Abstract
Single-stranded DNA or RNA libraries used in SELEX experiments usually include primer-annealing sequences for PCR amplification. In genomic SELEX, these fixed sequences may form base pairs with the central genomic fragments and interfere with the binding of target molecules to the genomic sequences. In this study, a method has been developed to circumvent these artificial effects. Primer-annealing sequences are removed from the genomic library before selection with the target protein and are then regenerated to allow amplification of the selected genomic fragments. A key step in the regeneration of primer-annealing sequences is to employ thermal cycles of hybridization-extension, using the sequences from unselected pools as templates. The genomic library was derived from the bacteriophage fd, and the gene 5 protein (g5p) from the phage was used as a target protein. After four rounds of primer-free genomic SELEX, most cloned sequences overlapped at a segment within gene 6 of the viral genome. This sequence segment was pyrimidine-rich and contained no stable secondary structures. Compared with a neighboring genomic fragment, a representative sequence from the family of selected sequences had about 23-fold higher g5p-binding affinity. Results from primer-free genomic SELEX were compared with the results from two other genomic SELEX protocols.
INTRODUCTION
The SELEX (Systematic Evolution of Ligands by EXponential enrichment), or in vitro selection, technique was originally developed to identify nucleic acid sequences or structures, termed aptamers, that bind tightly to target molecules (1,2). The procedure mimics natural evolution by employing an iterative process of selection and amplification of sequences from a combinatorial nucleic acid library. The library is usually created by chemical synthesis with a central random sequence region, which defines the scope and complexity of the library, and two flanking constant regions for PCR amplification. Results from SELEX generally are sequences having common motifs, and SELEX using double-stranded DNA (dsDNA) libraries is particularly useful for defining consensus sequences for dsDNA binding proteins (3). In contrast, libraries of single-stranded DNA (ssDNA) or RNA, which can fold into tertiary structures, can be used to select sequences that bind with high affinity and specificity to a variety of targets, including small molecules and proteins that are not thought of as nucleic acid binders (4–7). Therefore, aptamers can be used like antibodies for different applications, such as inhibition of enzymatic activity, protein detection and purification, and diagnostics (8–10). Catalytic RNA aptamers bearing specific activity, or ribozymes, can also be selected (11), and several types of deoxyribozymes (DNA ribozymes) have been identified (12), even though they have not been found in nature.
In a living cell, a protein may bind tightly and specifically to a site on the genome or to RNA transcripts to perform its functions. One way to identify such sequences for potential protein–nucleic acid interactions is by means of genomic SELEX (13). In genomic SELEX, the central random region of the library is substituted by DNA fragments derived from the whole genome of interest, and, if desired, the library can be transcribed to RNA for RNA SELEX selection with target proteins. Genomic RNA SELEX has been performed with bacteriophage MS2 coat protein (14) and Drosophila pre-mRNA splicing factor B52 (15). A similar procedure has been applied to identify ribosomal RNA fragments that bind to specific ribosomal proteins (16).
A problem with genomic SELEX is that the primer-annealing sequences of the library may base-pair with the central genomic fragments to form structures that are selected as sites for target binding (14). Interactions involving non-genomic, primer-binding sequences greatly limit the significance of results from genomic SELEX. To address this problem, primer-annealing sequences can be blocked by complementary oligonucleotides or switched to different sequences midway during the rounds of SELEX, as demonstrated by Gold and co-workers (14). In addition, Vater et al. (17) have shown that the primer-annealing sequences on the 5′ and 3′ ends can be trimmed to only 6 and 4 nt, respectively. However, non-naturally occurring sequences are still present in the library during selection with these approaches.
In this study, a method, termed primer-free genomic SELEX, has been developed to efficiently solve the problems caused by primer-annealing sequences. Under this experimental design, the primer-annealing sequences are completely removed from the library before selection with the target protein and are then regenerated to allow amplification of the selected genomic fragments. We chose the bacteriophage Ff gene 5 protein (g5p) to test this idea, because this protein has previously been used for regular SELEX selection of ssDNA sequences from a random library (18,19), and the size of the Ff genome is relatively small (6408 nt for fd and 6407 nt for f1 and M13), such that the genomic library is of minimal complexity for the development of this method.
The genome of the Ff viruses is a circular ssDNA comprising 11 tightly packed and overlapping genes and a 508 nt intergenic region (IG) (20,21). The IG contains five functional hairpin structures, denoted hairpins A–E (22), which include some regulatory elements, such as the morphogenetic signal for packaging (hairpin A), the complementary strand origin (involving hairpins B and C) and the initiation site for viral strand synthesis (on hairpin D) (23).
M13, f1 and fd of the Ff viruses encode an identical g5p (20). The role of g5p in viral DNA replication is to cooperatively bind to and thus sequester nascent ssDNA genomes from serving as templates for complementary DNA strand synthesis, and to package the genomes into complexes prior to assembly of the DNA into virions (24). Therefore, the g5p is usually considered to be a non-sequence-specific DNA binding protein. In addition to morphogenetic functions, the g5p has been shown to regulate mRNA translation of viral genes 1, 2, 3, 5 and 10 (25–28). Leader sequences on the 5′ end untranslated regions of the mRNAs are required for the regulation (25,26). It is most likely that the regulation is through g5p binding to the leader sequences to block the mRNA from being translated by ribosomes. The leader sequence from the gene 2 mRNA, and its DNA analog, forms G-quadruplex structures in vitro, and the structured forms are preferred over unstructured ones for g5p binding (29,30).
Therefore, it is likely that some regions in the viral genome or RNA transcripts are more preferred for g5p binding, and this binding is related to biological functions. Results from previous regular SELEX showed that the selected sequences (from a random library) folded to form G-quadruplex and hairpin structures (18,19,31), but were not homologous to those of the viral genome. Here, by using primer-free genomic SELEX, we identified an unexpected major fd genomic site that showed a higher affinity for g5p than did a neighboring sequence.
