Abstract
A β-sheet mini-protein based on the FBP11 WW1 domain sequence has been redesigned for the molecular recognition of ssDNA. A previous report showed that a β-hairpin peptide dimer, (WKWK)2, binds ssDNA with low micromolar affinity but with little selectivity over duplex DNA. This report extends those studies to a three-stranded β-sheet mini-protein designed to mimic the OB-fold. The new peptide binds ssDNA with low micromolar affinity and shows about 10-fold selectivity for ssDNA over duplex DNA. The redesigned peptide no longer binds its native ligand, the polyproline helix, confirming that the peptide has been redesigned for the function of binding ssDNA. Structural studies provide evidence that this peptide consists of a well structured β-hairpin made of Strands 2&3 with a less structured first strand that provides affinity for ssDNA but does not improve the stability of the full peptide. These studies provide insight into protein-DNA interactions as well as a novel example of protein-redesign.
Keywords: Protein Redesign, Protein-DNA interactions, beta-sheet, WW Domain
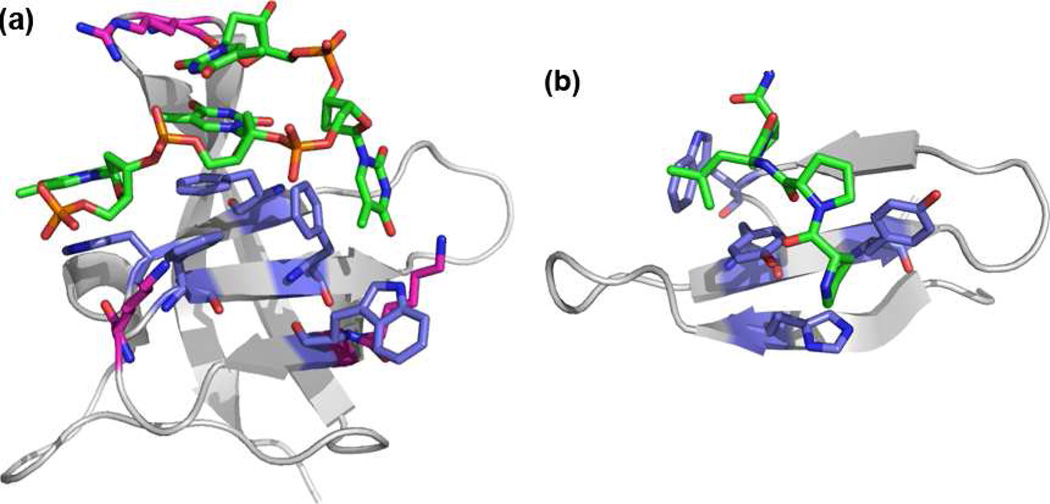
Protein-DNA interactions play a crucial role in many biological processes. Interactions involving protein recognition of single-stranded DNA (ssDNA) are essential in processes such as DNA recombination, replication and repair (1–4), telomere regulation (5–6), and cold shock response (7–12). Proteins recognize ssDNA primarily through an oligonucleotide/oligosaccharide binding motif (OB-fold, Figure 1a). The binding site of the OB-fold is a solvent-exposed beta-sheet surface composed of two three-stranded anti-parallel β-sheets (3, 4). These proteins interact with ssDNA through a combination of electrostatic and aromatic interactions on the exposed β-sheet surface as well as hydrogen-bonding interactions.
Figure 1.
(a) Structure of a single OB-fold in cold shock protein B from Bacillus subtilis bound to dT6 (pdb: 2es2)3b; (b) Structure of the FBP11 WW1 domain bound to a polyproline helix (pdb: 2dyf).
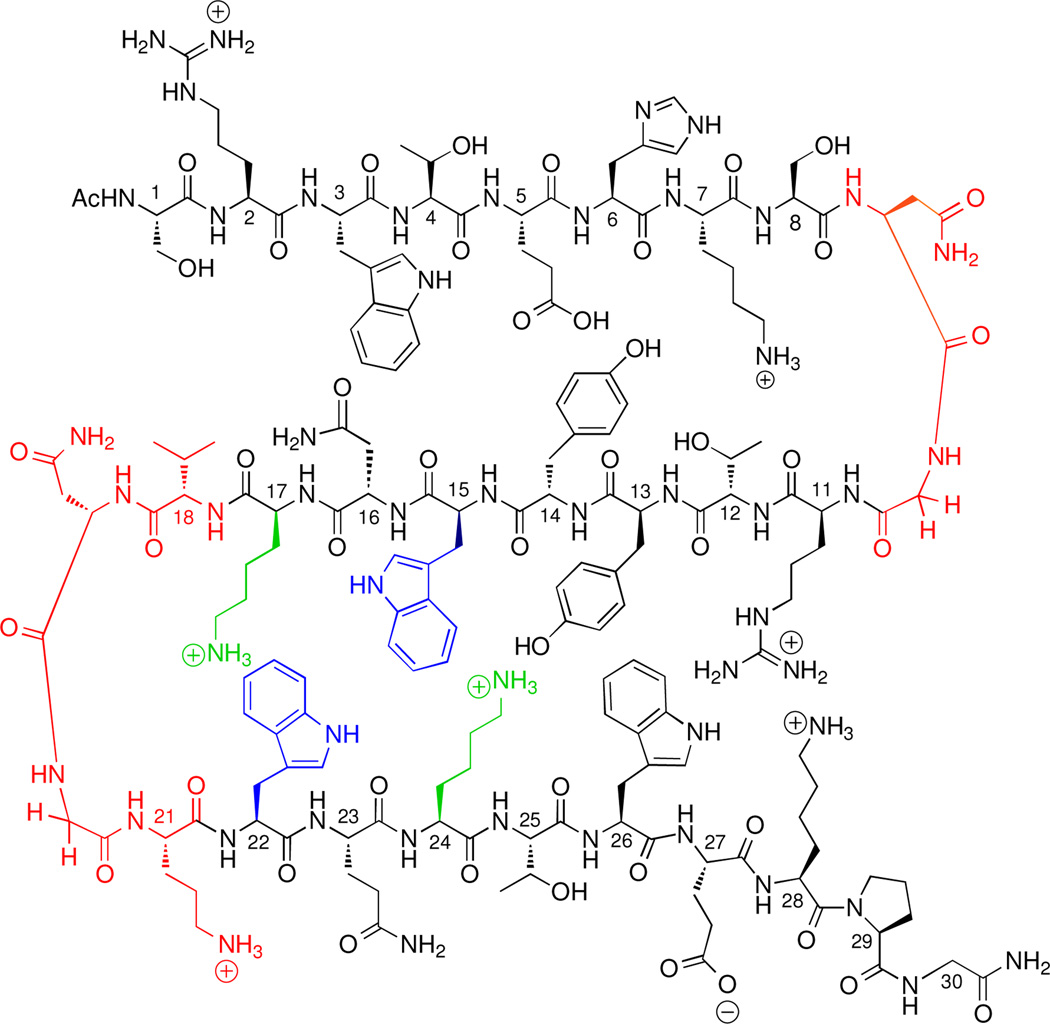
Although much research has been performed to determine the mechanisms by which proteins interact with dsDNA, the interactions between β-sheet proteins and ssDNA are less well studied. This laboratory previously reported a β-hairpin peptide dimer designed as a minimalist OB-fold mimic. The peptide (WKWK)2 consists of two well folded beta-hairpins, each with a binding cleft made up of two diagonal Trp sidechains and two Lys residues, which binds ssDNA with a Kd of 3 µM (13). This is comparable to the 6 µM Kd of cold shock protein A with ssDNA, which consists of a single OB-fold (9). (WKWK)2 loosely mimics an OB-fold in that it uses a combination of aromatic and electrostatic interactions on the face of a β-sheet to bind to the unpaired nucleotides in ssDNA (13–15) However, (WKWK)2 was found to bind dsDNA with a similar affinity, albeit primarily through electrostatic interactions (13). Structure-function studies demonstrated additional differences in the mechanism of binding to ss- and dsDNA (16). In particular, these studies indicated that binding to duplex DNA may be occurring via groove-binding. Thus, we hypothesized that addition of a third strand may inhibit binding to duplex DNA while maintaining or increasing affinity for ssDNA. To this end, a three-stranded β-sheet peptide based on the FBP11 WW1 domain peptide (Figure 1b) has been designed for binding to ssDNA. While WW domains have been studied extensively in the areas of protein folding and protein design (17–26), the potential for DNA binding has never been explored. The WW domain proteins are three-stranded β-sheet mini-proteins known for their conserved tryptophan residues. The natural ligands for WW domains are proline-rich sequences which often form polyproline helices (27–38) with the FBP11 WW1 domain ligand being PPLP (27–32). Knowledge of the structural features of this class of proteins combined with data from previous ssDNA binding peptides from this laboratory led to this redesign of a WW domain mutant (Figure 2) which binds ssDNA in the low micromolar range with about 10-fold selectivity over duplex DNA. This manuscript describes the redesign of a WW domain peptide as a molecular receptor selective for ssDNA, mimicking the natural OB-fold domain. These model systems provide a method to reveal factors contributing to protein-nucleic acid recognition.
Figure 2.
WW domain Mut1. Turn mutations are shown in red and the WKWK binding cleft is shown in blue and green.
Experimental Procedures
Peptide Synthesis and Purification
Peptides were synthesized via automated solid phase peptide synthesis using an Applied Biosystems Pioneer Peptide Synthesizer. Fmoc protected amino acids were used with a PEGPAL-PS resin. Amino acid residues were activated with HBTU (O-benzotriazole-N,N,N’,N’,-tetramethyluronium hexafluorophosphate) and HOBT (N-hydroxybenzotriazole) along with DIPEA (diisopropylethylamine) in DMF (N,N-dimethylformamide). Amino acids were deprotected with 2% DBU (1,8-diazabicyclo[5.4.0]undec-7-ene) and 2% piperidine in DMF for approximately 10 minutes. Each amino acid was coupled on an extended cycle of 75 minutes to improve coupling. The N-terminus of each peptide was acetylated using 5% acetic anhydride and 6% lutidine in DMF for 30 minutes. Cleavage of the peptides from the resin as well as side chain deprotection was performed in 95% trifluoroacetic acid (TFA), 2.5% H2O, and 2.5% triisopropylsilane (TIPS) for three hours. TFA was evaporated by bubbling with nitrogen, and ether was added to the resulting product. The peptide was then extracted with water and lyophilized to a powder.
