the supernatant was discarded and 1ml absolute ethanol was added to precipitate and stored at the room temperature for 10 min and centrifuged at 14,000rpm for one minute. The supernatant was discarded. This process was repeated by adding 70% ethanol, same as the previous steps. Finally, supernatant was discarded and Microtubes were placed at 65C° for 5 min to vaporize the ethanol residue and the pellet was collected and used in DNA extraction (Habibian et al.,2013).
DNA extraction
High pure PCR template preparation kit (Roche, Germany) was used for extraction of DNA, according to the manufacturer’s instruction. The extracted DNA was stored at -70C°until PCR amplification.
Nested PCR
Based on detection of EBNA-2 region of EBV genome, the typing of EBV is determined (Rocío.,2006). The nested-PCR was performed in two rounds, for first round the following primers, EBNA-2F:TGGAAACCCGTCACTCTC(48572-89) and anti - sense EBNA-2I TAATGGCATAGGTGGAATG(49355-73) was used. 25μl reaction mixture, containing 7μl of the extracted DNA, 2.5μl PCR buffer 10X (Roche), 0.5 μl deoxynucleotide triphosphate 10nM (Roche), 1U Taq Polymerase (Roche), 20μM of each primer and distilled water up to 25µl was subjected to thermocycler(Techne TC-5000,UK) and programmed for 1cycle : 94C° for 5 min followed by 35 cycles: 94C° for 45 s, 50.5C° for 45 s, 72C° for 45 s and final extension 72C° for 10 min. The second round, 4μl of the first round PCR product, with forward primers EBNA-2C, AGGGATGCCTGGACACAAGA (48810-29) and anti sense EBNA- 2G, GCCTCGGTTGTGACAGAG (49048-65), was used for EBV type1 detection. For type 2 determination EBNA-2B anti sense TTGAAGAGTATGTCCTAAGG was used. Following thermal condition was programed : 1 cycle, 94C° for 5 min: followed by 35 cycles:94C° for 45 s, 53.5C° for 45 s, 72C° for 45 s and final extension 72C° for 10 min (Rocío et al., 2006).
Gel electrophoresis
The final PCR products were subjected to electrophoresis in 2% agarose gel, stained with safe stain and visualized under ultraviolet transilluminator. Cell line B95.8 was used as a positive control to detect EBV(29 Rocío et al., 2006). Expected PCR product for EBV type 1 was 250 bp and for EBV type2 (300 bp) (2 Rocío et al., 2006).
Sequencing
To determine EBV genotyping the PCR products of 14 positive samples were sequenced (ABI, Bioneer company, South Korea) and blasted using NCBI and EBV database.
Statistics
The variable such as mean, standard deviation, were analyzed using SPSS version 20, the frequency of EBVDNA among the males and in WHO Histology Types were analyzed by chi-square and Fisher`s exact tests.
Results
38/50(76%) of patients with Nasopharyngeal Carcinoma (NPC) were males and 12/50(24%) females. High Frequency of 41/50(82%) NPC were observed among the patients > 50 years while lower cases of 9/50(18%) NPC were found among the patients <50 years (Table 1). Among 50 NPC cases 14 (28%) showed positive for EBVDNA including 3(6%)females and 11(22%) males(OR= 1.222, 95% CI = 0.277 -- 5.384, p = 0.791). All the detected positive EBVDNA were belonged to EBV type1, but EBV type 2 was detected among the patients with NPC. Fig. 1 shows the results of EBVDNA by PCR. 3/50 (6%) of females and 11/50 (22%) of males samples NPC samples showed positive for EBVDNA type1. EBVDNA was found in 2/14(22.22%)WHO histological type II while 12/41(29.26%) of WHO histological type III showed positive for EBVDNA. (OR= 0.690, 95% CI= 0.125-- 3.815, p = 0.670). The EBVDNA was not found in WHO histological type 1 (Table 2).
Table 1.
Characteristics of Patients with Nasopharyngeal Carcinoma
| Age group | Gender | Total | |
|---|---|---|---|
| Female | Male | ||
| <50 | 3(6%) | 6(12%) | 9(18%) |
| >50 | 9(18%) | 32(64%) | 41(82%) |
| Total | 12(24%) | 38(76%) | |
Table 1 shows 82% of NPC patients were above 50 years while 18% were below 50 years old
Figure 1.