MATERIALS AND METHODS
DNA sequences
The DNA sequences used in this report were purchased from Midland (Midland, TX) and desalted using gel filtration chromatography without further purification, unless otherwise noted. The sequences were 5′P [5′ end primer, d(GTGATCATGCACGAGA)], 5′P-N9 [5′ end primer for library construction, d(GTGATCATGCACGAGA-N9), where N9 stands for random incorporation of nine A, G, C or T nucleotides at the 3′ end during synthesis], 5′P-Fok [d(GTGATCATGCggatGA), where changes from 5′P are shown in lower case and the Fok I recognition site is underlined], c5′P [sequence complementary to 5′P, d(TCTCGTGCATGATCAC)], 3′P [3′ end primer, d(GCGAATTCTGCCTCTT)], 3′P-N9 [3′ end primer for library construction, d(GCGAATTCTGCCTCTT-N9)], 3′P-Fok [d(GCGAATTCTGgaTgTT)], 5′P2 [second 5′ end primer, d(GTAAGCTTCGTGCACA)], 5′P2-OH [d(GTAAGCTTCGTGCACA-2′OH), where the sugar moiety at the 3′ end is a ribose], c5′P2 [sequence complementary to the last 15 nt of 5′P2, d(TGTGCACGAAGCTTA)], AU-6 [an example of a selected sequence from regular genomic SELEX, purified by trityl-selective perfusion high-performance liquid chromatography (HPLC), d(GTGATCATGCACGAGA-CTGTCTCCGGCCTTTCTCACCCGTTTGAATCTTTGCCTACTCATTACTCCGGC-AAGAGGCAGAATTCGC), where the 53mer genomic fragment is underlined, named AU-6c53], AU-6c53 (purified by trityl-selective perfusion HPLC), I-7c26 [sequence from previous work (18) and used as a competitor, d(GTGCCACCCTCCTCTCTTGTTCTTGT); this sequence was purchased from Oligos Etc. (Wilsonville, OR) and gel-purifed], BA-8 (an example of a selected sequence from primer-free genomic SELEX, purified by trityl-selective perfusion HPLC, d(TGCCAGTTCTTTTGGGTATTCCGTTATTATTGCGTTTCCTCGGTTTCCTTCTGGTAA)] and BA-8-C [sequence from fd genomic DNA and used as a control, purified by trityl-selective perfusion HPLC, d(CTTTGTTCGGCTATCTGCTTACTTTCCTTAAAAAGGGCTTCGGTAAGATAGCTATTG)]. All the sequences are shown 5′ to 3′.
Denaturing gel electrophoresis
Polyacrylamide gels (acrylamide/bis = 19/1) with specified percentages (w/v) were prepared in TBE buffer (90 mM Tris-borate, pH 8.3, 2 mM EDTA) containing 7.6 M urea. The gels were usually pre-run at 500 V in TBE buffer for 10 min. 32P-labeled DNA samples were prepared in 33–50% formamide and heated at >70°C for 3 min before loading into the gel. Electrophoresis was performed at 450 V for 50 min.
Electrophoretic mobility shift assay (EMSA)
EMSAs were performed on agarose gels with TAE buffer (40 mM Tris-acetate, pH 8.3, 1 mM EDTA), as described previously (18).
Construction of the fd genomic library
The method for the construction of the fd genomic DNA library was modified from that used by Gold and co-workers (13). The procedure is described below.
First (complementary) strand synthesis. An aliquot of 1 μg of fd genomic DNA (0.51 pmol, or 3 × 1011 copies) was mixed with 80 pmol of primer 3′P-N9, and H2O was added to make a volume of 30 μl. The mixture was heated at 95°C for 3 min and immediately put on ice. dNTPs (0.5 mM each of dATP, dGTP, dCTP and dTTP) and 0.2 U/μl of DNA polymerase I large fragment (Klenow fragment, with 3′ exonuclease activity; Promega, Madison, WI) were added on ice. The final volume was 50 μl in a buffer of 50 mM Tris–HCl, pH 7.2, 10 mM MgSO4 and 0.1 mM DTT. The mixture was incubated on ice for 5 min, at room temperature for 25 min, and then at 50°C for 5 min. The reaction was stopped by adding 10 mM EDTA and heating at 75°C for 10 min. To reduce the occurrence of constructs having identical primers on both tails, the mixture of the above extension reaction was purified using QIAquick PCR Purification Kit (Qiagen, Valencia, CA) to remove the unreacted primers (the removal efficiency was ∼90% in this case).
Second (genomic) strand synthesis. The products from the first strand synthesis were used as templates for the second strand synthesis. Similar procedures were followed, except that 160 pmol of primer 5′P-N9 was used in this extension reaction.
Library isolation. The products from the second strand synthesis were resolved on an 8% denaturing polyacrylamide gel. 32P-labeled ssDNA markers of φX174 DNA/Hinf I (24–762 nt; Promega) were run in parallel. In this study, the bands corresponding to the 82mer and 118mer markers were respectively isolated and DNAs were eluted into 300 μl TE buffer (10 mM Tris–HCl, pH 7.4, 1 mM EDTA) at 4°C overnight. The preliminary DNA genomic libraries were recovered by ethanol precipitation.
Refinement of the library. To refine the preliminary library, PCR was performed with primers 5′P and biotin-conjugated 3′P (both lacking the 9 random nucleotides at the 3′ ends) under the following conditions: (i) 95°C for 2 min, (ii) 18 cycles at 95°C for 1 min, 55°C for 1 min and 72°C for 2 min, and (iii) final extension at 72°C for 10 min. The PCR products were extracted with phenol/chloroform and then incubated with immobilized streptavidin agarose beads (Pierce, Rockford, IL) in SA buffer (10 mM Tris–HCl, pH 7.4, 400 mM NaCl and 1 mM EDTA) at room temperature for 40 min, followed by washing the beads three times with SA buffer. The bound DNA was denatured in 120 mM NaOH at 37°C for 15 min to elute the genomic strands from the beads. The genomic strands were ethanol precipitated and finally subjected to purification on an 8% denaturing gel. The libraries with about 82mer and 118mer sequences were named FDG-82 and FDG-118, respectively.
Regular genomic SELEX
The procedure for regular genomic SELEX applied in this experiment was similar to that used for SELEX with a chemically synthesized random library in our previous report (18). About 60 pmol of ssDNA from the FDG-82 library was used for the initial round of selection. The g5p was added such that the selection ratios (bound DNA/total DNA) were 3.3–8% for the first and subsequent rounds. To approximate intracellular conditions, the reaction was performed in a buffer containing 10 mM Tris–HCl, pH 7.4, 200 mM KCl and 1.5 mM MgCl2. The binding mixture was separated on agarose gels in TAE buffer. The g5p-bound DNA was extracted from the gel and ethanol-precipitated before being subjected to PCR. PCR and the following ssDNA isolation were performed as described above for library refinement. Finally, denaturing gel-purified ssDNA was subjected to the next round of selection. After four rounds, the selected DNA was cloned and sequenced.