Peptides were purified by reversed-phase HPLC. A Vydac C-18 semi-preparative column was used for separation with a gradient of 5–35% solvent B over 25 minutes with solvent A 95:5 water:acetonitrile, 0.1% TFA and solvent B 95:5 acetonitrile:water, 0.1% TFA. Peptides were then lyophilized and peptide sequence was confirmed by MALDI mass spectrometry. After purification, all peptides were desalted with a Pierce D-Salt Polyacrylamide 1800 desalting column.
DNA Sample Preparation
DNA sequences were purchased from IDT (Integrated DNA Technologies). All DNA samples were dissolved in 10mM Na2HPO4, 100 mM NaCl, adjusted to pH 7.0. Concentrations of both DNA strands were determined using a Perkin Elmer Lambda 35 UV/Vis Spectrometer. Absorbance values were determined at 260 nm, and concentrations were calculated using the extinction coefficients of the two DNA strands (ε260, ssDNA = 95,500 M−1·cm−1 and ε260, dsDNA = 112600 M−1·cm−1). Equal concentrations of the two strands (in sodium phosphate buffer, pH 7.0) were pooled in a final concentration of 100 mM NaCl. The solution was heated at 95 °C for 5 minutes to anneal the strands and was then allowed to cool to room temperature before storing at −20 °C.
Fluorescence Titrations
To determine the recognition of single-stranded and double-stranded oligonucleotides by the peptides, fluorescence titrations were performed which followed the Trp quenching with increasing oligonucleotide concentration. Peptide and nucleotide samples were prepared in 10 mM sodium phosphate buffer, 100 mM NaCl, pH 7.0. Peptide concentrations were determined in 5 M guanidine hydrochloride by recording the absorbance of the Trp residues at 280 nm (ε = 5690 M−1cm−1) by UV/vis spectroscopy. Concentrations of nucleotides were determined by UV/vis spectroscopy by observing the absorbance at 260 nm. Fluorescence scans were obtained on a Cary Eclipse Fluorescence Spectrophotometer from Varian. The experiments were performed at 298 K using an excitation wavelength of 297 nm. Fluorescence emission intensities of the Trp residues at 348 nm were fit as a function of nucleotide concentration to the binding equation (Equation 1) on Kaleidagraph using non-linear least squares fitting (39).
| Equation 1 |
where I is the observed fluorescence intensity, Io is the initial fluorescence intensity of the peptide, I∞ is the fluorescence intensity at binding saturation, [L] is the concentration of added nucleotide, and Kd is the dissociation constant. Oligonucleotides have an observable absorbance at the excitation wavelength of Trp (297 nm), and therefore there is an inner filter effect for which one must take account. The absorbance of the oligonucleotides at 297 nm was monitored at known concentrations and the extinction coefficient was determined. New absorbance values were determined for each oligonucleotide concentration. Corrected fluorescence values were determined from the following equations (Equation 2 and Equation 3) (40).
| Equation 2 |
| Equation 3 |
where Fc is the corrected fluorescence, Fo is the fluorescence observed, and Ci is the correction factor for each absorbance value (i). Ai is the new absorbance value for each concentration determined by the extinction coefficient.
Stoichiometry of Binding
The stoichiometry of binding was determined by the molar variation method following the quenching of tryptophan fluorescence (41). The peptide concentration was held constant and the amount of DNA was increased. The concentration of the peptide was above the Kd so that as the DNA is added, the majority of it is bound, maximizing the change in signal. Thus, when saturation is reached, there is no more change in signal, and saturation represents the binding stoichiometry. Peptide concentrations were in the range of 25–50 µM, depending on the maximum DNA concentrations used. The conditions were limited to low DNA concentrations (~60 µM for ssDNA and ~40 µM for duplex DNA) because of the inner filter effect. After correction for the inner filter effect, the fluorescence intensity was plotted against the ratio of DNA/peptide concentrations to give the stoichiometry of binding. The stoichiometry of binding is shown in plots (Figures S1 and S2) with the X-intercept of the dashed lines indicating the stoichiometry for each.
Oligoproline Binding
Fluorescence titrations using the oligoproline ligand were performed using the same procedures as with DNA, but the increase in Trp fluorescence with increased oligoproline was measured. This increase in Trp fluorescence has been ascribed to the increased hydrophobic environment upon oligoproline binding. Fluorescence scans were obtained on a Cary Eclipse Fluorescence Spectrophotometer from Varian. The experiments were performed at 298 K using an excitation wavelength of 297 nm. Fluorescence emission intensities of the Trp residues at 340 nm were fit as a function of polyproline concentration to the following equation (Equation 4) on Kaleidagraph using non-linear least squares fitting (28).
| Equation 4 |
where Ffree is the fluorescence intensity without ligand added and Fsat is the fluorescence intensity of a saturating concentration of ligand titrated. [WL] is the fraction of WW domain bound to ligand and is obtained by Equation 5. [W0] is the total WW domain concentration used.
| Equation 5 |
Fluorescence Anisotropy
A fluorophore, 5’-Bodipy 630/650-X NHS Ester, was attached to the ssDNA sequence for all binding studies. For duplex DNA binding, the labeled DNA was annealed to the complementary sequence as above stated. The fluorophore has an absorbance maximum at 638 nm and an emission maximum at 653 nm. Its molar extinction coefficient is 101000 /M·cm. Fluorescence anisotropy was measured using a Varian Cary Eclipse Fluorescence Spectrophotometer with a temperature controller. Bodipy-labeled DNA samples (low micromolar concentrations) were titrated with peptide samples (0-low millimolar concentrations) in 10 mM Na2HPO4, 100 mM NaCl, pH 7.0. Fluorescence samples were analyzed at 298 K and were excited at 638 nm with excitation and emission slit widths of 2.5 nm. Fluorescence was observed at 653 nm, and the anisotropy was determined by the software that came with the instrument. The anisotropy was fit to the following equation (Equation 6) using Kaleidagraph to determine the binding constant
| Equation 6 |
where F is the fluorescence anisotropy, Io is the initial fluorescence intensity of the peptide, I∞ is the fluorescence intensity at binding saturation, [L] is the concentration of added nucleotide, [P] is the peptide concentration for each fraction, and Kd is the dissociation constant. Equation 6 was derived from equations given by Wang and coworkers (42).
Another fluorophore employed was 5-(and -6)-Carboxytetramethylrhodamine, mixed isomers (TAMRA), which was purchased from Biotium. TAMRA was coupled onto the peptide Mut1 at Orn21. The synthesis was completed by coupling Lys(ivDde) in the original ornithine position (Orn21Lys). The ivDde protecting group was orthogonally deprotected by treatment with 2% hydrazine in DMF. Manual coupling of TAMRA was performed with four equivalents of HOBT, HBTU, and DIPEA in DMF. Cleavage from the resin and side chain deprotection was completed as with all other peptides. The resulting peptide was purified by HPLC and its sequence and purity determined by mass spectrometry. Peptide concentrations were determined by UV-Vis using TAMRA’s extinction coefficient of 91000 M−1·cm−1 at wavelength 559 nm. This extinction coefficient was supplied by Integrated DNA Technologies, www.idtdna.com. The excitation wavelength used in the experiments is 559 nm, and the observed emission wavelength is 583 nm. Anisotropy experiments were performed using the same methods as with the Bodipy-labeled DNA experiments.
Circular Dichroism
CD measurements were obtained using an AVIV 62 DS Circular Dichroism spectrometer or an Applied Photophysics Chirascan Plus spectrometer. CD data was obtained for the WW Domain peptides at 30 µM from 260–185 nm. The peptides were dissolved in 10 mM Na2HPO4, pH 7.0. Wavelength scans were performed at 25 °C. Thermal denaturations were performed by measuring the ellipticity at 227 nm from 4 to 96 °C, with 4 degree temperature steps. Equilibration times were ten minutes at each step.
CD measurements for thermal denaturation and renaturation of Mut1 were obtained using an Applied Photophysics Chirascan Plus spectrometer at 30 µM in a 10 mM Na2HPO4, pH 7.0 buffer. CD data was obtained from 235–210 nm at 4 to 95°C with 4 degree temperature increments. The sample was equilibrated at each increment for 5 minutes. Spectra from 260–185 nm were obtained at 4, 24, 64, and 95°C for both denaturation and renaturation of Mut1.
NMR spectroscopy
NMR experiments were carried out on either a Varian Inova 600 MHz or Bruker Ultrashield 600 MHz Plus spectrometer. 2D TOCSY data was acquired with a 1 mM concentration sample prepared in 50 mM KD2PO4, 0.5mM DSS in 90% H2O and 10% deuterium oxide adjusted to pH 7.0 with sodium deuteroxide. TOCSY spectra were acquired using 36 scans per increment and 128 increments in the indirect dimension with a mixing time, D9, of 0.0200 sec. Solvent suppression was applied with the Varian software.
2D NOESY data was acquired with a 1 mM concentration sample prepared in 50 mM KD2PO4, 0.5 mM DSS and buffered to pD 7.0 (uncorrected) with sodium deuteroxide. NOESY spectra were acquired using 36 scans per increment and 128 increments in the indirect dimension with a mixing time, D8, of 0.0750 sec. Solvent suppression was applied with the Varian software.
Peptide proton assignments of Mut1 S12 and S23 were determined using standard methods (43). The proton assignments of Mut1 were determined using the 90% H2O 2D TOCSY and 2D TOCSY (D2O only buffer) spectra, using the 2D TOCSY and NOESY spectra of Mut1 S12 and S23 as a guide. Additionally, the 2D NOESY spectrum of Mut1 was used to confirm the assignments (See Supporting Information).