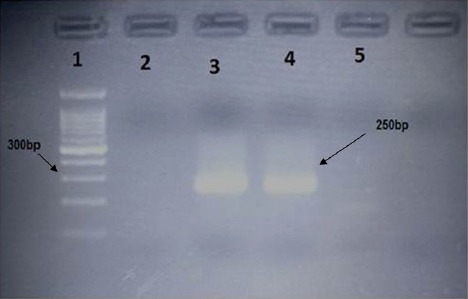
Shows the Results of EBNA2 Amplification Lane 1:100-bp size marker; Lane 2: Negative control; Lane 3:Positive control (positive band 250 bp (B95.8 cell line);Lanes 4-: positive EBV DNA sample.
Table 2.
Distribution of EBVDNA among Gender and WHO Histology Type.
| Feature | Positive | Negative | P value |
|---|---|---|---|
| n=50 | |||
| Sex | |||
| Male = 38 | 11 (28.9%) | 27 (71.5%) | 0.791 |
| Female= 12 | 3 (25.0%) | 9 (75.0%) | |
| WHO Histology Type | |||
| WHO Type II =9 | 2 (22.2%) | 7 (77.8%) | 0.670 |
| WHO Type III =41 | 12 (29.3%) | 29 (70.7%) |
The distribution of EBV DNA among males, females, (p=791), and in WHO histology type II, III (0.671).
Discussion
Appropriate diagnosis of NPC is crucial for patients’ survival. The association of viral infection with nasopharyngeal cancer is important strategy for future treatment and prevention. The role of EBV in NPC is most prominent and well documented (Pathmanathan et al., 1995;Mancao et al., 2005; Izumi et al., 1999; Borthakur et al.,2016; Fish et al; 2014; Bieging et al., 2009; Allen et al., 2005; Chen et al., 2005; Iwakiri et al.,2013; Parikhit et al., 2016). The detection of EBVDNA have been reported in NPC (Mirzamani., 2006; Hassan., 2006). In our study, 38(76 %) males and 12(24%) were females patients with NPC which inconsistent with Safavi-Naini et al finding in Iran (Safavi-Naini et al.,2015). With regard to a total collected 50 NPC samples during 10 years (2005-2015), it seems low rate of nasopharyngeal carcinoma exist in our region (Safavi-Naini et al.,2015). In our study most of patients with NPC were above 50 years which is consistent with Wang et al finding (2002). Based on the EBNA2, EBNA-3a, and EBNA-3c latent genes, EBV has been classified into two major strains, which are called as EBV type 1 (EBV-1) and type 2 (EBV-2) (Rowe et al., 1989;Cancian et al., 2011; Lucchesi et al., 2008;Sample et al., 1990;Rocío et al., 2006; Dolan et al., 2006). The two strains not only differ in their genotypes, but also they have functional differences in their transforming capacities. EBV-1 easily transforms B cells in culture, leading to immortalized lymphoblastoid cell lines (LCL), while EBV-2 unable to transform B cells (Dolan et al., 2006; Rowe et al., 1989). Type EBV type 1 is commonly associated with NPC tumor worldwide(Klumb et al.,2004; Bao-Lin et al., 2012; Xiao-Hui et al., 2015; Habibian et al.,2013; Yap et al., 2007; Kwok et al., 2014; Parviz et al., 2014) while EBV type 2 can infect and persist latent in CD8+ T cells(Coleman et al., 2015). In our study, EBVDNA type 1 was only detected in NPC tumor and is consistent with other findings (Habibian et al., 2013; Bao-Lin et al., 2012; Xiao-Hui et al., 2015). In our survey all the NPC samples were negative for EBV DNA type 2 strain. The expression of EBNA-1, LMP -1 and LMP-2 proteins’ have been exhibited to have an important role in NPC (Pathmanathan et al., 1995; Mancao et al., 2005; Izumi et al., 1999; Borthakur et al.,2016; Fish et al; 2014; Bieging et al., 2009; Allen et al., 2005; Chen et al., 2005; Iwakiri et al., 2013; Parikhit et al., 2016). In our study the expressions of EBNA-1, and LMP -1 and LMP-2 were not studied but requires further investigation. EBV infection commonly detected in NPC tumors with WHO histology type II and III subtype NPC (Chua et al 2003; Pakkirmasthan et al., 2015). In our study EBVDNA was found in 2 WHO histological type II while 12 EBV DNA were detected in WHO histological type III NPC subtypes. The prevalence of EBVDNA among 3(6 %) female and 11(22%) male patients with NPC was not found significant (P=0.791).