Primer-blocking genomic SELEX
The FDG-82 library was also used for this protocol, and the same procedures as in regular genomic SELEX were applied, except that the two primer regions of the library were hybridized with two complementary oligonucleotides before performing g5p binding reactions in each round. To achieve the highest degree of hybridization, the library was first incubated (37°C for 1 h) with c5′P-SS-biotin, which was complementary to the 5′ primer region of the library, and the 3′ end was linked to a biotin tag through a disulfide bond. Immobilized streptavidin beads were added to the mixture and then washed with SA buffer, such that only the library sequences that annealed to c5′P-SS-biotin were retained on the beads. The hybrids were eluted by incubating with 50 mM DTT at 42°C for 30 min (32). The yield for this step was ∼55%. The other complementary oligonucleotide (biotin-SS-3′P) was subsequently added, and hybrids with bound 3′P were purified in a similar way (yield ∼30%). In this protocol, SELEX was performed for four rounds before cloning and sequencing.
Primer-free genomic SELEX
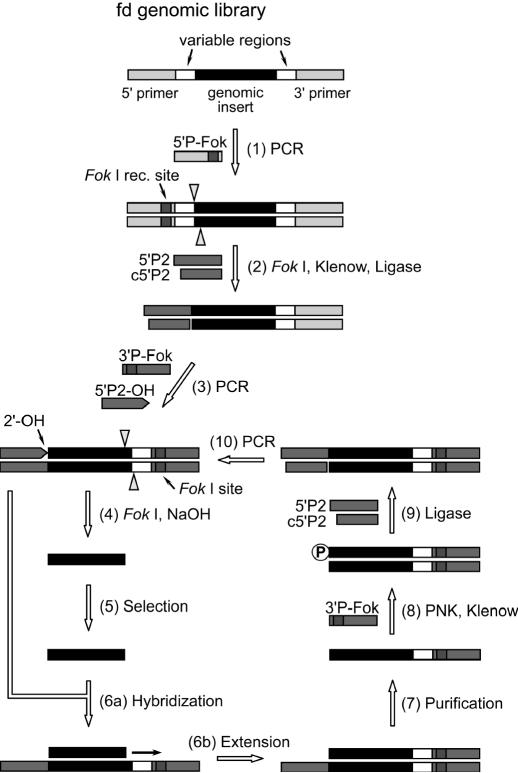
The FDG-118 library was used as a precursor library for this protocol. The whole procedure is schematically shown in Figure 1.
Figure 1.
A schematic procedure for primer-free genomic SELEX. Different regions of the genomic library are indicated and their lengths are not shown in proportion. Restriction cleavage sites of Fok I are approximately indicated by arrow heads. For details, see Materials and Methods.
Step 1. To remove the variable region on the 5′ end of the FDG-118 library (adjacent to the primer-annealing site and consisting of about 9 nt that were derived from the random tails of the primers used for library construction), a Fok I recognition site was introduced to the 5′ primer region by PCR in the presence of primers 5′P-Fok and 3′P. Fok I is a class 2 restriction endonuclease that cuts 9 nt downstream from a GGATG recognition site on that strand and 13 nt upstream on the complementary strand. A small amount of [α-32P]dCTP (ICN Biomedicals, Irvine, CA) was included in the reaction to internally label and track the products.
Step 2. The PCR products were digested by Fok I (15–20 U; Promega) at 37°C for 2 h, and the recessed DNA ends were filled by the Klenow fragment (10 U; Promega) at 37°C for 15 min. After phenol/chloroform extraction and ethanol precipitation, the DNA (∼3 pmol) was purified from a 6% denaturing polyacrylamide gel. To ligate a new 5′ end primer sequence to the library, 50 pmol of the pre-annealed 5′P2:c5′P2 duplex was added in the presence of 3–6 U of T4 DNA ligase (Promega). The reaction mixture was incubated for 16 h at 15°C and was then subjected to a 6% denaturing polyacrylamide gel. The ligated library was isolated from the gel. All the enzymatic reactions in this step were carried out in the buffers provided by the manufacturers. Steps 1 and 2 were modified from Shtatland et al. (14).
Step 3. As in step [1], PCR was performed to introduce a Fok I recognition site to the 3′ primer region of the library (primers: 5′P2 and 3′P-Fok). Subsequently, a ribose linkage was added to the junction of the 5′ primer and genomic inserts by another PCR reaction (primers: 5′P2-OH and 3′P-Fok). At least 1 ml of internally 32P-labeled PCR products (see above) was obtained at this step, which was then concentrated and ethanol precipitated. 5′P2, instead of 5′P2-OH, was included in a parallel PCR reaction (0.2 ml) to prepare dsDNA templates used at Step 6.
Step 4. To remove primer regions from both ends, the library was incubated with 15 U of Fok I in a volume of 20 μl and digested at 37°C. After 2 h, NaOH was directly added to the reaction to a final concentration of 0.2 M. Alkaline hydrolysis was performed ∼24 h at 37°C or 10 min at 95°C. The reaction was neutralized with acetic acid, ethanol precipitated and purified on an 8% denaturing polyacrylamide gel. The genomic insert produced at this step was ∼57 nt in length and contained no primer-annealing sequences; it could be easily separated from the cleaved primers (16–27mers) and complementary strands (∼87mer) on denaturing gels. Typically, 30–40 pmol of primer-free library was recovered.
Step 5. For the selection reaction, the g5p was added to the primer-free library and incubated at 37°C for 15 min. The reaction was performed in the same buffer as for regular SELEX, except that 20 mM Tris–HCl was used in this experiment. Selection ratios (bound DNA/total DNA) were 2.4–10.5%. The g5p-bound DNA was separated by EMSA and recovered as described previously (18).