Deviations in alpha hydrogen chemical shifts from random coil values, ΔδHα, were calculated according to Equation 7,
| Equation 7 |
where δHα,obs is the observed chemical shift of a given alpha hydrogen in the peptide, and δHα,RC is the random coil chemical shift of the corresponding proton determined from unstructured control peptides, Mut1-S1, Mut1-S2, and Mut1-S3 (Figure 3)
Figure 3.
Sequences of native FBP11 WW1 domain, WW domain mutants and controls, polyproline helix, and DNA sequences used in binding studies. Residues in the turn sequences and in the binding pocket are highlighted as in Figure 2, with turn sequences in red, Trp residues in the binding pocket in blue, and Lys residues in the binding pocket in green. Other mutations are shown in bold.
The extent of folding at the turn for each peptide can be determined by calculating the fraction folded from the Gly splitting from Gly residues in the turns (residues 10 and 20), as in equation 8 (54),
| Equation 8 |
where ΔδGlyobs is the observed glycine diastereotopic protons splitting for the peptide, and ΔδGly100 is the glycine splitting for the fully folded control peptide that is presumed to take on a 100% fold. The following disulfide-linked cyclic peptides were used as the fully folded control peptides, as is precedented in the literature (44). The underlined residues indicate the position of cyclization. The bold residue is the Gly in the turn for which the splitting was measured. The ΔδGly100 values were found to be 0.41 ppm and 0.70 ppm for Mut1-S12-cyc and Mut1-S23-cyc, respectively.
Mut1-S12-cyc: Ac-C-R-W-T-E-H-K-S-N-G-R-T-Y-Y-W-N-K-C-NH2
Mut1-S23-cyc: Ac-C-T-Y-Y-W-N-K-V-N-G-O-W-Q-K-T-W-E-C-NH2
Aggregation Studies
To assess possible aggregation, 1D spectra of Mut1 were acquired using 36 scans with solvent presaturation at four concentrations: 212 µM, 500 µM, 1000 µM, and 1317 µM. Concentrations were determined in 5 M guanidine hydrochloride by recording the absorbance of Trp (ε = 5690 M−1cm−1) and Tyr (ε = 1280 M−1cm−1) residues at 280 nm by UV/vis on a Thermoscientific Nanodrop 2000 spectrophotometer. Peak widths were determined for five distinct peaks and line broadening was not observed over these concentrations (see Supporting Information).
Results
Sequence Design
The FBP11 WW1 domain peptide (Figure 3a) used in these studies corresponds to residues 15–42 of the native peptide (residues 144–171 of the full mammalian FBP11 WW1 protein) (32–33), with an additional glycine residue coupled to the C-terminus for ease in synthesis. Using the same principles as with WKWK and (WKWK)2 (13–16), the FBP11 WW1 domain was mutated to form a putative binding cleft for ssDNA to give Mut1 (Figure 2; 3b). The C-terminal β-hairpin was mutated such that a Trp binding pocket was placed on the binding face with two flanking Lys residues cross-strand to the two tryptophans. The turn sequences were changed to Asn-Gly turns since these gave well folded turns in the hairpins used previously for nucleotide binding studies (13–16) and the mutations taken together gave the peptide a +5 net charge. The C-terminal β-hairpin sequence mimics the binding face of the β-hairpin peptide, WKWK. Position 16 (Position 2 in Mut1 (45)) is varied in several known FBP WW domains (32–33). Arginine was placed in that position for Mut1 since Arg is known to provide favorable contacts in DNA binding (46–49). Residues 2 and 4 are cross-strand from the binding pocket in strands 2 and 3 and may make favorable contacts to the DNA to improve binding affinity and selectivity. These additional interactions were not present in the WKWK monomer and dimer.
Two additional mutants were designed to further understand the characteristics that drive the interactions of interest. Mut2 (Figure 3c) was designed with no proline on the C-terminus. This residue is conserved in many different WW domains and has been shown to be crucial in the folding of native WW domain peptides (27). To determine if this residue is still important for folding and ssDNA recognition in Mut1, structure and binding studies of Mut2 were performed.
Research by Kelly, et al., has shown that the native turn between strands 1 and 2 is important for structure and stability (17) leading to another mutant made with the native turn one sequence replacing the Asn-Gly turn that was introduced in the original mutant. The other mutations remained the same to give Mut3 (Figure 3d).
The isolated hairpins made up of strands 1 and 2 (Mut1-S12) or strands 2 and 3 (Mut1-S23) as well as single strands (Mut1-S1, Mut1-S2, and Mut1-S3; Figure 3e–i) were also characterized as additional means to understand the role of each strand in folding and binding to ssDNA. An additional hairpin composed of strands 2 and 3 with an E27Q mutation, Mut1-S23-E27Q (Figure 3j), was also characterized to better understand the role of stability and electrostatic interactions in ssDNA recognition by Mut1. Structural and binding studies of Mut1-S23 and Mut1-S23-E27Q were also compared to WKWK monomer (Figure 3k).
A polyproline sequence based on a peptide used in studies as an FBP11 WW1 domain ligand (Figure 3l) (28) was synthesized to determine if binding to this sequence is affected by the mutations made. This polyproline helix contains the PPLP motif known as the ligand specific to the FBP11 WW1 domain (28, 34, 35).
CD Characterization of Folding
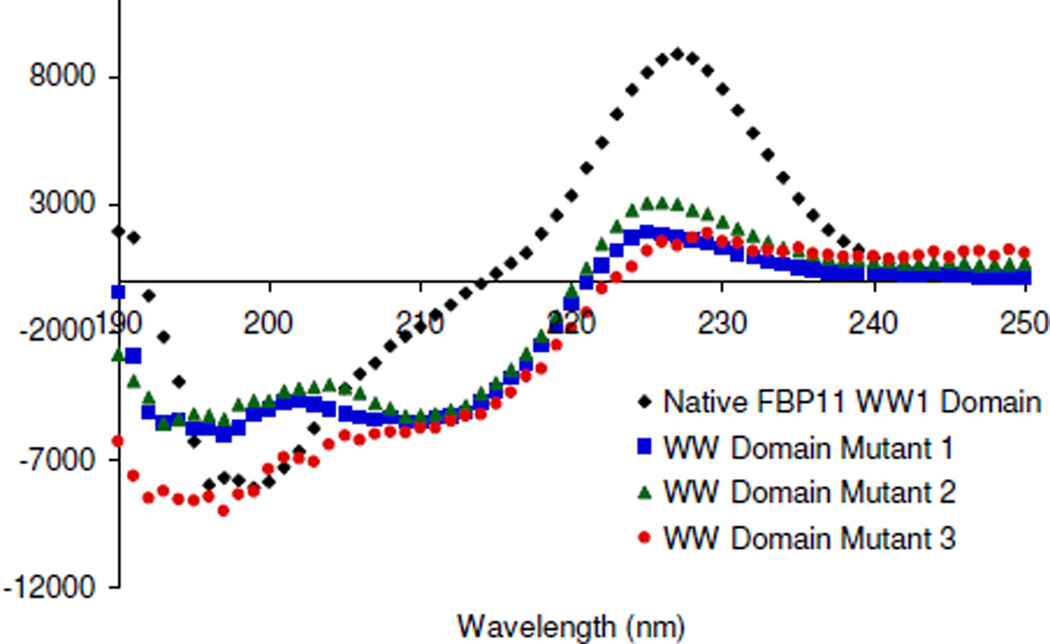
Structural studies on Mutants 1–3 were performed by CD and compared to the native FBP11 WW1 domain to determine the effect of the mutations on β-sheet structure (Figure 4). The CD spectra of the mutants differ from that of the native WW domain. In particular, the native protein displays a larger exciton coupling peak at 227 nm than the mutants. This is likely due to differences in orientations of the aromatic sidechains. Nonetheless, the mutants clearly exhibit β-sheet structure, as indicated by the minima at 210 nm. Although β-sheets typically have minima at about 215 nm, the shift to 210 nm is likely due to the contributions of the Trp residues, and the differences in these spectra could be due to the number and relative conformations of tryptophans in each peptide. The minimum near 195 nm may represent random coil due to some degree of fraying or some extent of polyproline helix character due to the KPG sequence at the C-terminus of the peptide, as a minimum is also observed in that region for the native protein. Mut3 displays a more significant minimum near 195 nm, which may be attributed to the more flexible turn sequence between strands 1 and 2.
Figure 4.
CD spectra for the native WW domain and WW domain mutants. Data was recorded at 30 µM peptide, 10 mM Na2HPO4, 25 °C, pH 7.0.