NPC is a complex disease and several factors such as environmental, genetic susceptibility, consumption of salted fish, preserved vegetables, cigarette smoking and family history may contribute to development of NPC(Ruan et al., 2010; Jin et al., 2016).
The concentration of plasma EBV DNA would be able to reflect the tumor load in patients with NPC (Lin et al., 2004; Chai et al.,2012). Following surgical resection of NPC tumor has been cleared EBV DNA in plasma within short time(Chan et al.,2008), but treated NPC patients by radiotherapy/chemoradiotherapy positive for EBV DNA resulted in development of metastasis (Leung et al., 2014). The recurrence of NPC positive for EBVDNA was described in patients with post-treatment EBV (Chan et al.,2002).
In the light of mentioned data to improve treatment and management, the screening of EBV DNA should be implemented for patients with NPC.
In summary, the frequency of EBVDNA among the patients with NPC was 14 (28%) cases. EBV type I is dominant in NPC tumors. EBVDNA was found in 2/9 (22.22%)WHO histological type II while 12/41 (29.26%) of WHO histological type III showed positive for EBVDNA. To improve treatment and management the screening of EBV DNA should be implemented for plasma or biopsy of patient with NPC before chemotrophy.
Acknowledgements
This study was done as a research project with 92143 registration number in Health Research Institute, Infectious and Tropical Disease Research Center, Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran. Our great appreciate goes to NiloofarNeisi to provide positive control for this study.
Authors’ Contributions
Study concept and design:ManoochehrMakvandi,Ali Teimoori, Nader saki, Soheila Nikakhlagh and Nastran Ranjbar;acquisitionof data:Toran Shahani;analysis and interpretation of data: ManoochehrMakvandi and Toran shahani; drafting of the manuscript: Toran Shahani, Zeinab Hosseini and NiloofarNeisi;critical revision of the manuscript for portant intellectual content: ManoochehrMakvandi;statisticalanalysis:ManoochehrMakvandi and Toran SHahani and Zeinab Hosseini; Administrative, technical, and material support:Toran Shahani,NiloofarNeisi,Ali Teimoori, Samira Pourrezaei,Zeinab Hosseini, Hashem Radmehr, AbdolnabiShabani, HamidehMehravaran,Azadeh haghi and,HadisKiani,; Study supervision:ManoochehrMakvandi and Ali Teimoori.
Funding/Support
This studywas supported byHealth Research Institute, infectious and Tropical diseaseResearch Center, Ahvaz Jundishapur Universityof Medical Sciences, Ahvaz, Iran.
Financial Disclosures
Authors have no financial interest related to the material in the manuscript.
References
- Aitken C, Sengupta S, Aedes C, et al. Heterogeneity within the Epstein-Barr virus nuclear antigen 2 gene in different strains of Epstein-Barr virus. J Gen Virol. 1994;75:95–100. doi: 10.1099/0022-1317-75-1-95. [DOI] [PubMed] [Google Scholar]
- Allen MD, Young LS, Dawson CW. The Epstein-Barr virus-encoded LMP2A and LMP2B proteins promote epithelial cell spreading and motility. J Virol. 2005;79:1789–1802. doi: 10.1128/JVI.79.3.1789-1802.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao-Lin H, Xiang-Ying X, Chun-Zhi Z, et al. Systematic review on Epstein-Barr virus (EBV) DNA in diagnosis of nasopharyngeal carcinoma in Asian populations. Asian Pacific J Cancer Prev. 2012;13:2577–81. doi: 10.7314/apjcp.2012.13.6.2577. [DOI] [PubMed] [Google Scholar]
- Bieging KT, Amick AC, Longnecker R, et al. Epstein–Barr virus LMP2A bypasses p53 inactivation in a MYC model of lymphomagenesis. Proc Natl Acad Sci USA. 2009;106:17945–50. doi: 10.1073/pnas.0907994106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borthakur P, Kataki K, Keppen C, et al. Expression of Epstein Barr virus encoded EBNA1 and LMP1 oncoproteins in nasopharyngeal carcinomas from northeast India. Asian Pac J Cancer Prev. 2016;17:3411–6. [PubMed] [Google Scholar]