Step 6. The following steps were dedicated to regenerating primer-annealing regions for the selected genomic sequences. To synthesize a 3′ primer region, the selected primer-free sequences were combined with 2.5 U of Taq, dNTPs (0.2 mM each of dATP, dGTP, dCTP and dTTP), and at least 2-fold amounts of full-length dsDNA templates from Step 3. The mixture was denatured at 94°C for 1 min and incubated at 72°C for 3 min for hybridization and extension. Analogous to a PCR reaction, this process was repeated for 20 cycles to maximize the yield (63–86%).
Step 7. The 3′-primer-bearing sequences (∼94mer) had to be purified on denaturing gels to remove the template strands (∼110mer), which might interfere with the following reactions.
Step 8. The sequence was then converted to blunt end duplexes before ligation of a 5′ primer sequence. The purified products from Step 7 were phosphorylated at the 5′ end by T4 polynucleotide kinase (Promega). The reaction (20 μl) was carried out for 30 min at 37°C and stopped by heating 2 min at 95°C. To the same tube, 100 pmol of 3′P-Fok was added to hybridize to the 3′ primer region of the library (95°C for 1 min, 60°C for 10 min and 25°C for 10 min), and then dNTPs (0.25 mM each of dATP, dGTP, dCTP and dTTP) and Klenow fragment (5 U) were added to give a final volume of 40 μl. The reaction was performed at 37°C for 25 min and 50°C for 5 min. After phenol/chloroform extraction and ethanol precipitation, the sample was resuspended in water and, without further purification, was ready for ligation.
Steps 9 and 10. The ligation reaction was performed with the pre-annealed 5′P2:c5′P2 duplexes, and the ligated products were purified from denaturing gels as described in Step 2. The overall yields for Steps 8 and 9 were 63–76%. The library at this point had acquired both the 5′ and 3′ primer regions and could be amplified by PCR, with primers 5′P2-OH and 3′P-Fok, to begin another round of SELEX. In our experiments, a total of five rounds were performed, and selected DNA from the fourth and fifth rounds was cloned and sequenced.
RESULTS
Construction and characterization of genomic library
A series of ssDNA libraries derived from the bacteriophage fd genome were constructed, based on the protocol used by Gold and co-workers (13). FDG-82 (∼82mer) and FDG-118 (∼118mer) were two libraries obtained using that protocol, and respectively carried about 50 and 86 nt of genomic sequence in the central region (called genomic inserts). The FDG-82 library was characterized by sequencing, statistical analysis, PCR and genomic insert overlapping analysis, and the results showed that the library essentially contained randomly distributed genomic inserts that covered the entire genome, including the hairpins from the IG (data not shown). We assume that FDG-118 as well as FDG-82 was a similarly representative genomic library for SELEX experiments.
Regular genomic SELEX
FDG-82 was first used for SELEX selection of high-affinity genomic sequences for binding to g5p. The procedure was similar to that previously used for SELEX with g5p and a chemically synthesized random library (18). To approximate physiologically relevant conditions, the selection reaction was performed at 37°C in a pH 7.4 buffer containing 200 mM KCl and 1.5 mM MgCl2. Table 1 shows 28 cloned sequences after four rounds of selection. These sequences represented two major (in genes 2 and 6) and one minor (in gene 3) genomic sites and were respectively grouped into three families (I, II and III). The sequences within each family largely overlapped. Two common aspects of these sequences were observed. First, they were located in pyrimidine-rich regions of the genome; this was consistent with the known binding preference of g5p for pyrimidines (33,34). Second, a few mutations were found in most sequences and were generally located near the extreme ends of the variable regions; these mutations presumably originated from mismatches of the primers with the hybridized genomic sequences during construction of the library (13).
Table 1. Twenty-eight DNA sequences from the fourth round of regular genomic SELEX.
aOnly the sequences of the genomic inserts are shown. Mutated nucleotides are presented in lower cases. A short segment of the fd genomic sequence is shown for comparison; the numbers above the sequence indicate the start or end positions of the sequence in the genome (GI:215394).
bThe number in parentheses is the number of clones with identical sequences.
It has been reported that primer-annealing sites might be involved in base pairing with the genomic inserts (14), and this could lead to artifacts if the resulting structures play a role in binding with the target protein. We chose a representative sequence AU-6 from family I to test the influence of primer-annealing sites on the DNA structures and g5p-binding affinities. AU-6 had no mutations in the genomic insert and was an 85mer containing the 5′ and 3′ primer-annealing sites. A truncated 53mer sequence, AU-6c53, was also synthesized without the primer-binding tails for comparison. Ultraviolet (UV) melting experiments (data not shown) indicated that AU-6c53 at 200 KCl had no apparent structures (melting temperature, Tm < 15°C), whereas the Tm of AU-6 was ∼35°C, close to the reaction temperature (37°C) for selection, suggesting that some DNA structures might be involved when binding with g5p. Further EMSA experiments supported this argument. As shown in lane 1 of Figure 2A, two complexes, intermediate and saturated complexes, appeared on the gel when AU-6 was bound by g5p. This EMSA pattern was similar to those of g5p binding to DNA structures of G-quadruplexes and hairpins, as reported previously (18,19,31). Possible AU-6 structures were calculated by mfold (35), and a few stem-loop foldings were predicted, in all of which one or both of the primer-annealing sites were extensively involved in base pairing (data not shown).
Figure 2.
Effects of primer hybridization of AU-6 on g5p binding. (A) EMSA. 32P-labeled AU-6 (1 μM) was heated at 95°C for 3 min in the absence (lanes 1–5) or presence (lanes 6–10) of 1.5 μM 3′P (3′ primer). The mixtures were cooled slowly at room temperature for 15 min to allow primer hybridization. Various concentrations (0–16 μM) of a DNA competitor (I-7c26) were added before adding 12.8 μM of g5p. The binding reaction was performed at 37°C for 30 min and subjected to a 2.5% agarose gel. Shown is a phosphor image of the dried gel. (B) Quantitation of complex formation inhibited by the competitor in the absence (open circle) or presence (solid circle) of 3′P. Percentages of g5p·AU-6 complexes (intermediate plus saturated) retained in each lane of (A) were quantitated and normalized to the samples without competitors (lane 1 or 6).