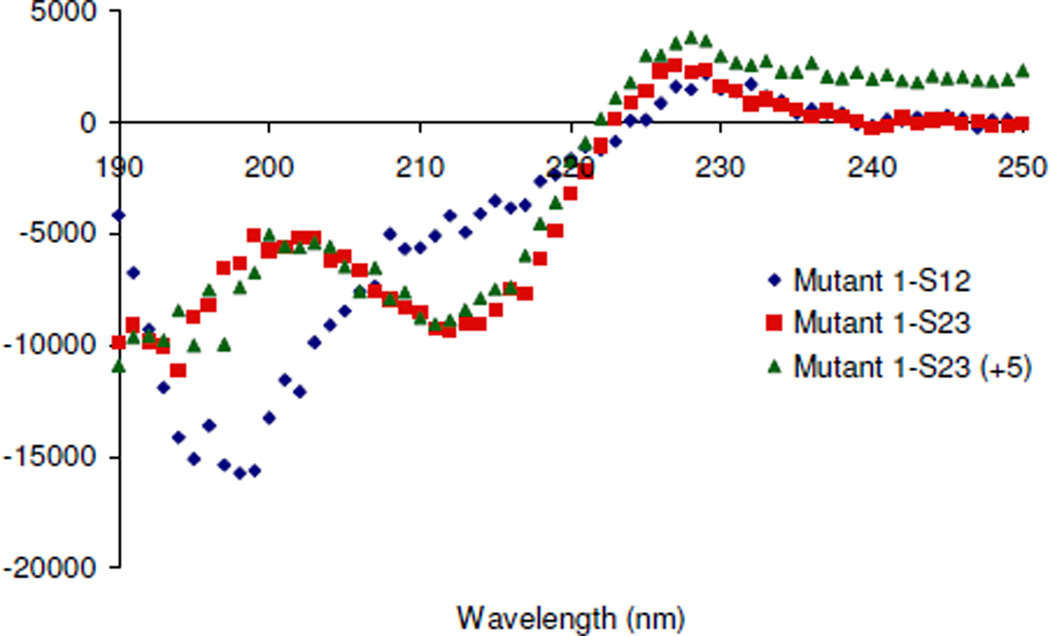
The Mut1 control peptides Mut1-S12 and Mut1-S23 were also analyzed by CD (Figure 5). Mut1-S12 is primarily unstructured with a minimum at 198 nm corresponding to random coil but with a shoulder at 215 nm, which is indicative of β-sheet structure. Mut1-S23 gives a minimum CD signal at about 215 nm, which is consistent with beta-sheet peptides and proteins. This peptide also has a minimum at about 195 nm, which may be due to fraying or the KPG tail at the C-terminus. Thus, it appears that the random coil nature of the three-stranded sheets primarily comes from strand one. Each of the three control peptides, Mut1-S1, Mut1-S2, and Mut1-S3, produced CD wavelength data consistent with random coil peptides, as expected (Figure S9).
Figure 5.
CD data for WW domain mutant control peptides. Data was recorded at 30 µM peptide, 10 mM Na2HPO4, 25°C, pH 7.0.
Thermal denaturation studies were conducted for the native WW domain and for Mutants 1–3 to determine the stability of the mutants compared to the native peptide. Thermal denaturations were performed following the Trp exciton coupling peak at 227 nm over a range of temperatures. The native FBP11 WW1 domain has a melting temperature similar to that of the hPin1 and FBP28 WW domains (17). Mutants 1 and 3 have similar stability to that of the native WW domain, but Mut2 appears to be less stable (Figure 6). The reduced stability of Mut2 verifies the role of Pro27 in stabilizing the mutants, as has been observed in the native protein (27).
Figure 6.
Thermal denaturation plots for the Native WW domain and the mutant peptides. Thermal denaturations were followed by CD at 227 nm from 4–96 °C with 4°/step. Data was recorded at 30 µM peptide, 10 mM Na2HPO4, pH 7.0. Lines are meant to guide the eye.
A thermal renaturation study was also performed for Mut1 to investigate the folding behavior of the mutant. Although Mut1 showed no sign of aggregation up to 1.3 mM by NMR (see Experimental Section and Supporting Information, Figure S12), thermal renaturation did not reproduce the same CD spectrum as was initially observed (see Supporting Information, Figure S10), indicating that the mutations have resulted in a peptide that is not as well behaved as the native protein. Nonetheless, this finding does not impact the results described below.
Characterization of the Peptide-ssDNA Interactions
The stoichiometry of binding was determined for Mut1 as well as Mut1-S23 and ssDNA by fluorescence quenching using the molar variation method (see Experimental Section). A 1:1 binding stoichiometry for the interaction between Mut1 and the 11-mer ssDNA sequence (Figure 3m) was determined, in agreement with the stoichiometry reported for the (WKWK)2 interaction with dA5 and with the same 11-mer ssDNA (see Supporting Information, Figure S1) (14–16). The binding stoichiometry for the interaction between Mut1-S23 and ssDNA was also determined to be 1:1 (see Supporting Information, Figure S2).
Binding of ssDNA to each of the peptides was determined by quenching of the tryptophan fluorescence as described in the Experimental Procedures. A correction for the inner filter effect arising from absorbance of the nucleobases at the excitation wavelength of Trp was performed for all binding data (see Experimental Procedures).
As a reference point, the dissociation constant of the WKWK monomer to the 11-mer ssDNA sequence was determined to be 39.3 µM (Table 1). This affinity is much weaker than that of (WKWK)2, presumably due to the lack of the second DNA binding pocket provided by the dimer as well as a lower net charge. WKWK can be compared to Mut1 and Mut1-S23 to gain information regarding structural and sequence related aspects of ssDNA binding for each.
Table 1.
Dissociation constants for the binding interaction between WW domain peptides and ssDNAa
| Peptide | Kd, (error) |
µM |
|---|---|---|
| Native WW domain | 12500b | |
| WKWK | 39 (2) | |
| WKWK dimer | 3.5 (0.2)c | |
| Mut1 | 17 (2) | |
| Mut1-S23 | 32 (2) | |
| Mut1-S23-E27Q | 23 (1) | |
| Mut2 | 34 (1) | |
| Mut3 | 46 (3) |
Conditions: 10 mM sodium phosphate buffer, 100 mM NaCl, pH 7.0, 25 °C. Each value is the average of at least two measurements. The error is from the fitting.
This data was collected using fluorescence anisotropy. The conditions are the same as with the fluorescence quenching binding measurements.
Mut1 was designed with two strands mimicking the binding pocket of the β-hairpin WKWK and has a net charge of +5. The additional N-terminal strand was intended to allow additional contacts to improve the binding to ssDNA. Fluorescence quenching experiments determined that Mut1 binds ssDNA with an affinity of 16.6 µM (Table 1). This peptide does not bind ssDNA as well as the WKWK dimer, which has a net charge of +8 and two aromatic binding pockets (14–16). However, Mut1 displays more than 2-fold tighter binding than the WKWK hairpin monomer, which has a similar aromatic binding cleft as Mut1 and a +4 charge. This suggests that strand 1 contributes to the binding affinity and provides some additional contacts favorable for binding. Whether these additional contacts are electrostatic or not was explored further, as described below.
The C-terminal proline was removed in Mut2 to determine if the residue is important in binding since it has been shown to be critical for folding of the native peptide (27). Pro37 in the native WW domain (Pro29 in Mut1) interacts with a small hydrophobic pocket made up of the N-terminal Trp12 as well as Tyr24, which stabilizes the structure (27). Thermal denaturations suggest that it contributes to the stability of the mutants as well. Binding of Mut2 to ssDNA is weaker than Mut1, with a Kd of 34.2 µM (Table 1). This indicates that stability of the three-stranded sheet influences binding, confirming the importance of the folded structure.
Replacing the Asn-Gly turn sequence in turn 1 of Mut1 with the native turn sequence in Mut3 also weakened the binding as compared to the original mutant by about three-fold (Kd = 45.8 µM, Table 1). It may be that strand 1 is less folded in Mut3, which would explain the more negative peak at 195 nm in the CD spectrum for Mut3 relative to Mut1 (Figure 4).
To further explore the role of strand 1 in Mut1, the binding of Mut1-S23 to ssDNA was analyzed by fluorescence quenching. The affinity was reduced by about two-fold (Kd = 32.4 µM, Table 1). The overall charge of the peptide is +4 versus the +5 charge for the full peptide, which could explain the difference in binding. However, Mut1-S23 binds ssDNA more strongly than does WKWK even though the charge is the same. Mut1-S23 has a longer sequence and additional contacts may enhance binding.
To determine whether charge is the only factor affecting the difference in binding of ssDNA for Mut1 versus Mut1-S23, a glutamic acid in strand 3 of Mut1-S23 was mutated to a glutamine (E27Q), resulting in a peptide with the same net charge as Mut1. The binding affinity of Mut1-S23-E27Q was intermediate to the full peptide and strands 2&3, with a Kd of 22.9 µM (Table 1). This indicates that the increased net positive charge improves the binding affinity, but the data suggests that strand 1 in Mut1 also contributes to binding in some way other than simply net charge.
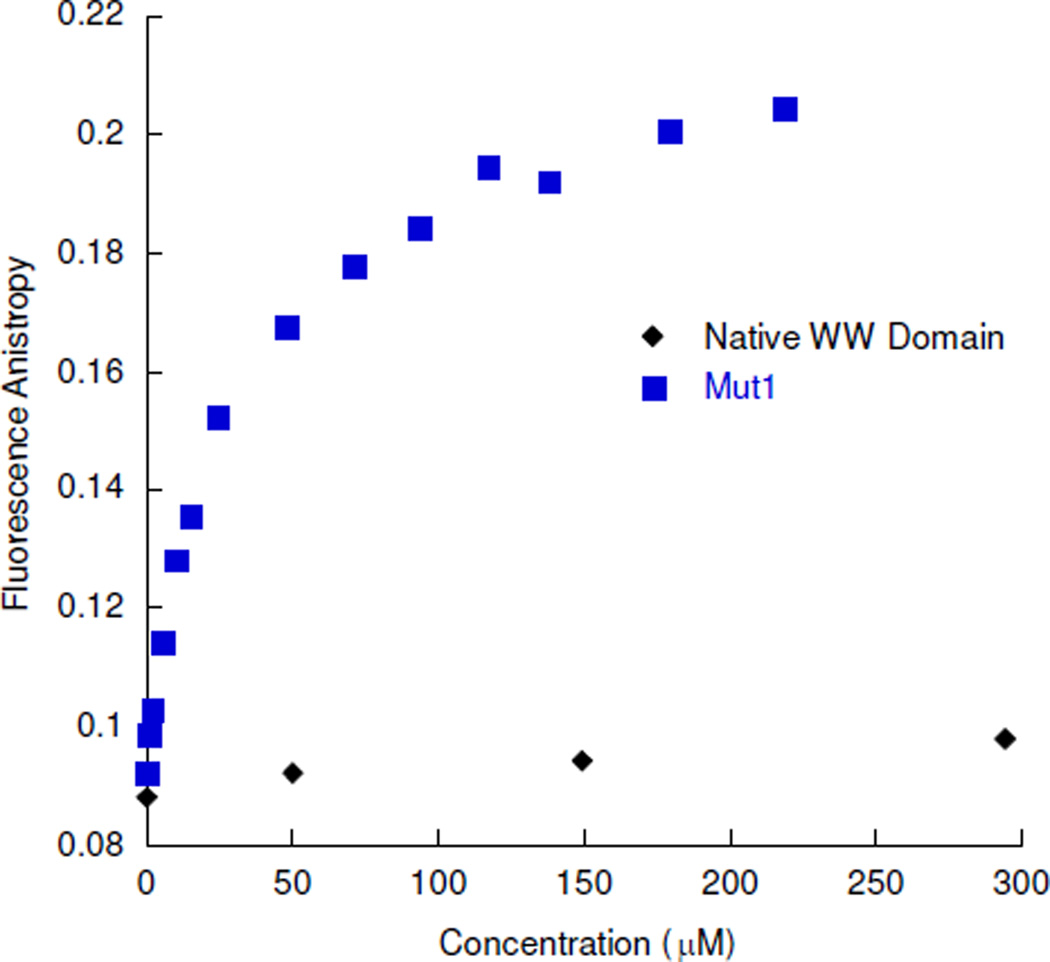
As a control, we measured the binding of the native FBP11 WW1 domain to ssDNA. While all of the data for the mutants binding to ssDNA and duplex DNA were obtained by fluorescence quenching, the binding affinity for the native peptide to these DNA sequences was too weak to be determined using that method due to inner filter effects. Fluorescence anisotropy was therefore employed to determine the binding of the native peptide to the DNA sequences. A fluorophore, 5’-Bodipy 630/650-X NHS Ester, was attached to the ssDNA sequence. Fluorescence anisotropy data gave a Kd in the millimolar range, providing evidence that the native peptide has little affinity for ssDNA (Figure 7, Table 1). Mut1 binds Bodipy-labeled ssDNA with a Kd of about 26 µM, similar to that determined by fluorescence quenching. This provides further evidence that the native WW domain has been redesigned to bind ssDNA (Figure 7). Control experiments were performed to confirm that the peptide does not bind to the fluorophore directly.