- Cancian L, Bosshard R, Lucchesi W, et al. C-Terminal region of EBNA-2 determines the superior transforming ability of type 1 Epstein-Barr virus by enhanced gene regulation of LMP-1 and CXCR7. PLoS Pathog. 2011;7:e1002164. doi: 10.1371/journal.ppat.1002164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai SJ, Pua KC, Saleh A, et al. Clinical significance of plasma Epstein-Barr Virus DNA loads in a large cohort of Malaysian patients with nasopharyngeal carcinoma. J Clin Virol. 2012;55:34–9. doi: 10.1016/j.jcv.2012.05.017. [DOI] [PubMed] [Google Scholar]
- Chan KC, Leung SF, Yeung SW, et al. Quantitative analysis of the transrenal excretion of circulating EBV DNA in nasopharyngeal carcinoma patients. Clin Cancer Res. 2008;14:4809–13. doi: 10.1158/1078-0432.CCR-08-1112. [DOI] [PubMed] [Google Scholar]
- Chan AT, Lo YM, Zee B, et al. Plasma Epstein-Barr virus DNA and residual disease after radiotherapy for undifferentiated nasopharyngeal carcinoma. J Natl Cancer Inst. 2002;94:1614–9. doi: 10.1093/jnci/94.21.1614. [DOI] [PubMed] [Google Scholar]
- Chen F, Liu C, Lindvall C. Epstein-Barr virus latent membrane 2A (LMP2A) down-regulates telomerase reverse transcriptase (hTERT) in epithelial cell lines. Int J Cancer. 2005;113:284–89. doi: 10.1002/ijc.20594. [DOI] [PubMed] [Google Scholar]
- Chua DT, Sham JS, Kwong DL, et al. Treatment outcome after radiotherapy alone for patients with Stage I-II nasopharyngeal carcinoma. Cancer. 2003;98:74–80. doi: 10.1002/cncr.11485. [DOI] [PubMed] [Google Scholar]
- Coleman CB, Wohlford EM, Smith NA, et al. Epstein-Barr virus type 2 latently infects T cells, inducing an atypical activation characterized by expression of lymphotactic, cytokines. J Virol. 2015;89:2301–12. doi: 10.1128/JVI.03001-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson CW, Port RJ, Young LS. The role of the EBV-encoded latent membrane proteins LMP1 and LMP2 in thepathogenesis of nasopharyngeal carcinoma (NPC), Semin. Cancer Biol. 2012;22:144–53. doi: 10.1016/j.semcancer.2012.01.004. [DOI] [PubMed] [Google Scholar]
- Dolan A, Addison C, Gatherer D, et al. The genome of Epstein-Barr virus type 2 strain AG876. J Virol. 2006;350:164–70. doi: 10.1016/j.virol.2006.01.015. [DOI] [PubMed] [Google Scholar]
- Fish K, Chen J, Longnecker R. Epstein-Barr virus latent membrane protein 2A enhances MYC-driven cell cycle progression in a mouse model of B lymphoma. Blood. 2014;123:530–40. doi: 10.1182/blood-2013-07-517649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habibian A, Makvandi M, Samarbafzadeh A, et al. Epstein-Barr Virus DNA frequency in paraffin embedded tissues of Non-Hodgkin lymphoma patients from Ahvaz, Iran. Jundishapur J Health Res. 2013;4:315–20. [Google Scholar]
- IARC. Working group on the evaluation of carcinogenic risks to humans: Epstein-Barr virus and Kaposi’s sarcoma herpesvirus/human herpesvirus 8 lyon, international agency for research on cancer. 1997:47–373. [Google Scholar]
- Izumi KM, Cahir McFarl ED, Riley EA, et al. The residues between the two transformation effector sites of Epstein–Barr virus latent membrane protein 1 are not critical for B-lymphocyte growth transformation. J Virol. 1999;73:9908–16. doi: 10.1128/jvi.73.12.9908-9916.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwakiri D, Minamitani T, Samanta M. Epstein–Barr virus latent membrane protein 2A contributes to anoikis resistance through ERK activation. J Virol. 2013;87:8227–34. doi: 10.1128/JVI.01089-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin-Xin B, Xiao-Yu Z, Wen-Sheng L, et al. Genetic susceptibility to the endemic form of NPC. Chin Clin Oncol. 2016;5:15. doi: 10.21037/cco.2016.03.11. [DOI] [PubMed] [Google Scholar]