To further explore the effects of primer-annealing sites and the resulting structures on g5p-binding affinity, g5p was added to samples containing AU-6, in which the 3′ primer-annealing site was free or blocked by a complementary oligonucleotide, and various amounts of an unrelated single-stranded competitor DNA (I-7c26) were present. As shown in Figure 2, the g5p-binding affinity for AU-6 was apparently reduced so that the competitor was more effective when the 3′ end of AU-6 was inhibited from intrastrand base pairing. Therefore, the primer-annealing sites of the fd genomic library may play an important role in g5p binding during this regular SELEX experiment, and the selected genomic inserts alone may not be high-affinity g5p binders.
Primer-blocking genomic SELEX
To reduce or eliminate the interactions between primer-annealing sites and genomic inserts, the primer-binding tails of the same original genomic library (FDG-82) were blocked by hybridization with complementary oligonucleotides before the selection reactions with the g5p were performed in each round; otherwise the same experimental procedures were followed as for regular genomic SELEX. Table 2 shows the cloned sequences after four rounds of selection. Out of 21 clones, 19 (family IV) overlapped in the coding region of gene 2 and were almost identical to the family I sequences from regular genomic SELEX (see Table 1). Apparently, this region of gene 2 represented by the family IV sequences was a preferred g5p binding site, even though the affinity of a similar sequence was reduced by blocking the primer-annealing sites (see Figure 2). In contrast, we did not identify any sequences homologous to those in family II, the other major group of sequences from regular genomic SELEX. Therefore, it was likely that the g5p-binding affinity was more significantly inhibited for the sequences in family II than those in family I (≈family IV) when the primer-annealing sites were blocked.
Table 2. Twenty-one DNA sequences from the fourth round of primer-blocking genomic SELEX.
aOnly the sequences of the genomic inserts are shown. Mutated nucleotides are presented in lower cases. A short segment of the fd genomic sequence is shown for comparison; the numbers above the sequence indicate the start or end positions of the sequence in the genome (GI:215394).
bAZ-4 and AZ-14 are located at genomic positions 5570–5620 (in the intergenic region) and 1757–1806 (in gene 3), respectively. The shaded region of AZ-4 is from part of the predicted hairpin A in the intergenic region.
In the primer-blocking protocol, non-genomic (primer-annealing) sequences are still present, even though they are hybridized and are inhibited from base pairing with the genomic inserts. Some unforeseen factors associated with the tails still could be affecting target protein binding. To completely eliminate those potential problems, we designed a new SELEX protocol, termed primer-free genomic SELEX, in which the primer-annealing sites were removed prior to the g5p selection step and then regenerated prior to amplification.
Primer-free genomic SELEX
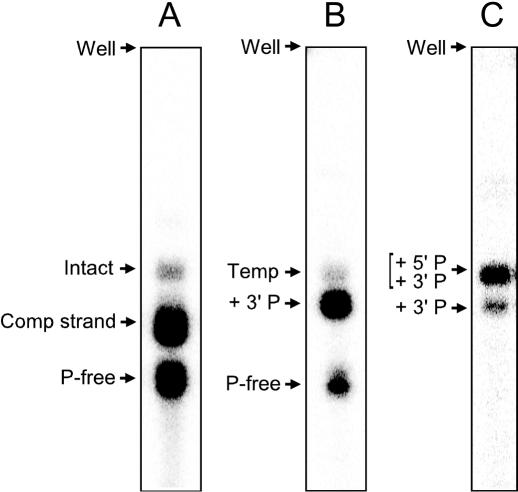
A primer-free genomic library was generated by introducing a restriction site to the 3′ end and a ribose linkage to the 5′ end primer-annealing regions, which were removed by enzymatic and alkaline treatments (Figure 1, Steps 1–4; ‘P-free’ band in Figure 3A). The variable regions, which usually contained a few point mutations due to the library construction (see Tables 1 and 2), were also concomitantly removed by this procedure. Since a considerable length of the DNA strands had to be deleted, we chose a longer genomic library (FDG-118, ∼118mers) for the starting material, which was finally trimmed to ∼57mers, containing only genomic sequences, for the selection reaction with g5p (Figure 1, Step 5). To regenerate primer-annealing sites, we hybridized the selected sequences to the unselected dsDNA templates under very stringent conditions (Figure 1, Step 6a). Success of this step relied on the fact that the PCR-amplified dsDNA templates (after Step 3) should contain many copies of each unique sequence, including the ones whose genomic inserts were selected by g5p binding. Thus, the selected sequences should be able to find complementary strands within a mixture of the templates. The hybridized genomic inserts were then elongated by a thermal-stable DNA polymerase to synthesize the 3′ primer-annealing sequence (Figure 1, Step 6b). The elongated products were quite homogenous in length and formed a discrete band on denaturing gels; this band was well separated from the templates and unreacted DNA, and thus could be unambiguously isolated (‘+ 3′P’ band in Figure 3B). This key step was usually accomplished with high yields (63–86%) in our experiments. The following ligation of the 5′ primer-annealing tail was relatively straightforward (Figure 1, Steps 7–9; ‘+ 5′P + 3′P’ band in Figure 3C). Finally, the selected sequences were amplified by PCR for another round of SELEX.
Figure 3.
Examples of gels used to monitor products from each round of primer-free genomic SELEX. (A) Isolation of primer-free DNA after restriction enzyme and alkaline treatments (Figure 1, Step 4). (B) Isolation of samples after synthesis of the 3′ primer-annealing sequence (Figure 1, Steps 6–7). Note that the amount of the templates was at least 2-fold higher than that of the primer-free DNA; the templates were intentionally reduced in radioactivity to give a lighter band on the gel. (C) Isolation of samples after ligation of the 5′ primer-annealing sequence (Figure 1, Step 9). Shown are phosphor images of 8% denaturing polyacrylamide gels. Intact, dsDNA PCR product; Comp strand, complementary strand; P-free, primer-free DNA; Temp, template DNA; + 3′P, DNA containing the 3′ primer-annealing sequence; + 5′P and + 3′P, DNA containing both 5′ and 3′ primer-annealing sequences.
Table 3 shows 26 cloned sequences after four rounds of selection with our current protocol. One major (family V) and one minor (family VI) groups of sequences were identified, both different from those in the previous two experiments. Because the variable regions were removed, mutations at the ends of the selected sequences were greatly reduced, except in the very end nucleotides. (The underlined regions in Table 3 covered the variable regions and were synthesized during regeneration of the 3′ primer.) UV melting experiments showed that the Tm of a representative sequence, BA-8, from family V was ∼28°C in 200 mM KCl (data not shown), suggesting that sequences in this pyrimidine-rich family did not form stable DNA structures at 37°C, the temperature used for the g5p binding reaction during SELEX.