Figure 7.
Bodipy-ssDNA titrated with the native WW domain and Mut1. Fluorescence monitored by fluorescence anisotropy; 1.5 µM Bodipy-ssDNA for Mut1; 50 µM Bodipy-ssDNA for native WW domain; 10 mM sodium phosphate buffer, 100 mM NaCl, pH 7.0, 25 °C. The error for Mut1 is less than 8% for each point, with the exception of the point at 150 µM, for which the error is 15%. The error for the Native WW domain was not determined.
Based on previous studies of (WKWK)2, which does not exhibit any sequence selectivity, the sequence-selectivity of Mut 1 for ssDNA was not investigated. We expect that Mut1 will not be sequence selective, as binding appears to be driven by aromatic interactions and electrostatic interactions, as is the case with the OB fold.
Characterization of the Mut1-dsDNA Interaction
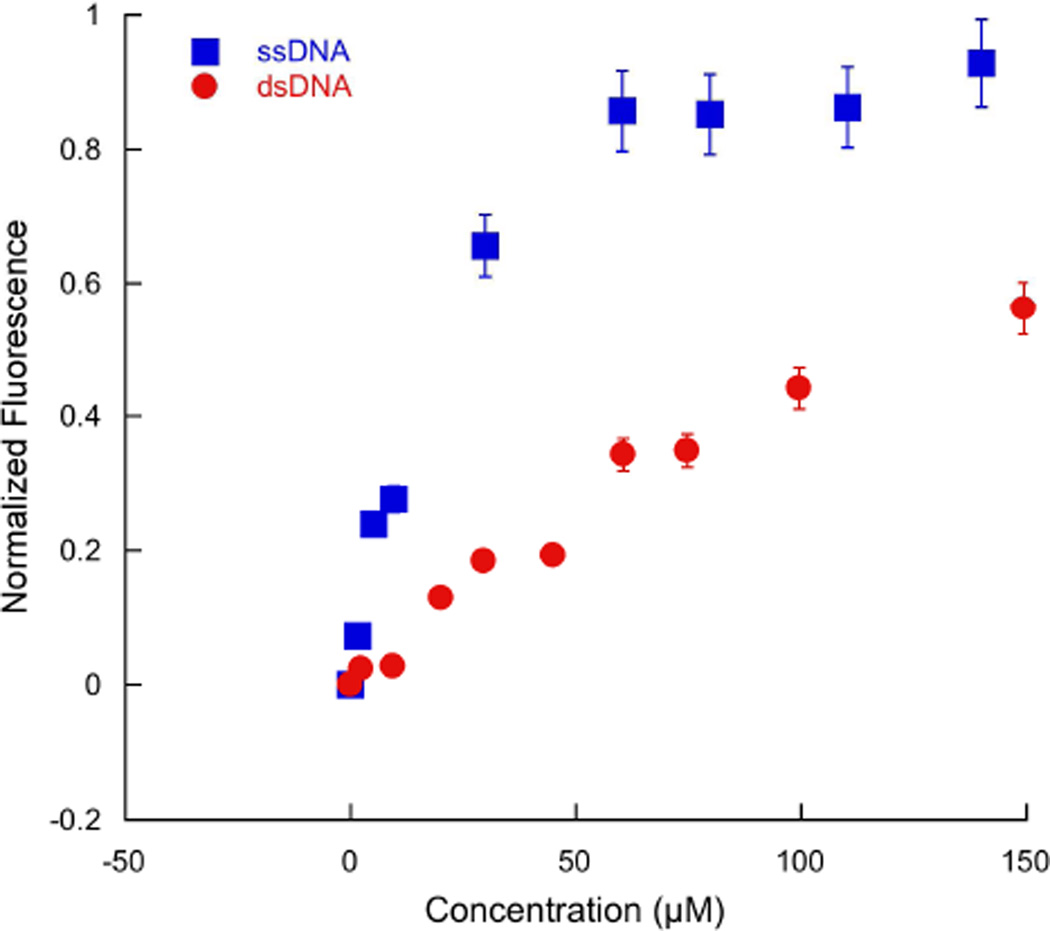
Because of inner filter effects due to DNA absorption at the excitation wavelength of Trp, fluorescence quenching of Trp could not be used for the duplex DNA binding studies with Mut1. Instead, fluorescence anisotropy was utilized to determine a more accurate dissociation constant for the binding interaction between Mut1 and duplex DNA. (5-(and -6)-Carboxytetramethylrhodamine (TAMRA) was coupled onto the peptide Mut1 at position 21 for these binding experiments via attachment to a Lys sidechain (see Experimental Procedures). This amino acid substitution has been shown to have minimal effects on stability and folding in model β-hairpins (50). As a control, fluorescence anisotropy was also used to measure the binding of Mut1 to ssDNA. The fluorescence anisotropy data for binding to ssDNA is consistent with the fluorescence quenching data, giving a dissociation constant of 20.4 µM (Table 2). The anisotropy data shows that Mut1 binds ssDNA with about 10-fold selectivity over duplex DNA (Kd = 189 µM, Table 2; Figure 8). This selectivity for ssDNA versus duplex DNA is much greater than that observed for WKWK dimer (13, 16). This is consistent with earlier studies that suggested WKWK dimer is a groove binder (16). The three-stranded sheet in Mut1 is likely too big to function as a groove binder and results in significantly greater selectivity for ssDNA.
Table 2.
Comparison of dissociation constants for the binding interaction between WW domain peptides and ssDNA and dsDNA sequences and Polyproline Helixa
| Peptide | ssDNA Kd, µM (error) |
dsDNA Kd, µM (error) |
Polyproline Helix Kd, µM (error) |
|---|---|---|---|
| Native FBP11 WW1 domain | >6000b | >500b | 60 (6) |
| WKWK dimer | 3.5 (0.2)c | 4.6 (0.4)c | n. d.d |
| TAMRA-Mut1 | 20 (4) | 190 (20) | No binding observed |
Conditions: 10 mM sodium phosphate buffer, 100 mM NaCl, pH 7.0, 25 °C. Each value is the average of at least two measurements. The error is from the fitting. n.d. denotes a measurement that was not determined.
This data was collected using fluorescence anisotropy with Bodipy-labeled DNA.
n. d. = not determined.
Figure 8.
Fluorescence anisotropy titrations of TAMRA-WW domain Mut1 titrated with unlabeled ssDNA (blue) and duplex DNA (red); 2 and 20 µM peptide, respectively, 10 mM sodium phosphate buffer, 100 mM NaCl, pH 7.0, 25 °C.
Control experiments showed that binding of the native WW domain to duplex DNA was very weak, as is the case with ssDNA. This provides further evidence that Mut1 has been successfully redesigned to bind DNA and that the native peptide has little affinity for any DNA sequence.