- Khabir A, Karray H, Rodriguez S, et al. EBV latent membrane protein 1 abundance correlates with patient age but not with metastatic behavior in North African nasopharyngeal carcinomas. J Virol. 2005;2:39. doi: 10.1186/1743-422X-2-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klumb CE, Hassan R, Oliveria DE, et al. Geographic Variation in Epstein-Barr virus-associated Burkitt’s lymphoma in children from Brazil. Int J Cancer. 2004;108:66–70. doi: 10.1002/ijc.11443. [DOI] [PubMed] [Google Scholar]
- Kwok H, Wu CW, Palser AL, et al. Genomic diversity of Epstein-Barr virus genomes isolated from primary nasopharyngeal carcinoma biopsy samples. J Virol. 2014;88:10662–72. doi: 10.1128/JVI.01665-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung SF, Chan KC, Ma BB, et al. Plasma Epstein-Barr viral DNA load at midpoint of radiotherapy course predicts outcome in advanced-stage nasopharyngeal carcinoma. Ann Oncol. 2014;25:1204–8. doi: 10.1093/annonc/mdu117. [DOI] [PubMed] [Google Scholar]
- Lin JC, Wang WY, Chen KY, et al. Quantification of plasma Epstein-Barr virus DNA in patients with advanced nasopharyngeal carcinoma. N Engl J Med. 2004;350:2461–70. doi: 10.1056/NEJMoa032260. [DOI] [PubMed] [Google Scholar]
- Lucchesi W, Brady G, Dittrich-Breiholz O, et al. Differential gene regulation by Epstein-Barr virus type 1 and type 2 EBNA2. J Virol. 2008;82:7456–66. doi: 10.1128/JVI.00223-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancao C, Altmann M, Jungnickel B, et al. Rescue of ‘crippled’ germinal center B cells from apoptosis by Epstein–Barr virus. Blood. 2005;106:4339–44. doi: 10.1182/blood-2005-06-2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markus K, Christine G, Wolfgang H. The lytic phase of Epstein-Barr virus requires a viral genome with 5-Methylcytosine residues in CpG sites. J Virol. 2012;86:447–58. doi: 10.1128/JVI.06314-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirzamani N, Salehian P, Farhadi M, et al. Detection of EBV and HPV in nasopharyngeal carcinoma by in situ hybridization. Exp Mol Pathol. 2006;81:231–34. doi: 10.1016/j.yexmp.2006.04.006. [DOI] [PubMed] [Google Scholar]
- Pakkirmasthan A, Kurakula S. Nasopharyngeal cancers: a retrospective comparative analysis of radiotherapy alone versus chemo-radiation (Benghazi experience) Indian J Cancer. 2015;52:391–5. doi: 10.4103/0019-509X.176718. [DOI] [PubMed] [Google Scholar]
- Parikhit B, Kangkana K, Chenole K, et al. Expression of Epstein Barr virus encoded EBNA1 and LMP1 oncoproteins in nasopharyngeal carcinomas from northeast India. Asian Pac J Cancer Prev. 2016;17:3411–41. [PubMed] [Google Scholar]
- Deyhimi P, Kalantari M. Study of Epstein-Barr virus expression in Burkitt’s lymphoma by polymerase chain reaction and in situ hybridization: A study in Iran. Dent Res J (Isfahan) 2014;11:380–85. doi: 10.4103/1735-3327.135917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathmanathan R, Prasad U, Sadler F, et al. Clonal proliferations of cells infected with Epstein-Barr virus in preinvasive lesions related to nasopharyngeal carcinoma. N Engl J Med. 1995;333:693–98. doi: 10.1056/NEJM199509143331103. [DOI] [PubMed] [Google Scholar]