Table 3. Twenty-six DNA sequences from the fourth round of primer-free genomic SELEX.
aOnly the sequences of the genomic inserts are shown. Mutated nucleotides are shown in lower cases. Underlined are 11 nt (including the 9mer variable region) that were absent in protein-binding reactions but were synthesized during regeneration of the 3′ primer (see Figure 1, Step 6b). A short segment of the fd genomic sequence is shown for comparison; the numbers above the sequence indicate the start or end positions of the sequence in the genome (GI:215394).
bThe number in parentheses is the number of clones with identical sequences.
cThe shaded region is from part of the predicted hairpin A in the intergenic region.
dBA-19, BA-6 and BB-8 are located at genomic positions 1990–2058 (in gene 3), 2540–2605 (in gene 3) and 4917–4987 (in gene 4), respectively.
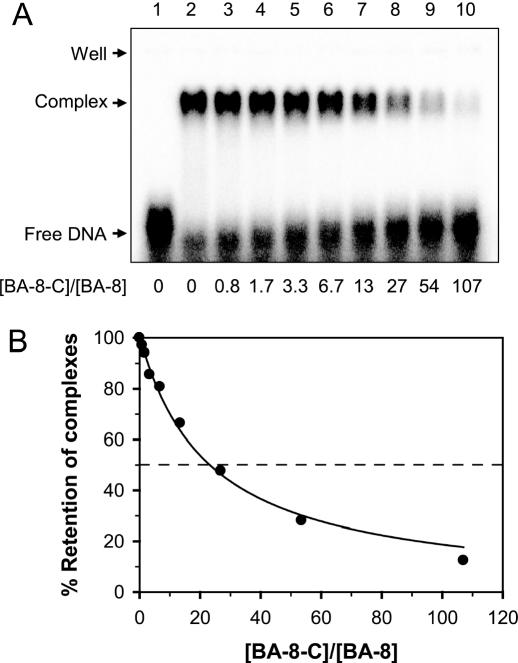
Future work is needed to assess whether interactions of g5p with BA-8 (or other homologs in family V) play a role in the cell. For this work, we made a preliminary determination of the relative binding affinities of BA-8 and a downstream control sequence BA-8-C (both 57mers) for g5p. As shown in Figure 4, a ∼23-fold higher concentration of BA-8-C than BA-8 was required to dissociate 50% of the g5p·BA-8 complexes, demonstrating that, in the current assay system, the selected genomic sequences in family V had higher affinity for g5p binding than a neighboring sequence. However, this competitive binding assay was performed with two pure, synthesized sequences. Actual SELEX selections are carried out in the presence of thousands of different genomic sequences, where it is possible that one sequence is selectively bound by g5p because it forms an interstranded structure with another sequence. Therefore, criteria other than the relative binding affinities of individual sequences may be needed to assess the significance of selected sequences.
Figure 4.
Relative g5p-binding affinity of BA-8 versus BA-8-C. (A) EMSA. A fixed amount of 32P-labeled BA-8 (1 μM) was mixed with increasing concentrations of unlabeled BA-8-C (0–107 μM) in a buffer of 20 mM Tris–HCl, pH 7.4, 200 mM KCl and 1.5 mM MgCl2. After adding 11.2 μM of g5p, the mixtures were incubated at 37°C for 15 min and subjected to a 2.5% agarose gel. Shown is a phosphor image of the dried gel. Lane 1, 32P-labeled BA-8 only. (B) Quantitation of complex formation inhibited by BA-8-C. Percentages of g5p·BA-8 complexes retained in lanes 2–10 of (A) were quantitated and normalized to the sample without BA-8-C (lane 2).
On the other hand, sequences in family VI were derived from the intergenic region of the genome, and included a downstream segment of hairpin A, the morphogenetic signal for viral packaging (22,23). Interestingly, one cloned sequence from the primer-blocking protocol (AZ-4, see Table 2) was similar to this family, though it contained a much shorter hairpin A segment and was not redundant. Accordingly, we performed one more round of primer-free genomic SELEX selection to see whether family VI would be enriched. Out of 27 cloned sequences from the fifth round of selection, 23 were similar to the sequences in family V, and only two were homologous to those in family VI (data not shown). Therefore, with our protocol and the FDG-118 genomic library, sequences of family V were still the best g5p binders.
DISCUSSION
A new primer-free method
Genomic SELEX is a useful tool for identifying genomic DNA or RNA transcripts that bind tightly to target proteins in vitro, and an expectation is that identified protein–nucleic acid interactions will give insight into in vivo biological events. To yield significant results, the binding between target proteins and the genomic library should not be influenced by artificial factors, such as the presence of primer-annealing sequences of the library. In this study, we first employed regular genomic SELEX with the bacteriophage Ff g5p and showed that the primer-annealing sites did affect protein binding. The primer-binding tails of a typical sequence AU-6 appeared to form base pairs with genomic inserts of the selected sequences, and the affinity for g5p was decreased when the tails were blocked by complementary oligonucleotides (see Figure 2). We next employed a primer-blocking protocol, which gave a family of sequences homologous to AU-6, so an unforeseen influence of the primer-annealing sequences could still not be excluded. Therefore, a primer-free genomic SELEX procedure was developed to circumvent problems related to the presence of primer-annealing sequences.