Binding to the Native Polyproline Helix
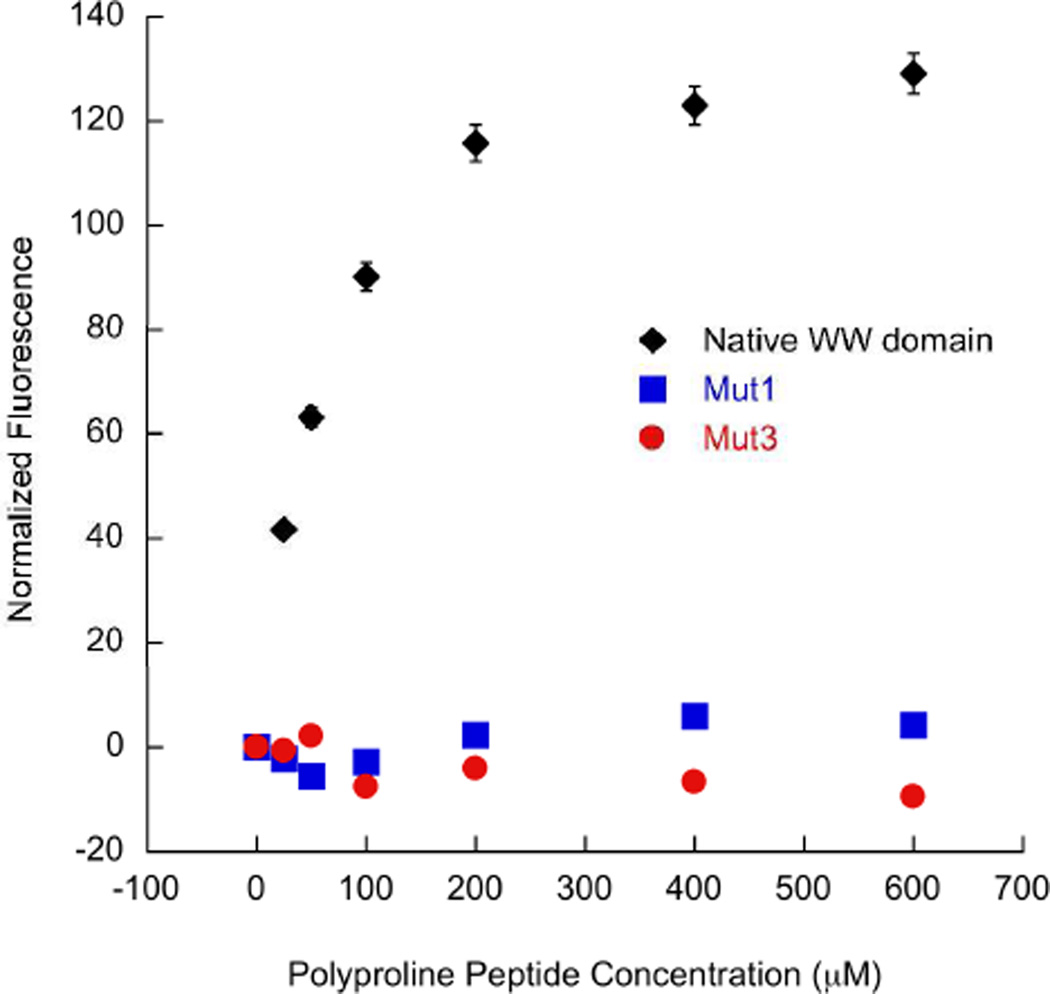
The ability of Mut1 to bind the natural ligand of FBP11 WW1 domain was investigated by conducting fluorescence experiments with a polyproline helix containing a PPLP consensus sequence (Figure 3l) and comparing that to binding of the consensus sequence to the native WW domain peptide. The binding affinity for the polyproline helix with the native WW domain was 59.6 µM (Table 2, Figure 9) (51). In contrast, no measurable binding was observed between Mut1 or Mut3and the polyproline helix (Table 2, Figure 9). This demonstrates the loss of function for its natural ligand and verifies that the WW domain has been redesigned to bind ssDNA.
Figure 9.
Fluorescence titrations of the native WW domain, Mut1, or Mut3 with oligoproline peptide (Figure 3l) following the increase in Trp fluorescence upon binding to the oligoproline peptide; 15 µM WW domain peptide, 0–600 µM polyproline helix; 10 mM sodium phosphate buffer, 100 mM NaCl, pH 7.0, 25 °C. This data was normalized by subtracting the initial peptide observed fluorescence from each fluorescence value. The error is less than 3% for each data point.
NMR Characterization of Mut1
NMR experiments were conducted to more fully characterize the folding of Mut1 and the role of strand 1 in stability and binding. Mut1, Mut1-S12, and Mut1-S23 were studied by one- and two-dimensional NMR. TOCSY experiments were used to assign peaks and NOESY experiments verified correct strand register (see Supporting Information, Figure S11). Interestingly, NOEs also clearly indicated the interaction of Pro29 with the aromatic pocket on strands 1 and 2, as is found in the native protein.
Glycine splitting and α-hydrogen (Hα) chemical shift values were determined to characterize the full peptide, both hairpins, the three individual strands, and their respective stabilities. The Hα chemical shifts of the hairpins relative to the random coil peptides indicate the degree of β-sheet structure for each residue along the peptide backbone. Downfield shifting of Hα protons is evidence of increased hairpin population, with a chemical shift difference of greater than 0.1 ppm taken to indicate β-sheet structure (52–53). A second method to determine the extent of folding of each peptide is to determine the Gly Hα splitting values for Gly residues in the turns (54). As folding of the hairpin increases, the splitting of the hydrogens increases as well, giving a measure of stability at the turn. A comparison of the Gly splitting in the β-hairpin with that of a fully folded cyclic control peptide gives the extent of folding and overall stability for the peptide (See Experimental Procedures and Supporting Information).
NMR data revealed that the N-terminal β-hairpin, Mut1-S12, has a glycine splitting value of 0.11 for Gly10, giving a fraction folded of 26% (Table 3). The C-terminal β-hairpin, Mut1-S23, is significantly more stable, with a glycine splitting value of 0.67 for Gly20 and a fraction folded of 89% (Table 3). This is similar to the WKWK peptide, which has a reported percent folding value of 96% (14–15).
Table 3.
Fraction folded for Mut1 and the beta-hairpins Mut1-S12 and Mut1-S23.a
| Peptide | Gly chemical shifts, ppmb | ΔδGly, ppmb | Fraction Folded (Gly Splitting)c |
|---|---|---|---|
| Mut1-S12 | 3.91, 4.02 | 0.11 | 0.26 |
| Mut1-S23 | 3.45, 4.12 | 0.67 | 0.89 |
| Mut1 | 3.82, 4.08 (turn 1) | 0.26 | 0.62 |
| 3.38, 4.06 (turn 2) | 0.68 | 0.91 |
Conditions: Values calculated from data obtained at 25 °C, 50 mM potassium dideuterium phosphate, pH 7.0 (uncorrected), referenced to DSS.
Error is ± 0.01 ppm, determined by as determined by the acquired points and spectral width.
Error is ± 0.02, based on the error in ΔδGly.
Comparison of Gly splitting for the full Mut1, Mut1-S12, and Mut1-S23 indicate that while the hairpin formed by strands 1&2 alone is poorly folded, addition of strand 3 in the full-length peptide adds stability to this hairpin, exhibiting an increase in structure from 26% to 62% folded for Gly10 (Table 3). This data suggests some degree of cooperative folding of the three-stranded sheet. This type of cooperativity is not unprecedented (55–60). For example, Searle et al., found that the N-terminal strand of their three-stranded beta sheet (peptide 1–24) cooperatively stabilized the C-terminal hairpin of the peptide (55–57). Likewise, Kelly and coworkers have shown that the hPin1 WW domain and various mutants exhibit cooperative unfolding (59–60). In contrast to Mut1-S12, strands 2&3 alone are as well folded in Mut1-S23 as in the full peptide, with a percent folded of 89% and 91% for Gly20, respectively (Table 3).
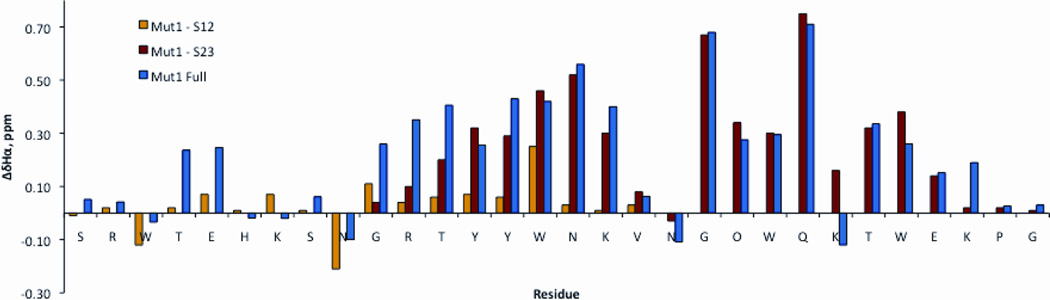
Hα chemical shift differences for the two hairpins and the full peptide are consistent with both the CD data and the NMR glycine splitting data (Figure 10). This data indicates that Mut1-S12 is only marginally folded at best, but when incorporated into the full length peptide, strand 1 exhibits modestly increased downfield shifting, corresponding to an increase in folding. In contrast, strands 2 and 3 are well folded in both Mut1 and Mut1-S23.
Figure 10.
NMR chemical shift differences for Mut1, Mut1-S12, and Mut1-S23 relative to unfolded controls (Mut1-S1, Mut1-S2, and Mut1-S3). Downfield shifting of > 0.1 ppm indicate beta-sheet structure. Upfield shifting of Asn9 and Asn19 is consistent with a beta-turn conformation. Upfield shifting of K24 is due to ring current effects from the cross-strand Trp15. Conditions: 25 °C, 50 mM potassium dideuterium phosphate, pD 7.4 (uncorrected), referenced to DSS. Error is ± 0.01 ppm, as determined by the acquired points and spectral width.
Discussion & Conclusions
We have found that introduction of a WKWK binding pocket into a three-stranded β-sheet via mutation of a native WW domain leads to a change in function of the native protein from a polyproline helix binder to a ssDNA receptor. In contrast, the native WW domain exhibits no binding to ssDNA. Structure/function studies indicate that Pro29 helps to stabilize the folded state of the mutant, as has been observed for the native protein, and this also impacts ssDNA binding affinity. In addition, Mut1, which has an Asn-Gly turn between strands 1 and 2, was determined to have a stronger binding affinity to ssDNA than did Mut3, which has the native turn sequence at that position. CD studies suggest that Mut3 has a higher random coil population than does Mut1, presumably due to a less structured turn region and less-well folded strand 1, which may explain the weaker binding of Mut3. Comparison of Mut1 to the truncated peptides, Mut1-S23 and Mut1-S23-E27Q, confirms that strand 1 contributes to ssDNA binding, albeit weakly. NMR studies indicate that Mut1 is well folded at strands 2 and 3, but that strand 1 is less well folded. Nonetheless, strand 1 influences binding to ssDNA.