- Ruan HL, Qin HD, Shugart YY, et al. Developing genetic epidemiological models to predict risk for nasopharyngeal carcinoma in high-risk population of China. PLoS One. 2013;8:e56128. doi: 10.1371/journal.pone.0056128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rechsteiner MP, Berger C, Zauner L, et al. Latent membrane protein 2B regulates susceptibility to induction of lytic Epstein-Barr virus infection. J Virol. 2008;82:1739–47. doi: 10.1128/JVI.01723-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocío H, Lídia RW, Claudio GS, et al. Epstein-Barr Virus (EBV) detection and typing by PCR: a contribution to diagnostic screening of EBV-positive Burkitt’slymphoma. Diag Pathol. 2006;1:17. doi: 10.1186/1746-1596-1-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rovedo M, Longnecker R. Epstein–Barr virus latent membrane protein 2B (LMP2B) modulates LMP2A activity. J Virol. 2007;81:84–94. doi: 10.1128/JVI.01302-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe M, Young LS, Cadwallader K, et al. Distinction between Epstein-Barr virus type A (EBNA 2A) and type B (EBNA 2B) isolates extends to the EBNA 3 family of nuclear proteins. J Virol. 1989;63:1031–39. doi: 10.1128/jvi.63.3.1031-1039.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safavi-Naini A, Raad N, Ghorbani J, et al. Incidence trends and geographical distribution of nasopharyngeal carcinoma in Iran. Iran J Cancer Prev. 2015;8:24–8. [PMC free article] [PubMed] [Google Scholar]
- Sample J, Young L, Martin B, et al. Epstein-Barr virus types 1 and 2 differ in their EBNA-3A, EBNA-3B, and EBNA-3C genes. J Virol. 1990;64:4084–92. doi: 10.1128/jvi.64.9.4084-4092.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma TD, Singh TT, Laishram RS, et al. Nasopharyngeal carcinoma-a clinico-pathological study in a regional cancer centre of northeastern India. Asian Pac J Cancer Prev. 2011;12:1583–7. [PubMed] [Google Scholar]
- Scholle F, Bendt KM, Raab-Traub N. Epstein-Barr virus LMP2A transforms epithelial cells, inhibits cell differentiation, and activates Akt. J Virol. 2000;74:10681–89. doi: 10.1128/jvi.74.22.10681-10689.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su-Mei C, Malcolm JS, Chao-Nan Q. The prevalence and prevention of nasopharyngeal carcinoma in China. Chin J Cancer. 2011;30:114–19. doi: 10.5732/cjc.010.10377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temple RM, Zhu J, Budgeon L, et al. Efficient replication of Epstein-Barr virus in stratified epithelium in vitro. Proc Natl Acad Sci U S A. 2014;111:16544–49. doi: 10.1073/pnas.1400818111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya S. Diagnosis of Epstein-Barr virus-associated diseases. Crit Rev Oncol/Hematol. 2002;44:227–38. doi: 10.1016/s1040-8428(02)00114-2. [DOI] [PubMed] [Google Scholar]
- Tsao SW, Tsang CM, Pang PS, et al. The biology of EBV infection in human epithelial cells. Semin Cancer Biol. 2012;22:137–43. doi: 10.1016/j.semcancer.2012.02.004. [DOI] [PubMed] [Google Scholar]
- Wang WY, Chien YC, Jan JS, et al. Consistent sequence variation of Epstein-Barr virus nuclear antigen 1 in primary tumor and peripheral blood cells of patients with nasopharyngeal carcinoma. Clin Cancer Res. 2002;8:2586–90. [PubMed] [Google Scholar]
- Wang Y, Zhang Y, Ma S. Racial differences in nasopharyngeal carcinoma in the United States. Cancer Epidemiol. 2013;37:793–802. doi: 10.1016/j.canep.2013.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao-Hui Z, Li-Xia L, Xi-Zhao L, et al. Quantification of Epstein–Barr virus DNA load in nasopharyngeal brushing samples in the diagnosis of nasopharyngeal carcinoma in southern. China Cancer Sci. 2015;106:1196–1201. doi: 10.1111/cas.12718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap YY, Hassan S, Chan M, et al. Epstein-Barr virus DNA detection in the diagnosis of nasopharyngeal carcinoma. Otolaryngol Head Neck Surg. 2007;136:986–91. doi: 10.1016/j.otohns.2006.11.027. [DOI] [PubMed] [Google Scholar]
- Young LS, Rickinson AB. Epstein-Barr virus: 40 years on. Nat Rev Cancer. 2004;4:757–68. doi: 10.1038/nrc1452. [DOI] [PubMed] [Google Scholar]