A major challenge in the design of a primer-free genomic SELEX protocol is how to efficiently regenerate the primer-annealing sites for ssDNA or RNA libraries. (In contrast, ligation of primer-annealing sequences to dsDNA libraries with cohesive or blunt ends is more straightforward.) In preliminary experiments, we tried using T4 RNA ligase from various suppliers to ligate primers to single-stranded genomic fragments, but the yields were very low and not consistent (usually <10%; data not shown). In addition, optimal conditions for ligation include low temperatures (<20°C), which may selectively reduce the ligation efficiency for sequences that form structures. In our final protocol, we performed a thermal cycle reaction to synthesize the 3′ primer-annealing site. DNA structures including templates (dsDNA, having both 5′ and 3′ primer-annealing sites) were denatured at 94°C, following which the selected genomic fragments were hybridized to complementary templates and elongated by the Taq polymerase at 72°C to synthesize the 3′ primer-annealing site. The denaturation-hybridization-polymerization process was repeated several times to increase the yields. This protocol relies on successful hybridization, and thus libraries or selection pools that are more complex (diverse) than the fd genomic library will have to be tested for efficiency of hybridization at this step. The fd genomic DNA is only 6408 nt in length, and it is theoretically represented by about the same number of unique sequences in the library. The size is extremely small, compared with the random chemical library of 26 nt (comprising ∼1015 sequences) used in our previous work. If a more complex starting library is used, a few initial rounds of regular or other types of SELEX could be performed to reduce the diversity of the selection pool, and help decrease the chance of preferential selection of sequences with repeated elements, before the primer-free protocol is applied.
SELEX with libraries lacking primer-annealing sites, similar to our primer-free genomic SELEX, has been used by Pagratis et al. (36). To regain PCR-amplifiable sequences, the selected sequences in their procedure were biotinylated and immobilized to capture (‘fish’) complementary template strands (having both primer-annealing sites) by hybridization. This is straightforward, but template sequences that are highly, but not exactly, complementary to the selected genomic fragments may also be captured and amplified to yield false-positive results. In our system, PCR-amplifiable sequences are directly derived from the selected genomic fragments, and thus false-positive results are less likely.
Selected genomic sequences
In the present work, two major groups of sequences from two loci of the fd genome were selected by regular genomic SELEX (see Table 1), and the genomic fragments were predicted to form structures with the primer-annealing sites. Results from genomic SELEX with a primer-blocking protocol showed that sequences homologous to AU-6 were still selected and dominated when the same starting library was used (family IV in Table 2 was homologous to family I in Table 1). It was not likely that these sequences were selected from unblocked strands, since we employed an immobilization-elution method to maximize the population of primer-annealed sequences in the selection pool (for details see Materials and Methods). In addition, the g5p seemed not to displace the primers when binding to primer-annealed sequences, because the EMSA patterns for AU-6 were different in the presence and absence of complementary oligomers (Figure 2A, compare lanes 1 and 6). Therefore, the genomic inserts in family I may indeed be preferred g5p binding sites, but the influence of hybridized primer-annealing tails on g5p binding to the central genomic sequences is still difficult to assess, especially when it was recently found that the g5p is able to bind to dsDNA more strongly than previously recognized (37). Moreover, the selected sequences by our primer-free protocol did not include those of family I, although it should be pointed out that a different library was used for the primer-free protocol. In any case, SELEX with genomic fragments containing primer-annealing tails, being intact or blocked, is not the best strategy for confident, unambiguous selection of biologically significant sequences.
As described in the introduction, the g5p performs its biological functions by binding to the viral genomic DNA and some RNA transcripts (24–28). The whole genomic DNA is saturated when bound by g5p, whereas in the case of RNA transcripts the primary binding sites are in the leader sequences, probably leaving the rest of the RNA unbound. Therefore, the control of viral protein expression by g5p may involve more specific protein–nucleic acid interactions than does the control of viral DNA replication and assembly. In this respect, selection of RNA sequences by genomic RNA SELEX may be more relevant in terms of the sequences having a biological function. The genomic DNA sequences selected in this study may have no function other than perhaps a fortuitous role in initiating cooperative binding of g5p to the viral genome. A more elaborate set of experiments, beyond the scope of this paper, will be required to test for any biological function of the selected sequences. Nevertheless, our primer-free genomic SELEX offers a method to identify potentially biologically important nucleic acid sequences for target molecules while simultaneously reducing artificial effects.
Acknowledgments
ACKNOWLEDGEMENTS
Support was provided by grant AT-503 from the Robert A. Welch Foundation and in part by grant 009741-0021-1999 from the Texas Advanced Technology Program.
REFERENCES
- 1.Ellington A.D. and Szostak,J.W. (1990) In vitro selection of RNA molecules that bind specific ligands. Nature, 346, 818–822. [DOI] [PubMed] [Google Scholar]
- 2.Tuerk C. and Gold,L. (1990) Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science, 249, 505–510. [DOI] [PubMed] [Google Scholar]
- 3.Mohibullah N., Donner,A., Ippolito,J.A. and Williams,T. (1999) SELEX and missing phosphate contact analyses reveal flexibility within the AP-2[alpha] protein: DNA binding complex. Nucleic Acids Res., 27, 2760–2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.James W. (2001) Nucleic acid and polypeptide aptamers: a powerful approach to ligand discovery. Curr. Opin. Pharmacol., 1, 540–546. [DOI] [PubMed] [Google Scholar]
- 5.Feigon J., Dieckmann,T. and Smith,F.W. (1996) Aptamer structures from A to zeta. Chem. Biol., 3, 611–617. [DOI] [PubMed] [Google Scholar]