This three-stranded motif provides substantial (approximately 10-fold) selectivity for ssDNA over dsDNA, unlike the (WKWK)2, which has less than 2-fold selectivity for ssDNA (13). Previous studies suggest that (WKWK)2 binds to duplex DNA via groove binding (16). Thus, the selectivity observed for Mut1 may be due to inhibition of groove binding due to the additional strand (strand 1).
These studies provide insight into features that can provide structure-selectivity in protein-DNA interactions. This model system represents a conceptual mimic of the OB-fold, which consists of a beta-sheet surface that binds ssDNA. As such, it provides insight into the minimal features responsible for ssDNA binding in OB-fold-containing proteins, such as replication protein A, which is involved in DNA replication and repair (1–4). This study also provides a novel example of protein redesign, conceptually similar to the protein grafting approach developed by Schepartz (61), but in this case grafting a designed peptide motif onto a native protein. It is also one of the few examples of a redesigned functional β-sheet (62). Further mutation studies are underway to optimize binding affinity.
Supplementary Material
Acknowledgements
We gratefully acknowledge funding from the NIH General Medical Sciences (Grant # 1R01GM072691) for funding of this work. ALS acknowledges support from a GAANN fellowship.
Abbreviations
- FBP11 WW1 domain
forming binding protein 11 WW1 domain
- ssDNA
single-stranded deoxyribonucleic acid
- dsDNA
double-stranded DNA or duplex DNA
- WKWK
peptide designated by two tryptophan residues cross-strand from two lysine residues on the binding face of the peptide
- OB-fold
oligonucleotide/oligosaccharide binding fold
- Mut1-S1
Mutant 1 Strand 1
- Mut1-S2
Mutant 1 Strand 2
- Mut1-S3
Mutant 1 Strand 3
- Mut1-S12
hairpin composing Mutant 1 Stands 1 and 2
- Mut1-S23
hairpin composing Mutant 1 Strands 2 and 3
- CD
circular dichroism
- NMR
nuclear magnetic resonance
- TAMRA
5-(and -6)-Carboxytetramethylrhodamine, mixed isomers
- DMF
N,N-dimethylformamide
- TFA
trifluoroacetic acid
- DIPEA
Diisopropylethylamine
- TIPS
Triisopropylsilane
- HOBT
N-hydroxybenzotriazole
- HBTU
O-benzotriazole-N,N,N’,N’,-tetramethyluronium hexafluorophosphate
Footnotes
This research was supported by a grant from the National Institutes of Health (1R01 GM072691).
ALS acknowledges support from a GAANN fellowship.
Supporting Information Available. Molar variation plots, Fluorescence spectra, CD wavelength data, NMR assignments, and NOEs. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Bochkarev A, Pfuetzner RA, Edwards AM, Frappier L. Structure of the single-stranded-DNA-binding domain of replication protein A bound to DNA. Nature. 1997;385:176–181. doi: 10.1038/385176a0. [DOI] [PubMed] [Google Scholar]
- 2.Wold MS. Replication Protein A: A Heterotrimeric, Single-Stranded DNA-Binding Protein Required for Eukaryotic DNA Metabolism. Annu. Rev. Biochem. 1997;66:61–92. doi: 10.1146/annurev.biochem.66.1.61. [DOI] [PubMed] [Google Scholar]
- 3.Theobald DL, Mitton-Fry RM, Wuttke DS. Nucleic Acid Recognition by OB-Fold Proteins. Annu. Rev. Biophys. Biomol. Struct. 2003;32:115–133. doi: 10.1146/annurev.biophys.32.110601.142506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bochkarev A, Bochkareva E. From RPA to BRCA2: lessons from single-stranded DNA binding by the OB-fold. Curr. Opin. Struct. Biol. 2004;14:36–42. doi: 10.1016/j.sbi.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 5.Anderson EM, Halsey WA, Wuttke DS. Site-Directed Mutagenesis Reveals the Thermodynamic Requirements for Single-Stranded DNA Recognition by the Telomere-Binding Protein Cdc13. Biochemistry. 2003;42:3751–3758. doi: 10.1021/bi027047c. [DOI] [PubMed] [Google Scholar]
- 6.Mitton-Fry RM, Anderson EM, Hughes TR, Lundblad V, Wuttke DS. Conserved Structure for Single-Stranded Telomeric DNA Recognition. Science. 2002;296:145–147. doi: 10.1126/science.1068799. [DOI] [PubMed] [Google Scholar]
- 7.Max KEA, Zeeb M, Bienert R, Balbach J, Heinemann U. Common mode of DNA binding to cold shock domains: Crystal structure of hexathymidine bound to the domain-swapped form of a major cold shock protein from Bacillus caldolyticus. FEBS J. 2007;274:1265–1279. doi: 10.1111/j.1742-4658.2007.05672.x. [DOI] [PubMed] [Google Scholar]
- 8.Max KEA, Zeeb M, Bienert R, Balbach J, Heinemann U. T-rich DNA Single Strands Bind to a Preformed Site on the Bacterial Cold Shock Protein Bs-CspB. J. Mol. Biol. 2006;360:702–714. doi: 10.1016/j.jmb.2006.05.044. [DOI] [PubMed] [Google Scholar]
- 9.Hillier BJ, Rodriguez HM, Gregoret LM. Coupling protein stability and protein function in Escherichia coli CspA. Folding Des. 1998;3:87–93. doi: 10.1016/S1359-0278(98)00014-5. [DOI] [PubMed] [Google Scholar]
- 10.Newkirk K, Feng W, Jiang W, Tejero R, Emerson SD, Inouye M, Montelione GT. Solution NMR structure of the major cold shock protein (CspA) from Escherichia coli: Identification of a binding epitope for DNA. Proc. Natl. Acad. Sci. USA. 1994;91:5114–5118. doi: 10.1073/pnas.91.11.5114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schindelin H, Marahiel MA, Heinemann U. Universal nucleic acid-binding domain revealed by crystal structure of the B. subtilis major cold-shock protein. Nature. 1993;364:164–168. doi: 10.1038/364164a0. [DOI] [PubMed] [Google Scholar]
- 12.Schnuchel A, Wiltscheck R, Czisch M, Herrier M, Willimsky G, Graumann P, Marahiel MA, Holak TA. Structure in solution of the major cold-shock protein from Bacillus subtilis. Nature. 1993;364:169–171. doi: 10.1038/364169a0. [DOI] [PubMed] [Google Scholar]
- 13.Butterfield SM, Cooper WJ, Waters ML. Minimalist protein design: a beta-hairpin peptide that binds ssDNA. J. Am. Chem. Soc. 2005;127:24–25. doi: 10.1021/ja045002o. [DOI] [PubMed] [Google Scholar]
- 14.Butterfield SM, Waters ML. A designed beta-hairpin peptide for molecular recognition of ATP in water. J. Am. Chem. Soc. 2003;125:9580–9581. doi: 10.1021/ja0359254. [DOI] [PubMed] [Google Scholar]
- 15.Butterfield SM, Sweeney MM, Waters ML. The recognition of nucleotides with model beta-hairpin receptors: investigation of critical contacts and nucleotide selectivity. J. Org. Chem. 2005;70:1105–1114. doi: 10.1021/jo0491105. [DOI] [PubMed] [Google Scholar]
- 16.Stewart AL, Waters ML. Structural effects on ss- and dsDNA recognition by a beta-hairpin peptide. ChemBioChem. 2009;10:539–544. doi: 10.1002/cbic.200800524. [DOI] [PubMed] [Google Scholar]
- 17.Jager M, Zhang Y, Bieschke J, Nguyen H, Dendle M, Bowman ME, Noel JP, Gruebele M, Kelly JW. Structure–function–folding relationship in a WW domain. Proc. Natl. Acad. Sci. USA. 2006;103:10648–10653. doi: 10.1073/pnas.0600511103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fernandez-Escamilla AM, Ventura S, Serrano L, Jimenez MA. Design and NMR conformational study of a β-sheet peptide based on Betanova and WW domains. Protein Sci. 2006;15:2278–2289. doi: 10.1110/ps.062186506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karanicolas J, Brooks CL. The structural basis for biphasic kinetics in the folding of the WW domain from a formin-binding protein: Lessons for protein design? Proc. Natl. Acad. Sci. USA. 2003;100:3954–3959. doi: 10.1073/pnas.0731771100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kraemer-Pecore CM, Lecomte JTJ, Desjarlais JR. A de novo redesign of the WW domain. Protein Sci. 2003;12:2194–2205. doi: 10.1110/ps.03190903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang X, Kowalski J, Kelly JW. Increasing protein stability using a rational approach combining sequence homology and structural alignment: Stabilizing the WW domain. Protein Sci. 2001;10:1454–1465. doi: 10.1110/ps.640101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jager M, Nguyen H, Crane JC, Kelly JW, Gruebele M. The folding mechanism of a β-sheet: the WW domain. J. Mol. Biol. 2001;311:373–393. doi: 10.1006/jmbi.2001.4873. [DOI] [PubMed] [Google Scholar]
- 23.Macias MJ, Gervais V, Civera C, Oschkinat H. Structural analysis of WW domains and design of a WW prototype. Nat. Struct. Biol. 2000;7:375–379. doi: 10.1038/75144. [DOI] [PubMed] [Google Scholar]
- 24.Koepf EK, Petrassi M, Ratnaswamy G, Huff ME, Sudol M, Kelly JW. Characterization of the Structure and Function of W→ F WW Domain Variants: Identification of a Natively Unfolded Protein That Folds upon Ligand Binding. Biochemistry. 1999;38:14338–14351. doi: 10.1021/bi991105l. [DOI] [PubMed] [Google Scholar]
- 25.Nguyen H, Jager M, Moretto A, Gruebele M, Kelly JW. Tuning the free-energy landscape of a WW domain by temperature, mutation, and truncation. Proc. Natl. Acad. Sci. USA. 2003;100:3948–3953. doi: 10.1073/pnas.0538054100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jager M, Dendle M, Fuller AA, Kelly JW. A cross-strand Trp–Trp pair stabilizes the hPin1 WW domain at the expense of function. Protein Sci. 2007;16:2306–2313. doi: 10.1110/ps.072904107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kato Y, Miyakawa T, Kurita J, Tanokura M. Structure of FBP11 WW1-PL Ligand Complex Reveals the Mechanism of Proline-rich Ligand Recognition by Group II/III WW Domains. J. Biol. Chem. 2006;281:40321–40329. doi: 10.1074/jbc.M609321200. [DOI] [PubMed] [Google Scholar]
- 28.Pires JR, Parthier C, do Aido-Machado R, Wiedemann U, Otte L, Bohm G, Rudolph R, Oschkinat H. Structural Basis for APPTPPPLPP Peptide Recognition by the FBP11WW1 Domain. J. Mol. Biol. 2005;348:399–408. doi: 10.1016/j.jmb.2005.02.056. [DOI] [PubMed] [Google Scholar]
- 29.Ball LJ, Kuhne R, Schneider-Mergener J, Oschkinat H. Recognition of proline-rich motifs by protein-protein-interaction domains. Angew. Chem. Int. Ed. 2005;44:2852–2869. doi: 10.1002/anie.200400618. [DOI] [PubMed] [Google Scholar]
- 30.Kato Y, Nagata K, Takahashi M, Lian L, Herrero JJ, Sudol M, Tanokura M. Common Mechanism of Ligand Recognition by Group II/III WW Domains: Redefining their functional classification. J. Biol. Chem. 2004;279:31833–31841. doi: 10.1074/jbc.M404719200. [DOI] [PubMed] [Google Scholar]
- 31.Bedford MT, Chan DC, Leder P. FBP WW domains and the Abl SH3 domain bind to a specific class of proline-rich ligands. EMBO J. 1997;16:2376–2383. doi: 10.1093/emboj/16.9.2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Otte L, Wiedemann U, Schlegel B, Pires JR, Beyermann M, Schmieder P, Krause G, Volkmer-Engert R, Schneider-Mergener J, Oschkinat H. WW domain sequence activity relationships identified using ligand recognition propensities of 42 WW domains. Protein Sci. 2003;12:491–500. doi: 10.1110/ps.0233203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zarrinpar A, Lim WA. Converging on proline: the mechanism of WW domain peptide recognition. Nat. Struct. Biol. 2000;7:611–613. doi: 10.1038/77891. [DOI] [PubMed] [Google Scholar]
- 34.Espinosa JF, Syud FA, Gellman SH. An autonomously folding β-hairpin derived from the human YAP65 WW domain: Attempts to define a minimum ligand-binding motif. Peptide Sci. 2005;80:303–311. doi: 10.1002/bip.20205. [DOI] [PubMed] [Google Scholar]
- 35.Dalby PA, Hoess RH, DeGrado WF. Evolution of binding affinity in a WW domain probed by phage display. Protein Sci. 2000;9:2366–2376. doi: 10.1110/ps.9.12.2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chan DC, Bedford MT, Leder P. Formin binding proteins bear WWP/WW domains that bind proline-rich peptides and functionally resemble SH3 domains. EMBO J. 1996;15:1045–1054. [PMC free article] [PubMed] [Google Scholar]
- 37.Macias MJ, Hyvonen M, Baraldi E, Schultz J, Sudol M, Saraste M, Oschkinat H. Structure of the WW domain of a kinase-associated protein complexed with a proline-rich peptide. Nature. 1996;382:646–649. doi: 10.1038/382646a0. [DOI] [PubMed] [Google Scholar]
- 38.Chen HI, Sudol M. The WW domain of Yes-associated protein binds a proline-rich ligand that differs from the consensus established for Src homology 3-binding modules. Proc. Natl. Acad. Sci. USA. 1995;92:7819–7823. doi: 10.1073/pnas.92.17.7819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lim WA, Fox RO, Richards FM. Stability and peptide binding affinity of an SH3 domain from the Caenorhabditis elegans signaling protein Sem-5. Protein Sci. 1994;3:1261–1266. doi: 10.1002/pro.5560030812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lohman TM, Mascotti DP. Nonspecific ligand-DNA equilibrium binding parameters determined by fluorescence methods. Methods Enzymol. 1992;212:424–458. doi: 10.1016/0076-6879(92)12027-n. [DOI] [PubMed] [Google Scholar]
- 41.C Y Huang CY. Determination of Binding Stoichiometry by the Continuous Variation Method: The Job Plot. Meth. Enzymol. 1982;87:509–525. doi: 10.1016/s0076-6879(82)87029-8. [DOI] [PubMed] [Google Scholar]
- 42.Wang Y, Hamasaki K, Rando RR. Specificity of Aminoglycoside Binding to RNA Constructs Derived from the 16S rRNA Decoding Region and the HIV-RRE Activator Region. Biochemistry. 1997;36:768–779. doi: 10.1021/bi962095g. [DOI] [PubMed] [Google Scholar]
- 43.Wuthrich K. NMR of Proteins and Nucleic Acids. New York: Wiley-Interscience; 1986. [Google Scholar]
- 44.Syud FA, Espinosa JF, Gellman SH. NMR-Based Quantification of β-Sheet Populations in Aqueous Solution through Use of Reference Peptides for the Folded and Unfolded States. J. Am. Chem. Soc. 1999;121:11577–11578. [Google Scholar]
- 45.The amino acid numbering for the mutant peptides is 1–30.
- 46.Allen MD, Yamasaki K, Ohme-Takagi M, Tateno M, Suzuki M. A novel mode of DNA recognition by a b-sheet revealed by the solution structure of the GCC-box binding domain in complex with DNA. EMBO J. 1998;17:5484–5496. doi: 10.1093/emboj/17.18.5484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ohki I, Shimotake N, Fujita N, Jee J-G, Ikegami T, Nakao M, Shirakawa M. Solution Structure of the Methyl-CpG Binding Domain of Human MBD1 in Complex with Methylated DNA. Cell. 2001;105:487–497. doi: 10.1016/s0092-8674(01)00324-5. [DOI] [PubMed] [Google Scholar]
- 48.Luscombe NM, Laskowski RA, Thornton JM. Amino acid–base interactions: a three-dimensional analysis of protein–DNA interactions at an atomic level. Nucleic Acids Res. 2001;29:2860–2874. doi: 10.1093/nar/29.13.2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moodie SL, Mitchell JBO, Thornton JM. Protein Recognition of Adenylate: An Example of a Fuzzy Recognition Template. J. Mol. Biol. 1996;263:486–500. doi: 10.1006/jmbi.1996.0591. [DOI] [PubMed] [Google Scholar]
- 50.Cooper WJ, Waters ML. Turn residues in beta-hairpin peptides as points for covalent modification. Org. Lett. 2005;7:3825–3828. doi: 10.1021/ol0510116. [DOI] [PubMed] [Google Scholar]
- 51.Pires, et al., found that the interaction between FBP11 WW1 domain and a similar polyproline helix lacking a Tyr tag had a Kd of 145 µM. See reference 28.
- 52.Wishart DS, Sykes BD, Richards FM. Relationship between Nuclear Magnetic Resonance Chemical Shift and Protein Secondary Structure. J. Mol. Biol. 1991;222:311–333. doi: 10.1016/0022-2836(91)90214-q. [DOI] [PubMed] [Google Scholar]
- 53.Maynard AJ, Sharman GJ, Searle MS. Origin of β-Hairpin Stability in Solution: Structural and Thermodynamic Analysis of the Folding of a Model Peptide Supports Hydrophobic Stabilization in Water. J. Am. Chem. Soc. 1998;120:1996–2007. [Google Scholar]
- 54.Searle MS, Griffiths-Jones SR, Skinner-Smith H. Energetics of Weak Interactions in a β-hairpin Peptide: Electrostatic and Hydrophobic Contributions to Stability from Lysine Salt Bridges. J. Am. Chem. Soc. 1999;121:11615–11620. [Google Scholar]
- 55.Sharman GJ, Searle MS. Dissecting the effects of cooperativity on the stabilisation of a de novo designed three stranded anti-parallel β-sheet. Chem. Commun. 1997:1955–1956. [Google Scholar]
- 56.Sharman GJ, Searle MS. Cooperative Interaction between the Three Strands of a Designed Antiparallel β-Sheet. J. Am. Chem. Soc. 1998;120:5291–5300. [Google Scholar]
- 57.Griffiths-Jones SR, Searle MS. Structure, Folding, and Energetics of Cooperative Interactions between the β-Strands of a de Novo Designed Three-Stranded Antiparallel β-Sheet Peptide. J. Am. Chem. Soc. 2000;122:8350–8356. [Google Scholar]
- 58.Kortemme T, Ramirez-Alvarado M, Serrano L. Design of a 20-Amino Acid, Three-Stranded β-Sheet Protein. Science. 1998;281:253–256. doi: 10.1126/science.281.5374.253. [DOI] [PubMed] [Google Scholar]
- 59.Kaul R, Angeles AR, Jager M, Powers ET, Kelly JW. Incorporating β-Turns and a Turn Mimetic out of Context in Loop 1 of the WW Domain Affords Cooperatively Folded β-Sheets. J. Am. Chem. Soc. 2001;123:5206–5212. doi: 10.1021/ja0102890. [DOI] [PubMed] [Google Scholar]
- 60.Nguyen H, Jager M, Kelly JW, Gruebele M. Engineering a beta-sheet protein toward the folding speed limit. J. Phys. Chem. B. 2005;109:15182–15186. doi: 10.1021/jp052373y. [DOI] [PubMed] [Google Scholar]
- 61.Zondlo NJ, Schepartz A. Highly Specific DNA Recognition by a Designed Miniature Protein. J. Am. Chem. Soc. 1999;121:6938–6939. [Google Scholar]
- 62.Smith TJ, Stains CI, Meyer SC, Ghosh I. Inhibition of β-Amyloid Fibrillization by Directed Evolution of a β-Sheet Presenting Miniature Protein. J. Am. Chem. Soc. 2006;128:14456–14457. doi: 10.1021/ja065557e. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.