- 6.Gold L., Polisky,B., Uhlenbeck,O. and Yarus,M. (1995) Diversity of oligonucleotide functions. Annu. Rev. Biochem., 64, 763–797. [DOI] [PubMed] [Google Scholar]
- 7.Wilson D.S. and Szostak,J.W. (1999) In vitro selection of functional nucleic acids. Annu. Rev. Biochem., 68, 611–647. [DOI] [PubMed] [Google Scholar]
- 8.Pileur F., Andreola,M.L., Dausse,E., Michel,J., Moreau,S., Yamada,H., Gaidamakov,S.A., Crouch,R.J., Toulme,J.J. and Cazenave,C. (2003) Selective inhibitory DNA aptamers of the human RNase H1. Nucleic Acids Res., 31, 5776–5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murphy M.B., Fuller,S.T., Richardson,P.M. and Doyle,S.A. (2003) An improved method for the in vitro evolution of aptamers and applications in protein detection and purification. Nucleic Acids Res., 31, e110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Golden M.C., Collins,B.D., Willis,M.C. and Koch,T.H. (2000) Diagnostic potential of PhotoSELEX-evolved ssDNA aptamers. J. Biotechnol., 81, 167–178. [DOI] [PubMed] [Google Scholar]
- 11.Bittker J.A., Phillips,K.J. and Liu,D.R. (2002) Recent advances in the in vitro evolution of nucleic acids. Curr. Opin. Chem. Biol., 6, 367–374. [DOI] [PubMed] [Google Scholar]
- 12.Li Y. and Breaker,R.R. (1999) Deoxyribozymes: new players in the ancient game of biocatalysis. Curr. Opin. Struct. Biol., 9, 315–323. [DOI] [PubMed] [Google Scholar]
- 13.Singer B.S., Shtatland,T., Brown,D. and Gold,L. (1997) Libraries for genomic SELEX. Nucleic Acids Res., 25, 781–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shtatland T., Gill,S.C., Javornik,B.E., Johansson,H.E., Singer,B.S., Uhlenbeck,O.C., Zichi,D.A. and Gold,L. (2000) Interactions of Escherichia coli RNA with bacteriophage MS2 coat protein: genomic SELEX. Nucleic Acids Res., 28, e93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim S., Shi,H., Lee,D.K. and Lis,J.T. (2003) Specific SR protein-dependent splicing substrates identified through genomic SELEX. Nucleic Acids Res., 31, 1955–1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stelzl U., Spahn,C.M. and Nierhaus,K.H. (2000) Selecting rRNA binding sites for the ribosomal proteins L4 and L6 from randomly fragmented rRNA: application of a method called SERF. Proc. Natl Acad. Sci. USA, 97, 4597–4602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vater A., Jarosch,F., Buchner,K. and Klussmann,S. (2003) Short bioactive Spiegelmers to migraine-associated calcitonin gene-related peptide rapidly identified by a novel approach: tailored-SELEX. Nucleic Acids Res., 31, e130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wen J.-D., Gray,C.W. and Gray,D.M. (2001) SELEX selection of high-affinity oligonucleotides for bacteriophage Ff gene 5 protein. Biochemistry, 40, 9300–9310. [DOI] [PubMed] [Google Scholar]
- 19.Wen J.-D. and Gray,D.M. (2004) Ff gene 5 single-stranded DNA-binding protein assembles on nucleotides constrained by a DNA hairpin. Biochemistry, 43, 2622–2634. [DOI] [PubMed] [Google Scholar]
- 20.Beck E. and Zink,B. (1981) Nucleotide sequence and genome organisation of filamentous bacteriophages fl and fd. Gene, 16, 35–58. [DOI] [PubMed] [Google Scholar]
- 21.Rapoza M.P. and Webster,R.E. (1995) The products of gene I and the overlapping in-frame gene XI are required for filamentous phage assembly. J. Mol. Biol., 248, 627–638. [DOI] [PubMed] [Google Scholar]
- 22.Schaller H. (1979) The intergenic region and the origins for filamentous phage DNA replication. Cold Spring Harb. Symp. Quant. Biol., 43, 401–408. [DOI] [PubMed] [Google Scholar]
- 23.Zinder N.D. and Horiuchi,K. (1985) Multiregulatory element of filamentous bacteriophages. Microbiol. Rev., 49, 101–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Model P. and Russel,M. (1988) In Calendar,R. (ed.), The Bacteriophages. Plenum Press, NY, Vol. 2, pp. 375–390. [Google Scholar]
- 25.Michel B. and Zinder,N.D. (1989) Translational repression in bacteriophage f1: characterization of the gene V protein target on the gene II mRNA. Proc. Natl Acad. Sci. USA, 86, 4002–4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zaman G., Smetsers,A., Kaan,A., Schoenmakers,J. and Konings,R. (1991) Regulation of expression of the genome of bacteriophage M13. Gene V protein regulated translation of the mRNAs encoded by genes I, III, V and X. Biochim. Biophys. Acta, 1089, 183–192. [DOI] [PubMed] [Google Scholar]
- 27.Yen T.S. and Webster,R.E. (1982) Translational control of bacteriophage f1 gene II and gene X proteins by gene V protein. Cell, 29, 337–345. [DOI] [PubMed] [Google Scholar]
- 28.Model P., McGill,C., Mazur,B. and Fulford,W.D. (1982) The replication of bacteriophage f1: gene V protein regulates the synthesis of gene II protein. Cell, 29, 329–335. [DOI] [PubMed] [Google Scholar]
- 29.Oliver A.W. and Kneale,G.G. (1999) Structural characterization of DNA and RNA sequences recognized by the gene 5 protein of bacteriophage fd. Biochem. J., 339, 525–531. [PMC free article] [PubMed] [Google Scholar]
- 30.Oliver A.W., Bogdarina,I., Schroeder,E., Taylor,I.A. and Kneale,G.G. (2000) Preferential binding of fd gene 5 protein to tetraplex nucleic acid structures. J. Mol. Biol., 301, 575–584. [DOI] [PubMed] [Google Scholar]
- 31.Wen J.-D. and Gray,D.M. (2002) The Ff gene 5 single-stranded DNA-binding protein binds to the transiently folded form of an intramolecular G-quadruplex. Biochemistry, 41, 11438–11448. [DOI] [PubMed] [Google Scholar]
- 32.Soukup G.A., Cerny,R.L. and Maher,L.J.,III (1995) Preparation of oligonucleotide-biotin conjugates with cleavable linkers. Bioconjug. Chem., 6, 135–138. [DOI] [PubMed] [Google Scholar]
- 33.Bulsink H., Harmsen,B.J. and Hilbers,C.W. (1985) Specificity of the binding of bacteriophage M13 encoded gene-5 protein to DNA and RNA studied by means of fluorescence titrations. J. Biomol. Struct. Dyn., 3, 227–247. [DOI] [PubMed] [Google Scholar]
- 34.Mou T.-C., Gray,C.W. and Gray,D.M. (1999) The binding affinity of Ff gene 5 protein depends on the nearest- neighbor composition of the ssDNA substrate. Biophys. J., 76, 1537–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.SantaLucia J. Jr (1998) A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics. Proc. Natl Acad. Sci. USA, 95, 1460–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pagratis N., Gold,L., Shtatland,T. and Javornik,B. (2001) Truncation SELEX method. USA Patent 6261774.
- 37.Mou T.-C., Shen,M.C., Terwilliger,T.C. and Gray,D.M. (2003) Binding and reversible denaturation of double-stranded DNA by Ff gene 5 protein. Biopolymers, 70, 637–648. [DOI] [PubMed] [Google Scholar]